Abstract
BACKGROUND & AIM: p53 activation by cellular stresses induces the transcription of hundreds of its target genes. To elucidate the entire picture of its downstream pathway, we screened a cDNA microarray dataset of adriamycin-treated HCT116 p53−/− or p53+/+ cells and identified EPSIN 3 as a novel p53 target. METHODS: Potential p53 binding sequences in the EPSIN 3 locus were evaluated by reporter and CHIP assays. To investigate the role of EPSIN 3 in the p53 downstream pathway, we assessed DNA damage-induced apoptosis in EPSIN 3-knockdown HCT116 cells or Epsin 3-deficient mice. In addition, we evaluated EPSIN 3 expression levels in various tissues, including gastric adenocarcinoma, human gastric mucosa with or without Helicobacter pylori infection, and mouse acute gastritis tissues induced by indomethacin. RESULTS: In response to DNA damage, p53 induced the expression of EPSIN 3 through the p53 binding elements in the EPSIN 3 promoter and the first intron. Knockdown of EPSIN 3 resulted in resistance to DNA damage-induced apoptosis both in vitro and in vivo. EPSIN 3 expression was down-regulated in gastric cancer tissues compared with normal tissues. In addition, Helicobacter pylori infection and indomethacin-induced acute gastritis repressed EPSIN 3 expression in gastric mucosa. CONCLUSIONS: EPSIN 3 is a novel p53 target and a key mediator of apoptosis. Chronic or acute mucosal inflammation as well as p53 inactivation induced down-regulation of EPSIN 3 and subsequently caused apoptosis resistance, which is a hallmark of cancer cells.
Introduction
p53 is one of the most intensively studied tumor suppressor genes [1], [2], [3]. p53 activation by cellular stresses induces the transcription of hundreds of its target genes, which affect various processes, such as cell cycle arrest, senescence, apoptosis, and posttranscriptional modification [4], [5], [6]. Previously, our group identified a number of p53 target genes: p53AIP1, p53R2, p53RDL, XEDAR, PADI4, ISCU, and Cystatin C [6], [7], [8], [9], [10], [11], [12], [13], [14]. Despite research efforts, the full picture of the p53 downstream pathway remains unclear.
EPSINs are accessory proteins implicated in clathrin-mediated endocytosis by binding to ubiquitin moieties on the cytoplasmic regions of membrane proteins. In vertebrates, three family members (EPSIN 1–3) have been identified to date. EPSIN 1 and EPSIN 2 are ubiquitously expressed in most type of tissues and are involved in the internalization of membrane proteins such as Notch, the transferrin receptor, EGFR, protease-activating receptor 1 and VEGFR2 [15], [16]. Deletion of both Epsin 1 and Epsin 2 results in embryonic lethality, owing to a defective vascular phenotype, thus suggesting their important roles during development [17]. In cancer tissues, EPSIN 1 and EPSIN 2 are up-regulated and have been implicated in endocytosis of VEGFR2 and the Notch-Notch ligand complex, thus resulting in vascularization, proliferation, invasion and cancer metastasis [18], [19]. In contrast, EPSIN 3 is only expressed in parietal cells in the stomach and in wounded keratinocytes in the skin epithelium [20], [21], and its function in both normal and cancerous tissues has not yet been characterized. Here, through a transcriptome analysis of HCT116 p53+/+ and HCT116 p53−/− cells treated with adriamycin, we identified EPSIN 3 as a novel p53 target that is involved in the apoptotic pathway and gastric carcinogenesis.
Results
Induction of EPSIN 3 by Cellular Stress
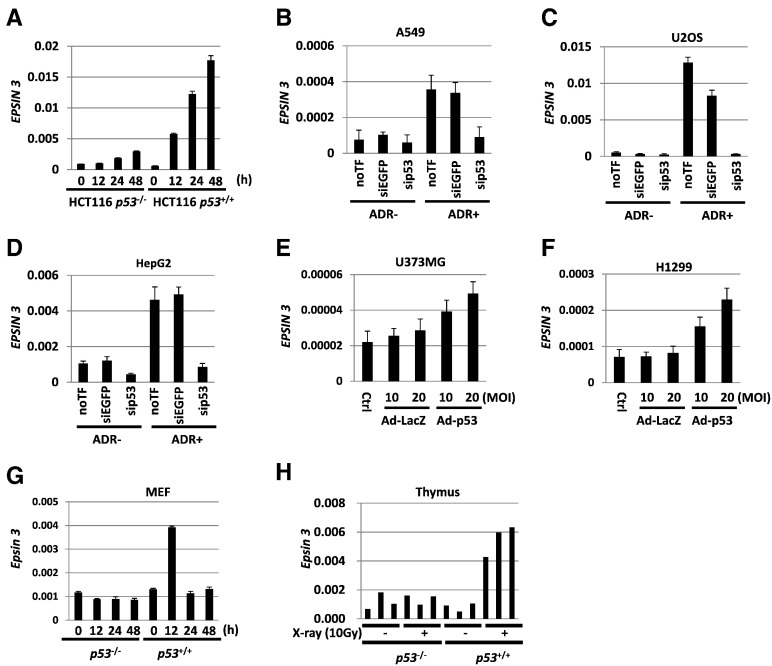
To identify novel p53 targets, we conducted a cDNA microarray analysis of mRNA isolated from HCT116 p53+/+ and HCT116 p53−/− cells that were treated with 2 μg/ml of adriamycin (ADR). Among the 22,310 human genes, we selected EPSIN 3 for further analysis because EPSIN 3 was markedly induced by ADR treatment only in HCT116 p53+/+ cells, and the other Epsin family proteins have not been reported to be p53 targets. The results of the cDNA microarray analysis were validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis (Figure 1A). We confirmed that the mRNA expression of EPSIN 3 was induced by ADR in other cell lines harboring wild-type p53: A549, U2OS and HepG2, and this induction was inhibited by knockdown of p53 (Figure 1, B–D and Supplementary Figure S1, A–C). We also found that EPSIN 3 mRNA expression was induced in U373 MG (p53-mutant) and H1299 (p53-deficient) cells infected with Ad-p53 but not in Ad-LacZ-infected cells (Figure 1, E and F).
Figure 1.
(A) qRT-PCR analysis of EPSIN 3 in HCT116 p53−/− and p53+/+ cells. After incubation with medium containing 2 μg/ml of ADR for 2 h, HCT116 p53−/− and p53+/+ cells were incubated with culture medium that did not contain ADR for the indicated hours. (B - D) A549, U2OS and HepG2 cells were transfected with sip53 24 h before treatment with ADR. siEGFP and no transfection (noTF) were used as controls. At 48 h after treatment with 2 μg/ml of ADR, total RNA was subjected to qRT-PCR. (E, F) qRT-PCR analysis of EPSIN 3 in U373 MG (p53 mutant) and H1299 (p53 null) cells infected with adenovirus expressing either p53 (Ad-p53) or LacZ (Ad-LacZ) at a multipli2city of infection (MOI) of 10 or 20. (F) qRT-PCR analysis of Epsin 3 expression in MEF cells obtained from p53−/− and p53+/+ mice. The cells were harvested at the indicated times after 2 μg/ml ADR treatment for 2 h. Error bars represent the S.D. (n = 3). Gapdh was used to normalize the expression levels. (H) qRT-PCR analysis of Epsin 3 expression in the thymus of X-ray-irradiated p53−/− and p53+/+ mice (10 Gy) (n = 3 per group). Mice were sacrificed 24 h after irradiation with 10 Gy of X-rays. β-actin was used to normalize the expression levels in all qRT-PCR analyses unless otherwise noted.
