Abstract
Escherichia coli Dps (DNA-binding proteins from starved cells) is the prototype of a DNA-protecting protein family expressed by bacteria under nutritional and oxidative stress. The role of the lysine-rich and highly mobile Dps N-terminus in DNA protection has been investigated by comparing the self-aggregation and DNA-condensation capacity of wild-type Dps and two N-terminal deletion mutants, DpsΔ8 and DpsΔ18, lacking two or all three lysine residues, respectively. Gel mobility and atomic force microscopy imaging showed that at pH 6.3, both wild type and DpsΔ8 self-aggregate, leading to formation of oligomers of variable size, and condense DNA with formation of large Dps–DNA complexes. Conversely, DpsΔ18 does not self-aggregate and binds DNA without causing condensation. At pH 8.2, DpsΔ8 and DpsΔ18 neither self-aggregate nor cause DNA condensation, a behavior also displayed by wild-type Dps at pH 8.7. Thus, Dps self-aggregation and Dps-driven DNA condensation are parallel phenomena that reflect the properties of the N-terminus. DNA protection against the toxic action of Fe(II) and H2O2 is not affected by the N-terminal deletions either in vitro or in vivo, in accordance with the different structural basis of this property.
INTRODUCTION
Bacteria are faced with constantly changing habitats and, most importantly, with a variety of assaults due to nutrient limitation, high osmolarity, low pH and oxidative stress. Therefore, all bacteria have developed a large number of signal transduction and regulatory mechanisms that enable them to survive the stress conditions in the environment. Among the best-characterized ones are the systems operating in Escherichia coli. Under growth-limiting and potentially lethal conditions, E.coli cells express and accumulate a non-specific dodecameric DNA-binding protein named Dps, (DNA-binding protein from starved cells) (1). In the stationary phase, Dps becomes the most abundant nucleoid component (180 000 molecules per cell) and causes the compaction of DNA characteristic of this growth phase (2,3). Dps protects DNA effectively, both in vitro and in vivo, against oxidative cleavage, nucleases, UV light (4) and thermal shock (5). In E.coli K-12 and O157:H7, Dps also contributes to acid tolerance (6). Both oxidative and acid stress are associated with the production of hydroxyl radicals (OH•) via Fenton-type reactions:
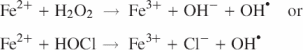
which lead mainly to DNA single and double strand breaks by oxidizing the sugar and base moieties.
Interaction between Dps and DNA, both in the purified state and in starved, stationary phase E.coli cells overexpressing Dps, is reported to result in the rapid formation of stable Dps–DNA co-crystals (7). However, the actual mode of the Dps–DNA interaction has not been fully elucidated. The surface of the Dps dodecamer does not display canonical DNA-binding motifs, and is dominated by negative charges just as the DNA molecule (8). On this basis, Frenkiel-Krispin et al. (9) proposed that Dps dodecamers cannot bind DNA directly and that complex formation is mediated through ion bridges formed by Mg2+ in a particular range of cation concentrations. The electrostatic potential calculations of Grant et al. (8) however, did not consider the contribution of the disordered, lysine-containing N-terminus, not visible in the X-ray structure, which contains three lysine residues in positions 5, 8 and 10. Nevertheless, Grant et al. (8) proposed a model of Dps–DNA interaction in which a major role in complex formation is attributed precisely to the N-terminal lysine residues. Given the remarkably similar packing of Dps dodecamers in protein crystals and of Dps–DNA complexes in electron microscopy preparations, Dps crystallization in vitro and binding of DNA were hypothesized to promote the same type of protein–protein interactions. Thus, in the hexagonal Dps crystal lattice, three adjacent dodecamers define holes that are lined by the lysine-rich N-termini. In the Dps–DNA complex, DNA is assumed to be threaded through such holes and to interact with the lysine residues. In accordance with this hypothesis, those members of the Dps family that do not possess a positively charged N-terminus do not appear to bind DNA based on agarose gel mobility assays. This is the case of the so-called Listeria innocua ferritin (10), Dlp-1 and Dlp-2 from Bacillus anthracis (11), and HP-NAP from Helicobacter pylori (12). Agrobacterium tumefaciens Dps does not alter DNA mobility in spite of the presence of a lysine-containing, albeit shorter than usual, N-terminus. However, this N-terminus was shown to be tightly bound to the protein surface (13).
Irrespective of the occurrence of interaction, Dps proteins afford DNA protection against oxidative damage by inhibiting formation of the toxic hydroxyl radicals. This is achieved by removal of Fe(II) from solution in a multi-step process that comprises rapid binding of Fe(II) at the highly conserved intersubunit ferroxidase center identified in the L.innocua protein (14), Fe(II) oxidation by H2O2, which is reduced to water, and sequestration of Fe(III) within the protein cavity (13,15) where a microcrystalline iron core is formed (16).
In the study (15) that unveiled the H2O2 detoxification mechanism operative in E.coli Dps, the family prototype, self-aggregation of this protein became immediately apparent. Self-aggregation leads to formation of oligomers of variable size, which tend to precipitate as indicated by the onset of turbidity. In fact, at pH values around neutrality, E.coli Dps requires at least 150 mM NaCl to be maintained in solution, whereas Dps proteins that lack the positively charged N-terminus like the L.innocua protein are fully soluble even in the absence of added salt.
In order to inquire on the role of the positively charged N-terminus in DNA condensation and Dps self-aggregation, two deletion mutants were constructed, DpsΔ8 and DpsΔ18. Given the N-terminal sequence of the wt protein (MSTAKLVKSKATNLLYTRND), DpsΔ8 lacks Lys-5 and Lys-8 while DpsΔ18 lacks Lys-5, Lys-8, Lys-10 as well as Arg-18. The DNA-binding and self-aggregation properties of DpsΔ8 and DpsΔ18 were compared to those of the wild-type protein in gel mobility assays and atomic force microscopy (AFM) experiments. In addition, the effect of the N-terminal deletions on the capacity to protect DNA against the radicals resulting from the reaction of Fe(II) and H2O2 was assessed in vitro and in vivo.
MATERIALS AND METHODS
Construction of pet-11a-dps
Plasmid pet-11a-dps contains the E.coli dps gene cloned under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG) inducible promoter of pet-11a (Novagen). The dps gene was amplified by PCR from plasmid pJE106 with primers Dps-f (5′-GAGGATATGAACATATGAGTACCGCT-3′) and Dps-r (5′-AAAGTTCTGGATCCTCAGCG ATGG-3′). The restriction sites for NdeI and BamHI are underlined. The amplified DNA fragment (547 bp) was digested with NdeI and BamHI, purified with the QIAquick PCR purification kit (Qiagen) and cloned into the expression vector pet-11a (Novagen) linearized with NdeI and BamHI. The construct was introduced into E.coli BL21DE3 cells and sequenced by E.coli strain BL21DE3 dideoxy sequencing to confirm the presence of the correct gene.
