Abstract
Objective:
Whereas few adenomas become cancer, most colorectal cancers arise from adenomas. Telomere length is a recognized biomarker in multiple cancers, and telomere maintenance mechanisms (TMM) are exploited by malignant cells. We sought to determine whether telomere length and TMM distinguish cancer-associated adenomas from those that are cancer-free.
Methods:
Tissues were identified as cancer-adjacent polyp (CAP)—residual adenoma contiguous with cancer—and cancer-free polyp (CFP)—adenomas without malignancy. Telomere length, TMM, and expression were measured in 102 tissues including peripheral blood leukocytes (PBLs), normal colon epithelium, adenoma, and cancer (in CAP cases) from 31 patients. Telomere length was measured in a separate cohort of 342 PBL from CAP and CFP patients.
Results:
The mean differences in telomere length between normal and adenoma were greater in CAP than in CFP cases, P=0.001; telomere length in PBL was 91.7 bp greater in CAP than in CFP, P=0.007. Each 100 bp telomere increase was associated with a 1.14 (1.04–1.26) increased odds of being a CAP, P=0.0063. The polyp tissue from CAP patients had shorter telomeres and higher Telomerase reverse transcriptase (hTERT) expression compared with polyps from CFP patients, P=0.05. There was a greater degree of alternative lengthening of telomere (ALT) level difference in CFP polyps than in CAP polyps. The polyp telomere lengths of aggressive CAPs were significantly different from the polyps of non-aggressive CAPs, P=0.01.
Conclusions:
Adenomas that progress to cancer exhibit distinct telomere length and TMM profiles. We report for the first time that PBL telomeres differ in patients with polyps that become malignant, and therefore may have clinical value in adenoma risk assessment and management.
Introduction
As the majority of colorectal cancers (CRCs) arise from adenomatous polyps but only a small fraction of adenomas become malignant, the clinical conundrum is to predict which adenomas will progress. One approach to this challenge has been to classify an adenoma as “advanced” if it has high-grade dysplasia, is larger than 1 cm, or has villous architecture. Among all individuals undergoing colonoscopy, 25–30% will have at least one polyp and 5% will have a lesion determined to be at increased risk for malignant transformation.1, 2, 3, 4 Even so, adenomas matched by degree of dysplasia, size, and villous architecture have differing outcomes.1
A recent editorial listing five big questions in CRC placed “what distinguishes malignant from benign polyps” at number two.5 As only a small percentage of adenomatous polyps will develop into cancer, identification of molecular features that predict the malignant potential of the polyp is a major clinical step in individualizing polyp patient management. Expanded understanding of polyp biology and pathology stands to inform physicians and patients of optimal surveillance requirements. Colonoscopy follow-up intervals for patients are currently determined based on a polyp's histology, size, quantity, and degree of dysplasia. Surveillance colonoscopies are prescribed in a range from every 2 to 3 months for patients with higher-risk polyps to annually, every 3 years, or as long as once in 5 years for patients with polyps deemed less risky for progression. Given the costs and potential morbidity associated with colonoscopy surveillance, patients stand to benefit when surveillance intervals are tailored based on defined molecular features of polyps and risk for malignancy.
CRC involves the transformation of normal colon tissue to precancerous polyps and then to a malignant neoplasm. Genetic instability either at the nucleotide or at the chromosomal level is implicated in CRC.6 Chromosomal stability is maintained partly through telomeres, which are the caps of linear chromosomes.7, 8, 9, 10, 11, 12, 13 Telomere length is a recognized biomarker in multiple cancers and is associated with patient survival.7 For CRC, survival is adversely affected if the tumor telomere length is shorter than that of corresponding normal colon tissue.13 In upto two-thirds of CRC cases, tumor telomere length has been reported to be 3–5 kb shorter than the telomeres of adjacent normal colon epithelium, which far exceeds the natural rate of telomere attrition expected in healthy tissues.12, 14 In other studies, peripheral blood leukocytes (PBLs) and cancers have exhibited both long and short telomeres.13, 15, 16, 17, 18
Cancers exploit two telomere maintenance mechanisms (TMM) to regulate telomere length in malignant cells: telomerase activation (TA) and alternative lengthening of telomeres (ALT). TA and ALT are mechanistically and clinically distinct19 including adverse effects for CRC with TA20 and chemoresistance in cancer cells exhibiting ALT.21 ALT had not been identified as arising in CRC until our first report of this process.9 Our finding that rectal cancers engage ALT, along with a report that rectal cancers without TA (but untested for ALT)22 have a better prognosis than cancers with TA, invites further investigation to discern the role of TMM with differences in survival among CRC patients. In addition, there are reports that TA and ALT are not mutually exclusive in CRC.9, 23 Currently, there is no consensus on the role of telomere length and TMM in neoplastic transformation of CRC (i.e., from normal to polyp to tumor, and in those polyps that do not become cancer).
A key gap in our current understanding of CRC includes determination of mechanistic features in the processes that lead to neoplastic transformation—normal colon to polyp to CRC. Our understanding of this process has been limited largely to the study of the association of risk for developing cancer in large population-based studies that follow the clinical behavior of cancer-free polyps (CFPs), without directly evaluating the molecular transformation of normal colon through polyp to cancer in the same person. In the current study, we measured telomere length and TMM in biospecimens from cancer-adjacent polyp (CAP) patients—those with residual adenomatous polyps contiguous with the primary cancer—and CFP patients—those with only polyp tissue without malignancy. For each group, assessments were made in PBL, normal colon, adenoma, and, when present, carcinoma.
