Abstract
Calorie restriction can extend life span in a variety of species including mammals, flies, nematodes, and yeast. Despite the importance of this nearly universal effect, little is understood about the molecular mechanisms that mediate the life-span-extending effect of calorie restriction in metazoans. Sir2 is known to be involved in life span determination and calorie restriction in yeast mother cells. In nematodes increased Sir2 can extend life span, but a direct link to calorie restriction has not been demonstrated. We now report that Sir2 is directly involved in the calorie-restriction life-span-extending pathway in Drosophila. We demonstrate that an increase in Drosophila Sir2 (dSir2) extends life span, whereas a decrease in dSir2 blocks the life-span-extending effect of calorie reduction or rpd3 mutations. These data lead us to propose a genetic pathway by which calorie restriction extends life span and provides a framework for genetic and pharmacological studies of life span extension in metazoans.
Keywords: aging, life span, Rpd3, histone deacetylase, Drosophila melanogaster
It has been known for ≈70 years that calorie restriction can dramatically extend the life span of rodents (1). In primates calorie restriction causes a number of physiological changes with positive health benefits (2, 3). Although calorie restriction has been the subject of intense investigation, little is understood about the molecular and cellular mechanisms by which a reduction in calorie intake effects life span extension. Recently it has been shown that the calorie-restriction life-span-extending effect is conserved across distant species from yeast to mammals (2, 4–12). Powerful molecular genetic techniques and the relatively short life span of model organisms such as yeast, nematodes, and flies provide the opportunity to uncover the molecular and cellular mechanisms underlying this universal effect on life span. Indeed, the use of these model organisms has implicated insulin-signaling, nutrient-sensing, and chromosome-remodeling proteins in either triggering calorie restriction or contributing to its life-span-extending effect (13–19).
The Rpd3/Sir2 histone deacetylases have been implicated in both life span determination and calorie restriction in yeast (15). Rpd3 and Sir2 can effect the activity of a variety of genes and physiological systems by deacetylating histones and other proteins such as p53 (12, 20). A decrease in Rpd3 or an increase in Sir2 extends mother cell life span in yeast (11, 21), and the effect of Sir2 on yeast life span is linked to calorie restriction (22). A similar mechanism may operate in metazoans, because an increase in Sir2 extends life span in nematodes (23), and a decrease in Rpd3 extends life span in flies (16). The increase in life span associated with decreased Rpd3 in flies is thought to occur through a mechanism related to calorie restriction (16). The finding of an increase in Drosophila Sir2 (dSir2) transcription in both long-lived rpd3 mutant flies and long-lived calorie-restricted normal flies implicates dSir2 as a potential member of the calorie-restriction life-span-extending pathway (16). Further evidence of a role for Sir2 in the determination of life span is the finding that the Sir2 agonist resveratrol extends life span in yeast, nematodes, and flies in a Sir2- and calorie-restriction-dependent manner (24, 25). These data suggest that Sir2 may be one of the primary elements of the calorie-restriction-induced life span extension in flies and other metazoans.
Materials and Methods
Fly Strains. The dSir24.5- and dSir25.26-null mutant lines were obtained from S. Smolik (Oregon Health & Science University, Portland); the dSir217-null mutant line was obtained from S. Astrom (Stockholm University, Stockholm); the ELAV-GeneSwitch line was obtained from H. Keshishian (Yale University, New Haven, CT); the Canton-S, armadillo-GAL4 driver, tubulin-GAL4 driver, and ELAV-GAL4 driver and the dSir2EP2300, dSir2EP2384, and dSir2KG00871 mutant lines were obtained from the Bloomington Drosophila Stock Center at Indiana University; the dSir2EYO3602 line was obtained from H. Bellen (Baylor College of Medicine, Houston); and the D42 driver was obtained from G. Boulianne (Hospital for Sick Children Research Institute, Toronto).
Genetic Crosses. The GAL4 and UAS binary system was used to drive overexpression of dSir2 (26, 27). To generate flies that ubiquitously overexpressed dSir2, >90 tubulin-GAL4/TM3 male flies were crossed to >100 virgin dSir2EP2300/CyO, dSir2EP2384/CyO, or dSir2EYO3602/CyO female flies. To generate flies that ubiquitously overexpressed dSir2 at a lower level, >90 armadillo-GAL4 male flies were crossed to >100 virgin dSir2EP2300/+ females. dSir2EP2300/+; tubulin-GA L4/+, Sir2EP2384/+; tubulin-GAL4/+, dSir2EYO3602/+; tubulin-GAL4/+, armadillo-GAL4/dSir2EP2300 progeny were used for the experimental life spans or semiquantitative RT-PCR. Control flies were obtained by mating >50 virgin F1 female flies and >50 F1 male flies from each condition to obtain white-eyed flies lacking balancer chromosomes, the tubulin-GAL4 driver chromosome, or chromosomes containing dSir2EP2300, dSir2EP2384, or dSir2EYO3602. A similar set of crosses was performed to obtain experimental and control flies for the neuronal overexpression of dSir2 in the ELAV-GAL4 studies, except that the ELAV-GAL4 driver stock was homozygous (ELAV-GAL4 is on the X chromosome), and >50 virgin F1 females flies were backcrossed to the ELAV-GAL4 stock so that control flies had an ELAV-GAL4 chromosome but no UAS-dSir2.
