Abstract
Background
BCDIN3D is a member of the Bin3 methyl-transferase family that targets the 5' mono-phosphate of nucleic acids. Although BCDIN3D has been shown to increasetumorigenic phenotypes and invasiveness in MDA-MB-231 cells, its the clinical implications in breast cancer remain unclear.
Methods
We screened for BCDIN3D using tissue microarrays constructed from 250 patients who were histologically confirmed to have invasive ductal breast carcinoma at the Fudan University Shanghai Cancer Center.
Results
The survival analysis by Kaplan-Meier and Cox regression showed that BCDIN3D expression level served as a prognostic factor for disease-free survival (P = 0.042). The prognostic value of BCDIN3D was most significant in triple-negative breast cancer (TNBC) patients (P = 0.007).
Conclusions
BCDIN3D might serve as an important prognostic factor for TNBC patients.
Keywords: BCDIN3D, breast cancer, TNBC
BACKGROUND
Breast cancer is the most common cancerin women, and accountfor approximately 23% of allcancer cases and approximately 14% of cancer deaths[1]. It is a heterogeneous disease embracing several different phenotypes [2], including luminal A, luminal B, human epidermal growth factor receptor 2 (HER2)-enriched, and triple-negative breast cancer (TNBC) [3]. Breast cancer results from the accumulation of genetic and epigenetic alterations[4, 5]. Epigenetic alterations, as defined by modifications of DNA, histones and coding/noncoding RNAs [6, 7], also contribute to various phases of neoplastic development including initiation, promotion, invasion, metastases.
RNA modification is a kind of epigenetic regulation of gene expression, analogous to DNA methylation and histone modification. [8]. Methylation is a ubiquitous modification that affects several residues/sites in molecules [9]. Methyl-transfer reactions to RNA nucleotides are catalyzed by a variety of RNA-MTases that include more than 60 members with hundreds of homologs, and which have so far been divided into four super-families[10]. BCDIN3D, a member of the Bin3 methyltransferase family, share homology within their putative S-adenosyl Methionine (SAM) binding motif from S. pombe to human. SAM (S-adenosylmethionine) is well known as the methyl donor for methyl-transferases that modify DNA, RNA, histones[11].
BCDIN3D is a methyltransferase that targets the 5'mono-phosphate of nucleic acids. As depletion of BCDIN3D in the MDA-MB-231 cells abolishes anchorage-independent growth and decreases invasiveness in MDA-MB-231 cells, but not growth ormigration [12], its prognostic value may be of interestHerein, we investigated BCDIN3D expression and its association with tumor progression and clinical outcome in a cohort of 250 patients who had undergone surgery for breast cancer in eastern Chinese women population.
RESULTS
Patient characteristics
Clinical-pathological characteristics of the study cohort are summarized in Table 1. Patients' median age at diagnosis was 52.02 years. After a mean follow-up time of 83.36 months, 55 of the 250 patients showed recurrence of disease. In univariate analysis, histological grade (HR, 1.761; 95% CI, 1.041-2.980; P = 0.035), tumor size (HR, 1.715; 95% CI, 1.086-2.708; P = 0.021), metastatic nodes (HR, 2.231;95% CI, 1.304-3.819; P = 0.003), were significantly associated with higher risk of recurrence and reached statistical significance as expected, however, only the association with tumor size and metastatic nodes remained statistically significant inmultivariate analysis (Table 3).
Table 1. Clinical-pathological characteristics of the study cohort.
| Clinical-pathological characteristics | No. | Percentage (%) |
|---|---|---|
| Age (years) (mean 52.02, SD 9.642, median 51, range 29-85) | ||
| ≤50 | 120 | 48.0 |
| >50 | 130 | 52.0 |
| Menopausal status | ||
| Pre | 108 | 43.2 |
| Post | 142 | 56.8 |
| TNM Stage | ||
| I | 74 | 29.6 |
| II | 132 | 52.8 |
| III | 42 | 16.8 |
| Unknown | 2 | 0.8 |
| Histological grade | ||
| I | 5 | 2.0 |
| II | 184 | 73.6 |
| III | 61 | 24.4 |
| Tumor size | ||
| T1 (≤2cm) | 115 | 46.0 |
| T2 (>2-5cm) | 120 | 48.0 |
| T3 (>5cm) | 13 | 5.2 |
| Unknown | 2 | 0.8 |
| Node status | ||
| Negative | 151 | 60.4 |
| Positive | 97 | 38.8 |
| Unknown | 2 | 0.8 |
| ER status | ||
| Negative | 143 | 57.2 |
| Positive | 106 | 42.4 |
| Unknown | 1 | 0.4 |
| PR status | ||
| Negative | 184 | 73.6 |
| Positive | 63 | 25.2 |
| Unknown | 3 | 1.2 |
| HER-2 status | ||
| Negative | 148 | 59.2 |
| Positive | 101 | 40.4 |
| Unknown | 1 | 0.4 |
| Molecular subtypea | ||
| Luminal like | 106 | 42.4 |
| HER-2 Positive | 42 | 16.8 |
| Triple Negative | 100 | 40.0 |
| Unknown | 2 | 0.8 |
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2; SD, standard deviation;
Definition of breast cancer molecular subtypes: luminal like (ER and/or PR positive, any HER-2 status), HER-2 Positive (ER and PR negative, HER-2 positive) and triple negative (ER negative, PR negative and HER-2 negative)
Table 3. Univariate and multivariate analysis of factors related to disease-free survival (DFS) in breast cancer patients.
