Abstract
Incorporation of nucleosides with novel base-constraining oxetane (OXE) modifications [oxetane, 1-(1′,3′-O-anhydro-β-d-psicofuranosyl nucleosides)] into antisense (AS) oligodeoxyribonucleotides (ODNs) should greatly improve the gene silencing efficiency of these molecules. This is because OXE modified bases provide nuclease protection to the natural backbone ODNs, can impart Tm values similar to those predicted for RNA–RNA hybrids, and not only permit but also accelerate RNase H mediated catalytic activity. We tested this assumption in living cells by directly comparing the ability of OXE and phosphorothioate (PS) ODNs to target c-myb gene expression. The ODNs were targeted to two different sites within the c-myb mRNA. One site was chosen arbitrarily. The other was a ‘rational’ choice based on predicted hybridization accessibility after physical mapping with self-quenching reporter molecules (SQRM). The Myb mRNA and protein levels were equally diminished by OXE and PS ODNs, but the latter were delivered to cells with approximately six times greater efficiency, suggesting that OXE modified ODNs were more potent on a molar basis. The rationally targeted molecules demonstrated greater silencing efficiency than those directed to an arbitrarily chosen mRNA sequence. We conclude that rationally targeted, OXE modified ODNs, can function efficiently as gene silencing agents, and hypothesize that they will prove useful for therapeutic purposes.
INTRODUCTION
The development of mRNA targeted gene silencing drugs is driven by their potential for exquisite specificity, and the promise of low toxicity (1–3). Of the various strategies under investigation, antisense (AS) oligodeoxyribonucleotides (ODNs) are among the most intensively studied, and the most mature with regard to clinical development (4). Still, it is widely perceived that the efficiency of these molecules can be improved (5,6), especially for clinical use (7–9), and it was this goal that prompted the studies reported herein.
The AS ODNs silence gene expression by hybridizing with single-stranded sequence within the targeted mRNA molecule. The resulting ODN–RNA duplex can block translation of the targeted mRNA by steric inhibition of the ribosomal complex, and/or by forming a substrate for RNase H, a naturally occurring enzyme that digests the RNA component of an ODN–RNA duplex (10). Highly efficient and predictable mRNA cleavage occurs when AS ODN molecules are employed in a cell free system, but reliability erodes when these molecules are employed for actual gene silencing in living cells. The rapid degradation of DNA by endo- and exonucleases is an important contributing factor to this problem, and as a result many chemical modifications of natural DNA have been investigated in hopes of improving AS ODN stability in vivo (3,11,12). Alterations that affect the phosphodiester bridges between nucleosides are the most commonly employed strategies. Sulfur substitution of the oxygen atom, leading to the formation of a phosphorothioate (PS) backbone, is arguably the most successful of these modifications because in addition to making the ODNs relatively nuclease resistant, they are water soluble and recruit RNase H, a property that is thought to be critical for in vivo gene silencing utility (13–15). Nonetheless, these highly desirable properties are offset by the often significant binding of these negatively charged molecules to plasma, as well as intracellular proteins (16,17), which can lead to coagulopathies, complement activation, and non-specific cytotoxicity (18–20). Other commonly employed backbone modifications, such as peptide nucleic acids (PNAs) or morpholino ODNs do not recruit RNase H, and because they are neutral in charge, do not easily pass cellular membranes (21,22). These attributes would be predicted to significantly diminish their utility for clinical applications.
Recently, much attention has been focused on AS ODN molecules with sugar-modified nucleosides (23). Among these, ODNs modified with ‘North’ (3′-endo-2′-exo) conformationally constrained nucleosides have attracted considerable attention (24). Upon incorporation of such a constrained nucleoside into an ODN, the ODN–RNA duplex formed after hybridization is driven to an RNA–RNA type conformation with much higher thermodynamic stability. However, this and other such modifications do not support RNase H mediated cleavage of the mRNA strand that is involved in the heteroduplex (25). To solve this problem, and to keep the otherwise desirable properties of sugar modified molecules, we developed a novel oxetane (OXE) modification of nucleosides that, when incorporated into ODNs, would impart enhanced stability against endo- and exonucleases, allow tight binding affinity with RNA, support RNase H activity, and accelerate catalysis of the mRNA target (26–30).
In this study, we report the use of an OXE modified AS ODN with natural phosphate backbone (26–30) to silence gene expression in an intact cellular system and compare its effectiveness with an ODN of identical sequence but with a PS backbone. Additionally, we report a direct comparison of silencing efficiency between an ODN that was arbitrarily targeted versus one that was ‘rationally’ directed.
MATERIALS AND METHODS
ODN synthesis
The PS ODNs were synthesized in the University of Pennsylvania Cancer Center Nucleic Acid Facility (UPCC NAF) on an Expedite™ 8909 Nucleic Acid Synthesis System (Applied Biosystems, Foster City, CA) by using standard phosphoramidite chemistry. The ODNs were then deprotected with concentrated ammonia and purified by reversed-phase high-performance liquid chromatography (HPLC). Concentrations of the full-length ODNs were determined by ultraviolet (UV) spectroscopy at 260 nm by using calculated extinction coefficients.
Self-quenching reporter molecules (SQRM), DNA hairpin molecules with self-complementary 5′ and 3′ ends to which a fluorophor and a quencher respectively are conjugated, were synthesized with phosphodiester backbones in the UPCC NAF in following manner. The 5′-fluorescein and 3′-C7-DABCYL groups were synthesized by using 3′-C7-DABCYL CPG (Glen Research Corporation, Sterling, VA) as support and fluorescein phosphoramidite (Cruachem Limited, Glasgow, Scotland, UK) for 5′-end modification with standard phosphoramidite chemistry on the Expedite™ 8909 Nucleic Acid Synthesis System. The SQRMs were deprotected and purified as described for ODNs above. The concentrations of the SQRMs were determined by UV spectroscopy.
