Abstract
During fungal diversity surveys of the order Eurotiales in Korea, two fungal strains, EML-DG33-1 and EML-NCP50, were isolated from samples of rat dung and fig tree leaf collected at a garden located in Gwangju in 2014. To complete the National Species List of Korea, it is a prerequisite to verify whether many questionable species, which were previously recorded but not confirmed, indeed present in Korea. Herein, the isolates were confirmed as undescribed species, Paecilomyces variotii and Talaromyces amestolkiae based on the combination of morphological and phylogenetic analyses of multigenes including the rDNA internal transcribed spacer, β-tubulin, and RNA polymerase II subunit 2.
Keywords: Fig tree leaf, Morphology, Multigene phylogenetic analysis, Paecilomyces variotii, Rat dung, Talaromyces amestolkiae
The order Eurotiales described by Beny and Kimbrough [1] consists of three families: Trichocomaceae, Thermoascaceae, and Eremomycetaceae. Especially, the Trichocomaceae contains species which are important to both industry and medicine and as mycotoxin producer on various foods [2]. The most well-known genera include Aspergillus, Penicillium, Paecilomyces, and Talaromyces [2,3,4].
The genus Paecilomyces (teleomorph, Byssochlamys) in the Eurotiales was originally described by Bainier [5], based on a single species, Paecilomyces variotii. This species was characterized by verticillate conidiophores with divergent whorls of phialides, which have a cylindrical or inflated base, tapering to a long and distinct neck. The genus was revised by Brown and Smith [6] and modified by Samson [7], who defined 31 species and divided the genus into two sections, Paecilomyces and Isarioidea. To date, more than 100 species of the genus Paecilomyces have been recognized [8]. The type species of Paecilomyces, P. variotii, has a sexual Byssochlamys state [9]. This species is frequently found in soils, animals, indoor environments, and food products [7,10]. Some species of Paecilomyces have been isolated from insects, and some can even cause infections in humans [11,12,13]. Most members of the genus Paecilomyces have optimum growth temperatures ranging from 30~37℃ [9]. Several studies have demonstrated the importance of Paecilomyces species in various biotechnological applications, including the production of tannase [6,7,8,9,10] and secondary metabolites, some of which have useful biological activities [14,15].
On the other hand, the genus Talaromyces in the Eurotiales was described by Benjamin [16] with Talaromyces vermiculatus as the type species. This genus was characterized by soft cleistothecial ascomata with a wall of interwoven hyphae. It was re-defined and restricted to species producing asci in chains by Stolk and Samson [17]. Samson et al. [18] successfully transferred species of Penicillium subgenus Biverticillium to Talaromyces by following the one fungus one name concept. On the other hand, Yilmaz et al. [19] provided a monograph and accepted 88 species of Talaromyces. Based on internal transcribed spacer (ITS), β-tubulin, and RBP2 multigene phylogeny, Yilmaz et al. [19] divided those species in seven sections including: Bacillispori, Helici, Islandici, Purpurei, Subinflati, Talaromyces, and Trachyspermi. Especially, several species in Talaromyces such as T. thermophilus, T. funiculosum, and T. marneffei show biotechnological and medical important properties [20].
In Korea, seven species of Paecilomyces and seventeen species of Talaromyces have been reported in Korea, of which only three Paecilomyces and five Talaromyces species have been well described, excluding P. variotii and T. amestolkiae [21]. In the Korean fungal species list published by the National Institute of Biological Resources (NIBR), the Paecilomyces variotii species is listed as an undescribed record which does not match any references.
During fungal diversity surveys of the order Eurotiales in Korea, two fungal strains, EML-DG33-1 as a dung fungus and EML-NCP50 as an endophyte, were isolated from samples of rat dung and fig tree leaf collected at a garden located in Gwangju in 2014. The objective of the present study was to clarify the phylogenetic status of the polyphyletic species of P. variotii and T. amestolkiae and to confirm them as undescribed species in Korea based on the morphological and multigene phylogenetic analyses.
MATERIALS AND METHODS
Isolation of fungal strains from rat dung and fig tree leaves
Rat dung samples were collected using sterile forceps, from the garden of Chonnam National University, Gwangju, Korea, in 2014. The samples were transferred to the laboratory in plastic bags, placed on sterile moist Whatman’s filter paper in Petri dishes, and incubated in a moist chamber at 25℃ for 7 days. Hyphal tips were transferred to potato dextrose agar (PDA; Difco, Franklin Lakes, NJ, USA) plates using a stereomicroscope. To isolate pure cultures, individual colonies of varied morphologies were transferred to PDA plates. On the other hand, leaf samples of fig tree (Ficus sp.) were collected from the campus of Chonnam National University, Gwangju, Korea, in 2014. Collected samples were stored in sterile polyethylene bags where they were cleaned under running tap water to remove debris before use, air dried, and then processed for isolation of endophytic fungi. The leaves were cut into small, 1 cm long and 0.5 cm wide pieces. Tissue pieces were surface-sterilized with 2% sodium hypochlorite for 3 min and with 70% ethanol for 1min, and then washed three times with sterile distilled water. The surface-sterilized samples were allowed to dry on sterile paper towel in a laminar airflow chamber. Ten fragments from each leaf were placed onto PDA and Rose Bengal chloramphenicol agar (Difco) supplemented with the antibiotic, streptomycin sulfate (0.4 mg/mL; Sigma-Aldrich, Munich, Germany), to suppress bacterial growth. After incubation at 25℃ for 5 days, individual hyphal tips of the developing fungal colonies were removed and placed onto in PDA medium to be incubated for 5~10 days.
To isolate pure cultures, individual colonies of varied morphologies were transferred to PDA plates. Pure isolates of P. variotii (EML-DG33-1 and EML-DG33-2) and T. amestolkiae (EML-NCP50) were maintained on PDA slant tubes and also stored in 20% glycerol at – 80℃ at the Environmental Microbiology Laboratory Fungarium, Chonnam National University, Gwangju. Dry cultures (EML-DG33-1 and EML-NCP50) were preserved at Chonnam National University Fungal Collection (CNUFC), Division of Food Technology, Biotechnology & Agrochemistry, College of Agriculture & Life Sciences, Gwangju. Duplicates of EML-DG33-1 and EML-NCP50 were also deposited as glycerol stock at – 80℃ at the Culture Collection of NIBR, Incheon, as KOSPFGC000002024 and KOSPFGC000002029, respectively.
