Abstract
Super-enhancers (SEs) are key transcriptional drivers of cellular, developmental, and disease states in mammals, yet the conservational and regulatory features of these enhancer elements in nonmammalian vertebrates are unknown. To define SEs in zebrafish and enable sequence and functional comparisons to mouse and human SEs, we used genome-wide histone H3 lysine 27 acetylation (H3K27ac) occupancy as a primary SE delineator. Our study determined the set of SEs in pluripotent state cells and adult zebrafish tissues and revealed both similarities and differences between zebrafish and mammalian SEs. Although the total number of SEs was proportional to the genome size, the genomic distribution of zebrafish SEs differed from that of the mammalian SEs. Despite the evolutionary distance separating zebrafish and mammals and the low overall SE sequence conservation, ∼42% of zebrafish SEs were located in close proximity to orthologs that also were associated with SEs in mouse and human. Compared to their nonassociated counterparts, higher sequence conservation was revealed for those SEs that have maintained orthologous gene associations. Functional dissection of two of these SEs identified conserved sequence elements and tissue-specific expression patterns, while chromatin accessibility analyses predicted transcription factors governing the function of pluripotent state zebrafish SEs. Our zebrafish annotations and comparative studies show the extent of SE usage and their conservation across vertebrates, permitting future gene regulatory studies in several tissues.
The identification of transcriptional regulators is central for understanding tissue-specific expression programs. Enhancers are cis-regulatory elements able to recruit transcription factors (TFs) and the transcriptional apparatus to activate their target gene expression (Smith and Shilatifard 2014; Heinz et al. 2015; Ren and Yue 2015). Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) has been a frequently used strategy to generate genome-wide enhancer annotations (Visel et al. 2009; Bernstein et al. 2010; Creyghton et al. 2010; Rada-Iglesias et al. 2011; Kieffer-Kwon et al. 2013; Vermunt et al. 2014; Prescott et al. 2015; Villar et al. 2015). ChIP-seq-based approaches have shown that a subset of mammalian enhancers are found in close sequence proximity to one another, forming large regions of hyperactive chromatin referred to as super-enhancers (SEs) or stretch enhancers (Lovén et al. 2013; Parker et al. 2013; Whyte et al. 2013). This structure distinguishes them from shorter, more compacted regions referred to as typical enhancers.
SEs are characterized by their high level of histone H3 lysine 27 acetylation (H3K27ac) density, a mark associated with active enhancers and promoters (Creyghton et al. 2010; Rada-Iglesias et al. 2011), and the binding of a high abundance of TFs, transcriptional coactivators, and chromatin remodelers (Hnisz et al. 2013; Whyte et al. 2013). Analyses of the SE dynamics during lineage commitment of specific cell types have shown that SEs are remodeled during differentiation, having crucial roles in cell fate determination (Adam et al. 2015; Thakurela et al. 2015; Vahedi et al. 2015). Moreover, SEs are enriched for single nucleotide polymorphisms (SNPs) associated with a broad spectrum of diseases including but not limited to cancers, type 1 diabetes, Alzheimer's disease, and multiple sclerosis (Hnisz et al. 2013; Parker et al. 2013; Vahedi et al. 2015). For example, a fraction of human T-cell acute lymphoblastic leukemia cases exhibits somatic mutations that create MYB TF binding sites that generate a SE adjacent to the TAL1 oncogene (Mansour et al. 2014). Despite a basic understanding of the features and functions of mammalian SEs and a recently published catalog of SEs in nonvertebrates (Wei et al. 2016), the extent to which the defining characteristics of mammalian SEs also apply to similar regulatory regions in species outside of the mammalian clade is not known.
Comparative analyses of enhancers in different species have been invaluable for our understanding of their evolution (for review, see Domené et al. 2013; Rubinstein and de Souza 2013). Here, we employed the zebrafish model as an exemplar to define SE biology in vertebrates (Howe et al. 2013; Kaufman et al. 2016). Previous studies of zebrafish have successfully identified stage-specific enhancers involved in early development and have highlighted their general low sequence conservation (Aday et al. 2011; Bogdanović et al. 2012; Lee et al. 2015). Although these enhancer annotations open the possibility to gain fundamental insights into gene regulation during embryonic development, they do not address the tissue-specificity of enhancers in zebrafish.
To identify cell- and tissue-specific enhancers, in particular SEs, we analyzed the distribution of H3K27ac in zebrafish pluripotent cells and four adult tissues. Our comparative analyses of zebrafish, mouse, and human SEs highlight their differences and similarities and advance the study of gene regulation in zebrafish by identifying a set of SE candidates involved in cellular identity.
