Abstract
In mammalian cells, early defenses against infection by pathogens are mounted through a complex network of signaling pathways shepherded by immune-modulatory pattern-recognition receptors. As obligate parasites, the survival of viruses is dependent upon the evolutionary acquisition of mechanisms that tactfully dismantle and subvert the cellular intrinsic and innate immune responses. Here, we review the diverse mechanisms by which viruses that accommodate DNA genomes are able to circumvent activation of cellular immunity. We start by discussing viral manipulation of host defense protein levels by either transcriptional regulation or protein degradation. We next review viral strategies used to repurpose or inhibit these cellular immune factors by molecular hijacking or by regulating their post-translational modification status. Additionally, we explore the infection-induced temporal modulation of apoptosis to facilitate viral replication and spread. Lastly, the co-evolution of viruses with their hosts is highlighted by the acquisition of elegant mechanisms for suppressing host defenses via viral mimicry of host factors. In closing, we present a perspective on how characterizing these viral evasion tactics both broadens the understanding of virus-host interactions and reveals essential functions of the immune system at the molecular level. This knowledge is critical in understanding the sources of viral pathogenesis, as well as for the design of antiviral therapeutics and autoimmunity treatments.
Keywords: Viral immune evasion, virus-host interactions, intrinsic immunity, innate immunity, proteasomal degradation, post-translational modification, apoptosis, viral mimicry
Introduction
Interactions between hosts and pathogens involve intricate processes of adaptation and co-evolution. While host cells have acquired core mechanisms of defense, viruses have evolved effective means to subvert or inhibit these defense mechanisms. One class of prominent human pathogens is that of DNA viruses. These viruses include poxviruses, herpesviruses, adenoviruses, polyomaviruses and papilloma viruses, and are connected with a diverse range of human diseases and even with increased mortality. The progression and spread of an infection depends on the “chess game”-like battle between host cell defenses and DNA virus evasion mechanisms.
The ability of a cell to elicit intrinsic and innate immune responses against DNA viruses is an essential component of host defense, acting immediately upon infection with these viruses. These host defenses act to inhibit viral infection, as well as to prime the adaptive immune responses and promote a global antiviral state within the host organism. As such, the function of these immune processes is essential for the survival of the host and the maintenance of a healthy system. The misregulation of immune responses is linked to the development of devastating autoimmune diseases, including type I diabetes, multiple sclerosis, and lupus erythematosus (reviewed in).
Intrinsic immune responses during viral infection utilize host cell transcriptional machinery to inhibit viral gene expression. Specialized proteins that bridge intrinsic and innate immunity are pattern recognition receptors (PRRs). These proteins are responsible for the early detection of the virus by recognizing specific viral molecular moieties, including nucleic acids, proteins, and lipids (reviewed in (Thompson et al., 2011)). This recognition elicits a temporal cascade of immune signaling pathways. For example, recognition of viral DNA by cellular DNA sensors triggers the signaling cascade that leads to the production of type I interferons (IFNs), usually IFNβ. This expression further induces the amplification of IFNs and interferon-stimulated genes (ISGs), which occurs several steps after the initial sensing event (reviewed in (Haller, Kochs, and Weber, 2006). The presence of viral DNA can be recognized by PRRs in multiple subcellular compartments, including the cytosol, endosomes, and the nucleus (as reviewed in (Crow, Javitt, and Cristea, 2015)). This sensing event signals through adaptor molecules in a sensor-dependent manner and leads to one of several outcomes—either transcription of type I and type II interferons or other cytokines (through translocation of IRFs and NF-κB), or cleavage of cytokines to produce their mature forms as exemplified by the function of inflammasomes (reviewed in (Crow, Javitt, and Cristea, 2015)). Cell death that occurs prematurely, e.g. before viral particle packaging and egress, limits the ability of the virus to spread. Therefore, another host defense mechanism utilizes cell death pathways as a mean to inhibit virus infection.
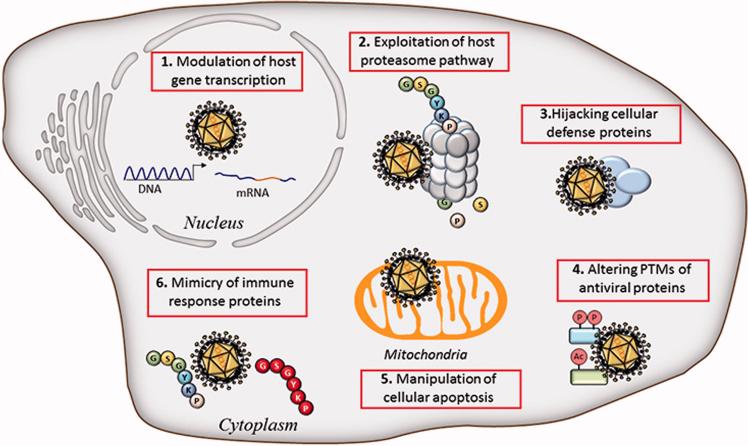
In response to these diverse mechanisms of host defense, viruses have evolved a plethora of tactics to ensure their replication and spread. The co-evolution of viruses with their hosts is in fact reflected by the sophisticated mechanisms of counter-defense to host antiviral strategies. Here, we review the current knowledge of the mechanisms DNA viruses use to inhibit or evade early host defense responses (Figure 1). Specifically, we start in chapter 1 by discussing viral means to inhibit host defense gene transcription. In chapter 2, we present an overview of the virus exploitation of the host proteasome pathway for the degradation of cellular defense factors. We next review the ability of viruses to hijack host defense proteins, to regulate immune effectors by post-translational modifications, and to inhibit cellular apoptosis for efficient replication (chapters 3, 4, and 5). In chapter 6, we present the striking ability of viruses to mimic host factors in order to act as signaling sinks of cellular antiviral components. Altogether, these strategies reveal examples of convergent and divergent evolution across DNA viruses, highlighting the efficacy of viral immune evasion to secure successful replication and spread. In closing, we highlight emerging themes in viral immune evasion strategies and the value of understanding these strategies for the development of antiviral therapies.
Figure 1.
Strategies used by DNA viruses to evade early host immune responses. Shown is an overview of viral strategies discussed in this review. Nuclear viral strategies include modulation of host gene expression while other strategies involve utilizing proteasome and apoptosis pathways, modulation of protein functions via PTM or hijacking, and viral mimicry of host immune proteins.
1. Antagonism of host defense gene transcription by multiple mechanisms
Viruses have effective means to inhibit de novo transcription of critical host antiviral genes, as well as fast-acting strategies to antagonize the stability of host RNA already present within the cell upon infection. Of the latter, the capacity of viruses to advantageously regulate cellular gene expression can be met through mechanisms of host shutoff, viral micro RNAs, and selective targeting of RNA, all of which ultimately suppress a significant degree of cellular protein synthesis. This subsection presents an overview of these mechanisms. Additional post-transcriptional modulatory mechanisms employed by viruses, including RNA editing and RNA splicing, is reviewed elsewhere (Hogg, 2016).
1.1. Virus induced destabilization of mRNA
An efficacious host shutoff approach used by viruses is the promotion of global RNA decay (Covarrubias et al., 2009; Glaunsinger and Ganem, 2004; Kwong and Frenkel, 1987; Lin et al., 2004; Read and Frenkel, 1983; Rowe et al., 2007; Sato et al., 2002). Notably, several α- and γ-herpesviruses have been well studied for their modulation of mRNA stability through viral ribonuclease-induced mRNA degradation (Figure 2, left). As an example, the human herpes simplex virus-1 (HSV-1) protein UL41, or virion host shutoff (vhs), is an mRNA-specific endonuclease that is packaged within the tegument protein layer of mature virion particles (Elgadi, Hayes, and Smiley, 1999; Everly et al., 2002; Schek and Bachenheimer, 1985). Upon cell entry, vhs is dispersed throughout the cytoplasm, rapidly degrading host and viral mRNAs prior to de novo viral gene expression, seizing the synthesis of cellular protein, and disaggregating preexisting polyribosomes (Smiley, Elgadi, and Saffran, 2001). These roles serve to prevent the expression of cellular immune factors and divert host machinery for viral replication. Importantly, the temporal degradation of host and viral mRNA levels promotes a shift in the cascade of viral gene expression from early to late viral genes. Mutations in vhs display reductions in viral titers as a result of increased levels of early gene transcripts and delays in late gene expression (Oroskar and Read, 1987; Oroskar and Read, 1989; Read and Frenkel, 1983).
Figure 2.
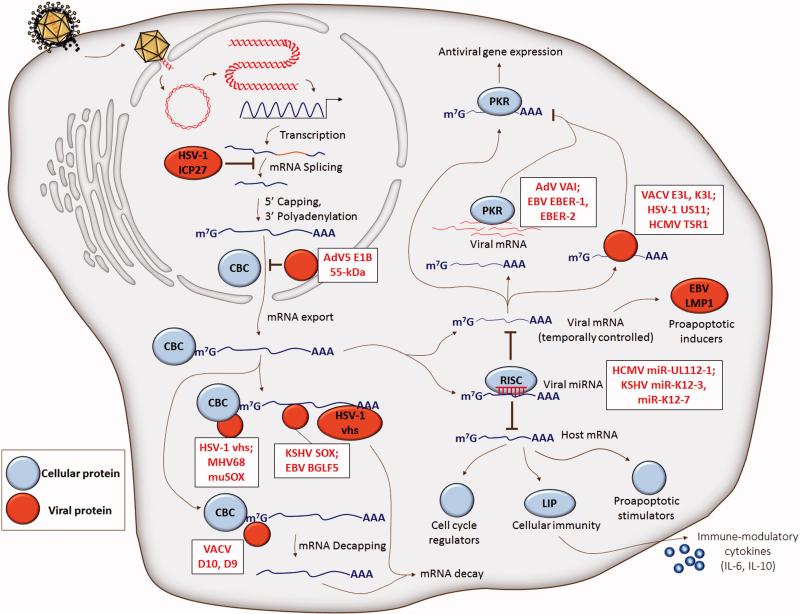
DNA virus immune evasion by antagonism of host defense gene transcription. Cellular (blue) mRNA is targeted for destabilization before and after the generation of mature mRNA that is exported to the cytoplasm with CBC. Viral (red) ribonuclease-induced fragmentation of RNA polymerase II-derived transcripts is initiated at different primary cleavage sites, including the 5’ 7-methylguanosine cap, 3’ poly-adenylated tail, and 3’ untranslated region. Decapping and destabilization of mRNA is facilitated by VACV D10 and D9 proteins. Adenovirus E1B 55-kDa associates with E4 Orf6 to selectively promote the export of viral late mRNAs to the cytoplasm, while inhibiting the export of cellular mRNA. Viral mRNA can act as a sequestration sink for host defense factors like PKR, ultimately suppressing antiviral gene expression. Viral microRNAs target specific cellular genes to either downregulate host defense gene expression, or attenuate antigen presentation through cytokine-dependent means of inhibiting immune cell maturation. Cap-binding protein complex, CBC; herpes simplex virus-1, HSV-1; Epstein-Barr virus, EBV; vaccina virus, VACV; adenovirus, AdV; human cytomegalovirus, HCMV; Kaposi's sarcoma virus, KSHV; protein kinase R, PKR.
Viruses may exhibit convergent evolution as evidenced by mRNA degradation through multiple means of mRNA targeting and different positions of primary cleavage. Viral RNases, like HSV vhs, Kaposi's sarcoma-associated herpesvirus (KSHV) ORF37 (SOX), murine herpesvirus 68 (MHV68) ORF37 (muSOX), and Epstein-Barr virus (EBV) ORF37 homolog (BGLF5), cleave mRNA into fragments. In a study exploring the patterns of RNA cleavage by the aforementioned DNA viral factors, it was determined that, in addition to evidence for global mRNA degradation (Rivas, Schmaling, and Gaglia, 2016), these viral RNases displayed preferential cleavage of RNA polymerase II-derived transcripts that contain a 7-methylguanosine 5’ cap and 3’ poly-adenylated tail (Gaglia et al., 2012). Through one of its RNA destabilizing mechanisms, vhs associates preferentially with the 5’ end of transcripts through the cap-binding protein complex, whereas the methods of mRNA targeting by SOX, muSOX, and BGLF5 await future characterization (Figure 2, left). Endonucleolytic cleavage of mRNA subsequently occurs near the cap for muSOX, or at internal positions for vhs, SOX, and BGLF5. Ultimately, all of these viral factors require cellular Xrn1 5’ – 3’ exonuclease activity to degrade the cleaved mRNA body (Covarrubias et al., 2011). Vhs was also shown to specifically target RNA containing AU-rich elements within the 3’ untranslated region of mRNA (Rivas, Schmaling, and Gaglia, 2016; von Roretz et al., 2011). Vhs directly binds the cellular protein tristetraprolin, which itself binds to AU-rich elements (Shu, Taddeo, and Roizman, 2015), representing a possible procedure through which vhs mediates targeted degradation. Of particular relevance to viral immune evasion, AU-rich elements are prevalent within mRNAs encoded by innate immune response genes (Schott and Stoecklin, 2010), which may highlight an evolved viral mean to specifically dampen host antiviral immunity in tandem with mechanisms of global mRNA destabilization. It has been further elucidated that an immediate-early protein of HSV-1, the multifunctional ICP27, contributes to the inhibition of mRNA processing by being necessary for and sufficient to impair proper splicing of premature mRNAs containing introns (Hardy and Sandri-Goldin, 1994). A staggering percentage of cellular genes contain introns, compared to no more than 5% of HSV-1 genes, of which a majority are immediately-early genes (Smiley, Elgadi, and Saffran, 2001). In this way, HSV-1 may indirectly avoid interferences with gene expression by not having an obligation to undergo splicing.
In addition to its characterized functions in viral replication, the involvement of vhs in preventing the expression of critical host defense genes supports a model of host shutoff as a means of immune evasion. Mice infected with nonfunctional vhs displayed elevated innate immune responses activated by the host (Duerst and Morrison, 2004; Murphy et al., 2003; Pasieka et al., 2008). Specifically, vhs counteracts host immune factors by facilitating the degradation of antiviral interferon stimulated genes (ISG), including ZAP and viperin (Shen et al., 2014; Su, Zhang, and Zheng, 2015). It was determined that the ectopic expression of ZAP prevented the infection and progression of a mutant HSV-1 virus strain lacking vhs, while overexpression of vhs promoted the degradation of ZAP mRNA (Su, Zhang, and Zheng, 2015). Similarly, wild-type HSV-1 infection was capable of reducing levels of viperin mRNA, whereas UL41-null HSV-1 infection did not (Shen et al., 2014). Given that ISGs can restrict the replication of DNA and RNA viruses (Chin and Cresswell, 2001; Shen et al., 2014), these findings point to the evolution of an effective HSV-1 immune evasion tactic, and whether other viruses also have mechanisms of viperin inhibition remains to be determined.
The regulation of mRNA turnover allows a cell to rapidly adjust gene expression in an adaptive manner. Vaccinia virus (VACV), the prototype for the laboratory study of poxviruses, modulates both viral and host gene expression during the course of infection (Parrish, Resch, and Moss, 2007). VACV D10 protein was identified to contain mRNA decapping activity, an enzymatic activity not previously shown to be encoded by a DNA virus. This discovery provides a mechanism for control of both host and viral gene expression. The conservation of D10 proteins in all sequenced poxviruses suggests an evolved mechanism of immune system subversion that is an active area of research. In fact, it was subsequently established by the same laboratory that the decapping activity of D10 is dependent on a Nudix hydrolase motif that is also present in the VACV D9 protein, which shares 25% sequence identity with D10 (Parrish and Moss, 2007).
A viral strategy for destabilizing cellular mRNA by preventing its nuclear export has also been reported. Adenovirus 5 was shown to inhibit cellular mRNA export, while promoting the selective export of viral late mRNAs to the cytoplasm (Gonzalez et al., 2006). This selectivity is achieved by the adenovirus E1B 55-kDa protein, which interacts with the viral E4 Orf6 and four cellular proteins (cullin 5, Rbx, and elongins B and C) to generate an infection-specific E3 ubiquitin ligase (Harada et al., 2002). It was also established that the export of viral late mRNAs occurs through the Nxf1/Tap pathway. However, it remains unclear if the associated substrates of the E3 ligase are targeted for proteasome-dependent degradation.
1.2 Translational inhibition by viral microRNAs
Alternative to inducing active mRNA degradation, DNA viruses encode and express non-coding RNAs, or microRNAs (miRNA), approximately 19-23 nucleotides long that can target specific cellular genes (Figure 2, right). Although the roles of a majority of viral miRNAs have yet to be fully understood, early assessments of viral miRNA target genes in immune and non-immune cells suggests that viral miRNAs are critical for subverting the host antiviral immune response (Boss and Renne, 2010). For example, the microRNA miR-UL112-1 of the human cytomegalovirus (HCMV) was shown to downregulate the expression of a ligand involved in the killing of NK cells, the major histocompatibility complex class I-related chain B (Stern-Ginossar et al., 2007). Aside from direct miRNA inhibition of host defense genes, Qin et al. provide the first report of viral miRNA-dependent induction of cytokines by targeting a host defense gene (Qin et al., 2010). It was determined that in human myelomonocytic cell lines, KSHV miR-K12-3 and miR-K12-7 target an isoform of the immune-stimulatory transcription factor C/EBPb, LIP, culminating in the expression of interleukins IL-6 and IL-10. As IL-6 and IL-10 have roles in inhibiting the maturation of dendritic cells, thereby preventing antigen presentation, the targeting of miR-K12-3 and miR-K12-7 to LIP represents an indirect immune evasion strategy mounted during KSHV infection. Aside from directly targeting the cellular immune pathway, viral miRNAs also target proapoptotic genes. As an example, the EBV miRNAs miR-BART16, miR-BART17-5p, and miR-BART1-5p were shown to target and attenuate the expression of LMP1, an EBV transforming factor that facilitates cellular growth and survival through the stimulation of NF-κB (Lo et al., 2007). Temporal control of the downregulation of LMP1 by EBV miRNAs ensures cell longevity by preventing LMP1-induced apoptosis and NF-κB inhibition.
1.3. Viral RNA as a sequestration sink for host defense factors
To hinder the production of virion particles during infection, cells possess an army of pattern recognition receptors and interferon-induced antiviral molecules that bind foreign nucleic acid and arrest viral protein synthesis. Of these antiviral effectors, the double stranded RNA (dsRNA)-dependent cellular protein kinase R (PKR) is critically involved in translational inhibition and stasis, as well as apoptosis induction, of infected cells. PKR binds viral dsRNA and phosphorylates the cellular translation initiation factor, eIF2α, thereby stalling protein synthesis. Many protein antagonists of PKR, derived from DNA viruses, have been identified (Figure 2, right). These include VACV E3L and K3L, HCMV TSR1, and HSV-1 US11 (Davies et al., 1993; Marshall et al., 2009; Peters et al., 2002). As an example, VACV E3L sequesters dsRNA, thus preventing viral mRNA-dependent activation of PKR or other dsRNA receptors (Griffiths and Coen, 2005). From the perspective of non-protein antagonists, several viruses express non-coding, double-stranded, or structured RNA molecules, which competitively inhibit host dsRNA receptors involved in cellular immunity. An area of active research is the identification of viral factors that use self-RNA as a proxy to subvert the role of PKR in translational inhibition by binding to PKR and preventing activation of the catalytic domain. These include RNA derived from adenovirus and EBV (Langland et al., 2006; Launer-Felty, Wong, and Cole, 2015). The adenovirus RNA molecule, virus-associated RNA-I, or VAI, which is an approximately 160 nucleotide dsRNA molecule, is one such self-RNA decoy that binds to PKR. Structural analyses, including small angle x-ray scattering and analytical ultracentrifugation, established that VAI adopts a flat duplex conformation stabilized by a pseudoknot, facilitating the formation of an apical stem and several loop regions. The association of the apical stem with the central domain of VAI generates a monomeric PKR binding site, the interaction of which not only occurs with high affinity and precision, but also in a fashion that avoids kinase activation (Dzananovic et al., 2014; Launer-Felty, Wong, and Cole, 2015). The constitutively expressed EBER-1 and EBER-2 RNAs of EBV are composed of 167 and 172 nucleotides, respectively (Glickman, Howe, and Steitz, 1988), and similarly sequester PKR from RNA products of viral replication (Laing, Matys, and Clemens, 2001; Launer-Felty, Wong, and Cole, 2015). Stem-loop IV of EBER-1 binds PKR at two different regions, providing the theoretical capability of sequestering two binding domains of PKR with one EBER-1 molecule (Vuyisich, Spanggord, and Beal, 2002).