Methods
Sample characteristics and preparation
Through an IRB-approved Biobank for Gastrointestinal Health Research (BGHR; IRB 622–00, PI LA Boardman), polyps with adjacent normal and tumor specimens were harvested following surgical resection and snap-frozen in liquid nitrogen and maintained in a −70 freezer. All polyps are villous subtypes with low-grade dysplasia. Upto three 1 cm2 full-thickness specimens from the center of the cancer and upto three 1 cm2 full-thickness specimens of the polyp were used via this mechanism. In addition to polyp and cancer tissues, three 1 cm2 normal colonic epithelium full-thickness specimens at least 8 cm from the polyp/tumor margin were harvested. All tissues were macrodissected using a hematoxylin and eosin -stained tissue slide with areas of normal epithelium, polyp, or cancer reviewed and identified by a pathologist. CFPs and normal colonic epithelium at least 8 cm from the polyp were collected at the time of colonoscopic resection. Peripheral blood was obtained when possible, before removal of the CFP polyp or CAP polyp/cancer and neo-adjuvant treatment. The adenomatous polyps without cancer (CFPs) were matched to the CAP for the features of polyp size (categorical size: 1–2, 2–5, and >5 cm); histologic category (villous adenoma); and degree of dysplasia (low grade in these cases). More detailed information is included in the Supplementary Materials and Methods section.
Patient outcome
CFP cases were classified as aggressive if the polyp recurred on subsequent colonoscopic examination following a complete resection of the polyp. Surveillance colonoscopies were performed at 3–6 month intervals for 1 year and every 3–5 years thereafter. CAP patients were classified as aggressive if CRC recurred or if a Stage IV patient was never cancer-free. More detailed information as well as clinical and epidemiological annotations are included in the Supplementary Materials and Methods section.
DNA and RNA extraction
DNA was extracted with the PureGene method based on our previous report that the DNA extraction method has an impact on accurate measurements of telomere length.24, 25 RNA was extracted using the MiRNeasy mini kit (Qiagen, Hilden, Germany).
Telomere length assessment
Telomere length was measured using a modified monochrome quantitative PCR (MMQPCR) method as previously described, with no deviation in preparation of mastermix, use of primers, etc. from Rode et al.16, 26, 27, 28 Real-time measurements were carried out on the ABI Viia-7 machine and all base pair calculations were made based on K562 cell DNA as a calibrator, using the ΔΔCT method as previously described.28
Telomere length using Universal STELA
Universal STELA was performed using the protocol and oligonucleotides as described previously.29, 30 In brief, Universal STELA was performed as described in Bendix et al., with the exception that double-stranded panhandle DNA with 5′ TA overhang at one end was used for ligation at chromosome and subtelomeric regions to avoid the fill-in step before PCR reactions.
ALT assessment
Telomeric C-Circle DNA is partially single-stranded telomere DNA circles specific to cells exhibiting ALT, and was assayed at Capital Biosciences (Gaithersburg, MD), using a modified C-circle-qPCR protocol.9, 31 ALT values greater than 1 are standardly used as ALT+ however, for our analyses we used all values and reported them as differences between tissues—not based on a binary ALT+ or ALT−.
RNA sequencing (stranded)
Total RNA was transferred into library preparation, which was an automated variant of the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA). Oligo dT beads were used to select mRNA from the total RNA sample. Resultant cDNA went through library preparation using broad designed indexed adapters substituted in for multiplexing. Flowcell cluster amplification and sequencing were performed according to the manufacturer's protocols using either the HiSeq 2000 or HiSeq 2500. Each run was a 101 bp paired-end with barcoding. Data were analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation. More detailed information as well as processing of RNA sequencing (RNA-seq) data are included in the Supplementary Materials and Methods section.
Assessment of telomerase by hTERT expression
hTERT expression was used as a surrogate for telomerase activity32 and was determined using RNA-seq. Normalized expression values as described above for hTERT were used in all subsequent analyses (genes and values in Supplementary Table 2).
Statistical Analysis
Tests of significance on telomere length and TMM data (hTERT expression and ALT values) were performed using appropriate statistical methods. Statistical significance was determined between matched tissues using the Wilcoxon signed-rank test. Wilcoxon rank-sum test was used to test for differences between CAP and CFP cases. A difference was considered significant if the P value was <0.05. Boxplots, histograms, and density plots were processed in R 2.15.1.33 Correlations were performed using the cor function in R, using default parameters and using Pearson method. Normalized values as described above from our RNA-seq data for the 183 genes reported in Lafferty–Whyte were used to generate the heatmap in Figure 2 (genes and values in Supplementary Table 3). Heatmaps were generated using default parameters using the heatmap and hclust functions in R. Age-adjusted telomere length was calculated as base pair differences between the actual telomere length and predicted telomere length by age, using a general linear model.
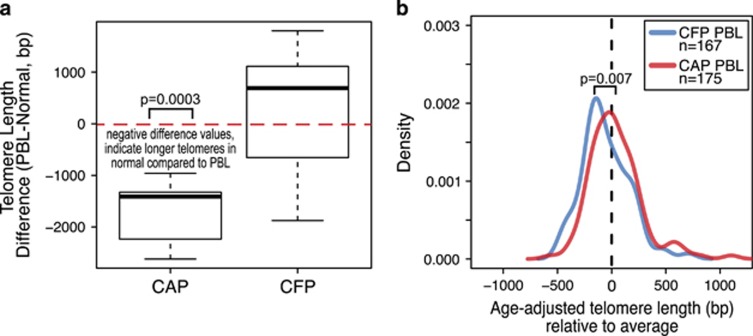
Figure 2.
Peripheral blood leukocyte telomeres are longer in cancer-adjacent polyp (CAP) patients. (a) Telomere length difference in base pairs between normal epithelium and PBL; from left to right, PBL minus normal for CAP and CFP cases. Red, dashed line drawn at zero shows positive and negative difference trends. The negative difference in the CAP cases indicates longer telomeres in the normal compared with the matched PBL. The differences between the PBL and normal in the CAP cases are significant, P=0.0003. (b) Density plots for age-adjusted PBL telomere length in 167 CAP (blue line) and 175 CFP (red line) cases. Dashed line at zero indicates the average telomere length for the population (n=342), with x axis-negative values indicating shorter telomeres than average and positive values being longer telomere length than average. There is a significant difference in the PBL telomere lengths between CAP and CFP cases.
