Abstract
Computer-controlled microfluidics would advance many types of cellular assays and microscale tissue engineering studies wherever spatiotemporal changes in fluidics need to be defined. However, this goal has been elusive because of the limited availability of integrated, programmable pumps and valves. This paper demonstrates how a refreshable Braille display, with its grid of 320 vertically moving pins, can power integrated pumps and valves through localized deformations of channel networks within elastic silicone rubber. The resulting computerized fluidic control is able to switch among: (i) rapid and efficient mixing between streams, (ii) multiple laminar flows with minimal mixing between streams, and (iii) segmented plug-flow of immiscible fluids within the same channel architecture. The same control method is used to precisely seed cells, compartmentalize them into distinct subpopulations through channel reconfiguration, and culture each cell subpopulation for up to 3 weeks under perfusion. These reliable microscale cell cultures showed gradients of cellular behavior from C2C12 myoblasts along channel lengths, as well as differences in cell density of undifferentiated myoblasts and differentiation patterns, both programmable through different flow rates of serum-containing media. This technology will allow future microscale tissue or cell studies to be more accessible, especially for high-throughput, complex, and long-term experiments. The microfluidic actuation method described is versatile and computer programmable, yet simple, well packaged, and portable enough for personal use.
Keywords: pump, valve, bioreactor, mixer, perfusion
Advanced microfluidic cellular assays (1–6) and microscale tissue engineering studies (7–10) would benefit from robust and convenient methods to computer-control accurate spatiotemporal patterns of microfluidic flows in arrays of fluidic networks. In the past, fluidic control included syringe pumps (11), hydrogel valves (12), gravity-driven pumps (13), evaporation-based pumps (14, 15), acoustic pumps (16), gas-generation-based pumps (17, 18), and centrifugal force in CD chips (19). Electrokinetic flow (20–22), thermopneumatic (23, 24), pneumatic/hydraulic (25), or mechanical (26, 27) valves and pumps have a high degree of control, but require external connection lines to larger equipment for actuation. A few fully integrated and self-packaged systems (23, 28) have been developed, but lack the reconfigurability inherent with numerous active valves and pumps.
Here, we report a method to precisely control fluid flow inside elastomeric capillary networks by using multiple (tens to hundreds) computer-controlled, piezoelectric, movable pins. These pins are positioned as a grid on a refreshable Braille display (e.g., F. H. Papenmeier, Schwerte, Germany), which is a tactile device used by the visually impaired to read computer text. Each pin can act as a valve and be shifted upward to push against channels contained in silicone rubber and completely shut the channel. Three sequential valves of this type, moved in certain patterns, can be effectively used as a peristaltic pump. Synchronous control of multiple pins through Braille display software in conjunction with a program that refreshes a line of text results in integrated, in situ microfluidic pumps and valves. We demonstrate this method first by switching between laminar flow and mixed flow of reagents in the same cell culture channel through programming, then by seeding, compartmentalizing, and sustaining C2C12 mouse myoblast proliferation and differentiation within microfluidic chips under different flow patterns over extended time periods (up to 3 weeks). The versatility and reliability of an accessible and portable multiactuator array for this fluidic actuation process opens windows for the study of cells and other microfluidic application where active pumping and valving are required.
Materials and Methods
Device Fabrication. Each chip is composed of poly(dimethylsiloxane) (PDMS) and fabricated by using soft lithography (29). Prepolymer (Sylgard 184, Dow-Corning) at a 1:10 curing agent-to-base ratio was cast against positive relief features to form a flat, ≈1-mm-thick, negative replica for channel features. The relief features were composed of SU-8 (MicroChem, Newton, MA) and fabricated on a thin glass wafer (200 μm thick) by using backside diffused-light photolithography (30). The prepolymer was then cured at 60°C for 60 min, and holes were punched in it by a sharpened 14-gauge blunt needle. Channel indentations in the negative replica were sealed by a flat PDMS sheet. This sheet was 140 μm thick and formed by spin coating (200 rpm, 4 min) of a 1:10 ratio prepolymer onto glass wafers that were silanized with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane (United Chemical Technologies, Bristol, PA). This sheet was cured at 150°C for 60 min before sealing channels. An additional reservoir level was added only for cell culture devices. This layer was formed by adding prepolymer on machined brass molds to ≈1 cm. This component was then cured at 60°C for 60 min. To irreversibly seal all cured components, they were oxidized for 30 s in oxygen plasma and sealed together. The final assembly was incubated at 60°C for 10 min, injected with PBS that readily wicked into the channels, then incubated in 37°C incubators overnight.