In addition to protein and RNA products that prevent PKR activation, it is possible that DNA viruses encode other competitive inhibitory factors of cellular dsRNA receptors. While such viral factors remain to be discovered for DNA viruses, a genomic region of the RNA virus, Hepatitis C virus, contains a structured internal ribosomal entry site (IRES) that is also a competitive inhibitor of PKR-mediated dsRNA binding. Considering the larger genome sizes of many DNA viruses relative to RNA viruses, it is possible that the genomic luxury of many DNA viruses affords the capacity to encode a competitive IRES element within polycistronic mRNA. Current knowledge of existing IRES elements in DNA viruses is limited to that in the thymidine kinase gene of HSV-1 (Griffiths and Coen, 2005), upstream of the simian virus 40 vp3 coding region (Yu and Alwine, 2006), and upstream of the shrimp pathogen White Spot Syndrome Virus (Han and Zhang, 2006). Despite this, it is conceivable that these and other as-of-yet uncovered IRES elements in DNA viruses similarly act as competitive inhibitors of cellular dsRNA receptors.
2. Viruses exploit the proteasome pathway to destroy host antiviral proteins
The proteasome pathway is critical for maintaining cellular homeostasis. This pathway is responsible for regulating protein turnover for the majority of intracellular proteins ((Rock et al., 1994) and reviewed in (Lecker, Goldberg, and Mitch, 2006)). These proteins are post-translationally modified by a chain of ubiquitin (Ub) molecules, which triggers their recognition by the 26S subunit of the proteasome. Via a stepwise process, the multisubunit proteasome complex degrades the proteins into small peptides. Among its roles in maintaining cellular homeostasis, the proteasome pathway is critical for the regulation of the immune responses. For example, this pathway is responsible for generating peptides for antigen presentation by major histocompatibility complex (MHC) class I and initiation of adaptive immune responses (Rock et al., 1994). Furthermore, the proteasome-mediated degradation of polyubiquitinated IκB is an important step in innate immune response, necessary for the translocation of NF-κB to the nucleus for transcriptional activation of type I IFNs (reviewed in (Karin and Ben-Neriah, 2000)).
Given the importance of this pathway in regulating immune response proteins, viruses have evolved mechanisms that exploit this pathway to specifically inhibit host defense factors. In particular, many DNA viruses have acquired the property to encode for viral E3 ubiquitin ligases (Randow and Lehner, 2009). One example is ICP0 from HSV-1 that functions as an E3 ligase through its RING finger domain (Boutell, Sadis, and Everett, 2002). Another viral strategy is the recruitment of host E3 ligases. This section provides an overview of the viral proteins shown to target host proteins for degradation via the proteasome pathway. Specifically, this three-part section reviews the virus-induced degradation of proteins involved in intrinsic defense, linking intrinsic to innate immunity, and innate immune responses.
2.1. Virus-induced degradation of intrinsic immunity proteins
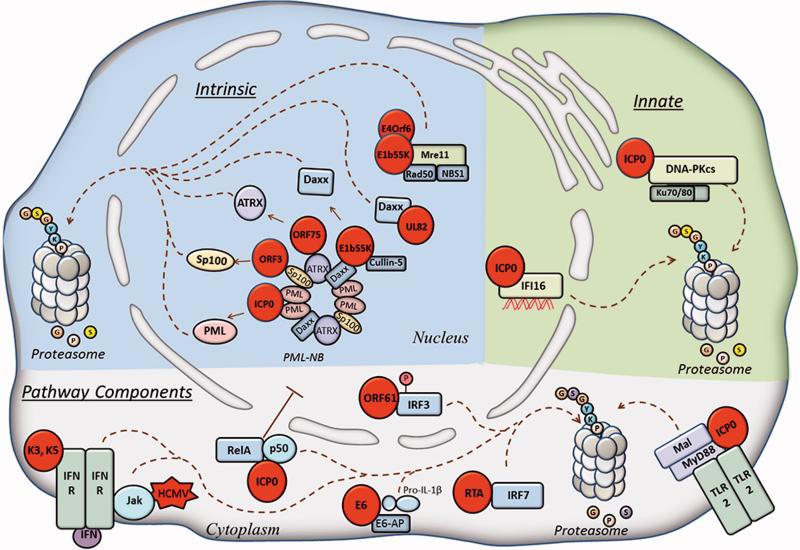
A major reported target of viral E3 ubiquitin ligases is PML, the main constituent of PML nuclear bodies (PML-NBs). PML-NBs are composed of several transiently-interacting proteins that form distinct puncta in the nuclear matrix, and have been shown to have antiviral functions (reviewed in (Everett and Chelbi-Alix, 2007) and (Scherer and Stamminger, 2016)). PML-NBs were reported to suppress a number of herpesviruses, including HSV-1(Everett et al., 2006), (HCMV) (Ahn and Hayward, 2000), EBV (Sivachandran, Wang, and Frappier, 2012) and KSHV (Marcos-Villar et al., 2009), as well as other DNA viruses, including adenovirus (Doucas et al., 1996), human papilloma virus (HPV) (Stepp, Meyers, and McBride, 2013), and parvovirus (Mitchell et al., 2014). The cell intrinsic defenses of PML-NBs are due in part to the epigenetic silencing of viral genomes (reviewed in (Scherer and Stamminger, 2016)). In agreement with its antiviral activity, PML knockdown leads to increased HSV-1 titers following infection in human fibroblasts (Diner et al., 2015). Several viral E3 ligases were reported to target PML for proteasomal degradation. The HSV-1 protein ICP0 was shown to directly target PML (Boutell, Orr, and Everett, 2003) (Figure 3, left). The details surrounding PML degradation are complicated, as studies have shown both SUMO-dependent (most isoforms of PML)(Boutell et al., 2011) and -independent (PML.I) (Cuchet-Lourenço et al., 2012) modes of degradation by ICP0. However, both virus-mediated modes of PML degradation lead to inhibition of intrinsic antiviral defenses. Similar to ICP0, the protein encoded by ORF75c of murine γherpesvirus-68 directly targets PML for degradation, an event found essential for efficient virus propagation (Ling et al., 2008).
Figure 3.
Viruses exploit host proteasome pathway for immune evasion. Intrinsic immune response (blue) include proteins of PML-NBs or the DDR that are targeted for proteasome-mediated degradation by several viruses (see text). Innate immune (pink) DNA sensors DNAPKcs and IFI16 are both targeted for degradation by HSV-1 viral protein ICP0. Immune response pathway components (within grey shading of cell) are targeted for degradation by several viruses. Viral proteins are represented by red circles. Dotted lines represent movement.
In addition to PML, other components of PML-NBs, including hDaxx, ATRX, and Sp100, were also shown to have antiviral properties, either collectively (as PML-NBs) or individually (Lukashchuk et al., 2008; Saffert and Kalejta, 2006; Stepp, Meyers, and McBride, 2013; Wagenknecht et al., 2015). Therefore, it may not be surprising that all viruses mentioned above as inhibited by PML-NBs have acquired mechanisms for targeting its diverse components either for degradation or dispersal (the latter of which will be discussed in chapter 3). For example, during infection with HCMV, hDaxx is recruited to the viral major immediate early promoter (MIEP) to suppress transcription of viral immediate-early (IE) genes. This function is counteracted by the HCMV tegument protein pp71 (UL82) (Saffert and Kalejta, 2006), which induces hDaxx degradation in a proteasome-dependent manner (Hwang and Kalejta, 2007; Saffert and Kalejta, 2006). However, it is interesting that this event seems to occur in an ubiquitin-independent manner (Hwang and Kalejta, 2007). This is not unprecedented for pp71, as this protein was also shown to induce the degradation of retinoblastoma (Rb) family members in a proteasome-dependent, ubiquitin-independent manner (Kalejta and Shenk, 2003). Another example is ORF75 of the γ-herpesvirus KSHV, which encodes a tegument protein that leads to the relocalization of hDaxx, dispersal of sp100, and degradation of ATRX (Full et al., 2014). This protein is homologous in sequence to phosphoribosylformylglycineamide amidotransferase (FGARAT), a cellular biosynthesis enzyme (Full et al., 2014). Indeed, another γ-herpesviruses that encodes a viral FGARAT is the herpesvirus saimiri from ORF3 that targets sp100 for degradation ((Full et al., 2012) and reviewed in (Scherer and Stamminger, 2016)). Lastly, the adenovirus (Ad5) protein E1B-55K, in complex with cellular cullin-5, has also been shown to target hDaxx for proteasomal-dependent degradation (Schreiner et al., 2010).
Host intrinsic immune responses also include the inhibition of viral gene transcription by using the host DNA damage response (DDR) pathway. Ad5 has been shown to target the DNA damage response protein Mre11 for degradation. Mre11, Rad50 and NBS1 form the MRN complex, which localizes to sites of DNA damage as part of the dsDNA break repair system. E4 mutant Ad5 infection elicits a cellular DDR that results in concatemerization of viral genomes (Weiden and Ginsberg, 1994). The consequence is that the viral genomes are too large to be packaged into virions (Boyer, Rohleder, and Ketner, 1999). To counteract this, the wild-type Ad5 virus has the ability to disperse the Mre11 complex, targeting Mre11 for degradation (Stracker, Carson, and Weitzman, 2002). This event relies on the viral proteins E4orf6 and E1b55K that together form a complex with E3 ligase activity (reviewed in (Weitzman and Ornelles, 2005)).
2.2. Degradation of host proteins that bridge intrinsic and innate immune responses
The recognition of viral DNA by host PRRs constitutes a core mechanism through which cells initiate innate immune responses (Crow, Javitt, and Cristea, 2015). Not surprisingly, viruses have acquired means to target PRRs for degradation to inhibit their DNA sensing functions, i.e., binding to viral DNA and stimulation of IFNs or pro-inflammatory cytokines. ICP0 of HSV-1 was shown to target the DNA sensor DNA-PK (Figure 3, right). DNA-PK is a protein complex that consists of the catalytic subunit (DNA-PKcs) and the heterodimer Ku70/80. Although this complex is more commonly known for its role in DNA repair processes, such as its contribution to non-homologous end-joining after DNA damage (Lieber et al., 2003), it was more recently reported as a sensor of viral DNA (Ferguson et al., 2012). Lees-Miller et al. found that ICP0 is required for a reduction in the half-life of DNA-PKcs (Lees-Miller et al., 1996). Shortly after, Roger Everett's group determined that ICP0 is necessary and sufficient for the proteasome-mediated degradation of DNA-PKcs (Parkinson, Lees-Miller, and Everett, 1999). Furthermore, it was determined that this activity by ICP0 is advantageous to HSV-1 replication, as the virus replicated more efficiently in DNA-PKcs-depleted cells (Parkinson, Lees-Miller, and Everett, 1999). HSV-1 infection induces a DNA damage response in cells. Therefore, it is tempting to speculate that the degradation of DNA-PKcs is beneficial to the virus in two ways: inhibition of a DNA damage response, and debilitating innate immune DNA sensing by DNA-PK. The role of ICP0 in inhibiting DNA sensing proteins is further exemplified by the degradation of the nuclear DNA sensor IFI16. Orzalli et al. showed that early during HSV-1 infection there is a requirement for ICP0 E3 ligase activity for the prompt degradation of IFI16 (Orzalli, DeLuca, and Knipe, 2012). Another study showed that ICP0 alone is not sufficient for inducing IFI16 degradation, suggesting that this degradation is also dependent on the function of another viral immediate-early protein yet to be determined (Cuchet-Lourenço et al., 2013). In agreement with these observations, another study showed that IFI16 degradation was slower and only partial during infection with a mutant virus devoid of all IE genes except for ICP0 (d106) (Diner et al., 2015). Additionally, degradation did not occur during infection with an ICP0-RING finger mutant virus, suggesting that ICP0 is necessary, but not sufficient for IFI16 degradation. Further investigations will be needed to determine which other viral or host proteins may be involved in targeting IFI16 for degradation. Nevertheless, the reduction in protein levels of IFI16 was shown to be favorable for viral replication. Altogether, the ability of viruses to target DNA sensors for degradation highlights an effective mean through which viruses can inhibit host immune defenses (Figure 3).
2.3. Virus-mediated degradation of innate immune pathway components
Many innate immune pathways exist as a compilation of events carried out by distinct protein interactions and post-translational modifications. These pathways are triggered by PRRs and signal through adaptor molecules to initiate the nuclear translocation of transcriptional activators; e.g. NF-κB, and interferon regulatory factor -3 and -7 (IRF3 and IRF7). IRF3 is a cytoplasmic protein that translocates to the nucleus upon its activation and dimerization, an event induced by its phosphorylation by Tank binding kinase 1 (TBK1) (Fitzgerald et al., 2003). During virus infection, the function of IRF3 is to stimulate the transcription of type I interferons (John Hiscott, 1999). ORF61 of varicella zoster virus (VZV) encodes for a phosphoprotein that contains a RING finger domain homologous to the equivalent domain in ICP0 of HSV-1 (Moriuchi et al., 1992). Activation of IRF3 was found to be inhibited during infection with VZV (Zhu et al., 2011). ORF61 targets activated IRF3, leading to its degradation in a proteasome-dependent manner; the RING finger domain of ORF61 was required for this activity (Zhu et al., 2011). As only active (phosphorylated) IRF3 was targeted by ORF61, this suggests a mechanism through which viral proteins recognize immune activation, avoiding the unnecessary degradation of an inactive protein that may increase cellular stress. Interestingly, the bovine ICP0 (bICP0) from bovine herpesvirus was also shown to trigger IRF3 degradation (Saira, Zhou, and Jones, 2007). However, ICP0 of HSV-1 does not target IRF3 for degradation, but instead inhibits it through an alternative mechanism (discussed in the virus hijacking section of this review). It is rather remarkable that several alpha-herpesviruses that encode homologous ICP0 genes have been shown to inhibit IRF3, yet the manner in which this inhibition ensues can differ.
Similar to IRF3, IRF7 is a transcriptional regulator of type I interferons and interferon-stimulated genes (ISGs). IRF7 is activated upon virus infection, which causes its dimerization in the cytoplasm, and subsequent translocation to the nucleus to bind to the interferon-stimulated response element (ISRE) on the promoters of interferon genes (Marié et al., 2000). The immediate-early transcriptional regulator RTA of KSHV was shown to directly target IRF7 for degradation in a proteasome-dependent manner (Yu, Wang, and Hayward, 2005) (Figure 3, bottom). Interestingly, although RTA is not known to have a traditional HECT or RING domain, it was found that the N-terminal domain of RTA was required for IRF7 ubiquitination and subsequent degradation in vitro, suggesting that this region encodes a non-canonical E3 ligase domain (Yu, Wang, and Hayward, 2005).
Toll-like receptor 2 (TLR2) is a PRR that is expressed on the cell membrane (Figure 3, bottom). Although primarily known for its function against bacterial, fungal and parasitic infection (Akira, Takeda, and Kaisho, 2001), TLR2 was recently shown to be activated in response to virus infection ((Compton et al., 2003; Kurt-Jones et al., 2004) and reviewed in (Akira, Uematsu, and Takeuchi, 2006)). During virus infection, its role in host defense is to recognize viral proteins at the cell surface and induce a signaling cascade that leads to an innate immune response. Within this signaling cascade are adaptor proteins, such as myeloid differentiation factor 88 (MyD88) and MyD88 adaptor-like (Mal), which initiate the activation of NF-κB that in turn results in the expression of pro- and anti-inflammatory cytokines (e.g., IL-6, reviewed in (Akira, Uematsu, and Takeuchi, 2006)). HSV-1 infection was shown to inhibit TLR2 downstream signaling and decrease of IL-6 levels (van Lint et al., 2010). This group went on to discover that MyD88 and Mal proteins are degraded through the proteasome in a manner dependent on the E3 ligase activity of ICP0 (van Lint et al., 2010).
The HSV-1 ICP0 protein was also shown to be responsible for the degradation of the p50 component of the NF-κB heterodimer (Zhang et al., 2013). Although there are several NF-κB family members, the predominant form of NF-κB exists as a heterodimer consisting of the p65/RelA and p50 subunits (Albert S. Baldwin, 1996). NF-κB –mediated interferon transcription requires the nuclear translocation of NF-κB. Zhang et al. showed that the p65/RelA subunit was inhibited from translocating to the nucleus as a direct result of the RING finger domain of ICP0 binding to p65/RelA. Interestingly, although the viral inhibition of NF-κB subunits p65/RelA and p50 both require the ICP0 RING finger domain, only the p50 subunit was found to be degraded (Zhang et al., 2013).
The downstream effect of type I IFN expression is the induction of ISGs. The signaling cascade is through the well-established Jak-STAT pathway; thus, Jak1 is an important regulator of ISG expression via the concerted efforts of Jak-STAT signaling (as reviewed in (Amsler, Verweij, and DeFilippis, 2013)). HCMV has acquired a mechanism to perturb this signaling, and Jak1 was shown to be degraded during infection. Although the specific viral protein mediating this activity has not yet been identified, the use of DNA polymerase inhibitors had minimal effect on the degradation event, suggesting the involvement of a tegument, immediate-early or early viral protein (Miller et al., 1998). The initiation of Jak-STAT signaling is through ligand binding of IFN receptors. Thus, another way viruses can inhibit ISG expression is by directly targeting IFN receptors. Indeed, KSHV proteins K3 and K5 were found to specifically target IFNγ receptor 1 for ubiquitination and subsequent degradation (Li et al., 2007).
In addition to the majority of PRRs that are known to trigger signaling cascades leading to transcriptional activation of antiviral cytokines, PRRs that trigger inflammasome formation culminate in the maturation of pro-inflammatory cytokines. For example, upon binding to viral DNA, the DNA sensor absent in melanoma 2 (AIM2) recruits apoptosis-associated speck-like containing a CARD (ASC) protein and pro-caspase-1. This interaction leads to the auto-proteolytic cleavage of caspase-1 and concludes with the cleavage of pro-interleukin-1β and secretion of the mature interleukin-1β (IL-1β). This, and related pro-inflammatory cytokines, have important roles in cell differentiation, cell death pathways, and priming adaptive immune responses (Lamkanfi and Dixit, 2014; Stienstra et al., 2010). From the virus perspective, the secretion of these cytokines can impede virus production at multiple levels. Although many routes of virus inhibition of inflammasomes have been described (see viral mimicry section), thus far one virus, HPV, has been shown to inhibit inflammasome function using a proteasome-dependent approach. The E6 protein of HPV has pleiotropic functions. One function of this protein is to commandeer the cellular protein E6-associated protein (E6-AP), an E3 ubiquitin ligase, a process extensively studied in the deregulation and degradation of the pro-apoptotic factor p53 (Huibregtse, Scheffner, and Howley, 1991). However, this interaction between E6 and cellular E6-AP was recently shown to inhibit downstream inflammasome activity (Niebler et al., 2013). The association of E6 with E6-AP has proven beneficial in HPV transformed keratinocytes, where they can target pro-IL-1β for proteasomal degradation, thus inhibiting the action of the NALP3 inflammasome (Niebler et al., 2013).
Of note, DNA viruses can also use viral factors to regulate viral protein levels. For example, the HSV-1 protein ICP0 was shown to regulate the degradation of the tegument protein pUL46 (Lin et al., 2013). pUL46 can impact the initial transcription of immediate early viral genes, and the temporal control of its protein levels was proposed as important for the progression through the virus life cycle. It remains to be determined whether this is a virus-mediated mechanism for limiting the levels of viral transcripts within the cell, which could lead to premature cell death. Altogether, it has become evident that DNA viruses, either by encoding their own E3 ligases or by taking over cellular ones, have efficient mechanisms for inhibiting host responses at multiple points within intrinsic and innate immune response pathways.
3. Viral hijacking of host defense proteins
A compelling strategy employed by DNA viruses to subvert cellular immune surveillance is to hijack host defense and immune-stimulatory proteins by means of sequestration and functional inhibition. This subsection highlights prominent protein classes within antiviral response pathways targeted by DNA viruses during early stages of infection (Figure 4).
Figure 4.