Results
H3K27ac marks hundreds of SEs in zebrafish
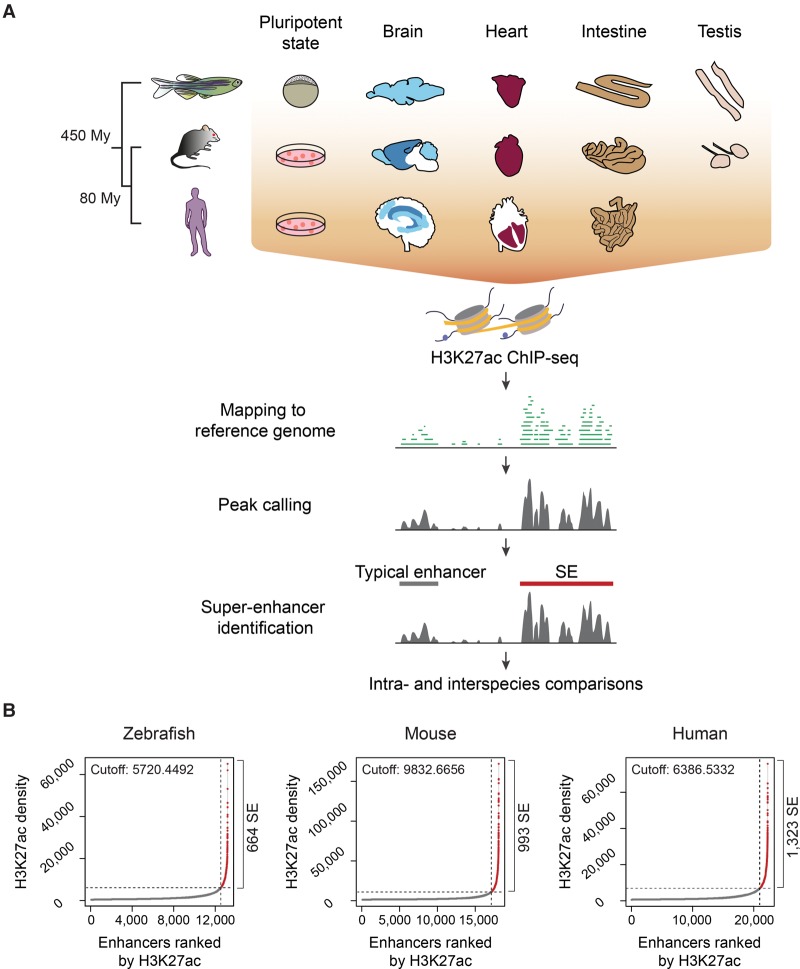
To assess characteristic features of vertebrate SEs, we identified enhancer regions in zebrafish (Danio rerio), mouse, and human brain, heart, intestine, testis, and pluripotent cells. For zebrafish, we used the early embryonic dome stage as a comparative stage to the pluripotent state of mouse and human ESCs (Schier and Talbot 2005). All mouse and human enhancer annotations, as well as zebrafish pluripotent state enhancer annotations, were based on publicly available data sets of the H3K27ac mark, whereas those of the zebrafish adult brain, heart, intestine, and testis were performed using in-house generated H3K27ac ChIP-seq data sets (Fig. 1A; Supplemental Table S1; Bernstein et al. 2010; Rada-Iglesias et al. 2011; Bogdanović et al. 2012; Chadwick et al. 2012; Mouse ENCODE Consortium 2012; Nord et al. 2013; Yue et al. 2014). To identify typical enhancers and SEs, H3K27ac-enriched regions were identified with SICER (Zang et al. 2009), filtered to discard active promoters, and stitched by the ROSE software (Fig. 1A; Lovén et al. 2013; Whyte et al. 2013). We identified an average of 743 and 1183 SEs for zebrafish and mammals, respectively (Fig. 1B; Supplemental Table S1; Supplemental Data set S1). Similar to mammalian SEs, most zebrafish SEs were longer than typical enhancers, although the length parameter was not explicitly considered for their identification (Supplemental Fig. S1A–C; examples of typical enhancers and SEs are shown in Supplemental Fig. S2A).
Figure 1.
Identification of typical enhancers and SEs in vertebrate genomes. (A) Workflow for the identification of vertebrate typical enhancers and SEs. Schematic representations depict the cells and tissues analyzed. (B) Saturation curves of H3K27ac density across brain data sets (whole brain for zebrafish, olfactory bulb for mouse, and middle frontal lobe for human). The number of ranked typical enhancers and SEs by H3K27ac density (x-axis) and their densities (y-axis) are plotted. Horizontal dotted lines represent density cutoffs used for the classification of SEs and vertical dotted lines demark SEs from typical enhancers. The total number of predicted SEs is noted on the right side of each graph.
Genomic distribution of zebrafish typical enhancers and SEs differs from that of mammalian regions
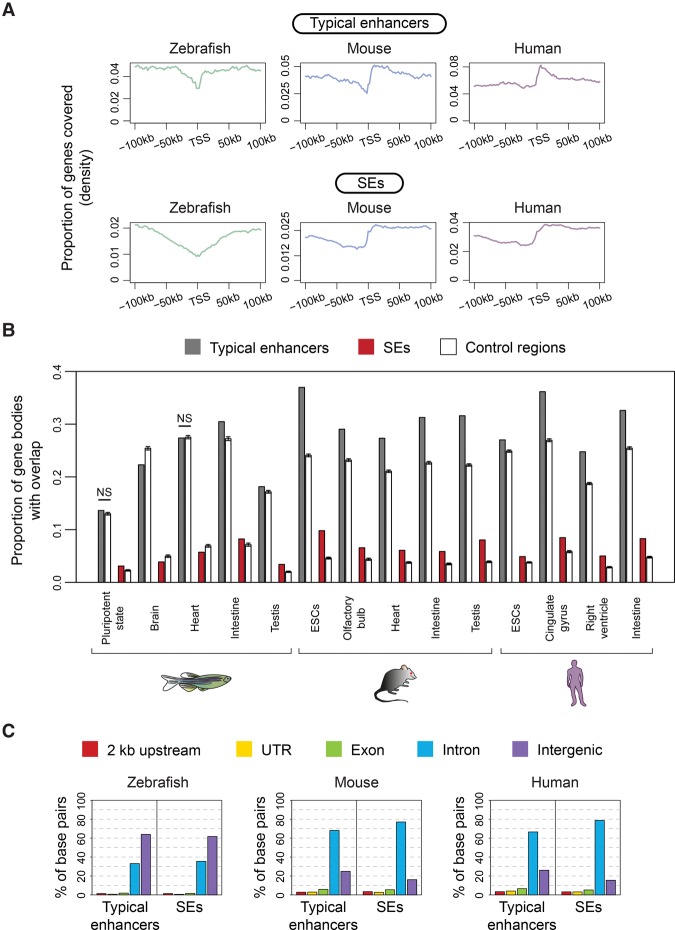
In contrast to mammalian SEs, which tend to overlap with gene bodies (Lovén et al. 2013; Whyte et al. 2013), neither zebrafish typical enhancers nor zebrafish SEs were preferentially enriched in the TSS downstream regions in any tissue or at any embryonic stage analyzed (Fig. 2A; Supplemental Fig. S2B). To assess if zebrafish typical enhancers and SEs were enriched in gene bodies, the proportion of genes covered by typical enhancers and SEs was calculated and compared to the proportion of genes covered by random control regions. As expected, mouse and human typical enhancers and SEs from all analyzed samples showed significant enrichments in gene bodies (P-values from z-scores ≤4.71 × 10−18), whereas gene-body enrichment of zebrafish typical enhancers and SEs showed variation among the different cells and tissues analyzed (Fig. 2B). Furthermore, we found that on average for all cells and tissues analyzed, ∼65% and ∼73% of mouse and ∼70% and ∼80% of human typical enhancer and SE sequences, respectively, overlapped introns (Fig. 2C). In zebrafish, only ∼28% of typical enhancer and ∼29% of SE sequences overlapped introns, and the majority of zebrafish typical enhancer and SE sequences (∼67% and ∼66%, respectively) overlapped intergenic regions in all zebrafish cells and adult tissues (Fig. 2C; Supplemental Fig. S2C). These drastic differences in genomic distribution cannot be solely explained by differences in the global genome composition of the three species, as >50% of the zebrafish, mouse, and human genomes correspond to intergenic sequences (Supplemental Fig. S2D).