The conditional ELAV-GeneSwitch driver was combined with the dSir2EP2300 line to drive overexpression of dSir2. A single cohort of F1 dSir2EP2300/+; ELAV-GeneSwitch/+ male and female adult flies was collected and placed on a diet of either food with RU-486 (mifepristone, Sigma) at a concentration of 200 μM or food with only diluent from the first day after eclosion.
Flies heteroallelic for dSir2-null mutations, dSir24.5/dSir25.26 flies, were generated by crossing dSir24.5/CyO to dSir25.26/CyO. Flies heterozygous for the null allele of rpd3 (rpd3def24) and the null allele dSir217 (28) or the hypomorphic allele dSir2EP2300 were crossed to Canton-S flies to generate a matched genetic background. dSir217/dSir2+; rpd3def24/rpd3+ flies were generated by crossing dSir2/CyO and rpd3/TM6Sb flies. dSir2EP2300/dSir2+; rpd3def24/rpd3+ flies possessing a copy of the dSir2-hypomorphic allele dSir2EP2300 and a copy of the rpd3-null allele rpd3def24 were generated in the same way. Control flies, generated in a similar manner, were obtained by mating >50 F1 CyO; Sb (without dSir2EP2300 or rpd3def24) male flies and >50 F1 virgin CyO; Sb (without dSir2EP2300 or rpd3def24) female flies to obtain flies with no balancer chromosome.
Life Span Determinations. Newly eclosed adults were collected, and ≈20 males and 20 females were placed into each vial with normal cornmeal sucrose food (per refs. 16 and 17) or high-calorie (15% yeast, 15% sucrose, and 2% agar) or low-calorie (5% yeast, 5% sucrose, and 2% agar) food (per ref. 7). Studies with the RU-486-ELAV-GeneSwitch driver used food in which RU-486, dissolved in 100% EtOH, was added during preparation when the food had cooled to 50°C, at a final concentration of 200 μM. Control food for the RU-486 experiments was made by adding the same amount of EtOH without RU-486 when the food had cooled to 50°C. Flies were maintained in a humidified temperature-controlled environmental chamber at 25°C. Every 2 days, flies were passed into new vials, and the number of dead flies was counted as described in ref. 16. Median and mean life spans and statistical log rank analyses were performed with statview (SAS Institute, Cary, NC). Maximum life spans were calculated as the mean of 10% survival, except for the studies with RU-486, in which 1% maximum life span was used.
Results
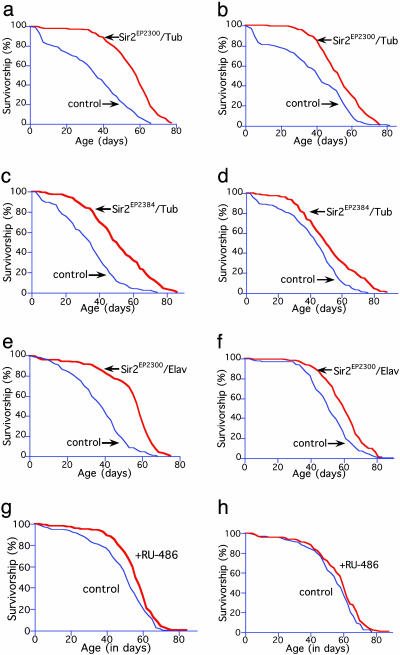
Increasing Sir2 Expression Extends Life Span in Drosophila. To test whether dSir2 is involved in longevity determination in the fly, we examined the life span of flies in which the level of dSir2 had been increased by using molecular genetic techniques. Flies were constructed that ubiquitously overexpressed dSir2 by combining, in individual flies, the Drosophila tubulin promoter fused to the gene for the yeast GAL4 activator protein (tubulin-GAL4 driver) with a native dSir2 gene that has a P element with GAL4-binding sites (EP-UAS) inserted just upstream (26, 27). Flies carrying the tubulin-GAL4 driver and each of the different EP-UAS-dSir2 genes, dSir2EP2300, dSir2EP2384, or dSir2EYO3602, had a >4-fold increase in dSir2 mRNA expression over the endogenous level (Fig. 5, which is published as supporting information on the PNAS web site). Consistent with our hypothesis that an increase in dSir2 in flies will increase life span, up to a 57% increase in average life span was seen in the tubulin-GAL4/dSir2EP2300, tubulin-GAL4/dSir22384, and tubulin-GAL4/dSir2EYO3602 flies, with an increase across all lines of 29% for females and 18% for males (Fig. 1 a–d and Table 1; see also Figs. 6 and 7, which are published as supporting information on the PNAS web site).