Construction of Dps deletion mutants Δ8 and Δ18
The dpsΔ18 gene lacking 18 amino acids at the N-terminus was amplified by PCR from the pJE106 plasmid carrying wt dps, using primers Δ18-f (5′-GCTTTATACCCATATGGATGTCTCC) and Dps-r (5′-AAAGTTCTGGATCCTCAGCGATGG). The dpsΔ8 gene lacking eight amino acids at the N-terminus was amplified by PCR from the pJE106 plasmid using primers Δ8-f (5′-CCGCTAAATTACATATGTCAAAAGCG) and Dps-r (5′-AAAGTTCTGGATCCTCAGCGATGG). The restriction sites for NdeI and BamHI are underlined. The amplified fragments for dpsΔ18 and dpsΔ8 (479 and 509 bp, respectively) were digested with NdeI and BamHI, purified with the QIAquick PCR purification kit and cloned into the expression vector pet-11a linearized with NdeI and BamHI. The constructs were introduced into E.coli BL21DE3 cells and sequenced by E.coli strain BL21DE3 dideoxy sequencing to confirm the presence of the correct genes.
Bacterial strains and media
E.coli strain BL21DE3 was transformed with plasmid pet-11a containing the dps wt, the dpsΔ8 or the dpsΔ18 genes. E.coli strain ZK126 dps::kan, knock-out for the dps gene (1), was kindly provided by Dr R. Kolter. Bacteria were grown on Luria–Bertani (LB) liquid medium (10 g/l tryptone, 5 g/l yeast extract, 5 g/l NaCl) or on the minimal medium M63 (0.2% glucose) and LB or M63 plates at 37°C containing 30 μg/ml kanamycin (Sigma Aldrich) or 50 μg/ml ampicillin.
Expression, purification and characterization of E.coli Dps
Dps was expressed and purified as described in Zhao et al. (15). In order to ensure complete removal of Mg2+, the purified protein was dialyzed extensively against 30 mM Tris–HCl buffer, 0.15 M NaCl at pH 7.8, containing Chelex (Bio-Rad). The characterization of the wt protein and of the deletion mutants described below was performed as in Ref. (8) by means of UV–visible spectroscopy, circular dichroism (CD) and analytical ultracentrifugation. Protein concentration was determined spectrophotometrically at 280 nm using a molar extinction coefficient (on a dodecamer basis) of 2.59 × 10−5 M−1 cm−1.
The possible onset of turbidity, reflecting self-aggregation, was assessed by monitoring absorbance at 230–400 nm of protein solutions at concentrations ranging from 0.06 to 2.7 μM in 10 mM Tris–HCl, 20 mM NaCl buffer, pH 7.0 in the presence or absence of 0.4 mM EDTA or in TAE (40 mM Tris-acetate and 2 mM EDTA) and BAE buffers (40 mM BisTris-acetate and 2 mM EDTA) in the pH range 6.3–8.7.
Expression and purification of DpsΔ18
E.coli BL21DE3 cells harboring the recombinant plasmid were grown in 1 l of liquid LB medium containing ampicillin (50 μg/ml) at 37°C to an optical density of 0.6 at 600 nm. The dps gene was induced by addition of 0.5 mM IPTG and the culture was incubated further for 3–4 h.
Cells were harvested (15 000 g for 20 min) and suspended in 10 ml of buffer A (50 mM Tris–HCl at pH 7.5, 0.5 mM DTT, 1 mM EDTA, 500 mM NaCl), and disrupted by sonication. The lysate was centrifuged at 15 000 g for 45 min and the supernatant was precipitated using two ammonium sulfate cuts at 30% and 60% saturation (w/v). At 60%, DpsΔ18 remains in solution after centrifugation (15 000 g for 45 min); the supernatant was dialyzed overnight at 4°C against 20 mM 2-(N-morpholino)ethanesulfonic acid (MES) at pH 6.0 and loaded onto a DEAE cellulose column (DE52) equilibrated with the same buffer. Dps Δ18 was eluted with 300 mM MES at pH 6.0, pooled and stored at −75°C. The purity of the preparation was probed by Coomassie blue staining of SDS–15% PAGE gels.
Expression and purification of DpsΔ8
Expression and purification of DpsΔ8 were performed as described for DpsΔ18 with one modification. The protein, which remains in solution at 60% saturation (w/v) ammonium sulfate, was dialyzed overnight at 4°C against 20 mM MES at pH 6.0. Under these conditions DpsΔ8 precipitates, and can be collected by centrifugation at 15 000 g for 15 min. DpsΔ8 was then resuspended in 30 mM Tris–HCl, 0.2 M NaCl at pH 8.0.
Gel retardation assays
The DNA-binding activity of wt Dps, DpsΔ8 and DpsΔ18 was assessed in gel shift assays using supercoiled pet-11a DNA (5600 bp, 20 nM) or pUC9-5S (3115 bp, 50 nM) as a probe. The plasmids were purified with the Qiaprep spin plasmid miniprep kit or with the Qiaquick Gel Extraction kit (Qiagen, Chatsworth, CA), which ensures removal of impurities and salts. DNA was incubated for 15 min at room temperature with the Dps proteins (3 μM) in TAE or BAE buffers at pH values over the range 6.3–8.7. In order to resolve the complexes, electrophoresis was carried out on 1% agarose gels in the same buffer used for incubation of the Dps–DNA mixture. The gels were stained with ethidium bromide and imaged by ImageMaster VDS (Amersham Biosciences).
In order to determine the stoichiometry of the interaction between wt Dps and DNA in BAE–TAE buffers, a 500 bp dsDNA fragment (10 nM) was used and gel mobility shift assays were performed as described above. The amounts of free DNA were estimated from the band intensities using the LisCap software (Amersham Biosciences) after imaging the gels by ImageMaster VDS.
Dps self-aggregation assays in agarose gels
Dps self-aggregation was monitored by following protein migration in agarose gels. The agarose gel and the buffers used were as those employed in the gel retardation assays described above. The gels were stained with Coomassie blue and imaged by ImageMaster VDS.
DNA protection assays in vitro
DNA protection from oxidative damage was assessed in vitro using pet-11a plasmid DNA (5600 bp, 20 nM). The reaction was carried out in 15 μl of 20 mM Tris–HCl, pH 7.3, containing 0.15 M NaCl. Plasmid DNA was allowed to interact with E.coli wt Dps or DpsΔ18 (3 μM) for 10 min prior to the addition of 50 μM FeSO4. After 2 min, H2O2 was added at a final concentration of 10 mM and the mixtures were incubated for 3 min at room temperature to allow complete consumption of Fe(II) present in solution (15). Thereafter, 2% SDS was added to the reaction mixture, which was incubated at 85°C for 5 min. Plasmid DNA was resolved by electrophoresis on 1% agarose gel in TAE. The gel was stained with ethidium bromide and imaged by ImageMaster VDS.
The protective effect of either E.coli Dps or DpsΔ18 was verified using the fluorimetric assay described by Yamamoto et al. (17) and Ceci et al. (13), which is based on the Fe(II)-mediated production of hydroxyl radicals that degrade deoxyribose to a chromogen. The chromogen fluorescence was used to determine hydroxyl radicals formation in potassium phosphate buffer 10 mM at pH 7.4, NaCl 63 mM, Dps 50–400 nM (data not shown).