Device Control. A Braille display (Navigator, Telesensory, Sunnyvale, CA) provided a grid of microactuator pins that reflected text displayed on a computer screen. These pins are actuated by piezoelectric bimorphs that can deliver up to 280 kPa of localized pressure. The most accessible method of control was to use a text editor (Boxer) to autoscroll through a prearranged text designed to actuate the correct Braille pins. More flexible programs, coded in Visual Basic, continually updated a line of characters corresponding to desired pin actuation. Braille screen reading software (hal, Dolphin Computer Access, Worcester, U.K.) directly controlled pin actuation based on the refreshing line of characters. The software was set to read only one line of displayed text that was determined by a program taking user input. This text was represented by a series of eight-pin (4 × 2) Braille cells.
General Cell Culture. Cell culture reagents were obtained from Invitrogen unless otherwise stated. C2C12 cells (31) were cultured under 5% CO2 on plastic tissue culture dishes (Fisher) in DMEM containing 15% FBS, 100 μg/ml streptomycin, and 100 units/ml penicillin. Cells were washed in PBS and detached from dishes by using trypsin/EDTA. These cells were injected into capillary channels and maintained with the same media.
Cell Seeding and Culture in Microchannels. Microchannels for culturing cells were loaded with PBS during the manufacturing process (see above). Fibronectin (100 μg/ml in PBS) was injected along the seed channel and incubated at 25°C for 10 min to coat the channel surface. This process involved placing a 30-gauge needle as a vent on one end of the injection channel and using positive pressure to inject contents on the other end while side channels were valved shut by the Braille pins. After a 15-min fibronectin incubation, media with 15% FBS were placed into the reservoir and circulated in all flow loops for 15 min. Cells were injected in a similar manner to fibronectin. After cells were inside the seed channel, two valves near the cell seeding channel entrance and exit were closed to stop any further flow and facilitate cell attachment. A 37°C, 5% CO2 incubator was used to house cells and the Braille display during cell attachment and culture. The attachment involved consecutive average flow rates of 0, 2, and 9 nl/min for 15 min each.
Results and Discussion
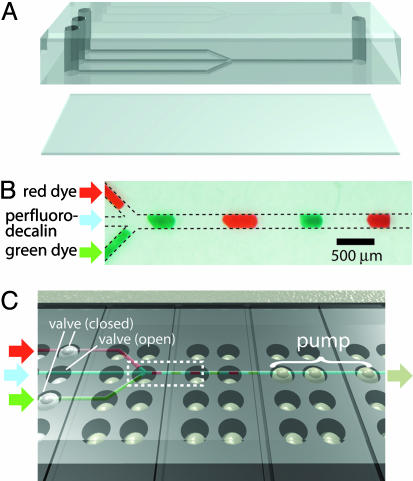
In a typical procedure, as shown in Fig. 1, we align and secure a network of channels onto a grid of Braille pin actuators such that valve positions are directly above the pins. Channel replicas in PDMS, created through soft lithography, are permanently sealed with a thin 140-μm PDMS sheet that separates channels from protruding Braille pins (Fig. 1A). Because of the elasticity of PDMS, the vertical force generated by a 0.9-mm activated Braille pin (≈0.2 N) can deform the PDMS sheet and areas directly adjacent to the channel above, constricting that particular channel section. A thicker PDMS sheet (e.g., 500 μm) or a high channel (e.g., 100 μm) will result in partial channel constriction. Deactivation of a Braille pin below a constricted channel will allow the elastomeric channel to regain its original, open shape. As previously reported, three valves can serve as a peristaltic pump (25). With a grid of pins on each Braille display, multiple valves and pumps can be used to generate versatile fluid flow. Fig. 1B shows one example where one pump and three valves (six Braille pins) can be used to generate complex, segmented flow. Here, two food dyes and an immiscible fluid (perfluorodecalin) are loaded. As shown in Fig. 1C, one outlet pump draws fluid while three valves, located at each inlet, determine which inlet fluid is moved. It is straightforward to meter varying combinations of each fluid at specific quantities for analytical and other purposes. It is also possible to create new pump and valve positions by changing only the channel design, or even by using the same channel design aligned in different configurations.
Fig. 1.
Schematic representation of Braille display-based microfluidics. (A) A typical two-layer design for use with Braille displays. Both layers were composed of PDMS. Channels were face down and separated from the Braille display surface by a thin (140 μm) sheet of PDMS. Holes were punched with a 14-gauge blunt needle. (B) Segmented flow generated by integrating pumps and valves. The three inlets on the left, from top to bottom, contain red food dye, perfluorodecalin, and green food dye. The three components alternatively move toward the outlet on the right. (C) Overview of the typical experimental setup. The multilayer assembly is positioned on top of the Braille display so that channels are aligned above Braille pins, facing down. Three potential valves are on the left, and one pump is on the right. Retracted pins are situated directly below channels, and actuated pins are displaced upward against the PDMS sheet, closing channels directly above it. The dotted box refers to what is shown in B.