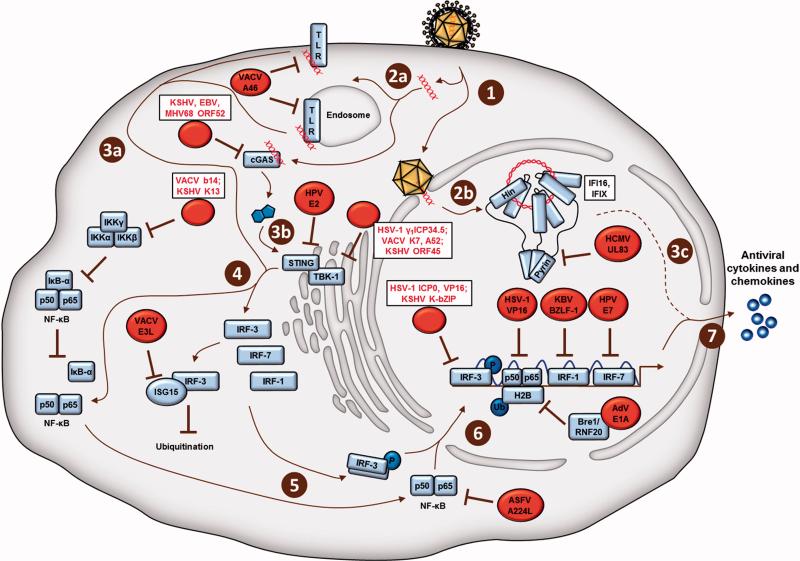
DNA virus hijacking and sequestration of host defense proteins. Depicted are viral proteins (red) shown simplistically as targeting the most downstream relevant host proteins (blue) involved in each of the following steps of cellular antiviral immune signaling. Detailed viral immune subversion mechanisms are described in the text. Upon early stages of infection of a cell with a DNA virus (1), the viral DNA genome is detected by cytoplasmic DNA sensors (2a), or nuclear DNA sensors (2b). Plasma membrane- or endosomal-localized TLRs (3a), as well as cGAS (3b) bind cytoplasmic viral DNA and ultimately activate several protein components of cellular immune signaling (4), including the transcription factors NF-KB, IRF-3, IRF-7, and IRF-1. Upon cytoplasmic activation, the transcription factors translocate to the nucleus (6), bind to regions of the host genome, and upregulate the expression of antiviral cytokines and chemokines (7). Herpes simplex virus-1, HSV-1; Epstein-Barr virus, EBV; vaccina virus, VACV; adenovirus, AdV; human cytomegalovirus, HCMV; Kaposi's sarcoma virus, KSHV; African swine fever virus, ASFV; Toll-Like receptor, TLR; histone 2B, H2B; phosphorylation, P; monubiquitination, Ub.
Immune-stimulatory proteins, such as those canonically associated with the STING-TBK-1-IRF-3 cytosolic signaling pathway, as well as with NF-κB and IRF transcriptional activity, have characterized functions in promoting the expression of antiviral cytokines. The immune signaling pathways initiated by DNA virus infections indeed intersect at the stage of transcription factor stimulation, making transcription factors amenable targets for virus factors to dampen and dismantle cellular immunity (Unterholzner and Bowie, 2008). As such, it is not surprising and well-documented that many viruses contain factors to prevent their activation. As IRF-3 has additionally been identified as an inhibitor of proapoptotic gene expression, it would stand to reason that viruses may target IRF-3 for inactivation. This would enable the establishment and maintenance of persistent and/or latent infections. The HSV-1 immediately-early transactivator ICP0 binds IRF-3, sequestering it within nuclear bodies alongside its co-activating proteins, CbP and p300 (Melroe et al., 2007). This sequestration event prevents transcription factor association with host gene binding sites that are required for type I IFN induction. A trans-inducing tegument factor, VP16, is one of the latest HSV-1 factors characterized to both inhibit NF-κB and hinder IRF-3 recruitment to its cofactor, CbP (Xing et al., 2013). It has been posited that a critical function of the interferon effector protein ISG15 is to directly counteract the proteolysis of IRF-3 during viral infection. ISG15 has been found to covalently bind to and stabilize IRF-3, preventing its ubiquitination during infection. Of note, VACV E3L was determined to target and dismantle ISG15 (Guerra et al., 2008), which speaks to the impressive repertoire of immune evasion strategies used by viruses.
In addition to sequestering IRF-3 and NF-κB, as described in chapter 2, the transcription factors IRF-7 and IRF-1 are known viral targets for immune evasion. As a protein with a relatively short half-life, IRF-7 protein stability is highly regulated in a cell type-specific manner (Prakash and Levy, 2006). During infection, IRF-7 destabilization is accelerated in many cell types in a proteasome-dependent fashion, as reviewed earlier. Alternative to proteasome-mediated degradation, the transcription factor K-bZIP of KSHV competes with IRF-3 for binding sites within the IFN-β promoter, thereby blocking promoter activation (Lefort et al., 2007). An immediate-early protein of EBV, BZLF-1, directly binds IRF-7 to inhibit its association with host genes (Unterholzner and Bowie, 2008). The elucidation of oncoprotein-mediated inhibition of transactivating IRF activity during HPV infection has reinforced the conceived supposition that immune response and viral immune evasion may be intimately tied to cancer (Park et al., 2000; Ronco et al., 1998; Um et al., 2002). IRF-1 has been characterized as a tumor suppressor based on the presence of low IRF-1 levels in a number of human cancers, its function in promoting apoptosis and suppressing cell proliferation in vitro, as well as the correlation between loss of IRF-1 and oncogene deregulation in vivo (Mboko et al., 2016; Nozawa et al., 1999; Savitsky et al., 2010). Furthermore, epidemiological studies have proposed that viral evasion factors of cellular immunity can be etiological agents of oncogenesis (Um et al., 2002). An early viral gene and implicated oncoprotein, E6, has been shown to bind IRF-3 and attenuate its ability to induce a type I IFN immune response following infection with the RNA virus, Sendai virus (Ronco et al., 1998). The HPV E7 gene encodes another oncoprotein that has been strongly implicated in the malignancy of cervical cancer (Park et al., 2000; Um et al., 2002). Upon exploring the effect of E7 expression on IFN signaling, researchers established that the amino terminal of E7 directly binds IRF-1 and attenuates the induction of IFN-β (Um et al., 2002).
Despite the contribution of NF-κB to the generation of antiviral chemokines and type I interferon, it has also been implicated in inhibiting the onset of virus infection-dependent apoptosis and thus facilitating the proliferation of host cells. This latter function seems functionally divergent from its role in intrinsic and innate immunity because apoptosis is a host defense mechanism that can annihilate virus-infected cells and prevent viral spread to neighboring cells (see chapter 5). In fact, several viruses promote NF-κB function to avoid apoptosis. VACV b14 prevents the phosphorylation, and thus activation of a NF-κB inhibitor, IKKB (Chen et al., 2008). KSHV protein K13 interacts with the IKKa-IKKb complex to prevent NF-κB inhibition (Matta et al., 2007). To reconcile these NF-κB functions from a pro-viral perspective, viruses can temporally control NF-κB in a biphasic manner. During early stages of infection, viruses can prevent NF-κB immune-activating roles through direct inhibition or proteasome-targeted degradation of NF-κB. As an example, the African Swine Fever Virus (ASFV) protein A238L is a homologue of a direct NF-κB cellular inhibitor, Ikbα, which sequesters NF-κB in the cytoplasm during early stages of infection (Tait et al., 2000). As the infection progresses, the ASFV protein A224L is temporally expressed later to activate NF-κB and inhibit caspases for the prevention of apoptosis. Evidently, the regulation of NF-κB activity during viral infection can be highly complex and temporally manipulated in response to both viral and cellular interests.
Aside from transcription factor inhibition by DNA viruses, several viral factors act on components upstream of either the STING-TBK-1-IRF-3 pathway or of transcription factor activity. The HSV-1 leaky-late protein γ1ICP34.5 was shown to bind directly to TBK-1, sequestering the kinase from interacting with and activating IRF-3 (Verpooten et al., 2009). Studies characterizing viral immune evasion factors have substantiated the functional relevance of cellular immune modulatory mechanisms. In a genome-wide expression screen of primary keratinocytes expressing the early HPV protein E2, STING and IFN-κ were two of 92 genes associated with innate immunity and were found to be suppressed (Sunthamala et al., 2014). In addition, the early HPV protein E7 and the adenovirus protein E1A were found to suppress IFN responses in immortalized HEK293 and HeLa cells (Lau et al., 2015). By immunoaffinity purification, both of these viral proteins were shown to interact with STING (Lau et al., 2015), suggesting that these viruses specifically target the STING pathway to inhibit early immune responses. A cogent demonstration of how understanding viral immune evasion can provide insight into mechanisms of cellular signaling is the exploration of VACV K7 protein inhibition of the TBK-1-IKKε complex (Schroder, Baran, and Bowie, 2008; Soulat et al., 2008). The authors determined that K7 targeted the human RNA helicase, DEAD-box protein 3 (DDX3), whereupon the TBK-1-IKKε complex was inhibited from upregulating IRFs and thus IFN-β levels. It was established for the first time that DDX3 functions downstream of PRRs, comprised of both RNA receptors and cytoplasmic DNA receptors, and indeed associates with the TBK-1-IKKε complex to stimulate IFN induction.
The multitude of examples by which DNA viruses inhibit components of the STING-TBK-1-IRF-3 signaling axis underscores the functional relevance of this pathway in intrinsic and innate immune defense, as well as the conservation of viral evasion strategies. The nuclear-replicating herpesvirus HCMV was found to prevent the activation of the nuclear-localized PRRs IFI16 and IFIX (Diner et al., 2015) in a non-degradative manner. The major tegument protein of HCMV, pUL83, was shown to specifically target the pyrin domain of IFI16 and IFIX, suppressing homotypic pyrin oligomerization and attenuating PRR-mediated antiviral immune responses (Li, Chen, and Cristea, 2013). In agreement with the involvement of pUL83 in suppressing immune responses, primary fibroblasts infected with a mutant HCMV strain lacking UL83 were able to induce over 10-fold greater levels of antiviral cytokines compared to wild-type HCMV infection. So, HSV-1 and HCMV evolved different means to suppress IFI16 functions, one destructive and one just inhibitory. Although these findings highlight the importance of IFI16 in antiviral response, it is puzzling that HCMV would evolve to maintain IFI16 following infection. This may be in part due to the roles of IFI16 in transcriptional regulation, and may represent the repurposing of IFI16 functions for the benefit of viral replication (Cristea et al., 2010). More studies will be needed to elucidate the diverse functions of IFI16 and IFIX in the context of infection. Another bona-fide DNA-binding PRR and known to function upstream of the STING-TBK-1-IRF-3 axis, cyclic GMP-AMP synthase (cGAS), was recently found to be inhibited by a tegument protein with homologues in the gammaherpesviruses KSHV, EBV, and MHV68 (Wu et al., 2015). In this study, it was established that the KSHV tegument protein ORF52 inhibited cGAS enzymatic activity of generating STING-activating cyclic dinucleotides by binding both to cGAS and DNA. The inhibitory titration of IFI16 and IFIX molecules by pUL83, as well as of cGAS by ORF52, reflects a “safety in numbers” approach to evade cellular immunity, considering that tegument proteins such as pUL83 are present in thousands of copies per virion particle (Varnum et al., 2004).
Alternative targets for DNA viruses outside of the STING-TBK1-IRF axis similarly act by binding and inhibiting host defense proteins. Of these immune modulatory pathways, toll-like receptor signaling is actively targeted by DNA viruses. The VACV protein A46 has been found to bind and sequester the TLR adaptor proteins Myd88, TRIF, Mal, and TRAM (reviewed in (Bowie and Unterholzner, 2008). In addition, VACV A52 binds the TBK-1-IKK stimulatory kinase, IRAK-2, and TNFR-associated factor TRAF6, ultimately preventing cellular immune signaling (Bowie and Unterholzner, 2008). The deficiency of either of these two proteins within virion particles resulted in lower infectivity in mouse models (Harte et al., 2003; Stack et al., 2005). Considering that VACV retains other factors that repress host defense (i.e., D9, D10, E3L, K7, b14), an evolved viral necessity may be a multi-step inhibition strategy of the same cellular immune response pathways. In lieu of this, viruses may activate particular viral immune evasion factors in a cell type-specific manner. Regardless, the final goal of such viral subversion is to effectively disrupt the partially redundant cellular immune pathways, which converge at the level of transcription factor activation, as well as antiviral cytokine and chemokine expression.
While still an emerging area of research, the study of viral infection on the posttranslational modification (PTM) state of virus and host factors has revealed dynamic and modification-specific regulation of proviral strategies and host defense mechanisms (discussed in chapter 4). Specific to viral hijacking, an uncommon example of PTM-mediated viral antagonism of a host immune protein is provided by the adenovirus E1A protein, the most immediately expressed viral protein upon viral infection (Fonseca et al., 2012). E1A had been well established as a critical viral transactivator for both manipulating gene transcription, as well as reprogramming cells to maintain active cell cycle state. It was subsequently determined that E1A achieved reprogramming of quiescent cell replication by altering histone 3 acetylation (Horwitz et al., 2008). An interaction screen identified an association between E1A and hBre1/RNF20, a protein involved in the monoubiquitination of histone 2B and initiating ISG expression upon viral infection (Fonseca et al., 2012). To inactivate components of ISG induction and to promote viral replication, E1A was found to bind and dissociate the hBre1 complex, thereby preventing histone 2B monoubiquitionation and subsequent expression of ISGs. The establishment of a key transcriptional effector, E1A, as a regulator of cellular histone PTM highlights the essentiality of protein modification in both viral immune evasion and host defense. More recently, ORF45, the immediate-early protein found in the tegument of KSHV virion particles, was shown to compete with IRF-7 for phosphorylation by acting as an alternative substrate for the kinases IKKε and TBK-1 (Liang et al., 2012). While it is tempting to conjecture that viral immune-inhibitory factors that have been identified to target an IRF may correspondingly target other IRFs, evidence exists for cases of substrate specificity. For example, ORF45 phosphorylation inhibition of IRF-7 was not recapitulated for IRF-3, signifying a kinase-indirect mechanism of ORF45 immune subversion (Liang et al., 2012). The exploration of dynamic PTM regulation during viral infection warrants future research and is further discussed in chapter 4.
4. Viruses can regulate antiviral proteins via post-translational modification
Another important mechanism through which viruses can inhibit host defenses involves the regulation of host protein post-translational modifications (PTM). One of these modifications, ubiquitination, was reviewed in the section on virus induced proteasome-dependent degradation. However, recent years have demonstrated that other PTMs are also dynamically modulated during viral infections, including phosphorylation, acetylation, and SUMOylation (Figure 5).
Figure 5.
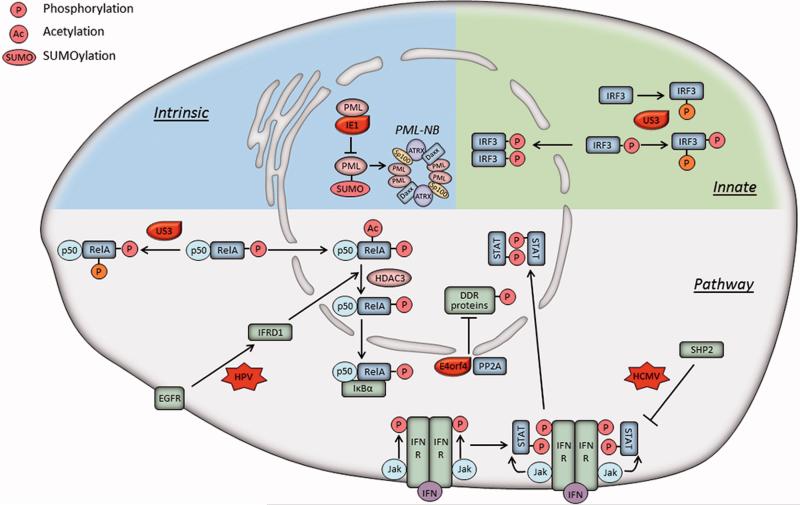
Viral regulation of host proteins via post translational modifications. HSV-1 US3 protein can hyperphosphorylate IRF3 and RelA to prevent their nuclear localization and transcription activation functions. HCMV can inhibit the Jak-mediated STAT1 phosphorylation in an SHP2-dependent manner, preventing STAT1-mediated induction of IFN-inducing genes. E4orf4 of adenovirus, together with PP2A, reduces phosphorylation of DDR proteins to inhibit DNA damage response. HPV infection enhances HDAC3-mediated RelA deacetylation by upregulating IFRD1 via the EGFR pathway, which decreases the duration of NF-κB signaling. HCMV IE1 binds to PML and limits its SUMOylation, repressing the formation of PML-NBs. Phosphorylation, P; acetylation, Ac; SUMOylation, SUMO.
Induction of protein phosphorylations has been linked to virus regulation of host innate immune response and downstream immune pathways. It is well established that the phosphorylation of IRF3 at Ser396 and its subsequent dimerization are required for its nuclear translocation and induction of type I IFN. During HSV-1 infection, the viral tegument Ser/Thr kinase US3 acts in immune evasion by targeting IRF3 (Wang et al., 2013). Specifically, US3 hyperphosphorylates IRF3 at an atypical site, Ser175. This inhibits its dimerization and nuclear translocation, thereby blocking IFN-β production. Consistently, the infection with a US3 kinase-dead (KD) mutant virus induces increased levels of IFN-β when compared to a wild type virus. The HSV-1 US3 kinase also targets the NF-κB signaling pathway by interacting with the NF- κB transcription factor subunit RelA (p65) (Wang et al., 2014). Activation of NF-κB regulates genes involved in production of antiviral cytokines and regulation of apoptotic processes (Chen and Greene, 2004). Similar to the IRF3 inhibition mechanism, US3 triggers the hyperphosphorylation of RelA at Ser75, blocking its nuclear translocation and repressing the expression of inflammatory cytokines (Wang et al., 2014). The US3 KD mutants do not block the nuclear translocation of RelA, indicating that the hyperphosphorylation is sufficient to prevent its nuclear accumulation.
Viral infections can also induce the phosphorylation of other host proteins, including histone deacetylases (HDACs). During infection with DNA viruses, the viral genome is rapidly associated with histones upon its entry into the nucleus. Therefore, HDACs can have important functions, as the histone acetylation status impacts viral gene transcription (Guise et al., 2013). The HCMV pUL97 kinase triggers the phosphorylation of HDAC1, thereby disrupting its binding to the major immediate early promoter of HCMV and aiding viral immediate early gene expression (Bigley et al., 2013). The HSV-1 US3 kinase also mediates phosphorylation of HDAC1 and HDAC2, thus blocking the silencing of viral genes by HDACs (Poon, Gu, and Roizman, 2006; Walters et al., 2010). In addition, ORF66p of VZV, homologous to HSV-1 US3, hyperphosphorylates HDAC1 and HDAC2 to release the viral genome from HDAC inhibition (Walters et al., 2009). The phosphorylation of HDACs may not be directly linked to viral immune evasion; however, as HDACs provide transcriptional regulation for a wide range of genes, it remains to be determined whether immune response genes are also affected.
Viral infections can also regulate cellular proteins by inhibiting phosphorylation events necessary for host defense. The phosphorylation of the signal transducer and activator of transcription 1 (STAT1) is required for its function as a transcription factor for IFN-inducing genes. This represents a step in the signal relay downstream of receptors for IFNs. Binding of IFNs to their receptors induces IFN receptor oligomerization and phosphorylation via Janus tyrosine kinases (Jaks) (Figure 5). Phosphorylation of IFN receptors leads to the subsequent recruitment and Jak-mediated phosphorylation of STAT proteins, allows STATs to dimerize and translocate into the nucleus, where they function as transcription factors. During HCMV infection, the Jak mediated STAT1 phosphorylation is inhibited (Baron and Davignon, 2008). The cellular tyrosine phosphatase SHP2 was proposed to be involved in the dephosphorylation of STAT1 and Jak1 (Baron and Davignon, 2008; You, Yu, and Feng, 1999), and this process was shown to be dependent on HCMV replication (Baron and Davignon, 2008).
Another host defense pathway regulated by phosphorylation is the cellular DDR. During infection with DNA viruses, the viral replication intermediates and the linear double stranded DNA are interpreted as DNA damage, thereby activating DDR. Viral replication can be inhibited by misrepair of viral DNA. Many DNA viruses have evolved mechanisms to inhibit the DDR, thus promoting viral replication (Weitzman, Lilley, and Chaurushiya, 2010). The adenovirus E4orf4 protein, in conjunction with the host phosphatase PP2A, was shown to reduce the phosphorylation of DDR proteins from the ataxia-telangiectasia mutated (ATM) and ATM- and Rad3-related (ATR) pathways, including ATM, 53BP1, Smc1, Nbs1, Chk1, and Chk2 (Brestovitsky et al., 2016). This decreased phosphorylation reduces DDR signaling, increasing the presence of damaged DNA. Although not a traditional DDR protein, another DNA binding protein that is phosphorylated by a viral protein is Barrier to Autointegration Factor (BAF). Unphosphorylated BAF acts to inhibit VACV replication; this antiviral function is circumvented by the VACV B1 kinase via phosphorylation of BAF (Wiebe and Traktman, 2007).
Viral regulation of protein acetylation was also shown to be relevant during infection. The small E1A protein of adenovirus has the ability to modulate histone H3 acetylation, thereby regulating host gene transcription and maintaining an active cell cycle (Horwitz et al., 2008). E1A was also reported to bind the tumor suppressor RB and the cellular acetyltransferase p300 (Ferrari et al., 2014). This interaction promotes p300 acetylation of K873/K874 in RB and K239 in E1A, which ultimately leads to chromatin condensation and the transcriptional repression of host defense genes (e.g., THBS1 and CTGF).