Fig. 1.
Increased expression of dSir2 extends life span in the fly. Life span extension is seen in female (a, c, e, and g) and male (b, d, f, and h) flies in which dSir2 expression is increased (red line) in a ubiquitous manner (a–d) or in the nervous system (e–h) compared with genetically matched controls (blue line). Survivorship curves for genetically matched controls and tubulin-GAL4 driver/dSir2EP2300 flies (a and b), tubulin-GAL4 driver/dSir2EP2384 flies (c and d), ELAV-GAL4 driver/dSir2EP2300 flies (e and f), and RU-486-ELAV-GeneSwitch driver/dSir2EP2300 flies (g and h). Genetically matched controls are flies derived from a second-generation cross of the offspring from the cross that generated the experimental flies in a–f. Controls in a–f are flies containing a mix of the chromosomes found in the experimental flies but which do not contain the GAL4 chromosome, the UAS-containing chromosome, or any balancer chromosome. Controls in g and h are flies from the same cohort fed only diluent and not RU-486. Each life span included at least 149 male and 159 female flies (16).
Table 1. Life span is extended when Sir2 expression is increased.
| Gender | Genotype | n | Median life span (% change) | χ2 | Maximal life span (% change) |
|---|---|---|---|---|---|
| F | Tub/dSir2EP2300 | 196 | 58 (57) | 139.754 | 73.62 (19) |
| F | Control | 171 | 37 | 62.1 | |
| M | Tub/dSir2EP2300 | 194 | 54 (32) | 37,024 | 73.57 (14) |
| M | Control | 179 | 41 | 65.43 | |
| F | Tub/dSir2EP2384 | 185 | 52 (44) | 84.391 | 78.96 (25) |
| F | Control | 159 | 36 | 62.75 | |
| M | Tub/dSir2EP2384 | 180 | 50 (14) | 24.597 | 81.7 (21) |
| M | Control | 149 | 44 | 67.53 | |
| F | ELAV/dSir2EP2300 | 208 | 59 (52) | 135.190 | 70 (19) |
| F | Control | 191 | 39 | 59.08 | |
| M | ELAV/dSir2EP2300 | 179 | 61 (20) | 28.347 | 80.56 (8) |
| M | Control | 202 | 51 | 75.29 | |
| F | Tub/dSir2EY03602 | 193 | 41 (-15) | 37.803 | 55.25 (-11) |
| F | Control | 183 | 47 | 62.24 | |
| M | Tub/dSir2EY03602 | 184 | 47 (9) | 24.461 | 74.26 (25) |
| M | Control | 161 | 43 | 58.79 | |
| F | ELAV-GS/dSir2EP2300 | 232 | 56 (12) | 19.10 | 84 (9) |
| F | Control | 202 | 51 | 77 | |
| M | ELAV-GS/dSir2EP2300 | 234 | 60 (-5) | 0.319* | 86 (-1) |
| M | Control | 187 | 63 | 87 | |
| F | ELAV-GS/dSir2EP2300 | 186 | 60 (5) | 6.260** | 88 (16) |
| F | Control | 200 | 56 | 76 | |
| M | ELAV-GS/dSir2EP2300 | 187 | 62 (-3) | 2.659*** | 84 (11) |
| M | Control | 185 | 64 | 76 |
The median and maximal life spans of male (M) and female (F) flies with increased expression of dSir2 ubiquitously (tubulin-GAL4/dSir2EP2300, tubulin-GAL4/dSir2EP2384, and tubulin-GAL4/dSir2EYO3602), expression restricted to neurons throughout life (ELAV-GAL4/dSir2EP2300), or expression restricted to adult neurons (ELAV-GeneSwitch/dSir2EP2300) compared with their genetically matched controls. n, Number of flies in each life span; Tub, tubulin-GAL4 driver; ELAV, ELAV-GAL4 driver; ELAV-GS, ELAV-GeneSwitch driver. (For example, Tub/dSir2EP2300 is a fly that possesses one copy of the tubulin-GAL4 driver and one copy of the dSir2EP2300 chromosome.) Controls are described in Materials and Methods. Control flies for ELAV-GeneSwitch driver flies were from the same cohort and fed only diluent, without RU-486. Statistical log rank analyses were performed with statview. Maximum life span is the mean of 10% survival, except for ELAV-GeneSwitch flies, which have a 1% maximum life span. P < 0.0001 for each experiment except where noted. *, P = 0.57; **, P = 0.01; ***, P = 0.10.