Bacterial survival in H2O2 gradients
E.coli strain ZK126 dps::kan lacking the dps gene, E.coli BL21DE3 transformed with plasmid pet-11a containing the dps wt gene and E.coli BL21DE3 transformed with plasmid pet-11a containing dpsΔ18 were grown on minimal M63 agar plates (0.2% glucose) containing ampicillin or kanamycin and a linear concentration gradient of H2O2 from 0 to 15 mM. Early stationary phase LB cultures (A600nm = 1.2) of the three strains were induced with 0.2 mM IPTG and after 15 min transferred to minimal M63-agar plates (0.2% glucose) containing 0.2 mM IPTG, the H2O2 gradient and the appropriate antibiotic. The plates were incubated at 37°C for 20 h and imaged by ImageMaster VDS. Gradient plate assays were carried out in triplicate. The expression levels of the proteins were assessed by SDS gel electrophoresis and N-terminal sequencing.
Atomic force microscopy
Dps–DNA complexes were prepared by incubating Dps at concentrations in the range 1–20 nM with DNA (1 nM) at 20°C for 5 min, in TAE or BAE at different pH values containing 5 mM NiCl2. Purified plasmid DNA (pUC9-5S) was used in the experiments. The samples were deposited onto freshly cleaved ruby mica (Mica New York, NY), incubated for about 1 min, rinsed with 10 drops of water and dried with a weak stream of nitrogen. AFM images were collected in air with a Nanoscope III microscope (Digital Instruments Inc., Santa Barbara, CA) operating in tapping mode. All operations were carried out at room temperature. Commercial diving board silicon cantilevers (Nanosensor or Olympus) were used. The microscope was equipped with a type E scanner (12 mm × 12 mm). Images (512 × 512 pixels) were collected with a scan size of 2 or 4 μm at a scan rate varying between two and four scan lines per second. Water was purified in a Milli-Q water purification apparatus (Millipore).
To verify the strength of the interaction with the mica surface, aggregates of wt Dps and Dps–DNA complexes at pH 6.3 and 8.0 were deposited onto the mica surface and imaged. Thereafter, the surface was rinsed sequentially and imaged for three times using either water or a 5 mM NiCl2 solution. No significant differences between water- and NiCl2-washed samples were observed.
RESULTS
Characterization of DpsΔ8 and DpsΔ18
Circular dichroism and analytical ultracentrifugation were used to test the conformation and state of association of the DpsΔ8 and DpsΔ18 mutants. No detectable structural changes are produced by the N-terminal deletions.
Dps self-aggregation at different pH values
The self-aggregation phenomenon of wt Dps and its N-terminal mutants was monitored in low ionic strength buffers (30–50 mM) in the pH range 6.3–8.7 by means of agarose gel electrophoresis, turbidity measurements and AFM imaging. Although it is under debate whether such extreme cytoplasmic pH values may be encountered in vivo (18,19), these experiments are still valuable in terms of discovering the roles of particular amino acid residues on various Dps functions.
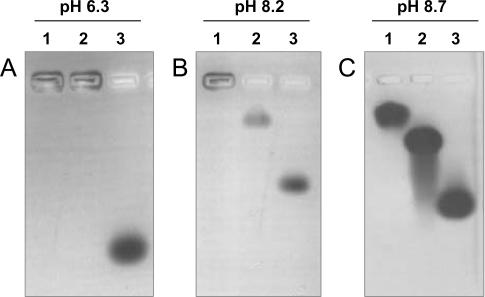
In BAE–TAE buffers at pH values below 8.2, wt Dps precipitates and does not enter the agarose gels, as evidenced by Coomassie blue staining (Figure 1A and B, lane 1). In TAE buffer at pH 8.7, the protein is soluble and migrates into the gel (Figure 1C, lane 1). Precipitation occurring at low pH values is reversible. DpsΔ8 self-aggregates and precipitates over a more limited pH range relative to the wt protein, namely below pH 7.3 (Figure 1A–C, lane 2 and data not shown). DpsΔ18 is fully soluble over the whole pH range studied (Figure 1A–C, lane 3). At any given pH, the increase in mobility follows the order wt Dps < DpsΔ8 < DpsΔ18, an indication that mobility reflects mainly the charge on the molecule.
Figure 1.
Self-aggregation of E.coli Dps at different pH values, monitored by agarose gel electrophoresis. In each panel, lane 1 refers to wt Dps, lane 2 to DpsΔ8, lane 3 to DpsΔ18. Conditions: BAE or TAE buffers, Coomassie blue staining.
Self-aggregation is reflected also by the onset of turbidity. Under conditions where the Dps proteins do not enter the agarose gel, the UV/visible spectrum shows clear evidence of turbidity. In the case of wt Dps at pH 6.3, for example, turbidity can be detected at protein concentrations as low as 60 nM.
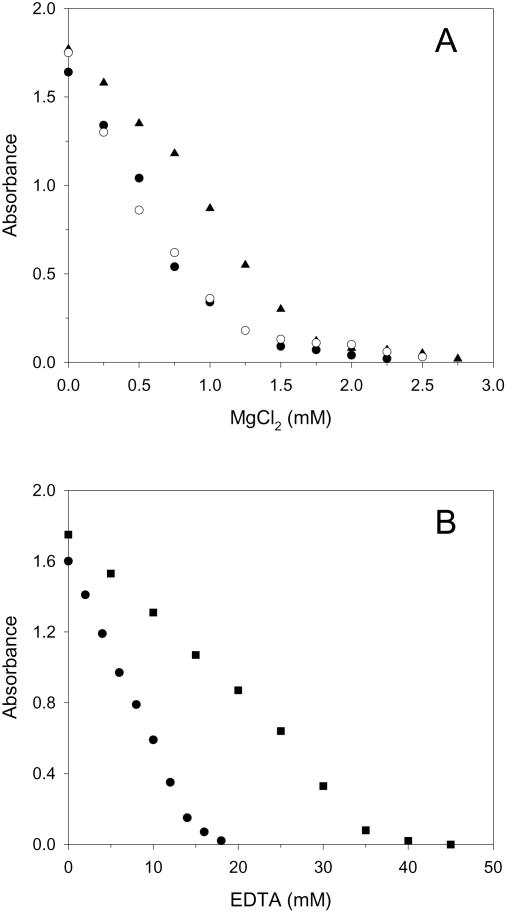
Such turbidity measurements lend themselves to assess the dependence of Dps self-aggregation on the presence of Mg2+ ions and EDTA. Frenkiel-Krispin et al. (9) proposed that Mg2+ ions may remain bound to Dps during its purification and that the presence of such cations within a particular concentration range is crucial for the formation of Dps–DNA co-crystals. Turbidity measurements were carried out under the same conditions used by these authors in their in vitro DNA–Dps co-crystallization experiments, namely 10 mM Tris–HCl, 20 mM NaCl, at pH 7.0. Figure 2A shows that in the absence of added Mg2+, the protein solutions have the same turbidity irrespective of the treatment with Chelex. Upon addition of Mg2+, turbidity decreases with increase in cation concentration and is abolished at about 1.5 mM Mg2+. In the presence of 0.4 mM EDTA, the Mg2+ concentration required to abolish turbidity increases to about 2.0 mM. Importantly, treatment of the protein with Chelex has no effect, indicating that the purification procedure used in the present work effectively removes Mg2+. Figure 2B shows the effect of EDTA additions to 10 mM Tris–HCl, 20 mM NaCl, at two different pH values, 7.0 and 7.8. The reduction of turbidity promoted by EDTA is indicative of a pH-dependent interaction with the protein. In particular, 18 and 40 mM EDTA are required to clarify the protein solutions at pH 7.8 and 7.0, respectively.