Morphological studies
To obtain samples for microscopic examination and growth rate determination, EML-DG33-1 and EML-NCP50 were cultured on each of the three different media. The media used were malt extract agar (MEA; 30 g malt extract, 1 g peptone, 20 g glucose, 0.005 g CuSO4 · 5H2O, 0.01 g ZnSO4 · 7H2O, and 20 g agar, in 1 L of deionized water), Czapeck yeast autolysate agar (CYA; 3 g NaNO3, 5 g yeast extract, 30 g sucrose, 1.3 g K2HPO4 · 3H2O, 0.5 g MgSO4 · 7H2O, 0.5 g KCl, 0.01 g FeSO4 · 7H2O, 0.005 g CuSO4 · 7H2O, 0.01 g ZnSO4 · 7H2O, and 20 g agar, in 1 L of deionized water), and yeast extract sucrose agar (YES; 20 g yeast extract, 150 g sucrose, 0.5 g MgSO4 · 7H2O, 0.005 g CuSO4 · 7H2O, 0.01 g ZnSO4 · 7H2O, and 20 g agar, in 1 L of deionized water). Plates were incubated at 20℃, 25℃, 30℃, 35℃, and 40℃ in the dark for 7 days. Samples were mounted in lactophenol solution (Junsei Chemical Co. Ltd., Tokyo, Japan) and examined under a light microscope (DFC290; Leica Microsystems, Wetzlar, Germany). Fine fungal structures were observed using scanning electron microscopy (SEM, Hitachi S4700; Hitachi, Tokyo, Japan). Samples were cultured in PDA medium in the dark at 27℃ for 7 days, fixed in 2.5% paraformaldehyde-glutaraldehyde in 0.05M phosphate buffer (pH 7.2) for 1 hr, and then washed with cacodylate buffer (Junsei Chemical Co. Ltd.). Cellular membranes were preserved by fixing the samples in 1% osmium tetroxide (Electron Microscopy Sciences, Hatfield, PA, USA) diluted with cacodylate buffer for 1 hr, washing again in cacodylate buffer, dehydrating in graded ethanol (Emsure, Darmstadt, Germany) and isoamyl acetate (Junsei Chemical Co. Ltd.), and drying in a fume hood. Finally, samples were sputter-coated with gold and observed under a Hitachi S4700 field emission scanning electron microscope at the Korea Basic Science Institute, Gwangju.
DNA extraction, PCR, and sequencing
Total genomic DNA was extracted directly from the mycelia using the HiGene Genomic DNA Prep Kit (BIOFACT Corp., Daejeon, Korea). The gene sequences of EML-DG33-1 and EML-NCP50-1, consisting of the ITS, Tub2, and RPB2 genes, were amplified with the primer pairs ITS1, ITS4 [22]; Bt2a, Bt2b [23]; and RPB2-5F, RPB2-7Cr [20], respectively. The PCR amplification mixture (total volume of 20 µL) contained 10 ng of fungal genomic DNA template, 5 pmol/µL of each primer, and Accupower PCR Premix (Taq DNA polymerase, dNTPs, buffer, and a tracking dye; Bioneer Corp., Daejeon, Korea). PCR products were purified using the Accuprep PCR Purification Kit (Bioneer Corp.) according to the manufacturer’s instructions. DNA sequencing was performed on an ABI 3700 Automated DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA).
Phylogenetic analysis
Sequences (Table 1) were subjected to phylogenetic analysis using Clustal_X v.1.83 [24] and Bioedit v. 5.0.9.1 software [25]. Phylogenies were assessed using MEGA 6 software [26]. Maximum likelihood (ML) phylogenetic trees were constructed for combined datasets of the ITS rDNA, Tub2, and RPB2 gene sequences. The nearest-neighbor-interchange was selected for the ML heuristic method, and the initial ML tree was set automatically. Sequences of Byssochlamys verrucosa and Trichocoma paradoxa and Coccidioides immiti were used as outgroups. The percentage of sequence identity was obtained using the basic local alignment search tool (BLAST) of the National Center for Biotechnology Information (NCBI) for each isolate.
Table 1. Sequences used in this study, including GenBank accession numbers.
Bold letters indicate accession numbers determined in our study.
ITS, internal transcribed spacer; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; HT, holotype; IsoT, isotype; T, ex-type strain; NT, neotype; EML, Environmental Microbiology Laboratory, Fungarium, Chonnam National University, Gwangju, South Korea; NRRL, ARS Culture Collection, Peoria, Illinois, USA.