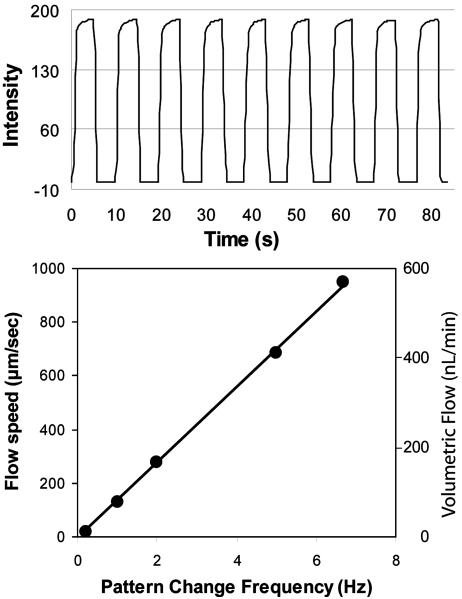
Fig. 2 indicates the performance of Braille pin-based valves and pumps. Valving efficiency and reproducibility are shown in Fig. 2 Upper by allowing a fluorescent solution to flow through the middle inlet of a three-inlet channel and a nonfluorescent solution to flow from the two side inlets. A fully sealed valve in this method depends on a cross-sectional channel shape that is bell-curved as opposed to rectangular. We developed a method to readily fabricate such cross-sectional shapes by using backside diffused light photolithography and confirmed full sealing by visual inspection and electrical conduction through valved channels (30). Fig. 2 Lower shows the linear performance of a three-pin peristaltic pump in relation to the frequency of pin-actuation pattern change. We typically looped the pattern, such as XXO, OXX, OOX, XOX in repetition, where X is a closed position and O is an open position, to pump toward the right. The resultant fluid flow is pulsatile, with transient movements in both directions. The net movement can be predicted by its linear relationship to the pattern change frequency, and flow direction can be switched by reversing the pattern of actuation.
Fig. 2.
Typical pump and valve characteristics. (Upper) A valve characterization. Fluorescein was flowed through the middle of three inlets, which were either switched open or closed by one valve while side inlets remained open. (Lower) A flow calibration for a three-pin pump used for the cell culture device shown in Fig. 4. Speeds are obtained by averaging the net translation of multiple fluorescent beads taken from the center of the channel. The pump pattern is XXO, OXX, OOX, XOX, where X represents a raised pin (valve closed) and O represents a lowered pin (valve open). The fastest reliable pump speed is an ≈120-ms delay between pin changes, which is taken as the fastest consistent actuation rate.
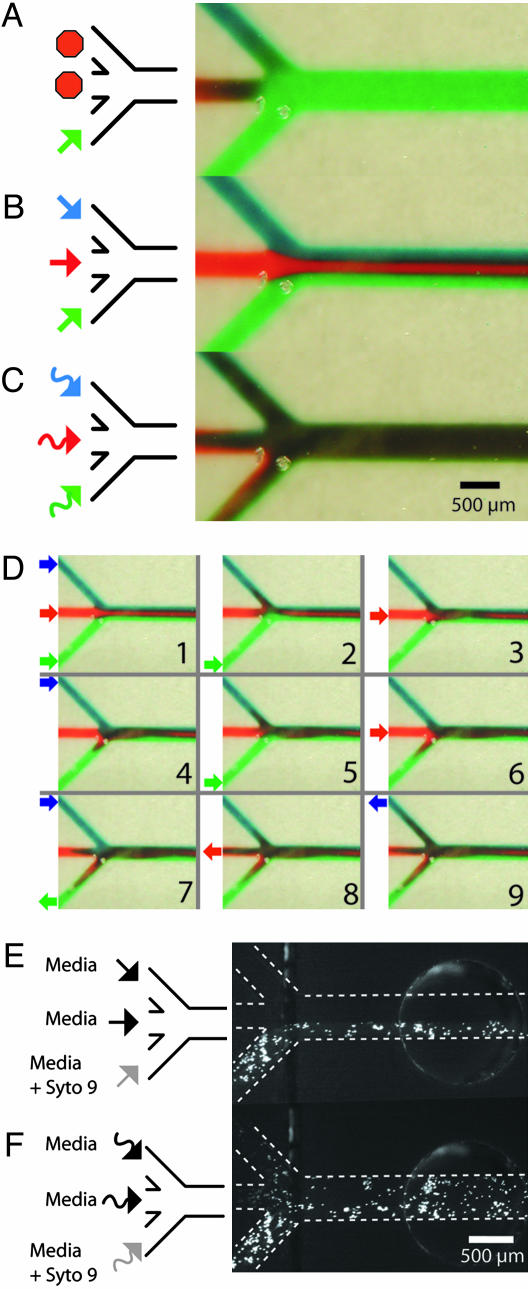
In vitro cell-based studies often involve delivery of reagents in various combinations and concentrations. Two challenges occur when translating this basic method to microfluidics. First, mixing of reagents is slow because it relies mainly on diffusion in the laminar flow regime, meaning there is no turbulence. Many past efforts have focused solely on solving this microfluidic mixing problem, often using complex channel geometries (32–35). Second, active fluid control components are difficult to integrate into microfluidics, thus limiting the flexibility of fluidic handling in terms of reagent metering and combination. Microfluidics using Braille displays amend both limitations. A grid of actuators can change dynamic flow patterns within the same channel between multiple, unmixed, laminar streams and actively mixed flows by switching between two pump algorithms (Fig. 3). The change was shown by a three-inlet, one-outlet channel architecture with one peristaltic pump controlling each inlet. When all three pumps were actuated in-phase toward the same direction, each with the same pin sequence at any given time, pseudo laminar flow was established (Fig. 3B). A switch in software input to a mixer algorithm, in which each pump progressed out-of-phase (36) and pumped one full cycle in reverse for every two cycles forward, caused the same nine pump pins to actively mix the three inlet streams at their intersection (Fig. 3C). In effect, active mixing caused segments of each stream to move toward or back into different inlets, and finally forward again significantly more mixed as shown stepwise in Fig. 3D. Mixing between the sequentially injected inlet fluids is also facilitated through the interlayering that occurs because of parabolic flow (37). This dynamic flow control process and a more rigorous analysis of mixing using both laminar and mixer algorithms can be seen in Movie 1, which is published as supporting information on the PNAS web site. Other algorithms can be used to mix and meter the fluidic input for different mixing ratios and channel architectures. The advantage of having numerous actuators becomes apparent when requiring an array of programmable mixers. For 18 inlet pumps for mixing, 54 independent actuators are needed (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site, for details).