Results
Telomere length distinguishes patients with polyps adjacent to cancer from those with CFPs
In this study we utilize a human tissue model of neoplastic transformation that captures the transition from normal colon to premalignant polyp and, in some cases, the transformation to cancer. These cases of neoplastic transformation are classified as CAP and CFP patients, with both groups including PBLs and/or normal colon epithelium as references (Figure 1a). It is important to note that CAP patient cases are different from other studies that compare polyps at different sites of the colon that are present when cancer is removed. CAP patient cases enable the unique opportunity to study the normal colon to polyp to cancer transition because the polyp tissue is the actual residual polyp of origin that is still in physical contact with the cancer (i.e., the polyp from which the cancer arose). In contrast, CFP patients were those who had a polyp removed and had no cancer present at the time of colonoscopy or after follow-up.
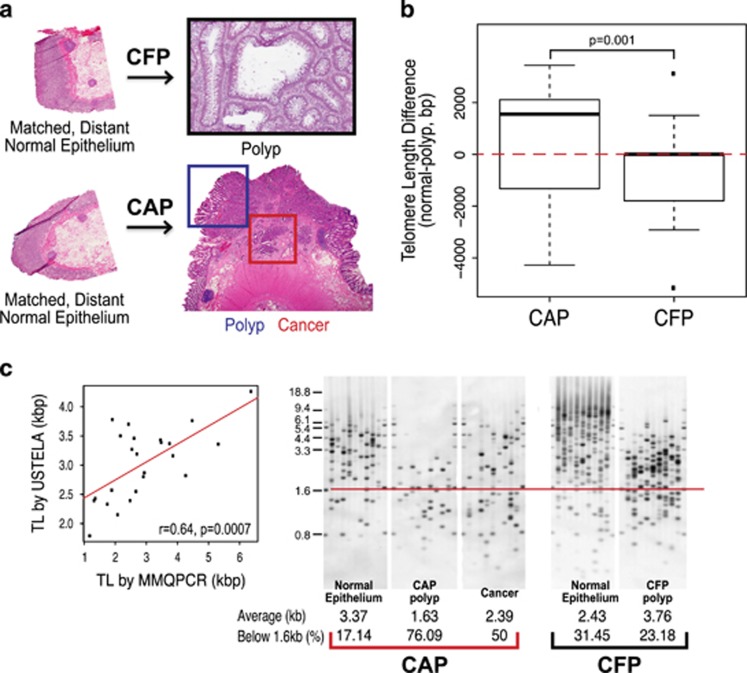
Figure 1.
Telomere length distinguishes cancer-adjacent polyps (CAPs) from cancer-free polyps (CFPs). (a) Corresponding tissue of CAP and CFP models that are used in this study. CFP cases include matched, distant normal colon epithelium, and the villous adenoma (polyp). All polyp cases used in the study were matched by histology and degree of dysplasia—villous adenomas with low-grade dysplasia. CFP cases are those that have had polyps present and removed that have not gone on to cancer. CAP cases include matched, distant normal colon epithelium, the polyp (residual polyp of origin) and the corresponding cancer that arose from the polyp. The cancer specimens from CAP cases represent a range of all stages. (b) Boxplots showing the values for the difference in telomere length in the polyp from the matched normal colon epithelium, for both CAP and CFP polyp cases from left to right, respectively. First, the telomere length for each polyp was subtracted from its matched normal telomere length to obtain a difference in telomere length value. Positive difference values indicate that the telomere length in the polyp tissue was shorter than the matched normal, and the inverse for the negative difference values (positive values, shorter polyp telomeres than normal; negative values, longer polyp telomeres than normal). Red, dashed line is drawn at zero for ease in determining positive and negative differences. The telomere length difference between CAP and CFP polyps is significant, P=0.001. (c) Scatterplot (far left) showing the correlation in telomere lengths in kilo base pairs (kbp) as determined from the modified monochrome quantitative PCR (MMQPCR) method and Universal STELA (USTELA) method, showing a strong, positive correlation. The other panels show results from USTELA from left to right, an exemplar CAP case and CFP case, showing normal, polyp, and cancer for the CAP and normal and polyp for the CFP. The horizontal red line indicates the threshold at which the load of short telomeres is determined (below 1.6 kb). In addition, the average telomere lengths in kilobases (kbs) and the percentage of telomeres below the 1.6 kb threshold (%) were both quantified and are included at the bottom of each electropherogram. There is an increasing amount of short telomeres from normal to polyp to cancer in the CAP case, with a slight telomere length increase in the cancer relative to the polyp (but still overall shorter compared with normal). Overall short telomeres occur to a lesser degree in the CFP polyp compared with normal as well as with the CAP tissues.
We measured telomere length in the PBL, normal colon, villous polyp with low-grade dysplasia, and tumor tissue of 15 CAP patients and the PBL, normal colon, and villous polyp with low-grade dysplasia tissue of 16 CFP patients (Supplementary Table 1,Supplementary Figure 1A and B). Distinct patterns of telomere length across tissue between the CAP and CFP tissues were evident. When the villous polyp tissues were compared directly with their matched normal colon tissue, the CAP cases showed a significantly greater degree of difference in telomere length than the CFP cases, P=0.001 (Figure 1b).
We found a high, positive correlation (r=0.64, P=0.0007) between the average telomere length determined using the MMQPCR method and that determined using Universal STELA (USTELA),29 which is a method for determining the presence of short telomeres on a chromosome-by-chromosome basis. (Figure 1d). In addition, we were able to confirm that the polyps with average short telomeres using MMQPCR were also among the cases showing the shortest telomeres per chromosome using USTELA (Figure 1c). The load of short telomeres in polyps in both CAP and CFP patients compared with the normal epithelium is recapitulated using USTELA, with the dramatic shortening visible by USTELA in the CAP polyps and cancer.