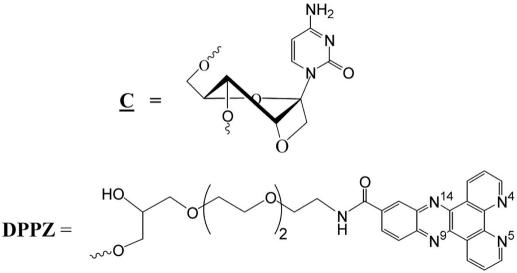
The AS and scrambled (SCR) sequence ODNs that contain OXE modified cytidine nucleosides were synthesized in the Department of Bioorganic Chemistry at University of Uppsala, Sweden, as previously reported (26–28,30). Dipyridophenazine (DPPZ) was conjugated to the 3′ end of each oxetane ODN sequence to increase stability against nucleases and to enhance mRNA target affinity (Figure 1). DPPZ derivatized CPG was used to conjugate the DPPZ at the 3′-end (31). All ODNs were purified by 20% denaturing (7 M urea) PAGE. The bands were visualized by UV shadowing, cut from the gel, and the ODNs were extracted with 0.3 M sodium acetate solution. The ODNs were desalted by using Sep-Pack C-18 cartridges (Waters, Milford, USA) and the concentrations were determined by UV spectroscopy.
Figure 1.
Chemical structure of OXE modification and the DPPZ moiety.
The sequences of ODNs employed in the study are given in Table 1. The 24mer sequence corresponds to c-myb codons 2–9 and the 30mer sequence was chosen as a result of physical mapping of c-myb mRNA.
Table 1. Sequences of OXE and PS modified ODNs employed.
| Name | Sequence |
|---|---|
| OXE 1 | d (TAT GCT GTG CCG GGG TCT TCG GGC) |
| OXE 2 | d (ACA GAC CAA CGT TTC GGA CCG TAT TTC TGT) |
| OXE 3 | d (ACA GAC CAA CGT TTC GGA CCG TAT TTC TGT) |
| PS 1 | d (TAT GCT GTG CCG GGG TCT TCG GGC) |
| PS 2 | d (ACA GAC CAA CGT TTC GGA CCG TAT TTC TGT) |
| Scr 1 | d (TCT TCA GAG ACT GCG ACA TAG CGC) |
| Scr 2 | d (ACC GTC CAT CGT GTA GCA ACC TTA GCA AGT) |
The oxetane modified cytosines are indicated in bold font and underlined. Each OXE molecule contains the DPPZ moiety at the 3′end for increased stability.
Cell culture and transfections
K562 cells were cultured at 37°C, in a fully humidified incubator (95% humidity) at 5% CO2 concentration. They were maintained in RPMI 1640 media that is supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine 0.5% penicillin/streptomycin. As required, the cells were removed from the culture and counted by means of a hemocytometer.
ODN transfections
The ODN transfections were carried out by nucleoporation using an Amaxa Nucleofector (Cologne, Germany) under conditions suggested by the manufacturer. In brief, 1 × 106 K562 cells were suspended in 100 μl of nucleofection buffer solution with 5 μg of ODNs (final concentrations of ODNs during the nucleofection procedure were 5.2 μM for the 24mers, and 4.3 μM for 30mers). Post transfection cell viability was determined by trypan blue exclusion. When calculating the amount (μg) of AS ODNs that is to be utilized for the nucleofection procedure, the molecular weight of DPPZ tail was subtracted. Immediately after the nucleoporation, cells were put into 1.5 ml of fresh RPMI media with 10% FBS and cultured for an additional 24 h or 48 h, after which time, the cells were harvested and employed for subsequent analyses.
Quantitative real time PCR (QRT-PCR)
Total RNA was isolated from cells by using Qiagen kits (Valencia, CA) as directed by the manufacturer. RNA (300 ng) was reverse transcribed by using random hexamers (Invitrogen, Carlsbad, CA) as primers. The resulting cDNA was employed for QRT–PCR reactions. These were carried out in triplicate with 1 μl of cDNA from the reverse transcription mixture. An ∼300 bp c-myb PCR product was obtained with the forward primer: d(TAG TCA ATG TCC CTC AGC CAG C) (starts at position 919 in c-myb) and reverse primer: d(GGA CGA TCATGC ACC TTG CT) (starts at position 1189). The c-myb probe was labeled with reporter dye: 6 carboxyfluorescein (FAM) at the 5′end, and the quencher (TAMRA) at the 3′end. The sequence of the probe was as follows: d(AGA ACA CCA CTC CAC TCC ATC TCT GCC A).
The 18S was used as a reporter gene for the QRT–PCR reaction. The product was obtained with forward primer: d(GGA CAT CTA AGG GCA TCA CAG ACC) and reverse primer: d(TGA CTC AAC ACG GGA AAC CTC AC). The 18S probe sequence was as follows: d(TGG CTG AAC GCC ACT TGT CCC TCT AA), labeled with FAM at the 5′ end and TAMRA at the 3′ end. The total reaction mix of 15 μl was preincubated at 50°C for 2 min. The protocol for PCR consisted of denaturation at 95°C for 10 min, followed by 36 cycles of 15 s denaturation at 92°C and 1.5 min of annealing and elongation at 60°C. The expected ∼300 bp c-myb PCR product was resolved on 1% agarose gel to confirm the specificity of the reaction. The negative control was carried out on RNA (no RT reaction).
Western blotting
The control and transfected cells were harvested after 24 or 48 h, washed twice in phosphate-buffered saline (PBS) and then pelleted. The pellet was lysed in 50 μl of triple-detergent lysis buffer (50 mM Tris, 150 mM NaCl, 0.02% sodium azide, 0.1% SDS and 1% NP-40) for 30 min. on ice. After centrifugation at 4°C, the supernatant containing total cellular proteins was collected on ice. A total amount of 100 μg of protein extract was resolved on 10% polyacrylamide gel, transferred to poly vinylidene fluoride (PVDF) membrane and probed overnight at 4°C with antibody against c-myb, clone 1-1 (Upstate, NY) at 1:1000 dilution. The membrane was then washed three times with TBS-T buffer and probed with secondary HRP conjugated antibody anti-mouse (Amersham), at 1:3000 dilution for 1 h at room temperature. Bands were visualized with enhanced chemiluminescence (ECL+) reagent. The same PVDF membrane was then stripped with stripping buffer and probed with antibody against beta actin (1:3000 dilution).
