Abstract
The abundant nuclear complex termed FACT affects several DNA transactions in a chromatin context, including transcription, replication, and repair. Earlier studies of yeast FACT, which indicated the apparent dispensability of conserved sequences at the N terminus of the FACT subunit Cdc68/Spt16, prompted genetic and biochemical studies reported here that suggest the domain organization for Spt16 and the other FACT subunit Pob3, the yeast homolog of the metazoan SSRP1 protein. Our findings suggest that each FACT subunit is a multidomain protein, and that FACT integrity depends on Pob3 interactions with the Spt16 Mid domain. The conserved Spt16 N-terminal domain (NTD) is shown to be without essential function during normal growth, but becomes important under conditions of replication stress. Genetic interactions suggest that some functions carried out by the Spt16 NTD may be partially redundant within FACT.
INTRODUCTION
FACT (Facilitates Chromatin Transcription) is an abundant complex that functions at the interface between chromatin and several enzymes that affect DNA. FACT has been identified biochemically in metazoans and yeast (Saccharomyces cerevisiae), and the genes encoding FACT subunits are conserved in the eukaryotic lineage (1–4). For yeast-cell viability the subunits of FACT are essential (5,6), almost certainly due to important chromatin-related processes that FACT mediates. These processes include transcription, DNA replication and DNA repair.
FACT (7) was initially identified as an agent that facilitates the passage of the RNA polymerase II (RNAPII) enzyme along a simple nucleosomal DNA template in vitro (2,7). FACT counteracts negative effects of the nucleosomal configuration of DNA on transcription elongation in vitro, most likely through its activities as a histone chaperone involved in the reversible reconfiguration of the nucleosome [reviewed in (8,9)]. FACT binds nucleosomes and H2A•H2B dimers in vitro, an activity correlated with the transcription of nucleosomal DNA (2,4,10,11). FACT also destabilizes interactions between H2A•H2B dimers and (H3•H4)2 tetramers in vitro (11), and may therefore mediate transcription by facilitating H2A•H2B displacement from the nucleosome. Displacement of an H2A•H2B histone dimer facilitates transcription in vitro (12,13), and is also seen upon transcription activity in vivo (14,15). Genetic studies in yeast support a role for FACT in nucleosome reconfiguration (16), and highlight the involvement of FACT in transcription (4,17). In vivo, FACT is distributed along transcribed regions in parallel with RNAPII (18–20) and can be found physically associated with the transcriptionally engaged form of RNAPII, further evidence for a role for FACT in transcription elongation (21). FACT also has chromatin-independent positive effects on transcription in vitro that are exerted at a post-initiation step (22), and FACT interaction with at least one sequence-specific DNA-binding protein is important for transcription in vivo (10).
FACT also has a negative role in vivo, mediating the repressive effects of chromatin on transcription (5,17,19,23–27). This activity of FACT is seen in the chromatin repression that normally prevents transcription when activators are absent or ineffective; FACT mutations can disrupt this repression, perhaps by impairing the reestablishment of proper nucleosome organization after the passage of RNAPII (19,27).
FACT is also involved in DNA replication. Immunodepletion of FACT from Xenopus oocytes compromises in vitro DNA synthesis (3), and mutations affecting yeast FACT show genetic interactions with a pol1/cdc17 mutation affecting DNA polymerase α, the catalytic subunit of the lagging-strand DNA polymerase α complex (6,28). Indeed, yeast FACT can be affinity-purified using immobilized Pol1/Cdc17 protein (6), and copurified with the 4-subunit polymerase α complex (29). Other evidence comes through the use of hydroxyurea (HU), a free-radical scavenger that causes replication stress by inhibiting the enzyme ribonucleotide reductase and limiting the synthesis of deoxyribonucleotides. Sensitivity to HU is conferred by many mutations that affect replication, as well as by mutations that impair the responses to replication stress. Several mutations affecting yeast FACT cause sensitivity to HU (4,28).
A DNA-repair role for FACT is also evident. In human cells, DNA damage caused by UV irradiation is potentiated in part through transcription-activation functions of p53 [reviewed in (30,31)]; one facet of this potentiation is the phosphorylation of serine 392 at the C terminus of p53 (32–34). This phosphorylation can be carried out by a complex of FACT and casein kinase II (CK2), with FACT providing p53 specificity (35,36). Yeast FACT also associates with CK2 (37).
The subunits of FACT, encoded by single genes in the genomes of eukaryotes examined so far, are highly conserved. The smaller FACT subunit is named Pob3 (yeast) or SSRP1 (human). Pob3 and SSRP1 are closely related sequences, but SSRP1 has an additional domain at its C terminus that is not found in Pob3. This domain, a high mobility group (HMG) fold, can bind damaged and distorted DNA (38,39). Pob3 lacks this HMG domain; however, the small HMG protein Nhp6 associates with yeast FACT and facilitates the binding of FACT to nucleosomes and the resulting nucleosome reorganization (4,40,41). A Pob3–Nhp6 fusion protein that mimics the structure of SSRP1 provides all essential Pob3 functions and many of the functions of Nhp6, including the restoration of chromatin repression (40). Thus Pob3 and Nhp6 may comprise a bipartite version of SSRP1. This difference between yeast and metazoan FACT may reflect additional roles for Nhp6 that do not involve FACT.
The larger subunit of FACT has been variously termed Cdc68 or Spt16, for its original discoveries in the yeast system (5,17). Despite end-to-end conservation of the Spt16 protein in a wide variety of eukaryotes, the N-terminal 30% of the yeast Spt16 polypeptide can be deleted without loss of essential functions, as shown by the cdc68-Δ922 (spt16-Δ922) mutation (26). However, chromatin repression is compromised by this mutation, and spt16-Δ922 mutant cells are also temperature sensitive for cell proliferation, indicating that essential functions are compromised at elevated growth temperatures (26).
The apparent dispensability of conserved sequences at the N terminus of FACT subunit Spt16 has prompted genetic and biochemical investigations of the domain organization for yeast FACT. Our data suggest that each FACT subunit is a multidomain protein and that the N-terminal domain (NTD) of Spt16 is indeed dispensable during normal growth, in part through redundant function with the rest of FACT. Previously reported temperature-sensitive mutations in the Spt16 NTD were found to destabilize the entire Spt16 polypeptide, accounting for the temperature sensitivity of mutations in this non-essential domain. The Spt16 NTD does become important under conditions of replication stress, where it mediates adaptation to stress conditions.
MATERIALS AND METHODS
Strains and plasmids
The spt16-Δ(6–435) mutation was identified in strain DE4B-17b (26). Haploid spt16Δ::KanMX strains (42) maintained by spt16 plasmids were derived from diploid strains BM64 (5,24) and W303. Plasmid pJW212, a gift from L. Howe and J. L. Workman, encodes full-length Spt16 with a C-terminal flag epitope; p314-M68 that carries a 3.5 kbp BamHI fragment encoding spt16-Δ(6–435). The plasmid-borne spt16-ΔNTD mutant allele was created from pJW212, after destruction of the SacII polylinker site, by cleavage at the open reading frame (ORF) N-terminal NcoI site in pJW212 and at the downstream BtgI site, followed by blunt-end ligation.
Partial proteolysis
Whole cell extracts were prepared (40) from cells grown to ∼5 × 107 cells/ml in YM1 rich medium, and FACT containing Pob3-(V5+His6) was purified by metal affinity chromatography (40). Size-exclusion chromatography was carried out in 50 mM Tris–HCl, pH 7.4, 100 mM NaCl, 2% glycerol, 1 mM 2-mercaptoethanol, 0.5 mM EDTA as described previously (1). FACT composed of Pob3-(V5+His6), and either Spt16-flag or HA-Spt16 (43) was digested with trypsin, endoprotease Glu-C, or chymotrypsin (all Sigma) at a ratio of 150:1 (w/w) in extraction buffer containing 50 mM NH4 acetate, 50 mM MgCl2 and 100 mM NaCl. For Pob3 analysis, partially purified FACT containing Spt16-flag was digested with trypsin or Glu-C at 25:1 ratio (w/w). Trypsin and chymotrypsin digestions were stopped with a 5-fold excess of soybean trypsin inhibitor (Sigma), while Glu-C digestions were stopped by heat denaturation in SDS loading buffer at 95°C for 10 min.