Figure 2.
Turbidity of wt Dps solutions as a function of MgCl2 (A) and EDTA (B) concentration. Turbidity, expressed in arbitrary units, was measured at 400 nm. Conditions: protein concentration 1 μM. In (A), purified Dps (open circles) and Chelex-treated Dps in 10 mM Tris–HCl, 20 mM NaCl, pH 7.0 (black circles); Chelex-treated Dps in the same buffer containing 0.4 mM EDTA (black triangles). In (B), Chelex-treated Dps in 10 mM Tris–HCl, 20 mM NaCl at pH 7.0 (black squares) and pH 7.8 (black circles).
To visualize Dps self-aggregation, wt protein and mutants in BAE–TAE buffers covering the pH range 6.3–8.7 were deposited onto freshly cleaved mica. For imaging purposes, protein concentration in AFM experiments was usually 10 nM, roughly one and two orders of magnitude lower than in turbidity and gel mobility measurements, respectively. At this low protein concentration, wt Dps appears as individual dodecamers and small aggregates at pH 6.3; at higher pH no aggregates were observed (see below). Neither Dps mutant exhibits aggregates at any pH value. This finding is of interest in comparison with the results obtained in the presence of DNA under similar conditions.
Dps-dependent condensation of DNA at different pH values
To compare wt and mutant Dps in their ability to interact with DNA, agarose gel mobility shift assays were performed using supercoiled pet-11a DNA as a probe. Reaction between 20 nM DNA and the Dps proteins in the concentration range 1–5 μM was allowed to proceed in BAE–TAE buffers, pH 6.3–8.7. As shown in Figure 3A, interaction of either wt Dps or DpsΔ8 with DNA at pH 6.3 generates complexes too large to migrate into the agarose gel. Conversely, in the presence of DpsΔ18 migration of DNA occurs. However, the significant, albeit very small, decrease in mobility of the fastest moving band and the blurring of the slower moving ones point to a weak interaction of the protein with DNA. At pH 7.7–8.2 (Figure 3B and C), the wt protein forms complexes with DNA that do not enter the agarose gel. Over the same pH range, DpsΔ18 does not affect DNA mobility, whereas DpsΔ8 alters slightly the pattern of DNA bands at pH 7.7, but not at pH 8.2. When pH is increased to 8.7 (Figure 3D), none of the Dps proteins interferes with DNA mobility. Using plasmid pUC9-5S (3115 bp) or a linear 500 bp dsDNA as probes, or increasing Dps concentration to 30 μM while keeping the DNA concentration constant, did not change the results of the experiments (data not shown).
Figure 3.
DNA gel retardation assays at different pH values. In panels (A–D), lane 1 refers to plasmid DNA with wt Dps, lane 2 to plasmid DNA with DpsΔ8, lane 3 to plasmid DNA with DpsΔ18, lane 4 to plasmid DNA alone. In panel (E), lane 1 refers to plasmid DNA, lanes 2–4 to plasmid DNA with Chelex-treated wt Dps in the presence of 10, 7.5, 3.75 mM MgCl2, respectively, lane 5 to plasmid DNA with Chelex-treated wt Dps, lane 6 to plasmid DNA with purified wt Dps. Conditions: in panels A–D, BAE or TAE buffers; in panel E, 10 mM Tris–HCl, 20 mM NaCl at pH 7.0; ethidium bromide staining.
The influence of MgCl2 on Dps–DNA complex formation was explored under the same conditions employed for turbidity measurements, namely in 10 mM Tris–HCl buffer, 20 mM NaCl, pH 7.0, using 1 μM wt Dps and 50 nM pUC9-5S (Figure 3E). When incubation is carried out in the absence of MgCl2, irrespective of the treatment with Chelex to remove divalent cations, all wt Dps forms large complexes with DNA that do not enter the agarose gel. When incubation is carried out in the presence of 3.75 mM MgCl2, the gel mobility shift assays indicate the presence of large complexes together with unbound DNA. Dps–DNA interaction is abolished at 7.5 mM MgCl2, in agreement with the observations of Frenkiel-Krispin et al. (9). Parallel experiments showed that neither NaCl (40–100 mM) nor NiCl2 (up to 20 mM) affect complex formation between wt Dps and DNA. The effect of NiCl2 was analyzed because this salt was used to deposit DNA onto the mica surface in AFM experiments (see below). In addition, Mg2+ in the concentration range 0–10 mM does not promote DpsΔ18–DNA interaction, in contrast with the hypothesis that the doubly charged cation is involved in stabilization of the protein–nucleic acid complex (6).
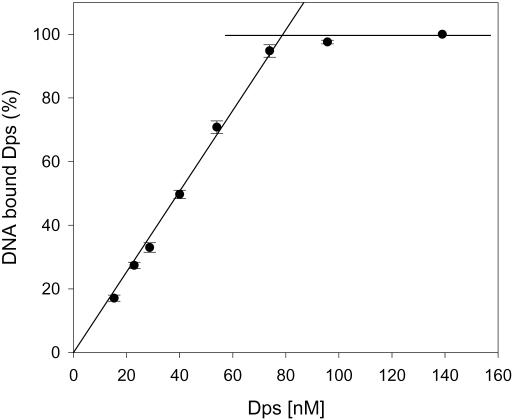
In a different set of gel retardation experiments that aimed at determining the average number of wt Dps molecules involved in DNA-containing complexes, 10 nM dsDNA (500 bp) was titrated with protein in the pH range 6.3–8.7. At pH 6.3, DNA is saturated by 80 nM Dps; the corresponding Dps:DNA molar ratio of 8:1 indicates that about one Dps dodecamer is bound per 60 bp of DNA (Figure 4). At higher pH values, the affinity and apparent binding stoichiometry progressively decrease (data not shown).
Figure 4.
Determination of the E.coli wt Dps–DNA binding stoichiometry at pH 6.3. The data are presented as the mean (±SD) of three independent experiments using 10 nM DNA (500 bp). Saturation of 10 nm DNA is achieved upon addition of 80 nM Dps, i.e. at a molar ratio of 8:1, which corresponds to binding of 1 Dps dodecamer per 62 DNA bp (on average).
To visualize the Dps–DNA interaction by AFM imaging, wt and mutant Dps, at the concentration of 10 nM, were mixed with 1 nM plasmid DNA (pUC9-5S) at different pH values in the range 6.3–8.0. The mixtures were incubated for a few minutes at room temperature and deposited onto freshly cleaved ruby mica in the presence of 5 mM NiCl2.