Slot blotting
Control cells, and cells nucleofected with 5 μg of OXE or PS modified AS ODNs, were collected at 5 min, and at 3 h after nucleofection and washed twice in PBS. The cell pellet was lysed in 400 μl of lysis buffer (50 mM HEPES pH 7.5; 85 mM KCL and 0.5% NP-40) for 20 min on ice. Thereafter cells were homogenized with a dounce and centrifuged at 5000 r.p.m. for 10 min. The supernatant was removed and the pellet was resuspended in 120 μl of diethyl pyrocarbonate (DEPC) water. Then, 0.4 M NaOH and 10 mM EDTA were added, and the samples were heated at 100°C for 10 min to denature DNA. The samples were applied on the Zeta-Probe membrane, which was first rehydrated with water, using Bio-Dot SF Microfiltration Apparatus (Bio-Rad). After the samples have been filtered through, 500 μl of 0.4 M NaOH was added and pulled through, until the wells were empty. The blotted membrane was removed from the apparatus and air-dried. Subsequently, DNA was UV cross linked to the membrane, and the membrane was pre-hybridized at 65°C by incubation with pre-warmed hybridization solution for 30 min. C-myb sense sequence, complementary to the sequence that was used for treating cells, was radiolabeled with T4 polynucleotide kinase (New England Bio Labs), and 0.075 mCi P-32 gamma ATP (Amersham). The probe was purified with Quick Spin (TE) Columns G-25 sephadex (Boehringer Mannheim). The pre-hybridized membrane was incubated with radiolabeled probe at 65°C for 12 h. Then the membrane was washed twice with buffer containing 0.2% sodium chloride-sodium citrate (SSC) and 0.2% SDS and put into the autoradiography cassette and incubated with film for 6 h at −80°C, after which time it was developed.
In vitro transcription
RNA was transcribed from a full-length, c-myb cDNA cassette cloned into a pcDNA3 plasmid (Invitrogen Corporation, Carlsbad, CA). The plasmid was linearized with XhoI (Promega Corporation, Madison, WI; New England Biolabs, Inc., Beverly, MA) and then transcribed with T7 RNA Polymerase by using the RiboMax™ Kit (Promega Corporation, Madison, WI). The RNA was purified by extraction with phenol/chloroform followed by ethanol precipitation or it was passed over an RNeasy column (QIAgen, Inc., Valencia, CA) and eluted in RNase-free water.
Mapping hybridization accessible regions of mRNA
We employed SQRM to identify potential hybridization accessible regions within the c-myb mRNA(32). A computer algorithm was developed to scan the cDNA sequence of interest for inverted repeats of 4–5 bases that were separated by an intervening sequence of 18–20 bases, to direct the SQRM rationally. It was hypothesized that such regions might self anneal by looping out a single strand available for hybridization. This was tested by synthesizing SQRM complementary to these sequences. They were dissolved in SQRM buffer to a final concentration of 100 nM and added to 1 μM of in vitro transcribed c-myb RNA target. Fluorescence emission was monitored by using a Packard FluoroCount Microplate Fluorometer (Packard Instrument Company, Meriden, CT). c-myb SQRMs and the SCR 1 control sequence were incubated with [1 μM] c-myb RNA for 30 min at 37°C. SQRMs were incubated in a separate well with DNase I, fast performance liquid chromatography (FPLC)-pure (Roche Diagnostics Corp., Indianapolis, IN) for 30 min at 37°C to obtain maximum fluorescence.
RESULTS
Effect of PS versus OXE modified ODNs on c-myb mRNA and protein expression
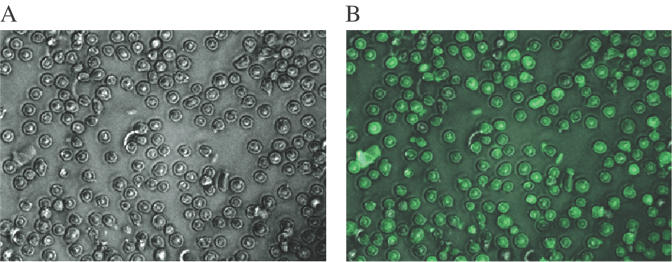
We compared the effectiveness of OXE versus PS modified ODNs by determining their respective abilities to silence the expression of Myb, an obligate transcription factor in hematopoietic cells (33,34). The ODNs were delivered by nucleoporation with an efficiency that exceeded 90% (Figure 2). Control cells were subjected to the identical procedure but in the absence of ODNs.
Figure 2.
Delivery of fluorescein labeled unmodified ODNs into K562 cells with the nucleoporation technique: phase (A) and fluorescent (B) low power (200×) photomicrographs.
We first examined the effectiveness of a PS ODN (PS 1) and an OXE ODN (OXE 1) targeted to the same arbitrarily chosen sequence within the c-myb mRNA. The sequence examined corresponded to c-myb codons 2–9. A PS AS ODN targeting the identical sequence has been employed in a previously reported clinical study (35). The effect of the various ODNs was examined at 6, 24, and 48 h after nucleoporation. No effect on mRNA or protein was seen at 6 h (data not shown), and effects at 24 and 48 h were equivalent. The 24 h mRNA data is shown (Figure 3). In these experiments, PS 1 appeared more effective than OXE1 since the former gave an ∼40% reduction of c-myb mRNA, whereas treatment with OXE 1 gave only an ∼20% reduction, a value not dissimilar from that obtained with an OXE ODN SCR sequence (Figure 3).
Figure 3.