Purification of FACT to homogeneity
A 3 l overnight culture at 7 × 107 cells/ml was diluted with 0.3 vol fresh YM1 medium and incubated for 6 h; cells were then harvested, washed with 0.9% NaCl, resuspended in 150 ml extraction buffer containing 50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 2 mM 2-mercaptoethanol, 4 mM EDTA, 0.1% Tween-20, and protease inhibitor mix (43), and broken using a French pressure cell (15 000 psi). The lysate was clarified by centrifugation (8000 g, 10 min), and the resulting pellet was re-extracted by sonication in 50 ml extraction buffer made to 0.5 M NaCl and centrifuged again. Pooled extract (∼300 ml) was adjusted to 150 mM NaCl, pH 7.4 and loaded onto a 50 ml DE-52 column, which was washed with 150 ml extraction buffer and eluted with a 120 ml 0.15–0.5 M NaCl gradient at 1.7 ml/min. Fractions containing FACT were pooled (∼24 ml) and loaded (3×) onto a 0.15 ml anti-flag–agarose (Sigma) column, which was washed with extraction buffer and eluted with 0.3 ml extraction buffer containing flag peptide (0.16 mg/ml; Sigma). The final yield of FACT was ∼60 μg, as determined by Coomassie staining.
GST–Pob3 fusion proteins
Plasmids for the production of glutathione S-transferase (GST)–Pob3 fusion proteins were constructed from PCR-amplified fragments of POB3, using Pfx Taq (Invitrogen) and primers containing BamHI and EcoRI restriction enzyme recognition sequences for directional, in-frame cloning into the expression vector pGEX2T (Pharmacia Biotech). Resulting plasmids were transformed into Escherichia coli cells (strain Top10), which were grown at 37°C to an optical density (600) of ∼0.5, at which point expression of GST polypeptides was induced by a 4 h incubation in 37°C in a medium containing 1 mM isopropyl-1-thio-β-D-galactopyranoside (Invitrogen); cells were then pelleted and stored at −20°C. Cell extracts were prepared as described previously (44). Glutathione–agarose beads (Sigma) as a 50% slurry in PBS were added and incubated at 4°C for 1 h with gentle agitation; bead-bound material was then treated as described previously (45). For quantitation, Coomassie-stained samples, resolved electrophoretically on denaturing 10% polyacrylamide gels, were compared to known quantities of BSA. Amounts of 50% GST-bead slurry were adjusted to make bound proteins equimolar for GST-pulldown assays.
Recombinant Spt16 polypeptides
Fragments of the SPT16 gene were PCR-amplified using Pfx Taq (Invitrogen) and primers that contained BamHI and SstI restriction enzyme recognition sequences, for directional cloning into vector pET32-LIC (Novagen). Expression from pET-Spt16 plasmids, encoding Spt16 polypeptide sequences fused N-terminally to thioredoxin, a His6 tag, and an S-tag was induced in E.coli (strain BL21-DE3) as described above. Recombinant proteins were purified under denaturing conditions essentially as described in Novagen's His•Bind Kits Inclusion Body Purification and Resin Chromatography sections. Urea was removed by sequential dialysis against buffer A (44) containing 4 M urea, then 2 M, and finally no urea. Samples were quantified as above, and centrifuged to remove any particulate matter before use.
GST-pulldown assays
Equimolar amounts of GST–Pob3 proteins and purified recombinant Spt16 polypeptides were incubated in a 50% glutathione–agarose bead slurry along with BSA (150 μg/ml) in buffer A containing protease inhibitors, or for high-stringency conditions in buffer B (buffer A made to 1% NP-40 and 0.5% sodium deoxycholate, plus protease inhibitors). Mixtures were rocked gently at 23°C for 1 h and then beads and supernate were separated. Pelleted beads were next washed for 15 min (five times) at 23°C in either PBS with 2% NP-40 and protease inhibitors, or buffer B (for higher stringency), suspended in an equal volume of 2× Laemmli loading buffer and boiled. Half of each solubilized sample and 5% of supernate were assessed by western blotting (43), using S-protein conjugated to HRP (Novagen) to detect recombinant Spt16 polypeptides.
RESULTS
Non-essential Spt16 N-terminal sequences
The cdc68-Δ922 (spt16-Δ922) mutation (Figure 1A) is an in-frame internal deletion that eliminates residues 6–306 from the 1035-residue Spt16 protein. Cells with this mutation are temperature-sensitive for growth and compromised for chromatin repression (26). Derivatives of temperature-sensitive spt16-Δ922 cells were selected that had become temperature-resistant due to the acquisition of a suppressor mutation. One suppressor mutation was found, by segregation analysis, to be genetically linked to the spt16-Δ922 locus, and was subjected to further analysis.
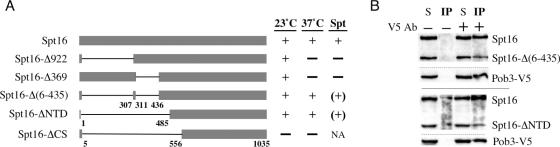
Figure 1.
Spt16 polypeptides. (A) Structures and biological activities of polypeptides deleted for N-terminal sequences. The in-frame deletions in spt16-Δ(6–435) are separated by Spt16 sequence (with one substitution due to codon joining). NA, not assessable. (B) Spt16 polypeptides lacking N-terminal sequences interact effectively with the FACT subunit Pob3. Anti-V5 antibody was used to immunoprecipitate Pob3-(V5+His6) from cells harboring Pob3-(V5+His6), wild-type full-length Spt16, and either Spt16-Δ(6–435) or Spt16-ΔNTD. Spt16 polypeptides and Pob3 were identified in the immunoprecipitate (IP) and supernate (S) fractions by western blotting. Different exposures of the Spt16 blots (15 s for supernates versus 60 s for IPs) are shown to emphasize relative abundances.
Sequencing of the spt16 locus from the spt16-Δ922 isolate containing the linked dominant suppressor mutation showed that the suppressor mutation is an additional in-frame deletion within the spt16-Δ922 ORF, removing 369 nucleotides downstream from the original spt16-Δ922 internal deletion. The resulting polypeptide lacks residues 6–435, with the residues IDPSED in their place (Figure 1A). This truncated Spt16 protein, termed Spt16-Δ(6–435), provides essential functions even at elevated temperatures and thus is more effective that the original Spt16-Δ922 protein, which misses only residues 6–306. Improved function due to the intragenic suppressing deletion in the spt16-Δ(6–435) mutant allele is also seen with respect to chromatin repression of transcription, which is compromised by the spt16-Δ922 mutation (26). Chromatin repression is measured by the degree of functional expression of the ‘Spt’ reporter genes his4-912δ and lys2-128δ; more expression of these reporter genes confers the ability to grow on media lacking histidine and/or lysine (the Spt− phenotype), and is indicative of impaired chromatin repression (46,47). Whereas the spt16-Δ922 mutant allele allows the growth of cells on these deficient media (26), the spt16-Δ(6–435) allele significantly decreases growth under these conditions, and thus restores a significant degree of chromatin repression at these reporter genes (Figure 1A). Moreover, spt16-Δ(6–435) in the homozygous diploid state also supports sporulation (data not shown), which the original spt16-Δ922 allele does not allow (26). Thus the additional internal deletion in spt16-Δ(6–435) suppresses the deleterious effects of the less extensive spt16-Δ922 internal deletion while maintaining effective essential Spt16 function.
The spt16-Δ369 and spt16-Δ(2–484) mutations suggest an NTD
Two more spt16 mutant genes harboring large deletions were created and tested for function. Fragment replacement was used to modify the spt16-Δ(6–435) mutant allele, thus creating the mutant allele spt16-Δ369. This gene lacks the deletion characteristic of spt16-Δ922, but retains the in-frame internal deletion (ORF basepairs 936–1304, encoding amino acids 312–435) that suppresses the phenotype of the spt16-Δ922 internal deletion, with all other sequences intact (Figure 1A). Like the original deletion allele spt16-Δ922, the internal deletion spt16-Δ369 causes temperature sensitivity and allows (weak) growth on His− and Lys− media, indicating that chromatin repression of the Spt reporter genes his4-912δ and lys2-128δ is compromised (Figure 1A). Thus, the combined deletion of two adjacent segments of the Spt16 protein, residues 6–306 and 312–435 [resulting in the Spt16-Δ(6–435) protein described above], has only limited effects under the conditions tested, while the presence of either of the N-terminal polypeptide segment in the absence of the other causes problems related to chromatin regulation (summarized in Figure 1A).
A more extensively deleted mutant allele was created using convenient restriction sites. This allele, spt16-ΔNTD (Figure 1A), lacks codons 2–484 but provides all essential functions of Spt16. Cells in which the Spt16-ΔNTD polypeptide is the only version of Spt16, grow at high temperature and maintain a significant degree of chromatin repression at the his4-912δ and lys2-128δ reporter genes (Figure 1A). These findings suggested that the Spt16-ΔNTD protein, and the Spt16-Δ(6–435) protein described above, maintain effective interactions with the other subunit of yeast FACT, Pob3. This conjecture was tested directly by competitive co-immunoprecipitation experiments. Anti-V5 antibody was used to immunoprecipitate epitope-tagged Pob3-(V5+His6) protein (40) from cells harboring two versions of Spt16 protein, full-length Spt16 and either Spt16-Δ(6–435) or Spt16-ΔNTD. Under these conditions, in which the total amount of Spt16 polypeptide was in excess, the truncated versions of Spt16 co-immunoprecipitated with Pob3, although somewhat less effectively than full-length Spt16 (Figure 1B). This in vivo Pob3 binding by N-terminally truncated Spt16 polypeptides, and their ability to provide essential functions, indicate that sequences downstream from residue 484 fold properly in the absence of upstream sequences, and begin to suggest a domain organization for Spt16. The more extensively deleted Spt16-ΔCS polypeptide [Cdc68-ΔCS, (26)] (Figure 1A) lacking N-terminal residues down to position 556 cannot supply essential function (26), consistent with a domain boundary in the 485–555 region.