PBL telomeres are longer in CAP patients
The telomere lengths of the PBL and the normal colon epithelium in the CAP cases were significantly different, P=0.0003 (Figure 2a). The normal colon epithelium had significantly longer telomeres than PBL in these CAP patients (Supplementary Figure 2A). For CFP cases, the telomere length of PBL and normal colon epithelium was not significantly different.
PBL telomere length was measured in a total of 342 PBL cases (167 PBL from patients with CAPs and 175 PBL from CFP patients). In both groups, there was a significantly negative correlation between age and telomere length. Per unit increase in age is associated with a significant telomere length decrease in both CFP and CAP groups (Supplementary Figure 2B). Interaction analysis suggested the association between age and telomere length is not conditioned on the CAP/CFP status (P=0.77). After age adjustment, CAP patients have on average 91.7 bp longer PBL telomere length than CFP patients (P=0.007; Figure 2b).
We evaluated telomere length based on the clinical behavior of the CFP and CAP cases. CAP and CFP patients were categorized as non-aggressive if the polyp in the CFP cases or cancer in the CAP cases did not recur, and as aggressive if the polyp recurred in the CFP case or if the cancer recurred in a CAP patient. We found that the aggressive PBL CAP group had longer telomeres than the aggressive PBL CFP group, with marginal significance (P=0.06; Supplementary Figure 2C). The patient outcome analyses were carried out with 126 and 84 for CAP and CFP cases, in which follow-up was available, respectively.
To investigate the ability of PBL telomere length to predict whether a case is a CAP or a CFP, we performed a logistic regression analysis in the 334 total CAP and CFP cases, and found that a 100 bp telomere length increase is associated with a 1.14 (1.04–1.26) increased odds of being a CAP case (P=0.0063), with a positive likelihood ratio of 1.33. We determined that with an increasing telomere length the positive predictive value increased for predicting the CAP status (Supplementary Figure 4A). Within the CAP cases, there was an increasing positive predictive value for predicting aggressive status among the CAP cases with the shortest telomeres (Supplementary Figure 4B). A similar trend was observed for predicting aggressive status based on telomere length within CFP cases (Supplementary Figure 4C).
CAPs and CFPs exhibit distinct TMMs
The TMM that is engaged in tumors has been shown to correlate with a distinct pattern of telomere length differences in the three tissue compartments of the PBL, normal colon, and rectal cancer.9 In the CAP and CFP patients whose telomere lengths were shown in Figure 1, we assessed telomerase activity through hTERT expression and ALT by C-circles across all tissues.22, 32, 34, 35, 36, 37, 38
The CAP polyps, which showed the most extreme short telomeres, had higher expression of hTERT when compared with the CFP polyps, P=0.05. The degree of difference in hTERT expression between the polyps and their matched normal colon tissue was significant for the CAP cases (P=0.02), but not for the CFP cases (P=0.15; Figure 3a). The hTERT differences in polyp tissue compared with their normal is not attributable to differences in the normal tissues, as the hTERT expression levels were not significantly different between CAP and CFP normal tissues (P=0.18). In contrast, the CFP polyps showed an overall higher level of ALT than in the CAP polyps (Figure 3b). In addition, there was a greater degree of observable ALT level difference in the CFP polyps to their normal when compared with CAP polyp cases.
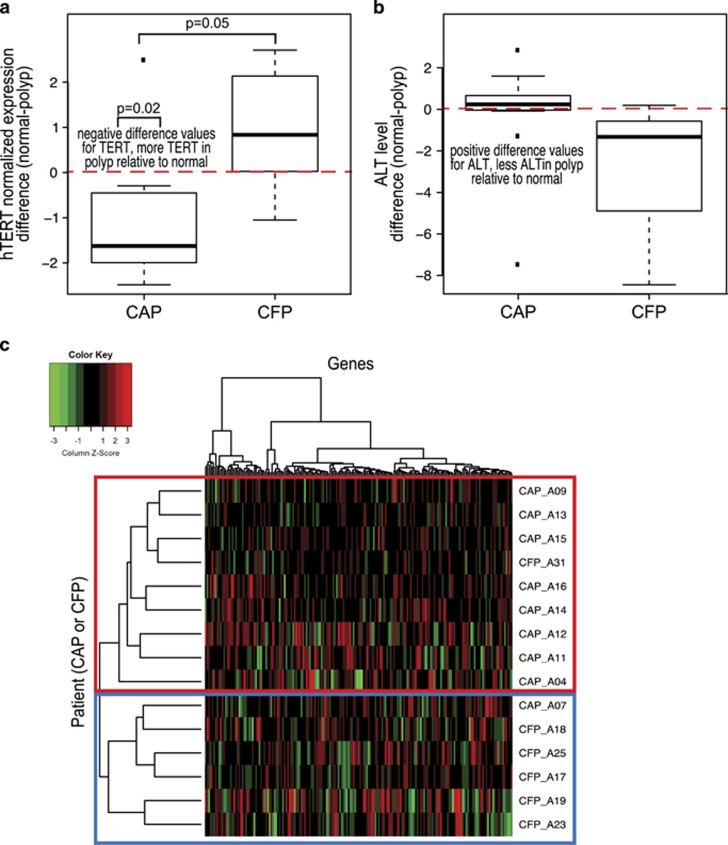
Figure 3.