Fig. 3.
Examples of reconfiguration between nonmixing and mixing flow. Three entering streams on the left merge together and exit to the right. Each entering stream is moved by its own three-pin peristaltic pump. (A) Fluidic switching allows only the green stream to flow. (B) Pseudo multiple laminar flows was achieved by running each pump in-phase. (C) Mixing was achieved with an algorithm that runs each pump out-of-phase and reverses a pump cycle once for every two cycles forward. (D) A timewise sequence of the mixing algorithm. Pictures were taken after significant fluidic movements. The arrows represent the direction and magnitude of flow just before this snapshot. Because each pump has sequences to move fluid forward and backward, the time a band of liquid will have had to undergo diffusive mixing will be different even at the same positions along the length of a channel depending on the phase of the pump cycle, with the additional band-broadening caused by backward momentum being largest closer to where the inlets merge. (E) Laminar flow was shown to stain cells only in a portion of the channel after 5.5 min. The bottom entering channel (left bottom) contained Syto 9, which stained C2C12 cells on one side of the main channel. (F) The mixing algorithm was subsequently used to stain all cells for 3.5 min. The arrows represent the mixing algorithm.
In Fig. 3 E and F, the same laminar and mixed patterns of flow were repeated for fluorescent Syto 9 staining of C2C12 cells in a manner similar to previous work (1, 2). Although cells were adherent throughout the width and length of the channel, laminar flow restricted the Syto 9 stream and staining to a third of the channel width (Fig. 3E). A subsequent switch to the mixing program caused all cells downstream of the inlet intersection to stain, as shown in Fig. 3F. Partially stained cells in the two media inlets show that contents from the Syto 9 stream have a residence time in the other inlets and indicates lateral redistribution of reagent across the width of the main channel. Although we have shown two specific cellular treatments, it is straightforward to automate combinations of different reagents, either mixed or unmixed, to treat cells or adjacent subpopulations of cells.
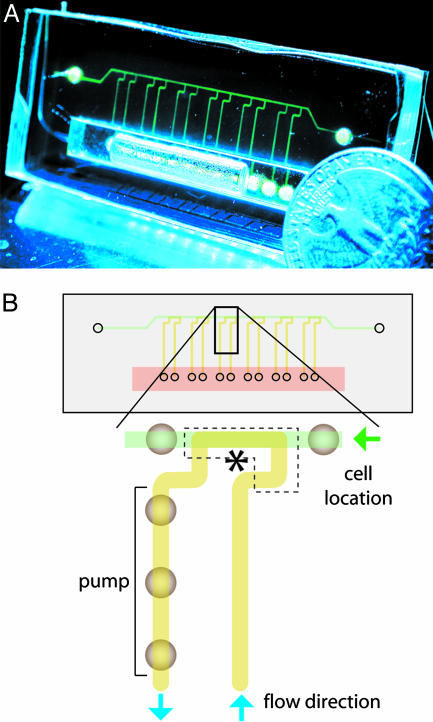
Complex fluidic control is also applicable to extended cellular studies and tissue culture where parallel yet individually tailored trials are needed. We designed a PDMS chip with microfluidic channels, shown in Fig. 4A, that allows a portable Braille display to automate parallel perfusion of cells. Such perfusion could sustain the C2C12 mouse myoblast cell line for up to 3 weeks in microfluidic channels. These cells are refreshed with recirculating media from a reservoir that is >3 orders of magnitude greater by volume than a single perfusion channel. The reservoir integrates as an additional PDMS layer while the entire device remains as a closed system to avoid the need for tubing and possible associated contamination. Reagents can be added or subtracted without leakage by using 30-gauge needles because PDMS self-seals. Fig. 4B shows the design of the same chip and a close-up of one of the perfusion loops. This design was made to reconfigure by compartmentalization between a cell seeding state and a cell culture state. During the seeding procedure, valves restrict the 12 longitudinal channels so that the top channel remains as the only open fluidic pathway. Cells are seeded through this channel with a cell injection needle, providing positive pressure and a vent needle at the two ends. This seeding procedure assigns precise locations for cells to be seeded and requires only one loading event to seed multiple sites with cells. During culture, seeded cells are broken into different compartments by valves, with each compartment being a part of an independent perfusion flow loop. Each flow loop has a different independent pump that brings media both to and from the on-chip reservoir and through the segments of seeded cells.