| Variables | DFS | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR(95% CIs) | P | HR(95% CIs) | P | |
| Age | 0.815(0.552-1.534) | 0.939 | 0.971(0.554-1.700) | 0.917 |
| Histological grade | 1.761(1.041-2.980) | 0.035 | 1.749(0.965-3.170) | 0.065 |
| Tumor Size | 1.715(1.086-2.708) | 0.021 | 1.648(1.042-2.606) | 0.033 |
| Metastatic nodes | 2.231(1.304-3.819) | 0.003 | 2.158(1.236-3.769) | 0.007 |
| ER | 0.786(0.456-1.356) | 0.388 | 1.491(0.713-3.117) | 0.288 |
| PR | 0.408(0.185-0.903) | 0.027 | 0.372(0.144-0.960) | 0.041 |
| HER-2 | 0.936(0.548-1.600) | 0.809 | 0.754(0.412-1.378) | 0.358 |
| BCDIN3D | 1.754(1.012-3.039) | 0.045 | 1.904(1.081-3.354) | 0.026 |
Abbreviations: HR, hazard ratio; CIs, confidence intervals; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2;
Expression patterns of BCDIN3D in breast cancer patients
In this cohort of 250 patients, TMAswere immunostained for BCDIN3D (representative images, Figure 1). The specificity of the antibody against BCDIN3D was confirmed by Western blot of human cell lysis. Positive staining of BCDIN3D was detected in 49.6% of tumors according to the scoring criterion described above (n = 124; 49.6% positive, 50.4% negative; Figure 1a-1d). We further analyzed relationships between clinicopathologic features and the expression level of BCDIN3D. The percentages of positive staining of protein were consistent across all subsets of patients (Table 2).
Figure 1. Representative image of immunohistochemical BCDIN3D staining were shown in both small pictures (×100 magnification) and large (×400 magnification).
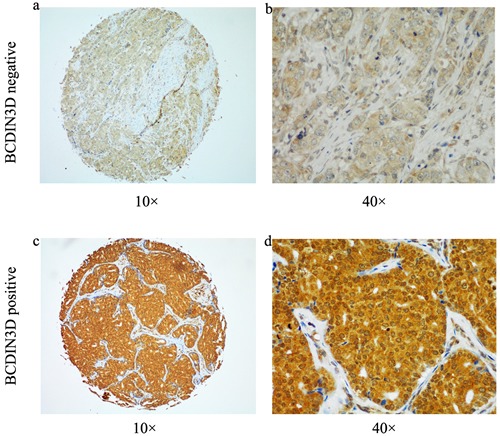
a.-b. Negative for BCDIN3D. c.-d. Positive for BCDIN3D.
Table 2. Correlations between patients' characteristics and expression status of BCDIN3D.