Figure 2.
Genomic distribution of typical enhancers and SEs. (A) Density plots representing the proportion of genes (y-axis) covered by typical enhancers and SEs in the vicinity of TSSs (x-axis) in zebrafish brain, mouse cerebellum, and human angular gyrus. (B) Proportion of gene bodies overlapping with typical enhancers, SEs, and control regions (y-axis) in different zebrafish, mouse, and human cells and tissues (x-axis). The mean and the standard deviation (black bars) calculated from bootstrap analyses of control regions are shown. All comparisons between typical enhancers and SEs and their controls have significant differences (P-values from z-scores ≤3 × 10−4), with the exception of zebrafish pluripotent state and heart typical enhancers. (NS) Not significant. (C) Distribution of typical enhancer and SE sequences across genomic features. The y-axis shows the percentage of total brain typical enhancer or SE base pairs overlapping the different genomic features represented in the legend. Adult brain data sets for mouse and human correspond to olfactory bulb and cingulate gyrus, respectively.
Vertebrate SEs are more cell- and tissue-specific than typical enhancers
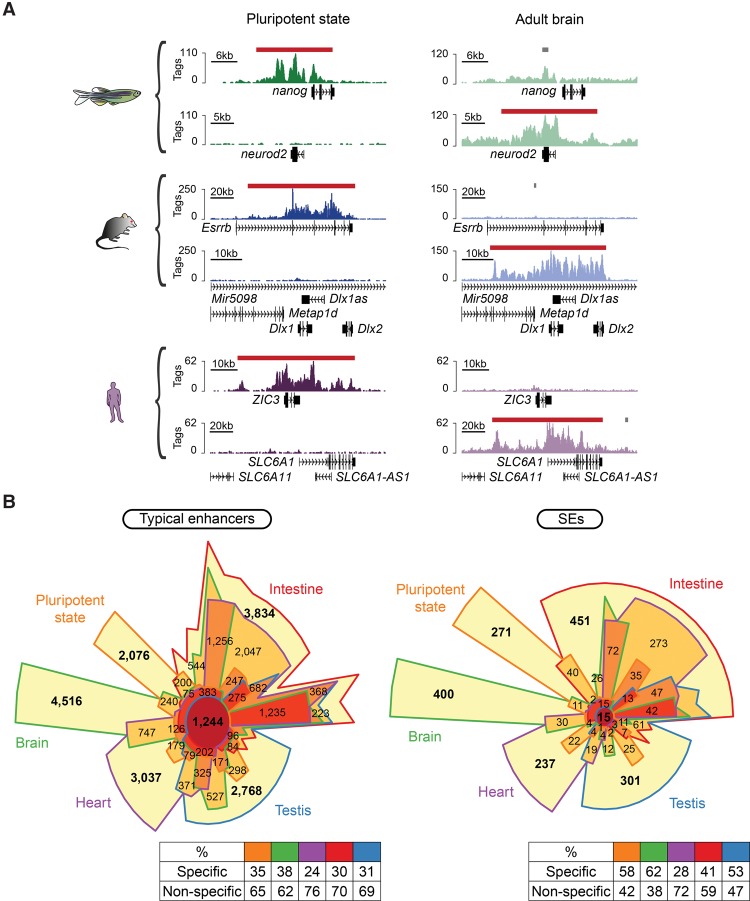
A notable characteristic of mammalian SEs is their association with key cellular identity genes (Fig. 3A; Hnisz et al. 2013; Whyte et al. 2013). Similar to mouse and human SEs, gene ontology (GO) annotations of the zebrafish SEs in pluripotent state, brain, heart, intestine, and testis showed enriched terms related to early development and pluripotency, neuronal components, signal transduction, immune pathways, and chromatin organization, respectively (Supplemental Fig. S3). In addition, our intra-species comparisons showed that, similar to mammals (Hnisz et al. 2013), zebrafish SEs exhibit higher cell- and tissue-specificity than typical enhancers (P-values from G-tests of independence ≤8.5 × 10−13, with the exception of zebrafish heart) (Fig. 3B; Supplemental Fig. S4).
Figure 3.
Cell and tissue specificity of vertebrate typical enhancers and SEs. (A) Distribution of H3K27ac at selected genes (genomic position represented on the x-axis) in both pluripotent state and adult brain of zebrafish, mouse, and human (raw tag counts represented on the y-axis). Typical enhancers and SEs are denoted by gray bars and red bars, respectively. (B) Chow-Ruskey diagrams representing the overlap between pluripotent state (orange), brain (green), heart (purple), intestine (red), and testis (blue) typical enhancers and SEs in zebrafish. Color-coded tables show the percentages of cell- or tissue-specific and nonspecific regions for each data set.