Fig. 4.
A typical PDMS-based perfusion cell culture device and its working scheme. (A) The device with fluorescein-loaded channels. The channel cross section is approximately an isosceles trapezoid with a height of 30 μm and an upper and lower base of 180 and 402 μm, respectively. (B) A scheme of the same device and a close-up of a single flow loop. Each of the six loops has a left and right valve separating each other, as well as a pump circulating fluid counterclockwise along the path colored yellow. Cells are seeded only in the top channel colored green. The dotted box with an asterisk appears in Fig. 5A.
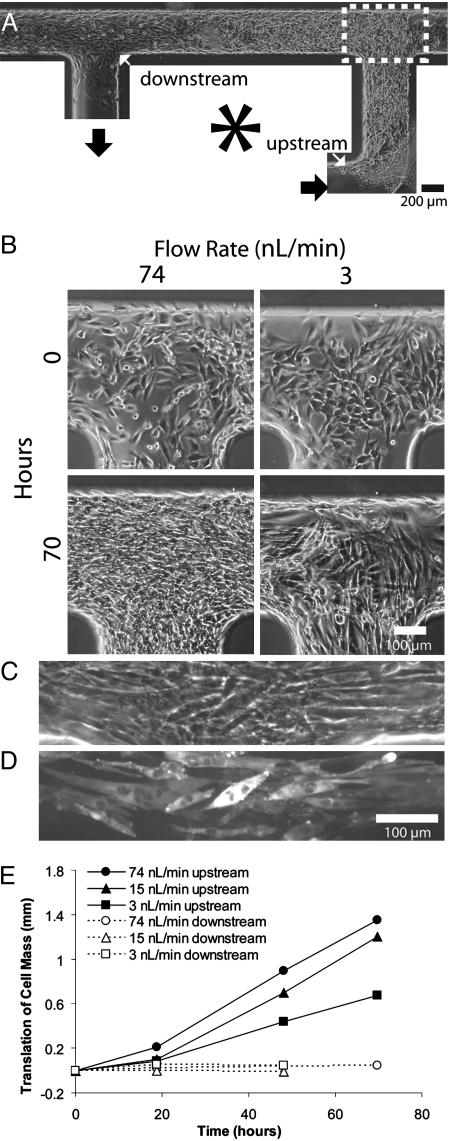
Fig. 5A shows a typical result of perfusion at 15 nl/min 39 h after initial seeding. Two indicators show that cells proliferate and/or migrate preferentially upstream or against the movement of fluid flow. First, whereas initial cell seeding results in a uniform cell density, subsequent perfusion culture results in higher cell density upstream. Second, cellular outgrowth from the initial boundaries is observed toward the upstream direction. Although we have not formally quantified proliferation or individual cell migration, we note that these observations are the opposite of what would be expected if cells were simply being dislocated downstream by fluid flow. Fig. 5B shows micrographs of cells cultured in different flow conditions. Whereas physiologically active muscle is perfused up to 25 times more than passive muscle (38) in humans, the range of this device was a 500-fold difference and could have been potentially larger. At flow rates that were too high (370 nl/min), most cells were dislodged, and the morphology of remaining cells was larger and more spread out. Within a range of intermediate flow rates (3–74 nl/min), cell density within channels increased with increasing flow. Flow rates at 74 nl/min resulted in a general increase of cell density throughout the length of the cell-seeded channel segment. At 15 nl/min, the density varied steadily along the channel length, and at the slowest flow rate (3 nl/min) no density change was prevalent except at the upstream region. With no flow, cells died after an overnight incubation, presumably because of lack of nutrients or growth factors and buildup of waste. Fig. 5C shows differentiated cells located at the downstream end of a longer circulatory path, and Fig. 5D shows immunohistostaining of the myosin heavy chain by active pumping of standard staining reagents and fusion as an indication of terminal muscle differentiation. Fig. 5E quantifies the expansion of cellular boundaries at the upstream and downstream locations of the original cellular mass when perfused at different intermediate flow rates. Throughout all flow rates, the downstream cellular boundaries remained relatively constant, whereas boundaries at the opposite end expand further upstream at a constant rate with higher flow rates, leading to faster outgrowth and shorter lag times before growth. However, subsequent reversal of the flow direction reversed the direction of cellular outgrowth (data not shown).