RESULTS
Phylogenetic analysis
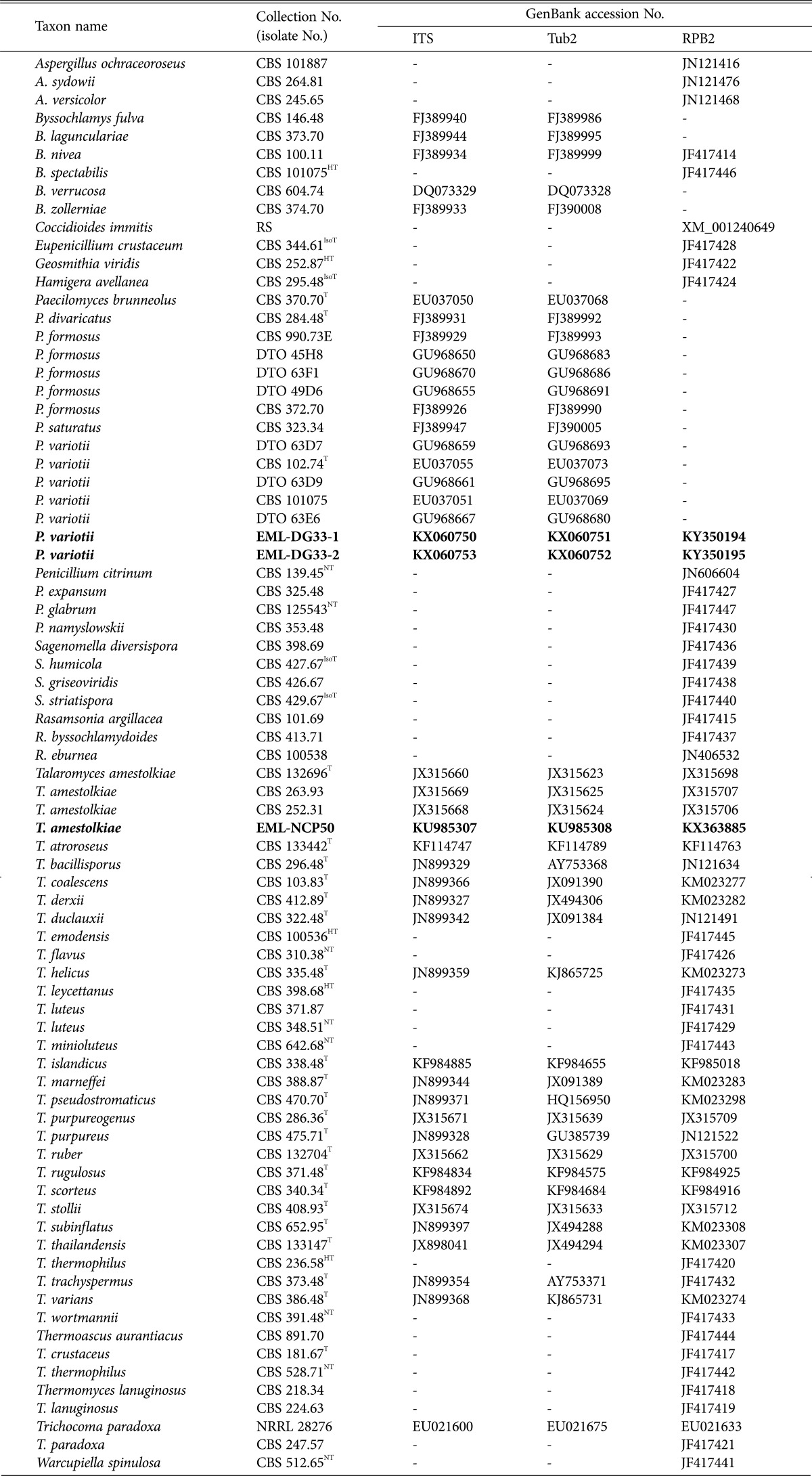
The groups of Eurotiales within ascomycete fungi has been known to be polyphyletic. In this study, the phylogenetic status of EML-DG33-1 and-2 isolates belonging to Paecilomyces & Byssochlamys clade and EML-NCP50 isolate belonging to Talaromyces s. str. and Penicillium subg. Biverticillium clade within Eurotiales are shown in the ML tree based on RPB2 sequence analysis (Fig. 1). The RPB2 sequence of EML-DG33-1 had 99.8% (971/973 bp) sequence similarity with the known sequence of a teleomorph of Paecilomyces, Byssochlamys spectabilis (GenBank accession No. JF417446).
Fig. 1. Phylogenetic relationships within genera Paecilomyces (teleomorph, Byssochlamys) and Talaromyces of the Trichocomaceae (Eurotiales) based on Maximum likelihood analysis of RPB2 sequence data. The corresponding sequence of Coccidioides immiti was used as the outgroup. Bootstrap support values > 50% are indicated at the nodes. The bar indicates the number of substitutions per position. Classification system presented by Houbraken et al. [3].
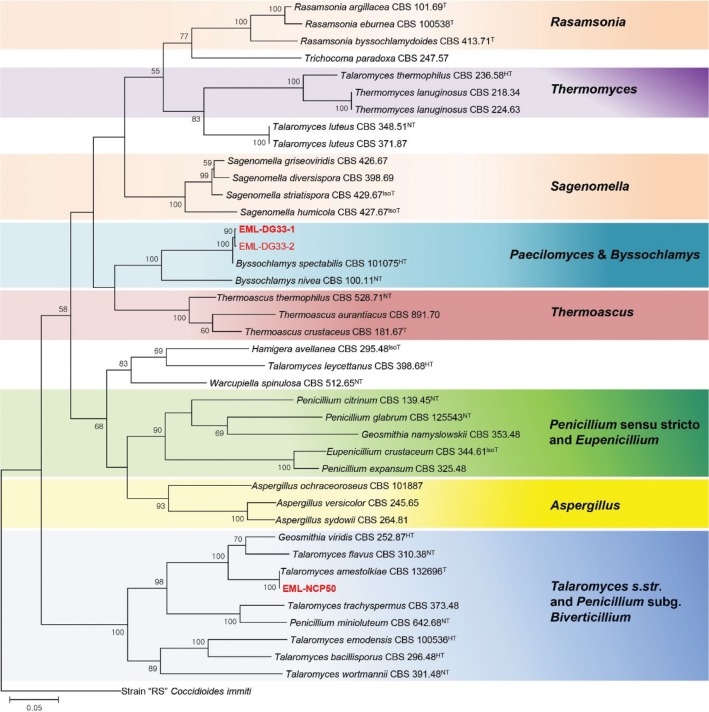
A BLAST search of ITS sequences via the NCBI database indicated that the ITS sequence of EML-DG33-1 is most closest to P. variotii DTO 63E (GenBank accession No. GU968667), with 98.7% (473/479 bp) homology. The Tub2 sequence of P. variotii DTO 63D9 (GenBank accession No. GU968695) showed 99.1% (447/451 bp) homology to that of EML-DG33-1. Based on this multigene analysis, the isolate was identified as P. variotii (Fig. 2). The ITS, Tub2, and RPB2 sequences of EML-DG33-1 and EML-DG33-2 were deposited in the NCBI database. The accession numbers for ITS, Tub2 and RPB2 sequences were KX060750, KX060751 and KY350194 for EML-DG33-1 isolate, and KX060753, KX060752, and KY350195 for EML-DG33-2 isolate, respectively. Analysis of the multigene genes placed the strains of EML-DG33-1 and EML-DG33-2 within the variotii group (Fig. 2).
Fig. 2. Phylogenetic tree of the Paecilomyces variotii EML-DG33-1 and EML-DG33-2 within the variotii group, based on maximum likelihood analysis of combined datasets for internal transcribed spacer rDNA, and Tub2. The corresponding sequence of Byssochlamys verrucosa was used as the outgroup. Bootstrap support values > 50% are indicated at the nodes. The bar indicates the number of substitutions per position. Classification presented by Houbraken et al. [27]. *Asterisk indicates classification system suggested by authors.