NF-κB pathway is regulated by inhibition of acetylation during HPV infection. RelA (p65) is known to be acetylated by p300/CBP and deacetylated by HDAC3 (Chen et al., 2001). This deacetylation promotes the binding of RelA, p50 and IκBα, leading to the nuclear export of the NF-κB complex. Thus, this reversible acetylation controls the duration of nuclear NF-κB action. During HPV infection, the RelA Lys310 acetylation is impaired. HPV induces the overexpression of interferon-related developmental regulator 1 (IFRD1) by upregulating epidermal growth factor receptor (EGFR). IFRD1 upregulation enhances the HDAC3-mediated RelA Lys310 deacetylation, thereby suppressing antiviral cytokines expression (Tummers et al., 2015).
Changes in protein SUMOylation have also been connected to viral infection. HSV-1 infection was shown to lead to the decrease in a large number of protein SUMOylation sites (Sloan et al., 2015). Given the requirement for PML SUMOylation in the formation of PML-NBs, this modification has been of particular interest. PML-NB function to silence viral gene expression, and as previously mentioned in this review, viruses have acquired different mechanisms to antagonize PML-NBs (Scherer and Stamminger, 2016). HSV-1 ICP0 was shown to use both SUMO-targeted and SUMO-independent mechanisms to induce the degradation of PML-NB. ICP0 acts as a SUMO-targeted ubiquitin ligase (STUbL), inducing the proteasomal degradation of SUMO-conjugated proteins, such as PML and other NB components (Boutell et al., 2011). In contrast, HCMV IE1 inhibits the PML-NB formation using a proteasome independent pathway. It binds to PML and limits its SUMOylation during infection, resulting in a dispersal and inactivation of PML-NBs. The central hydrophobic domain of HCMV IE1 was shown to be required for PML binding and deSUMOylation (Lee et al., 2004).
The regulation of the PTM status during the progression of a viral infection was not only shown to act in virus immune evasion, but also as a mechanism used by host cells to suppress infection. For example, the phosphorylation of the HCMV major tegument protein pUL83 was shown to impact its function in host immune evasion (Li, Chen, and Cristea, 2013). pUL83 phosphorylation at Ser364 was induced by cell host kinases. Importantly, this phosphorylation inhibited the ability of pUL83 to suppress the oligomerization of the interferon-inducible protein IFI16. IFI16 is a nuclear DNA sensor necessary for induction of IFN response upon infection with several herpesviruses. Therefore, this phosphorylation event shows that host cells can inhibit virus immune evasion and rescue IFI16 function by PTM of viral proteins (Li, Chen, and Cristea, 2013).
5. Manipulation of apoptosis by DNA viruses
Apoptosis is a well-characterized form of cell death, which plays a key role during viral infection. It constitutes one of the early host defense mechanisms that limits the availability of cells for viral infection, being able to eradicate viral infection early on. Premature cell death can effectively reduce viral replication and limit the progression of viral infection. As indicated in the sections above, viral propagation relies on the cellular machinery, modulating numerous biological processes. Upon entry, viruses disrupt normal cell functions, inducing stress responses that can stimulate the apoptosis pathway. Although most viral infections should trigger apoptosis, viruses have acquired multiple strategies for either suppressing or promoting apoptosis to avoid being eliminated by the host immune system (Roulston, Marcellus, and Branton, 1999; Shen and Shenk, 1995). The existence of viral products in diverse DNA viruses that modulate apoptosis indicates that the regulation of apoptosis is evolutionarily conserved and constitutes a core viral mechanism for inhibition of host defense.
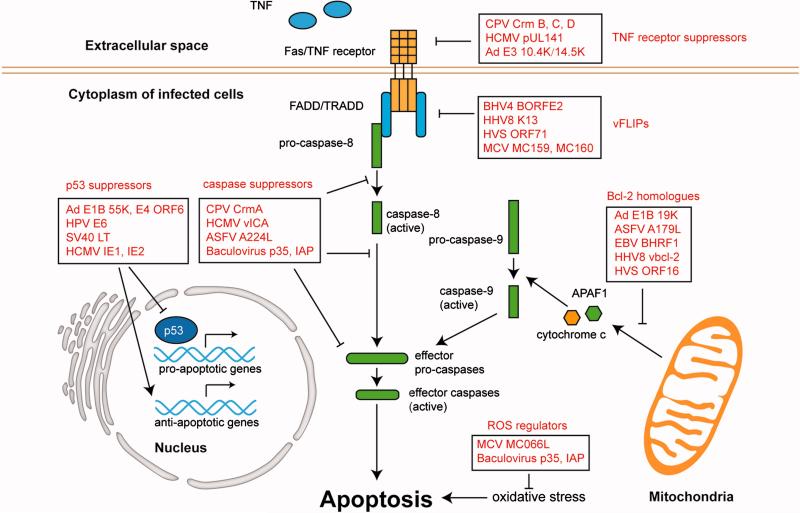
Apoptosis can be initiated through distinct mechanisms, being under the control of multiple cellular factors. The apoptosis inducing stimuli usually fall into two categories, extrinsic and intrinsic pathways, which both converge into the activation of effector caspases (e.g., caspase-3, -6, and -7), committing the cells to programmed cell death (Enari et al., 1998) (Figure 8). The extrinsic pathway begins at the cell membrane, where death receptors bind to signal factors, particularly tumor necrosis factor (TNF) and Fas ligands (Locksley, Killeen, and Lenardo, 2001). Upon binding of TNF or Fas to the TNF receptor, the Fas-associated death domain protein (FADD) and TNFR1-associated death domain protein (TRADD) form the death-inducing signaling complex (DISC), leading to the activation of caspase-8 and downstream effector caspases (Hsu, Xiong, and Goeddel, 1995; Kischkel et al., 1995). The intrinsic pathway is mediated by the mitochondria and held in check by Bcl-2 family members (Adams and Cory, 1998; Chao and Korsmeyer, 1998; Cory et al., 1994; Strasser, Huang, and Vaux, 1997) (Figure 8). Bcl-2 proteins play a pivotal role in deciding whether a cell will live or die by up- or down-regulating mitochondrial membrane permeability (MMP) (Crompton, 1999; Oltvai, Milliman, and Korsmeyer, 1993). Once pro-apoptotic Bcl-2 proteins are recruited to the mitochondrial membrane, MMP is altered, resulting in the release of cytochrome c, apoptosis inducing factor (AIF) and mitochondrial caspase (Susin et al., 1999). These released factors are essential components of the apoptosome, which activates effector caspases. Viruses have evolved strategies to manipulate both the extrinsic and intrinsic pathways. These viral strategies include the use of caspase suppressors, TNF receptor suppressors, viral FLICE (pro-caspase-8) inhibitory proteins (vFLIPs), and viral Bcl-2 homologues. Additionally, viruses inhibit apoptosis using p53 inhibitors, and reactive oxygen species (ROS) regulators. This section will review these strategies adopted by DNA viruses to block apoptosis pathways, as well as briefly describe examples of viral promotion of apoptosis.
5.1. DNA viruses suppress apoptosis
A common viral mechanism for suppressing apoptosis is via inhibition of caspases. Caspases, a group of cysteine proteases, are evolutionarily conserved executioners of the apoptotic response (Cohen, 1997; Shi, 2002; Thornberry and Lazebnik, 1998). All caspases are produced in the form of catalytically inactive pro-caspases, which are activated through cleavage at certain Aspartate residues (Riedl and Shi, 2004). Once the caspase cascade is initiated, the effector caspases will cleave a wide range of cellular proteins, inevitably leading to cell death. A number of viral proteins have been found to inhibit caspase protease activity. Cowpox virus (CPV) encodes a potent wide-spectrum caspase inhibitor, the cytokine response modifier A (CrmA), and homologues of this protein were also found in vaccinia virus as B13R and B22R gene products (Dbaibo and Hannun, 1998). The baculovirus protein p35 and the inhibitor of apoptosis (IAP) also target multiple caspases (Barry and McFadden, 1998; Miller, 1997). The African swine fever virus (ASFV) encodes an IAP homolog that was shown to specifically inhibit caspase-3 (Chacon et al., 1995; Nogal et al., 2001).
Viruses have also developed mechanisms to inhibit cell death by specifically targeting the extrinsic pathway. One strategy is to disrupt the binding between death receptors and TNF/Fas ligands on the cell membrane. An interesting example is provided by the adenovirus E3 gene products, 10.4K and 14.5K, which alter the presentation of Fas on the plasma membrane via its internalization and retention (Shisler et al., 1997), inhibiting TNF-induced apoptosis (Dimitrov et al., 1997; Krajcsi et al., 1996). Crm-B, -C, -D encoded by CPV, are TNFR-like glycoproteins that neutralize TNF (Hu, Smith, and Pickup, 1994; Loparev et al., 1998; Smith et al., 1996). Additionally, HCMV pUL141 restricts TNFR function by targeting TNF-related apoptosis-inducing ligand (TRAIL) to the endoplasmic reticulum, and dampening the stimulation of death receptors (Smith et al., 2013). Viruses also inhibit the extrinsic pathway downstream of TNFR. Some viruses encode viral FLICE (pro-caspase-8) inhibitory proteins (vFLIPs) that contain death-effector domains (DED). These vFLIPs block the interaction between pro-caspase-8 and death domain protein, thereby preventing the activation of caspase-8. MC159 and MC160 are two vFLIPs produced by molluscum contagiosum virus (MCV) that mimic cellular FADD and inhibit Fas-mediated cell death (Shisler and Moss, 2001). Similarly, another vFLIP protein from the bovine herpesvirus, BORFE2, suppresses Fas- and TNFR1-induced apoptosis (Wang et al., 1997). The HCMV UL36 gene encodes a multi-functional cell death suppressor, denoted viral inhibitor of caspase-8 activation (vICA) protein, which binds pro-caspase-8 and inhibits its activation by interference with its recruitment to DISC (McCormick et al., 2010). Similar vFLIPs from other herpesviruses have also been found, including herpesvirus saimiri protein ORF71 (Glykofrydes et al., 2000), equine herpesvirus 2 (EHV-2) protein E8 (Hu et al., 1997), and HHV-8 protein K13 (Tolani et al., 2014).
In addition to the extrinsic inhibitory mechanisms, viruses have also acquired means to suppress intrinsic apoptosis pathways. Many viruses encode Bcl-2 homologues, which in most cases counteract apoptosis triggered in response to abnormal cellular function, stress signals, and changes in mitochondria. One characteristic of the Bcl-2 family is that they can form heterodimers. Therefore, the Bcl-2 homologues encoded by viruses could interact with cellular Bcl-2 proteins and interfere with their functions. One of the first evidences showing that viruses inhibit apoptosis by modulating Bcl-2 function was reported for adenovirus. The adenovirus E1B 19K protein was demonstrated to form heterodimers with diverse Bcl-2 family members, thus maintaining the mitochondria membrane permeability (Rao et al., 1992). Lack of functional E1B 19K gene leads to severe cytopathogenic effect, featured by degradation of chromosomal DNA, premature cell death, and reduced yield of viral progeny (Granville et al., 1998; Rao, Modha, and White, 1997). This phenotype could be reversed by over-expression of host Bcl-2. Later, other virus families were also shown to encode homologues of the Bcl-2 family. ASFV expresses the Bcl-2 homolog A179L, which prevents apoptosis initiated by the interferon-induced double-stranded RNA-activated protein kinase (Brun et al., 1996). Anti-apoptotic viral factors encoded by herpesviruses have been extensively studied, and a number of Bcl-2 factors have been identified. Productive replication of HCMV depends on the expression of viral mitochondrial inhibitor of apoptosis (vMIA; pUL37 x1). vMIA, the smallest protein product of UL37, is localized to mitochondria early in infection (Goldmacher, 2002). It maintains MMP by neutralizing Bax, a pro-apoptotic Bcl-2 protein (Ma et al., 2012). Similarly, Epstein-Barr virus (EBV), human herpes virus 8 (HHV-8), and herpesvirus saimiri (HVS) encode Bcl-2 homologues that recruit Bcl-2 family proteins (Bax or Bak), keeping mitochondria from proapoptotic stimuli and stabilizing MMP (Derfuss et al., 1998; Khanim et al., 1997; Marshall et al., 1999; Nava et al., 1997; Niedobitek et al., 1991).
Aside from proteins, viral RNA of DNA viruses can also regulate the intrinsic apoptosis pathway. For example, HCMV encodes an abundant 2.7-kb non-coding RNA (β2.7), which interacts with the mitochondrial complex I, thereby maintaining adenosine triphosphate production necessary for the progression of the virus life cycle (Reeves et al., 2007) . This is a mean through which β2.7 can protect infected cells from mitochondrial stress, and subvert mitochondria-induced apoptosis.
A well-established viral mechanism for inhibiting apoptosis is the targeting of p53. As p53 controls the transcription of Bcl-2 family members (Miyashita et al., 1994), this also provides an indirect modulation of Bcl-2 proteins. Several DNA oncogenic viruses tackle p53-mediated apoptosis either by blocking or downregulating p53. In adenovirus, E1B-55K and E4 ORF6 products bind p53 and block its transcriptional activation (Dobner et al., 1996; Kratzer et al., 2000; Martin and Berk, 1998). Similarly, SV40 large T antigen (LT) interacts with p53, together with retinoblastoma tumor suppressor protein (Yanai and Obinata, 1994) (pRB), altering their control of cell death. Additionally, as stated in the above section describing virus-induced protein degradation, the HPV E6 product disrupts p53 DNA binding ability and targets p53 for rapid proteasome-mediated degradation (Scheffner et al., 1990). HCMV proteins were also reported to target p53. Specifically, the immediate early proteins IE1 and IE2 interact with p53 in the nucleus, reducing its tumor suppressor function and down-regulating the transcription of proapoptotic genes (Chiou et al., 2006; Sainz et al., 2005; Taylor and Bresnahan, 2005; Taylor and Bresnahan, 2006).
In addition to inhibiting extrinsic and intrinsic apoptosis pathways, viruses also suppress stimuli that trigger apoptosis. Elevated ROS levels, which are linked to apoptosis, have been noticed during viral infections (Harman, 1992; Schwarz, 1996). Some viral products function to maintain oxidative levels and prevent subsequent apoptosis. For example, the MCV MC066L protein exhibits homology to glutathione peroxidase, a key regulator of cellular reactive oxygen species (ROS). MC066L blocks apoptosis mediated by UV radiation or peroxide treatment (Shisler et al., 1998). The baculovirus p35 gene product and IAP also play roles in suppressing ROS-induced apoptosis (Sah et al., 1999).
5.2. DNA viruses can also benefit from apoptosis
The virus life cycle is a complex and finely-tuned process, during which the suppression and induction of apoptosis need to be balanced in order to achieve effective replication and dissemination. Although a growing number of viral proteins have been shown to block apoptosis, some viruses take advantage of the induction of apoptosis at certain stages of infection. For example, viruses can benefit from apoptosis during the lytic cycle once they are matured and ready to egress (Bideshi et al., 2005; Mi et al., 2001; Teodoro and Branton, 1997). Apoptosis at a late stage of infection facilitates the dissemination of matured viral particles. Ectromelia virus, a type of poxvirus that infects skin cells, upregulates Fas ligands in the suprabasal layer of conjunctiva, leading to apoptosis in epithelial cells. The shedding of the epithelial layer of conjunctiva may help the transmission of virus-containing apoptotic cells to the surrounding environment (Hu et al., 1997). Additionally, some viruses depend on the proteolytic activity of caspases to establish infection. Aleutian mink disease virus (AMDV), a member of parvoviruses, serves as a good example. It encodes NS1 protein that can be cleaved by caspase-3, generating five distinct NS1 products. These NS1 products localize to the nucleus and acts as transcriptional regulators. Therefore, apoptosis-induced caspase-3 activation is critical for productive viral replication (Best et al., 2003; Best, Wolfinbarger, and Bloom, 2002). Similar mechanisms have also been found for other DNA viruses, including adenoviruses (Grand et al., 2002), MCV (Shisler and Moss, 2001), and HSV-1 (Munger, Hagglund, and Roizman, 2003), but the contribution of these caspase-dependent cleavages to the respective virus life cycles remain to be explored.
In addition to apoptosis, programmed necrosis serves as an important alternate defense mechanism against viral infection. Recent studies demonstrate the importance of programmed necrosis pathways in antiviral response, as well as how viruses have evolved to countermeasure this frontline defense strategy (Chan et al., 2003; Cho et al., 2009; Upton, Kaiser, and Mocarski, 2010; Upton, Kaiser, and Mocarski, 2012). This topic has been elegantly reviewed by (Kaiser, Upton, and Mocarski, 2013).
6. Viral mimicry of host factors to inhibit immune responses
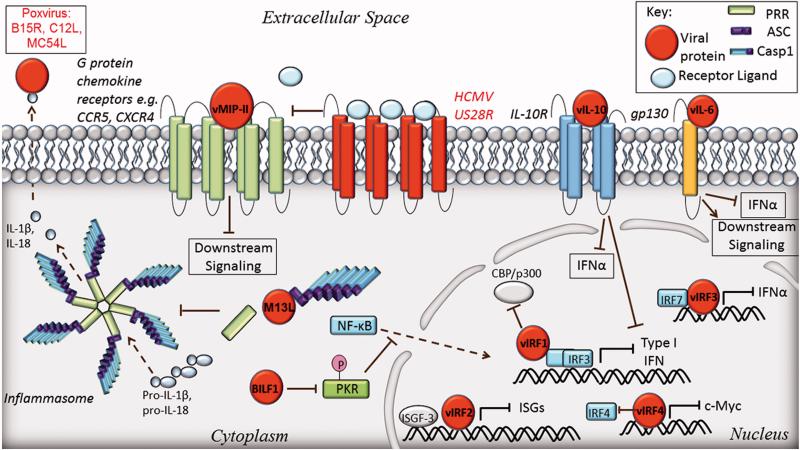
Innate immune signaling molecules, such as cytokines and chemokines, are essential for cellular protection against DNA viruses. These ligand molecules function in an autocrine and paracrine manner, circulating within the host to alert neighboring cells of the existence of danger, as well as to stimulate adaptive immune responses. In this way, they are the essence of global protection against DNA viruses. From the virus perspective, these signaling components are devastating to virus spread, and therefore to virus “survival”. Once cytokines are secreted from an infected cell, it may become more difficult for the virus to inhibit immune responses, as these proteins are now extracellular. Many DNA viruses have evolved an ingenious approach to inhibit these immune response effectors, which involves mimicking cellular cytokines and chemokines. These viral mimics can compete for immune receptor binding sites and inhibit downstream signaling. This section reviews the current knowledge of DNA viruses that mimic host defense proteins as a strategy to evade immune responses.
6.1. Cytokine mimics of herpesviruses
Herpesviruses offer prime examples of viral mimicry of host immune factors. Thus far, viral mimics have been characterized for the β-herpesvirus HCMV, and the γ-herpesviruses EBV and KSHV. HCMV encodes two evasin mimics: an IL-10 cytokine mimic (UL111a ORF, hereafter referred to as cmvIL-10) (Kotenko et al., 2000), and a chemokine receptor mimic encoded by US28 (Gao and Murphy, 1994) (Fig. 7, top). Although numerous cytokine and chemokine homologs have been identified for HCMV, only cmvIL-10 and US28 have been reported as having immuno-suppressive properties. Similarly, EBV also has an IL-10 mimic (BCRF1) (Liu et al., 1997), as well as a cysteine-X-cysteine motif receptor (CXCR) homolog (BILF1) (Beisser et al., 2005; Paulsen et al., 2005). The IL-12p40-like protein of EBV forms the heterodimer IL-27 and modulates adaptive immune responses. The late-acting aspect of protein is beyond the scope of this review, and we refer the reader to (Pflanz et al., 2002). KSHV has many mimics, which include three cysteine-cysteine (CC)-chemokines, an IL-6 mimic, and viral versions of IRF proteins (Burýšek and Pitha, 2001; Burýšek et al., 1999; Joo et al., 2007; Lee et al., 2014; Molden et al., 1997; Yamin et al., 2013).
Figure 7.
Viruses mimic host immune response proteins. Cell membrane: Chemokine receptors (e.g. CCR5, CXCR4) are blocked from binding their ligands by KSHV vMIP-II. HCMV US28 receptor binds promiscuously to a variety of chemokine ligands- leading to fewer molecules being available to bind their receptors. Viral IL-10 (HCMV, EBV) binds human IL-10 receptor inhibiting transcription of IFNα or γ, respectively. The cofactor of the IL-6 receptor gp130 binds viral IL-6 (KSHV) resulting in activation of downstream signaling, or inhibition of IFNα. Cytoplasm: myxoma virus protein M13L inhibits inflammasome formation by binding the pyrin domain of ASC thereby inhibiting the interaction of ASC with the pyrin domain of a PRR. Mature IL-1β resulting from inflammasome activity is inhibited by VACV B15R which acts as a soluble IL-1 receptor. Levels of secreted IL-18 is depleted by several poxvirus family proteins including MCV MC54L, VACV C12L, and variola virus vIL-18BP. EBV protein BILF1 inhibits NF-κB nuclear translocation by modifying PKR phosphorylation status. Nucleus: KSHV viral IRF homologues inhibit transcription of immune response genes by effecting host IRF binding promoter regions.