Fig. 5.
On-chip cell culture. (A) A typical cell culture loop at a flow rate of 15 nl/min. Fluid is pumped by pins on the left side in a counterclockwise direction. (B) C2C12 cells cultured in channels circulated with cell culture media at different flow rates. The flow rate was calibrated in Fig. 2B.(C) Phase-contrast images of muscle differentiated and undifferentiated cells aligned in the movement of flow. (D) Fluorescent micrograph of the same differentiated cells stained for the myosin heavy chain. Positive staining and multinucleated cells indicated differentiation. Staining was carried out by using the same pumping strategy as media circulation to dispense all reagents. (E) Upstream (solid lines) and downstream (dashed lines) growth as indicated by the length of the bulk movement of cells at the two regions. The flow speeds are 74, 15, and 3 nl/min. The respective average residence times of media in the seeded cell segment are 0.3, 1.5, and 7.7 min.
The observed graded cell proliferation or directional migration suggests the reduction of mitogens, chemotactic agents, nutrients, and/or oxygen, or the accumulation of cellular waste over the length of the seeded cells (39). The extracellular fluid-to-cell ratio (by volume) in the channels matches the physiological value of 0.5, whereas the same ratio is ≈200 in conventional Petri dish culture (8). The small ratio facilitates chemical gradients because cells can consume and secrete at appreciable levels so that compounds are depleted or accumulate along the channel length in each segment of entering fluid (40). Generation of gradients during perfusion is relevant to a variety of physiological cellular microenvironments in developing tissues where morphogenetic gradients are important in specifying cell fate (41).
Using Braille displays as active fluidic controllers allows the basic function of microfluidic systems to be readily accessible and programmable without the need for large and expensive laboratory facilities. A number of advantageous microfluidic pumps and valves have been demonstrated previously. Most notably, pneumatic/hydraulic pumps and valves using multilayer soft lithography (25) and micromachined pumps and valves (23, 27, 28) can be more closely spaced than Braille pins and arranged entirely arbitrarily. However, Braille display-based microfluidic systems have six characteristics that make them especially versatile. (i) Channel dimensions are ideal for eukaryotic cell handling and microscale tissue engineering. (ii) Portable Braille displays are commercially available and relatively inexpensive (one-time capital investment of approximately $10 per pin). (iii) The total fluid actuation system is compact in size (as small as 8 × 18 × 3.6 cm), can be powered by battery or Universal Serial Bus, and is easy to move among cell incubators, laminar flow cabinets, and microscopes. (iv) Each display holds 64 (4 × 16) to 3,072 (48 × 64) vertically movable pins arranged in a planar grid, so a multitude of distinct channel designs can be driven by the same pin grid as cassettes without the need for external tubing or wiring. The predefined actuator arrangement is also useful as a standard to allow facile transfer of designs between different chips. Pins are spaced ≈2.5 or 3.8 mm apart depending on whether they are on the same or between Braille cells. (v) As opposed to combinatorially controlled actuators, the ability to independently address actuators simultaneously enables a greater degree of freedom in fluidic movement. This flexibility is demonstrated in the continuous fluid mixing array (Fig. 3 and Fig. 7, which is published as supporting information on the PNAS web site). (vi) Although the currently used channel material (PDMS) has a number of advantages, such as optical transparency and disposability, this system can potentially be used with other elastic materials (42).
Compared to other microbioreactor systems (7–10), Braille display-based microfluidic bioreactors are more densely packed and not limited to linear and unidirectional perfusion. Fluidic direction can be switched or recycled to accommodate different stages of tissue growth. For example, one seeding event is able to prepare multiple compartments at precise locations, but compartments can be separated either for single-pass or looped circulation. Because most perfusion systems use a macroscopic pump (peristaltic or syringe) and reservoir for each cell segment, it would be difficult to recompartmentalize or redirect fluid flow. We believe that the flexibility and long-term cell maintenance capabilities of Braille display-based microfluidics will enable higher-throughput cellular studies where spatiotemporal cell and fluid patterns are required such as in study of autocrine/paracrine signaling, cell differentiation, and morphogenesis. These methods will also be useful in fabricating microfluidic systems for studies of metabolism, for screening by using arrays of cells, in mimicry of in vivo niches, and for development of cell-based sensors.