Cancer-adjacent polyps (CAPs) and cancer-free polyps (CFPs) exhibit distinct telomere maintenance mechanisms. (a) Boxplot of values for the difference in the normalized hTERT expression in the polyp from the matched normal colon epithelium. hTERT expression in the polyp was subtracted from that in the matched normal for each case to obtain a difference value. CAP and CFP case, left and right, respectively. Negative difference values indicate that the hTERT expression in the polyp tissue is higher than the matched normal, and the inverse for the negative difference values (negative values, higher TERT expression in polyp compared with matched normal; positive values, lower TERT expression in polyp compared with matched normal values). Red, dashed line is drawn at zero for ease in determining positive and negative differences. There is a difference in TERT expression between normal and polyp in the CAP cases (P=0.02), and not for the CFP cases. In addition, the differences in TERT expression between CAP and CFP polyp cases are different, P=0.05. (b) Boxplot showing the values for the difference in the normalized alternative lengthening of telomere (ALT) levels in the polyp from the matched normal colon epithelium, left boxplot is the CAP cases and right boxplot is the CFP cases. These boxplots were generated by the same method as in part A, but for ALT levels. This plot is to show the ALT trend in differences, P>0.05. (c) Heatmap generated for genes reported by Lafferty–Whyte as defining telomerase-positive and ALT-positive cell lines. Plotted in this heatmap are 183 genes from our RNA-seq data. Hierarchical clustering was performed for genes and polyp cases. The left axis is the clustering for the CAP and CFP polyp cases, and the top axis is the clustering for the genes. The top 10 cases for hTERT expression and top 10 cases for ALT expression are shown. Five of the cases overlapped between the groups as having both hTERT expression and ALT level. The red box shows the CAP group that clusters together on the basis of expression and the blue box represents the CFP case cluster.
It has been previously reported that telomerase-positive and ALT-positive tumor cell lines exhibit distinct gene expression signatures.39 This gene expression signature was refined to a panel of 297 genes that have a significant association with TMM in liposarcoma tissues. From the CAP and CFP cases, we identified the 10 cases in which the polyp tissue had the highest TERT expression and the 10 cases with the highest ALT level in the polyp. We expected that the majority of cases with the highest TERT expression would be CAPs and those with the highest ALT level would be CFPs, which was the case. We then performed hierarchical clustering on the genes reported in Lafferty–Whyteet al., and found that the cases clustered in two distinct groups with all CAPs except one clustering together and all CFPs grouped together in a second cluster (Figure 3c). These results indicate that the CAP and CFP polyps exhibit distinct gene expression signatures based on their TMM status.
Telomere dynamics have an impact on patient outcome based on severity of the case
We characterized both CAP and CFP cases on their clinical behavior, and categorized them as aggressive or non-aggressive (as first shown in Figure 2). CAP cases were determined to be clinically aggressive if a Stage I–III cancer recurred or presented at Stage IV, whereas CFP cases were classified as aggressive if there was a recurrence of the polyp in the same anatomical location as identified by the previous polypectomy scar and/or submucosal tattoo marking. We found that aggressive polyps had shorter telomeres, regardless of their association with cancer (Figure 4a). The aggressive CAPs exhibited the shortest range of telomere lengths, with the polyp tissue exhibiting the shortest of telomere lengths. The aggressive CAP polyp telomere lengths were significantly different from the non-aggressive CAP polyp telomere lengths, P=0.01. Aggressive CFP polyps had a larger range of telomere lengths, but also exhibited overall shorter telomeres compared with both non-aggressive CAP and CFP polyps.
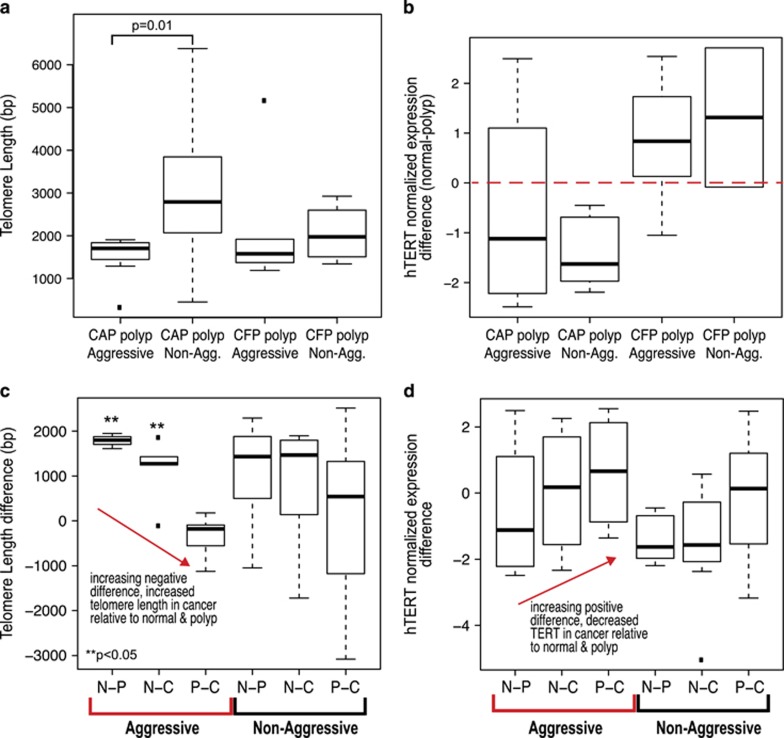
Figure 4.
Telomere dynamics are associated with patient outcome and malignant potential. (a) Boxplot showing the range of telomere length values in base pairs (bp), from left to right, aggressive CAP polyp, non-aggressive CAP polyp, aggressive CFP polyp, and non-aggressive CFP polyp cases. The aggressive CAP polyps are different from the non-aggressive CAP polyps, P=0.01. (b) Boxplot showing the values for the difference in the normalized hTERT expression in the polyp from the matched normal colon epithelium, from left to right, aggressive CAP polyp, non-aggressive CAP polyp, aggressive CFP polyp, and non-aggressive CFP polyp cases. hTERT expression in the polyp was subtracted from that in the matched normal for each case to obtain a difference value. Negative difference values indicate higher hTERT expression in the polyp tissue relative to matched normal, and the inverse for the negative difference values (negative values, higher TERT expression in polyp compared with matched normal; positive values, lower TERT expression in polyp compared with matched normal values). Red, dashed line is drawn at zero for ease in determining positive and negative differences. (c) Telomere length difference values in base pairs for aggressive (left, red brackets) and non-aggressive cases (right, black brackets). The x axis shows the difference that was taken to obtain the differences in telomere length. N-P, CAP polyp subtracted from normal colon epithelium; N-C, cancer subtracted from normal colon epithelium; P-C, cancer subtracted from polyp. **P<0.05 indicates a significant difference. (d) hTERT expression differences for aggressive (left, red brackets) and non-aggressive cases (right, black brackets). N-P, CAP polyp subtracted from normal colon epithelium; N-C, cancer subtracted from normal colon epithelium; P-C, cancer subtracted from polyp.