QTR-PCR assay performed on RNA isolated from K562 cells transfected with OXE and PS antisense (AS) ODN molecules. Data are presented as a function of c-myb mRNA copies relative to 18S RNA. As noted above, 1 × 106 K562 cells were suspended in 100 μl of nucleofection buffer solution with 5 μg of ODNs (final concentrations of ODNs during the nucleofection procedure were 5.2 μM for the 24mers, and 4.3 μM for 30mers). Control cells were subjected to nucleoporation, but in the absence of ODNs. OXE1 and PS 1 have identical sequence (24mer). OXE2, OXE3 and PS 2 are also sequence identical. Based on the mapping procedure described, these ODNs were predicted to hybridize with their target with high efficiency. OXE 2 and OXE 3 differ in number of oxetane modified nucleosides per molecule (four and five, respectively). Scrambled sequences (SCR 1 and 2) have the same nucleotide composition as OXE1 and OXE 2 respectively but in a random order.
In contrast to these results, both PS and OXE ODNs, targeted to a region that was identified as accessible with the mapping procedure, were much more efficient at extinguishing mRNA expression (Figure 3). PS 2 performed somewhat better than the corresponding OXE molecules (OXE 2 and 3) yielding mRNA reduction, measured by QRT–PCR of ∼85 versus 70% with the OXE ODNs. It should be noted that although OXE 2 and OXE 3 were of identical sequence, OXE 2 contained four oxetane-modified cytidines, whereas OXE 3 contained five such modifications. Nevertheless, their biological activity, as determined by the ability to diminish c-myb mRNA levels did not appear to differ significantly when measured by QRT–PCR.
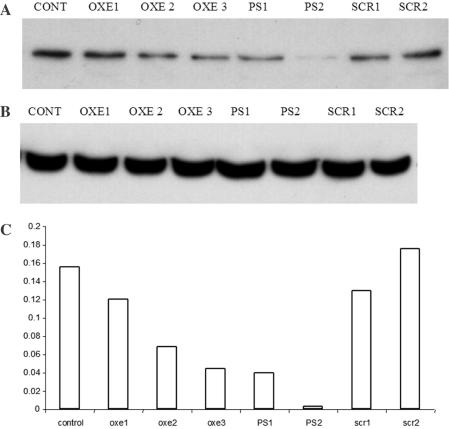
The correlation between c-myb mRNA extinction and protein diminution was also examined (Figure 4). The diminution of Myb protein after treatment with OXE 1, ∼20% when compared with untreated cells, correlated well with its effect on the mRNA, and did not appear to differ significantly from a SCR sequence control. The corresponding PS 1 molecule, which suppressed c-myb mRNA levels by ∼40%, effected a substantial reduction of the protein as well. Again, in agreement with their effects on mRNA levels, the AS 30mers were highly efficient in decreasing protein levels in the treated cells. OXE 2 and 3 suppressed Myb protein ∼70% when compared with untreated controls. Only trace protein was detectable in cells treated with PS 2.
Figure 4.
Western blot on lysates from K562 control and transfected cells. About 1 × 106 K562 cells were suspended in 100 μl of nucleofection buffer solution with 5 μg of ODNs (final concentrations of ODNs during the nucleofection procedure were 5.2 μM for the 24mers, and 4.3 μM for 30mers). Cells were lysed and 100 μg of total protein extract was resolved on polyacrylamide gel. The proteins were than transferred to PVDF membrane and probed first with antibody against c-myb (A), then the same membrane was stripped and probed again with antibody against beta actin (B). The intensity of bands was measured with a densitometer and the ratio of c-myb versus beta actin is presented in the graph (C).
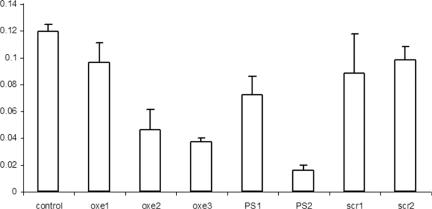
Effect of ODNs on cell proliferation
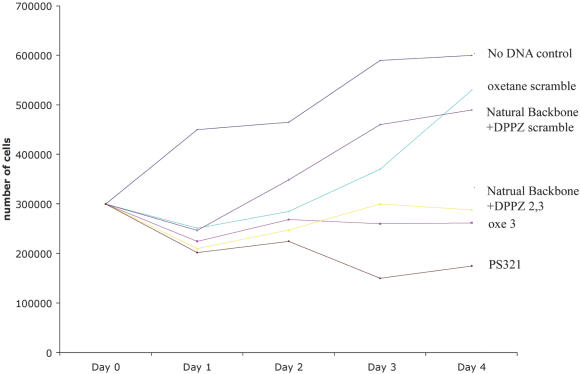
We next examined the effect of the various ODN modifications on K562 cell proliferation (Figure 5). We expected to see that inhibition of Myb expression would correlate with effects on cell growth and this was in fact observed. When compared with the proliferation of untreated control cells, a scrambled sequence OXE ODN, and a scrambled natural backbone ODN with a 3′-DPPZ terminator, had minimal effects on cell growth. These results indicate that the DPPZ moiety is not toxic to cells and does not degrade sequence specificity. In contrast, PS, OXE, and natural backbone-3′-DPPZ targeted to the previously identified hybridization accessible region within the c-myb mRNA all gave significant growth inhibition over the course of the four-day experiment. In accord with its apparently greater inhibition of c-myb mRNA and protein levels, the PS ODNs inhibit cell proliferation to a greater extent than the other ODNs evaluated. Nevertheless, the possibility that ‘non-specific’ effects of the PS backbone were contributing to this effect cannot be ruled out.
Figure 5.
Cell proliferation after electroporation of OXE and PS modified ODNs. About 3 × 105 K562 cells were suspended in 100 μl of nucleofection buffer solution with 5 μg of the respective ODNs. Following electroporation cells were washed and then transferred to 12 well costar plates (Corning Inc., Corning, NY), each well containing 1.5 ml of RPMI with 10% FBS. An aliquot of cells were removed every 24 h over 4 days and counted with a hemocytometer.