As mentioned earlier in this review, IL-10 is primarily an anti-inflammatory cytokine, inhibiting the production of pro-inflammatory cytokines as a regulatory mechanism for a controlled immune response. Among the cytokines inhibited by IL-10 are IL-1β, and type I and II IFNs. Viruses use these IL-10 properties to hamper host defenses via pro-inflammatory cytokine expression. cmvIL-10 is a gene that was likely pirated from the host genome (Kotenko et al., 2000) and binds with high affinity to the IL-10 receptor (Jones et al., 2002). The effects of cmvIL-10 binding the IL-10 receptor have shown to be effective (Spencer et al., 2002), despite only having 27% sequence homology to human IL-10 (hIL-10) (Jones et al., 2002). A major producer of type I interferons during infection are plasmacytoid dendritic cells; however, the expression of IFNα was severely abrogated in these cells and shown to be directly due to cmvIL-10 (Chang et al., 2009). The fact that cmvIL-10 has relatively little overlap in sequence to hIL-10 may exemplify an adaptation the virus has acquired to promote the anti-inflammatory actions of IL-10, while not sharing the cell proliferative events stimulated by this cytokine (Spencer et al., 2002). The EBV vIL-10 encoded by BCRF1 has 80% sequence homology to hIL-10 (Vieira et al., 1991). Despite having such high homology to human IL-10, BCRF1 has a much lower affinity for the IL-10 receptor (Liu et al., 1997). This is in contrast to cmvIL-10, which has lower sequence homology but higher affinity for the IL-10 receptor. Despite these differences, both viral IL-10 homologues seem to have acquired many inhibitory functions of hIL-10 whilst not acquiring the stimulatory functions. Indeed, BCRF1 has been shown to inhibit IFNγ early during infection in human peripheral blood mononuclear cells (Swaminathan et al., 1993), but does not share the function of hIL-10 in stimulating MHC II expression on B cells (Go et al., 1990). It is remarkable how two herpesvirus mimics of hIL-10 have evolved different means of procuring anti-inflammatory properties of hIL-10, while not acquiring its pro-stimulatory properties. Interestingly, the parapoxvirus Orf, that infects sheep, goat, and humans, also encodes an IL-10 mimic (Fleming et al., 1997). However, in ovine cells, orfIL-10 retained both the anti-inflammatory and immuno-stimulatory functions of IL-10 (Haig et al., 2002). Although not a herpesvirus, the fact that Orf virus also encodes a viral IL-10 portrays how viruses can adopt similar strategies to evade host immunity.
Contrasting the soluble cytokine cmvIL-10, HCMV also has a chemokine receptor mimic, US28 (Figure 7). US28 is expressed early in the virus life cycle (Zipeto et al., 1999) and has been implicated in having oncogenic and immunoevasive roles (reviewed in (McSharry, Avdic, and Slobedman, 2012)). Here, we focus on the latter activity of US28. A 30% sequence homology exists between US28 and β-chemokine receptor CCR1, and limited homology with CCR2 and CCR5 is within the ligand binding domain (Gao and Murphy, 1994). US28 is rather promiscuous in its binding to chemokines, which includes its binding to CCL2 and CCL5 that are secreted during infection (Bodaghi et al., 1998; Gao and Murphy, 1994). It is postulated that US28 acts as a sink by sequestering these chemokines which would inhibit the chemotaxis of immune cells by outcompeting host CCR binding. Indeed, Bodaghi et al. showed that US28 bound CCL2 and CCL5 in infected fibroblasts, which resulted in reduced extracellular levels of the chemokines (Bodaghi et al., 1998). The promiscuity of receptor-ligand binding is also represented by KSHV vMIP-II except in this case, vMIP-II is the ligand. vMIP-II (a CCL mimic) was shown to bind CCR-1, -2, -5, as well as CXCR4 in COS-7 cells (Kledal et al., 1997). Competition assays utilizing conditioned media containing radiolabeled vMIP-II showed the ability of vMIP-II to displace CCL2 from CCR2 and CCL3 from CCR-1 and -3 (Kledal et al., 1997). Receptor activity was measured by calcium mobilization, where endogenous ligands can increase intracellular calcium, an activity that was blocked by vMIP-II (Kledal et al., 1997). It could be hypothesized that the blocking of intracellular calcium by vMIP-II may indicate that the chemotactic action of these receptors then is also blocked. Indeed, Yamin et al. showed that vMIP-II antagonizes CX3CR1 and CCR5 in naïve natural killer (NK) cells leading to inhibited migration of NK cells (Yamin et al., 2013). They went on to show that KSHV vMIP-I and -3 did not share this inhibitory function (Yamin et al., 2013).
EBV encodes an α-chemokine CXC receptor BILF1 (Paulsen et al., 2005). In contrast to the receptor ligand requisitioning technique used by HCMV US28, BILF1 has not yet displayed the ability to bind CCLs or CXCLs (Beisser et al., 2005). Instead, BILF1 alters the phosphorylation of PKR, an RNA-dependent antiviral protein that aids in apoptosis during viral infection (Beisser et al., 2005). Among PKR functions is the ability to activate NF-κB, a transcription factor that regulates type I IFNs (Williams, 1995). Thus, perhaps the function of BILF1 is to inhibit early interferon signaling during EBV infection.
The cytokine IL-6 has both pro- and anti-inflammatory properties. One vital function of IL-6 is in the transition from innate to adaptive immune responses. IL-6 downregulates chemokines related to neutrophil function while upregulating chemokines that attract monocytes (reviewed in (Scheller et al., 2011)). KSHV encodes an IL-6 mimic (vIL-6) that has ~25% homology to human IL-6 (hIL-6), but has high sequence homology within the binding domain (Neipel et al., 1997) and retains the overall structure of a 4-helix bundle (Chow et al., 2001). The resulting protein is able to bypass the receptor binding requirements that hIL-6 is restricted by. vIL-6 needs only the gp130 cofactor (Figure 7), while hIL-6 requires gp130 in complex with gp80 for effective signaling to occur (Chow et al., 2001; Molden et al., 1997). vIL-6 has the ability to block IFNα signaling during infection by inhibiting Jak-STAT signaling mediated by IFN receptor tyk2 phosphorylation (Chatterjee et al., 2002). Interestingly, the promoter region of vIL-6 contains two sequences that resemble ISREs that were shown to be responsive to IFNα-mediated transcriptional activation (Chatterjee et al., 2002). Since vIL-6 can be induced by IFNα and can also suppress IFNα induction, vIL-6 has cleverly acquired control to enforce a negative feedback loop (Chatterjee et al., 2002).
KSHV has yet another mimicking strategy to inhibit host interferon responses. KSHV encode four genes that have some homology to human IRF genes (Moore et al., 1996). The proteins, termed vIRF-1, -2, -3, and -4, have all been found to have pro-viral roles. However, the mechanisms by which each inhibits the host differ. vIRFs 1-3 have all been shown to inhibit IFN response during infection (Joo et al., 2007; Mutocheluh et al., 2011; Zimring, Goodbourn, and Offermann, 1998). vIRF1 was found to bind both cellular IRF3 and IRF7; however its ability to block IFN responses was limited to its interaction with IRF3 (Lin et al., 2001). Furthermore, vIRF1 did not inhibit the dimerization or translocation of IRF3, but rather blocked its IFN induction function by inhibiting its ability to recruit its cofactors CBP/p300 (Lin et al., 2001). vIRF2 was found to have multiple functions during infection that inhibit IFN expression. First, vIRF2 was shown to inhibit PKR by preventing its autophosphorylation and therefore, blocking its activation (Burýšek and Pitha, 2001). Later, vIRF2 was found to exploit cellular IRF3 turnover mediated by caspase-3 (Aresté, Mutocheluh, and Blackbourn, 2009). Lastly, vIRF2 restricts ISG expression by inhibiting the interferon stimulating gene factor complex (ISGF-3) (Mutocheluh et al., 2011). The broad activity of vIRF2 displayed by the ability to act early and late highlights the importance of this viral factor in promoting viral “survival”. vIRF3 also has multiple immune inhibitory functions. IRF7-mediated IFNα production was shown to be inhibited by vIRF3 (Joo et al., 2007) and also restricts IRF5-mediated apoptosis signaling (Wies et al., 2009). vIRF4 has recently been found to reduce cellular IRF4 expression as well as to compete with IRF4 for c-Myc promoter binding, thereby inhibiting c-Myc transcription (Lee et al., 2014). c-Myc has many roles in the cell, but of particular relevance is its role in apoptosis regulation (Hoffman and Liebermann, 2008). Since the function of vIRF4 was found to be important for efficient lytic cycle of KSHV, this may suggest that the antiviral nature of c-Myc is to promote apoptosis during infection, an event that is inhibited by vIRF4. Altogether, although these IRF mimics from KSHV may differ in their modes of action, it is clear that the viral mimicry of these important IFN inducers aids in KSHV immune evasion.
6.2 Mimics from Poxvirus target inflammasomes
Poxviruses are somewhat unique in their replication cycles, as unlike most other known DNA viruses, they replicate in the cytoplasm. Therefore, it may be more difficult for the virus to evade host innate immune responses, as most known DNA sensors reside in the cytoplasm. Several cytoplasmic PPRs stimulate the formation of an inflammasome complex (Figure 7), and such complexes were shown to be induced upon infection with poxviruses. To counteract this host response, poxviruses have developed immune evasion strategies based on either inhibition of cytoplasmic inflammasome assembly or on mimicking inflammasome signaling molecules. The inflammasome typically consists of a PRR, the adaptor protein ASC, and caspase-1. The PRR interacts with the ASC via interaction through their pyrin domains (PYD). This is a necessary step for the auto-proteolytic cleavage of pro-caspase-1 to its mature form. Similar to the nuclear HCMV strategy reviewed in chapter 3, Myxoma virus has devised a way to inhibit cytoplasmic PYD-PYD interactions. Myxoma virus is a poxvirus that infects rabbits. This virus produces a protein that shares homology with pyrin domain-containing protein, M13L (Johnston et al., 2005). M13L interacts directly with ASC, leading to decreases in caspase-1 cleavage and diminished IL-1β and IL-18 secretion (Johnston et al., 2005). This suggests that the function of M13L is to outcompete the PRR PYD binding to ASC.
Inhibition of inflammasome formation is not the only trick of mimicry that poxviruses have acquired. Indeed, poxviruses also utilize mechanisms similar to those discussed for herpesviruses, mimicking cytokines or their receptors. Vaccinia virus was found to encode two genes, B15R and B18R, that share homology to human and mouse IL-1 receptors (Smith and Chan, 1991). B15R is able to bind to murine IL-1β and inhibit B and T lymphocyte proliferative responses normally induced by IL-1β (Spriggs et al., 1992), suggesting that B15R is a soluble IL-1β receptor mimic. IL-18 is another product of inflammasomes, which induces IFNγ and stimulates NK cells (reviewed in (Dinarello et al., 2013). Since IL-18 stimulates inflammatory responses, and its misregulation is associated with autoimmune diseases (reviewed in (Dinarello et al., 2013)), strict regulation of IL-18 is required for optimum homeostasis. Cellular regulation of IL-18 involves an IL-18 binding protein (IL-18BP) that has higher affinity for IL-18 than the IL-18Rα does for IL-18 (Dinarello et al., 2013). Most poxviruses have been determined to have IL-18BP encoding genes (Krumm et al., 2008) suggesting that poxviruses intend to nullify the immune stimulatory functions of IL-18 by binding IL-18 before it can bind to its receptor. Several poxviruses have been shown to adopt this approach, including Molluscum antagiosum virus (MCV), variola virus (varv), and VACV. MCV is a poxvirus that causes small raised lesions in humans. Although in healthy individuals the virus is rather benign, in that there is little evidence for inflammation, the lesions can persist for many months or even years, suggesting a suppression of the local immune response (reviewed in (Smith et al., 1999)). MC54L of Molluscum antagiosum was found to bind human IL-18 (Xiang and Moss, 2001), which may contribute to the inhibition of a local immune response around the infected lesions. Varv was the virus responsible for the deadly smallpox disease, prior to its eradication by vaccination. Varv also inhibits IL-18 from binding its receptor by way of varvIL-18BP, which outcompetes hIL-18R for IL-18 binding (Meng, Leman, and Xiang, 2007). Lastly, vaccinia virus was also found to encode an IL-18BP homolog (Smith, Bryant, and Alcamí, 2000). This gene, C12L is thought to be responsible for inhibiting IL-18-mediated immune responses, since infection with a C12L mutant in mice displayed increased pro-inflammatory responses- including elevated IFNγ production and NK and T-cell cytotoxicity (Reading and Smith, 2003).
Closing remarks
Herein, we have highlighted shared and unique mechanisms by which DNA viruses evade cellular intrinsic and innate immune responses. We emphasize the presence of convergent and divergent viral strategies that function to dismantle host immunity. These strategies work either by preventing antiviral cytokine/chemokine expression or by preserving cellular viability in the interests of viral replication and spread. The current knowledge inspires many questions regarding evolution-driven viral designs that serve to abate cellular immune detection. Do DNA viruses have conserved means to exploit immune signaling pathways for the promotion of viral replication? Is there a precedent for the evolutionary loss of molecular signatures recognized by PRRs as foreign? For example, the gamma-herpesvirus MCMV contains an abundance of TLR9-stimulating CpG motifs, whereas such motifs are underrepresented in the related MHV68 genome (Pezda et al., 2011). The absence of CpG motifs in MHV68 begs the question of whether this virus has evolved to specifically evade the recognition by TLR9. Additionally, is the deficiency in lysine residues in many herpesvirus proteins demonstrative of an evolved means to evade cellular ubiquitin-mediated degradation? Furthermore, virus strategies that facilitate latency in hosts is an active area of research that is reviewed elsewhere (Lieberman, 2016). These questions and others expose the intricacies of the virus-host relationship, which warrant continued research.
While some of these immune evasion tactics are starting to be well defined, the knowledge of others remains limited because of the complex interrelationship between viruses and hosts. Insight into these viral subversion strategies not only sheds light upon fundamental cellular signaling pathways, but also provides therapeutic opportunities. To facilitate the generation of antiviral vaccines and treatments, ongoing efforts are focused on removing viral evasion factors while maintaining attenuated virus strains. The characterization of viral targets for immune evasion can also be leveraged as drug targets for autoimmune diseases that require suppression of host immunity. Hence, characterizing these mechanisms can afford the capacity to develop a multifaceted toolkit to regulate cellular immunity in personalized medicine against immunity-related afflictions.
Figure 6.
DNA viruses suppress apoptotic processes. Viruses exploit host extrinsic and intrinsic apoptotic pathways via distinct viral proteins (red). Initiation of the extrinsic pathway is inhibited by virus-encoded TNF receptor suppressors. Downstream of the Fas/TNF receptors, within the cytosol, the activation of caspase-8 is inhibited by vFLIPS. The inhibition of caspase-8 will shut off the caspase cascade. In addition, viruses target other caspases by caspase suppressors. To prevent the activation of the intrinsic pathway, viral proteins mimic host Bcl-2 proteins and regulate mitochondrial membrane potential by recruiting these proteins to the outer membrane of mitochondria. This recruitment will reduce the release of APAF1, cytochrome c, and other mitochondria apoptotic factors. Therefore, these viral Bcl-2 homologues can suppress the activation of caspase-9, and in turn, apoptosis. P53 is critically involved in the regulation of the expression of pro-apoptotic factors, thus it constitutes a target of some viruses. A number of viral proteins function as p53 suppressors that modulate the levels of pro-apoptotic and anti-apoptotic genes. Of note, oxidative stress can be controlled by viral products, such as MCV MC066L, and baculovirus p35 and IAP. These viral proteins prevent host cells from ROS mediated apoptosis, but the underlying mechanisms await further characterization.
Acknowledgments
We are grateful for funding from the National Institutes of Health, grants R01 [GM114141] and R21 [AI102187] to IMC and an F31 AI114240-01A1 to MSC.
Footnotes
Declaration of interest
The authors have no competing interests or conflicts of interest in this review.