Spt16 domains suggested by partial proteolysis of native FACT
In parallel with the above genetic studies, we assessed domain boundaries using limited proteolysis. Proteases with different cleavage specificities, namely trypsin, chymotrypsin and endoprotease Glu-C (V8 protease), were used to digest purified FACT, and the sizes of the resulting fragments were estimated by denaturing gel electrophoresis. FACT composed of epitope-tagged Spt16 and Pob3 proteins was used, which allowed direct identification of N-terminal and/or C-terminal polypeptides.
FACT containing N-terminally HA-tagged Spt16 supplies all essential functions, as does FACT containing C-terminally (V5+His6)-tagged Pob3 (40,43). A version of FACT composed of HA-Spt16 and Pob3-(V5+His6) is similarly functional in vivo (data not shown). This doubly tagged FACT was digested with trypsin for increasing times, proteolysis was halted with soybean trypsin inhibitor, and the digest was analyzed by western blotting. Anti-HA antibody detected a major 55 kDa N-terminal fragment of Spt16 (Figure 2A, NTD). This disjoined N-terminal fragment, the NTD, was stable to incubation with trypsin after all full-length Spt16 protein had been cleaved, indicating that the trypsin-generated NTD maintains a folded state. Glu-C also generated an HA-tagged Spt16 NTD of similar size (data not shown), consistent with preferential proteolytic cleavage in a boundary region defining the 55 kDa NTD (excluding the HA tag) as an independently folded domain.
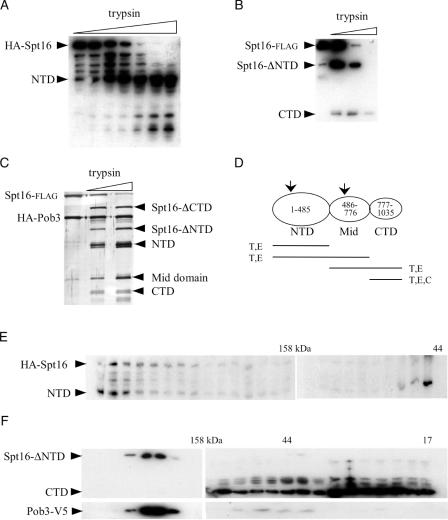
Figure 2.
Domain organization of the Spt16 subunit through FACT partial proteolysis. (A) Purified FACT (1) composed of epitope-tagged HA-Spt16 and Pob3-(V5+His6) was treated with trypsin for increasing times, resolved electrophoretically through a 10% polyacrylamide gel, and analyzed by western blotting using anti-HA antibody. N-terminally HA-tagged Spt16 fragments are indicated. (B) Purified FACT composed of Spt16-flag and Pob3-(V5+His6) was treated with trypsin or chymotrypsin and similarly analyzed using anti-flag antibody. C-terminally flag-tagged Spt16 fragments are indicated. (C) Purified FACT was trypsin-digested, resolved electrophoretically and silver-stained. Band identification was confirmed by western blotting (data not shown). (D) Spt16 domain organization, with likely subdomain boundaries indicated by arrows. Products of trypsin (T), Glu-C (E), and chymotrypsin (C) proteolysis are indicated. (E) Material from trypsin-digested FACT containing HA-Spt16 was resolved by exclusion chromatography, and the resulting fractions were resolved electrophoretically and analyzed by western blotting using anti-HA antibody. (F) Material from trypsin-digested FACT containing Spt16-flag was analyzed as in (E) using antibody against V5 (top) and flag (bottom). In this experiment, trypsin cleaved off the Spt16 NTD from all detectable FACT.
The above trypsin digestion of FACT that contained HA-tagged Spt16 also generated smaller HA-tagged polypeptide fragments, suggesting that the NTD may be folded into subdomains (Figure 2A; see also 2D). This possibility is supported by NTD sequence considerations (see Discussion).
Partial proteolysis also produced HA-tagged polypeptides larger than the NTD, highlighting downstream cleavage sites that suggested the presence of a C-terminal domain (CTD). To assess the presence of an independently folded CTD of Spt16, we used purified FACT that contained Spt16 tagged at its C terminus with the flag epitope. Spt16-flag provides all essential functions (data not shown), even when combined with Pob3-(V5+His6) or with Pob3 HA-tagged at its N terminus [(1); data not shown]. Both of these doubly tagged versions of FACT were separately subjected to limited digestion with trypsin, chymotrypsin, or Glu-C. All three enzymes generated a C-terminal Spt16 fragment of 28 kDa (Figure 2B, and data not shown), indicating that the boundary between this Spt16 CTD and the upstream ‘middle’ domain is highly accessible. Trypsin and Glu-C also produced a major Spt16-flag fragment of 62 kDa, consistent with Spt16 minus its NTD (Figure 2B, Spt16-ΔNTD); Glu-C, but not the other enzymes, also produced a 46 kDa C-terminal fragment (data not shown). These findings suggest that, as a subunit of FACT, Spt16 is folded into three major domains: a 55 kDa NTD, a 28 kDa CTD, and a 34 kDa middle (Mid) domain, which itself may comprise subdomains.
The 62 kDa fragment (Spt16 minus its NTD) generated by trypsin cleavage was analyzed by Edman sequencing, which showed that trypsin cleaved at arginine 485. This residue is at the downstream end of a ∼30-residue span that shows limited sequence conservation. This span is devoid of cleavage sites for chymotrypsin, consistent with the absence of a disjoined NTD fragment in chymotrypsin digests (data not shown). The NTD boundary suggested by partial proteolysis is consistent with the activity of the Spt16-ΔNTD mutant polypeptide created genetically as described above, which comprises the intact Mid and C-terminal domains.
The domain structure of Spt16 suggested above is supported by the trypsin cleavage pattern of FACT purified to homogeneity. The resistance of Pob3 to low levels of trypsin (see below) allowed the identification of silver-stained Spt16 fragments (Figure 2C, lanes 2 and 3). In this analysis the NTD and the combined Mid+CTD (Spt16-ΔNTD) were especially resistant to proteolysis, while the CTD was more labile (Figure 2C, and data not shown). Figure 2D summarizes Spt16 domain organization.
To assess the folded nature of the disjoined Spt16 NTD, affinity-purified FACT containing HA-Spt16 was treated lightly with trypsin to an extent that generated some Spt16 NTD cleavage product but which also left some FACT undigested (Figure 2A). This mixture was then resolved by exclusion chromatography to size-fractionate the NTD, FACT minus the NTD, and undigested FACT, which runs at >200 kDa [Figure 2E; (1)]. Western analysis of the column eluate showed that, while a portion of the NTD fractionated as a monomer, much of the disjoined NTD co-migrated with intact FACT as indicated by undigested, full-length Spt16 (Figure 2E), most likely as a result of the maintenance of NTD non-covalent interactions after trypsin cleavage.
A different result was obtained by similar analysis, using the same buffer conditions, of the Spt16 CTD. Size-fractionation of trypsin-digested FACT that contained Spt16-flag showed that the bulk of the disjoined CTD fractionated away from Pob3 (Figure 2F), indicating little non-covalent association with other domains of FACT.
The results in Figures 1B and 2F suggest that the Pob3 interacts with the Spt16 Mid domain. To learn more about this interaction, the blot in Figure 2F was reprobed with polyclonal anti-Spt16 antibody, which detected an untagged Mid domain co-migating with Pob3 (data not shown). Thus the Spt16 CTD is not necessary for Pob3 binding. This result was confirmed by Pob3 co-immunoprecipitation of tagged Spt16-ΔCTD polypeptide from trypsin-treated FACT, and by the purification of a complex of Pob3 and Mid domain from an extract of cells harboring a plasmid expressing the Mid-domain tagged with the flag epitope and the SV40 NLS (data not shown). These in vivo findings are also supported by the in vitro experiments described below. Pob3 interaction is thus provided mainly by the Spt16 Mid domain.