To further investigate the regulation of Epsin 3 by p53, we evaluated Epsin 3 expression in mouse embryonic fibroblast (MEF) cells of p53+/+ and p53−/− mice that were treated with 2 μg/ml of ADR. Epsin 3 mRNA expression was increased 12 h after ADR treatment in the MEFs of p53+/+ mice but not in the MEF cells of p53−/− mice (Figure 1G). Moreover, RNA purified from the thymi of p53+/+ and p53−/− mice that were irradiated with 10 Gy of X-rays was also subjected to qRT-PCR analysis. Epsin 3 mRNA expression was increased after X-ray irradiation in the thymi of p53+/+ mice but not in the thymi of p53−/− mice (Figure 1H). These results clearly demonstrated the regulation of EPSIN 3 by p53 in vitro and in vivo.
EPSIN 3 is a Direct Target of p53
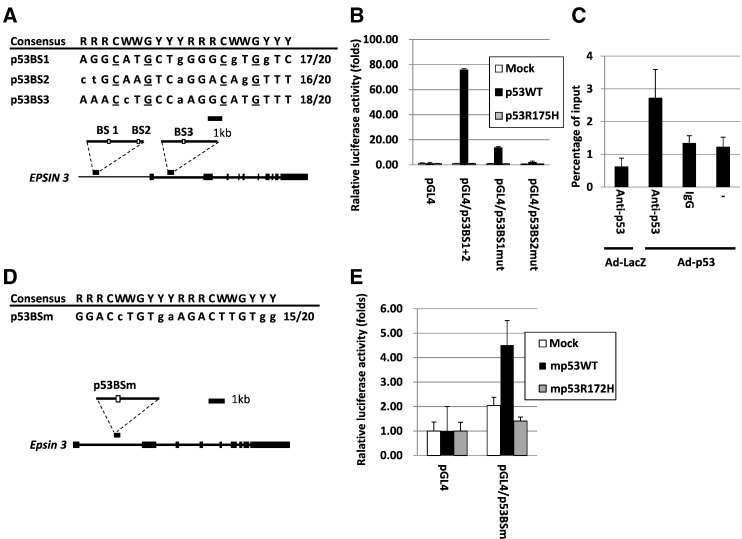
We surveyed the p53 binding sequence [22] within the EPSIN 3 locus and identified three potential binding sites (Figure 2A). A 449-base DNA fragment containing p53BS1 and p53BS2 and a 428-base DNA fragment containing p53BS3 were amplified and cloned into the pGL4.24 vector (pGL4.24/p53BS1+ 2 and pGL4.24/p53BS3, respectively). A reporter assay revealed that U373 MG cells transfected with pGL4.24/p53BS1+ 2 showed increased luciferase activity only in the presence of a plasmid designed to express wild-type p53 (Figure 2B and Supplementary Figure S2A). Mutant human p53 (p53R175H) was used as a negative control. However, base substitutions in either potential binding site (pGL4.24/p53BSmut) diminished the enhanced luciferase activity, particularly in the p53BS2 sites. To verify whether p53 directly binds to p53BS2, we performed a ChIP assay using U373 MG cells infected with either Ad-p53 or Ad-LacZ. After precipitation with an anti-p53 antibody, DNA fragments harboring p53BS2 were quantified with qRT-PCR assays. The results showed that p53 bound to the genomic region including the p53BS2 site in cells infected with Ad-p53 (Figure 2C). In addition, p53 was shown to bind to genomic region including p53BS3 in U-2 OS cells treated with etoposide [23], although p53BS3 exhibited lower transcriptional activity than p53BS1+ 2 (Supplementary Figure S2). These findings suggest that p53 would regulate EPSIN 3 expression through multiple p53 binding elements in the EPSIN 3 genome.
Figure 2.
(A, D) White boxes indicate the location of the potential p53 binding sequences (p53BS1, p53BS2, p53BS3 and p53BSm). Comparison of these potential binding sequences to the consensus p53 binding sequence. R, purine; W, A or T; Y, pyrimidine. Nucleotides identical to the consensus sequence are in capital letters. The underlined cytosine and guanine residues were substituted for thymine to examine the specificity of the p53 binding sequence. (B, E) Results of the luciferase assay of the genomic fragments containing p53BSs with or without amino acid substitutions at the p53BSs. The luciferase activity is indicated relative to the activity of the mock vector with S.D. (n = 3). Mutant p53 represents the plasmid expressing either human p53 with a missense mutation (R175H) or mouse p53 with a missense mutation (R172H) (C). U373 MG cells were used for these analyses.
We also found a potential p53-binding site (p53BSm) in the first intron of the mouse Epsin 3 gene (Figure 2D) and confirmed transactivation of the reporter gene by wild-type mouse p53 (Figure 2E and Supplementary Figure S2B). Mutant mouse p53 (mp53R172H) was used as a negative control. Thus, our findings clearly indicated that human and mouse Epsin 3 are novel p53 targets.
Cleavage of EPSIN 3 Protein
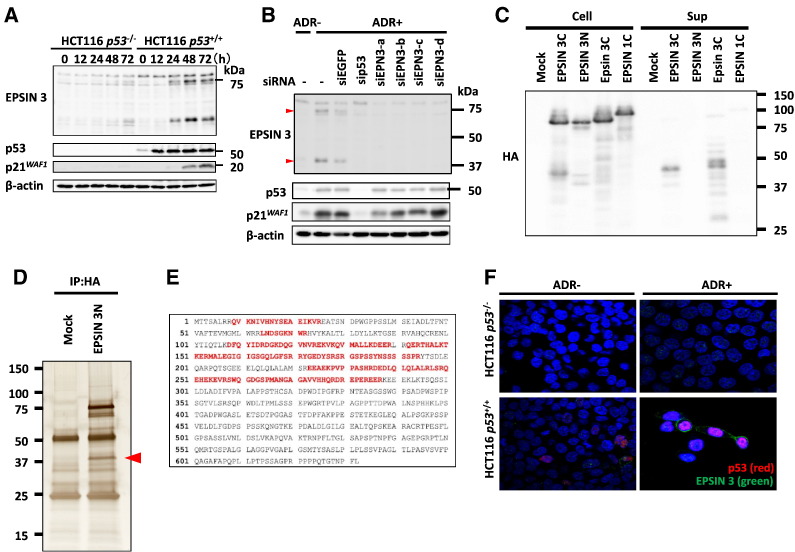
To investigate the induction of EPSIN 3 protein expression by p53, we performed western blot analysis using HCT116 p53−/− and HCT116 p53 +/+ cells that were treated with ADR. We found the induction of two protein bands at approximately 80 kDa and 40 kDa only in ADR-treated HCT116 p53+/+ cells (Figure 3A). Knockdown of EPSIN 3 by siRNAs (siEPN3-a-d) completely eliminated these two protein bands (Figure 3B).
Figure 3.