References
- 1.Adams JM, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–6. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 2.Ahn J- H, Hayward GS. Disruption of PML-Associated Nuclear Bodies by IE1 Correlates with Efficient Early Stages of Viral Gene Expression and DNA Replication in Human Cytomegalovirus Infection. Virology. 2000;274:39–55. doi: 10.1006/viro.2000.0448. [DOI] [PubMed] [Google Scholar]
- 3.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 4.Akira S, Uematsu S, Takeuchi O. Pathogen Recognition and Innate Immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Albert S, Baldwin J. THE NF-κB AND IκB PROTEINS: New Discoveries and Insights. Annual Review of Immunology. 1996;14:649–81. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 6.Amsler L, Verweij MC, DeFilippis VR. The Tiers and Dimensions of Evasion of the Type I Interferon Response by Human Cytomegalovirus. Journal of Molecular Biology. 2013;425:4857–71. doi: 10.1016/j.jmb.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aresté C, Mutocheluh M, Blackbourn DJ. Identification of Caspase-mediated Decay of Interferon Regulatory Factor-3, Exploited by a Kaposi Sarcoma-associated Herpesvirus Immunoregulatory Protein. Journal of Biological Chemistry. 2009;284:23272–85. doi: 10.1074/jbc.M109.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baron M, Davignon JL. Inhibition of IFN- -Induced STAT1 Tyrosine Phosphorylation by Human CMV Is Mediated by SHP2. The Journal of Immunology. 2008;181:5530–36. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- 9.Barry M, McFadden G. Apoptosis regulators from DNA viruses. Current opinion in immunology. 1998;10:422–30. doi: 10.1016/s0952-7915(98)80116-7. [DOI] [PubMed] [Google Scholar]
- 10.Beisser PS, Verzijl D, Gruijthuijsen YK, Beuken E, Smit MJ, Leurs R, Bruggeman CA, Vink C. The Epstein-Barr Virus BILF1 Gene Encodes a G Protein-Coupled Receptor That Inhibits Phosphorylation of RNA-Dependent Protein Kinase. Journal of Virology. 2005;79:441–49. doi: 10.1128/JVI.79.1.441-449.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Best SM, Shelton JF, Pompey JM, Wolfinbarger JB, Bloom ME. Caspase cleavage of the nonstructural protein NS1 mediates replication of Aleutian mink disease parvovirus. J Virol. 2003;77:5305–12. doi: 10.1128/JVI.77.9.5305-5312.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Best SM, Wolfinbarger JB, Bloom ME. Caspase activation is required for permissive replication of Aleutian mink disease parvovirus in vitro. Virology. 2002;292:224–34. doi: 10.1006/viro.2001.1238. [DOI] [PubMed] [Google Scholar]
- 13.Bideshi DK, Tan Y, Bigot Y, Federici BA. A viral caspase contributes to modified apoptosis for virus transmission. Genes Dev. 2005;19:1416–21. doi: 10.1101/gad.1300205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bigley TM, Reitsma JM, Mirza SP, Terhune SS. Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding. J Virol. 2013;87:7393–408. doi: 10.1128/JVI.02825-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bodaghi B, Jones TR, Zipeto D, Vita C, Sun L, Laurent L, Arenzana-Seisdedos F, Virelizier J- L, Michelson S. Chemokine Sequestration by Viral Chemoreceptors as a Novel Viral Escape Strategy: Withdrawal of Chemokines from the Environment of Cytomegalovirus-infected Cells. The Journal of Experimental Medicine. 1998;188:855–66. doi: 10.1084/jem.188.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boss IW, Renne R. Viral miRNAs: tools for immune evasion. Curr Opin Microbiol. 2010;13:540–45. doi: 10.1016/j.mib.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boutell C, Cuchet-Lourenco D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. Plos Pathog. 2011;7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boutell C, Cuchet-Lourenço D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence. PLoS Pathog. 2011;7:e1002245. doi: 10.1371/journal.ppat.1002245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boutell C, Orr A, Everett RD. PML Residue Lysine 160 Is Required for the Degradation of PML Induced by Herpes Simplex Virus Type 1 Regulatory Protein ICP0. Journal of Virology. 2003;77:8686–94. doi: 10.1128/JVI.77.16.8686-8694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boutell C, Sadis S, Everett RD. Herpes Simplex Virus Type 1 Immediate-Early Protein ICP0 and Its Isolated RING Finger Domain Act as Ubiquitin E3 Ligases In Vitro. Journal of Virology. 2002;76:841–50. doi: 10.1128/JVI.76.2.841-850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–22. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer J, Rohleder K, Ketner G. Adenovirus E4 34k and E4 11k Inhibit Double Strand Break Repair and Are Physically Associated with the Cellular DNA-Dependent Protein Kinase. Virology. 1999;263:307–12. doi: 10.1006/viro.1999.9866. [DOI] [PubMed] [Google Scholar]
- 23.Brestovitsky A, Nebenzahl-Sharon K, Kechker P, Sharf R, Kleinberger T. The Adenovirus E4orf4 Protein Provides a Novel Mechanism for Inhibition of the DNA Damage Response. Plos Pathog. 2016;12:e1005420. doi: 10.1371/journal.ppat.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brun A, Rivas C, Esteban M, Escribano JM, Alonso C. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology. 1996;225:227–30. doi: 10.1006/viro.1996.0592. [DOI] [PubMed] [Google Scholar]
- 25.Burýšek L, Pitha PM. Latently Expressed Human Herpesvirus 8-Encoded Interferon Regulatory Factor 2 Inhibits Double-Stranded RNA-Activated Protein Kinase. Journal of Virology. 2001;75:2345–52. doi: 10.1128/JVI.75.5.2345-2352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burýšek L, Yeow W-S, Lubyová B, Kellum M, Schafer SL, Huang YQ, Pitha PM. Functional Analysis of Human Herpesvirus 8-Encoded Viral Interferon Regulatory Factor 1 and Its Association with Cellular Interferon Regulatory Factors and p300. Journal of Virology. 1999;73:7334–42. doi: 10.1128/jvi.73.9.7334-7342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chacon MR, Almazan F, Nogal ML, Vinuela E, Rodriguez JF. The African swine fever virus IAP homolog is a late structural polypeptide. Virology. 1995;214:670–4. doi: 10.1006/viro.1995.0083. [DOI] [PubMed] [Google Scholar]
- 28.Chan FKM, Shisler J, Bixby JG, Felices M, Zheng LX, Appel M, Orenstein J, Moss B, Lenardo MJ. A role for tumor necrosis factor receptor-2 and receptor-interacting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–21. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 29.Chang WLW, Barry PA, Szubin R, Wang D, Baumgarth N. Human cytomegalovirus suppresses type I interferon secretion by plasmacytoid dendritic cells through its interleukin 10 homolog. Virology. 2009;390:330–37. doi: 10.1016/j.virol.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chao DT, Korsmeyer SJ. BCL-2 family: regulators of cell death. Annu Rev Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 31.Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS. Viral IL-6-Induced Cell Proliferation and Immune Evasion of Interferon Activity. Science. 2002;298:1432–35. doi: 10.1126/science.1074883. [DOI] [PubMed] [Google Scholar]
- 32.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–7. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 33.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 34.Chen RAJ, Ryzhakov G, Cooray S, Randow F, Smith GL. Inhibition of I kappa B kinase by vaccinia virus virulence factor b14. Plos Pathog. 2008;4 doi: 10.1371/journal.ppat.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. P Natl Acad Sci USA. 2001;98:15125–30. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chiou SH, Yang YP, Lin JC, Hsu CH, Jhang HC, Yang YT, Lee CH, Ho LL, Hsu WM, Ku HH, Chen SJ, Chen SS, Chang MD, Wu CW, Juan LJ. The immediate early 2 protein of human cytomegalovirus (HCMV) mediates the apoptotic control in HCMV retinitis through up-regulation of the cellular FLICE-inhibitory protein expression. Journal of immunology. 2006;177:6199–206. doi: 10.4049/jimmunol.177.9.6199. [DOI] [PubMed] [Google Scholar]
- 37.Cho Y, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FKM. Phosphorylation-Driven Assembly of the RIP1-RIP3 Complex Regulates Programmed Necrosis and Virus-Induced Inflammation. Cell. 2009;137:1112–23. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chow D-c, He X-l, Snow AL, Rose-John S, Garcia KC. Structure of an Extracellular gp130 Cytokine Receptor Signaling Complex. Science. 2001;291:2150–55. doi: 10.1126/science.1058308. [DOI] [PubMed] [Google Scholar]
- 39.Cohen GM. Caspases: the executioners of apoptosis. The Biochemical journal. 1997;326(Pt 1):1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, Finberg RW. Human Cytomegalovirus Activates Inflammatory Cytokine Responses via CD14 and Toll-Like Receptor 2. Journal of Virology. 2003;77:4588–96. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cory S, Strasser A, Jacks T, Corcoran LM, Metz T, Harris AW, Adams JM. Enhanced cell survival and tumorigenesis. Cold Spring Harbor symposia on quantitative biology. 1994;59:365–75. doi: 10.1101/sqb.1994.059.01.041. [DOI] [PubMed] [Google Scholar]
- 42.Covarrubias S, Gaglia MM, Kumar GR, Wong W, Jackson AO, Glaunsinger BA. Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1. Plos Pathog. 2011;7 doi: 10.1371/journal.ppat.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Covarrubias S, Richner JM, Clyde K, Lee YJ, Glaunsinger BA. Host Shutoff Is a Conserved Phenotype of Gammaherpesvirus Infection and Is Orchestrated Exclusively from the Cytoplasm. J Virol. 2009;83:9554–66. doi: 10.1128/JVI.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cristea IM, Moorman NJ, Terhune SS, Cuevas CD, O'Keefe ES, Rout MP, Chait BT, Shenk T. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J Virol. 2010;84:7803–14. doi: 10.1128/JVI.00139-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crompton M. The mitochondrial permeability transition pore and its role in cell death. The Biochemical journal. 1999;341(Pt 2):233–49. [PMC free article] [PubMed] [Google Scholar]
- 46.Crow MS, Javitt A, Cristea IM. A Proteomics Perspective on Viral DNA Sensors in Host Defense and Viral Immune Evasion Mechanisms. Journal of Molecular Biology. 2015;427:1995–2012. doi: 10.1016/j.jmb.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cuchet-Lourenço D, Anderson G, Sloan E, Orr A, Everett RD. The Viral Ubiquitin Ligase ICP0 Is neither Sufficient nor Necessary for Degradation of the Cellular DNA Sensor IFI16 during Herpes Simplex Virus 1 Infection. Journal of Virology. 2013;87:13422–32. doi: 10.1128/JVI.02474-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cuchet-Lourenço D, Vanni E, Glass M, Orr A, Everett RD. Herpes Simplex Virus 1 Ubiquitin Ligase ICP0 Interacts with PML Isoform I and Induces Its SUMO-Independent Degradation. Journal of Virology. 2012;86:11209–22. doi: 10.1128/JVI.01145-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davies MV, Chang HW, Jacobs BL, Kaufman RJ. The E3l and K3l Vaccinia Virus Gene-Products Stimulate Translation through Inhibition of the Double-Stranded Rna-Dependent Protein-Kinase by Different Mechanisms. J Virol. 1993;67:1688–92. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dbaibo GS, Hannun YA. Cytokine response modifier A (CrmA): a strategically deployed viral weapon. Clinical immunology and immunopathology. 1998;86:134–40. doi: 10.1006/clin.1997.4476. [DOI] [PubMed] [Google Scholar]
- 51.Derfuss T, Fickenscher H, Kraft MS, Henning G, Lengenfelder D, Fleckenstein B, Meinl E. Antiapoptotic activity of the herpesvirus saimiri-encoded Bcl-2 homolog: stabilization of mitochondria and inhibition of caspase-3-like activity. J Virol. 1998;72:5897–904. doi: 10.1128/jvi.72.7.5897-5904.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dimitrov T, Krajcsi P, Hermiston TW, Tollefson AE, Hannink M, Wold WS. Adenovirus E3-10.4K/14.5K protein complex inhibits tumor necrosis factor-induced translocation of cytosolic phospholipase A2 to membranes. J Virol. 1997;71:2830–7. doi: 10.1128/jvi.71.4.2830-2837.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dinarello C, Novick D, Kim S, Kaplanski G. Interleukin-18 and interleukin-18 Binding Protein. Frontiers in Immunology. 2013;4 doi: 10.3389/fimmu.2013.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. 2015 doi: 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diner BA, Li T, Greco TM, Crow MS, Fuesler JA, Wang J, Cristea IM. The functional interactome of PYHIN immune regulators reveals IFIX is a sensor of viral DNA. Molecular systems biology. 2015;11:787. doi: 10.15252/msb.20145808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diner BA, Lum KK, Javitt A, Cristea IM. Interactions of the Antiviral Factor Interferon Gamma-Inducible Protein 16 (IFI16) Mediate Immune Signaling and Herpes Simplex Virus-1 Immunosuppression. Molecular & Cellular Proteomics. 2015;14:2341–56. doi: 10.1074/mcp.M114.047068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dobner T, Horikoshi N, Rubenwolf S, Shenk T. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science. 1996;272:1470–73. doi: 10.1126/science.272.5267.1470. [DOI] [PubMed] [Google Scholar]
- 58.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes & Development. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 59.Duerst RJ, Morrison LA. Herpes simplex virus 2 virion host shutoff protein interferes with type I interferon production and responsiveness. Virology. 2004;322:158–67. doi: 10.1016/j.virol.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 60.Dzananovic E, Patel TR, Chojnowski G, Boniecki MJ, Deo S, McEleney K, Harding SE, Bujnicki JM, McKenna SA. Solution conformation of adenovirus virus associated RNA-I and its interaction with PKR. J Struct Biol. 2014;185:48–57. doi: 10.1016/j.jsb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 61.Elgadi MM, Hayes CE, Smiley JR. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–64. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Enari M, Sakahira H, Yokoyama H, Okawa K, Iwamatsu A, Nagata S. A caspase-activated DNase that degrades DNA during apoptosis, and its inhibitor ICAD. Nature. 1998;391:43–50. doi: 10.1038/34112. [DOI] [PubMed] [Google Scholar]
- 63.Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: Implications in antiviral defence. Biochimie. 2007;89:819–30. doi: 10.1016/j.biochi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 64.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML Contributes to a Cellular Mechanism of Repression of Herpes Simplex Virus Type 1 Infection That Is Inactivated by ICP0. Journal of Virology. 2006;80:7995–8005. doi: 10.1128/JVI.00734-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Everly DN, Feng PH, Mian IS, Read GS. mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: Genetic and biochemical evidence that Vhs is a nuclease. J Virol. 2002;76:8560–71. doi: 10.1128/JVI.76.17.8560-8571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ferguson BJ, Mansur DS, Peters NE, Ren H, Smith GL. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. 2012 doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK, Berk AJ. Adenovirus Small E1A Employs the Lysine Acetylases p300/CBP and Tumor Suppressor Rb to Repress Select Host Genes and Promote Productive Virus Infection. Cell Host & Microbe. 2014;16:663–76. doi: 10.1016/j.chom.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKK epsilon and TBK1 are essential components of the IRF3 signaling pathway. Nature Immunology. 2003;4:491–96. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 69.Fleming SB, McCaughan CA, Andrews AE, Nash AD, Mercer AA. A homolog of interleukin-10 is encoded by the poxvirus orf virus. Journal of Virology. 1997;71:4857–61. doi: 10.1128/jvi.71.6.4857-4861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J, Mymryk JS. Adenovirus Evasion of Interferon-Mediated Innate Immunity by Direct Antagonism of a Cellular Histone Posttranslational Modification. Cell Host & Microbe. 2012;11:597–606. doi: 10.1016/j.chom.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Full F, Jungnickl D, Reuter N, Bogner E, Brulois K, Scholz B, Stürzl M, Myoung J, Jung JU, Stamminger T, Ensser A. Kaposi's Sarcoma Associated Herpesvirus Tegument Protein ORF75 Is Essential for Viral Lytic Replication and Plays a Critical Role in the Antagonization of ND10-Instituted Intrinsic Immunity. PLoS Pathog. 2014;10:e1003863. doi: 10.1371/journal.ppat.1003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Full F, Reuter N, Zielke K, Stamminger T, Ensser A. Herpesvirus Saimiri Antagonizes Nuclear Domain 10-Instituted Intrinsic Immunity via an ORF3-Mediated Selective Degradation of Cellular Protein Sp100. Journal of Virology. 2012;86:3541–53. doi: 10.1128/JVI.06992-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaglia MM, Covarrubias S, Wong W, Glaunsinger BA. A Common Strategy for Host RNA Degradation by Divergent Viruses. J Virol. 2012;86:9527–30. doi: 10.1128/JVI.01230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao JL, Murphy PM. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. Journal of Biological Chemistry. 1994;269:28539–42. [PubMed] [Google Scholar]
- 75.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol Cell. 2004;13:713–23. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 76.Glickman JN, Howe JG, Steitz JA. Structural-Analyses of Eber1 and Eber2 Ribonucleoprotein-Particles Present in Epstein-Barr Virus-Infected Cells. J Virol. 1988;62:902–11. doi: 10.1128/jvi.62.3.902-911.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Glykofrydes D, Niphuis H, Kuhn EM, Rosenwirth B, Heeney JL, Bruder J, Niedobitek G, Muller-Fleckenstein I, Fleckenstein B, Ensser A. Herpesvirus saimiri vFLIP provides an antiapoptotic function but is not essential for viral replication, transformation, or pathogenicity. J Virol. 2000;74:11919–27. doi: 10.1128/jvi.74.24.11919-11927.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Go NF, Castle BE, Barrett R, Kastelein R, Dang W, Mosmann TR, Moore KW, Howard M. Interleukin 10, a novel B cell stimulatory factor: unresponsiveness of X chromosome-linked immunodeficiency B cells. The Journal of Experimental Medicine. 1990;172:1625–31. doi: 10.1084/jem.172.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Goldmacher VS. vMIA, a viral inhibitor of apoptosis targeting mitochondria. Biochimie. 2002;84:177–85. doi: 10.1016/s0300-9084(02)01367-6. [DOI] [PubMed] [Google Scholar]
- 80.Gonzalez R, Huang W, Finnen R, Bragg C, Flint SJ. Adenovirus E1B 55-Kilodalton Protein Is Required for both Regulation of mRNA Export and Efficient Entry into the Late Phase of Infection in Normal Human Fibroblasts. Journal of Virology. 2006;80:964–74. doi: 10.1128/JVI.80.2.964-974.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grand RJ, Schmeiser K, Gordon EM, Zhang X, Gallimore PH, Turnell AS. Caspase-mediated cleavage of adenovirus early region 1A proteins. Virology. 2002;301:255–71. doi: 10.1006/viro.2002.1586. [DOI] [PubMed] [Google Scholar]
- 82.Granville DJ, Carthy CM, Yang D, Hunt DW, McManus BM. Interaction of viral proteins with host cell death machinery. Cell death and differentiation. 1998;5:653–9. doi: 10.1038/sj.cdd.4400388. [DOI] [PubMed] [Google Scholar]
- 83.Griffiths A, Coen DM. An unusual internal ribosome entry site in the herpes simplex virus thymidine kinase gene. P Natl Acad Sci USA. 2005;102:9667–72. doi: 10.1073/pnas.0504132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. Vaccinia virus E3 protein prevents the antiviral action of ISG15. Plos Pathog. 2008;4 doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guise AJ, Budayeva HG, Diner BA, Cristea IM. Histone deacetylases in herpesvirus replication and virus-stimulated host defense. Viruses. 2013;5:1607–32. doi: 10.3390/v5071607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haig DM, Thomson J, McInnes CJ, Deane DL, Anderson IE, McCaughan CA, Imlach W, Mercer AA, Howard CJ, Fleming SB. A comparison of the anti-inflammatory and immuno-stimulatory activities of orf virus and ovine interleukin-10. Virus Research. 2002;90:303–16. doi: 10.1016/s0168-1702(02)00252-6. [DOI] [PubMed] [Google Scholar]
- 87.Haller O, Kochs G, Weber F. The interferon response circuit: Induction and suppression by pathogenic viruses. Virology. 2006;344:119–30. doi: 10.1016/j.virol.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Han F, Zhang XB. Internal initiation of mRNA translation in insect cell mediated by an internal ribosome entry site (IRES) from shrimp white spot syndrome virus (WSSV). Biochem Bioph Res Co. 2006;344:893–99. doi: 10.1016/j.bbrc.2006.03.229. [DOI] [PubMed] [Google Scholar]
- 89.Harada JN, Shevchenko A, Shevchenko A, Pallas DC, Berk AJ. Analysis of the Adenovirus E1B-55K-Anchored Proteome Reveals Its Link to Ubiquitination Machinery. Journal of Virology. 2002;76:9194–206. doi: 10.1128/JVI.76.18.9194-9206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hardy WR, Sandri-Goldin RM. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. Journal of Virology. 1994;68:7790–99. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Harman D. Role of free radicals in aging and disease. Annals of the New York Academy of Sciences. 1992;673:126–41. doi: 10.1111/j.1749-6632.1992.tb27444.x. [DOI] [PubMed] [Google Scholar]
- 92.Harte MT, Haga IR, Maloney G, Gray P, Reading PC, Bartlett NW, Smith GL, Bowie A, O'Neill LAJ. The poxvirus protein A52R targets toll-like receptor signaling complexes to suppress host defense. J Exp Med. 2003;197:343–51. doi: 10.1084/jem.20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–72. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 94.Hogg JR. Viral Evasion and Manipulation of Host RNA Quality Control Pathways. Journal of Virology. 2016;90:7010–18. doi: 10.1128/JVI.00607-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus Small e1a Alters Global Patterns of Histone Modification. Science (New York, NY) 2008;321:1084–85. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horwitz GA, Zhang KL, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;321:1084–85. doi: 10.1126/science.1155544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hsu H, Xiong J, Goeddel DV. The TNF receptor 1-associated protein TRADD signals cell death and NF-kappa B activation. Cell. 1995;81:495–504. doi: 10.1016/0092-8674(95)90070-5. [DOI] [PubMed] [Google Scholar]