Pob3 domains suggested by partial proteolysis
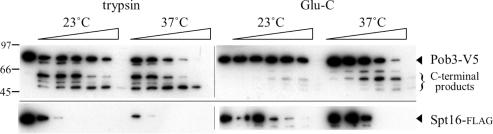
In the context of FACT, Pob3 is more resistant than Spt16 to proteolytic treatments (Figure 2C, and data not shown). However, a higher concentration of trypsin cleaved the Spt16-flag•Pob3-(V5+His6) version of FACT to generate two major C-terminal Pob3 fragments, of apparent sizes 62 and 50 kDa including the tag (Figure 3). Glu-C gave similar Pob3 fragments (Figure 3, and data not shown). [Electrophoretic mobilities of the 551-residue Pob3 protein and tagged derivatives are lower than that predicted by molecular mass; (1)]. Probing the same blots with anti-flag antibody (Figure 3) showed that the appearance of the Pob3-(V5+His6) fragments coincided with extensive cleavage of Spt16, suggesting that protease-sensitive sites in Pob3 are shielded by Spt16 protein. Assignment of Pob3 cleavage sites is complicated by the uncertainty in molecular weight estimates due to the anomalous electrophoretic mobilities noted above, and the instability of Pob3 fragments produced by FACT cleavage. Nonetheless, some insight was provided by the expression of Pob3-(V5+His6) in E.coli cells, which yielded C-terminal fragments due to cleavages at residues 214 and 221 (data not shown). Based on all of these findings, our preliminary prediction is that Pob3 has 23 kDa NTDs and 40 kDa CTDs that are stabilized by association with the Mid domain of Spt16. The resistance of Pob3 to cleavage suggests that this boundary region may be sequestered in FACT.
Figure 3.
Domain organization of the Pob3 subunit through FACT partial proteolysis. Purified FACT composed of Spt16-flag and Pob3-(V5+His6) was treated with Glu-C or trypsin, and samples removed at increasing times were analyzed as in Figure 2, using anti-V5 antibody to detect Pob3 C-terminal polypeptides. Membranes were stripped and reprobed with anti-flag antibody to assess Spt16-flag cleavage.
In vitro dissection of Spt16•Pob3 interactions
FACT subunit interactions were assessed directly using GST-pulldown assays with recombinant Spt16 and Pob3 polypeptides. Several purified recombinant Spt16 polypeptide fragments, each fused N-terminally to a cassette of thioredoxin, His6, and S-tag (THS; Figure 4A), were assayed for binding to GST–Pob3 fusions. Each recombinant THS–Spt16 peptide was mixed with equimolar amounts of GST–Pob3 or GST bound to glutathione–agarose beads, and bound material was assessed by western blotting for S-tagged Spt16 peptides. Results are summarized in Figure 4A.
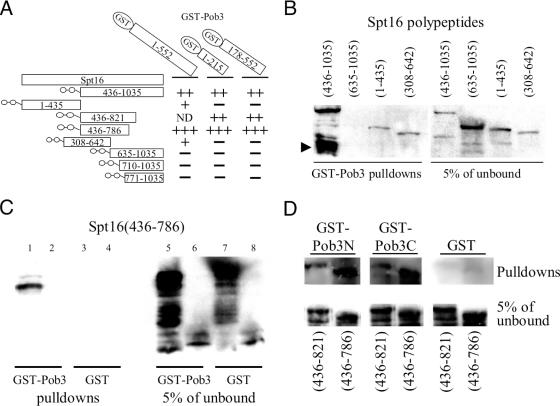
Figure 4.
Spt16–Pob3 interactions in vitro. (A) Schematic of recombinant Spt16 (side) and GST–Pob3 (top) polypeptides used, and the interactions indicated by western analysis of Spt16 polypeptides. The THS at the N terminus of each Spt16 polypeptide is represented by the double circle. ND, not determined. (B) GST–Pob3 pulldowns of S-tagged Spt16 polypeptides. None of the Spt16 polypeptides interacted with GST-bound beads, and the THS moiety itself did not interact with beads carrying GST or GST–Pob3 (data not shown). (C) GST pulldowns of the Spt16(436–786) Mid domain, using high-stringency buffer B. Lanes 1 and 3, S-tagged Spt16(436–786); lanes 2 and 4, the THS moiety; lanes 5–8, input for the experiments of lanes 1–4, respectively. (D) Interactions (buffer B) of the Mid-domains Spt16(436–786) and Spt16(463–821) with GST–Pob3 polypeptides bearing N- or C-terminal residues 1–215 and 178–552, respectively.
As expected, avid binding to GST–Pob3 was seen for THS–Spt16(436–1035) recombinant protein, containing the Mid and C-terminal domains (Figure 4B). Stronger binding to GST–Pob3 was repeatedly found for an ∼45 kDa S-tagged proteolytic fragment of THS–Spt16(436–1035), even though this fragment was not abundant in the input or unbound material (Figure 4B, arrowhead). The presence of the S-tag shows that this fragment contains Mid-domain sequences (436 to ∼700, taking into account the THS tag), indicating the expected strong Pob3 binding by the Mid domain.
The Mid and C-terminal domains of Spt16 were tested separately for Pob3 binding. The CTD polypeptides THS–Spt16(710–1035) and THS–Spt16(771–1035) each failed to associate with GST–Pob3 (data not shown), as expected from the results of partial proteolysis (Figure 2F). The Mid-domain polypeptides THS–Spt16(436–786) and THS–Spt16 (436–821) associated non-specifically with the GST-bound agarose beads (data not shown), but higher-stringency washes indicated specific and strong binding to GST–Pob3 by the Mid domain THS–Spt16(436–786) (Figure 4C). THS–Spt16 (635–1035) and THS–Spt16(308–642), containing Mid-domain fragments, associated far less avidly with GST–Pob3 (Figure 4B), suggesting that the Mid domain needs to be intact for effective Pob3 binding.
The NTD polypeptide THS–Spt16(1–435) bound weakly but reproducibly to GST–Pob3 (Figure 4B), analogously to the binding of the Spt16 NTD to the rest of FACT after partial proteolysis (Figure 2E). Thus the NTD may also be involved in Pob3 binding.
Experiments to determine the Spt16-interacting region(s) of Pob3 used GST fusions mimicking the domains suggested by partial proteolysis. GST–Pob3N, containing Pob3 residues 1–215, and GST–Pob3C, containing Pob3 residues 178–552, were incubated at equimolar amounts with purified recombinant THS–Spt16 peptides. The THS–Spt16(436–1035) Mid+CTD polypeptide bound specifically to both GST–Pob3N and GST–Pob3C polypeptides, and not to the GST-bound beads (data not shown), and at higher stringency the two Spt16 Mid-domain polypeptides interacted specifically with both GST–Pob3N and GST–Pob3C (Figure 4D). Pob3 may therefore have two binding surfaces for Cdc68.
NTD point mutations destabilize the Spt16 polypeptide
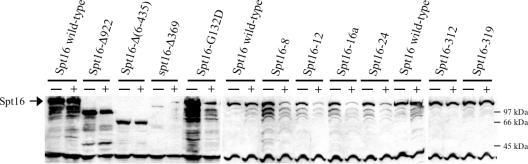
The finding that the Spt16 NTD is dispensable for essential functions appears inconsistent with the presence of point mutations in this domain that compromise essential functions. Several mutant versions of Spt16 with substitutions only in the NTD bring about temperature sensitivity for cell proliferation (4,17). For at least one Spt16 NTD mutation, causing a G132D substitution, temperature sensitivity is accompanied by destabilization of the mutant protein (43). Increased expression of the spt16-G132D mutant allele (cdc68-1), using a multicopy plasmid, alleviates much of this temperature sensitivity (24), consistent with temperature sensitivity due to an abnormally short Spt16 half-life. We therefore tested several other temperature-sensitive spt16 mutant alleles with point mutations affecting the NTD (4). Isogenic strains were created in which four mutant alleles encoding NTD point mutations, spt16-16a (R204W,A273V,C290V,D318N), spt16-8 (G369D,R373T), spt16-24 (T434I), and spt16-12 (A417T plus downstream substitutions), present on centromeric (low-copy) plasmids, supplied only the Spt16 protein. These mutant proteins, plus deleted proteins described here and analogously expressed, were evaluated for half-life as described in (43). Logarithmically growing cells were treated with the protein-synthesis inhibitor cycloheximide to stop the production of new polypeptides, and the relative abundance of pre-existing Spt16 protein was determined by western analysis. In each case the addition of cycloheximide immediately halted cell proliferation (data not shown). With the exception of the Spt16-Δ369 polypeptide, all mutant proteins were relatively stable at 23°C, as indicated by polypeptide abundance in the growing cells (Figure 5). In contrast, every tested protein with an NTD alteration, with the exceptions of Spt16-Δ922 (20) and Spt16-Δ(6-435), showed significant decreases in abundance during high-temperature incubation (Figure 5). The temperature sensitivities displayed by cells with these N-terminal spt16 mutations may therefore be related to the shorter half-lives of the mutant proteins at a restrictive temperature. Consistent with this idea, the spt16-16a, spt16-8, spt16-24 and spt16-12 mutant alleles, like spt16-G132D (24), allow high-temperature growth when present on multicopy plasmids (data not shown).