We observed overall higher TERT expression in both aggressive and non-aggressive CAP polyps, whereas in non-aggressive CAPs and CFP polyps there were lower levels of TERT relative to the matched normal epithelium (Figure 4b). The aggressive CFP polyps showed higher levels of polyp TERT compared with the non-aggressive CFP polyps; however, overall both aggressive and non-aggressive CAPs show strikingly higher TERT levels. These results suggest that telomere length may be used to distinguish cases based on patient outcome, whereas TERT expression levels appear more linked to the malignant potential of the polyp rather than overall patient outcome.
Patterns of telomere length and TMM across malignant transformation
Relative to the normal epithelium, the polyp tissue of the CAP cases had shorter telomeres, P=0.01 (Supplementary Figure 3A). Similar to the polyp tissue, the telomere length of the cancer tissue is significantly shorter than that of the normal colon (P=0.03). The trends observed in the CAP polyps for TMM (hTERT and ALT) were similarly observed in the cancer tissues, but to a lesser degree. hTERT expression was slightly lower in the cancer, and ALT levels were slightly higher in the cancer tissue as compared with the polyps (Supplementary Figure 3B). Interestingly, there was a wide range of hTERT expression overall in the cancer tissue relative to the normal and polyp tissues.
There was a greater degree of telomere length difference between the polyp and cancer tissue in the aggressive cases, with the telomere lengthening being more dramatic in the aggressive CAP cases (Figure 4c). The non-aggressive cases showed a similar trend for telomere length, but to a lesser degree than in the aggressive cases, especially for the polyps. Likewise, TMM showed distinct patterns between aggressive and non-aggressive cases. hTERT expression was highest in the polyp and increase in the cancer as compared with normal; when polyp and cancer are compared there are lower levels of TERT in the aggressive cancer cases (Figure 4d).
Discussion
This study presents a distinct profile of telomere dynamics that appear to be associated with adenoma risk of malignant progression. By identifying distinctions in the molecular features (in this case telomere dynamics) between cancer-transformed and CFPs, these studies may also yield markers present in the peripheral blood that may allow non-invasive risk assessment. The development of CRC is generally considered a linearly progressive disease from normal to polyp to cancer, similar to a model of aging, and indeed telomeres and their maintenance mechanisms are highly associated with aging and cancer progression. However, the dynamic telomere landscape of adenomatous polyps and the transition point to cancer have not been evaluated systematically in a group of polyps and their adjacent cancer from a collective set of individual patients, nor have these pathways been studied across multiple telomere-related features in comparison with similar CFPs. Characterizing the molecular profiles of polyps undergoing the transition to cancer that differentiate CFPs is an important contribution in identifying opportunities to provide optimal treatment choices for patient with polyps that carry the risk of CRC.
In this report, we present telomere length, TMM, and expression results across a total of 102 tissues (PBL, normal, polyp, and cancer) representing 31 patient cases. We have also measured telomere length in an additional 342 patient PBL samples. Continuing to identify telomere phenotypes across patient samples will improve our understanding of the role of telomere dynamics in neoplastic transformation. We utilized only one histologic and dysplasia category of polyps in this study so as not to confound the results with any other molecular or pathological feature of the tissue. Examining other histological and clinical subtypes of polyps in future studies will be important in further dissecting transition points in neoplastic transformation. In addition, we ensured that all the polyps used in this study resected at colonoscopy were removed by similar techniques so as not to introduce any predisposition for recurrence based on inconsistent polypectomy techniques. We have designed this model of colorectal polyp with or without progression to cancer carefully so that the results included in this study and any further studies using these cases will be solely attributable to biological phenomena.
The finding that both telomere length and TMM are associated with the malignant potential of polyps is an important and novel finding. It suggests that patients with telomere shortening in the polyp and tumor relative to normal may have a poorer outcome based on the association of shortened telomere lengths in the polyps and tumors of patients with aggressive CRC. This study also identified differences in telomere length and TMM between polyps with and without adjacent cancer, and related those differences to patient outcomes.
Although tissues start off with equivalent telomeres, changes in telomere length occur in parallel but at different rates during the lifespan of each individual.40, 41 In this study, we show that the telomere length in the normal colon epithelium is significantly longer than that in the PBL of CAP patients. This lengthening occurs more dramatically in the aggressive cases, suggesting that early transformation events may be occurring in the normal colon. Telomere dynamics in both the PBL and the normal tissue from which a cancer originates may inform the patient's prognosis. In this study we have found that a high normal colon epithelium to PBL telomere length ratio characterizes the cases associated with cancer.
As we observed an unexpected distinction between PBL and normal colon epithelium as well as differences in the PBL between CAP and CFP cases, it was necessary to clarify this finding in a larger set of patients. Following telomere length measurement in a set of over 300 patient PBL cases, we discovered that CAP cases exhibited longer telomeres than the CFP cases. Both long and short telomeres in the PBL are reported as being associated with a higher risk for developing many types of cancer, including CRC.13, 15, 16, 17, 18, 27, 28, 42, 43 PBL telomere length can be sustained or lengthened, providing opportunities for intervention.44, 45, 46 As no other study has examined the PBL telomere length between CAP and CFP cases (only cancer vs. non-cancer patients), it was previously not possible to know whether telomere length was associated with cancer or CFPs. This is a novel and important finding that PBL telomere length is longer in CAP patients and shows potential to be used as a marker to distinguish patients with polyps that have developed cancer from those that have remained cancer-free. In addition, longer telomeres in the blood of the CAP patients indicate that the blood could be used as a surrogate for the telomere length in the normal colon in CAP cases (which we showed were much longer than the telomere length of the normal colon of CFP cases).