(A) HCT116 p53−/− and HCT116 p53+/+ cells were treated with 2 μg/ml ADR for 2 h and then incubated in normal medium. At the indicated times after the treatment, whole cell extracts were subjected to immunoblotting with anti-EPSIN 3, anti-p53, anti-p21, or anti β-actin antibodies. (B) We designed four small interfering RNAs (siRNAs): siEPSIN 3-a-d. HCT116 cells were transfected with each siRNA 24 h before treatment with ADR. siEGFP was used as a control. At 48 h after ADR treatment, whole cell extracts were subjected to western blot analysis. Red arrows indicate the protein bands of EPSIN 3. (C) At 24 h after HEK293T cells were transfected with one of the following expression plasmids: EPSIN 3 tagged with HA at its C-terminus (EPSIN 3C), EPSIN 3 tagged with HA at its N-terminus (EPSIN 3 N), mouse Epsin 3 tagged with HA at its C-terminus (Epsin 3C) or EPSIN 1 tagged with HA at its C-terminus (EPSIN 1), the culture medium was replaced with serum-free medium. After another 24 h, the medium was collected, and acetone-precipitated protein samples were subjected to western blot analysis. (D) HEK293T cells were transfected with either EPSIN 3 N or mock vector. At 36 h after transfection, whole cell lysates were subjected to immunoprecipitation with an anti-HA agarose antibody, and the precipitants were analyzed using SDS-PAGE and subsequent silver staining. The red arrow indicates the N-terminal segment of EPSIN 3. (E) The whole amino acid sequence of EPSIN 3 is shown. Red letters indicate peptide sequences detected by mass spectrometry analysis. (F) After incubation with medium containing 0 or 2 μg/ml of ADR for 2 h, HCT116 p53−/− and p53+/+ cells were incubated for 48 h with medium that did not contain ADR. Adherent cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. EPSIN 3 (green) and p53 (red) were visualized using specific primary antibodies and fluorescein-conjugated secondary antibodies. Nuclei were stained with DAPI (blue).
To further characterize these two proteins, we constructed plasmids expressing human EPSIN 3 tagged with hemagglutinin (HA) at either the carboxyl or amino-terminus (EPSIN 3C and EPSIN 3 N, respectively). In addition to the 80 kDa protein, the anti-HA antibody detected several protein bands at approximately 40 kDa in cells transfected with either EPSIN 3C- or EPSIN 3 N–expressing plasmids (Figure 3C). These findings suggested that cleavage of EPSIN 3 protein generates an N-terminal and C-terminal fragment of approximately 40 kDa each.
Epsin 3 protein has been shown to be included in exosomes purified from human urine samples [24]. Similar to this finding, the C-terminus of the EPSIN 3 protein was detected in the culture medium of HEK293T cells transfected with plasmid expressing either human EPSIN 3C or mouse Epsin 3C. Together, these results suggest that EPSIN 3 is likely to be cleaved into two fragments, and its C-terminal fragment is secreted into the culture medium.
To determine the cleavage site of the EPSIN 3 protein, we purified an N-terminal, 37 kDa fragment from HEK293T cells that were transfected with an EPSIN 3 N-expressing plasmid (Figure 3D). Mass spectrometry analysis of this band identified several peptide fragments spanning amino acids 9–288 of the EPSIN 3 protein (Figure 3E), thus indicating that EPSIN 3 is likely to be cleaved near amino acid position 288. However, immunocytochemistry indicated that EPSIN 3 was expressed on the plasma membrane of ADR-treated HCT116 p53+/+ cells 48 h after ADR treatment (Figure 3F, and Supplementary Figure S3). Together, our results suggest that EPSIN 3 is primarily localized at the plasma membrane, whereas its C-terminal fragment is secreted into the extracellular space.
Regulation of Cancer Cell Growth and Apoptosis by EPSIN 3
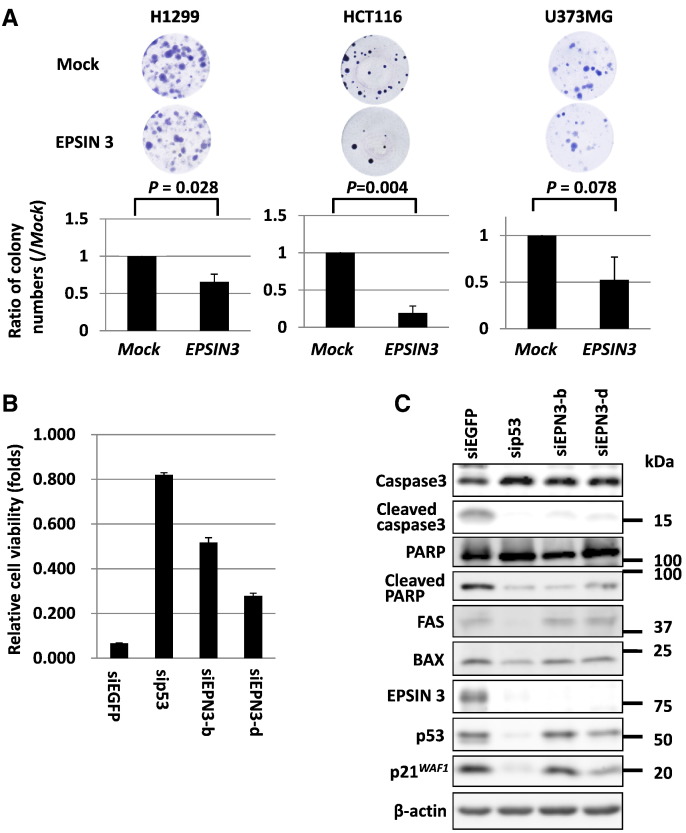
To explore the role of EPSIN 3 in the growth of cancer cells, we performed a colony formation assay using three cancer cell lines: H1299, HCT116, and U373 MG. The results showed that ectopic expression of EPSIN 3 significantly reduced tumor cell growth in H1299 and HCT116 cells compared with mock-transfected cells (Figure 4A). Then, we performed a cell proliferation assay using HCT116 cells transfected with either siEPN3-b or siEPN3-d. The results showed that knockdown of EPSIN 3 inhibited the ADR-induced suppression of growth (Figure 4B). We further examined the effect of EPSIN 3 on ADR-induced apoptosis. Interestingly, knockdown of EPSIN 3 in ADR-treated HCT116 cells reduced cleaved caspase-3 and cleaved PARP levels, thus indicating regulation of apoptosis by EPSIN 3 (Figure 4C).
Figure 4.
(A) H1299, HCT116 and U373 MG cells transfected with either an EPSIN 3-expressing plasmid or mock vector were cultured in medium containing 0.8, 0.5 and 0.5 mg/ml of geneticin, respectively, for 2–3 weeks. Error bars represent the S.D. (n = 3). The P value was calculated by using Student's t tests. (B) At 24 h after transfection of each siRNA, HCT116 cells were seeded and cultured on ultra-low-attachment plates. Then, 24 h after plating, HCT116 cells were treated with 1 μg/ml ADR for 48 h and subjected to cell proliferation analysis. Relative cell viability was calculated by dividing the absorbance of ADR-treated cells by that of untreated cells. Error bars represent the S.D. (n = 3). (C) Western blot analysis was performed using HCT116 cells treated as described above. siRNA against EGFP was used as a control.