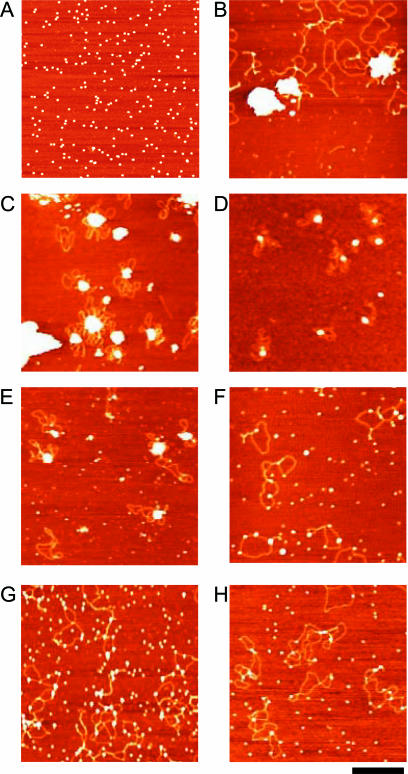
The wt protein alone appears as single dodecamers well spread over the mica surface at pH 7.6 (Figure 5A) even though some aggregates are formed at pH 6.3 (data not shown). In contrast, in the presence of DNA, all dodecamers are sequestered into large Dps–DNA condensates, an indication of strong macromolecular interactions. These large Dps–DNA condensates appear to contain one or more plasmid molecules looping out from a central Dps aggregate; their size decreases dramatically upon rising pH. In some cases, the Dps–DNA complexes are too large to permit visualization of the DNA molecules involved (Figure 5B–D). When the amount of protein is decreased 2- to 3-fold, numerous DNA molecules appear to be protein-free, but the average size of the protein–DNA condensates remains essentially the same (data not shown).
Figure 5.
AFM images of E.coli Dps–DNA complexes. (A) Wt Dps in TAE at pH 7.6. (B) Wt Dps and plasmid DNA in TAE at pH 7.6. (C) Wt Dps and plasmid DNA in BAE at pH 6.3. (D) Wt Dps and plasmid DNA in TAE at pH 8.0. (E) DpsΔ8 and plasmid DNA in BAE at pH 6.3. (F) DpsΔ8 and plasmid DNA in TAE at pH 7.8. (G) DpsΔ18 and plasmid DNA in BAE at pH 6.3. (H) Wt Dps and plasmid DNA in TAE at pH 8.0 in the presence of 5 mM MgCl2. The bar represents 400 nm. Dps dodecamers and DNA in (F) and (G) appear enlarged with respect to (C–E) and (H) because of a larger tip broadening effect.
The N-terminal deletion mutant DpsΔ8 forms aggregates at pH 6.3 (Figure 5E) whereas it binds to DNA with no condensation at pH 7.8 (Figure 5F), in agreement with the gel mobility data. Notably, at pH 6.3 DpsΔ8 aggregation is not complete as shown by the numerous single dodecamers spread over the mica surface, an indication of a weaker macromolecular interactions. DpsΔ18 does not give rise to condensates when mixed with DNA even at pH 6.3 (Figure 5G). Despite the absence of DNA condensation, Dps molecules are bound to the DNA plasmid randomly and appear as ‘beads on a string’. The DNA contour length of Dps-bound plasmids in Figure 5F–H is comparable with that of the protein-free plasmids indicating that dodecamer binding to DNA does not entail wrapping and other modes of DNA compaction.
The influence of MgCl2 on the wt Dps–DNA interaction was also explored. In accordance with the gel shift experiments, no Dps–DNA aggregates are obtained when 5 mM MgCl2 is added to the reaction buffer TAE, pH 8.0 (Figure 5H). Likewise, rinsing the surface with a 10 mM MgCl2 solution results in an almost complete disaggregation of the Dps–DNA complexes.
In vitro DNA protection against hydroxyl radical formation by wt Dps and DpsΔ18
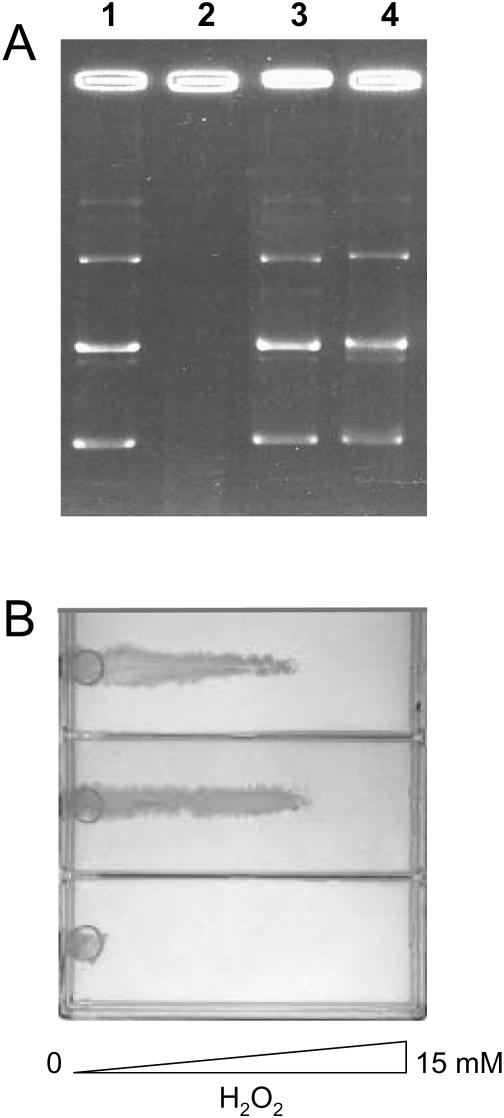
To establish whether the N-terminal deletion mutant DpsΔ18 is able to protect DNA from oxidative damage under conditions where no binding is observed, in vitro DNA damage assays were performed. E.coli wt Dps was used as a control. The combined effect of 50 μM Fe(II) and 10 mM H2O2 on the integrity of plasmid pet-11a was assessed in the absence and presence of the two Dps proteins in 20 mM Tris–HCl, 100 mM NaCl, pH 7.5. Both E.coli wt Dps and DpsΔ18 afford complete protection whereas DNA is fully degraded in the absence of protein (Figure 6A).
Figure 6.
DNA protection assay in vitro (A) and E.coli resistance to H2O2 challenge (B). In panel (A), lane 1 refers to plasmid DNA, lane 2 to plasmid DNA exposed first to 50 μM FeSO4 and then to 10 mM H2O2, lane 3 to plasmid DNA plus 3 μM E.coli Dps exposed first to 50 μM FeSO4 and then to 10 mM H2O2, lane 4 to plasmid DNA plus 3 μM DpsΔ18 exposed first to 50 μM FeSO4 and then to 10 mM H2O2. Panel (B) shows the survival of E.coli BL21DE3 harboring the dpsΔ18 gene (top) in comparison with that of E.coli BL21DE3 harboring the dps wt gene (middle) and that of E.coli strain ZK126 dps::kan, knock-out for the dps gene (bottom), grown on minimal M63 (0.2% glucose) plates containing a linear concentration gradient of H2O2.