Figure 5.
Spt16 polypeptide stabilities. Extracts of isogenic spt16Δ derivatives maintained by plasmid-borne spt16 mutant alleles, including several with NTD point mutations (4), were made from cells growing at 23°C (−) or incubated at 39°C for 1 h with the protein-synthesis inhibitor cycloheximide (+), resolved electrophoretically, and assessed by western analysis for Spt16 polypeptide. India ink staining (66) prior to anti-Spt16 antibody binding (data not shown) and the 40 kDa non-specific band verified equal loading. The resected polypeptides Spt16-Δ922, Spt16-Δ(6–435), and Spt16-Δ369 have higher mobilities than full-length Spt16.
Spt16-312 and Spt16-319, two novel temperature-sensitive proteins with substitutions in the Mid and C-terminal domains (A.F. O'Donnell, G.C. Johnston and R.A. Singer, in preparation) had longer half-lives at the restrictive temperature (Figure 5), and increased expression from multicopy plasmids did not alleviate temperature sensitivity (data not shown). Thus spt16 temperature sensitivity is not invariably accompanied by Spt16 polypeptide instability.
The Spt16 NTD influences essential FACT functions
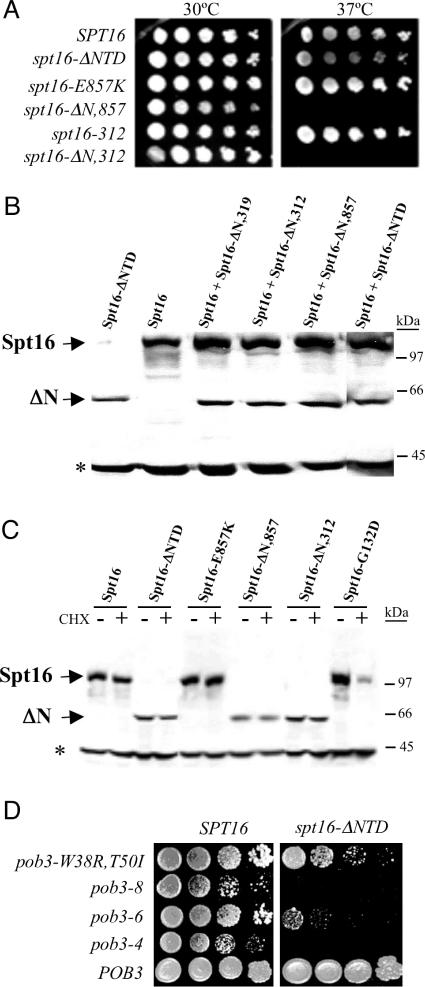
The NTD of Spt16, although dispensable under normal growth conditions, is conserved in sequence across the eukaryotic spectrum, suggesting an important function for this domain. Therefore, indications of an important function were sought through genetic means. Combining the deletion mutation of spt16-ΔNTD with the downstream point mutations characterizing the spt16-312 allele mentioned above, or with the E857K substitution mutation encoded by spt16-E857K (A.F. O'Donnell, unpublished data), yielded plasmid-borne mutant alleles spt16-ΔN,312 and spt16-ΔN,E857K that are unable to support growth at 37°C, in contrast to spt16-ΔNTD, spt16-312 and spt16-E857K (Figure 6A). It is even more striking to note that combining the spt16-ΔNTD deletion with the point mutations of spt16-319 created a version of Spt16 unable to support growth even at 30°C (data not shown). Polypeptide degradation may not contribute to these effects; the Spt16-ΔN,319, Spt16-ΔN,312 and Spt16-ΔN,E857K polypeptides are abundant and stable when expressed from a low-copy plasmid in wild-type cells (Figure 6B), and Spt16-ΔN,312 and Spt16-ΔN,E857K, as the sole Spt16 polypeptide in cells, are also stable at the restrictive temperature (Figure 6C). These findings suggest that the Spt16 NTD, although without essential function, can influence the essential functions(s) of downstream Spt16 domains.
Figure 6.
The spt16-ΔNTD mutation affects essential FACT activities. (A) Five-fold serial dilutions of isogenic spt16Δ derivatives maintained by the indicated low-copy plasmid-borne spt16 mutant alleles were spotted onto rich medium and incubated at the indicated temperature. (B) Extracts of 30°C wild-type cells harboring the same low-copy spt16 mutant plasmids were assessed by western analysis for Spt16 polypeptide. The asterisk indicates the non-specific 40 kDa band. (C) Isogenic spt16Δ derivatives with plasmid-borne spt16 mutant alleles were assessed for Spt16 polypeptide stability as in Figure 5. The asterisk indicates the non-specific 40 kDa band. (D) Serial dilutions of isogenic pob3Δ cells (SPT16) and spt16Δ pob3Δ cells with a low-copy spt16-ΔNTD plasmid (spt16-ΔNTD), maintained by the indicated low-copy pob3 mutant or POB3 plasmids, were spotted on rich medium and incubated at 35°C.
More insight was provided by the use of mutations altering the other FACT subunit, Pob3. Temperature-sensitive pob3 mutations affect cells in ways that implicate Pob3 in DNA replication (28). Combining temperature-sensitive pob3 mutant alleles with spt16-ΔNTD gave double-mutant cells with enhanced temperature sensitivities (Figure 6D). These findings suggest that the NTD of Spt16 influences replication-related function(s) of Pob3, and may have a role in replication.
A replication function for the Spt16 NTD
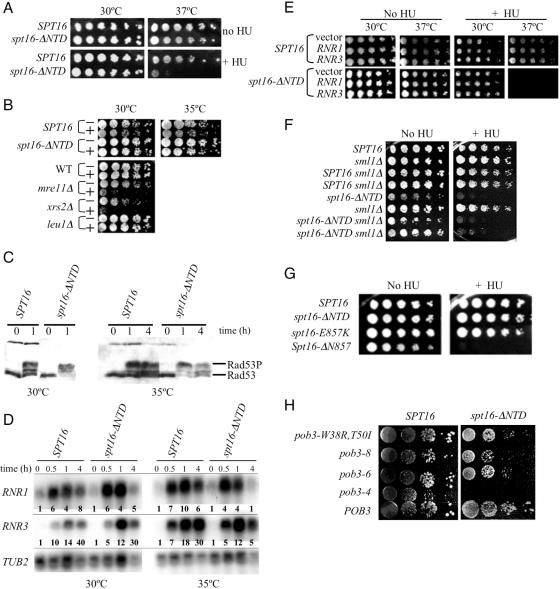
Involvement of the Spt16 NTD in replication was substantiated by investigations involving HU. This free-radical scavenger causes replication stress by inhibiting the enzyme ribonucleotide reductase and decreasing the synthesis of deoxyribonucleotides needed for DNA replication. Several FACT mutations cause sensitivity to HU (4,28). Cells lacking the Spt16 NTD also exhibit sensitivity to HU (Figure 7A). These cells also are sensitive to methylmethane sulfonate, another inducer of replication stress [data not shown; (48)]. Therefore the Spt16 NTD, although dispensable for normal DNA replication, becomes important during times of replication stress.
Figure 7.
The Spt16 NTD mediates HU sensitivity. (A) Five-fold serial dilutions of isogenic spt16Δ derivatives maintained by a plasmid-borne spt16-ΔNTD or SPT16 gene were spotted onto rich medium and incubated at the indicated temperature in the presence or absence of 50 mM HU. (B) Growing cells were transferred to rich medium with (+) or without (−) 200 mM HU, incubated for 8 h, and then serially diluted, transferred to HU-free solid medium, and incubated at the indicated temperature for colony formation. Isogenic mre11Δ and xrs2Δ cells (67) were positive controls for sensitivity, while isogenic leu1Δ and wild-type (WT) cells were negative controls. (C) Rad53 protein was detected by western analysis (using antibody from Santa Cruz Biotechnology) in extracts of growing cells incubated for the indicated times in rich medium containing 200 mM HU. Rad53P indicates phosphorylated Rad53. (D) RNR1 and RNR3 RNAs in extracts of growing cells incubated for the indicated times in rich medium containing 200 mM HU were detected by northern analysis, and quantified (bolded values in each lane) relative to TUB2 RNA values by densitometric analysis using multiple exposures and ImageJ software (US National Institutes of Health; available at http://rsb.info.nih.gov/nih-image/). (E) Serial dilutions of stationary-phase cultures of cells harbouring the RNR1 and RNR3 expression plasmids pBAD070 and pBAD079, or the control plasmid pBAD054, were spotted on rich medium with or without 50 mM HU and incubated at the indicated temperature. (F) Cells with the sml1Δ deletion (67) and isogenic derivatives with tagged SPT16 or spt16-ΔNTD at the chromosomal locus were spotted on rich medium with or without 50 mM HU and incubated at 30°C. Two segregants of sml1Δ derivatives are shown. (G) Five-fold serial dilutions of isogenic spt16Δ derivatives carrying the indicated plasmid-borne spt16 allele were spotted onto rich medium with or without 50 mM HU and incubated at 30°C. (H) Serial dilutions of cultures of the strains shown in Figure 6D were spotted on rich medium containing 100 mM HU and incubated at 30°C.