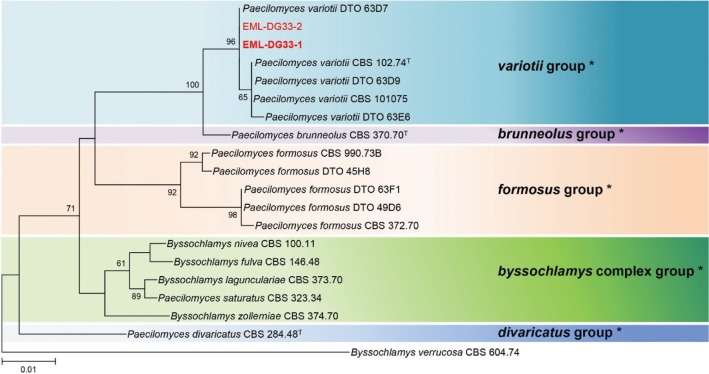
BLAST search analysis indicated that the ITS (accession No. KU985307), Tub2 (accession No. KU985308), and RPB2 (accession No. KX363885) sequences of EML-NCP50 had 99.8% (524/525 bp), 100% (427/427 bp), and 100% (1,038/1,038 bp) sequence similarity, respectively, with the known sequences of T. amestolkiae DTO179F5 (GenBank accession Nos. JX315660, JX315623, and JX315698, respectively). Analysis of the multigene genes placed the EML-NCP50 strain within the section Talaromyces (Fig. 3).
Fig. 3. Phylogenetic tree of the Talaromyces amestolkiae EML-NCP50 within the Sect. Talaromyces based on maximum likelihood analysis of combined datasets for internal transcribed spacer rDNA, Tub2, and RPB2. The corresponding sequence of Trichocoma paradoxa was used as the outgroup. Bootstrap support values > 50% are indicated at the nodes. The bar indicates the number of substitutions per position.
Taxonomy of EML-DG33-1
Paecilomyces variotii Bainier, Bull. Soc. Mycol. Fr. 23:27 (1907) (Table 2, Fig. 4)
Table 2. Morphological comparison of the isolate EML-DG33-1 and closely related species.
aFrom the description by Samson [7].
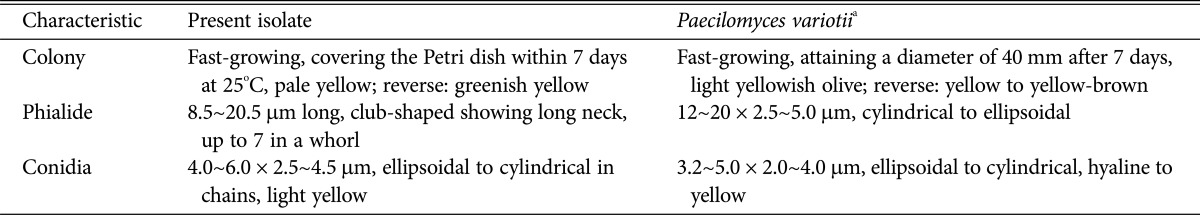
Fig. 4. Morphological and conidial characteristics of the Paecilomyces variotii isolate, EML-DG33-1. A, D, Colonies on malt extract agar; B, E, Colonies on yeast extract sucrose agar; C, F, Colonies on Czapeck yeast autolysate agar. A~C, Obverse view; D~F, Reverse view; G~J, Branched conidiophores and club-shaped phialides (arrows); K, Conidial chain (scale bars: G = 50 µm, H~J = 20 µm, K = 10 µm).
≡ Penicillium variotii (Bainier) Sacc., Syll. Fung. 22:1273 (1913)
=Penicillium aureocinnamomeum Biourge, La Cellule 33:213 (1923)
Description
The colonies of EML-DG33-1 exhibited fast growth on MEA, covering the Petri dish within 7 days at 25℃. The color of the colonies was pale yellow and white at the margins. The color of the colony reverse was greenish yellow (Fig. 4A and 4D). Mycelial growth on MEA was sparse; however, sporulation was extensive. Conidiophores were mostly short and irregularly branched (Fig. 4G and 4I). Phialides were club-shaped with a long neck, with up to seven in a whorl, and measured 8.5~20.5 µm in length (Fig. 3H). Conidia were ellipsoidal to cylindrical in chains, yellow in color, and measured 4.0~6.0 × 2.5~4.5 µm (Fig. 4K).
Colonies on YES grew faster than those on CYA, and attained a diameter of 53~56 mm after 7 days at 25℃. The color of the colonies was deep yellow, and the color of the colony reverse was dull brown. Conidia grown on YES were often slightly larger than those grown on CYA (Fig. 4B and 4E). Colonies on CYA grew very slowly, and attained a diameter of 40~42 mm after 7 days at 25℃. The color of the colonies was light yellow, and the color of the colony reverse was brownish yellow in the center and white at the margins (Fig. 4C and 4F).
Culture characteristics
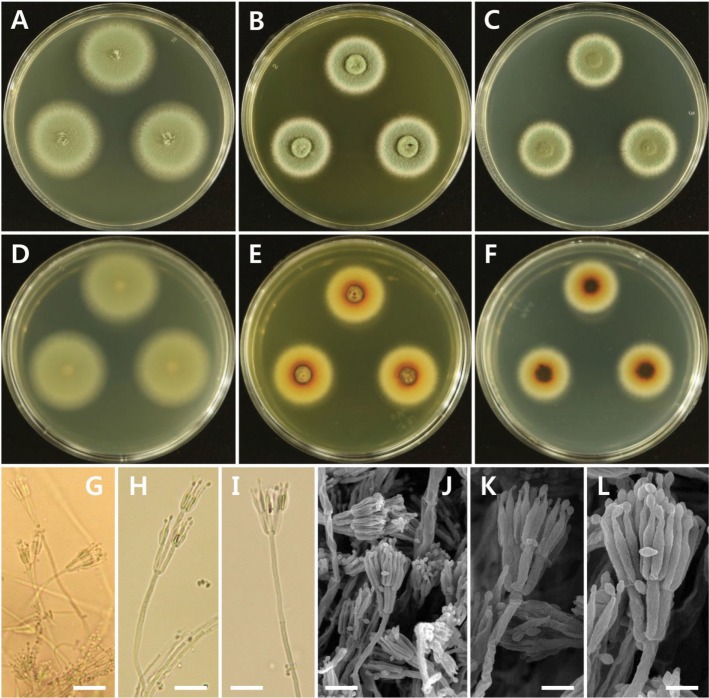
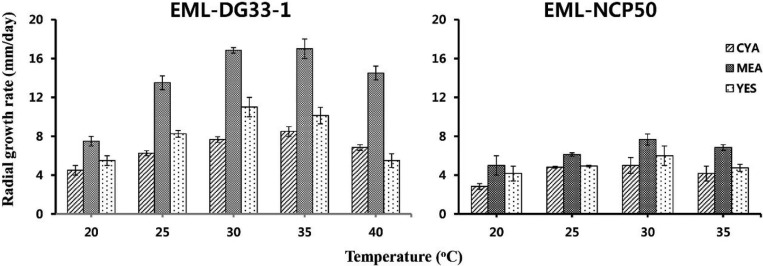
The isolate was observed to grow over a wide range of temperatures with varying growth rates on MEA, YES, and CYA. The average growth rates on MEA, YES, and CYA were 13.5, 8.0, and 6.0 mm/day, respectively. The optimum growth temperature range was 30~35℃, slow growth was observed at below 20℃, and could grow well at temperatures as high as 40℃.