To determine whether a threshold level of dSir2 expression is required for life span extension in the fly, we examined flies in which the armadillo-GAL4 driver was combined with the dSir2EP2300 chromosome. The armadillo-GAL4 driver is a weaker driver than the tubulin-GAL4 driver: compared with control flies, armadillo-GAL4/dSir2EP2300 flies showed only a 10–20% increase in dSir2 mRNA levels and no life span extension, suggesting that a significant increase in dSir2 mRNA is required to cause an extension in life span (data not shown).
Increasing Sir2 Expression in Neurons Extends Life Span. Knowing that ubiquitous overexpression of dSir2 increases life span, we wanted to determine which tissues normally express dSir2 in adults and whether an effect in a single tissue could mediate the life span extension caused by dSir2 overexpression. Using anti-dSir2 antibodies (28, 29), we found that, similar to embryos and larvae (29), in adults dSir2 protein is found at high levels in the nuclei of neurons and in the nuclei and cytoplasm of fat body cells (Fig. 8, which is published as supporting information on the PNAS web site). The finding of a prominent expression of dSir2 in the nervous system of normal animals led us to examine whether an increase in dSir2 in neurons may be one of the primary mediators of the Sir2-related life span extension. Neuronal dSir2 overexpression in flies carrying the pan-neuronal promoter ELAV-GAL4 driver and dSir2EP2300 extended the average life span by 52% in females and 20% in males (Fig. 1 e and f, respectively, and Table 1).
ELAV-GAL4 drives expression in embryos and larvae as well as adults. A different system for overexpressing dSir2 was used to test whether increased expression of dSir2 only in adult neurons might lead to life span extension. The RU-486 Gene-Switch system allows for the comparison of genetically identical animals from the same cohort, one group receiving RU-486, which induces expression of the EP-UAS gene, and the other group receiving only diluent (30, 31). In two independent trials the maximum life span of adult dSir2EP2300/ELAV-GeneSwitch flies receiving RU-486 was increased by 9% and 16%, respectively, in females and was decreased by 1% and increased by 10%, respectively, in males (Fig. 1 g and h, respectively, and Table 1). In females, respective 5% and 12% increases in median life span were also seen. The smaller increase in life span in flies with the ELAV-GeneSwitch driver, relative to those with the ELAV driver, is consistent with a lower level of dSir2 induction with the ELAV-GeneSwitch driver at the dose of RU-486 used (200 μM). In addition, preliminary studies suggest that RU-486 itself may have some mild deleterious effects on life span in flies, especially on males (unpublished results). The greater life span extension obtained by using the standard ELAV driver could be due partly to increased dSir2 activity before eclosion in adult ELAV-GAL4/dSir2EP2300 flies. Regardless, the results demonstrate that pan-neuronal overexpression of dSir2 during only the adult stage is sufficient to produce a modest extension of maximal life span.
To further explore the possibility that a subset of neurons may be important in dSir2-mediated life span extension, we examined the life span of flies containing the D42-GAL4 motoneuron-specific driver (32) and dSir2EP2300. No life span extension was seen in the D42/dSir2EP2300 flies (data not shown).
Decreases in physical activity, reproductive status (especially in females), or calorie intake are known to increase life span in the fly (reviewed in ref. 33). Although we did not perform quantitative studies, visual inspection suggested no obvious decrease in physical activity or fertility in the long-lived dSir2-overexpressing flies compared with their matched controls. Furthermore, the fact that life span was found to be significantly increased in both males and females suggests that a potential decrease in female reproduction is unlikely to be the primary cause of the observed life span extension.
Considered together, the results of experiments driving dSir2 demonstrate that overexpression of dSir2 correlates well with increased life span in flies. In four different driver-GAL4/UAS-dSir2 lines in which dSir2 was substantially overexpressed either ubiquitously or in neurons, the life span of flies was extended significantly. However, when the driver caused only a small ubiquitous increase in dSir2 or an increase only in motor neurons, life span was not extended. Furthermore, an intermediate increase in dSir2 in the adult nervous system caused by the ELAV-GeneSwitch driver caused an intermediate increase in life span.
