ABSTRACT
Cell suspensions of Pelosinus sp. strain UFO1 were previously shown, using spectroscopic analysis, to sequester uranium as U(IV) complexed with carboxyl and phosphoryl group ligands on proteins. The goal of our present study was to characterize the proteins involved in uranium binding. Virtually all of the uranium in UFO1 cells was associated with a heterodimeric protein, which was termed the uranium-binding complex (UBC). The UBC was composed of two S-layer domain proteins encoded by UFO1_4202 and UFO1_4203. Samples of UBC purified from the membrane fraction contained 3.3 U atoms/heterodimer, but significant amounts of phosphate were not detected. The UBC had an estimated molecular mass by gel filtration chromatography of 15 MDa, and it was proposed to contain 150 heterodimers (UFO1_4203 and UFO1_4202) and about 500 uranium atoms. The UBC was also the dominant extracellular protein, but when purified from the growth medium, it contained only 0.3 U atoms/heterodimer. The two genes encoding the UBC were among the most highly expressed genes within the UFO1 genome, and their expressions were unchanged by the presence or absence of uranium. Therefore, the UBC appears to be constitutively expressed and is the first line of defense against uranium, including by secretion into the extracellular medium. Although S-layer proteins were previously shown to bind U(VI), here we showed that U(IV) binds to S-layer proteins, we identified the proteins involved, and we quantitated the amount of uranium bound.
IMPORTANCE Widespread uranium contamination from industrial sources poses hazards to human health and to the environment. Herein, we identified a highly abundant uranium-binding complex (UBC) from Pelosinus sp. strain UFO1. The complex makes up the primary protein component of the S-layer of strain UFO1 and binds 3.3 atoms of U(IV) per heterodimer. While other bacteria have been shown to bind U(VI) on their S-layer, we demonstrate here an example of U(IV) bound by an S-layer complex. The UBC provides a potential tool for the microbiological sequestration of uranium for the cleaning of contaminated environments.
KEYWORDS: contamination, uranium sequestration, uranium-binding protein
INTRODUCTION
Uranium is highly toxic and is also a radionuclide with a long half-life (1). Uranium mining, milling, and other anthropogenic industrial activities have led to widespread environmental contamination with associated hazards to human health and to the environment (2). In the United States, the U.S. Department of Energy (DOE) oversees the monitoring and restoration of waste sites at 12 facilities contaminated with uranium (3). Uranium is present in oxic fresh water as highly soluble U(VI). At pH values lower than 5.0, U(VI) is predominantly found as the free uranyl ion (UO22+), and between pH values of 5.0 and 7.5, U(VI) is predominantly complexed with carbonate. At pH values higher than 7.5, UO2(OH)3− is thought to be the predominant uranyl species (1). U(VI) can also precipitate in the form of uranium phosphate minerals, such as autunite (4, 5). Under anoxic fresh water conditions, U(IV) is the main oxidation state of uranium, and U(IV) often precipitates as uraninite (UO2) (1). Uraninite readily oxidizes to U(VI) in the presence of O2, NO3−, and Fe(III) (6–8).
There is increasing interest in microbiological sequestration of uranium for the cleaning of contaminated environments (3, 4, 9). Here, we focused on the effects of uranium on Pelosinus sp. strain UFO1. This strictly anaerobic fermentative firmicute was isolated from an uncontaminated background well at the Oak Ridge Reservation (ORR) in Tennessee (10), one of the DOE facilities that has areas of uranium contamination (3). The genome of strain UFO1 has been sequenced (11). Although the organism was not grown in the presence of added uranium, cell suspensions of strain UFO1 were previously shown to sequester uranium in two different oxidation states (12). When incubated with uranyl acetate (U[VI]), approximately 17% of the uranium precipitated on the cell surface. Using transmission electron microscopy with energy-dispersive X-ray spectroscopy (TEM-EDS), it was identified as insoluble uranyl phosphate with a U/P ratio of 1:1. The remainder of the uranium was present as monomeric complexes associated with the biomass. Analysis by X-ray absorption spectroscopy showed that it was U(IV) bound by carboxylate and phosphoryl functional groups and was not present as UO2 (uraninite). When 100 μM anthraquinone-2,6-disulfonate (AQDS), a humic acid analog with electron shuttling properties, was added to the cell suspensions of strain UFO1, all of the uranium was sequestered as U(IV) (12). We hypothesized that this strain UFO1-bound U(IV) is associated with a specific protein or proteins. In this study, we grew strain UFO1 in the presence of uranium and analyzed the resulting biomass to identify and characterize uranium-binding proteins. We show that virtually all of the added uranium was bound by a heterodimeric protein located on the cell surface as well as released into the extracellular medium.