SEs associate with a conserved set of genes throughout vertebrate evolution
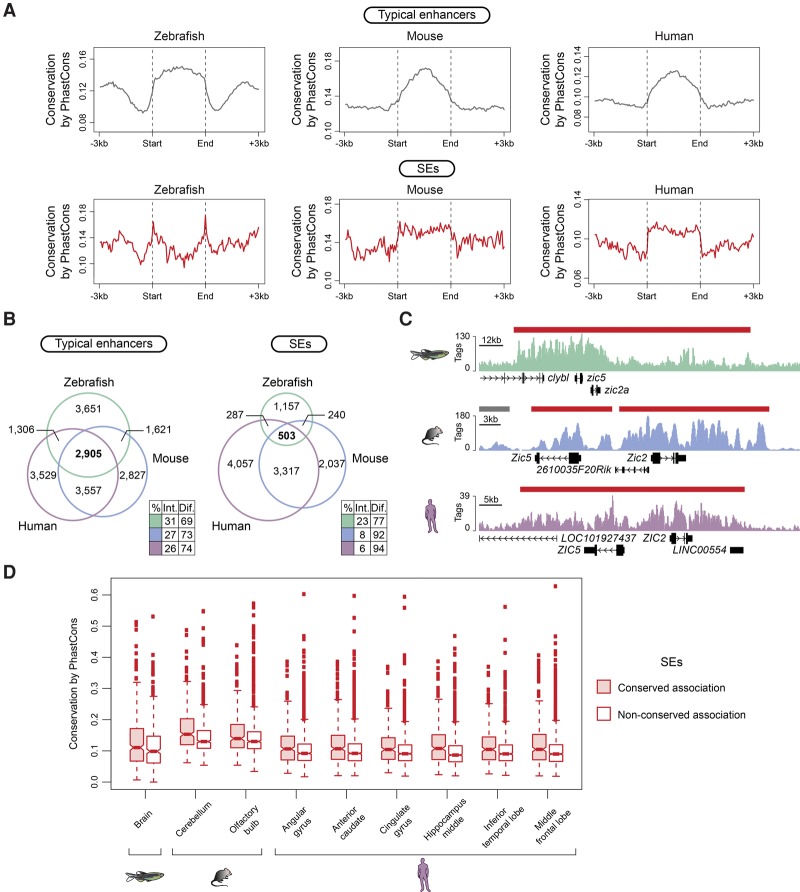
Collectively, typical enhancers and SEs showed higher sequence conservation than their immediate flanking regions (P-values from Wilcoxon rank-sum test ≤2.8 × 10−4, with the exception of typical enhancers from the right ventricle of the human heart) (Fig. 4A). While zebrafish SEs from most tissues analyzed had significantly higher sequence conservation than zebrafish typical enhancers (P-values from Wilcoxon rank-sum test ≤9.3 × 10−4), mouse and human sequence conservation differences were dependent on the tissue analyzed (Supplemental Fig. S5A). When we compared individual intergenic regions enriched for H3K27ac within typical enhancers and SEs, the higher conservation found for full-length SEs was diminished, and, for most of the data sets, typical enhancer regions were more conserved than SE regions (P-values from Wilcoxon rank-sum test ≤3.7 × 10−3) (Supplemental Fig. S5B). This observation is consistent with the fact that a higher proportion of SE constitutive regions overlaps intragenic sequences, which could artificially inflate the SE conservation estimate when analyzed as a whole unit (Supplemental Fig. S5C).
Figure 4.
SE conservation in vertebrates. (A) Metagenes of sequence conservation of typical enhancers and SEs from zebrafish whole brain, mouse olfactory bulb, and human middle frontal lobe. The x-axis depicts the start and end of typical enhancers and SEs flanked by 3 kb of adjacent sequence. The y-axis represents sequence conservation calculated by PhastCons. (B) Venn diagrams show the number of orthologous genes associated with brain typical enhancers (left) and SEs (right) in zebrafish (green), mouse (blue), and human (purple). Color-coded tables show the percentages of intersection and difference for each species. The observed differences in overlap between typical enhancers and SEs in the three species are significant (P-values ≤5.497 × 10−8) based on G-tests of independence. (C) ChIP-seq binding profiles for H3K27ac at the indicated loci in zebrafish, mouse, and human brain (raw tag counts represented on the y-axis). Typical enhancers and SEs are denoted by gray bars and red bars, respectively. Gene positions are noted along the x-axis. (D) Box plots depicting average sequence conservation of brain SEs with maintained orthologous association in zebrafish, mouse, and human and with no maintained orthologous association. The y-axis shows sequence conservation calculated by PhastCons. The box bounds the interquartile range divided by the median, and the notch approximates a 95% confidence interval for the median. All observed differences in conservation between SE categories are significant (P-value ≤9.1 × 10−3) based on Wilcoxon rank-sum tests.
Next, to determine if SEs tend to maintain their spatial association with orthologous genes throughout evolution, the genes associated with zebrafish, mouse, and human typical enhancers and SEs were compared based on homology annotations. The proportion of orthologous genes associated with typical enhancers in all three species was significantly larger than that associated with SEs (P-values from G-tests of independence ≤5.497 × 10−8) (Fig. 4B; Supplemental Fig. S6A–D; Supplemental Table S2). Approximately 42% of zebrafish SEs were associated with orthologous genes in mouse and human (pluripotent state = 110/473; brain = 321/664; heart = 325/850; intestine = 462/1145; testis = 362/581), and ∼27% and ∼21% of the mouse and human SEs, respectively, maintained their orthologous associations (examples are illustrated in Fig. 4C; Supplemental Fig. S6E–H). Importantly, mammalian SEs with conserved orthologous gene associations in the three species had higher sequence conservation than the nonassociated-SEs (P-values from Wilcoxon rank-sum test ≤4.7 × 10−3). Similar results were also observed for the zebrafish brain and testis SEs (P-values from Wilcoxon rank-sum test ≤9.1 × 10−3) (Fig. 4D; Supplemental Fig. 6I). Thus, despite overall low sequence conservation in vertebrates, SEs that maintained orthologous gene associations exhibited higher conservation at the sequence level than those lacking such associations.