- 98.Hu FQ, Smith CA, Pickup DJ. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology. 1994;204:343–56. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- 99.Hu SM, Vincenz C, Buller M, Dixit VM. A novel family of viral death effector domain-containing molecules that inhibit both CD-95- and tumor necrosis factor receptor-1-induced apoptosis. J Biol Chem. 1997;272:9621–24. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 100.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. The EMBO Journal. 1991;10:4129–35. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hwang J, Kalejta RF. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology. 2007;367:334–38. doi: 10.1016/j.virol.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 102.John Hiscott PP, Genin Pierre, Nguyen Hannah, Heylbroeck Christophe, Mamane Yael, Algarte Michele, Lin Rongtuan. Triggering the Interferon Response: The Role of IRF-3 Transcription Factor. Journal of Interferon & Cytokine Research. 1999;19(1):1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 103.Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricuttio D, Wang G, McFadden G. A Poxvirus-Encoded Pyrin Domain Protein Interacts with ASC-1 to Inhibit Host Inflammatory and Apoptotic Responses to Infection. Immunity. 2005;23:587–98. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 104.Jones BC, Logsdon NJ, Josephson K, Cook J, Barry PA, Walter MR. Crystal structure of human cytomegalovirus IL-10 bound to soluble human IL-10R1. Proceedings of the National Academy of Sciences. 2002;99:9404–09. doi: 10.1073/pnas.152147499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of Interferon Regulatory Factor 7 (IRF7)-Mediated Interferon Signal Transduction by the Kaposi's Sarcoma-Associated Herpesvirus Viral IRF Homolog vIRF3. Journal of Virology. 2007;81:8282–92. doi: 10.1128/JVI.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaiser WJ, Upton JW, Mocarski ES. Viral modulation of programmed necrosis. Curr Opin Virol. 2013;3:296–306. doi: 10.1016/j.coviro.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kalejta RF, Shenk T. Proteasome-dependent, ubiquitin-independent degradation of the Rb family of tumor suppressors by the human cytomegalovirus pp71 protein. Proceedings of the National Academy of Sciences. 2003;100:3263–68. doi: 10.1073/pnas.0538058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Karin M, Ben-Neriah Y. Phosphorylation Meets Ubiquitination: The Control of NF-κB Activity. Annual Review of Immunology. 2000;18:621–63. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 109.Khanim F, Dawson C, Meseda CA, Dawson J, Mackett M, Young LS. BHRF1, a viral homologue of the Bcl-2 oncogene, is conserved at both the sequence and functional level in different Epstein-Barr virus isolates. The Journal of general virology. 1997;78(Pt 11):2987–99. doi: 10.1099/0022-1317-78-11-2987. [DOI] [PubMed] [Google Scholar]
- 110.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, Peter ME. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995;14:5579–88. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kledal TN, Rosenkilde MM, Coulin F, Simmons G, Johnsen AH, Alouani S, Power CA, Lüttichau HR, Gerstoft J, Clapham PR, Clark-Lewis I, Wells TNC, Schwartz TW. A Broad-Spectrum Chemokine Antagonist Encoded by Kaposi's Sarcoma-Associated Herpesvirus. Science. 1997;277:1656–59. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 112.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proceedings of the National Academy of Sciences. 2000;97:1695–700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krajcsi P, Dimitrov T, Hermiston TW, Tollefson AE, Ranheim TS, Vande Pol SB, Stephenson AH, Wold WS. The adenovirus E3-14.7K protein and the E3-10.4K/14.5K complex of proteins, which independently inhibit tumor necrosis factor (TNF)-induced apoptosis, also independently inhibit TNF-induced release of arachidonic acid. J Virol. 1996;70:4904–13. doi: 10.1128/jvi.70.8.4904-4913.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber RH. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene. 2000;19:850–57. doi: 10.1038/sj.onc.1203395. [DOI] [PubMed] [Google Scholar]
- 115.Krumm B, Meng X, Li Y, Xiang Y, Deng J. Structural basis for antagonism of human interleukin 18 by poxvirus interleukin 18-binding protein. Proceedings of the National Academy of Sciences. 2008;105:20711–15. doi: 10.1073/pnas.0809086106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1315–20. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kwong AD, Frenkel N. Herpes-Simplex Virus-Infected Cells Contain a Function(S) That Destabilizes Both Host and Viral Messenger-Rnas. P Natl Acad Sci USA. 1987;84:1926–30. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Laing KG, Matys V, Clemens MJ. Analysis of the expression and function of the EBV-encoded small RNAs, the EBERs, in heterologous cells. Methods in molecular biology. 2001;174:45–66. doi: 10.1385/1-59259-227-9:45. [DOI] [PubMed] [Google Scholar]
- 119.Lamkanfi M, Dixit Vishva M. Mechanisms and Functions of Inflammasomes. Cell. 2014;157:1013–22. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 120.Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–10. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 121.Lau L, Gray EE, Brunette RL, Stetson DB. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science. 2015;350:568–71. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 122.Launer-Felty K, Wong CJ, Cole JL. Structural Analysis of Adenovirus VAI RNA Defines the Mechanism of Inhibition of PKR. Biophys J. 2015;108:748–57. doi: 10.1016/j.bpj.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lecker SH, Goldberg AL, Mitch WE. Protein Degradation by the Ubiquitin–Proteasome Pathway in Normal and Disease States. Journal of the American Society of Nephrology. 2006;17:1807–19. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- 124.Lee H- R, Doğanay S, Chung B, Toth Z, Brulois K, Lee S, Kanketayeva Z, Feng P, Ha T, Jung JU. Kaposi's Sarcoma-Associated Herpesvirus Viral Interferon Regulatory Factor 4 (vIRF4) Targets Expression of Cellular IRF4 and the Myc Gene To Facilitate Lytic Replication. Journal of Virology. 2014;88:2183–94. doi: 10.1128/JVI.02106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lee HR, Kim DJ, Lee JM, Choi CY, Ahn BY, Hayward GS, Ahn JH. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J Virol. 2004;78:6527–42. doi: 10.1128/JVI.78.12.6527-6542.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. Journal of Virology. 1996;70:7471–77. doi: 10.1128/jvi.70.11.7471-7477.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lefort S, Soucy-Faulkner A, Grandvaux N, Flamand L. Binding of Kaposi's sarcoma-associated herpesvirus K-bZIP to interferon-responsive factor 3 elements modulates antiviral gene expression. J Virol. 2007;81:10950–60. doi: 10.1128/JVI.00183-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Li Q, Means R, Lang S, Jung JU. Downregulation of Gamma Interferon Receptor 1 by Kaposi's Sarcoma-Associated Herpesvirus K3 and K5. Journal of Virology. 2007;81:2117–27. doi: 10.1128/JVI.01961-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li T, Chen J, Cristea IM. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell host & microbe. 2013;14:591–9. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li T, Chen J, Cristea Ileana M. Human Cytomegalovirus Tegument Protein pUL83 Inhibits IFI16-Mediated DNA Sensing for Immune Evasion. Cell Host Microbe. 2013;14:591–99. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liang Q, Fu B, Wu F, Li X, Yuan Y, Zhu F. ORF45 of Kaposi's Sarcoma-Associated Herpesvirus Inhibits Phosphorylation of Interferon Regulatory Factor 7 by IKKε and TBK1 as an Alternative Substrate. Journal of Virology. 2012;86:10162–72. doi: 10.1128/JVI.05224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human nonhomologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4:712–20. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 133.Lieberman Paul M. Epigenetics and Genetics of Viral Latency. Cell Host & Microbe. 2016;19:619–28. doi: 10.1016/j.chom.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lin AE, Greco TM, Döhner K, Sodeik B, Cristea IM. A Proteomic Perspective of Inbuilt Viral Protein Regulation: pUL46 Tegument Protein is Targeted for Degradation by ICP0 during Herpes Simplex Virus Type 1 Infection. Molecular & Cellular Proteomics : MCP. 2013;12:3237–52. doi: 10.1074/mcp.M113.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lin HW, Chang YY, Wong ML, Lin JW, Chang TJ. Functional analysis of virion host shutoff protein of pseudorabies virus. Virology. 2004;324:412–18. doi: 10.1016/j.virol.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 136.Lin R, Genin P, Mamane Y, Sgarbanti M, Battistini A, Harrington WJ, Barber GN, Hiscott J. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene. 2001;20:800–11. doi: 10.1038/sj.onc.1204163. [DOI] [PubMed] [Google Scholar]
- 137.Ling PD, Tan J, Sewatanon J, Peng R. Murine Gammaherpesvirus 68 Open Reading Frame 75c Tegument Protein Induces the Degradation of PML and Is Essential for Production of Infectious Virus. Journal of Virology. 2008;82:8000–12. doi: 10.1128/JVI.02752-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu Y, de Waal Malefyt R, Briere F, Parham C, Bridon JM, Banchereau J, Moore KW, Xu J. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. The Journal of Immunology. 1997;158:604–13. [PubMed] [Google Scholar]
- 139.Lo AKF, To KF, Lo KW, Lung RWM, Hui JWY, Liao GL, Hayward SD. Modulation of LMP1 protein expression by EBV-encoded microRNAs. P Natl Acad Sci USA. 2007;104:16164–69. doi: 10.1073/pnas.0702896104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 141.Loparev VN, Parsons JM, Knight JC, Panus JF, Ray CA, Buller RM, Pickup DJ, Esposito JJ. A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc Natl Acad Sci U S A. 1998;95:3786–91. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lukashchuk V, McFarlane S, Everett RD, Preston CM. Human Cytomegalovirus Protein pp71 Displaces the Chromatin-Associated Factor ATRX from Nuclear Domain 10 at Early Stages of Infection. Journal of Virology. 2008;82:12543–54. doi: 10.1128/JVI.01215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ma J, Edlich F, Bermejo GA, Norris KL, Youle RJ, Tjandra N. Structural mechanism of Bax inhibition by cytomegalovirus protein vMIA. Proc Natl Acad Sci U S A. 2012;109:20901–6. doi: 10.1073/pnas.1217094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marcos-Villar L, Lopitz-Otsoa F, Gallego P, Muñoz-Fontela C, González-Santamaría J, Campagna M, Shou-Jiang G, Rodriguez MS, Rivas C. Kaposi's Sarcoma-Associated Herpesvirus Protein LANA2 Disrupts PML Oncogenic Domains and Inhibits PML-Mediated Transcriptional Repression of the Survivin Gene. Journal of Virology. 2009;83:8849–58. doi: 10.1128/JVI.00339-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Marié I, Smith E, Prakash A, Levy DE. Phosphorylation-Induced Dimerization of Interferon Regulatory Factor 7 Unmasks DNA Binding and a Bipartite Transactivation Domain. Molecular and Cellular Biology. 2000;20:8803–14. doi: 10.1128/mcb.20.23.8803-8814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Marshall EE, Bierle CJ, Brune W, Geballe AP. Essential Role for either TRS1 or IRS1 in Human Cytomegalovirus Replication. J Virol. 2009;83:4112–20. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marshall WL, Yim C, Gustafson E, Graf T, Sage DR, Hanify K, Williams L, Fingeroth J, Finberg RW. Epstein-Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J Virol. 1999;73:5181–5. doi: 10.1128/jvi.73.6.5181-5185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Martin MED, Berk AJ. Adenovirus E1B 55K represses p53 activation in vitro. J Virol. 1998;72:3146–54. doi: 10.1128/jvi.72.4.3146-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Matta H, Mazzacurati L, Schamus S, Yang T, Sun Q, Chaudhary PM. Kaposi's sarcoma-associated herpesvirus (KSHV) oncoprotein K13 bypasses TRAFs and directly interacts with the I kappa B kinase complex to selectively activate NF-kappa B without JNK activation. J Biol Chem. 2007;282:24858–65. doi: 10.1074/jbc.M700118200. [DOI] [PubMed] [Google Scholar]
- 150.Mboko WP, Olteanu H, Ray A, Xin G, Darrah EJ, Kumar SN, Kulinski JM, Cui WG, Dittel BN, Gauld SB, Tarakanova VL. Tumor Suppressor Interferon-Regulatory Factor 1 Counteracts the Germinal Center Reaction Driven by a Cancer-Associated Gammaherpesvirus. J Virol. 2016;90:2818–29. doi: 10.1128/JVI.02774-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McCormick AL, Roback L, Livingston-Rosanoff D, St Clair C. The human cytomegalovirus UL36 gene controls caspase-dependent and -independent cell death programs activated by infection of monocytes differentiating to macrophages. J Virol. 2010;84:5108–23. doi: 10.1128/JVI.01345-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McSharry BP, Avdic S, Slobedman B. Human Cytomegalovirus Encoded Homologs of Cytokines, Chemokines and their Receptors: Roles in Immunomodulation. Viruses. 2012;4:2448–70. doi: 10.3390/v4112448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Melroe GT, Silva L, Schaffer PA, Knipe DM. Recruitment of activated IRF-3 and CBP/p300 to herpes simplex virus ICP0 nuclear foci: Potential role in blocking IFN-beta induction. Virology. 2007;360:305–21. doi: 10.1016/j.virol.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Meng X, Leman M, Xiang Y. Variola Virus IL-18 Binding Protein Interacts with Three Human IL-18 Residues That Are Part of a Binding Site for Human IL-18 Receptor Alpha Subunit. Virology. 2007;358:211–20. doi: 10.1016/j.virol.2006.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Mi J, Li ZY, Ni S, Steinwaerder D, Lieber A. Induced apoptosis supports spread of adenovirus vectors in tumors. Human gene therapy. 2001;12:1343–52. doi: 10.1089/104303401750270995. [DOI] [PubMed] [Google Scholar]
- 156.Miller DM, Rahill BM, Boss JM, Lairmore MD, Durbin JE, Waldman JW, Sedmak DD. Human Cytomegalovirus Inhibits Major Histocompatibility Complex Class II Expression By Disruption of the Jak/Stat Pathway. The Journal of Experimental Medicine. 1998;187:675–83. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Miller LK. Baculovirus interaction with host apoptotic pathways. Journal of cellular physiology. 1997;173:178–82. doi: 10.1002/(SICI)1097-4652(199711)173:2<178::AID-JCP17>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 158.Mitchell AM, Hirsch ML, Li C, Samulski RJ. Promyelocytic Leukemia Protein Is a Cell-Intrinsic Factor Inhibiting Parvovirus DNA Replication. Journal of Virology. 2014;88:925–36. doi: 10.1128/JVI.02922-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–805. [PubMed] [Google Scholar]
- 160.Molden J, Chang Y, You Y, Moore PS, Goldsmith MA. A Kaposi's Sarcoma-associated Herpesvirus-encoded Cytokine Homolog (vIL-6) Activates Signaling through the Shared gp130 Receptor Subunit. Journal of Biological Chemistry. 1997;272:19625–31. doi: 10.1074/jbc.272.31.19625. [DOI] [PubMed] [Google Scholar]
- 161.Moore PS, Boshoff C, Weiss RA, Chang Y. Molecular Mimicry of Human Cytokine and Cytokine Response Pathway Genes by KSHV. Science. 1996;274:1739–44. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 162.Moriuchi H, Moriuchi M, Smith HA, Straus SE, Cohen JI. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. Journal of Virology. 1992;66:7303–08. doi: 10.1128/jvi.66.12.7303-7308.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Munger J, Hagglund R, Roizman B. Infected cell protein No. 22 is subject to proteolytic cleavage by caspases activated by a mutant that induces apoptosis. Virology. 2003;305:364–70. doi: 10.1006/viro.2002.1728. [DOI] [PubMed] [Google Scholar]
- 164.Murphy JA, Duerst RJ, Smith TJ, Morrison LA. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J Virol. 2003;77:9337–45. doi: 10.1128/JVI.77.17.9337-9345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mutocheluh M, Hindle L, Aresté C, Chanas SA, Butler LM, Lowry K, Shah K, Evans DJ, Blackbourn DJ. Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor-2 inhibits type 1 interferon signalling by targeting interferon-stimulated gene factor-3. Journal of General Virology. 2011;92:2394–98. doi: 10.1099/vir.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 166.Nava VE, Cheng EH, Veliuona M, Zou S, Clem RJ, Mayer ML, Hardwick JM. Herpesvirus saimiri encodes a functional homolog of the human bcl-2 oncogene. J Virol. 1997;71:4118–22. doi: 10.1128/jvi.71.5.4118-4122.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Neipel F, Albrecht JC, Ensser A, Huang YQ, Li JJ, Friedman-Kien AE, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. Journal of Virology. 1997;71:839–42. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Niebler M, Qian X, Höfler D, Kogosov V, Kaewprag J, Kaufmann AM, Ly R, Böhmer G, Zawatzky R, Rösl F, Rincon-Orozco B. Post-Translational Control of IL-1? via the Human Papillomavirus Type 16 E6 Oncoprotein: A Novel Mechanism of Innate Immune Escape Mediated by the E3-Ubiquitin Ligase E6-AP and p53. PLoS Pathog. 2013;9:e1003536. doi: 10.1371/journal.ppat.1003536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Niedobitek G, Young LS, Lau R, Brooks L, Greenspan D, Greenspan JS, Rickinson AB. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. The Journal of general virology. 1991;72(Pt 12):3035–46. doi: 10.1099/0022-1317-72-12-3035. [DOI] [PubMed] [Google Scholar]
- 170.Nogal ML, Gonzalez de Buitrago G, Rodriguez C, Cubelos B, Carrascosa AL, Salas ML, Revilla Y. African swine fever virus IAP homologue inhibits caspase activation and promotes cell survival in mammalian cells. J Virol. 2001;75:2535–43. doi: 10.1128/JVI.75.6.2535-2543.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nozawa H, Oda E, Nakao K, Ishihara M, Ueda S, Yokochi T, Ogasawara K, Nakatsuru Y, Shimizu S, Ohira Y, Hioki K, Aizawa S, Ishikawa T, Katsuki M, Muto T, Taniguchi T, Tanaka N. Loss of transcription factor IRF-1 affects tumor susceptibility in mice carrying the Ha-ras transgene or nullizygosity for p53. Genes & Development. 1999;13:1240–45. doi: 10.1101/gad.13.10.1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74:609–19. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 173.Oroskar AA, Read GS. A Mutant of Herpes-Simplex Virus Type-1 Exhibits Increased Stability of Immediate-Early (Alpha) Messenger-Rnas. J Virol. 1987;61:604–06. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Oroskar AA, Read GS. Control of Messenger-Rna Stability by the Virion Host Shutoff Function of Herpes-Simplex Virus. J Virol. 1989;63:1897–906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci U S A. 2012;109:E3008–17. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Park JS, Kim EJ, Kwon HJ, Hwang ES, Namkoong SE, Um SJ. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein - Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275:6764–69. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- 177.Parkinson J, Lees-Miller SP, Everett RD. Herpes Simplex Virus Type 1 Immediate-Early Protein Vmw110 Induces the Proteasome-Dependent Degradation of the Catalytic Subunit of DNA-Dependent Protein Kinase. Journal of Virology. 1999;73:650–57. doi: 10.1128/jvi.73.1.650-657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Parrish S, Moss B. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol. 2007;81:12973–78. doi: 10.1128/JVI.01668-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Parrish S, Resch W, Moss B. Vaccinia virus D10 protein has mRNA decapping activity, providing a mechanism for control of host and viral gene expression. P Natl Acad Sci USA. 2007;104:2139–44. doi: 10.1073/pnas.0611685104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pasieka TJ, Lu B, Crosby SD, Wylie KM, Morrison LA, Alexander DE, Menachery VD, Leib DA. Herpes simplex virus virion host shutoff attenuates establishment of the antiviral state. J Virol. 2008;82:5527–35. doi: 10.1128/JVI.02047-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Paulsen SJ, Rosenkilde MM, Eugen-Olsen J, Kledal TN. Epstein-Barr Virus-Encoded BILF1 Is a Constitutively Active G Protein-Coupled Receptor. Journal of Virology. 2005;79:536–46. doi: 10.1128/JVI.79.1.536-546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Peters GA, Khoo D, Mohr I, Sen GC. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J Virol. 2002;76:11054–64. doi: 10.1128/JVI.76.21.11054-11064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Pezda AC, Penn A, Barton GM, Coscoy L. Suppression of TLR9 Immunostimulatory Motifs in the Genome of a Gammaherpesvirus. The Journal of Immunology. 2011;187:887–96. doi: 10.4049/jimmunol.1003737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pflanz S, Timans JC, Cheung J, Rosales R, Kanzler H, Gilbert J, Hibbert L, Churakova T, Travis M, Vaisberg E, Blumenschein WM, Mattson JD, Wagner JL, To W, Zurawski S, McClanahan TK, Gorman DM, Bazan JF, de Waal Malefyt R, Rennick D, Kastelein RA. IL-27, a Heterodimeric Cytokine Composed of EBI3 and p28 Protein, Induces Proliferation of Naive CD4+ T Cells. Immunity. 2002;16:779–90. doi: 10.1016/s1074-7613(02)00324-2. [DOI] [PubMed] [Google Scholar]
- 185.Poon AP, Gu H, Roizman B. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc Natl Acad Sci U S A. 2006;103:9993–8. doi: 10.1073/pnas.0604142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Prakash A, Levy DE. Regulation of IRF7 through cell type-specific protein stability. Biochem Bioph Res Co. 2006;342:50–56. doi: 10.1016/j.bbrc.2006.01.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Qin ZQ, Kearney P, Plaisance K, Parsons CH. Kaposi's sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukocyte Biol. 2010;87:25–34. doi: 10.1189/jlb.0409251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11:527–34. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 189.Rao L, Debbas M, Sabbatini P, Hockenbery D, Korsmeyer S, White E. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc Natl Acad Sci U S A. 1992;89:7742–6. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Rao L, Modha D, White E. The E1B 19K protein associates with lamins in vivo and its proper localization is required for inhibition of apoptosis. Oncogene. 1997;15:1587–97. doi: 10.1038/sj.onc.1201323. [DOI] [PubMed] [Google Scholar]