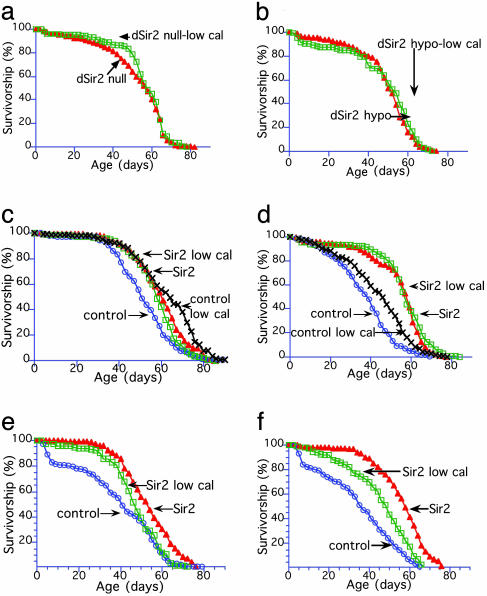
dSir2 Is Necessary for the Life-Span-Extending Effect of Calorie Restriction. Flies given low-calorie food, in addition to showing an increase in life span, showed an increase in dSir2 mRNA expression (16). To determine whether dSir2 is directly in the calorie-restriction life-span-extending pathway in flies, we compared the life span of flies that had reduced or no dSir2 expression on a diet of low-calorie food with that of genetically identical flies from the same cohort on a diet of normal or high-calorie food. Flies with either no dSir2 gene function [e.g., dSir24.5/dSir25.26 (29)] or with severely decreased dSir2 gene function (e.g., dSir2KG00871/dSir2KG00871) showed no life span extension on a diet of low-calorie food (Fig. 2 a and b) relative to genetically identical flies on a diet of normal or high-calorie food. The inability of dSir2 mutant flies to increase their life span in response to a low-calorie diet demonstrates that a sufficient level of dSir2 must be available for the activation of life span extension by calorie reduction and, furthermore, that dSir2 is an important element in the calorie-reduction life-span-extending pathway.
Fig. 2.
Calorie-reduction life span extension is mediated by dSir2. (a and b) The expected life span extension with calorie reduction was blocked by mutations in dSir2. Survivorship curves of dSir2-null [dSir24.5/dSir25.26 (29)] (a) and dSir2-hypomorphic (dSir2KG00871/dSir2KG00871) (b) female flies on normal (red filled triangles) and low-calorie (green open boxes) food. Male flies mutant for dSir2 also had no life span extension in response to caloric reduction. (c–f) The life span of long-lived dSir2-overexpressing flies was not further extended by calorie reduction. Survivorship curves of long-lived ELAV-GAL4/dSir2EP2300 (c and d) and tubulin-GAL4/dSir2EP2300 (e and f) male (c and e) and female (d and f) flies on normal (red filled triangles) and low-calorie (green open boxes) food and those of genetically matched controls (c and d) on normal (blue open circles) and low-calorie (black X's) food. Low-calorie food was as described in ref. 16 and Materials and Methods. Each life span included at least 179 male and 171 female flies (16).
Calorie Restriction Does Not Further Extend the Life Span of Long-Lived dSir2-Overexpressing Flies. If dSir2 mediates the effect of calorie-restriction-induced life span extension, we would also expect that calorie restriction would not further increase life span in flies in which dSir2 activity is already elevated. We examined the life span of two of the long-lived dSir2-overexpressing lines, tubulin-GAL4/dSir2EP2300 and ELAV-GAL4/dSir2EP2300, under normal and low-calorie food conditions. Under our husbandry conditions, a decrease in calorie content in the food typically increased the life span of normal flies by 35–40% or more (16). However, when the long-lived ELAV-GAL4/dSir2EP2300 and tubulin-GAL4/dSir2EP2300 flies were placed on a diet of low-calorie food, no further increase in life span was seen with the ELAV-GAL4/dSir2EP2300 flies, whereas the tubulin-GAL4/dSir2EP2300 flies showed a reduction in life span toward normal (Fig. 2 c–f). Control flies for ELAV-GAL4/dSir2EP2300 placed on a diet of low-calorie food demonstrated a 28% and 22% increase in median life span for males and females, respectively (Fig. 2 c and d). The lack of a cumulative effect of calorie reduction and dSir2 overexpression on life span suggests that life span extensions are mediated through similar or related pathways.