To investigate the molecular mechanism whereby EPSIN 3 regulates the apoptotic pathway, we explored EPSIN 3-interacting proteins by using immunoprecipitation and subsequent mass spectrometry analysis. The results indicated an interaction between EPSIN 3 and clathrin heavy chain (CHC) (Supplementary Figure S4A), a component of vesicular transport that also interacts with other Epsin family members and p53 [25], [26], [27]. We also identified an interaction between EPSIN 3 and p53 (Supplementary Figure S4B). However, the majority of EPSIN 3 protein did not co-localize with p53 or clathrin in ADR-treated HCT116 cells (data not shown). Moreover, knockdown of EPSIN 3 did not affect the expression of FAS and BAX, two important pro-apoptotic targets of p53 (Figure 4C). Therefore, p53 is unlikely to be a major mediator of EPSIN 3-related apoptotic pathway regulation.
Because EPSIN 3 is localized at the plasma membrane, we assumed that EPSIN 3, similar to other family members, is involved in the endocytosis of membrane proteins and subsequently represses anti-apoptotic responses. To screen membrane proteins regulated by EPSIN 3, we quantified the levels of membrane proteins using ADR-treated HCT116 cells transfected with siEPSIN 3 or siEGFP. Membrane proteins were purified by a glycopeptide-targeted labeling method and subjected to mass-spectrometric analysis. As a result, several anti-apoptotic membrane proteins such as CD44, KLK6, EPHA3, and ROR1 were shown to be highly expressed in EPSIN 3-knockdown cells compared with siEGFP-transfected cells (Supplementary Table 1). These findings suggest that EPSIN 3 may enhance apoptosis of cancer cells through endocytosis of oncogenic receptor proteins.
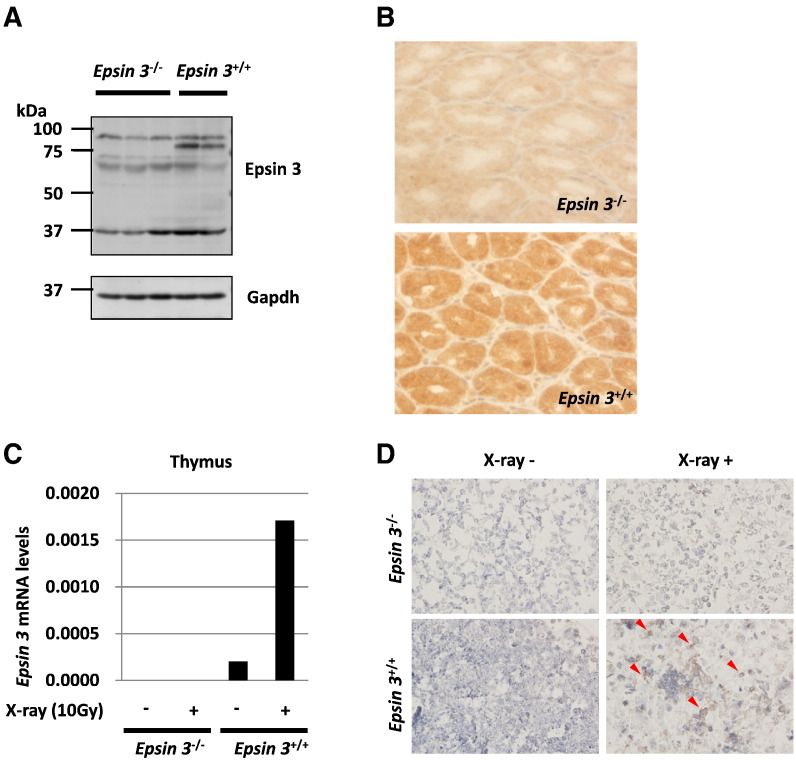
Then, we analyzed the involvement of Epsin 3 in DNA damage-induced apoptosis in vivo by using Epsin 3-deficient mice. We confirmed the lack of Epsin 3 expression in the stomachs of Epsin 3-knockout mice by western blotting and immunohistochemistry (Figure 5, A and B) because Epsin 3 has been shown to be highly expressed in gastric parietal cells. Consistent with the results of a previous report, Epsin 3-deficient mice showed no obvious phenotype [20]. Epsin 3 mRNA levels were markedly increased in the thymi of wild-type mice after X-ray irradiation, but these changes were not detectable in Epsin 3-deficient mice (Figure 5C). Next, we evaluated apoptotic cells by using a TUNEL assay and found that X-ray-induced apoptosis was decreased in the thymi of Epsin 3-deficient mice compared with wild-type mice (Figure 5D). Together, our results suggest that EPSIN 3 regulates the apoptotic pathway in response to genotoxic damage both in vitro and in vivo.
Figure 5.
(A) Western blot analysis of Epsin 3 protein expression in the stomachs of X-ray-irradiated Epsin 3−/− and Epsin 3+/+ mice (10 Gy) (n = 2–3 per group). The red arrow indicates the Epsin 3 protein band. Gapdh was used as the loading control. (B) Immunohistochemical analysis of Epsin 3 protein expression in the stomachs of X-ray-irradiated Epsin 3−/− and Epsin 3+/+ mice (10 Gy). All mice were sacrificed 24 h after irradiation. (C) qRT-PCR analysis of Epsin 3 in the thymi of X-ray-irradiated Epsin 3−/− and Epsin 3+/+ mice (10 Gy, 24 h) (n = 1 per group). β-actin was used to normalize the expression levels. (D) Apoptotic cells in the thymi of Epsin 3−/− and Epsin 3+/+ mice were evaluated using TUNEL assays 48 h after exposure to 3 Gy of X-ray irradiation. Red arrows indicate TUNEL-positive cells.
Dysregulation of EPSIN 3 Expression in Stomach Tissues
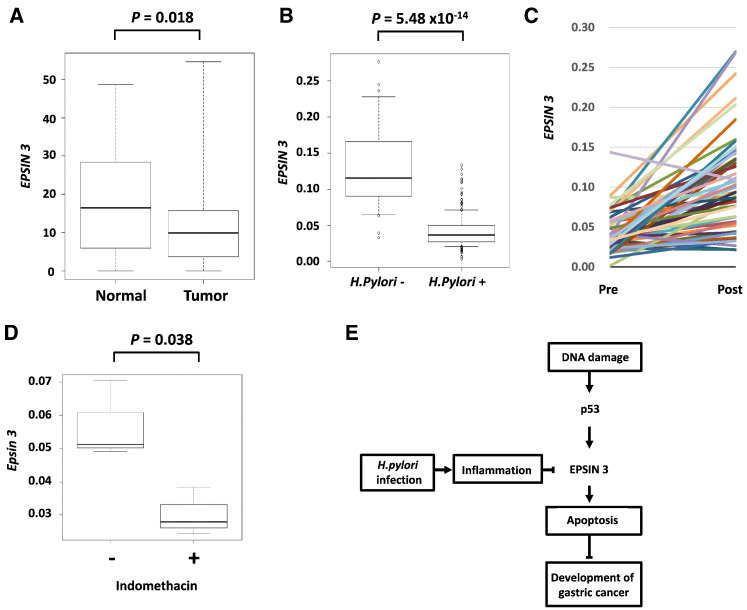
To investigate the role of EPSIN 3 in human carcinogenesis, we examined its expression by using RNA sequencing data of stomach adenocarcinoma tissues downloaded from the TCGA database [28]. Notably, the expression of EPSIN 3 was significantly decreased in stomach adenocarcinoma tissues compared with corresponding normal tissues (Figure 6A). However, EPSIN 3 expression was not significantly associated with overall survival (P = .37; log-rank test, Supplementary Figure S5). These results indicate that EPSIN 3 is mainly associated with the onset of disease but not with disease progression.
Figure 6.