Intracellular concentrations of PS and OXE ODNs
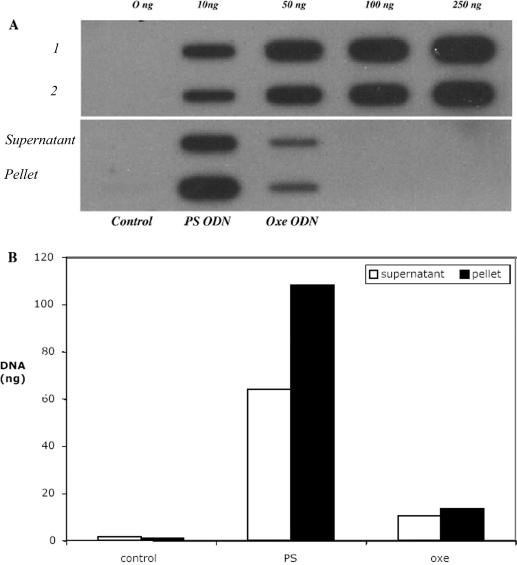
In the experiments described in Figures 3 and 4 above, the PS ODNs appeared to outperform the OXE modified ODNs. A number of factors could explain these results. One is that the ‘off target’ effects of PS ODNs contributed to the results observed. Another is the relatively simple possibility that the molecules were delivered intracellularly with differing efficiency despite the fact that the same delivery process was used. Since the latter was relatively easy to assess, we examined the intracellular concentration of the PS and OXE ODNs 5 min following nucleoporation, and then again 3 h later following the same procedure employed for the experiments described in Figure 3. Somewhat to our surprise, we found that the OXE modified ODNs were nucleoporated into cells with much less efficiency than the PS ODNs (Figure 6A). In the material analyzed 5 min after nucleoporation, it is clear from the intensity of the bands shown on the slot blot that considerably more PS ODNs were electroporated into the cells than OXE ODNs, despite the fact that the same number of cells (1 × 106) were suspended in identical amounts of ODN (5 μg). Also of interest, it appears that a significant portion of the PS ODNs was bound to intracellular structures since approximately twice the amount of material was found in the cell pellet as in the cell lysate. In contrast, the OXE ODNs appeared to be equally distributed between these two compartments.
Figure 6.
Quantitation of ODNs delivered to cells by nucleoporation. K562 cells were nucleoporated in buffer alone (Control), or with PS or OXE ODNs. Five minutes later, the cells were lysed and the supernatant was separated from the residual cellular material by centrifugation. The resulting supernatant and cell pellet was then transferred to the membrane, and the blots were probed with a radiolabeled complementary sequence. (A) Slot blot of ODNs present in K562 cell supernatants, and residual cell pellet. Rows 1 and 2 of the upper blot are, respectively, PS and OXE ODN standards applied to the membrane for quantitation purposes. Columns labeled on the lower half of the blot represent material found in the cell supernatant and pellet, obtained from untreated control cells and cells nucleoporated with PS and OXE ODNs. (B) Densitometry analysis of slot blot shown in (A) above. Open columns represent material present in the cell supernatant and black columns represent material present in the cell pellet of PS and OXE treated cells.
The amount of material specifically associated with the cell lysate and residual pellet was quantitated by densitometry (Figure 6B) by using known amounts of DNA spotted to the membrane for reference purposes (Figure 6A). By this method, we estimated that approximately six times more PS ODNs were delivered to cells than OXE ODNs. In addition, the visual impression that considerably more PS ODNs were bound to cellular structures than the OXE ODNs was confirmed. Very similar results, including the ratio of material found in cell lysate and residual pellet, were obtained at 3 h (not shown). The amount of ODNs actually delivered to cells was ∼1 and 5%, respectively, of the total OXE and PS ODNs to which the cells were exposed. These results suggest that, on a molar basis, the OXE were approximately six times more potent than the corresponding PS molecules. A credible explanation for this enhanced potency may be the fact that more OXE ODNs are bioavailable to hybridize with their intended mRNA target since they are not bound to intracellular membranes to the same extent as the PS molecules.
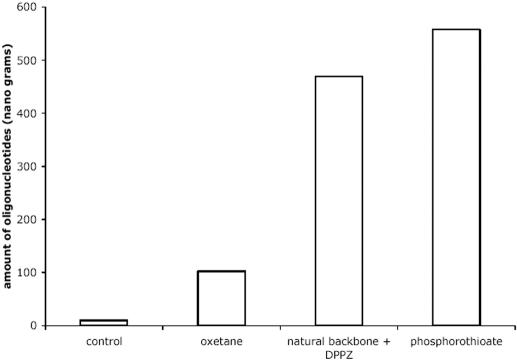
Finally, since the natural backbone conjugated to the 3′-DPPZ inhibited cell proliferation to the same extent as the OXE modified ODNs, we also examined the intracellular concentration of natural backbone ODNs with the 3′-DPPZ terminator, compared with OXE and PS ODNs of the same sequence, 5 min post nucleoporation. The densitometry measurements of total intracellular material blotted to a membrane reveals that delivery of the PS and natural backbone-3′DPPZ are essentially equivalent and approximately five to six times greater than that observed for the OXE modified ODNs (Figure 7). Accordingly, the DPPZ cannot be responsible for the decreased intracellular concentration of the OXE modified material, and at the same time, does not account for the apparent potency of the OXE modified sequence. Further, it is worth noting that the DPPZ terminator does not lead to a loss of specificity of the molecule (Figure 7).
Figure 7.
Effect of DPPZ moiety on intracellular delivery of ODNs. K562 cells were nucleoporated in buffer alone (control), or with PS, OXE, or natural backbone ODNs with a 3′-DPPZ moiety. Five minutes later, the cells were lysed and the resulting material was then slot blotted to a membrane, which was then probed with a radiolabeled complementary sequence. The densitometry measurement of DNA (ng) found on the slot blot corresponding to control, OXE, natural backbone 3′-DPPZ and PS treated cells are shown.