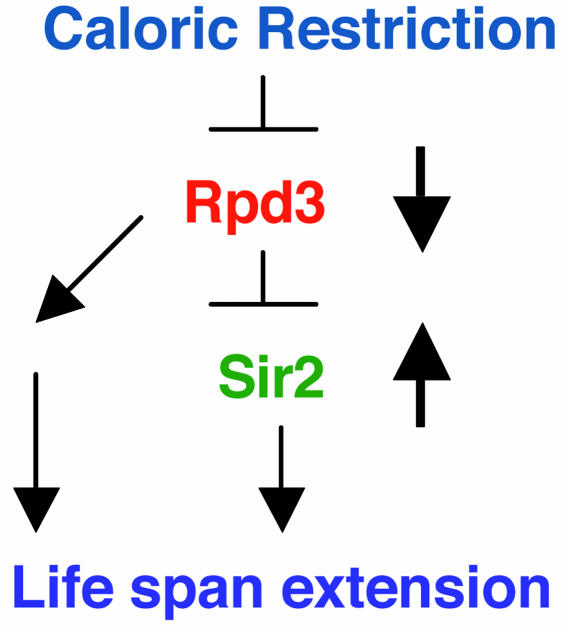
dSir2 and Rpd3 Are in the Same Calorie-Restriction Pathway. The data presented here on dSir2, along with previous work on life span and calorie reduction of rpd3 mutants (16), indicate that the life-span-extending effects of nutrient reduction in the fly are mediated through dSir2 and Rpd3. The observation of an increase in dSir2 mRNA in long-lived rpd3 mutants (16) led us to postulate that the effect of Rpd3 may depend partially on an increase in dSir2. Together these data suggest a model for how calorie reduction extends life span in the fly (Fig. 3). In this model, the stimulus of calorie reduction triggers a decrease in Rpd3 activity and a subsequent increase in dSir2 activity. The increase in dSir2, either alone or in conjunction with additional changes initiated by the decrease in Rpd3 activity, results in life span extension.
Fig. 3.
Calorie-restriction/Rpd3/Sir2 pathway for life span extension in the fly. In this model a stimulus of calorie reduction results in a decrease in Rpd3 (16). The reduction in Rpd3 activity decreases the inhibition of Sir2, causing an increase in Sir2 (16). The increase in Sir2, in conjunction with the decrease in Rpd3, effects life span extension.
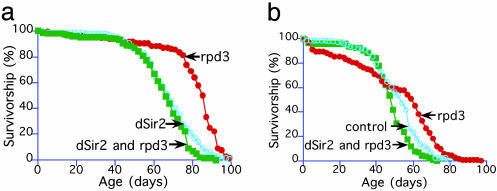
Further confirmation that dSir2 and Rpd3 are in the same life-span-extending pathway was obtained by examining the life span of flies carrying a long-lived rpd3 mutation (rpd3def24) and a dSir2 mutation (dSir217 or dSir2EP2300). dSir2 mutations do not reduce life span in otherwise normal flies (ref. 29 and data not shown). Therefore, if dSir2 were not in the same pathway as Rpd3, the life span would not be altered from the extended life span of rpd3 mutant flies. As predicted by our model, flies with both a dSir2 mutation and rpd3def24 mutation were not long-lived, whereas their counterparts, flies mutant for only rpd3def24, remained long-lived (Fig. 4). dSir2 and Rpd3, therefore, seem to be in the calorie-reduction life-span-extending pathway in flies. The finding that long-lived rpd3 mutations increase dSir2 levels (16) suggests that dSir2 is downstream of Rpd3.
Fig. 4.
Mutations in dSir2 block the life-span-extending effect of rpd3 mutants. The extension in life span seen with rpd3 mutation was prevented by mutations in dSir2. Flies labeled as “rpd3” are heterozygous for the rpd3def24-null allele of rpd3, whereas flies labeled as “dSir2 and rpd3” possess one copy of the life-span-extending rpd3-null mutation and one copy of either the dSir217-null (28) or dSir2EP2300-hypomorphic allele. (a) dSir217. Flies labeled “rpd3,” “dSir2 and rpd3,” and “control” were each backcrossed to a Canton-S background. (b) dSir2EP2300. Flies labeled “rpd3,” “dSir2 and rpd3,” and “control” were collected from the offspring of crosses between the rpd3-null allele, rpd3def24, and dSir2EP2300. Flies labeled “control” were derived from an additional cross to remove the rpd3 and dSir2EP2300 alleles and any balancer chromosomes. Each life span included at least 200 male and 200 female flies (16).
Discussion
The availability and utilization of nutrients seems to be a central feature of a variety of environmental and genetic interventions that extend life span in many different organisms. Calorie restriction, in particular, is one of the most well conserved interventions known, extending life span in organisms as diverse as yeast and mammals (2, 4–12). Despite the nearly universal effect on life span extension of calorie restriction, little is understood molecularly about how a decrease in nutrient availability triggers an extension in life span. Model organisms have proven useful for understanding the molecular and cellular mechanisms of other complex biological phenomena such as development. Organisms such as yeast, nematodes, and flies can rapidly provide information about causal relationships through the combined use of genetic and molecular approaches.