Sensitivity to HU can indicate several deficiencies, including impaired checkpoint signaling during DNA replication. When replication-fork activity stalls, as in response to a shortage of deoxynucleotide substrates, the replication checkpoint is activated. This checkpoint signaling has several effects, including inhibition of mitosis, relief of transcriptional repression for genes encoding ribonucleotide reductase, and ultimately the restoration of replication-fork function [reviewed in (49)]. Malfunction of this checkpoint signaling causes sensitivity to chronic exposure to HU, as in Figure 7A. A more direct measure of checkpoint function is the degree of resistance to acute, short-term HU exposure, which indicates whether checkpoint-mediated activities can spare a cell from irreversible damage related to replication-fork collapse (50–52). Control cells lacking the checkpoint proteins, Mre11 or Xrs2 were subjected to an 8 h exposure to a high concentration (200 mM) of HU and then transferred to solid HU-free medium to assess colony formation. As expected (53), these mutant cells with impaired checkpoint signaling failed to survive the acute HU treatment (Figure 7B). In marked contrast, the same HU treatment (at 30°C) of spt16-ΔNTD and spt16-Δ(6-435) mutant cells did not impair colony-forming ability (Figure 7B, and data not shown), and the treated cells were uniformly of the big-bud phenotype characteristic of checkpoint arrest (data not shown). This sensitivity to chronic HU treatment but normal resistance to acute HU treatment was seen in both the S288c and W303 genetic backgrounds. Similar assessment at 35°C, or with a 24 h HU exposure, also indicated that checkpoint signaling is intact in spt16-ΔNTD and spt16-Δ(6-435) mutant cells (Figure 7B, and data not shown). Thus the absence of the Spt16 NTD does not prevent this critical aspect of checkpoint signaling.
Checkpoint signaling was also assessed biochemically. Activation of checkpoint signaling leads to phosphorylation of the checkpoint protein Rad53, stimulation of the protein kinase activity of Rad53 for downstream signaling and decreasing Rad53 electrophoretic mobility (54). Figure 7C shows that Rad53 protein has shifted to a lower electrophoretic mobility upon HU treatment of spt16-ΔNTD and wild-type cells at both 30 and 35°C, indicating that Rad53 is phosphorylated effectively upon HU treatment of spt16-ΔNTD mutant cells.
One important consequence of replication-checkpoint signaling is an increase in ribonucleotide reductase expression (55). Direct assessment of endogenous mRNA levels for RNR1 and RNR3, which encode ribonucleotide reductase subunits, indicated that both genes were activated in spt16-ΔNTD mutant cells upon HU treatment (Figure 7D). Therefore the existence of at least two responses to checkpoint signaling, ribonucleotide reductase activation and maintenance of replication-fork stability as indicated by resistance to acute HU treatment, as well as Rad53 phosphorylation, indicate that checkpoint signaling is active in spt16-ΔNTD mutant cells.
Despite this checkpoint signaling, spt16-ΔNTD mutant cells are sensitive to HU (Figure 7A). We noted that during HU treatment high levels of RNR1 and RNR3 mRNAs were maintained in wild-type cells, whereas mRNA abundances had declined in spt16-ΔNTD mutant cells by the 4 h timepoint, especially at 35°C (Figure 7D). This decline paralleled the decline in Rad53 phosphorylation seen for spt16-ΔNTD mutant cells at this 35°C timepoint (Figure 7C). The Spt16 NTD may therefore facilitate sustained checkpoint signaling. The decline in checkpoint signaling and in RNR gene transcription in spt16-ΔNTD mutant cells upon extended HU exposure raised the possibility that ribonucleotide reductase activity in these mutant cells might become insufficient for the restart of stalled replication forks. However, two lines of evidence suggest that ribonucleotide reductase activity is not deficient in spt16-ΔNTD mutant cells. Ectopic expression of plasmid-borne RNR1 or RNR3 from the constitutive GAP promoter can alleviate HU sensitivity for checkpoint-deficient cells (50). However, overexpression of RNR1 or RNR3 from these plasmids did not alter the HU sensitivity of spt16-ΔNTD mutant cells (Figure 7E). We also eliminated the Sml1 protein, an inhibitor of ribonucleotide reductase (56). Checkpoint signaling causes the phosphorylation and degradation of Sml1, releasing preexisting ribonucleotide reductase enzyme from inhibition (57). A sml1Δ mutation makes cells resistant to HU and allows growth in the absence of checkpoint signaling (58). However, sml1Δ did not abrogate the HU sensitivity of spt16-ΔNTD cells (Figure 7F). These experiments suggest that the Spt16 NTD may mediate an activity downstream of checkpoint signaling itself, perhaps related to the resumption of replication by stalled replication forks.
Genetic indications of several replication functions for yeast FACT
Sensitivity to HU has been reported for yeast FACT mutants that have an intact Spt16 NTD (4,28). To determine whether these HU sensitivities are related to the actions of the NTD, genetic interactions between FACT mutations were assessed. These experiments showed that the spt16-ΔN,E857K mutant allele described above causes a degree of HU sensitivity greater than that shown by either spt16-ΔNTD or spt16-E857K themselves (Figure 7G). Similarly, several spt16-ΔNTD pob3 double-mutant derivatives exhibited synthetic HU sensitivity (Figure 7H). These findings suggest that FACT is involved in replication-fork activities in several ways, both dependent on and independent of the Spt16 NTD.
DISCUSSION
We report here an initial analysis, through genetic and biochemical analysis, of the domain organization of the subunits of yeast FACT. The domain interactions responsible for FACT integrity as determined here, and the functional aspects of the NTD of the Spt16 subunit, suggest that FACT functions may also be partially compartmentalized to domains.
Domain organization of the FACT subunits
The Spt16 protein may be folded into three major domains, as indicated by partial-proteolysis experiments, N-terminal sequence data and genetic findings (Figure 2D). The domain boundaries map to localized regions of low sequence conservation among Spt16 homologs from diverse species. Our findings suggest that Pob3 may be folded into two domains, separated by a short yeast-specific insert within the conserved Pob3/SSRP1 family. The SSRP1 members of the Pob3/SSRP1 protein family also have an HMG domain appended C-terminally. This HMG sequence is likely to be another independently folded portion of the polypeptide, given that this type of HMG domain is an independently folded entity in many contexts. The small HMG protein Nhp6 may provide this domain for yeast FACT (4,40).
FACT integrity
The Spt16 Mid domain is intact in Spt16 derivatives that bind Pob3 in vivo, and also binds Pob3 polypeptides in vitro. FACT is therefore stabilized through Pob3 interactions with the Spt16 Mid domain. Conversely, Spt16 binds in vitro to two different recombinant Pob3 polypeptides encompassing the NTDs and CTDs. Although these two Pob3 fragments have residues 178–215 in common, much of this ‘overlap’ sequence is absent from metazoan Pob3 homologs and most likely comprises a domain boundary. This 38-residue region is therefore unlikely to be responsible for the similar interactions of these two Pob3 fragments with Spt16. An internal duplication in Pob3/SSRP1 proteins has been reported that may produce similar polypeptide folds (59). This duplication spans Pob3 residues 4–104 and 374–475, placing one duplicated subdomain in each of the Pob3 domains that binds Spt16. This tandem duplication may provide binding surfaces for Spt16, one within each of the suggested Pob3 domains.
Analogous in vitro interaction studies have been carried out for human and Drosophila FACT (10,36). Fragments of human and Drosophila Spt16, corresponding to yeast residues 337–657 (for the human fragment), and ∼460–1035 and ∼630–1035 (for Drosophila fragments), were sufficient for in vitro SSRP1 binding. Thus in both metazoan cases, regions corresponding to the yeast Mid domain were involved. Note, however, that the 645–1035 fragment of yeast Spt16, analogous to the SSRP1-binding Drosophila fragment, did not bind Pob3 in vitro (Figure 4B), perhaps as a consequence of improper folding. The expected lack of interaction was also seen; human and Drosophila Spt16 polypeptide fragments that did not bind SSRP1 corresponding to N-terminal Spt16 sequences that are dispensable for yeast FACT integrity.
Correspondence between interactions within metazoan and yeast FACT is less evident for SSRP1/Pob3. Human SSRP1 was found to interact in vitro with hSpt16 through N-terminal sequences, whereas the downstream hSSRP1 fragments that were tested did not interact (36). For Drosophila FACT, the opposite pattern was seen, with C-terminal dSSRP1 sequences mediating dSpt16 interaction and N-terminal sequences remaining inert (10). Resolution of these differences awaits further study.