The same conclusion was reached in a different type of experiment, in which iron-dependent hydroxyl radical formation was assessed using a fluorimetric assay (13,17). The result of this experiment shows that E.coli Dps and DpsΔ18 inhibit hydroxyl radical formation with a similar efficiency. The extent of inhibition varies from 55% to 70% when the Fe(II) to Dps molar ratio decreases from 420 to 52 (data not shown).
In vivo DNA protection against hydroxyl radical formation by wt Dps and DpsΔ18
The contribution of DpsΔ18 and wt Dps to bacterial survival under H2O2 challenge was assessed in vivo by comparing the growth of the relevant strains to that of the Dps knockout strain ZK126-dps::kan monitoring bacterial growth in a H2O2 linear gradient between 0 and 15 mM. As shown in Figure 6B, the Dps knockout E.coli strain ZK126 dps::kan stops growing at H2O2 concentrations above 2 mM. In contrast, E.coli carrying the pet11a-Dps or the pet11a-DpsΔ18 plasmids grows up to 10 mM H2O2. No significant difference was observed in the protection efficiency of the DpsΔ18 mutant and wild-type proteins.
DISCUSSION
The structural determinants underlying the ability of E.coli Dps to bind and condense DNA are still elusive though all Dps proteins are named after this distinctive property of the family prototype that gives rise to large microcrystalline Dps–DNA complexes in starved E.coli cells (7). The present study shows that formation of such complexes is tightly linked to the capacity of E.coli Dps to self-aggregate and that both properties are correlated with the presence of the lysine-rich N-terminus.
Self-aggregation in solution is an unrecognized feature of E.coli Dps despite the fact that polymerization is apparent from the immediate onset of turbidity when protein solutions are brought to pH values around and below neutrality in buffers of ionic strength 30–50 mM. Formation of large aggregates is manifest also in the electrophoretic behavior of the wt protein, which is unable to enter the agarose gels below pH 8.2. In turn, the deletion mutants DpsΔ8 and DpsΔ18, lacking, respectively, two or all three N-terminus lysine residues, exhibit a behavior different from that of the wt protein as a function of pH. Thus, self-aggregation of the DpsΔ8 mutant is apparent only below pH 7.3, whereas the DpsΔ18 mutant does not self-aggregates even at pH 6.3, the lowest pH investigated (Figure 1). It follows that the cornerstone of the protein–protein interaction lies in the N-terminus, where the only titrating group(s) are the lysine residues in positions 5, 8 and 10. Given the pH range favoring self-aggregation, the pK values of these lysine residues must be anomalously low. Interestingly, the 23-symmetry (tetrahedral) of the Dps dodecamer brings the N-terminus at the edge of the 2-fold interfaces such that Lys-10, visible in one of the subunits, lies near the Arg-55 and His-112 residues of the symmetry related monomer (8). Thus, this positive patch, in the surrounding of the N-terminus lysine side chains, should lead to a significant decrease of the pK value. We propose that Dps self-aggregation is governed by the protonation state of the N-terminus lysines. When protonated, the N-terminus lysines are envisaged to interact with negatively charged carboxyl groups at the surface of neighboring Dps molecules. Though increased by this interaction, the pK values of the N-terminus lysines remain close to the value of physiological pH as indicated by the pH-dependence of self-aggregation (Table 1). This situation is not unprecedented. In cytochrome c, for example, lysine residues with pK values <7.0 are located within the positively charged recognition patch, which is involved in the interaction with physiological partners (20).
Table 1. Self-aggregation and DNA condensation of E.coli Dps and its deletion mutants DpsΔ8 and DpsΔ18 as a function of pH assessed in gel retardation and AFM experiments.
| pH | Dps | DpsΔ8 | DpsΔ18 | |||
|---|---|---|---|---|---|---|
| Self-aggregationa | DNA condensationb | Self-aggregationa | DNA condensationb | Self-aggregationa | DNA condensationb | |
| 6.3 | Yes | Yes | Yes | Yes | No | Noc |
| 7.3 | Yes | Yes | Yes | Yes | No | No |
| 7.7 | Yes | Yes | No | Noc | No | No |
| 8.2 | Yes | Yes | No | No | No | No |
| 8.7 | No | No | No | No | No | No |
aDps self-aggregation is defined as the capacity of dodecamers to form oligomers of variable size. The oligomers reach dimensions up to hundreds of nanometers as indicated by AFM imaging. They remain confined to the electrophoresis well and no migration is observed.
bDps-driven DNA condensation is defined as the capacity of the protein to form large Dps–DNA complexes containing many Dps molecules and one or more DNA plasmids. In the AFM images, the complexes appear as large protein aggregates surrounded by DNA loops from the condensed plasmids. The Dps–DNA condensates remain confined into the well during electrophoresis.
cDNA condensation as defined above is not observed under these specific conditions. However, the AFM images show that single Dps dodecamers bind to DNA as ‘beads on a string’. Migration of these small Dps–DNA complexes in agarose gels is retarded relative to free plasmid DNA.
When interaction with DNA is monitored by means of gel electrophoresis under the conditions used to follow Dps self-aggregation, it becomes apparent that whenever Dps proteins do not migrate in agarose gels, large complexes with DNA are formed that likewise do not enter agarose gels (Figures 1, 3 and Table 1). The DpsΔ8 and DpsΔ18 mutants, at pH values around 7.7–7.8 and 6.3, respectively, do not self-aggregate although interaction with DNA manifests itself in slight, albeit significant, changes in DNA mobility (Figure 3) indicative of DNA binding with no condensation.
The picture which emerges is that both Dps self-aggregation and condensation of DNA are processes correlated with the nature of the N-terminus. More specifically, the number of the N-terminus lysine residues appears to determine whether interaction with DNA will lead to formation of large complexes or not. The AFM images obtained with the DpsΔ8 mutant provide decisive information in this respect as they allow one to visualize protein binding to DNA distinctly from condensation. Either one of the two situations can be made visible, in full agreement with the gel electrophoresis experiments, by operating at the appropriate pH value. Thus, at pH values around 7.7–7.8 all plasmids contain few DpsΔ8 molecules bound as single dodecamers and resemble free plasmids in their morphology (Figure 5F), whereas at pH 6.3 condensation takes place and DNA appears bundled in folded loops in the periphery of large protein aggregates (Figure 5E). Similar DNA condensates are produced by the wt protein at all pH values investigated (Figure 5C and D). In contrast, the DpsΔ18 mutant binds DNA, albeit without formation of condensates, only at pH 6.3, the lowest pH value investigated (Figure 5G), in agreement with the gel electrophoresis data. The charge on the N-terminus therefore determines not only whether self-aggregation will occur, but also the mode of interaction with DNA. Self-aggregation and DNA condensation both require full protonation of at least one lysine residue (Lys-10) as shown by the DpsΔ8 mutant, whereas DNA binding can take place in the absence of the lysine-containing N-terminus as in DpsΔ18, at least at slightly acid pH values. This is probably due to the presence of other residues on the protein surface that ionize in this pH range (e.g. His-112 and His-117).