Specimen examined
Republic of Korea, Jeonnam Province, garden of the Chonnam national University located in Gwangju (35°10' N, 126°55' E), from a rat dung, 10 Sep 2014 (EML-DG33-1), deposited at the Culture Collection of NIBR, Incheon, as KOSPFGC000002024.
Taxonomy of EML-NCP50
Talaromyces amestolkiae Yilmaz, Houbraken, Frisvad & Samson, Persoonia 29: 48 (2012) (Table 3, Fig. 5)
Table 3. Morphological comparison of the isolate EML-NCP50 and closely related species.
aFrom the description by Yilmaz et al. [19].
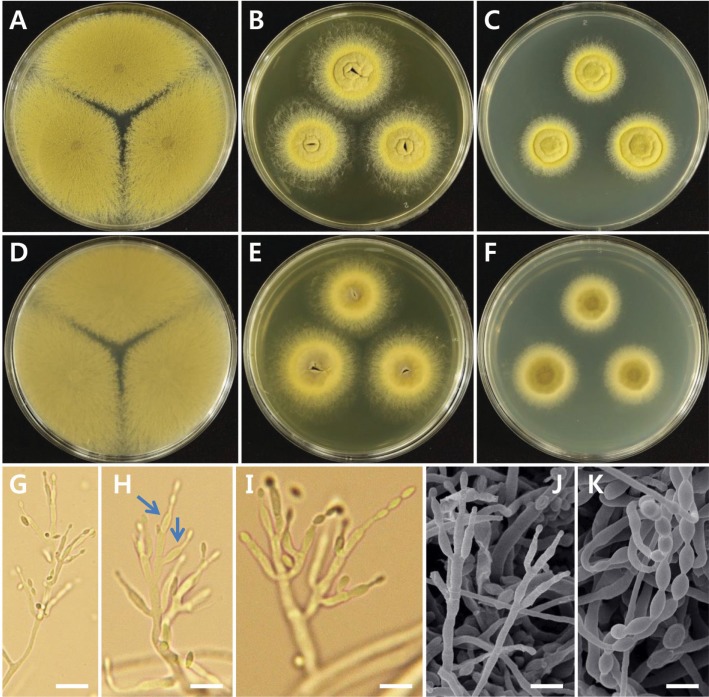
Fig. 5. Morphological characteristics of the Talaromyces amestolkiae isolate, EML-NCP50. Colonies on malt extract agar; B, E, Colonies on yeast extract sucrose agar; C, F, Colonies on Czapeck yeast autolysate agar. A~C, Obverse view; D~F, Reverse view; G~K, Conidiophores; L, Conidiophore and conidia (scale bars: G = 50 µm, H~J = 20 µm, K, L = 10 µm).
Description
Colonies were characterized after 7 days at 25℃. On the CYA agar, colonies were 21~24 mm in diameter and raised in the center. They had wide, regular margins (2~3 mm), white or yellow mycelium, which was reddish in the center, and moderately dense sporulation (Fig. 5C and 5F). On the YES agar, good sporulation on colonies was observed, whereas pigments and exudates were absent. The colonies were 34~36 mm in diameter. The mycelium on the YES agar was white; it exhibited moderately dense sporulation, narrow margins, and tufts (Fig. 5B and 5E). On the MEA agar, the mycelium was white, sometimes red in the center. Diameter of colonies that had regular wide margins varied within 34~53 mm. Sporulation varied from moderately dense to dense. Soluble pigment was absent (Fig. 5A and 5D).
The texture, conidiophores, conidia, metulae, phialide, and stripes were examined under a compound light microscope and SEM. Densely textured conidiophores were borne solitary or in small fascicles, simple or branched, smooth-walled, and pale to light brown in color (Fig. 5G and 5J). Conidia were smooth-walled, single-celled, ellipsoidal, and their size varied in the range of 2.5~4.0 × 1.0~3.5 µm (Fig. 5L). Metulae were arranged in verticils of 3~5; their length ranged from 9.5~13.0 µm and the width varied from 2.0~3.5 µm. There were 3~4 acerose phialides in total and their size varied in the range of 9.0~13.5 × 1.5~2.5 µm. Branches of stripes (2~3) were observed; they were smoothwalled, 86~201 µm long, and 2.0~3.0 µm wide.
Culture characteristics
The isolate was observed to grow over a wide range of temperatures with varying growth rates on MEA, YES, and CYA. The average growth rates on MEA, YES, and CYA were 6.5, 5.0, and 4.8 mm/day, respectively. Optimal growth was observed around 25~30℃, slow growth was observed at below 20℃, and no growth at 40℃ (Fig. 6).
Fig. 6. Effect of temperature and culture medium on mycelial growth of Paecilomyces variotii EML-DG33-1 and Talaromyces amestolkiae EML-NCP50. Mycelia were grown on malt extract agar (MEA), Czapeck yeast autolysate agar (CYA), and yeast extract sucrose agar (YES) at different temperatures, as indicated.
Specimen examined
Republic of Korea, Jeonnam Province, garden of the Chonnam national University located in Gwangju (35°10' N, 126°55' E), from a figs leaf, June 2014 (EML-NCP50), and deposited at the Culture Collection of NIBR, Incheon, as KOSPFGC000002029.