Overall, these results suggest that differences in telomere length in the PBL of patients can distinguish cases that have polyps with malignant potential from those that remain cancer-free. The lengthened telomere phenotype has the potential to develop concomitant CRC in a polyp, even if they were only found to have adenomatous tissue in a biopsy. PBL telomere length could be used to establish tailored surveillance intervals once limitations of all current techniques to measure telomere length are addressed so that telomere length measurements can be accurately compared between studies. In addition, incorporation of the telomere length with other telomere-related markers could improve the positive predictive value of detecting a CAP vs. CFP case. In order to utilize telomere length and TMM profiling in the clinic setting, additional standardization and optimization for the assay is necessary. We acknowledge that, although we present a comprehensive integration of telomere length and TMM in multiple patient tissues, improved strategies for detecting telomere length and TMM will improve the precision of telomere profiling in the clinical setting.
Altogether, the results presented here clarify the role of telomere dynamics in malignant transformation. Shortened telomeres and telomere maintenance engagement occur early in adenomatous polyp development, and are most dramatic with an increasing malignant potential of the polyp (CAP cases). This study shows the highly dynamic nature of both telomeres and their maintenance mechanisms across tissues from a single individual. This work indicates that additional molecular events are likely taking place at the polyp level, early in neoplastic progression, and are dynamic across tissue compartments. A particularly striking result is the changes in telomere length observed in the normal colon epithelium, indicating a very early feature of carcinogenesis. Telomere dynamics appear to be associated with the clinical patient outcome, and could serve as an important point of determining risk for recurrence in both polyp and CRC patients. Our findings of telomere lengthening in the PBL of cases associated with cancer suggest that telomere dynamics have the potential to serve as a blood-based marker of CRC risk. Defining specific telomere length measurements in combination with the TMM in the blood and tissues of a larger patient group with and without cancer may lead us closer to a molecular, individualized approach to the management of colorectal polyps and cancer.
Study Highlights
Acknowledgments
We thank Dr Stig Bojenson for invaluable help and expertise with the MMQPCR technique for telomere length measurement used here. The RNA-seq data used here were generated by the Genomics Platform, Broad Institute, and we thank Kristina Tracy and Donald Skifter for their expertise and review of this manuscript. We would also thank Dr Clifford Steer, Dr Khashayarsha Khazaie, and Dr David Ahlquist for their constructive review of this manuscript. Funding was generously provided to support this research by a grant from Eugene and Eva Lane and by the Mayo Foundation. We wish to thank Dr Mohamad Mouchli for his assistance in reviewing clinical records. This work was supported by the National Cancer Institute at the National Institutes of Health (Grant number R01 CA170357).
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Guarantor of the article: Lisa A. Boardman, MD.
Specific author contributions: Study concept and design: BRD and LAB; acquisition of data: BRD, RJ, TS, JS, and LAB; analysis and interpretation of data: BRD, XR, and LAB; drafting of the manuscript: BRD and LAB; critical revision of the manuscript for important intellectual content: all authors; statistical analysis: DG and AO; material and technical support: RJ, JA, and DD; study supervision: LAB. All authors have approved the final draft of this manuscript that was submitted.
Financial support: This work was supported by the National Cancer Institute at the National Institutes of Health (Grant number R01 CA170357). In addition, funding was generously provided by a grant from Eugene and Eva Lane through the Mayo Foundation.
Potential competing interests: None.
Supplementary Material
References
- Church J. Clinical significance of small colorectal polyps. Dis Colon Rectum 2004; 47: 481–485. [DOI] [PubMed] [Google Scholar]
- Heitman S, Ronksley P, Hilsden R et al. Prevalence of adenomas and colorectal cancer in average risk individuals: a systematic review and meta-analysis. Clin Gastroenterol Hepatol 2009; 7: 1272–1278. [DOI] [PubMed] [Google Scholar]
- Martinez M, Sampliner R, Marshall J et al. Adenoma characteristics as risk factors for recurrence of advanced adenomas. Gastroenterology 2001; 120: 1077–1083. [DOI] [PubMed] [Google Scholar]
- Winawer S, Zauber A, O'Brien MJ et al. Randomized comparison of surveillance intervals after colonoscopic removal of newly diagnosed adenomatous polyps. The National Polyp Study Workgroup. N Engl J Med 1993; 328: 901–906. [DOI] [PubMed] [Google Scholar]
- Chakradhar S. Colorectal cancer: 5 big questions. Nature 2015; 521: S16. [DOI] [PubMed] [Google Scholar]
- Carethers J, Jung B. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology 2015; 149: 1177–1190.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo E, Boardman L. Toward a molecular classification of colorectal cancer: the role of telomere length. Front Oncol 2014; 4: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu N, Skinner H, Litzelman K et al. Telomeres and telomere dynamics: relevance to cancers of the GI tract. Expert Rev Gastroenterol Hepatol 2013; 7: 733–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman L, Johnson R, Viker K et al. Correlation of chromosomal instability, telomere length and telomere maintenance in microsatellite stable rectal cancer: a molecular subclass of rectal cancer. PLoS One 2013; 8: e80015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman L, Litzelman K, Seo S et al. The association of telomere length with colorectal cancer differs by the age of cancer onset. Clin Transl Gastroenterol 2014; 5: e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojesen S. Telomeres and human health. J Intern Med 2013; 274: 399–413. [DOI] [PubMed] [Google Scholar]
- Engelhardt M, Drullinsky P, Guillem J et al. Telomerase and telomere length in the development and progression of premalignant lesions to colorectal cancer. Clin Cancer Res 1997; 3: 1931–1941. [PubMed] [Google Scholar]