RESULTS
Growth of strain UFO1 in the presence of uranium.
To characterize the sequestration of U(IV) by strain UFO1, the organism was grown in modified R2 medium supplemented with cysteine (0.5 g/liter). Cysteine was added as a reducing agent to remove O2 from the anaerobic growth medium and to act as an electron shuttle similar to AQDS (13, 14). AQDS was previously shown to result in the sequestration and accumulation of U(IV) in cell suspensions of strain UFO1 (12). The medium used to grow strain UFO1 in these experiments did not reduce AQDS chemically (see Fig. S1 in the supplemental material), but strain UFO1 reduces AQDS during growth (14). The enzymatic mechanism by which strain UFO1 reduces U(VI) is unknown, but it is aided by electron shuttles, such as AQDS and cysteine.
The oxidation state of the uranium associated with strain UFO1 grown in modified R2 broth was further confirmed using a 2-(5-bromo-2-pyridylazo)-5-diethylaminophenol (bromo-PADAP)-based colorimetric assay that specifically detects U(VI) (15). The U(VI)-specific assay showed that samples of strain UFO1 grown in the presence of 50 μM uranium and concentrated 10-fold contained 5.2 ± 3.6 μM U(VI). Inductively coupled plasma mass spectrometry (ICP-MS) showed that the same samples contained 357 ± 70 μM uranium. This indicates that the bulk of the uranium in the strain UFO1 samples was in the U(IV) oxidation state. It is possible that U(V) was also present, although U(V) disproportionates readily to U(VI) and U(IV), which, as opposed to U(V), were previously seen in association with cell suspensions of strain UFO1 (12, 16).
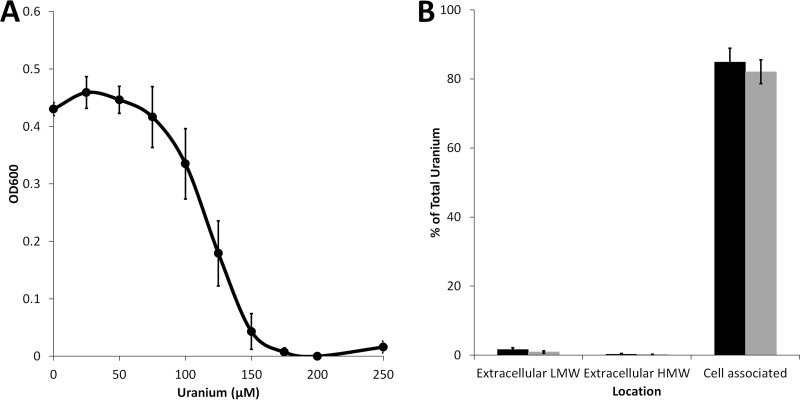
Strain UFO1 was quite sensitive to the presence of uranium in the growth medium. When grown in the presence of increasing concentrations of uranyl acetate (U[VI]) to stationary phase (20 h) (Fig. 1A), growth was inhibited at concentrations of uranium higher than 75 μM, and no growth was observed at a uranium concentration of 175 μM. The partitioning of uranium (using 25 and 50 μM in the medium) between fractions at late log phase of strain UFO1 is shown in Fig. 1B. Total uranium was measured (i) in the spent growth medium that passed through a 3-kDa filter (extracellular low molecular weight [LMW]), (ii) in the spent medium that could not pass through a 3-kDa filter (extracellular high molecular weight [HMW]), and (iii) in the cell pellet (cell associated). At concentrations of 25 and 50 μM uranium in the growth medium, more than 80% of the uranium was located in the cell-associated fraction at the end of growth, while less than 2% was found in the low-molecular-weight unbound fraction or in the extracellular HMW fraction. Approximately 20% of the uranium added to the cultures was unaccounted for, possibly due to adsorption to the 3-kDa filter or loss during digestion of the cell material before ICP-MS analysis. Nevertheless, the vast majority of the uranium added to growing cultures, even at the 50 μM concentration, was associated with the cells at the end of the growth phase.
FIG 1.
Growth and uranium partitioning of Pelosinus sp. Strain UFO1. (A) Endpoint growth after 20 h of Pelosinus sp. strain UFO1 grown in the presence of increasing concentrations of uranium. (B) Strain UFO1 was grown in the presence of 25 μM (black bars) or 50 μM (gray bars) uranium. After growth, cells were harvested and total uranium was measured in the supernatant that could flow through a 0.22-μm filter (extracellular low molecular weight [LMW]), in the supernatant that was retained from flowing through a 0.22-μm filter (extracellular high molecular weight [HMW]), and in the pellet (cell associated). Values are reported as the percentages of total uranium added to the original culture. Error bars represent the standard deviations of values from biological triplicates.