DISCUSSION
In this study, we combined a morphological description with a multigene analysis to assess the phylogenetic placement of a poorly reported species, Paecilomyces variotii in Korea. Although the species of Paecilomyces variotii has been only mentioned in several studies in Korea [15,21,28,29], there were no phylogenetic placement analyses or morphological descriptions or official record. Especially, the species of P. variotii has been used as a test microorganism in Korea, but it has not been described as undescribed species [30]. Because of lack of clear information about the species of P. variotii, confirmation of this species as undescribed species in Korea based on the detailed descriptions of the morphology and multigene phylogenetic analyses are required.
Dung is not only a rich source of nutrients, including carbohydrates, nitrogen, vitamins, minerals, and growth factors, but also contains a high amount of water with a neutral pH of around 6.5 [31,32]. Thus, it is considered a good substratum for fungal growth in a niche. A number of studies on the diversity of fungi have been reported with regard to different animal dung substrates [33,34,35]. Nyberg and Persson [36] observed 24 species in 14 genera in mouse dung, whereas Richardson [37] found 32 species in 17 genera from the dung of sheep, deer, cattle, rabbit, hare, and grouse. Although species of rat are among the most common animals worldwide, there is lack of information regarding the Paecilomyces species occurring in rat dung. Recently, the species P. variotii was isolated from the hair of golden hamster by Bagy et al. [38]. Notably, their study indicated that P. variotii and Aspergillus niger were the dominant groups of fungi recovered from the hair of golden hamsters. Furthermore, many species of fungi that have been isolated from dung produce secondary metabolites [32,39,40], such as appenolides A~C and coniochaetone-A, produced by Podospora appendiculata isolated from deer dung and Coniochaeta saccardoi isolated from lemming dung, respectively.
The ability of different species of the genus Paecilomyces to produce mycotoxin and other biological metabolites has been reported in previous studies [9]. According to Bokhari et al. [41], P. variotii produces the genotoxic mycotoxin, patulin. Peptide mycotoxins known as leucinostatins, with activity against fungi, are extracted from P. lilacinus and possess high toxicity to experimental animals [42]. In addition, Paecilomyces was used as a biological control. Perveen et al. [43] reported that the Paecilomyces species exhibit antifungal activity against Sclerotium rolfsii and Pythium aphanidermatum; whereas, P. lilacinus affects nematodes that attack plant roots [44].
In recent years, endophytes have received considerable scientific attention because they have been recognized as biological control agents. Vaz et al. [45] screened endophytic fungi from the leaves of Myrciaria floribunda, Alchornea castaneifolia, and Eugenia aff. bimarginata to examine their antimicrobial activity against pathogenic microorganisms. Thirty-eight fungal extracts exhibited antimicrobial activity against at least one of the target microorganisms tested. Similar results were obtained by Paul et al. [46,47], who showed that non-pathogenic endophytic fungi might reduce the growth of pathogenic C. acutatum, F. oxysporum, and Phytophthora capsici. In this study, the isolate EML-DG33-1 showed antifungal activity against A. alternata and F. oxysporum. The endophytic isolate EML-NCP50, weakly to moderately inhibited the growth of eight fungal pathogens (data not shown). This result is consistent with previous reports demonstrating that many endophytes exhibit antifungal activity against different pathogenic fungi [45]. Our study suggests that the strains, EML-DG33-1 and EML-NCP50, might be a source of biologically active secondary metabolites and biological control agents.
In addition, in the present study, we demonstrated the thermal tolerance of this EML-DG33-1 strain. In comparison with previous results [7], the colony morphology and cultural characteristics of the present isolate on MEA medium was similar to that of P. variotii.
Based on the sequences of the 18S and ITS rDNA regions, Luangsa-ard et al. [48] and Inglis and Tigano [49] demonstrated that Paecilomyces is polyphyletic across two Ascomycetes orders, the Eurotiales and the Hypocreales. Luangsa-ard et al. [48] analyzed the 18S rDNA and showed that the species P. variotii and its thermophilic relatives belong to the order Eurotiales (Trichocomaceae), and the mesophilic species related to P. farinosus are in the order Hypocreales (Clavicipitaceae and Hypocreaceae). In another aspect, Samson et al. [9] studied the genus Byssochlamys and its Paecilomyces anamorphs using a polyphasic approach to differentiate the species by analyzing each of ITS region, parts of the Tub2 and calmodulin gene. Our results showed that the strains EML-DG33-1 and EML-DG33-2 belonged to a variotii group containing P. variotii species. There was only two RPB2 sequences data in the Paecilomyces & Byssochlamys clade available in GenBank (Fig. 1). The results of phylogenetic analysis showed that this species belongs to the same clade presented by Houbraken et al. [27].
To our knowledge, the genus Talaromyces has been frequently isolated from soil and root aquatic in Korea [50,51]. Figs trees are rarely found in Korea. There have been no studies of fungal endophytes on the host in Korea. Thus, this finding suggest that figs tree may be useful source of fungal endophytics. Endophytic microorganisms have been discovered in all plant families, representing various species in different climatic regions of the world [52]. Species of fig tree (Ficus sp.) are native throughout the tropical region with several species extending into the semi-warm temperate zones. In 2001, Suryanarayanan and Vijaykrishna [53] isolated the endophytes from the aerial root of fig tree (Ficus benghalensis) in India. Wang et al. [54] investigated the fungal diversity on fallen leaves of Ficus in northern Thailand in 2008.
So far, several studies to evaluate and compare the phylogenetic relationships between some species belonging to the genus Talaromyces have been carried out, using the ITS region, β-tubulin, and RPB2 [18,19]. Phylogenetic analyses of three genes showed that EML-NCP50 belonged to the genus Talaromyces, including T. amestolkiae, T. stollii, and T. ruber. Morphological characteristics of the isolate EML-NCP50 were similar to those of T. amestolkiae described by Yilmaz et al. [19]. Therefore, these results confirmed that the isolate EML–NCP50 belongs to the species T. amestolkiae within sect. Talaromyces.
Some genera including Paecilomyces and Talaromyces belonging to Eurotiales within class Ascomycetes are polyphyletic (Fig. 1). Thus, to include unrecorded species in a national species list, it is very important to confirm the first records of poorly known species based on the detailed morphological descriptions and the current molecular phylogenetic placement analyses.