Analysis of accessible chromatin identifies differences between zebrafish typical enhancer and SE composition
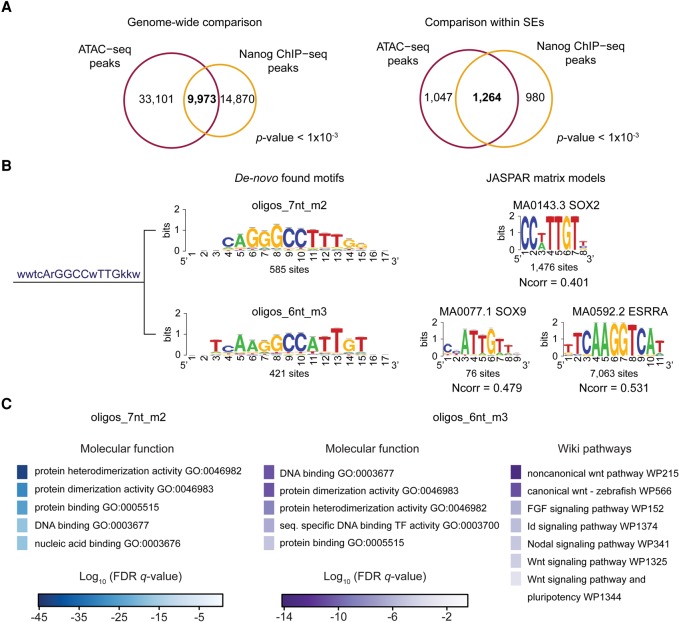
Within zebrafish SEs, we sought to demarcate transcription factor binding site (TFBS) hotspots or epicenters, defined as regions shorter than 1 kb bound by at least five TFs involved in cell identity (Siersbæk et al. 2014; Adam et al. 2015). To overcome the lack of zebrafish ChIP-seq data, we focused on the identification of accessible chromatin regions by ATAC-seq (Supplemental Fig. S7A; Buenrostro et al. 2013). To confirm that ATAC-seq data can be mined to identify TFBSs in zebrafish, we compared pluripotent state ATAC-seq (Kaaij et al. 2016) and Nanog ChIP-seq peaks (Xu et al. 2012). These comparisons showed significant overlap at both the genome-wide level and within SEs (P-value < 1 × 10−3) (Fig. 5A).
Figure 5.
Analysis of zebrafish SE composition by ATAC-seq. (A) Venn diagrams representing the overlap between ATAC-seq peaks (purple) and Nanog peaks (orange) genome-wide (left) and within pluripotent state SEs (right). (B) Cluster, consensus motif sequence, and logos of SOX-related de novo–found motifs in ATAC-seq peaks within SEs (left). JASPAR matrix models (right) of SOX2, SOX9, and ESRRA. (Ncorr) Normalized correlation between identified motifs and JASPAR models. (C) Top molecular function and wiki pathway GO terms enriched for the ATAC-seq peaks containing sites of the de novo identified oligos_7nt_m2 (left) and oligos_6nt_m3 (right) motifs shown in B. Binomial FDR q-values for the GO terms are displayed in a color-scale (q-values ≤6.7 × 10−4).
A differential analysis of ATAC-seq peaks within typical enhancers and SEs identified 12 clusters of overrepresented motifs within SEs (Supplemental Fig. S7B). Our set of consensus motifs included those with similarity to matrix models of pluripotency-associated TFs, such as SOX2, EOMES, and FOXD3 (Sutton et al. 1996; Hromas et al. 1999; Avilion et al. 2003; Kidder and Palmer 2010). The motif that correlated with the SOX2 matrix was the consensus of two motifs: one similar to the SOX2 matrix model and the second motif similar to the SOX9 and ESRRA matrix models (Fig. 5B). GO annotation of the SE ATAC-seq peaks containing sites of these two motifs showed enrichment for TF function and pluripotency terms that were not identified by the global analysis of pluripotent state SEs (Fig. 5C; Supplemental Fig. S3A). Thus, our results predict a set of TFs with enriched binding to accessible chromatin regions highly associated with pluripotency.
Dissections of vertebrate SEs identify functionally conserved elements
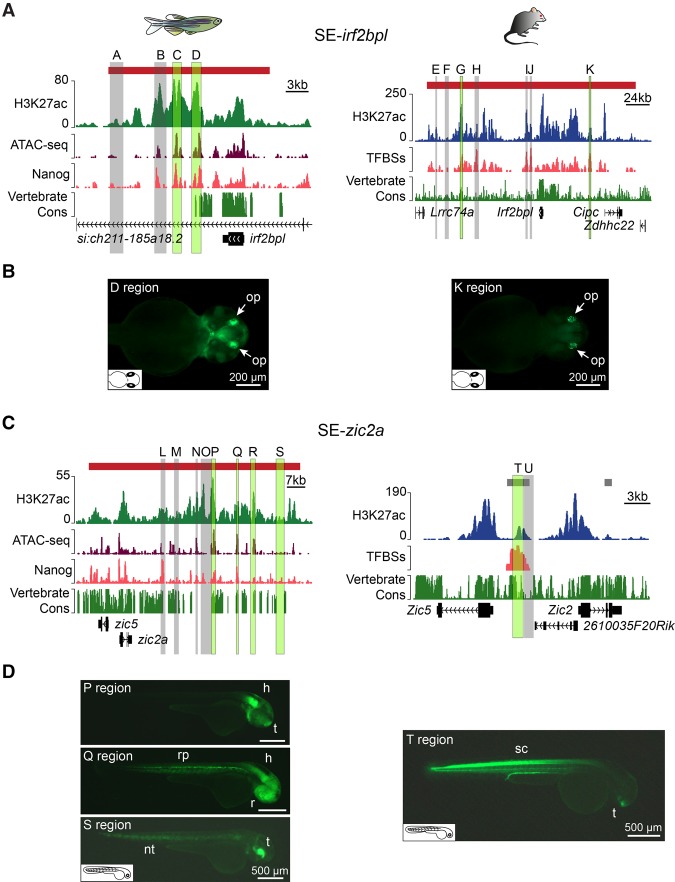
To determine the different contribution of regions within SEs, two SEs having conserved associations with irf2bpl and zic2a (hereafter referred to as SE-irf2bpl and SE-zic2a) (Fig. 4C; Supplemental Fig. S6A) were tested by GFP reporter assays in zebrafish embryos (Supplemental Fig. S8A). Twelve zebrafish gene distal regions were selected for the enhancer activity test based on their H3K27ac, ATAC-seq, and Nanog ChIP-seq profiles (Fig. 6; Supplemental Table S3). To evaluate the functional conservation of the equivalent mouse SEs, nine mouse regions, selected based on the presence or absence of TFBSs for 14 pluripotent state TFs, were tested (Supplemental Fig. S7A; Supplemental Table S3; Chen et al. 2008; Heng et al. 2010; Ma et al. 2011; Vella et al. 2012; Betschinger et al. 2013; Whyte et al. 2013). It should be noted that while the mouse Zic2–associated region is a typical enhancer at the pluripotent state (Fig. 6C), it is identified as a SE in the brain (Fig. 4C).