| Clinical-pathological characteristics | Cases | BCDIN3D | |||
|---|---|---|---|---|---|
| - | + | Pa | |||
| 250 | 126 | 124 | |||
| Percentage (%) | 50.40% | 49.60% | |||
| Age(years) | |||||
| ≤50 | 120 | 62 | 58 | 0.521 | |
| >50 | 130 | 64 | 66 | ||
| Menopausal status | |||||
| Pre | 108 | 54 | 54 | 1.000 | |
| Post | 142 | 72 | 70 | ||
| TNM Stage | |||||
| I | 74 | 32 | 42 | 0.250 | |
| II | 132 | 73 | 59 | ||
| III | 42 | 21 | 21 | ||
| Histological grade | |||||
| I | 5 | 1 | 4 | 0.334 | |
| II | 184 | 92 | 92 | ||
| III | 61 | 33 | 28 | ||
| Tumor size | |||||
| T1 (≤2cm) | 115 | 52 | 63 | 0.318 | |
| T2 (>2-5cm) | 120 | 66 | 54 | ||
| T3 (>5cm) | 13 | 8 | 5 | ||
| Node status | |||||
| Negative | 151 | 75 | 76 | 0.697 | |
| Positive | 97 | 51 | 46 | ||
| ER status | |||||
| Negative | 143 | 69 | 74 | 0.522 | |
| Positive | 106 | 56 | 50 | ||
| PR status | |||||
| Negative | 184 | 88 | 96 | 0.243 | |
| Positive | 63 | 36 | 27 | ||
| HER-2 status | |||||
| Negative | 148 | 78 | 70 | 0.368 | |
| Positive | 101 | 47 | 54 | ||
Abbreviations:ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2;
Pearson χ2 test
BCDIN3D was identified as a significant prognostic factor in breast cancer patients, especially in triple-negative breast cancers (TNBC)
Both univariate and adjusted multivariate survival analyses showed significant differences in DFS between the BCDIN3D positive and negative groups. BCDIN3D positive group having a significantly higher incidence of disease eventsin both univariate analysis (HR = 1.754; 95% CI: 1.012-3.039; P = 0.045) and multivariate analysis. (HR = 1.904; 95% CI: 1.081-3.354; P = 0.026) (Table 3). The BCDIN3D positive group also showed worse DFS in the Kaplan-Meier survival analysis (P = 0.042; Figure 2a). Thus, these results indicate that BCDIN3D expression isdirect associated with breast cancer recurrence. Then we analyzed the relationship between BCDIN3D expression and survival according to the different breast cancer subtypes. We found the prognostic value of BCDIN3D for DFS was most significant among patients with TNBC (P = 0.007; Figure 2b). In the TNBCsubset, patients with positive BCDIN3D staining showed a higher likelihood of occurence of disease events (HR = 3.584; 95% CI: 1.319-9.737; P = 0.012) in univariate analysis and remain the same trend in multivariate analysis. (HR = 3.719; 95% CI: 1.345-10.283; P = 0.011)(Table 4).
Figure 2. Elevated expression of BCDIN3D predictsworse clinical outcome in breast cancer patients, especially in TNBC.
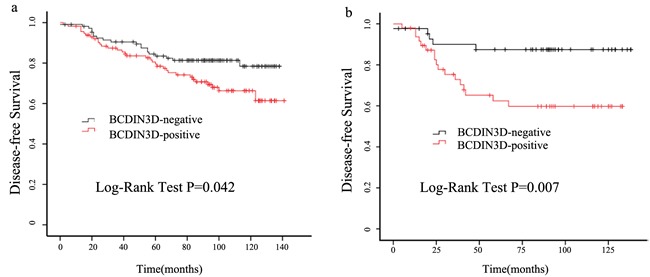
a. Cumulative disease-free survival curves of patients with positive or negative expression of BCDIN3D in breast cancer patients. b. Cumulative disease-free survival curves of patients with positive or negative expression of BCDIN3D in TNBC patients.
Table 4. Univariate and multivariate analysis of factors related to disease-free survival (DFS) in TNBC patients.
| Variables | DFS | |||
|---|---|---|---|---|
| Univariate Analysis | Multivariate Analysis | |||
| HR(95% CIs) | P | HR(95% CIs) | P | |
| Age | 0.967(0.417-2.238) | 0.937 | 1.192(0.503-2.824) | 0.690 |
| Histological grade | 1.273(0.553-2.927) | 0.570 | 0.953(0.392-2.315) | 0.915 |
| Tumor Size | 2.837(1.381-5.825) | 0.005 | 1.950(1.023-3.719) | 0.042 |
| Metastatic nodes | 3.195(1.364-7.486) | 0.007 | 3.157(1.265-7.880) | 0.014 |
| BCDIN3D | 3.584(1.319-9.737) | 0.012 | 3.719(1.345-10.283) | 0.011 |
Abbreviations: HR, hazard ratio; CIs, confidence intervals; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2;
DISCUSSION
Breast Cancer, is now known to involve epigenetic abnormalities along with genetic alterations [13]. Epigenetic modifications are early events in breast carcinogenesis and could be usefulfor early detection, prognosis, and targeted therapy of breast cancer [14]. Post-transcriptional gene regulation by noncoding RNA commonly referred as microRNAs is a kind of epigenetic regulation [15, 16]. It has become one of the most dynamic and fast-growing fields in science. Despite the rapid advance of RNA research, the enzymes that post-transcriptionally modify RNA have been less investigated[17].