(A) Using the RNA sequence data of the in human gastric cancer tissues obtained from the database of The Cancer Genome Atlas (TCGA), gene expression analysis was performed. Box-plots indicate EPSIN 3 expression in stomach tissues. The vertical axis indicates the normalized expression level of EPSIN 3, the top bar represents the maximum observed value, the lower bar represents the minimum observed value, the top of the box is the upper or third quartile, the bottom of the box is the lower or first quartile, and the middle bar is the median value. The P value was calculated by using Mann–Whitney U tests. (B) qRT-PCR analysis of EPSIN 3 mRNA levels in biopsy samples from the gastric mucosa in the absence (n = 28) or presence (n = 214) of H. pylori. The vertical axis indicates the expression level of EPSIN 3 normalized against β-actin expression, the top bar represents 90% of the maximum observed value, the lower bar represents 10% of the maximum observed value, the top of the box is the upper or third quartile, the bottom of the box is the lower or first quartile, and the middle bar is the median value. The P value was calculated by a Mann–Whitney U test. (C) Comparison of the expression levels of EPSIN 3 in each individual patient (n = 54) before and after H. pylori eradication. The vertical axis indicates the expression level of EPSIN 3 normalized against β-actin expression. (D) qRT-PCR analysis of Epsin 3 mRNA levels in the stomachs of mice that were sacrificed 24 h after treatment with 40 mg/kg of intraperitoneal indomethacin (n = 3). The vertical axis indicates the expression level of EPSIN 3 normalized against β-actin expression. The P value was calculated by using Student's t test. (E) Scheme of EPSIN 3 mediated gastric carcinogenesis. EPSIN 3 is inactivated in the stomach by p53 inactivation and H. pylori infection, and its inactivation causes apoptosis resistance, a hallmark of cancer cells.
Because substantial evidence has indicated that chronic infection with Helicobacter pylori (H. pylori) is associated with the development of gastric cancer, we evaluated whether H. pylori infection could alter EPSIN 3 expression. Interestingly, EPSIN 3 mRNA levels were decreased in the gastric mucosa from patients with H. pylori infection compared with the gastric mucosa from H. pylori-negative control subjects (Figure 6B). Moreover, EPSIN 3 expression levels were increased after eradication of H. pylori in 50 of 54 (92.6%) patients (Figure 6C). We also evaluated Epsin 3 expression in a mouse model of acute inflammation induced by intraperitoneal injection of indomethacin. As a result, Epsin 3 expression was significantly decreased in mouse stomachs after indomethacin treatment (Figure 6D). These findings indicated that either p53 inactivation or mucosal inflammation caused by H. pylori or indomethacin induces down-regulation of EPSIN 3 expression and subsequently causes apoptosis resistance, which is a hallmark of cancer cells (Figure 6E).
Discussion
Here, we found that EPSIN 3 is a mediator of p53-induced apoptosis. In addition, we observed that EPSIN 3 expression was significantly decreased in gastric cancer tissues and the gastric mucosa in people infected with H. pylori. Epsin family members share structural similarities: an ENTH domain at the N-terminus, 2 ubiquitin interacting motifs (UIMs), a DPW motif, 2 or 3 clathrin binding motifs in the central region, and an EH domain at the C-terminus. All of these structures work cooperatively and lead to membrane invagination at an initial step of endocytosis. Although several lines of evidence have suggested that EPSIN 1 and 2 promote carcinogenesis [18], [19], our results indicated a tumor suppressive function of EPSIN 3 in both humans and mice. Interestingly, both human and mouse Epsin 3 are cleaved in the central part the protein, and the C-terminal fragments (which no longer contain ENTH and UIM domains essential for initiation of endocytosis) are secreted into the extracellular space. EPSIN 3 is located at the plasma membrane, and a previous proteome analysis identified Epsin 3 in human urine exosomes, which contain various membrane proteins. We also evaluated the effect of cleaved EPSIN 3 on tumor cell growth by using conditioned medium from HEK293T cells transfected with EPSIN 3-expressing plasmid. However, EPSIN 3-containing medium did not show a growth suppressive effect (data not shown), indicating that the full-length intracellular EPSIN 3 protein likely plays a major role in EPSIN 3-mediated growth suppression.
In our analysis, we demonstrated that EPSIN 3 mediates apoptotic signaling in cancer cells. DNA damage induces the intrinsic apoptotic signaling cascade, in which a number of genes, including known p53 targets, form a complicated network [29]. EPSIN 3 is induced at later time points (48 h after adriamycin treatment) like the p53-induced pro-apoptotic protein p53AIP1 [12]. Notably, knockdown of EPSIN 3 abrogated the cleavage of caspase-3 and PARP to the same degree as knockdown of p53, thus suggesting that EPSIN 3 is involved in a key step in the p53-caspase-3 axis.
Our results indicated that EPSIN 3 regulates the expression of anti-apoptotic membrane proteins, such as CD44, KLK6, EPHA3, and ROR1. Among them, proteinase-activated receptor 1 (PAR1), a G protein-coupled receptor for thrombin, was shown to be up-regulated in cancer cells [30]. Aberrant endocytosis of the receptor causes persistent downstream signaling, promoting invasion of cancer cells, and silencing of PAR1 leads to apoptosis of tumor cells in a mouse xenograft model [31]. Taken together, EPSIN 3 may enhance apoptosis of cancer cells through endocytosis of oncogenic receptor proteins.
We also found that EPSIN 3 expression was decreased in gastric cancer tissues as well as in gastric mucosa infected with H. pylori. In addition, EPSIN 3 expression was increased after H. pylori eradication. Interestingly, EPSIN 3 expression was also reduced in a mouse model of acute inflammation caused by intraperitoneal indomethacin injection. Chronic inflammation caused by H. pylori infection may be a major cause of EPSIN 3 down-regulation in gastric mucosa. Because EPSIN 3 is a key mediator of DNA damage-induced apoptosis and p53 is the most frequently mutated gene in gastric cancer [28], [32], [33], [34], the p53-EPSIN 3 pathway might play an important role in gastric carcinogenesis.
Materials and Methods
cDNA Microarray
Gene expression analysis was performed using a SurePrint G3 Human GE 8x60K microarray (Agilent, Santa Clara, CA, USA) according to the manufacturer's protocol, as described previously [13]. Briefly, either HCT116 p53+/+ or HCT116 p53−/− cells were treated with ADR and incubated at 37°C until the time of harvest. Total RNA was isolated from the cells by using standard protocols. Each RNA sample was labeled and hybridized to the array slides.
Cell Culture and Transfection
The human embryonic kidney cell line HEK293T was obtained from Riken Cell Bank. The human cancer cell lines U373 MG (astrocytoma), H1299 (non-small cell lung cancer), and HCT116 (colorectal adenocarcinoma) were purchased from American Type Culture Collection. Human mammary epithelial cells MCF10A p53+/+ or MCF10A p53−/− were purchased from Sigma Aldrich (St. Louis, MO, USA). HCT116 p53+/+ and HCT116 p53−/− cell lines were generously provided by B. Vogelstein (Johns Hopkins University, Baltimore, MD, USA). HEK293T and U373 MG cells were transfected with plasmids by using Fugene6 (Promega, Madison, WI, USA) and Lipofectamine LTX (Invitrogen, Carlsbad, CA, USA), respectively. Small interfering RNA (siRNA) oligonucleotides, which were commercially synthesized by Sigma Genosys, were transfected with Lipofectamine RNAiMAX reagent (Invitrogen). Sequences of the siRNA oligonucleotides used are shown in Supplementary Table 2.