Figure 6.
Functional analysis of vertebrate SEs. (A) Genomic context and conservation of the zebrafish (left) and mouse (right) irf2bpl and Irf2bpl loci. Horizontal bars represent SEs (red). Raw H3K27ac ChIP-seq, ATAC-seq, and Nanog ChIP-seq profiles are shown in tag counts (y-axis). The TFBS track represents the TFBS enrichment along the mouse locus. The Vertebrate Cons tracks represent conservation scores calculated by PhastCons. Gray and green highlighted regions correspond to the regions tested in reporter assays. Regions driving specific GFP expression are indicated in green. (B) GFP expression driven by the zebrafish SE-irf2bpl D region (left) and the mouse K region (right) in transgenic zebrafish embryos at 48 hpf. White arrows indicate the olfactory placode (op). (C) Genomic context and conservation of the zebrafish and mouse zic2a and Zic2 loci as described in A. Horizontal bars represent typical enhancers (gray) and SEs (red). (D) GFP expression driven by the zebrafish P, Q, and S regions (left) and the mouse T region (right). (h) Hindbrain, (nt) notochord, (r) retina, (rp) roof plate, (sc) spinal cord, (t) telencephalon.
For zebrafish SE-irf2bpl, there was a strong concordance between enhancer activity and the presence of a high ATAC-seq signal (Fig. 6A,B; Supplemental Fig. S8B). Remarkably, the GFP expression pattern driven by the conserved zebrafish region D and the mouse region K (Fig. 6A) substantially overlapped within the olfactory placode (Fig. 6B). Similarly, the mouse region G (Fig. 6A) drove dim GFP expression in the olfactory placode at ∼24 h post-fertilization (hpf) with peak GFP expression in the roof plate at 48 hpf (Supplemental Fig. S8B).
For zebrafish SE-zic2a, 75% of SE-zic2a regions exhibiting enhancer activity also contained ATAC-seq peaks and displayed high sequence conservation (the P, Q, and R regions) (Fig. 6C,D; Supplemental Fig. S8C). Interestingly, the zebrafish S region, originally selected as a control region based on the lack of sequence conservation and the absence of H3K27ac and ATAC-seq signals, drove specific GFP expression in the notochord and telencephalon (Fig. 6D) similar to the spinal cord and telencephalon expression driven by the equivalent mouse T region (Fig. 6D). As the S region contained a mildly enriched Nanog peak (Fig. 6C) and predicted TFBSs (Supplemental Table S3), it likely corresponds to a redundant or “shadow” enhancer that is not active under homeostatic conditions and, consequently, is not found by ATAC-seq (Fig. 6C).
Taken together, our results confirm that SEs contain regions with evolutionary conserved enhancer functions and emphasize the importance of analyzing comprehensive hyperactive chromatin regions instead of isolated enhancers to allow the identification of enhancers with partially redundant activities.
Discussion
In this study, we identify tissue-specific enhancers in zebrafish, focusing on hyperactive chromatin regions or SEs. Our comparative analyses support a model in which SEs specify uniquely important cell- and tissue-specific regulatory regions across species (Hnisz et al. 2013; Saint-André et al. 2016) and highlight the difference in genomic distribution between zebrafish and mammalian SEs. While the majority of mammalian SEs overlap with their target genes (Whyte et al. 2013), zebrafish typical enhancers and SEs are mainly located within intergenic regions. Similarly, during early zebrafish development, differentially methylated DNA regions, ∼50% of which are enriched for enhancer-associated chromatin marks including H3K27ac, are mainly embedded within intergenic sequences (Lee et al. 2015). Future analyses incorporating the enhancer annotations of additional species may reveal if the intergenic distribution of zebrafish regulatory regions is a distinctive feature.
Similar to what has been shown for zebrafish and mammalian enhancers (Bogdanović et al. 2012; Lee et al. 2015; Villar et al. 2015), our PhastCons value-based sequence conservation analysis showed that both zebrafish typical enhancers and SEs have overall low sequence conservation and that SE intergenic constitutive regions do not display higher conservation than those of typical enhancers. However, the sequence conservation was detectably higher in the fraction of SEs that has maintained an association with orthologous genes in zebrafish, mouse, and human compared to the fraction lacking conserved orthologous associations. It remains to be determined if those SEs with orthologous gene associations have an evolutionary common origin, or if they independently evolved in the three species. Notably, enhancers shared between human and chimp also display higher sequence conservation than species-biased enhancers (Prescott et al. 2015).
Previous studies have reported enhancer regions with overlapping functions in phylogenetically distant species (Hare et al. 2008; Taher et al. 2011; Clarke et al. 2012). However, the genome-wide prediction of those regions is not trivial (Taher et al. 2011), as sequence conservation alone does not necessarily predict functional conservation, and regions with high sequence conservation can drive different patterns of expression in reporter assays (Goode et al. 2011). Thus, it is remarkable that we defined equivalent subregions in two SEs with conserved enhancer functions. Although the extent of enhancer redundancy is poorly understood, a recent study has shown the genome-wide pervasiveness of shadow enhancers during Drosophila development (Cannavò et al. 2016). Indeed, one of the zebrafish SE regions identified in this study likely represents a shadow enhancer with a conserved function. For these reasons, we propose that the future identification of shadow enhancers will benefit from the analysis of whole hyperactive chromatin regions rather than the analysis of isolated enhancers.
Our study reveals the genome-wide distribution of tissue-specific cis-regulatory elements in zebrafish and identifies the key SE complement in this important model system. Moreover, the characterized genomic distribution of zebrafish typical enhancers and SEs, together with our comparative analyses to those of mammals, solidifies our understanding of pervasive and conserved vertebrate transcriptional mechanisms.