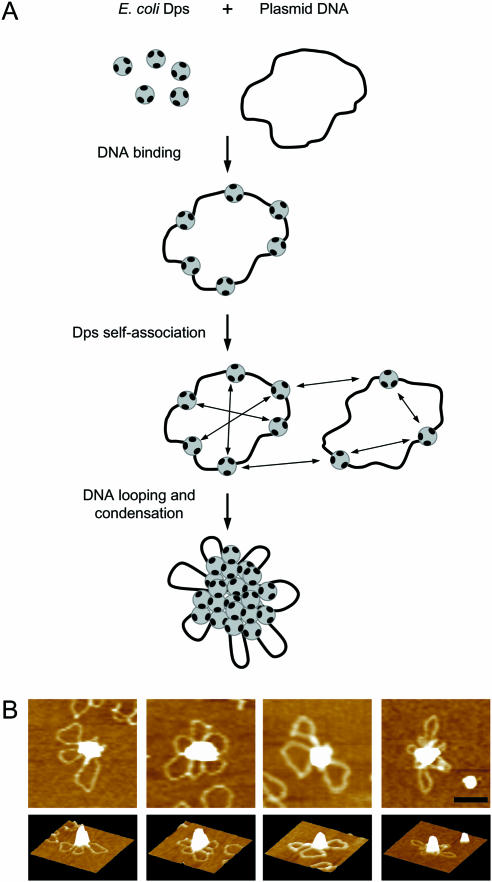
The positively charged N-termini have a regular disposition on the protein surface, which is dictated by the highly symmetrical assembly of the dodecamer. It follows that individual Dps dodecamers can interact simultaneously with other Dps and/or DNA molecules. The sequence of the events leading to DNA condensation is schematized in Figure 7A. Dps dodecamers bind DNA at random with no sequence specificity. Thereby the plasmid-bound Dps molecules become confined in space. The consequent increase in local protein concentration favors the mutual interaction between Dps molecules bound on the same or on neighboring DNA plasmids inducing DNA looping and condensation. In turn, protein–protein interactions between such relatively small Dps–DNA multimeric complexes (Figure 7B), can foster the formation of larger structures in a strongly cooperative fashion. A similar two-step mechanism has been proposed for other DNA-condensing proteins e.g. RIP60 (21). It may be argued that the highly symmetrical disposition of the N-termini on the surface of the Dps dodecamer lends itself to the formation of ordered structures such as those observed in the nucleoid of starved E.coli cells (3,9).
Figure 7.
DNA condensation driven by E.coli Dps self-aggregation. (A) Proposed model. Dps dodecamers are depicted as gray circles with dark patches representing the positively charged N-terminal tail of the monomers. For simplicity, the patches are drawn without correlation to the number and symmetry of the N-termini within the Dps dodecamer. Wt-Dps, DpsΔ8 and DpsΔ18 dodecamers bind DNA in a non-specific manner by interaction with the negatively charged phosphate groups. The binding of several Dps dodecamers to the same DNA plasmid confines the proteins in space. Thereby the local protein concentration is increased and mutual interactions between Dps molecules bound to the same plasmid or to closely spaced ones can take place. The different behavior of the wt and mutant proteins and the pH dependence of the DNA condensation process are determined by the number of lysine residues in the N-terminus and their pK values. In wt Dps, the interaction capacity of the N-terminus is at its highest due to the presence of three lysine residues. Thus, interaction with DNA is driven towards condensation even at pH values where the lysine residues are not fully protonated, e.g. at pH 8.0. In the DpsΔ8 mutant, the N-terminus has a lower interaction capacity since it carries only Lys-10. Thus, interaction with DNA is driven towards condensation only at pH values where full protonation of this lysine residue occurs, i.e. at pH < 7.7. At higher pH values, interaction is either limited to the binding step (i.e. over the range pH 7.7–8.0) or does not take place at all (e.g. at pH 8.2) due to the progressive deprotonation of Lys-10. The latter scenario applies also to DpsΔ18, which lacks the N-terminus lysines; the limited binding observed at pH 6.3 may be ascribed to other positively charged residues (e.g. histidines) on the protein surface. (B) AFM images of selected wt Dps–DNA condensates. Upper panels show top views of the condensates; lower panels show the corresponding lateral three-dimensional views. The bar represents 100 nm; the heights of the aggregates from left to right are 11.5, 10.3, 6.3 and 6.8 nm.
Due to their common structural basis, Dps self-aggregation and DNA condensation display a similar dependence on pH and on the presence of salts or small charged molecules capable of binding to the macromolecular partners. The latter aspect is of special relevance in view of the suggestion by Frenkiel-Krispin et al. (9) that the Dps dodecamer cannot interact directly with DNA, but only through bridges formed by Mg2+ within a particular concentration range. This conclusion was based on in vitro Dps–DNA co-crystallization experiments conducted in the presence of MgCl2 and EDTA at different concentrations. However, both MgCl2 and EDTA interact with Dps, as shown clearly by experiments in which the two molecules are studied separately under conditions of controlled ionic composition, and the interaction abolishes Dps self-aggregation and DNA condensation at appropriate concentrations (Figures 2 and 3E). In addition, given the parallelism between the two processes, Mg2+ should be able to promote self-aggregation yet this is not observed: neither in the wt protein nor in the DpsΔ18 mutant. A full analysis of the complex interplay between pH, Mg2+ and EDTA binding delineated by these results is beyond the scope of the present work.
A final important point concerns the effect of the N-terminal deletion on the ability of Dps to protect DNA from the reactive oxygen species generated during aerobic metabolism, e.g. the hydroxyl radicals produced from H2O2 and Fe(II) through Fenton chemistry. The N-terminal deletion mutant DpsΔ18, which is unable to bind and condense DNA at physiological pH values, exerts a protective effect comparable to that of the wt protein not only in vitro (Figure 6A), but also in vivo (Figure 6B). Thus, the bacterial cells expressing wt Dps or the DpsΔ18 mutant grow similarly and up to higher H2O2 levels than the strain that lacks the dps gene (∼10 mM versus ∼1 mM H2O2). This result is in accordance with the knowledge that protection of DNA against the toxic action of H2O2 depends on the capacity of Dps to sequester Fe(II) and convert it to Fe(III) using H2O2 as an oxidant (13,15). It is also in keeping with recent studies that assign to Dps an important role in the protection of several bacterial pathogens, like Salmonella enterica (22), Helicobacter pylori (12,23), Campylobacter jejuni (24), Porphyromonas gingivalis (25), Streptococcus suis (26), from H2O2 produced by the host phagocytes during infection. Among these pathogens only S.enterica Dps possesses a lysine-rich N-terminus (22).