DISCUSSION
The AS ODNs with ‘North’ conformationally constrained nucleotides have attracted considerable attention because of their ability to drive the ODN strand of the ODN–RNA hybrid duplex to an RNA type orientation, thereby increasing target affinity (3,25). Such molecules can be broadly classified into three groups. The first group comprises ODNs synthesized with various 2′-O-alky moieties (23), or electron withdrawing substituents like fluorine, in the ribo configuration. The second group of ODNs encompasses nucleic acids containing pyranose derivatives such as hexitol and cyclohexene nucleic acids (25). The ODN–RNA hybrid duplexes of this type also mimic RNA–RNA type helices. The third group consists of ODNs that contain conformationally constrained bicyclic or tricyclic monomer nucleotide units. The novel 1′,2′-oxetane modified nucleosides developed by the Chattopadhyaya laboratory are members of this group of so-called locked nucleic acids (LNA). Because of the conformational pre-organization that occurs upon incorporation of these ‘North’ constrained units into the ODNs, even partial modification of the ODN strand can drive the ODN–RNA duplex into a rigid RNA–RNA type duplex.
The RNA–RNA duplexes have higher Tms than corresponding DNA–RNA duplexes but they are not permissive of RNase H mediated target strand cleavage. The ability to support RNase H activity can be partially or fully restored by incorporating natural or PS bases back into these ODNs creating so-called mixed or chimeric ODNs (12,23,38,39,40). In some cases, for example with LNA modified ODNs, a run of 6–10 regular or PS nucleotides is necessary to obtain effective RNase H cleavage (36,41,42). This is undesirable because PS nucleotides can increase off target silencing, and other non-specific effects, whereas use of natural nucleosides that are less active in this regard, leaves the ODNs susceptible to endonuclease activity (43). The 1′,2′-oxetane modified nucleoside addresses many of these concerns, in particular the need to allow RNase H activation (28). It is true that each oxetane–cytidine modification will drop the Tm of an OXE ODN–RNA hybrid by ∼3°C (27), but this loss in thermodynamic stability can be at least partially, or even fully, compensated for (depending on the sequence composition and length) by the 3′-DPPZ group (29). In addition, the OXE modifications impart endonuclease stability to the ODNs and at the same time, they appear to actually accelerate the catalysis of the RNA target (27,29). Further, because they may be incorporated into an all-natural backbone, they are much less likely to produce the non-specific effects and attendant cytotoxicity of molecules that are stabilized by a PS modified backbone, such as LNA.
For all these reasons, we hypothesized that OXE modified AS ODNs with natural phosphodiester backbones would prove highly useful for in vivo gene targeting applications. The studies reported herein appear to provide direct confirmation of this hypothesis. In this regard, we evaluated three OXE modified molecules that were directed against the c-myb mRNA in K562 human leukemia cells and compared their efficacy with PS ODNs of identical length and sequence. With two of the three OXE modified ODNs that were evaluated (OXE 2 and 3), we observed a large decrease in c-myb mRNA (∼70%) and Myb protein in treated cells. The SCR sequence ODNs of identical base content were without effect, which suggests that the inhibition observed was sequence specific. The OXE 1 molecule suppressed c-myb mRNA to the same extent as its scrambled control (∼20%), which suggests that its effects were not sequence specific. In contrast, the OXE 2 and 3 ODNs were both effective. Whereas they differed in terms of number of OXE modified cytidines, these 30mers were of identical sequence. We hypothesize that their efficacy was due to the fact, that they were directed to a region of the c-myb mRNA, which our SQRM driven physical mapping studies suggested was available for hybridization. Virtually identical results were obtained in hamster fibroblast cells engineered to express c-myb (TKTS+), which suggests that these results were not specific to K562 cells (data not shown).
To specifically address whether the number of OXE modifications/molecule would correlate with an ODN's functional efficacy, we synthesized the specifically targeted ODNs with either four or five modified cytosines (OXE 2 and OXE 3, respectively). The effects of these molecules on c-myb mRNA expression, and cell growth were quite similar. These results indicate that, at least for this ODN, the incorporation of an additional OXE modified cytidine did not significantly increase activity, or cytotoxicity of a more general nature. These observations are of practical utility. Since only 3 OXE modifications per molecule are thought to give an ∼4-fold protection against endonucleases, and the 3′-DPPZ moiety lends additional protection against exonucleases, the cost of increasing the number of modified nucleosides beyond 4 or 5 bases may well be avoidable. Nonetheless, the issue of what constitutes a minimal number of modifications for any given ODN will need to be more systematically addressed.
With regard to the potential gene silencing efficiency of OXE modified ODNs, we observed that PS containing ODNs gave greater reduction in the targeted (c-myb) mRNA and protein than the corresponding oxetane molecule. However, this difference was likely to be more apparent than real because when measured, the ability to deliver these materials by nucleoporation varied with the backbone. In this regard, PS molecules had approximately six times the intracellular concentration as the OXE molecules. The reason for this difference is not clear but as shown in Figure 7, it is not the result of the DPPZ moiety. It is possible that efflux rates of PS and OXE molecules differ but we think this unlikely since the intracellular ratio of PS to OXE molecules was essentially the same immediately, and 3 h after nucleoporation. One must also consider the possibility that the Tm reduction caused by the oxetane modified cytidine residues could lead to a decreased ability to detect these molecules on the slot blot, though examination of Figure 6A would suggest that such a difference could not account for the results obtained. For all these reasons then, we believe that on a molar basis, the OXE molecules are in fact more effective gene silencing agents than their corresponding PS molecules. Accordingly, if the amount of OXE molecules delivered into cells could be augmented, their real efficacy in comparison to PS ODNs would be more apparent. Furthermore, since the OXE modified molecules do not bind to intracellular proteins to the same extent as PS ODNs (Figure 6B), it is likely that they are more available to hybridize with their intended mRNA target, and less likely to cause non-specific effects because of adhering to intracellular proteins and structures.