Supplementary Material
Acknowledgments
We thank Dr. Shian-Huey Chiang (University of Michigan, Ann Arbor) for the C2C12 cells and Prof. Marie E. Csete, Mai T. Lam, and Prof. Mark Burns for helpful review of this manuscript and use of clean-room facilities. This material is based on work supported by the U.S. Army Research Laboratory and the U.S. Army Research Office under Contract/Grant DAAD19-03-1-0168, National Science Foundation Grant BES-0238625, The Whitaker Foundation, a Nathan Shock Center for Aging Research Pilot Grant, and the National Aeronautics and Space Administration BioScience and Engineering Institute. W.G. thanks the University of Michigan Undergraduate Research Opportunities Program for the Summer Biomedical Research Fellowship.
Author contributions: W.G., X.Z., N.F., B.S.C., and S.T. designed research; W.G., X.Z., N.F., B.S.C., and S.T. performed research; W.G., X.Z., and N.F. contributed new reagents/analytic tools; W.G., X.Z., N.F., and S.T. analyzed data; and W.G. and S.T. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PDMS, poly(dimethylsiloxane).
References
- 1.Takayama, S., McDonald, J. C., Ostuni, E., Liang, M. N., Kenis, P. J. A., Ismagilov, R. F. & Whitesides, G. M. (1999) Proc. Natl. Acad. Sci. USA 96, 5545–5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takayama, S., Ostuni, E., LeDuc, P., Naruse, K., Ingber, D. & Whitesides, G. M. (2003) Chem. Biol. 10, 123–130. [DOI] [PubMed] [Google Scholar]
- 3.Jeon, N. L., Baskaran, H., Dertinger, S. K. W., Whitesides, G. M., Water, L. V. D. & Toner, M. (2002) Nat. Biotechnol. 20, 826–830. [DOI] [PubMed] [Google Scholar]
- 4.Wheeler, A. R., Throndset, W. R., Whelan, R. J., Leach, A. M., Zare, R. N., Liao, Y. H., Farrell, K., Manger, I. D. & Daridon, A. (2003) Anal. Chem. 75, 3581–3586. [DOI] [PubMed] [Google Scholar]
- 5.Leclerc, E., Sakai, Y. & Fujii, T. (2003) Biomed. Microdev. 5, 109–114. [Google Scholar]
- 6.Beebe, D., Wheeler, M., Zeringue, H., Walters, E. & Raty, S. (2002) Theriogenology 57, 125–135. [DOI] [PubMed] [Google Scholar]
- 7.Sikavitsas, V. I., Bancroft, G. N., Holtorf, H. L., Jansen, J. A. & Mikos, A. G. (2003) Proc. Natl. Acad. Sci. USA 100, 14683–14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sin, A., Chin, K. C., Jamil, M. F., Kostov, Y., Rao, G. & Shuler, M. L. (2003) Biotechnol. Prog. 20, 338–345. [DOI] [PubMed] [Google Scholar]
- 9.Powers, M. J., Domansky, K., Kaazempur-Mofrad, M. R., Kalezi, A., Capitano, A., Upadhyaya, A., Kurzawski, P., Wack, K. E., Stolz, D. B., Kamm, R., et al. (2002) Biotechnol. Bioeng. 78, 257–269. [DOI] [PubMed] [Google Scholar]
- 10.Allen, J. W. & Bhatia, S. N. (2003) Biotechnol. Bioeng. 82, 253–262. [DOI] [PubMed] [Google Scholar]
- 11.Hatch, A., Kamholz, A. E., Hawkins, K. R., Munson, M. S., Schilling, E. A., Weigl, B. H. & Yager, P. (2001) Nat. Biotechnol. 19, 461–465. [DOI] [PubMed] [Google Scholar]
- 12.Zhao, B., Moore, J. S. & Beebe, D. J. (2001) Science 291, 1023–1026. [DOI] [PubMed] [Google Scholar]
- 13.Cho, B. S., Schuster, T. G., Zhu, X., Chang, D., Smith, G. D. & Takayama, S. (2003) Anal. Chem. 75, 1671–1675. [DOI] [PubMed] [Google Scholar]
- 14.Effenhauser, C. S., Harttig, H. & Kramer, P. (2002) Biomed. Microdevices 4, 27–32. [Google Scholar]
- 15.Walker, G. M. & Beebe, D. J. (2002) Lab Chip 2, 57–61. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen, N. T. & White, R. M. (1999) Sens. Actuators A Phys. 77, 229–236. [Google Scholar]