The Spt16 CTD
The CTD of Spt16 does not make stable associations with Pob3 or upstream Spt16 domains (Figure 2F), and therefore may be tethered loosely to the rest of FACT. We found that an spt16-ΔCTD mutant allele cannot keep cells alive, but attempts to understand the biological effects of the CTD have so far been stymied by the in vivo instability of Spt16-ΔCTD polypeptide (J. R. Stevens, G.C.J. and R.A.S., unpublished observations). However, recombinant human Spt16 lacking C-terminal residues corresponding to most of the CTD also lacks the histone-binding activity of full-length hSpt16 (11), suggesting that the Spt16 CTD may function as a mobile histone-binding domain.
NTD fragments and FACT function
The phenotypes of the spt16-Δ(5-435) and spt16-ΔNTD alleles, encoding Spt16 polypeptides lacking the NTD, indicate that the NTD is not essential for growth at 37°C or for chromatin repression of transcription, and therefore these properties depend on Spt16 Mid and CTD domains. However, Spt16 polypeptides in which only a fragment of the NTD is appended to Mid+CTD, such as Spt16-Δ922 and Spt16-Δ369, are compromised for these functions. These observations suggest that the NTD fragments in these polypeptides may be misfolded to an extent that they interfere with proper Mid+CTD function or with Pob3 function.
Structure predictions support the notion that the NTD fragments in the Spt16-Δ922 and Spt16-Δ369 polypeptides may indeed be misfolded. The Spt16 NTD resembles members of the ‘pita-fold’ family of aminopeptidase enzymes (59,60), with a type-I fold (61) predicted for an aminopeptidase-like NTD subregion. Eukaryotic aminopeptidases have another domain, of heterogeneous structure (62). This added NTD of eukaryotic aminopeptidase enzymes is mimicked, in size and position, by a potential subdomain of the Spt16 NTD, so that the entire Spt16 NTD resembles a type-Ib enzyme structure. NTD subdomains are in fact suggested by partial-proteolysis results (Figure 2A). In any case, the Spt16 NTD fragments studied here, in the Spt16-Δ922 and Spt16-Δ369 polypeptides, each lack approximately half of the putative ‘pita-fold’ subdomain.
Spt16 NTD function
The experiments reported here, and related ones concerning metazoan FACT (10,36), suggest that the NTD of the FACT subunit Spt16 is not critical for FACT integrity. This domain is almost certainly, therefore, involved in interactions with other proteins. However, these interactions are not important for global transcription if the rest of FACT is intact. On the other hand, NTD interactions are important under conditions of replication stress, as evidenced by the phenotype of spt16-ΔNTD mutant cells. Even during replication stress these interactions may not affect transcription, because the Spt16 NTD is not necessary for transcriptional activation of the RNR genes (Figure 7D). The Spt16 NTD may therefore be important for the restart of stalled replication forks that accumulate upon replication stress.
The postulated NTD interactions during replication stress are not obvious. The NTD interacts with the Sas3 component of the NuA3 histone acetyltransferase complex (63). Although this interaction has certain transcription-related effects, an appreciation of the consequences of this interaction awaits further investigation. A more relevant interaction for replication stress may involve the protein kinase CK2. A fragment of human Spt16 encompassing a majority of the NTD interacts in vitro with the α′ catalytic subunit of CK2 (36) in a complex that phosphorylates and activates p53 (35,36). Activation of p53 takes place upon replication stress brought on by HU treatment (64), and p53 becomes localized to stalled replication forks (65). An analogous CK2•FACT association exists in yeast (37), but its significance is currently unclear.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Steve Elledge, Tim Formosa, LeAnn Howe and Jerry Workman for plasmids, Jeff Coles for preliminary experiments, David Carruthers and Kendra Gillis for technical assistance, and Chris Barnes for helpful discussions. This work was supported by a grant from the Canadian Institutes of Health Research. A.F.O. was supported by the Natural Sciences and Engineering Research Council of Canada, the Canadian Institutes of Health Research, the Killam Foundation and the Nova Scotia Health Research Foundation. J.K. was supported by the Canadian Institutes of Health Research; L.V.M. was supported by grants from the Dalhousie Cancer Biology Research Group.
REFERENCES
- 1.Brewster N.K., Johnston,G.C. and Singer,R.A. (1998) Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem., 273, 21972–21979. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G., Wu,W.-H., Lane,W.S., Hampsey,M. and Reinberg,D. (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature, 400, 284–288. [DOI] [PubMed] [Google Scholar]
- 3.Okuhara K., Ohta,K., Seo,H., Shioda,M., Yamada,T., Tanaka,Y., Dohmae,N., Seyama,Y., Shibata,T. and Murofushi,H. (1999) A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr. Biol., 9, 341–350. [DOI] [PubMed] [Google Scholar]
- 4.Formosa T., Eriksson,P., Wittmeyer,J., Ginn,J., Yu,Y. and Stillman,D.J. (2001) Spt16–Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J., 20, 3506–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone E.A., Clark,C.D., Chiang,A. and Winston,F. (1991) Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 5710–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wittmeyer J. and Formosa,T. (1997) The Saccharomyces cerevisiae DNA polymerase α catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol., 17, 4178–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orphanides G., LeRoy,G., Chang,C.-H., Luse,D.S. and Reinberg,D. (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell, 92, 105–116. [DOI] [PubMed] [Google Scholar]
- 8.Belotserkovskaya R. and Reinberg,D. (2004) Facts about FACT and transcript elongation through chromatin. Curr. Opin. Genet. Dev., 14, 139–146. [DOI] [PubMed] [Google Scholar]
- 9.Belotserkovskaya R., Saunders,A., Lis,J.T. and Reinberg,D. (2004) Transcription through chromatin: understanding a complex FACT. Biochim. Biophys. Acta, 1677, 87–99. [DOI] [PubMed] [Google Scholar]
- 10.Shimojima T., Okada,M., Nakayama,T., Ueda,H., Okawa,K., Iwamatsu,A., Handa,H. and Hirose,S. (2003) Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev., 17, 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belotserkovskaya R., Oh,S., Bondarenko,V.A., Orphanides,G., Studitsky,V.M. and Reinberg,D. (2003) FACT facilitates transcription-dependent nucleosome alteration. Science, 301, 1090–1093. [DOI] [PubMed] [Google Scholar]
- 12.Chang C.-H. and Luse,D.S. (1997) The H3/H4 tetramer blocks transcript elongation by RNA polymerase II in vitro. J. Biol. Chem., 272, 23427–23434. [DOI] [PubMed] [Google Scholar]
- 13.Kireeva M.L., Walter,W., Tchernajenko,V., Bondarenko,V., Kashlev,M. and Studitsky,V.M. (2002) Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell, 9, 541–552. [DOI] [PubMed] [Google Scholar]
- 14.Jackson V. (1990) In vivo studies on the dynamics of histone–DNA interaction: evidence for nucleosome dissolution during replication and transcription and a low level of dissolution independent of both. Biochemistry, 29, 719–731. [DOI] [PubMed] [Google Scholar]
- 15.Kimura H. and Cook,P.R. (2001) Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol., 153, 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formosa T., Ruone,S., Adams,M.D., Olsen,A.E., Eriksson,P., Yu,Y., Rhoades,A.R., Kaufman,P.D. and Stillman,D.J. (2002) Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics, 162, 1557–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowley A., Singer,R.A. and Johnston,G.C. (1991) CDC68, a yeast gene that affects regulation of cell proliferation and transcription, encodes a protein with a highly acidic carboxyl terminus. Mol. Cell. Biol., 11, 5718–5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders A., Werner,J., Andrulis,E.D., Nakayama,T., Hirose,S., Reinberg,D. and Lis,J.T. (2003) Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science, 301, 1094–1096. [DOI] [PubMed] [Google Scholar]
- 19.Mason P.B. and Struhl,K. (2003) The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol., 23, 8323–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim M., Ahn,S.-H., Krogan,N.J., Greenblatt,J.F. and Buratowski,S. (2004) Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J., 23, 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindstrom D.L., Squazzo,S.L., Muster,N., Burckin,T.A., Wachter,K.C., Emigh,C.A., McCleery,J.A., Yates,J.R.,III and Hartzog,G.A. (2003) Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol., 23, 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wada T., Orphanides,G., Hasegawa,J., Kim,D.-K., Shima,D., Yamaguchi,Y., Fukuda,A., Hisatake,K., Oh,S., Reinberg,D. and Handa,H. (2000) FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell, 5, 1067–1072. [DOI] [PubMed] [Google Scholar]