In conclusion, our studies clearly demonstrate that oxetane–cytidine modified molecules are able to hybridize with their intended target in living cells, and that they are efficient gene silencing agents. Our studies also suggest that rational targeting can significantly increase the apparent potency of these, and possibly other AS nucleic acids as well. Accordingly, we believe that appropriately targeted, OXE modified ODNs could well prove a significant addition to the armamentarium of anticancer drugs, and in other diseases where gene silencing is expected to lead to useful therapeutic consequences.
Acknowledgments
ACKNOWLEDGEMENTS
J.B.O. is supported by a grant from the Polish Research Council (2P05A 12326). A.M.G. is supported by grants from the NIH and the Doris Duke Charitable Foundation. Generous financial support from the Swedish Natural Science Research Council (Vetenskapsrådet), the Swedish Foundation for Strategic Research (Stiftelsen för Strategisk Forskning) and Philip Morris USA Inc. to J.C. is also gratefully acknowledged.
REFERENCES
- 1.Dykxhoorn D.M., Novina,C.D. and Sharp,P.A. (2003) Killing the messenger: short RNAs that silence gene expression. Nature Rev. Mol. Cell. Biol., 4, 457–467. [DOI] [PubMed] [Google Scholar]
- 2.Jansen B. and Zangemeister-Wittke,U. (2002) Antisense therapy for cancer—the time of truth. Lancet Oncol., 3, 672–683. [DOI] [PubMed] [Google Scholar]
- 3.Kurreck J. (2003) Antisense technologies. Improvement through novel chemical modifications. Eur. J. Biochem., 270, 1628–1644. [DOI] [PubMed] [Google Scholar]
- 4.Opalinska J.B. and Gewirtz,A.M. (2002) Nucleic-acid therapeutics: basic principles and recent applications. Nature Rev. Drug Discov., 1, 503–514. [DOI] [PubMed] [Google Scholar]
- 5.Matteucci M.D. and Wagner,R.W. (1996) In pursuit of antisense. Nature, 384, 20–22. [PubMed] [Google Scholar]
- 6.Stein C.A. (1995) Does antisense exist? Nature Med., 1, 1119–1121. [DOI] [PubMed] [Google Scholar]
- 7.Calogero A., Hospers,G.A. and Mulder,N.H. (1997) Synthetic oligonucleotides: useful molecules? A review. Pharm. World Sci., 19, 264–268. [DOI] [PubMed] [Google Scholar]
- 8.Gewirtz A.M. (2000) Oligonucleotide therapeutics: a step forward [editorial; comment]. J. Clin. Oncol., 18, 1809–1811. [DOI] [PubMed] [Google Scholar]
- 9.Lebedeva I. and Stein,C.A. (2001) Antisense oligonucleotides: promise and reality. Annu. Rev. Pharmacol. Toxicol., 41, 403–419. [DOI] [PubMed] [Google Scholar]
- 10.Crooke S.T. (1999) Molecular mechanisms of action of antisense drugs. Biochim. Biophys. Acta, 1489, 31–44. [DOI] [PubMed] [Google Scholar]
- 11.Akhtar S., Kole,R. and Juliano,R.L. (1991) Stability of antisense DNA oligodeoxynucleotide analogs in cellular extracts and sera. Life Sci., 49, 1793–1801. [DOI] [PubMed] [Google Scholar]
- 12.Matteucci M. (1997) Oligonucleotide analogues: an overview. Ciba Found Symp., 209, 5–14; discussion 14–18. [PubMed] [Google Scholar]
- 13.Bonham M.A., Brown,S., Boyd,A.L., Brown,P.H., Bruckenstein,D.A., Hanvey,J.C., Thomson,S.A., Pipe,A., Hassman,F., Bisi,J.E. et al. (1995) An assessment of the antisense properties of RNase H-competent and steric-blocking oligomers. Nucleic Acids Res., 23, 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gee J.E., Robbins,I., van der Laan,A.C., van Boom,J.H., Colombier,C., Leng,M., Raible,A.M., Nelson,J.S. and Lebleu,B. (1998) Assessment of high-affinity hybridization, RNase H cleavage, and covalent linkage in translation arrest by antisense oligonucleotides. Antisense Nucleic Acid Drug Dev., 8, 103–111. [DOI] [PubMed] [Google Scholar]
- 15.Stein C.A. (2000) Is irrelevant cleavage the price of antisense efficacy? Pharmacol Ther., 85, 231–236. [DOI] [PubMed] [Google Scholar]
- 16.Levin A.A. (1999) A review of the issues in the pharmacokinetics and toxicology of phosphorothioate antisense oligonucleotides. Biochim. Biophys. Acta, 1489, 69–84. [DOI] [PubMed] [Google Scholar]
- 17.Shoeman R.L., Hartig,R., Huang,Y., Grub,S. and Traub,P. (1997) Fluorescence microscopic comparison of the binding of phosphodiester and phosphorothioate (antisense) oligodeoxyribonucleotides to subcellular structures, including intermediate filaments, the endoplasmic reticulum, and the nuclear interior. Antisense Nucleic Acid Drug Dev., 7, 291–308. [DOI] [PubMed] [Google Scholar]
- 18.Henry S.P., Novotny,W., Leeds,J., Auletta,C. and Kornbrust,D.J. (1997) Inhibition of coagulation by a phosphorothioate oligonucleotide. Antisense Nucleic Acid Drug Dev., 7, 503–510. [DOI] [PubMed] [Google Scholar]
- 19.Henry S.P., Templin,M.V., Gillett,N., Rojko,J. and Levin,A.A. (1999) Correlation of toxicity and pharmacokinetic properties of a phosphorothioate oligonucleotide designed to inhibit ICAM-1. Toxicol. Pathol., 27, 95–100. [DOI] [PubMed] [Google Scholar]
- 20.Shaw D.R., Rustagi,P.K., Kandimalla,E.R., Manning,A.N., Jiang,Z. and Agrawal,S. (1997) Effects of synthetic oligonucleotides on human complement and coagulation. Biochem. Pharmacol., 53, 1123–1132. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen P.E. (2000) Antisense peptide nucleic acids. Curr. Opin. Mol. Ther., 2, 282–287. [PubMed] [Google Scholar]