Methods
ChIP-seq assays
Whole brains, hearts, intestines, and testis were dissected from same-age adult male AB zebrafish. Two biological replicates were prepared from each tissue. ChIP-seq was performed as previously described (Guenther et al. 2008) using Abcam H3K27ac antibody (ab4729, lot# GR259887-1). Purified chromatin was used for single-end library preparation following standard Illumina protocols. For more details, see Supplemental Material.
Identification of typical enhancers and SEs
H3K27ac ChIP-seq data sets were mapped to their corresponding reference genomes (Zv9 for zebrafish, mm10 for mouse, and hg38 for human) using Bowtie 2 version 2.1.0 (Langmead and Salzberg 2012). Peak calling was performed with SICER version 1.1 (Zang et al. 2009); if available, input libraries were used as controls for the peak calling (Supplemental Table S1). Identified peaks were filtered to discard peaks for which the main summit was within promoter regions and used as input for the ROSE algorithm version 0.1 to identify typical enhancers and SEs. For detailed parameters, see Supplemental Material and Supplemental Files S1,S2.
Computational analyses
The calculation of typical enhancer and SE distributions around TSSs was performed using Nebula (Boeva et al. 2012). Typical enhancer and SE enrichments over gene bodies were calculated with a customized script (Supplemental File S3) and control enrichments were obtained by bootstrap resampling with 100 iterations. To calculate the percentage of typical enhancer and SE sequences overlapping with genomic features, typical enhancer and SE annotations were compared to RefSeq Gene annotations (Rosenbloom et al. 2015) using BEDTools intersect function (Quinlan and Hall 2010). Sequence conservation scores were calculated based on the vertebrate conservation PhastCons tracks from UCSC (Siepel and Haussler 2005; Siepel et al. 2005) associated with each of the genome versions used for read mapping using hgWiggle (Kent et al. 2002) and a customized Python script (Supplemental File S4). For ortholog comparisons, typical enhancer and SE target genes were annotated based on gene proximity using Nebula. All gene names were converted to Ensembl ids and compared based on homology annotations from Ensembl (Genes 82) (Cunningham et al. 2015). Analysis of the ATAC-seq library was performed as previously described (Buenrostro et al. 2013). Overrepresented motifs in ATAC-seq peaks within SEs were identified using the RSAT peak-motifs tool (Thomas-Chollier et al. 2012a,b). For more details, see Supplemental Material.
Microinjections
Each of the vectors containing SE regions (for cloning details, see the Supplemental Material) was co-injected with Tol2 mRNA into one-cell stage zebrafish embryos. GFP expression was monitored during the first 3 d post-fertilization. All injection experiments were repeated at least twice (Supplemental Table S3). For more details, see Supplemental Material.
Data access
Zebrafish H3K27ac ChIP-seq data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) (Edgar et al. 2002) under accession number GSE75734.
Supplementary Material
Acknowledgments
We thank Igor Ulitsky, Matthew Guenther, and Violaine Saint-André for helpful comments on this manuscript. We also thank all members of the Shkumatava lab for help with zebrafish dissections and for useful discussions. High-throughput sequencing was performed by the ICGex NGS platform of Institut Curie supported by the grants ANR-10-EQPX-03 (Equipex) and ANR-10-INBS-09-08 (France Génomique Consortium) from the ANR (“Investissements d'Avenir” program) and by the Canceropole Île-de-France. This work was supported by grants from the European Research Council (FLAME-337440), ATIP-Avenir and La Fondation Bettencourt Schueller and Fondation pour la Recherche Medicale (FRM DBI201312285578). Y.A.P.-R. was partially funded by a scholarship from Secretaría de Ciencia, Tecnología e Innovación – Seciti, México.
Author contributions: A.S. conceived and designed the project. Y.A.P.-R., V.B., A.C.M., and A.S. designed experiments; A.C.M. and A.S. performed zebrafish ChIP-seq; Y.A.P.-R. performed computational analyses and prepared plasmid constructs; Y.A.P.-R. and A.B. performed microinjections and microscopy; S.M. assisted with experimental work; Y.A.P.-R., A.C.M., and A.S. wrote the manuscript. All authors reviewed and approved the manuscript; V.B., E.B., and A.S. supervised the project.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.203679.115.