Calorie Restriction Extends Life Span in Flies by Increasing dSir2. The search for elements that extend life span in metazoans has identified the involvement of the insulin-signaling, nutrient-sensing, and Sir2 pathways (13, 14, 17–19, 34). Although the Sir2 pathway has been linked to calorie availability in yeast, it has not been shown to function in the calorie restriction pathway in metazoans (22). The data we present here demonstrate a direct link between the life-span-extending effects of dSir2 and calorie restriction in the fly. We used five different GAL-4 drivers (tubulin, ELAV, armadillo, ELAV-Gene-Switch, and D42-motoneuron) to drive expression of endogenous dSir2 genes with three separate nearby insertions of UAS elements. In four strains in which dSir2 expression was increased substantially, either ubiquitously or in neuronal cells, the life span of the flies was extended substantially, up to 57% when dSir2 mRNA expression was increased 4-fold. Conversely, in two other similarly constructed strains in which dSir2 expression was not elevated or was only marginally elevated, life span was not altered relative to that of control flies. Thus, in six fly strains constructed by using different combinations of drivers and dSir2 responders, increased longevity correlated very well with elevation of dSir2. Furthermore, we show here that life span cannot be extended by calorie restriction in flies that lack dSir2 activity, nor can life span be further increased by calorie restriction in flies in which dSir2 activity is already raised. The recent findings that a Sir2 agonist, resveratrol (shown to increase the activity of yeast, nematode, fly, and human Sir2) extends life span in yeast, nematodes, and flies in a manner that is Sir2-dependent and associated with calorie restriction provide additional evidence for a primary role of Sir2 activity in determining life span in metazoans (24, 25). Together, these observations make a strong case that calorie restriction extends life span in flies by increasing dSir2 activity.
dSir2 Seems to Operate After Rpd3 in the Pathway Mediating Life Span Extension by Calorie Restriction. The data presented here, in conjunction with our previous work on Rpd3 (16), show that dSir2 and Rpd3 are important components in the calorie-restriction life-span-extending pathway of flies. A decrease in dSir2 prevents the life-span-extending effect of calorie restriction, and the life-span-extending effect of calorie restriction is not cumulative with the life-span-extending effect of increased dSir2. Similarly, the life-span-extending effect of Rpd3 mutations is not cumulative with the effect of calorie restriction (16). We reported previously (16) that long-lived flies with reduced Rpd3 activity have elevated dSir2 mRNA. We now show that, in flies with decreases in both Rpd3 and dSir2 activity, life span is not extended, indicating that an increase in dSir2 activity in response to a decrease in Rpd3 activity is necessary for life span extension. Together these data suggest that dSir2 is downstream of Rpd3 in the calorie-restriction life-span-extending pathway in flies. This model provides a useful framework and testable model for examining the relationship of Sir2, calorie reduction, and longevity by using genetic, molecular, and pharmaceutical approaches (Fig. 3). The documentation of a molecular genetic pathway responsible for effecting calorie-restriction-related life span extension will be useful for identifying biochemical mediators and drug interventions that can mimic calorie restriction. Given the conservation of elements of the calorie restriction/Rpd3/Sir2 pathway in extending life span in yeast and now flies, agents that stimulate the activity of Sir2 are potential tools for extending life span in metazoans (24, 25).
Supplementary Material
Acknowledgments
We thank Joseph Jack and Stewart Frankel for critical reading of the manuscript; Tyson Bross for help with the log rank analyses and mortality rate determinations; Suzanne Kowalski and Dianna Schwarz for technical support; Stefan Astrom (Stockholm University, Stockholm) for the dSir217 flies and an anti-dSir2 antibody; Sarah Smolik (Oregon Health & Science University, Portland) for the dSir24.5 and dSir25.26 flies and an anti-dSir2 antibody; Haig Keshishian (Yale University, New Haven, CT) for the ELAV-GeneSwitch flies; Hugo Bellen (Baylor College of Medicine, Houston) for the dSir2EYO3602 flies; Gabrielle Boulianne for the D42 flies (Hospital for Sick Children Research Institute, Toronto); and the Bloomington Drosophila Stock Center at Indiana University for the tubulin-GAL4, ELAV-GAL4, dSir2KG00871, dSir2EP2300, and dSir2EP2384 flies. This work was supported by National Institutes of Health Grants ES11463, AG20816, and AG23088 (to B.R.), and AG16667 (to S.L.H.), the Ellison Medical Foundation, and the Donaghue Foundation (S.L.H.). S.L.H. is an Ellison Medical Foundation Senior Investigator and a member of the Scientific Advisory Board of Elixir Pharmaceuticals, Inc. (Cambridge, MA).