- 22.Summerton J. and Weller,D. (1997) Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev., 7, 187–195. [DOI] [PubMed] [Google Scholar]
- 23.Manoharan M. (1999) 2′-carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta, 1489, 117–130. [DOI] [PubMed] [Google Scholar]
- 24.Marquez V.E., Siddiqui,M.A., Ezzitouni,A., Russ,P., Wang,J., Wagner,R.W. and Matteucci,M.D. (1996) Nucleosides with a twist. Can fixed forms of sugar ring pucker influence biological activity in nucleosides and oligonucleotides? J. Med. Chem., 39, 3739–3747. [DOI] [PubMed] [Google Scholar]
- 25.Herdewijn P. (1999) Conformationally restricted carbohydrate-modified nucleic acids and antisense technology. Biochim. Biophys. Acta, 1489, 167–179. [DOI] [PubMed] [Google Scholar]
- 26.Amirkhanov N.V., Pradeepkumar,P.I. and Chattopadhyaya,J. (2002) Kinetic analysis of the RNA cleavage of the conformationally-constrained oxetane-modified antisense-RNA hybrid duplex by RNase H. JCS. Perkin, 2, 976–984. [Google Scholar]
- 27.Pradeepkumar P.I., Amirkhanov,N.V. and Chattopadhyaya,J. (2003) Antisense oligonucleotides with oxetane-constrained cytidine enhances hetroduplex stability, elicits satisfactory RNase H response as well as show improved resistance to both exo and endonucleases. Org. Biomol. Chem., 1, 81–92. [DOI] [PubMed] [Google Scholar]
- 28.Pradeepkumar P.I. and Chattopadhyaya,J. (2001) Oxetane modified antisense oligonucleotides promote RNase H cleavage of the complementary RNA strand in the hybrid duplex as efficiently as the native, and offer improved endonuclease resistance. JCS. Perkin, 2, 2074–2083. [Google Scholar]
- 29.Pradeepkumar P.I., Cheruku,P., Plashkevych,O., Acharya,P., Gohil,S. and Chattopadhyaya,J. (2004) Synthesis, physicochemical and biochemical studies of 1′,2′-oxetane constrained adenosine and guanosine modified oligonucleotides, and their comparison with those of the corresponding cytidine and thymidine analogues. J. Am. Chem. Soc., 126, 11484–11499. [DOI] [PubMed] [Google Scholar]
- 30.Pradeepkumar P.I., Zamaratzki,E., Foldesi,A. and Chattopadhyaya,J. (2000) Transmission of the conformational information in the antisense–RNA hybrid duplex influences the pattern of the RNase H cleavage reaction. Tetrahedon Lett., 41, 8601–8607. [Google Scholar]
- 31.Ossipov D., Zamaratski,E. and Chattopadhyaya,J. (1999) Helv. Chim. Acta, 82, 2186–2200. [Google Scholar]
- 32.Sokol D.L., Zhang,X., Lu,P. and Gewirtz,A.M. (1998) Real time detection of DNA.RNA hybridization in living cells. Proc. Natl Acad. Sci. USA, 95, 11538–11543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gewirtz A.M. and Calabretta,B. (1988) A c-myb antisense oligodeoxynucleotide inhibits normal human hematopoiesis in vitro. Science, 242, 1303–1306. [DOI] [PubMed] [Google Scholar]
- 34.Mucenski M.L., McLain,K., Kier,A.B., Swerdlow,S.H., Schreiner,C.M., Miller,T.A., Pietryga,D.W., Scott,W.J., Jr and Potter,S.S. (1991) A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell, 65, 677–689. [DOI] [PubMed] [Google Scholar]
- 35.Luger S.M., O'Brien,S.G., Ratajczak,J., Ratajczak,M.Z., Mick,R., Stadtmauer,E.A., Nowell,P.C., Goldman,J.M. and Gewirtz,A.M. (2002) Oligodeoxynucleotide-mediated inhibition of c-myb gene expression in autografted bone marrow: a pilot study. Blood, 99, 1150–1158. [DOI] [PubMed] [Google Scholar]
- 36.Braasch D.A. and Corey,D.R. (2001) Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol., 8, 1–7. [DOI] [PubMed] [Google Scholar]
- 37.Orum H. and Wengel,J. (2001) Locked nucleic acids: a promising molecular family for gene-function analysis and antisense drug development. Curr. Opin. Mol. Ther., 3, 239–243. [PubMed] [Google Scholar]
- 38.Agrawal S., Jiang,Z., Zhao,Q., Shaw,D., Cai,Q., Roskey,A., Channavajjala,L., Saxinger,C. and Zhang,R. (1997) Mixed-backbone oligonucleotides as second generation antisense oligonucleotides: in vitro and in vivo studies. Proc. Natl Acad. Sci. USA, 94, 2620–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 40.Tidd D.M. (1992) Methylphosphonodiester/phosphodiester chimeric oligodeoxynucleotides. Biochem. Soc. Trans., 20, 746–749. [DOI] [PubMed] [Google Scholar]
- 41.Bondensgaard K., Petersen,M., Singh,S.K., Rajwanshi,V.K., Kumar,R., Wengel,J. and Jacobsen,J.P. (2000) Structural studies of LNA–RNA duplexes by NMR: conformations and implications for RNase H activity. Chemistry, 6, 2687–2695. [DOI] [PubMed] [Google Scholar]
- 42.Kurreck J., Wyszko,E., Gillen,C. and Erdmann,V.A. (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res., 30, 1911–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uhlmann E., Peyman,A., Ryte,A., Schmidt,A. and Buddecke,E. (2000) Use of minimally modified antisense oligonucleotides for specific inhibition of gene expression. Methods Enzymol., 313, 268–284. [DOI] [PubMed] [Google Scholar]