In conclusion, the recognition that Dps self-aggregation is the driving force in the formation of large Dps–DNA complexes represents yet another proof that protein–protein interactions play a major role in nucleoid structuring. In terms of the dual mechanism of DNA protection, the present data confirm that the highly conserved capacity of Dps proteins to oxidize and uptake iron provides a universal protection mechanism towards oxidative DNA damage. The physical protection afforded by Dps–DNA condensation appears to be restricted to those species whose Dps proteins possess a lysine-containing, freely mobile N-terminus. It may be hypothesized that the latter mechanism applies to DNA protection against detrimental factors other than hydroxyl radicals like nucleases, UV and γ radiation, high temperature and chemical mutagens. Interestingly, E.coli Dps has a higher affinity for DNA at acidic pH values that mimic the stress conditions typically encountered by enterobacteria during host-infection, prolonged periods of starvation and acid stress challenges.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Ministero Istruzione Università e Ricerca ‘Biologia strutturale e dinamica di proteine redox’ (to G.L.R and E.C.), ‘Centro di Eccellenza Biologia e Medicina Molecolare’ (to E.C.) and ‘FIRB 2001’ (to C.R.).
REFERENCES
- 1.Almiron M., Link,A.J., Deirdre,F. and Kolter,R. (1992) A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev., 6, 2646–2654. [DOI] [PubMed] [Google Scholar]
- 2.Azam T.A., Iwata,A., Nishimura,A., Ueda,S. and Ishihama,A. (1999) Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol., 181, 6361–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim J., Yoshimura,S.H., Hizume,K., Ohniwa,R,L., Ishihama,A. and Takeyasu,K. (2004) Fundamental structural units of the Escherichia coli nucleoid revealed by atomic force microscopy. Nucleic Acids Res., 32, 1982–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martinez A. and Kolter,R. (1997) Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol., 179, 5188–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nair S. and Finkel,S.E. (2004) Dps protects cells against multiple stresses during stationary phase. J. Bacteriol., 186, 4192–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi S.H., Baumler,D.J. and Kaspar,C.W. (2000) Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl. Environ. Microbiol., 66, 3911–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf S.G., Frenkiel,D., Arad,T., Finkel,S.E., Kolter,R. and Minsky,A. (1999) DNA protection by stress-induced biocrystallization. Nature, 400, 83–85. [DOI] [PubMed] [Google Scholar]
- 8.Grant R.A., Filman,D.J., Finkel,S.E., Kolter,R. and Hogle,J.M. (1998) The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nature Struct. Biol., 5, 294–303. [DOI] [PubMed] [Google Scholar]
- 9.Frenkiel-Krispin D., Levin-Zaidman,S., Shimoni,E., Wolf,S.G., Wachtel,E.J., Arad,T., Finkel,S.E., Kolter,R. and Minsky,A. (2001) Regulated phase transitions of bacterial chromatin: a non-enzymatic pathway for generic DNA protection. EMBO J., 20, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bozzi M., Mignogna,G., Stefanini,S., Barra,D., Longhi,C., Valenti,P. and Chiancone,E. (1997) A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem., 272, 3259–3265. [DOI] [PubMed] [Google Scholar]
- 11.Papinutto E., Dundon,W.G., Pitulis,N., Battistutta,R., Montecucco,C. and Zanotti,G. (2002) Structure of two iron-binding proteins from Bacillus anthracis. J. Biol. Chem., 277, 15093–15098. [DOI] [PubMed] [Google Scholar]
- 12.Zanotti G., Papinutto,E., Dundon,W.G., Battistutta,R., Seveso,M., Del Giudice,G., Rappuoli,R. and Montecucco,C. (2002) Structure of the neutrophil-activating protein from Helicobacter pylori. J. Mol. Biol., 323, 125–130. [DOI] [PubMed] [Google Scholar]
- 13.Ceci P., Ilari,A., Falvo,E. and Chiancone,E. (2003) The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J. Biol. Chem., 278, 20319–20326. [DOI] [PubMed] [Google Scholar]
- 14.Ilari A., Stefanini,S., Chiancone,E. and Tsernoglou,D. (2000) The dodecameric ferritin from Listeria innocua contains a novel intersubunit iron-binding site. Nature Struct. Biol., 7, 38–43. [DOI] [PubMed] [Google Scholar]
- 15.Zhao G., Ceci,P., Ilari,A., Giangiacomo,L., Laue,T.M., Chiancone,E. and Chasteen,N.D. (2002) Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells. A ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem., 277, 27689–27696. [DOI] [PubMed] [Google Scholar]
- 16.Ilari A., Ceci,P., Ferrari,D., Rossi,G.L. and Chiancone,E. (2002) Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J Biol Chem., 277, 37619–37623. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y., Poole,L.B., Hantgan,R.R. and Kamio,Y. (2002) An iron-binding protein, Dpr, from Streptococcus mutans prevents iron-dependent hydroxyl radical formation in vitro. J. Bacteriol., 184, 2931–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi H., Saito,H. and Kakegawa,T. (2000) Bacterial strategies to inhabit acidic environments. J. Gen. Appl. Microbiol., 46, 235–243. [DOI] [PubMed] [Google Scholar]
- 19.Slonczewski J.L., Rosen,B.P., Alger,J.R. and Macnab,R.M. (1981) pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl Acad. Sci., 78, 6271–6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crnogorac M.M., Ullmann,G.M. and Kostic,N.M. (2001) Effects of pH on protein association: modification of the proton-linkage model and experimental verification of the modified model in the case of cytochrome c and plastocyanin. J. Am. Chem. Soc., 123, 10789–10798. [DOI] [PubMed] [Google Scholar]
- 21.Montigny W.J., Houchens,C.R., Illenye,S., Gilbert,J., Coonrod,E., Chang,Y.C. and Heintz,N.H. (2001) Condensation by DNA looping facilitates transfer of large DNA molecules into mammalian cells. Nucleic Acids Res., 29, 1982–1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halsey T.A., Vazquez-Torres,A., Gravdahl,D.J., Fang,F.C. and Libby,S.J. (2004) The ferritin-like Dps protein is required for Salmonella enterica serovar Typhimurium oxidative stress resistance and virulence. Infect. Immun., 72, 1155–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooksley C., Jenks,P.J., Green,A., Cockayne,A., Logan,R.P. and Hardie,K.R. (2003) NapA protects Helicobacter pylori from oxidative stress damage, and its production is influenced by the ferric uptake regulator. J. Med. Microbiol., 6, 461–469. [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa T., Mizunoe,Y., Kawabata,S., Takade,A., Harada,M., Wai,S.N. and Yoshida,S. (2003) The iron-binding protein Dps confers hydrogen peroxide stress resistance to Campylobacter jejuni. J. Bacteriol., 185, 1010–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ueshima J., Shoji,M., Ratnayake,D.B., Abe,K., Yoshida,S., Yamamoto,K. and Nakayama,K. (2003) Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun., 71, 1170–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pulliainen A.T., Haataja,S., Kahkonen,S. and Finne,J. (2003) Molecular basis of H2O2 resistance mediated by Streptococcal Dpr. Demonstration of the functional involvement of the putative ferroxidase center by site-directed mutagenesis in Streptococcus suis. J. Biol. Chem., 278, 7996–8005. [DOI] [PubMed] [Google Scholar]