- 23.Prelich G. and Winston,F. (1993) Mutations that suppress the deletion of an upstream activating sequence in yeast: involvement of a protein kinase and histone H3 in repressing transcription in vivo. Genetics, 135, 665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Q., Johnston,G.C. and Singer,R.A. (1993) The Saccharomyces cerevisiae Cdc68 transcription activator is antagonized by San1, a protein implicated in transcriptional silencing. Mol. Cell. Biol., 13, 7553–7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lycan D., Mikesell,G., Bunger,M. and Breeden,L. (1994) Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol., 14, 7455–7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evans D.R.H., Brewster,N.K., Xu,Q., Rowley,A., Altheim,B.A., Johnston,G.C. and Singer,R.A. (1998) The yeast protein complex containing Cdc68 and Pob3 mediates core-promoter repression through the Cdc68 N-terminal domain. Genetics, 150, 1393–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan C.D., Laprade,L. and Winston,F. (2003) Transcription elongation factors repress transcription initiation from cryptic sites. Science, 301, 1096–1099. [DOI] [PubMed] [Google Scholar]
- 28.Schlesinger M.B. and Formosa,T. (2000) POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics, 155, 1593–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wittmeyer J., Joss,L. and Formosa,T. (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase α. Biochemistry, 38, 8961–8971. [DOI] [PubMed] [Google Scholar]
- 30.Ryan K.M., Phillips,A.C. and Vousden,K.H. (2001) Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol., 13, 332–337. [DOI] [PubMed] [Google Scholar]
- 31.Wahl G.M. and Carr,A.M. (2001) The evolution of diverse biological responses to DNA damage: insights from yeast and p53. Nature Cell Biol., 3, e277–e286. [DOI] [PubMed] [Google Scholar]
- 32.Sakaguchi K., Sakamoto,H., Lewis,M.S., Anderson,C.W., Erikson,J.W., Appella,E. and Xie,D. (1997) Phosphorylation of serine 392 stabilizes the tetramer formation of tumor suppressor protein p53. Biochemistry, 36, 10117–10124. [DOI] [PubMed] [Google Scholar]
- 33.Kapoor M. and Lozano,G. (1998) Functional activation of p53 via phosphorylation following DNA damage by UV but not gamma radiation. Proc. Natl Acad. Sci. USA, 95, 2834–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu H., Taya,Y., Ikeda,M. and Levine,A.J. (1998) Ultraviolet radiation, but not gamma radiation or etoposide-induced DNA damage, results in the phosphorylation of the murine p53 protein at serine-389. Proc. Natl Acad. Sci. USA, 95, 6399–6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keller D.M., Zeng,X., Wang,Y., Zhang,Q.H., Kapoor,M., Shu,H., Goodman,R., Lozano,G., Zhao,Y. and Lu,H. (2001) A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell, 7, 283–292. [DOI] [PubMed] [Google Scholar]
- 36.Keller D.M. and Lu,H. (2002) p53 Serine 392 phosphorylation increases after UV through induction of the assembly of the CK2•hSPT16•SSRP1 complex. J. Biol. Chem., 277, 50206–50213. [DOI] [PubMed] [Google Scholar]
- 37.Krogan N.J., Kim,M., Ahn,S.H., Zhong,G., Kobor,M.S., Cagney,G., Emili,A., Shilatifard,A., Buratowski,S. and Greenblatt,J.F. (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol., 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bruhn S.L., Pil,P.M., Essigmann,J.M., Housman,D.E. and Lippard,S.J. (1992) Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl Acad. Sci. USA, 89, 2307–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yarnell A.T., Oh,S., Reinberg,D. and Lippard,S.J. (2001) Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem., 276, 25736–25741. [DOI] [PubMed] [Google Scholar]
- 40.Brewster N.K., Johnston,G.C. and Singer,R.A. (2001) A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol., 21, 3491–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhoades A.R., Ruone,S. and Formosa,T. (2004) Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell. Biol., 24, 3907–3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wach A., Brachat,A., Pöhlmann,R. and Philippsen,P. (1994) New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast, 10, 1793–1808. [DOI] [PubMed] [Google Scholar]
- 43.Xu Q., Singer,R.A. and Johnston,G.C. (1995) Sug1 modulates yeast transcription activation by Cdc68. Mol. Cell. Biol., 15, 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahn Y.-T., Wu,X.-L., Biswal,S., Velmurugan,S., Volkert,F.C. and Jayaram,M. (1997) The 2 μm-plasmid-encoded Rep1 and Rep2 proteins interact with each other and colocalize to the Saccharomyces cerevisiae nucleus. J. Bacteriol., 179, 7497–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sengupta A., Blomqvist,K., Pickett,A.J., Zhang,Y., Chew,J.S.K. and Dobson,M.J. (2001) Functional domains of yeast plasmid-encoded Rep proteins. J. Bacteriol., 183, 2306–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winston F. (1992) Analysis of SPT genes: a genetic approach toward analysis of TFIID, histones, and other transcription factors of yeast. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Plainview, NY, pp. 1271–1293. [Google Scholar]
- 47.Winston F. and Carlson,M. (1992) Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet., 8, 387–391. [DOI] [PubMed] [Google Scholar]
- 48.Tercero J.A. and Diffley,J.F. (2001) Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature, 412, 553–557. [DOI] [PubMed] [Google Scholar]
- 49.Osborn A.J., Elledge,S.J. and Zou,L. (2002) Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol., 12, 509–516. [DOI] [PubMed] [Google Scholar]
- 50.Desany B.A., Alcasabas,A.A., Bachant,J.B. and Elledge,S.J. (1998) Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev., 12, 2956–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lopes M., Cotta-Ramusino,C., Pellicioli,A., Liberi,G., Plevani,P., Muzi-Falconi,M., Newlon,C.S. and Foiani,M. (2001) The DNA replication checkpoint response stabilizes stalled replication forks. Nature, 412, 557–561. [DOI] [PubMed] [Google Scholar]
- 52.Sogo J.M., Lopes,M. and Foiani,M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science, 297, 599–602. [DOI] [PubMed] [Google Scholar]
- 53.D'Amours D. and Jackson,S.P. (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev., 15, 2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pellicioli A., Lucca,C., Liberi,G., Marini,F., Lopes,M., Plevani,P., Romano,A., Di Fiore,P.P. and Foiani,M. (1999) Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J., 18, 6561–6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elledge S.J., Zhou,Z., Allen,J.B. and Navas,T.A. (1993) DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays, 15, 333–339. [DOI] [PubMed] [Google Scholar]
- 56.Chabes A., Domkin,V. and Thelander,L. (1999) Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J. Biol. Chem., 274, 36679–36683. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X., Chabes,A., Domkin,V., Thelander,L. and Rothstein,R. (2001) The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J., 20, 5344–3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao X., Muller,E.G.D. and Rothstein,R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell, 2, 329–340. [DOI] [PubMed] [Google Scholar]
- 59.Ponting C.P. (2002) Novel domains and orthologues of eukaryotic transcription elongation factors. Nucleic Acids Res., 30, 3643–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aravind L. and Koonin,E.V. (1998) Eukaryotic transcription regulators derive from ancient enzymatic domains. Curr. Biol., 8, R111–R113. [DOI] [PubMed] [Google Scholar]
- 61.Bradshaw R.A., Brickey,W.W. and Walker,K.W. (1998) N-terminal processing: the methionine aminopeptidase and Nα-acetyl transferase families. Trends Biochem. Sci., 23, 263–267. [DOI] [PubMed] [Google Scholar]
- 62.Lowther W.T. and Matthews,B.W. (2000) Structure and function of the methionine aminopeptidases. Biochim. Biophys. Acta, 1477, 157–167. [DOI] [PubMed] [Google Scholar]
- 63.John S., Howe,L., Tafrov,S.T., Grant,P.A., Sternglanz,R. and Workman,J.L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAFII30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)–FACT complex. Genes Dev., 14, 1196–1208. [PMC free article] [PubMed] [Google Scholar]
- 64.Nayak B.K. and Das,G.M. (2002) Stabilization of p53 and transactivation of its target genes in response to replication blockade. Oncogene, 21, 7226–7229. [DOI] [PubMed] [Google Scholar]
- 65.Sengupta S., Linke,S.P., Pedeux,R., Yang,Q., Farnsworth,J., Garfield,S.H., Valerie,K., Shay,J.W., Ellis,N.A., Wasylyk,B. and Harris,C.C. (2003) BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J., 22, 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hancock K. and Tsang,V.C.W. (1983) India ink staining of proteins on nitrocellulose paper. Anal. Biochem., 133, 157–162. [DOI] [PubMed] [Google Scholar]
- 67.Winzeler E.A., Shoemaker,D.D., Astromoff,A., Liang,H., Anderson,K., Andre,B., Bangham,R., Benito,R., Boeke,J.D., Bussey,H. et al. (1999) Functional characterization of the S.cerevisiae genome by gene deletion and parallel analysis. Science, 285, 901–906. [DOI] [PubMed] [Google Scholar]