ACKNOWLEDGEMENTS
This work was supported by the Project on Survey and Discovery of Indigenous Species of Korea funded by NIBR of the Ministry of Environment, and in part by a fund from the National Institute of Animal Science under Rural Development Administration, Republic of Korea. We are grateful to Dr. Paul Kirk from Royal Botanic Gardens, Kew, United Kingdom, for kind review of the manuscript.
References
- 1.Benny GL, Kimbrough JW. A synopsis of the orders and families of Plectomycetes with keys to genera. Mycotaxon. 1980;12:1–91. [Google Scholar]
- 2.Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houbraken J, Spierenburg H, Frisvad JC. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek. 2012;101:403–421. doi: 10.1007/s10482-011-9647-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geiser DM, Gueidan C, Miadlikowska J, Lutzoni F, Kauff F, Hofstetter V, Fraker E, Schoch CL, Tibell L, Untereiner WA, et al. Eurotiomycetes: Eurotiomycetidae and Chaetothyriomycetidae. Mycologia. 2006;98:1053–1064. doi: 10.3852/mycologia.98.6.1053. [DOI] [PubMed] [Google Scholar]
- 5.Bainier G. Mycothèque de l'École de Pharmacie. XI. Paecilomyces, genre nouveau de Mucédinées. Bull Trimest Soc Mycol Fr. 1907;23:26–27. [Google Scholar]
- 6.Brown AH, Smith G. The genus Paecilomyces Bainier and its perfect stage Byssochlamys Westling. Trans Br Mycol Soc. 1957;40:17–89. [Google Scholar]
- 7.Samson RA. Paecilomyces and some allied Hyphomycetes. Stud Mycol. 1974;6:1–119. [Google Scholar]
- 8.He J, Kang J, Lei B, Wen T. Paecilomyces wawuensis, a new species isolated from soil in China. Mycotaxon. 2011;115:303–310. [Google Scholar]
- 9.Samson RA, Houbraken J, Varga J, Frisvad JC. Polyphasic taxonomy of the heat resistant ascomycete genus Byssochlamys and its Paecilomyces anamorphs. Persoonia. 2009;22:14–27. doi: 10.3767/003158509X418925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houbraken J, Varga J, Rico-Munoz E, Johnson S, Samson RA. Sexual reproduction as the cause of heat resistance in the food spoilage fungus Byssochlamys spectabilis (anamorph Paecilomyces variotii) Appl Environ Microbiol. 2008;74:1613–1619. doi: 10.1128/AEM.01761-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou K, Zhao XL, Han LP, Cao MM, Chen C, Shi BZ, Luo DQ. Paecilomycines A and B, novel diterpenoids, isolated from insect-pathogenic fungi Paecilomyces sp. ACCC 37762. Helv Chim Acta. 2015;98:642–649. [Google Scholar]
- 12.Marzec A, Heron LG, Pritchard RC, Butcher RH, Powell HR, Disney AP, Tosolin FA. Paecilomyces variotii in peritoneal dialysate. J Clin Microbiol. 1993;31:2392–2395. doi: 10.1128/jcm.31.9.2392-2395.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor FJ, Guarro J. Clinical manifestations, treatment and outcome of Paecilomyces lilacinus infections. Clin Microbiol Infect. 2006;12:948–960. doi: 10.1111/j.1469-0691.2006.01481.x. [DOI] [PubMed] [Google Scholar]
- 14.Dong Q, Wang H, Xing X, Ji S. Identification and characterization of a special species of Paecilomyces. Ann Microbiol. 2012;62:1587–1592. [Google Scholar]
- 15.Liu J, Li F, Kim EL, Li JL, Hong J, Bae KS, Chung HY, Kim HS, Jung JH. Antibacterial polyketides from the jellyfishderived fungus Paecilomyces variotii. J Nat Prod. 2011;74:1826–1829. doi: 10.1021/np200350b. [DOI] [PubMed] [Google Scholar]
- 16.Benjamin CR. Ascocarps of Aspergillus and Penicillium. Mycologia. 1955;47:669–687. [Google Scholar]
- 17.Stolk AC, Samson RA. The genus Talaromyces: studies on Talaromyces and related genera II. Mycologia. 1973;65:1221–1223. [Google Scholar]
- 18.Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol. 2011;70:159–183. doi: 10.3114/sim.2011.70.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yilmaz N, Visagie CM, Houbraken J, Frisvad JC, Samson RA. Polyphasic taxonomy of the genus Talaromyces. Stud Mycol. 2014;78:175–341. doi: 10.1016/j.simyco.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- 21.Lee YS, Jung HY, Lee HB, Kim SH, Shin KS, Eom AH, Kim C, Lee SY Korean Society of Mycology. National list of species of Korea. Ascomycota, Glomeromycota, Zygomycota, Myxomycota, Oomycota. Incheon: National Institute of Biological Resources; 2015. [Google Scholar]
- 22.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and application. San Diego (CA): Academic Press; 1990. pp. 315–322. [Google Scholar]
- 23.Houbraken J, Due M, Varga J, Meijer M, Frisvad JC, Samson RA. Polyphasic taxonomy of Aspergillus section Usti. Stud Mycol. 2007;59:107–128. doi: 10.3114/sim.2007.59.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- 26.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houbraken J, Verweij PE, Rijs AJ, Borman AM, Samson RA. Identification of Paecilomyces variotii in clinical samples and settings. J Clin Microbiol. 2010;48:2754–2761. doi: 10.1128/JCM.00764-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim MJ, Lee H, Choi YS, Kim GH, Huh NY, Lee S, Lim YW, Lee SS, Kim JJ. Diversity of fungi in creosote-treated crosstie wastes and their resistance to polycyclic aromatic hydrocarbons. Antonie Van Leeuwenhoek. 2010;97:377–387. doi: 10.1007/s10482-010-9416-6. [DOI] [PubMed] [Google Scholar]