- 191.Read GS, Frenkel N. Herpes-Simplex Virus Mutants Defective in the Virion-Associated Shutoff of Host Polypeptide-Synthesis and Exhibiting Abnormal Synthesis of Alpha-(Immediate Early) Viral Polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Reading PC, Smith GL. Vaccinia Virus Interleukin-18-Binding Protein Promotes Virulence by Reducing Gamma Interferon Production and Natural Killer and T-Cell Activity. Journal of Virology. 2003;77:9960–68. doi: 10.1128/JVI.77.18.9960-9968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reeves MB, Davies AA, McSharry BP, Wilkinson GW, Sinclair JH. Complex I Binding by a Virally Encoded RNA Regulates Mitochondria-Induced Cell Death. Science. 2007;316:1345–48. doi: 10.1126/science.1142984. [DOI] [PubMed] [Google Scholar]
- 194.Riedl SJ, Shi Y. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol. 2004;5:897–907. doi: 10.1038/nrm1496. [DOI] [PubMed] [Google Scholar]
- 195.Rivas HG, Schmaling SK, Gaglia MM. Shutoff of Host Gene Expression in Influenza A Virus and Herpesviruses: Similar Mechanisms and Common Themes. Viruses. 2016;8 doi: 10.3390/v8040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Rock KL, Gramm C, Rothstein L, Clark K, Stein R, Dick L, Hwang D, Goldberg AL. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–71. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 197.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes & Development. 1998;12:2061–72. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Roulston A, Marcellus RC, Branton PE. Viruses and apoptosis. Annual review of microbiology. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 199.Rowe M, Glaunsinger B, van Leeuwen D, Zuo JM, Sweetman D, Ganem D, Middeldorp J, Wiertz EJHJ, Ressing ME. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. P Natl Acad Sci USA. 2007;104:3366–71. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Saffert RT, Kalejta RF. Inactivating a Cellular Intrinsic Immune Defense Mediated by Daxx Is the Mechanism through Which the Human Cytomegalovirus pp71 Protein Stimulates Viral Immediate-Early Gene Expression. Journal of Virology. 2006;80:3863–71. doi: 10.1128/JVI.80.8.3863-3871.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Sah NK, Taneja TK, Pathak N, Begum R, Athar M, Hasnain SE. The baculovirus antiapoptotic p35 gene also functions via an oxidant-dependent pathway. Proc Natl Acad Sci U S A. 1999;96:4838–43. doi: 10.1073/pnas.96.9.4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sainz B, LaMarca HL, Garry RF, Morris CA. Synergistic inhibition of human cytomegalovirus replication by interferon-alpha/beta and interferon-gamma. Virol J. 2005;2 doi: 10.1186/1743-422X-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Saira K, Zhou Y, Jones C. The Infected Cell Protein 0 Encoded by Bovine Herpesvirus 1 (bICP0) Induces Degradation of Interferon Response Factor 3 and, Consequently, Inhibits Beta Interferon Promoter Activity. Journal of Virology. 2007;81:3077–86. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sato H, Callanan LD, Pesnicak L, Krogmann T, Cohen JI. Varicella-zoster virus (VZV) ORF17 protein induces RNA cleavage and is critical for replication of VZV at 37 degrees C but not 33 degrees C. J Virol. 2002;76:11012–23. doi: 10.1128/JVI.76.21.11012-11023.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the IRF transcription factor family. Cancer Immunol Immun. 2010;59:489–510. doi: 10.1007/s00262-009-0804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 Oncoprotein Encoded by Human Papillomavirus Type-16 and Type-18 Promotes the Degradation of P53. Cell. 1990;63:1129–36. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 207.Schek N, Bachenheimer SL. Degradation of Cellular Messenger-Rnas Induced by a Virion-Associated Factor during Herpes-Simplex Virus-Infection of Vero Cells. J Virol. 1985;55:601–10. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2011;1813:878–88. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 209.Scherer M, Stamminger T. Gem: Emerging role of PML nuclear bodies in innate immune signaling. Journal of Virology. 2016 doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Scherer M, Stamminger T. Gem: Emerging role of PML nuclear bodies in innate immune signaling. Journal of virology. 2016 doi: 10.1128/JVI.01979-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schott J, Stoecklin G. Networks controlling mRNA decay in the immune system. Wires Rna. 2010;1:432–56. doi: 10.1002/wrna.13. [DOI] [PubMed] [Google Scholar]
- 212.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. Proteasome-Dependent Degradation of Daxx by the Viral E1B-55K Protein in Human Adenovirus-Infected Cells. Journal of Virology. 2010;84:7029–38. doi: 10.1128/JVI.00074-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Schroder M, Baran M, Bowie AG. Viral targeting of human DEAD-box protein 3 reveals its novel role in IRF3/7 activation. Cytokine. 2008;43:313–13. doi: 10.1038/emboj.2008.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Schwarz KB. Oxidative stress during viral infection: A review. Free Radical Bio Med. 1996;21:641–49. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 215.Shen GH, Wang KZ, Wang S, Cai MS, Li ML, Zheng CF. Herpes Simplex Virus 1 Counteracts Viperin via Its Virion Host Shutoff Protein UL41. J Virol. 2014;88:12163–66. doi: 10.1128/JVI.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shen Y, Shenk TE. Viruses and apoptosis. Current opinion in genetics & development. 1995;5:105–11. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 217.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–70. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 218.Shisler J, Yang C, Walter B, Ware CF, Gooding LR. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J Virol. 1997;71:8299–306. doi: 10.1128/jvi.71.11.8299-8306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shisler JL, Moss B. Molluscum contagiosum virus inhibitors of apoptosis: The MC159 v-FLIP protein blocks Fas-induced activation of procaspases and degradation of the related MC160 protein. Virology. 2001;282:14–25. doi: 10.1006/viro.2001.0834. [DOI] [PubMed] [Google Scholar]
- 220.Shisler JL, Senkevich TG, Berry MJ, Moss B. Ultraviolet-induced cell death blocked by a selenoprotein from a human dermatotropic poxvirus. Science. 1998;279:102–5. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 221.Shu MF, Taddeo B, Roizman B. Tristetraprolin Recruits the Herpes Simplex Virion Host Shutoff RNase to AU-Rich Elements in Stress Response mRNAs To Enable Their Cleavage. J Virol. 2015;89:5643–50. doi: 10.1128/JVI.00091-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sivachandran N, Wang X, Frappier L. Functions of the Epstein-Barr Virus EBNA1 Protein in Viral Reactivation and Lytic Infection. Journal of Virology. 2012;86:6146–58. doi: 10.1128/JVI.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Sloan E, Tatham MH, Groslambert M, Glass M, Orr A, Hay RT, Everett RD. Analysis of the SUMO2 Proteome during HSV-1 Infection. Plos Pathog. 2015;11:e1005059. doi: 10.1371/journal.ppat.1005059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Smiley JR, Elgadi MM, Saffran HA. Herpes simplex virus vhs protein. Method Enzymol. 2001;342:440–51. doi: 10.1016/s0076-6879(01)42565-1. [DOI] [PubMed] [Google Scholar]
- 225.Smith CA, Hu FQ, Smith TD, Richards CL, Smolak P, Goodwin RG, Pickup DJ. Cowpox virus genome encodes a second soluble homologue of cellular TNF receptors, distinct from CrmB, that binds TNF but not LT alpha. Virology. 1996;223:132–47. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- 226.Smith GL, Chan YS. Two vaccinia virus proteins structurally related to the interleukin-1 receptor and the immunoglobulin superfamily. Journal of General Virology. 1991;72:511–18. doi: 10.1099/0022-1317-72-3-511. [DOI] [PubMed] [Google Scholar]
- 227.Smith KJ, Col, Usa MC, Yeager J, Cdr, Usn MC, Skelton H. Molluscum contagiosum: its clinical, histopathologic, and immunohistochemical spectrum. International Journal of Dermatology. 1999;38:664–72. doi: 10.1046/j.1365-4362.1999.00677.x. [DOI] [PubMed] [Google Scholar]
- 228.Smith VP, Bryant NA, Alcamí A. Ectromelia, vaccinia and cowpox viruses encode secreted interleukin-18-binding proteins. Journal of General Virology. 2000;81:1223–30. doi: 10.1099/0022-1317-81-5-1223. [DOI] [PubMed] [Google Scholar]
- 229.Smith W, Tomasec P, Aicheler R, Loewendorf A, Nemcovicova I, Wang ECY, Stanton RJ, Macauley M, Norris P, Willen L, Ruckova E, Nomoto A, Schneider P, Hahn G, Zajonc DM, Ware CF, Wilkinson GWG, Benedict CA. Human Cytomegalovirus Glycoprotein UL141 Targets the TRAIL Death Receptors to Thwart Host Innate Antiviral Defenses. Cell Host & Microbe. 2013;13:324–35. doi: 10.1016/j.chom.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Soulat D, Buerckstuemmer T, Westermayer S, Goncalves A, Bauch A, Stefanovic A, Hantschel O, Bennett KL, Decker T, Superti-Furga G. The DEAD-box helicase DDX3X is a critical component of the TANK-binding kinase 1-dependent innate immune response. Wien Klin Wochenschr. 2008;120:21–21. doi: 10.1038/emboj.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Spencer JV, Lockridge KM, Barry PA, Lin G, Tsang M, Penfold MET, Schall TJ. Potent Immunosuppressive Activities of Cytomegalovirus-Encoded Interleukin-10. Journal of Virology. 2002;76:1285–92. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Spriggs MK, Hruby DE, Maliszewski CR, Pickup DJ, Sims JE, Buller RML, VanSlyke J. Vaccinia and cowpox viruses encode a novel secreted interleukin-1-binding protein. Cell. 1992;71:145–52. doi: 10.1016/0092-8674(92)90273-f. [DOI] [PubMed] [Google Scholar]
- 233.Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. Vaccinia virus protein Toll-like-interleukin-1 A46R targets multiple receptor adaptors and contributes to virulence. J Exp Med. 2005;201:1007–18. doi: 10.1084/jem.20041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Stepp WH, Meyers JM, McBride AA. Sp100 Provides Intrinsic Immunity against Human Papillomavirus Infection. mBio. 2013;4 doi: 10.1128/mBio.00845-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Stienstra R, Joosten LAB, Koenen T, van Tits B, van Diepen JA, van den Berg SAA, Rensen PCN, Voshol PJ, Fantuzzi G, Hijmans A, Kersten S, Müller M, van den Berg WB, van Rooijen N, Wabitsch M, Kullberg B-J, van der Meer JWM, Kanneganti T, Tack CJ, Netea MG. The Inflammasome-Mediated Caspase-1 Activation Controls Adipocyte Differentiation and Insulin Sensitivity. Cell Metabolism. 2010;12:593–605. doi: 10.1016/j.cmet.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Stracker TH, Carson CT, Weitzman MD. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature. 2002;418:348–52. doi: 10.1038/nature00863. [DOI] [PubMed] [Google Scholar]
- 238.Strasser A, Huang DC, Vaux DL. The role of the bcl-2/ced-9 gene family in cancer and general implications of defects in cell death control for tumourigenesis and resistance to chemotherapy. Biochimica et biophysica acta. 1997;1333:F151–78. doi: 10.1016/s0304-419x(97)00019-x. [DOI] [PubMed] [Google Scholar]
- 239.Su CH, Zhang J, Zheng CF. Herpes simplex virus 1 UL41 protein abrogates the antiviral activity of hZAP by degrading its mRNA. Virol J. 2015;12 doi: 10.1186/s12985-015-0433-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Sunthamala N, Thierry F, Teissier S, Pientong C, Kongyingyoes B, Tangsiriwatthana T, Sangkomkamhang U, Ekalaksananan T. E2 Proteins of High Risk Human Papillomaviruses Down-Modulate STING and IFN-kappa Transcription in Keratinocytes. Plos One. 2014;9 doi: 10.1371/journal.pone.0091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost MC, Alzari PM, Kroemer G. Mitochondrial release of caspase-2 and -9 during the apoptotic process. J Exp Med. 1999;189:381–94. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Swaminathan S, Hesselton R, Sullivan J, Kieff E. Epstein-Barr virus recombinants with specifically mutated BCRF1 genes. Journal of Virology. 1993;67:7406–13. doi: 10.1128/jvi.67.12.7406-7413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Tait SWG, Reid EB, Greaves DR, Wileman TE, Powell PP. Mechanism of inactivation of NF-kappa B by a viral homologue of I kappa B alpha - Signal-induced release of I kappa B alpha results in binding of the viral homologue to NF-kappa B. J Biol Chem. 2000;275:34656–64. doi: 10.1074/jbc.M000320200. [DOI] [PubMed] [Google Scholar]
- 244.Taylor RT, Bresnahan WA. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J Virol. 2005;79:3873–7. doi: 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Taylor RT, Bresnahan WA. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J Virol. 2006;80:10763–71. doi: 10.1128/JVI.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Teodoro JG, Branton PE. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–46. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Thompson MR, Kaminski JJ, Kurt-Jones EA, Fitzgerald KA. Pattern Recognition Receptors and the Innate Immune Response to Viral Infection. Viruses. 2011;3:920–40. doi: 10.3390/v3060920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Thornberry NA, Lazebnik Y. Caspases: Enemies within. Science. 1998;281:1312–16. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 249.Tolani B, Matta H, Gopalakrishnan R, Punj V, Chaudhary PM. NEMO Is Essential for Kaposi's Sarcoma-Associated Herpesvirus-Encoded vFLIP K13-Induced Gene Expression and Protection against Death Receptor-Induced Cell Death, and Its N-Terminal 251 Residues Are Sufficient for This Process. Journal of Virology. 2014;88:6345–54. doi: 10.1128/JVI.00028-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tummers B, Goedemans R, Pelascini LP, Jordanova ES, van Esch EM, Meyers C, Melief CJ, Boer JM, van der Burg SH. The interferon-related developmental regulator 1 is used by human papillomavirus to suppress NFkappaB activation. Nature communications. 2015;6:6537. doi: 10.1038/ncomms7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Um SJ, Rhyu JW, Kim EJ, Jeon KC, Hwang ES, Park JS. Abrogation of IRF-1 response by high-risk HPV E7 protein in vivo. Cancer Lett. 2002;179:205–12. doi: 10.1016/s0304-3835(01)00871-0. [DOI] [PubMed] [Google Scholar]
- 252.Unterholzner L, Bowie AG. The interplay between viruses and innate immune signaling: Recent insights and therapeutic opportunities. Biochem Pharmacol. 2008;75:589–602. doi: 10.1016/j.bcp.2007.07.043. [DOI] [PubMed] [Google Scholar]
- 253.Upton JW, Kaiser WJ, Mocarski ES. Virus Inhibition of RIP3-Dependent Necrosis. Cell Host & Microbe. 2010;7:302–13. doi: 10.1016/j.chom.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Upton JW, Kaiser WJ, Mocarski ES. DAI complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host & Microbe. 2012;11:290–97. doi: 10.1016/j.chom.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.van Lint AL, Murawski MR, Goodbody RE, Severa M, Fitzgerald KA, Finberg RW, Knipe DM, Kurt-Jones EA. Herpes Simplex Virus Immediate-Early ICP0 Protein Inhibits Toll-Like Receptor 2-Dependent Inflammatory Responses and NF-κB Signaling. Journal of Virology. 2010;84:10802–11. doi: 10.1128/JVI.00063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, Wang D, Camp DG, Rodland K, Wiley S, Britt W, Shenk T, Smith RD, Nelson JA. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome (vol 78, pg 10960, 2004). J Virol. 2004;78:13395–95. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Verpooten D, Ma YJ, Hou SW, Yan ZP, He B. Control of TANK-binding Kinase 1-mediated Signaling by the gamma(1)34.5 Protein of Herpes Simplex Virus 1. J Biol Chem. 2009;284:1097–105. doi: 10.1074/jbc.M805905200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Vieira P, de Waal-Malefyt R, Dang MN, Johnson KE, Kastelein R, Fiorentino DF, deVries JE, Roncarolo MG, Mosmann TR, Moore KW. Isolation and expression of human cytokine synthesis inhibitory factor cDNA clones: homology to Epstein-Barr virus open reading frame BCRFI. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:1172–76. doi: 10.1073/pnas.88.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.von Roretz C, Di Marco S, Mazroui R, Gallouzi IE. Turnover of AU-rich-containing mRNAs during stress: a matter of survival. Wires Rna. 2011;2:336–47. doi: 10.1002/wrna.55. [DOI] [PubMed] [Google Scholar]
- 260.Vuyisich M, Spanggord RJ, Beal PA. The binding site of the RNA-dependent protein kinase (PKR) on EBER1 RNA from Epstein-Barr virus. Embo Rep. 2002;3:622–27. doi: 10.1093/embo-reports/kvf137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Wagenknecht N, Reuter N, Scherer M, Reichel A, Müller R, Stamminger T. Contribution of the Major ND10 Proteins PML, hDaxx and Sp100 to the Regulation of Human Cytomegalovirus Latency and Lytic Replication in the Monocytic Cell Line THP-1. Viruses. 2015;7:2751. doi: 10.3390/v7062751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Walters MS, Erazo A, Kinchington PR, Silverstein S. Histone deacetylases 1 and 2 are phosphorylated at novel sites during varicella-zoster virus infection. J Virol. 2009;83:11502–13. doi: 10.1128/JVI.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Walters MS, Kinchington PR, Banfield BW, Silverstein S. Hyperphosphorylation of histone deacetylase 2 by alphaherpesvirus US3 kinases. J Virol. 2010;84:9666–76. doi: 10.1128/JVI.00981-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang GH, Bertin J, Wang Y, Martin DA, Wang J, Tomaselli KJ, Armstrong RC, Cohen JI. Bovine herpesvirus 4 BORFE2 protein inhibits Fas- and tumor necrosis factor receptor 1-induced apoptosis and contains death effector domains shared with other gamma-2 herpesviruses. J Virol. 1997;71:8928–32. doi: 10.1128/jvi.71.11.8928-8932.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang K, Ni L, Wang S, Zheng C. Herpes simplex virus 1 protein kinase US3 hyperphosphorylates p65/RelA and dampens NF-kappaB activation. J Virol. 2014;88:7941–51. doi: 10.1128/JVI.03394-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wang S, Wang K, Lin R, Zheng C. Herpes simplex virus 1 serine/threonine kinase US3 hyperphosphorylates IRF3 and inhibits beta interferon production. J Virol. 2013;87:12814–27. doi: 10.1128/JVI.02355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Weiden MD, Ginsberg HS. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:153–57. doi: 10.1073/pnas.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Weitzman MD, Lilley CE, Chaurushiya MS. Genomes in conflict: maintaining genome integrity during virus infection. Annual review of microbiology. 2010;64:61–81. doi: 10.1146/annurev.micro.112408.134016. [DOI] [PubMed] [Google Scholar]
- 269.Weitzman MD, Ornelles DA. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene. 2005;24:7686–96. doi: 10.1038/sj.onc.1209063. [DOI] [PubMed] [Google Scholar]
- 270.Wiebe MS, Traktman P. Barrier to Autointegration Factor is a Host Defense Against Poxvirus Replication that is Overcome by the Viral B1 Kinase. Cell host & microbe. 2007;1:187–97. doi: 10.1016/j.chom.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wies E, Hahn AS, Schmidt K, Viebahn C, Rohland N, Lux A, Schellhorn T, Holzer A, Jung JU, Neipel F. The Kaposi's Sarcoma-associated Herpesvirus-encoded vIRF-3 Inhibits Cellular IRF-5. Journal of Biological Chemistry. 2009;284:8525–38. doi: 10.1074/jbc.M809252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Williams BRG. The role of the dsRNA-activated kinase, PKR, in signal transduction. Seminars in Virology. 1995;6:191–202. [Google Scholar]
- 273.Wu JJ, Li WW, Shao YM, Avey D, Fu BS, Gillen J, Hand T, Ma SM, Liu X, Miley W, Konrad A, Neipel F, Stuerzl M, Whitby D, Li H, Zhu FX. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host & Microbe. 2015;18:333–44. doi: 10.1016/j.chom.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Xiang Y, Moss B. Correspondence of the Functional Epitopes of Poxvirus and Human Interleukin-18-Binding Proteins. Journal of Virology. 2001;75:9947–54. doi: 10.1128/JVI.75.20.9947-9954.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Xing JJ, Ni LW, Wang S, Wang KZ, Lin RT, Zheng CF. Herpes Simplex Virus 1-Encoded Tegument Protein VP16 Abrogates the Production of Beta Interferon (IFN) by Inhibiting NF-kappa B Activation and Blocking IFN Regulatory Factor 3 To Recruit Its Coactivator CBP. J Virol. 2013;87:9788–801. doi: 10.1128/JVI.01440-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Yamin R, Kaynan NS, Glasner A, Vitenshtein A, Tsukerman P, Bauman Y, Ophir Y, Elias S, Bar-On Y, Gur C, Mandelboim O. The Viral KSHV Chemokine vMIP-II Inhibits the Migration of Naive and Activated Human NK Cells by Antagonizing Two Distinct Chemokine Receptors. PLoS Pathog. 2013;9:e1003568. doi: 10.1371/journal.ppat.1003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yanai N, Obinata M. Apoptosis Is Induced at Nonpermissive Temperature by a Transient Increase in P53 in Cell-Lines Immortalized with Temperature-Sensitive Sv40 Large T-Antigen Gene. Exp Cell Res. 1994;211:296–300. doi: 10.1006/excr.1994.1090. [DOI] [PubMed] [Google Scholar]
- 278.You M, Yu DH, Feng GS. Shp-2 tyrosine phosphatase functions as a negative regulator of the interferon-stimulated Jak/STAT pathway. Molecular and Cellular Biology. 1999;19:2416–24. doi: 10.1128/mcb.19.3.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Yu YJ, Alwine JC. 19S Late mRNAs of simian virus 40 have an internal ribosome entry site upstream of the virion structural protein 3 coding sequence. J Virol. 2006;80:6553–58. doi: 10.1128/JVI.00517-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yu YX, Wang SZE, Hayward GS. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity. 2005;22:59–70. doi: 10.1016/j.immuni.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 281.Zhang J, Wang K, Wang S, Zheng C. Herpes Simplex Virus 1 E3 Ubiquitin Ligase ICP0 Protein Inhibits Tumor Necrosis Factor Alpha-Induced NF-κB Activation by Interacting with p65/RelA and p50/NF-κB1. Journal of Virology. 2013;87:12935–48. doi: 10.1128/JVI.01952-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Zhu H, Zheng C, Xing J, Wang S, Li S, Lin R, Mossman KL. Varicella-Zoster Virus Immediate-Early Protein ORF61 Abrogates the IRF3-Mediated Innate Immune Response through Degradation of Activated IRF3. Journal of Virology. 2011;85:11079–89. doi: 10.1128/JVI.05098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zimring JC, Goodbourn S, Offermann MK. Human Herpesvirus 8 Encodes an Interferon Regulatory Factor (IRF) Homolog That Represses IRF-1-Mediated Transcription. Journal of Virology. 1998;72:701–07. doi: 10.1128/jvi.72.1.701-707.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zipeto D, Bodaghi B, Laurent L, Virelizier JL, Michelson S. Kinetics of transcription of human cytomegalovirus chemokine receptor US28 in different cell types. Journal of General Virology. 1999;80:543–47. doi: 10.1099/0022-1317-80-3-543. [DOI] [PubMed] [Google Scholar]