A post-transcriptional modification enzyme BCDIN3D O-methylates the 5′ terminal mono-phosphate group of the precursor of some miRNA, which increasedtumorigenic phenotypes and cells invasiveness[12]. To our knowledge, no one else has explored its clinical significance. Our study is the first to evaluate the prognostic value of BCDIN3D in breast cancer patients. Our finding that patients with BCDIN3D positive tumors had worse DFS than the BCDIN3D negative group supports previous biological function study results from other researchers. Interestingly, this association was found to be most significant among patients with TNBC. TNBC breast tumors lack ER, PR, and HER-2 expression and occupy 15-20% of all breast cancers. It is generally more aggressive, has higher rates of relapse and decreased overall survival [18]. As few biomarkers are widely considered to be predictive for TNBC prognosis, and thus predictive factors for TNBC are urgently need. Our findings suggest that BCDIN3D might serve as an important prognostic factor for TNBC patients.
Our study had several limitations, including the small data set, few cases of stage I patients, lack of validation in an independent series of cases, and the composition of the study cohort which did not exactly represent that of the breast cancer population. Howeverour study was based on the use of TMAs, which can guarantee the consistency and coherence of these factors.
CONCLUSIONS
In conclusion, our results associate, for the first time, higher BCDIN3D levels with worse DFS, especially in triple-negative breast cancer population, which suggests its potential use as a predictive biomarker. Furthermore, as more future basic and clinical studies uncover the underlying mechanism in this pattern, BCDIN3D could emerge as a desperately needed therapeutic targetin triple-negative breast cancer.
MATERIALS AND METHODS
Patients and specimens
250 primary breast cancer samples of stage I to III invasive ductal carcinoma cases were collected randomly at the Department of Breast Surgery in Fudan University Shanghai Cancer Center (FDUSCC, Shanghai, P.R. China) between August 2001 and March 2006. Their clinical-pathological characteristics and the systemic therapies are presented in Table 1. In this retrospective cohort study, they have been followed regularly, and 227 cases obtained the clinical outcome, with the last update in September 2013. Their median follow-up time was 96 months.
Ethics statement
This study was approved by the Ethics Committee of FDUSCC, and each participant signed an informed consent document.
Tissue microarrays (TMAs)
TMAs were constructed from formalin-fixed, paraffin-embedded samples of carcinomas obtained from the250 breast cancer patients. A hematoxylin- and eosin-stained section of each tumor block was used to mark representative tumor regions. Tissue cylinders with a diameter of 2 mm were punched from the above regions and transferred to recipient array blocks using a Tissue Micro Arrayer (Beecher Instruments, Sliver Springs, MD, USA). TMAs werecomposed of duplicate cores from different areas of the same tumor to compare staining patterns.
IHC experimental procedures
The tissue micro arrays were subjected to immunohistochemical staining for BCDIN3D, using a 2-step protocol (GTVisionTMIII). The primary antibodies used were monoclonal BCDIN3D antibody (Santa Cruz Biotechnology, CA, USA sc-390348). The specificity of the BCDIN3D antibody was validated by western blot (Figure S1). The TMAs were deparaffinized with xylene, and rehydrated with an ethanol gradientThe sections were then rinsed with phosphate buffer solution (PBS) for immunohistochemical staining. For antigen retrieval the sections were immersed in 0.01 M tris sodium citrate pH 6.0 and boiled at 121 °C for 5 min followed by 2 min simmering. All slides were incubated with nonspecific staining blocking agent for 20 minutes to quench endogenous peroxidase activity and then with anti-BCDIN3D (1:200) at 4 °C overnight. Primary antibodies were detected by HRP-conjugated secondary antibodies followed by colorimetric detection with 3, 3-diaminobenzidine (DAB). The TMAs were then counterstained with Gill hematoxylin and dehydrated in an ascending ethanol series before clearing with xylene and mounting under a coverslip.
Evaluation of immunohistochemical variables
TMAs were stained and scored semi-quantitatively. The score used in all subsequent analysis was the average across the available cores. Staining was graded for intensity of staining (0, no staining; 1+, faint/equivocal; 2+, moderate; 3+, strong) and percentage of cells stained (0, no staining; 1+, < 10% of cells stained; 2+, 10% -50% of cells; and 3+, >50% of cells stained). For this study, SI ≥3 was defined as positive staining, and SI ≤2 was defined as negative staining. Scoring was reviewed in parallel by two experienced breast disease pathologistswho were blinded to all clinical data.