Cell Treatments
We generated and purified replication-deficient recombinant viruses expressing either p53 (Ad-p53) or LacZ (Ad-LacZ), as described previously [12]. U373 MG (p53-mutant) and H1299 (p53-null) cells were infected with viral solutions at various amounts of multiplicity of infection (MOI) and incubated at 37°C until the time of harvest. To induce genotoxic stress, cells were incubated with 2 μg/ml of adriamycin (ADR) for 2 h.
Antibodies
Anti-actin monoclonal antibody (AC15), anti-EPSIN 3 polyclonal antibody (HPA055546), and anti-HA agarose monoclonal antibody (HA-7) were purchased from Sigma-Aldrich. Anti-p21 monoclonal antibody (OP64) and anti-p53 monoclonal antibody (OP43) were purchased from Merck Millipore. Anti-caspase-3 monoclonal antibody (8G10) and anti-cleaved caspase-3 monoclonal antibody (5A1E) were purchased from Cell Signaling Technology (Beverly, MA, USA).
Knockout Mice
p53-deficient mice were provided by the RIKEN BioResource Center (Ibaraki, Japan) [35]. Epsin 3-deficient mice were generated by in vitro fertilization using Epsin 3-deficient sperm purchased from KOMP (Knockout Mouse Project: https://www.komp.org/). Genotypes were confirmed by PCR analysis. The primer sequences used are indicated in Supplementary Table 2. All mice were maintained under specific pathogen-free conditions and were handled in accordance with the Guidelines for Animal Experiments of the Institute of Medical Science (University of Tokyo, Tokyo, Japan). Mice were irradiated using an X-ray irradiation system (MBR-1520R-3, Hitachi, Tokyo, Japan).
Quantitative Real-Time PCR
Total RNA was isolated from human cells and mouse tissues by using RNeasy Plus Mini Kits (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Complementary DNAs were synthesized using Super Script III reverse transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) was conducted using SYBR Green Master Mix on a Light Cycler 480 (Roche, Basel, Switzerland). Absolute copy numbers were calculated using serial dilutions of a plasmid including a cDNA fragment as a standard. Expression of each mRNA was normalized against that of β-actin or Gapdh. The primer sequences used are shown in Supplementary Table 2.
Gene Reporter Assay
DNA fragments, including the potential p53-binding sites (p53BS), were amplified and cloned into the pGL4.24-promoter vector (Promega). The primers used for amplification are shown in Supplementary Table 2. Site-directed mutagenesis was performed to introduce “T” point mutations replacing the “C”s at the 4th and the 14th nucleotide and the “G”s at the 7th and the 17th nucleotide of the p53BS (Supplementary Table 2). Reporter assays were performed using a Dual Luciferase assay system (Promega), as described previously [8].
Chromatin Immunoprecipitation Assay
ChIP assays were performed using an EZ-Magna ChIP G - Chromatin Immunoprecipitation Kit (Merck Millipore, Darmstadt, Germany) by following the manufacturer's protocol. In brief, U373 MG cells infected with either Ad-p53- or Ad-LacZ at an MOI of 10 were cross-linked with 1% formaldehyde for 10 min, washed with PBS, and lysed in nuclear lysis buffer. The lysate was then sonicated using a Bioruptor UCD-200 (CosmoBio, Tokyo, Japan) to shear the DNA to approximately 200 to 1000 bps. The supernatant from 1 × 106 cells was used for each immunoprecipitation with either anti-p53 antibody (OP140, Merck Millipore) or normal mouse IgG (sc-2025, Santa Cruz, Santa Cruz, CA, USA). Before immunoprecipitation, 2% of the supernatant was removed and designated “input”. Column-purified DNA was quantified by qRT-PCR. The primer sequences used are shown in Supplementary Table 2.
Plasmid Construction
The entire coding sequence of the EPSIN 3 cDNA was amplified by PCR using KOD-Plus DNA polymerase (Toyobo, Osaka, Japan) and inserted into the EcoRI and XhoI sites of the pCAGGS vector. The construct sequence was confirmed by DNA sequencing analysis. The primers used for amplification are shown in Supplementary Table 2.
Western Blot Analysis
To prepare whole cell extracts, cells were collected and lysed in chilled RIPA buffer (50 mmol/L Tris–HCl at pH 8.0, 150 mmol/L NaCl, 0.1% SDS, 0.5% sodium deoxycholate, and 1% NP40) containing 1 mM phenylmethylsulfonyl fluoride (PMSF), 0.1 mM DTT and 0.1% Calbiochem Protease Inhibitor Cocktail Set III, EDTA-Free (EMD Chemicals Inc., Merck KGaA, Darmstadt, Germany). The samples were sonicated for 15 min with a 30-sec on/30-sec off cycle using a Bioruptor UCD-200. To prepare the tissue extracts, frozen tissues were placed in a 2 ml tube containing 14 K beads with the lysis buffer described above and homogenized twice for 30 sec at 6000 rpm in a Precellys 24 Dual system (Bertin Technologies, France). After centrifugation at 16000 × g for 15 min, the supernatants were collected and boiled in SDS sample buffer (Bio-Rad, Hercules, CA, USA). SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed for each sample, and the proteins were then transferred to a nitrocellulose membrane (Hybond™ ECL™, Amersham, Piscataway, NJ, USA). Protein bands on the western blots were visualized by chemiluminescent detection (ECL, Amersham).
Condensation of Secreted Proteins in Media
At 24 h after transfection of the expression plasmid into HEK293T cells, the culture medium was replaced with serum-free medium, and after another 24 h, the medium was collected. The same volume of cold acetone was added to the medium and incubated at −80°C for 2 h. After centrifugation, the precipitant was suspended with SDS sample buffer (Bio-Rad).
Immunoprecipitation
At 36 h after transfection of the expression plasmid into HEK293T cells, cells were collected and lysed in chilled lysis buffer (50 mmol/L Tris–HCl (pH 8.0), 150 mmol/L NaCl, and 0.5% NP40). Lysates were precleared by incubation with protein G-Sepharose 4B (Invitrogen) and mouse IgG at 4°C for 1 h. Then, precleared cell extracts were incubated overnight with an anti-HA agarose antibody at 4°C. The beads were washed four times with 1 ml of ice-cold lysis buffer, and the immunoprecipitated proteins were released from the beads by boiling in sample buffer for 2 min.
Quantification of Membrane Proteins by Mass Spectrometric Analysis
The siEPN3-b or siEPN3-d vector was transfected into HCT116 cells. After 24 h, cells were treated with 2 μg/ml ADR for 2 h. Then, cells were lysed with a solution of 8 M urea and 50 mM ammonium bicarbonate, reduced in 20 mM Tris(2-carboxyethyl)phosphine with 50 mM ammonium bicarbonate for 30 min at 37°C and alkylated in 50 mM iodoacetamide with 50 mM ammonium bicarbonate for 45 min in the dark at 25°C. Trypsin GOLD solution was added at a ratio of 1/50 (w/w) enzyme to protein and incubated at 37°C for 16 h. The resulting peptides were desalted with an Oasis HLB 1 ml cartridge (Waters) and dried with a vacuum spin dryer. To enrich glycopeptides, peptides were dissolved in PBS and incubated with 50 μl of Con-A agarose beads (Seikagaku Corporation, Japan) at 25°C for 60 min. Following 4 washes with PBS, N-glycosylated peptides were specifically eluted by incubation with 1 unit of PNGase-F solution (Sigma) at 37°C for 6 h. The released de-glycosylated peptides were finally analyzed with an LTQ-Orbitrap-Velos LC/MS system and subjected to database search analysis in which amino acid substitution (Asn > Asp) was added to the dynamic modification setting in Proteome Discoverer software to determine de-glycosylation sites generated by PNGase-F.