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Masoro, E. J. (2000) Exp. Gerontol. 35, 299–305. [DOI] [PubMed] [Google Scholar]
- 2.Lane, M. A., Mattison, J., Ingram, D. K. & Roth, G. S. (2002) Microsc. Res. Tech. 59, 335–338. [DOI] [PubMed] [Google Scholar]
- 3.Mattison, J. A., Lane, M. A., Roth, G. S. & Ingram, D. K. (2003) Exp. Gerontol. 38, 35–46. [DOI] [PubMed] [Google Scholar]
- 4.Wanagat, J., Allison, D. B. & Weindruch, R. (1999) Toxicol. Sci. 52, 35–40. [DOI] [PubMed] [Google Scholar]
- 5.Weindruch, R. (1996) Toxicol. Pathol. 24, 742–745. [DOI] [PubMed] [Google Scholar]
- 6.Kayo, T., Allison, D. B., Weindruch, R. & Prolla, T. A. (2001) Proc. Natl. Acad. Sci. USA 98, 5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman, T. & Partridge, L. (1996) Proc. R. Soc. London Ser. B 263, 755–759. [DOI] [PubMed] [Google Scholar]
- 8.Good, T. P. & Tatar, M. (2001) J. Insect Physiol. 47, 1467–1473. [DOI] [PubMed] [Google Scholar]
- 9.Lakowski, B. & Hekimi, S. (1998) Proc. Natl. Acad. Sci. USA 95, 13091–13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin, S. J., Defossez, P. A. & Guarente, L. (2000) Science 289, 2126–2128. [DOI] [PubMed] [Google Scholar]
- 11.Jiang, J. C., Wawryn, J., Shantha Kumara, H. M. & Jazwinski, S. M. (2002) Exp. Gerontol. 37, 1023–1030. [DOI] [PubMed] [Google Scholar]
- 12.Guarente, L. (2000) Genes Dev. 14, 1021–1026. [PubMed] [Google Scholar]
- 13.Kenyon, C. (2001) Cell 105, 165–168. [DOI] [PubMed] [Google Scholar]
- 14.Tatar, M., Bartke, A. & Antebi, A. (2003) Science 299, 1346–1351. [DOI] [PubMed] [Google Scholar]
- 15.Hekimi, S. & Guarente, L. (2003) Science 299, 1351–1354. [DOI] [PubMed] [Google Scholar]
- 16.Rogina, B., Helfand, S. L. & Frankel, S. (2002) Science 298, 1745. [DOI] [PubMed] [Google Scholar]
- 17.Rogina, B., Reenan, R. A., Nilsen, S. P. & Helfand, S. L. (2000) Science 290, 2137–2140. [DOI] [PubMed] [Google Scholar]
- 18.Hwangbo, D. S., Gersham, B., Tu, M. P., Palmer, M. & Tatar, M. (2004) Nature 429, 562–566. [DOI] [PubMed] [Google Scholar]
- 19.Kapahi, P., Zid, B. M., Harper, T., Koslover, D., Sapin, V. & Benzer, S. (2004) Curr. Biol. 14, 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunet, A., Sweeney, L. B., Sturgill, J. F., Chua, K. F., Greer, P. L., Lin, Y., Tran, H., Ross, S. E., Mostoslavsky, R., Cohen, H. Y., et al. (2004) Science 303, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 21.Kaeberlein, M., McVey, M. & Guarente, L. (1999) Genes Dev. 13, 2570–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, S. J., Kaeberlein, M., Andalis, A. A., Sturtz, L. A., Defossez, P. A., Culotta, V. C., Fink, G. R. & Guarente, L. (2002) Nature 418, 344–348. [DOI] [PubMed] [Google Scholar]
- 23.Tissenbaum, H. A. & Guarente, L. (2001) Nature 410, 227–230. [DOI] [PubMed] [Google Scholar]
- 24.Howitz, K. T., Bitterman, K. J., Cohen, H. Y., Lamming, D. W., Lavu, S., Wood, J. G., Zipkin, R. E., Chung, P., Kisielewski, A., Zhang, L. L., et al. (2003) Nature 425, 191–196. [DOI] [PubMed] [Google Scholar]
- 25.Wood, J. G., Rogina, B., Lavu, S., Howitz, K., Helfand, S. L., Tatar, M. & Sinclair, D. (2004) Nature 430, 686–689. [DOI] [PubMed] [Google Scholar]
- 26.Brand, A. H. & Perrimon, N. (1993) Development 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 27.Rorth, P. (1996) Proc. Natl. Acad. Sci. USA 93, 12418–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Astrom, S. U., Cline, T. W. & Rine, J. (2003) Genetics 163, 931–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman, B. L., Lundblad, J. R., Chen, Y. & Smolik, S. M. (2002) Genetics 162, 1675–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. (2001) Proc. Natl. Acad. Sci. USA 98, 12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roman, G., Endo, K., Zong, L. & Davis, R. L. (2001) Proc. Natl. Acad. Sci. USA 98, 12602–12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkes, T. L., Elia, A. J., Dickinson, D., Hilliker, A. J., Phillips, J. P. & Boulianne, G. L. (1998) Nat. Genet. 19, 171–174. [DOI] [PubMed] [Google Scholar]
- 33.Helfand, S. L. & Rogina, B. (2003) Bioessays 25, 134–141. [DOI] [PubMed] [Google Scholar]
- 34.Guarente, L. & Kenyon, C. (2000) Nature 408, 255–262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.