- Valls C, Pinol C, Rene J et al. Telomere length is a prognostic factor for overall survival in colorectal cancer. Colorectal Dis 2011; 13: 1265–1272. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Furugori E, Esaki Y et al. Correlation of telomere lengths in normal and cancers tissue in the large bowel. Cancer Lett 2000; 158: 179–184. [DOI] [PubMed] [Google Scholar]
- Riegert-Johnson DL, Boardman L, Crook J et al. Shorter peripheral blood telomeres are a potential biomarker for patients with advanced colorectal adenomas. Int J Biol Markers 2012; 27: e375–e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode L, Nordestgaard B, Bojesen S. Peripheral blood leukocyte telomere length and mortality among 64,637 individuals from the general population. J Natl Cancer Inst 2015; 107: djv074. [DOI] [PubMed] [Google Scholar]
- Segui N, Guino E, Pineda M et al. Longer telomeres are associated with cancer risk in MMR-proficient hereditary non-polyposis colorectal cancer. PLoS One 2014; 9: e86063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K, Codd V, Rice T et al. Longer genotypically-estimated leukocyte telomere length is associated with increased adult glioma risk. Oncotarget 2015; 6: 42468–42477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hills M, Conomos D et al. Telomere extension by telomerase and ALT generates variant repeats by mechanistically distinct processes. Nucleic Acids Res 2014; 42: 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertler R, Rosenberg R, Stricker D et al. Prognostic potential of the telomerase subunit human telomerase reverse transcriptase in tumor tissue and nontumorous mucosa from patients with colorectal carcinoma. Cancer 2002; 95: 2103–2111. [DOI] [PubMed] [Google Scholar]
- Flynn R, Cox K, Jeitany M et al. Alternative lengthening of telomeres renders cancer cells hypersensitive to ATR inhibitors. Science 2015; 347: 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi L, Cech T. Inventory of telomerase components in human cells reveals multiple subpopulations of hTR and hTERT. Nucleic Acids Res 2014; 42: 8565–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaurreta M, Maestro M, Rafael S et al. Telomerase activity in colorectal cancer, prognostic factor and implications in the microsatellite instability pathway. World J Gastroenterol 2007; 13: 3868–3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boardman L, Skinner H, Litzelman K. Telomere length varies by DNA extraction method: implications for epidemiologic research—response. Cancer Epidemiol Biomarkers Prev 2014; 23: 1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham J, Johnson R, Litzelman K et al. Telomere length varies by DNA extraction method: implications for epidemiologic research. Cancer Epidemiol Biomarkers Prev 2013; 22: 2047–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon R. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res 2009; 37: e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rode L, Bojesen S, Weischer M et al. Short telomere length, lung function and chronic obstructive pulmonary disease in 46,396 individuals. Thorax 2013; 68: 429–435. [DOI] [PubMed] [Google Scholar]
- Weischer M, Bojesen S, Cawthon R et al. Short telomere length, myocardial infarction, ischemic heart disease, and early death. Arterioscler Thromb Vasc Biol 2012; 32: 822–829. [DOI] [PubMed] [Google Scholar]
- Bendix L, Horn P, Jensen U et al. The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell 2010; 9: 383–397. [DOI] [PubMed] [Google Scholar]
- Holohan B, Hagiopian M, Lai T et al. Perifosine as a potential novel anti-telomerase therapy. Oncotarget 2015; 6: 21816–21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau L, Dagg R, Henson J et al. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res 2013; 41: e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolquist K, Ellisen L, Counter C et al. Expression of TERT in early premalignant lesions and a subset of cells in normal tissues. Nat Genet 1998; 19: 182–186. [DOI] [PubMed] [Google Scholar]
- Team RDC.. R: A language and environment for statistical computing. R Foundation for Statistical Computing 2010.
- Aschacher T, Wolf B, Enzmann F et al. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene 2015; 35: 94–104. [DOI] [PubMed] [Google Scholar]
- Jones A, Beggs A, Carvajal-Carmona L et al. TERC polymorphisms are associated both with susceptibility to colorectal cancer and with longer telomeres. Gut 2012; 61: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan T, Englezou A, Gupta J et al. Telomere elongation in immortal human cells without detectable telomerase activity. EMBO J 1995; 14: 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henson J, Cao Y, Huschtscha L et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol 2009; 27: 1181–1185. [DOI] [PubMed] [Google Scholar]
- Henson J, Reddel R. Assaying and investigating alternative lengthening of telomeres activity in human cells and cancers. FEBS Lett 2010; 584: 3800–3811. [DOI] [PubMed] [Google Scholar]
- Lafferty-Whyte K, Cairney C, Will M et al. A gene expression signature classifying telomerase and ALT immortalization reveals an hTERT regulatory network and suggests a mesenchymal stem cell origin for ALT. Oncogene 2009; 28: 3765–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie N, Dempster M, Dunlop M et al. Telomere reduction in human colorectal carcinoma and with ageing. Nature 1990; 346: 866–868. [DOI] [PubMed] [Google Scholar]
- Kim H, Kim Y, Kim H et al. Telomere length changes in colorectal cancers and polyps. J Korean Med Sci 2002; 17: 360–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Doherty J, Burgess S et al. Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum Mol Genet 2015; 24: 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plentz R, Wiemann S, Flemming P et al. Telomere shortening of epithelial cells characterises the adenoma-carcinoma transition of human colorectal cancer. Gut 2003; 52: 1304–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettleton J, Diez-Roux A, Jenny N et al. Dietary patterns, food groups, and telomere length in the Multi-Ethnic Study of Atherosclerosis (MESA). Am J Clin Nutr 2008; 88: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornish D, Lin J, Chan J et al. Effect of comprehensive lifestyle changes on telomerase activity and telomere length in men with biopsy-proven low-risk prostate cancer: 5-year follow-up of a descriptive pilot study. Lancet Oncol 2013; 14: 1112–1120. [DOI] [PubMed] [Google Scholar]
- Tiainen A, Mannisto S, Blomstedt P et al. Leukocyte telomere length and its relation to food and nutrient intake in an elderly population. Eur J Clin Nutr 2012; 66: 1290–1294. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.