References
- Adam RC, Yang H, Rockowitz S, Larsen SB, Nikolova M, Oristian DS, Polak L, Kadaja M, Asare A, Zheng D, et al. 2015. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521: 366–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND. 2011. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol 357: 450–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. 2003. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev 17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. 2010. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol 28: 1045–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Nichols J, Dietmann S, Corrin PD, Paddison PJ, Smith A. 2013. Exit from pluripotency is gated by intracellular redistribution of the bHLH transcription factor Tfe3. Cell 153: 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Lermine A, Barette C, Guillouf C, Barillot E. 2012. Nebula—a web-server for advanced ChIP-seq data analysis. Bioinformatics 28: 2517–2519. [DOI] [PubMed] [Google Scholar]
- Bogdanović O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJC, Gomez-Skarmeta JL. 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22: 2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavò E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, Ciglar L, Korbel JO, Furlong EE. 2016. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol 26: 38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick LH. 2012. The NIH Roadmap Epigenomics Program data resource. Epigenomics 4: 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Clarke SL, VanderMeer JE, Wenger AM, Schaar BT, Ahituv N, Bejerano G. 2012. Human developmental enhancers conserved between deuterostomes and protostomes. PLoS Genet 8: e1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2015. Ensembl 2015. Nucleic Acids Res 43: D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domené S, Bumaschny VF, de Souza FSJ, Franchini LF, Nasif S, Low MJ, Rubinstein M. 2013. Enhancer turnover and conserved regulatory function in vertebrate evolution. Philos Trans R Soc Lond B Biol Sci 368: 20130027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode DK, Callaway HA, Cerda GA, Lewis KE, Elgar G. 2011. Minor change, major difference: divergent functions of highly conserved cis-regulatory elements subsequent to whole genome duplication events. Development 138: 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Lawton LN, Rozovskaia T, Frampton GM, Levine SS, Volkert TL, Croce CM, Nakamura T, Canaani E, Young RA. 2008. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev 22: 3403–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare EE, Peterson BK, Iyer VN, Meier R, Eisen MB. 2008. Sepsid even-skipped enhancers are functionally conserved in Drosophila despite lack of sequence conservation. PLoS Genet 4: e1000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Romanoski CE, Benner C, Glass CK. 2015. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol 16: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JCD, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. 2010. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Stem Cell 6: 167–174. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, Hoke HA, Young RA. 2013. Super-enhancers in the control of cell identity and disease. Cell 155: 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hromas R, Ye H, Spinella M, Dmitrovsky E, Xu D, Costa RH. 1999. Genesis, a Winged Helix transcriptional repressor, has embryonic expression limited to the neural crest, and stimulates proliferation in vitro in a neural development model. Cell Tissue Res 297: 371–382. [DOI] [PubMed] [Google Scholar]
- Kaaij LJ, Mokry M, Zhou M, Musheev M, Geeven G, Melquiond AS, de Jesus Domingues AM, de Laat W, Niehrs C, Smith AD, et al. 2016. Enhancers reside in a unique epigenetic environment during early zebrafish development. Genome Biol 17: 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. 2016. A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351: aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidder BL, Palmer S. 2010. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res 20: 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al. 2013. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 155: 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lowdon RF, Maricque B, Zhang B, Stevens M, Li D, Johnson SL, Wang T. 2015. Developmental enhancers revealed by extensive DNA methylome maps of zebrafish early embryos. Nat Commun 6: 6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153: 320–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. 2011. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol 18: 120–127. [DOI] [PubMed] [Google Scholar]
- Mansour MR, Abraham BJ, Anders L, Berezovskaya A, Gutierrez A, Durbin AD, Etchin J, Lawton L, Sallan SE, Silverman LB, et al. 2014. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science 346: 1373–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse ENCODE Consortium. 2012. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol 13: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, Black BL, et al. 2013. Chromatin stretch enhancer states drive cell-specific gene regulation and harbour human disease risk variants. Proc Natl Acad Sci 110: 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott SL, Srinivasan R, Marchetto MC, Grishina I, Narvaiza I, Selleri L, Gage FH, Swigut T, Wysocka J. 2015. Enhancer divergence and cis-regulatory evolution in the human and chimp neural crest. Cell 163: 68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B, Yue F. 2015. Transcriptional enhancers: bridging the genome and phenome. Cold Spring Harb Symp Quant Biol 80: 17–26. [DOI] [PubMed] [Google Scholar]
- Rosenbloom KR, Armstrong J, Barber GP, Casper J, Clawson H, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2015. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 43: D670–D681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M, de Souza FSJ. 2013. Evolution of transcriptional enhancers and animal diversity. Philos Trans R Soc Lond B Biol Sci 368: 20130017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-André V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, Bradner JE, Young RA. 2016. Models of human core transcriptional regulatory circuitries. Genome Res 26: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A, Talbot WS. 2005. Molecular genetics of axis formation in zebrafish. Annu Rev Genet 39: 561–613. [DOI] [PubMed] [Google Scholar]
- Siepel A, Haussler D. 2005. Phylogenetic hidden Markov models. In Statistical methods in molecular evolution (ed. Nielsen R), pp. 325–351. Springer, New York. [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. 2005. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res 15: 1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbæk R, Rabiee A, Nielsen R, Sidoli S, Traynor S, Loft A, La Cour Poulsen L, Rogowska-Wrzesinska A, Jensen ON, Mandrup S. 2014. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep 7: 1443–1455. [DOI] [PubMed] [Google Scholar]
- Smith E, Shilatifard A. 2014. Enhancer biology and enhanceropathies. Nat Struct Mol Biol 21: 210–219. [DOI] [PubMed] [Google Scholar]
- Sutton J, Costa R, Klug M, Field L, Xu D, Largaespada DA, Fletcher CF, Jenkins NA, Copeland NG, Klemsz M, et al. 1996. Genesis, a winged helix transcriptional repressor with expression restricted to embryonic stem cells. J Biol Chem 271: 23126–23133. [DOI] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, Ovcharenko I. 2011. Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res 21: 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakurela S, Sahu SK, Garding A, Tiwari VK. 2015. Dynamics and function of distal regulatory elements during neurogenesis and neuroplasticity. Genome Res 25: 1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Herrmann C, Defrance M, Sand O, Thieffry D, van Helden J. 2012a. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res 40: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M, Darbo E, Herrmann C, Defrance M, Thieffry D, van Helden J. 2012b. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat Protoc 7: 1551–1568. [DOI] [PubMed] [Google Scholar]
- Vahedi G, Kanno Y, Furumoto Y, Jiang K, Parker SCJ, Erdos MR, Davis SR, Roychoudhuri R, Restifo NP, Gadina M, et al. 2015. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature 520: 558–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella P, Barozzi I, Cuomo A, Bonaldi T, Pasini D. 2012. Yin Yang 1 extends the Myc-related transcription factors network in embryonic stem cells. Nucleic Acids Res 40: 3403–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermunt MW, Reinink P, Korving J, de Bruijn E, Creyghton PM, Basak O, Geeven G, Toonen PW, Lansu N, Meunier C, et al. 2014. Large-scale identification of coregulated enhancer networks in the adult human brain. Cell Rep 9: 767–779. [DOI] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Zhang S, Shang S, Zhang B, Li S, Wang X, Wang F, Su J, Wu Q, Liu H, et al. 2016. SEA: a super-enhancer archive. Nucleic Acids Res 44: D172–D179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fan ZP, Müller P, Fogley R, DiBiase A, Trompouki E, Unternaehrer J, Xiong F, Torregroza I, Evans T, et al. 2012. Nanog-like regulates endoderm formation through the Mxtx2-nodal pathway. Dev Cell 22: 625–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, Sandstrom R, Ma Z, Davis C, Pope BD, et al. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature 515: 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. 2009. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 25: 1952–1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.