- 17.Munyan, J. W., Fuentes, H. V., Draper, M., Kelly, R. T. & Woolley, A. T. (2003) Lab Chip 3, 217–220. [DOI] [PubMed] [Google Scholar]
- 18.Hong, C.-C., Murugesan, S., Kim, S., Beaucage, G., Choi, J.-W. & Ahn, C. H. (2003) Lab Chip 3, 281–286. [DOI] [PubMed] [Google Scholar]
- 19.Lai, S., Wang, S., Luo, J., Lee, J., Yang, S. & Madou, M. J. (2004) Anal. Chem. 76, 1832–1837. [DOI] [PubMed] [Google Scholar]
- 20.Broyles, B. S., Jacobson, S. C. & Ramsey, J. M. (2003) Anal. Chem. 75, 2761–2767. [DOI] [PubMed] [Google Scholar]
- 21.Harrison, D. J., Fluri, K., Seiler, K., Fan, Z., Effenhauser, C. S. & Manz, A. (1993) Science 261, 895–897. [DOI] [PubMed] [Google Scholar]
- 22.Emrich, C. A., Tian, H., Medintz, I. L. & Mathies, R. A. (2002) Anal. Chem. 74, 5076–5083. [DOI] [PubMed] [Google Scholar]
- 23.Liu, R. H., Yang, J., Lenigk, R., Bonanno, J. & Grodzinski, P. (2004) Anal. Chem. 76, 1824–1831. [DOI] [PubMed] [Google Scholar]
- 24.Handique, K., Burke, D. T., Mastrangelo, C. H. & Burns, M. A. (2001) Anal. Chem. 73, 1831–1838. [DOI] [PubMed] [Google Scholar]
- 25.Unger, M. A., Chou, H.-P., Thorsen, T., Scherer, A. & Quake, S. R. (2000) Science 288, 113–116. [DOI] [PubMed] [Google Scholar]
- 26.Schabmueller, C. G. J., Koch, M., Mokhtari, M. E., Evans, A. G. R., Brunnschweiler, A. & Sehr, H. (2002) J. Micromech. Microeng. 12, 420–424. [Google Scholar]
- 27.Yang, X., Grosjean, C., Tai, Y.-C. & Ho, C.-M. (1998) Sens. Actuators A Phys. 64, 101–108. [Google Scholar]
- 28.Burns, M. A., Johnson, B. N., Brahmasandra, S. N., Handique, K., Webster, J. R., Krishnan, M., Sammarco, T. S., Man, P. M., Jones, D., Heldsinger, D., et al. (1998) Science 282, 484–487. [DOI] [PubMed] [Google Scholar]
- 29.Duffy, D. C., McDonald, J. C., Schueller, O. J. A. & Whitesides, G. M. (1998) Anal. Chem. 70, 4974–4984. [DOI] [PubMed] [Google Scholar]
- 30.Futai, N., Gu, W. & Takayama, S. (2004) Adv. Mater. 16, 1320–1323. [Google Scholar]
- 31.Yaffe, D. & Saxel, O. (1977) Differentiation 7, 159–166. [DOI] [PubMed] [Google Scholar]
- 32.Stroock, A. D., Dertinger, S. K. W., Ajdari, A., Mezic, I., Stone, H. A. & Whitesides, G. M. (2002) Science 295, 647–651. [DOI] [PubMed] [Google Scholar]
- 33.Liu, R. H., Stremler, M. A., Sharp, K. V., Olsen, M. G., Santiago, J. G., Adrian, R. J., Aref, H. & Beebe, D. J. (2000) J. Microelectromech. Syst. 9, 190–197. [Google Scholar]
- 34.Jen, C., Wu, C., Lin, Y. & Wu, C. (2003) Lab Chip 3, 77–81. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, T. J., Ross, D. & Locascio, L. E. (2002) Anal. Chem. 74, 45–51. [DOI] [PubMed] [Google Scholar]
- 36.Glasgow, I. & Aubry, N. (2003) Lab Chip 3, 114–120. [DOI] [PubMed] [Google Scholar]
- 37.Handique, K. & Burns, M. A. (2001) J. Micromech. Microeng. 11, 548–554. [Google Scholar]
- 38.Guyton, A. C. (1986) in Textbook of Medical Physiology, ed. Dreibelbis, D. (Saunders, Philadelphia), pp. 336–337.
- 39.Corti, S., Salani, S., Bo, R. D., Sironi, M., Strazzer, S., D'Angelo, M.G., Comi, G. P., Bresolin, N. & Scarlato, G. (2001) Exp. Cell Res. 268, 36–44. [DOI] [PubMed] [Google Scholar]
- 40.Ledezma, G. A., Folch, A., Bhatia, S. N., Balis, U. J., Yarmush, M. L. & Toner, M. (1999) J. Biomech. Eng. 121, 58–64. [DOI] [PubMed] [Google Scholar]
- 41.Tabata, T. & Takei, Y. (2004) Development (Cambridge, U.K.) 131, 703–712. [DOI] [PubMed] [Google Scholar]
- 42.Rolland, J. P., Van Dam, R. M., Schorzman, D. A., Quake, S. R. & DeSimone, J. M. (2004) J. Am. Chem. Soc. 126, 2322–2323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.