- 29.Kim HR, Kim JH, Bai DH, Ahn BH. Identification and characterization of useful fungi with α-amylase activity from the Korean traditional Nuruk. Mycobiology. 2011;39:278–282. doi: 10.5941/MYCO.2011.39.4.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee S, Xu S, Bivila CP, Lee H, Park MS, Lim YW, Yamamoto N. Triazole susceptibilities in thermotolerant fungal isolates from outdoor air in the Seoul capital area in South Korea. PLoS One. 2015;10:e0138725. doi: 10.1371/journal.pone.0138725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webster J. Presidential address: coprophilous fungi. Trans Br Mycol Soc. 1970;54:161–180. [Google Scholar]
- 32.Sarrocco S. Dung-inhabiting fungi: a potential reservoir of novel secondary metabolites for the control of plant pathogens. Pest Manag Sci. 2016;72:643–652. doi: 10.1002/ps.4206. [DOI] [PubMed] [Google Scholar]
- 33.Thilagam L, Nayak BK, Nanda A. Studies on the diversity of coprophilous microfungi from hybrid cow dung samples. Int J PharmTech Res. 2015;8:135–138. [Google Scholar]
- 34.Nguyen TT, Lee SH, Bae S, Jeon SJ, Mun HY, Lee HB. Characterization of two new records of zygomycete species belonging to undiscovered taxa in Korea. Mycobiology. 2016;44:29–37. doi: 10.5941/MYCO.2016.44.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Alvarado P, Alves-Silva G, Ammirati JF, Ariyawansa HA, et al. Fungal diversity notes 253-366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 2016;78:1–237. [Google Scholar]
- 36.Nyberg Å, Persson IL. Habitat differences of coprophilous fungi on moose dung. Mycol Res. 2002;106:1360–1366. [Google Scholar]
- 37.Richardson MJ. Diversity and occurrence of coprophilous fungi. Mycol Res. 2001;105:387–402. [Google Scholar]
- 38.Bagy MM, Abdel-Mallek AY, El-Sahanawany AA, Morsi GA. Studies on fungi associated with laboratory animal ‘golden hamster’ and antibiotic effects of aloe sap, garlic extract and onion oil. J Islam Acad Sci. 1997;10:3–12. [Google Scholar]
- 39.Wang Y, Gloer JB, Scott JA, Malloch D. Appenolides A-C: three new antifungal furanones from the coprophilous fungus Podospora appendiculata. J Nat Prod. 1993;56:341–344. doi: 10.1021/np50093a005. [DOI] [PubMed] [Google Scholar]
- 40.Wang H, Gloer JB, Scott JA, Malloch D. Coniochaetones A and B: new antifungal benzopyranones from the coprophilous fungus Coniochaeta saccardoi. Tetrahedron Lett. 1995;36:5847–5850. [Google Scholar]
- 41.Bokhari F, Gherbawy Y, Najjar A. Detection of the patulinproducing potential of some Paecilomyces variotii strains isolated from the air samples of Jeddah City, Saudi Arabia, using the RAPD-PCR technique. Aerobiologia. 2009;25:49–54. [Google Scholar]
- 42.Mikami Y, Fukushima K, Arai T, Abe F, Shibuya H, Ommura Y. Leucinostatins, peptide mycotoxins produced by Paecilomyces lilacinus and their possible roles in fungal infection. Zentralbl Bakteriol Mikrobiol Hyg A. 1984;257:275–283. [PubMed] [Google Scholar]
- 43.Perveen Z, Ramzan N, Noreen N, Rajpuit AQ, Shahzad S. In vitro evaluation of biocontrol potential of Paecilomyces species against Sclorotium rolfsii and Pythium aphanidermatum. Int J Biol Biotechnol. 2015;12:407–411. [Google Scholar]
- 44.Anastasiadis IA, Giannakou IO, Prophetou-Athanasiadou DA, Gowen SR. The combined effect of the application of a biocontrol agent Paecilomyces lilacinus, with various practices for the control of root-knot nematodes. Crop Prot. 2008;27:352–361. [Google Scholar]
- 45.Vaz AB, Brandão LR, Vieira ML, Pimenta RS, Morais PB, Sobral ME, Rosa LH, Rosa CA. Diversity and antimicrobial activity of fungal endophyte communities associated with plants of Brazilian savanna ecosystems. Afr J Microbiol Res. 2012;6:3173–3185. [Google Scholar]
- 46.Paul NC, Kim WK, Woo SK, Park MS, Yu SH. Fungal endophytes in roots of Aralia species and their antifungal activity. Plant Pathol J. 2007;23:287–294. [Google Scholar]
- 47.Paul NC, Deng JX, Sang HK, Choi YP, Yu SH. Distribution and antifungal activity of endophytic fungi in different growth stages of chili pepper (Capsicum annuum L.) in Korea. Plant Pathol J. 2012;28:10–19. [Google Scholar]
- 48.Luangsa-ard JJ, Hywel-Jones NL, Samson RA. The polyphyletic nature of Paecilomyces sensu lato based on 18S-generated rDNA phylogeny. Mycologia. 2004;96:773–780. doi: 10.1080/15572536.2005.11832925. [DOI] [PubMed] [Google Scholar]
- 49.Inglis PW, Tigano MS. Identification and taxonomy of some entomopathogenic Paecilomyces spp. (Ascomycota) isolates using rDNA-ITS sequences. Genet Mol Biol. 2006;29:132–136. [Google Scholar]
- 50.Adhikari M, Yadav DR, Kim S, Um YH, Kim HS, Lee HB, Lee YS. Discovery of two new Talaromyces species from crop field soil in Korea. Mycobiology. 2015;43:402–407. doi: 10.5941/MYCO.2015.43.4.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.You YH, Park JM, Park JH, Kim JG. Diversity of endophytic fungi associated with the roots of four aquatic plants inhabiting two wetlands in Korea. Mycobiology. 2015;43:231–238. doi: 10.5941/MYCO.2015.43.3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fisher PJ, Petrini O. Location of fungal endophytes in tissues of Suaeda fruiticosa: a preliminary study. Trans Br Mycol Soc. 1987;89:246–249. [Google Scholar]
- 53.Suryanarayanan TS, Vijaykrishna D. Fungal endophytes of aerial roots of Ficus benghalensis. Fungal Divers. 2001;8:155–161. [Google Scholar]
- 54.Wang HK, Hyde KD, Soytong K, Lin FC. Fungal diversity on fallen leaves of Ficus in northern Thailand. J Zhejiang Univ Sci B. 2008;9:835–841. doi: 10.1631/jzus.B0860005. [DOI] [PMC free article] [PubMed] [Google Scholar]