Statistical analyses
Disease-free survival(DFS) was defined as the time between the date of the primary surgery to the date of relapse/breast cancer specific death or September 2013. The first recurrence of disease at a local, regional, or distant site; diagnosis of contralateral breast cancer; and the breast cancer specific death were considered DFS events. Patients with study end date and loss of follow-up were considered to be censored. Correlations between clinical-pathological parameters and BCDIN3D were tested using the Chi-squared test. Survival outcomes were estimated using the Kaplan-Meier method and were compared between the groups using log-rank statistics. Univariate and multivariate analyseswere carried out using the Cox risk proportion model. Statistics was analyzed using SPSS (version 13.0; SPSS Company). All P values are two-sided; P value less than 0.05 was considered significant. All analyses were based on the observed data with the assumption that missing data were completely at random.
SUPPLEMENTARY MATERIAL FIGURE
Acknowledgments
This research was supported by grants from the National Natural Science Foundation of China ( 81202082). These funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
CONFLICTS OF INTERESTS
The authors declare that they have no competing interests.
Authors' contributions
ZS, JW conceived and designed the study. LY, YC and XH performed the experiments and analyzed the data. LS and FQ contributed reagents, materials, and analysis tools. All authors read and approved the final manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, Shalon D, Brown PO, Botstein D. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, Senn HJ. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24:2206–2223. doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kanwal R, Gupta S. Epigenetic modifications in cancer. Clin Genet. 2012;81:303–311. doi: 10.1111/j.1399-0004.2011.01809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Virani S, Colacino JA, Kim JH, Rozek LS. Cancer epigenetics: a brief review. ILAR J. 2012;53:359–369. doi: 10.1093/ilar.53.3-4.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xhemalce B. From histones to RNA: role of methylation in cancer. Brief Funct Genomics. 2013;12:244–253. doi: 10.1093/bfgp/els064. [DOI] [PubMed] [Google Scholar]
- 7.Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S. Epigenetics in breast cancer: what's new? Breast Cancer Res. 2011;13:225. doi: 10.1186/bcr2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu N, Pan T. RNA epigenetics. Transl Res. 2014 doi: 10.1016/j.trsl.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sibbritt T, Patel HR, Preiss T. Mapping and significance of the mRNA methylome. Wiley Interdiscip Rev RNA. 2013;4:397–422. doi: 10.1002/wrna.1166. [DOI] [PubMed] [Google Scholar]
- 10.Frankel AD, Smith HO. Restriction and modification enzymes detect no allosteric changes in DNA with bound lac repressor or RNA polymerase. J Mol Biol. 1981;146:611–619. doi: 10.1016/0022-2836(81)90049-8. [DOI] [PubMed] [Google Scholar]
- 11.Loenen WA. S-adenosylmethionine: jack of all trades and master of everything? Biochem Soc Trans. 2006;34:330–333. doi: 10.1042/BST20060330. [DOI] [PubMed] [Google Scholar]
- 12.Xhemalce B, Robson SC, Kouzarides T. Human RNA methyltransferase BCDIN3D regulates microRNA processing. Cell. 2012;151:278–288. doi: 10.1016/j.cell.2012.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23:781–783. doi: 10.1101/gad.1787609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dworkin AM, Huang TH, Toland AE. Epigenetic alterations in the breast: Implications for breast cancer detection, prognosis and treatment. Semin Cancer Biol. 2009;19:165–171. doi: 10.1016/j.semcancer.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006;5:60. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanwal R, Gupta S. Epigenetics and cancer. J Appl Physiol (1985) 2010;109:598–605. doi: 10.1152/japplphysiol.00066.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He C. Grand challenge commentary: RNA epigenetics? Nat Chem Biol. 2010;6:863–865. doi: 10.1038/nchembio.482. [DOI] [PubMed] [Google Scholar]
- 18.Bastien RR, Rodriguez-Lescure A, Ebbert MT, Prat A, Munarriz B, Rowe L, Miller P, Ruiz-Borrego M, Anderson D, Lyons B, Alvarez I, Dowell T, Wall D, Segui MA, Barley L, Boucher KM, et al. PAM50 breast cancer subtyping by RT-qPCR and concordance with standard clinical molecular markers. BMC Med Genomics. 2012;5:44. doi: 10.1186/1755-8794-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