Identification of the Cleavage Site of EPSIN 3
Immunoprecipitates bound to the anti-HA antibody-conjugated agarose beads were separated on SDS-PAGE and silver stained. The excised EPSIN 3 bands were reduced in 10 mM Tris(2-carboxyethyl)phosphine (Sigma-Aldrich) with 50 mM ammonium bicarbonate (Sigma-Aldrich) for 30 min at 37°C and alkylated in 50 mM iodoacetamide (Sigma-Aldrich) with 50 mM ammonium bicarbonate for 45 min in the dark at 25°C. Trypsin GOLD (Promega) solution was added at a ratio of 1/50 (w/w) enzyme to protein and incubated at 37°C for 16 h. The resulting peptides were extracted from the gel and analyzed with an LTQ-Orbitrap-Velos mass spectrometer (Thermo Scientific) combined with an UltiMate 3000 RSLC nanoflow HPLC system (Thermo Scientific). The MS/MS spectra were compared to a Homo sapiens protein sequence database in SwissProt by using Proteome Discoverer 1.4 software (Thermo Scientific), in which a false discovery rate of 1% was set for both peptide and protein identification filters.
Immunocytochemistry
Adherent cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. The cells were then submerged in blocking solution (3% BSA/PBS containing 0.2% Triton X-100) for 1 h at room temperature and incubated with either rabbit anti-EPSIN 3 antibody (diluted 1:500) or mouse anti-p53 antibody (diluted 1:1000) in blocking solution overnight at 4°C. Primary antibodies were stained with goat anti-rabbit or goat anti-mouse secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 594, respectively, (each diluted 1:2000) for 1 h at room temperature, stained with DAPI and visualized on a FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan).
Colony Formation Assay
Colony formation assays were performed in six-well culture plates. Cells transfected with either pCAGGS/EPSIN 3 or mock plasmid were cultured in the presence of geneticin (Invitrogen) for 2–3 weeks. The colonies were stained with crystal violet (Sigma) and scored using ImageJ software.
Cell Proliferation and Apoptosis Analysis
HCT116 cells were seeded on cell culture dishes coated with polyethyleneimine and transfected with siRNA. At 24 h after transfection, the cells were transferred to ultra-low-attachment plates (Corning, NY, USA). After another 24 h, the cells were treated with 1 μg/ml of ADR for 48 h. To assess cell viability, cells were subjected to an ATP measurement assay using a Cell Titer-Glo Luminescent Viability Assay (Promega) according to the manufacturer's protocol. For the detection of apoptosis, cells treated as described above were subjected to western blot analysis with antibodies against pro-caspase-3 and cleaved caspase-3.
Immunohistochemistry
Stomach sections from 6-week-old mice were formalin-fixed and paraffin-embedded and subjected to immunohistochemistry. After deparaffinization with xylene, antigens were retrieved at 125°C for 30 sec in DAKO Target Retrieval Solution (EDTA buffer, pH 6.0). Intrinsic peroxidases and proteins were blocked. Anti-Epsin 3 antibody diluted to 1/150 in DAKO Antibody Diluent was added to the samples and incubated for 1 h at 37°C. The samples were subsequently treated with Envision Plus anti-rabbit IgG as a secondary antibody for 1 h at 37°C. A chromogen substrate was added, and the specimens were counterstained with hematoxylin.
TUNEL Assay
Frozen sections of thymus tissues were obtained from either Epsin 3-deficient or wild-type mice 24 h after exposure to 3 Gy of X-ray irradiation, and apoptotic cells were visualized by in situ end labeling using an Apoptosis In Situ Detection Kit (Wako, Osaka, Japan) according to the manufacturer's protocol.
Database Analysis
Data on EPSIN 3 expression levels in gastric cancer samples and the related clinical information were downloaded from the TCGA project via a data portal on 15 May 2015. The expression levels in normal tissues and tumors were compared using Mann–Whitney U tests. The overall survival rate of patients with gastric cancer was compared using the Kaplan–Meier method between two groups stratified according to the expression levels of EPSIN 3 in gastric cancer tissues (below or above the median expression level among the cohort).
Collection of Biopsy Samples of Gastric Mucosa
Tissues samples were obtained from patients who underwent esophagogastroduodenoscopy (EGD) at the Toyoshima Endoscopy Clinic, Tokyo, Japan. We collected mucosal tissue from the gastric angulus via biopsy. H. pylori infection was determined by a positive result in the 13C–urea breath test (UBIT, Otsuka, Tokushima, Japan), stool antigen test, or a combination of serum immunoglobulin G antibody test (E-plate, Eiken, Tokyo, Japan) and pathological evaluation (hematoxylin and eosin staining). Participants infected with H. pylori received eradication therapy. At least 8 weeks after the completion of eradication therapy, successful eradication was confirmed by either a negative 13C–urea breath test or a negative stool antigen test [36]. Then, a second EGD was performed 6 months to 1 year after the first EGD performed at the time of enrollment, and mucosal tissues were again collected from the gastric angulus by biopsy. RNA purified from the remaining tissues were subjected to qRT-PCR analysis. This research project was approved by the institutional review board at the Institute of Medical Science, University of Tokyo. All participants provided written, informed consent.
Author Contributions
J. Mori, C. Tanikawa, Y. Funauchi, and K. Matsuda initiated the project. J. Mori conducted most of the experiments and data analysis. O. Toyoshima collected gastric mucosal samples. K. Ueda and N. Ohnishi conducted LC–MS/MS analysis. J Mori, C. Tanikawa, and K. Matsuda prepared the manuscript.
The following are the supplementary data related to this article.
List of membrane proteins increased in EPSIN 3-knockdown cells
Sequences of DNA and RNA oligonucleotides
Supplementary Figures
Acknowledgments
This work was supported in part by a grant from the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology of Japan to K. Matsuda and C. Tanikawa; a grant from the Japan Agency for Medical Research and Development to K. Matsuda and C. Tanikawa; a grant from the Ministry of Health, Labor and Welfare of Japan to K. Matsuda; and a grant from the Takeda Science Foundation to K. Matsuda and C. Tanikawa.
Footnotes
Disclosure of Potential Conflicts of Interest: The authors declare no potential conflicts of interest.
References
- 1.Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science. 1989;244:217–221. doi: 10.1126/science.2649981. [DOI] [PubMed] [Google Scholar]
- 2.Eliyahu D, Michalovitz D, Eliyahu S, Pinhasi-Kimhi O, Oren M. Wild-type p53 can inhibit oncogene-mediated focus formation. Proc Natl Acad Sci U S A. 1989;86:8763–8767. doi: 10.1073/pnas.86.22.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finlay CA, Hinds PW, Levine AJ. The p53 proto-oncogene can act as a suppressor of transformation. Cell. 1989;57:1083–1093. doi: 10.1016/0092-8674(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 4.Caron de Fromentel C, Soussi T. TP53 tumor suppressor gene: a model for investigating human mutagenesis. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 5.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanikawa C, Ueda K, Nakagawa H, Yoshida N, Nakamura Y, Matsuda K. Regulation of protein Citrullination through p53/PADI4 network in DNA damage response. Cancer Res. 2009;69:8761–8769. doi: 10.1158/0008-5472.CAN-09-2280. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 8.Tanikawa C, Matsuda K, Fukuda S, Nakamura Y, Arakawa H. p53RDL1 regulates p53-dependent apoptosis. Nat Cell Biol. 2003;5:216–223. doi: 10.1038/ncb943. [DOI] [PubMed] [Google Scholar]
- 9.Tanikawa C, Furukawa Y, Yoshida N, Arakawa H, Nakamura Y, Matsuda K. XEDAR as a putative colorectal tumor suppressor that mediates p53-regulated anoikis pathway. Oncogene. 2009;28:3081–3092. doi: 10.1038/onc.2009.154. [DOI] [PubMed] [Google Scholar]
- 10.Funauchi Y, Tanikawa C, Yi Lo PH, Mori J, Daigo Y, Takano A, Miyagi Y, Okawa A, Nakamura Y, Matsuda K. Regulation of iron homeostasis by the p53-ISCU pathway. Sci Rep. 2015;5:16497. doi: 10.1038/srep16497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mori J, Tanikawa C, Funauchi Y, Lo PH, Nakamura Y, Matsuda K. Cystatin C as a p53-inducible apoptotic mediator that regulates cathepsin L activity. Cancer Sci. 2016;107:298–306. doi: 10.1111/cas.12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 13.Koguchi T, Tanikawa C, Mori J, Kojima Y, Matsuda K. Regulation of myo-inositol biosynthesis by p53-ISYNA1 pathway. Int J Oncol. 2016;48:2415–2424. doi: 10.3892/ijo.2016.3456. [DOI] [PubMed] [Google Scholar]
- 14.Tanikawa C, Nakagawa H, Furukawa Y, Nakamura Y, Matsuda K. CLCA2 as a p53-inducible senescence mediator. Neoplasia. 2012;14:141–149. doi: 10.1593/neo.111700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazazic M, Bertelsen V, Pedersen KW, Vuong TT, Grandal MV, Rodland MS, Traub LM, Stang E, Madshus IH. Epsin 1 is involved in recruitment of ubiquitinated EGF receptors into clathrin-coated pits. Traffic. 2009;10:235–245. doi: 10.1111/j.1600-0854.2008.00858.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen B, Dores MR, Grimsey N, Canto I, Barker BL, Trejo J. Adaptor protein complex-2 (AP-2) and Epsin-1 mediate protease-activated receptor-1 internalization via phosphorylation- and ubiquitination-dependent sorting signals. J Biol Chem. 2011;286:40760–40770. doi: 10.1074/jbc.M111.299776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Ko G, Zatti A, Di Giacomo G, Liu L, Raiteri E, Perucco E, Collesi C, Min W, Zeiss C. Embryonic arrest at midgestation and disruption of Notch signaling produced by the absence of both Epsin 1 and Epsin 2 in mice. Proc Natl Acad Sci U S A. 2009;106:13838–13843. doi: 10.1073/pnas.0907008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tessneer KL, Pasula S, Cai X, Dong Y, Liu X, Yu L, Hahn S, McManus J, Chen Y, Chang B. Endocytic adaptor protein Epsin is elevated in prostate cancer and required for cancer progression. ISRN Oncol. 2013;2013:420597. doi: 10.1155/2013/420597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasula S, Cai X, Dong Y, Messa M, McManus J, Chang B, Liu X, Zhu H, Mansat RS, Yoon SJ. Endothelial Epsin deficiency decreases tumor growth by enhancing VEGF signaling. J Clin Invest. 2012;122:4424–4438. doi: 10.1172/JCI64537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko G, Paradise S, Chen H, Graham M, Vecchi M, Bianchi F, Cremona O, Di Fiore PP, De Camilli P. Selective high-level expression of Epsin 3 in gastric parietal cells, where it is localized at endocytic sites of apical canaliculi. Proc Natl Acad Sci U S A. 2010;107:21511–21516. doi: 10.1073/pnas.1016390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spradling KD, McDaniel AE, Lohi J, Pilcher BK. Epsin 3 is a novel extracellular matrix-induced transcript specific to wounded epithelia. J Biol Chem. 2001;276:29257–29267. doi: 10.1074/jbc.M101663200. [DOI] [PubMed] [Google Scholar]
- 22.el- Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 23.Smeenk L, van Heeringen SJ, Koeppel M, Gilbert B, Janssen-Megens E, Stunnenberg HG, Lohrum M. Role of p53 serine 46 in p53 target gene regulation. PLoS One. 2011;6:e17574. doi: 10.1371/journal.pone.0017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol. 2009;20:363–379. doi: 10.1681/ASN.2008040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drake MT, Downs MA, Traub LM. Epsin binds to clathrin by associating directly with the clathrin-terminal domain. Evidence for cooperative binding through two discrete sites. J Biol Chem. 2000;275:6479–6489. doi: 10.1074/jbc.275.9.6479. [DOI] [PubMed] [Google Scholar]
- 26.Rosenthal JA, Chen H, Slepnev VI, Pellegrini L, Salcini AE, Di Fiore PP, De Camilli P. The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J Biol Chem. 1999;274:33959–33965. doi: 10.1074/jbc.274.48.33959. [DOI] [PubMed] [Google Scholar]
- 27.Enari M, Ohmori K, Kitabayashi I, Taya Y. Requirement of clathrin heavy chain for p53-mediated transcription. Genes Dev. 2006;20:1087–1099. doi: 10.1101/gad.1381906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 30.Bar-Shavit R, Turm H, Salah Z, Maoz M, Cohen I, Weiss E, Uziely B, Grisaru-Granovsky S. PAR1 plays a role in epithelial malignancies: transcriptional regulation and novel signaling pathway. IUBMB Life. 2011;63:397–402. doi: 10.1002/iub.452. [DOI] [PubMed] [Google Scholar]
- 31.Booden MA, Eckert LB, Der CJ, Trejo J. Persistent signaling by dysregulated thrombin receptor trafficking promotes breast carcinoma cell invasion. Mol Cell Biol. 2004;24:1990–1999. doi: 10.1128/MCB.24.5.1990-1999.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kakiuchi M, Nishizawa T, Ueda H, Gotoh K, Tanaka A, Hayashi A, Yamamoto S, Tatsuno K, Katoh H, Watanabe Y. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 33.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 34.Lin Y, Wu Z, Guo W, Li J. Gene mutations in gastric cancer: a review of recent next-generation sequencing studies. Tumour Biol. 2015;36:7385–7394. doi: 10.1007/s13277-015-4002-1. [DOI] [PubMed] [Google Scholar]
- 35.Tsukada T, Tomooka Y, Takai S, Ueda Y, Nishikawa S, Yagi T, Tokunaga T, Takeda N, Suda Y, Abe S. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene. 1993;8:3313–3322. [PubMed] [Google Scholar]
- 36.Malfertheiner P, Megraud F, O'Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T. Management of Helicobacter pylori infection--the Maastricht IV/Florence Consensus Report. Gut. 2012;61:646–664. doi: 10.1136/gutjnl-2012-302084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of membrane proteins increased in EPSIN 3-knockdown cells
Sequences of DNA and RNA oligonucleotides
Supplementary Figures