A 15-MDa uranium-binding complex is present in the membranes of strain UFO1.
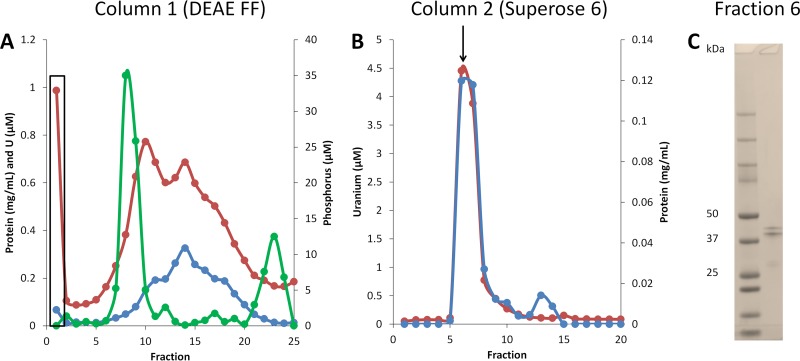
The combined membrane and cytoplasmic fractions of cells grown in the presence of 50 μM uranium were subjected to multistep column chromatography combined with ICP-MS analysis to purify and detect uranium binding proteins (Fig. 2). The uranium-binding proteins were likely either of high molecular weight, membrane associated, or both, as ultracentrifugation of the cell extract at 100,000 × g removed over 98% of the uranium from the soluble fraction. The uranium-binding proteins in the combined membrane and cytoplasmic fractions failed to bind to either an anionic DEAE fast flow (FF) column (pH 8.0) or a HiTrap sulfopropyl (SP) high performance (HP) cation exchange column (pH 7.0), as the majority of the uranium was located in the flowthrough fractions from both columns. However, this was highly advantageous, as most of the proteins in the membrane and cytoplasmic fractions bound to the ion exchange columns, thereby significantly purifying the uranium-binding proteins (see Table S1).
FIG 2.
Purification of the uranium-binding complex from combined membrane and cytoplasmic fractions. The uranium-binding complex (UBC) was purified from the combined membrane and cytoplasmic fractions by two column chromatography steps, anion exchange (DEAE FF) (A) and size exclusion (Superose 6) (B). Fractions were then analyzed for protein (blue lines), uranium (red lines), and phosphorus (green line). The flowthrough fraction from column 1 was used for the second purification step and is denoted by a box in panel A. (C) Denaturing gel image for the purified UBC from fraction 6 of the Superose 6 column (arrow in panel B).
After anion-exchange chromatography (with a DEAE FF column) (Fig. 2A), a portion of the uranium-containing flowthrough material was concentrated and subjected to Superose 6 size exclusion chromatography (Fig. 2B). At this point, the samples contained two major proteins identified by SDS electrophoresis corresponding to masses of 49.6 kDa and 45.4 kDa (Fig. 2C). These two proteins were identified by matrix-assisted laser desorption ionization (MALDI) mass spectrometry of tryptic digests of the SDS-PAGE gel bands as UFO1_4203 (AIF53746.1) and UFO1_4202 (AIF53745.1), respectively. This uranium-binding complex, which we termed UBC, was extremely large with an estimated size of 15 MDa. This is smaller than the exclusion limit (40 MDa) of the Superose 6 column, but larger than the fractionation range of the column (5 kDa to 5 MDa). Analysis for conserved domains (http://www.ncbi.nlm.nih.gov) showed that both proteins contained an S-layer-type domain (see Table S2). Additionally, UFO1_4202 contained an OprB or porin superfamily domain, and UFO1_4203 contained an outer membrane channel superfamily domain and a conserved domain of unknown function (DUF3373).
The calculated size of a single heterodimeric unit of UBC was 95 kDa (from the gene sequences), and based on this value, each heterodimer contained 3.3 atoms of uranium as determined by ICP-MS (see Table S1). Hence, the purified UBC of approximately 15 MDa in size was proposed to contain 150 heterodimers (UFO1_4203 and UFO1_4202) and about 500 uranium atoms. A faint band with a mass of 25 kDa was also observed on the SDS gel of the UBC sample, and this was identified as a flagellin domain protein encoded by UFO1_4112. However, this was assumed to be a contaminant and not part of the UBC. The two subunits of the UBC were analyzed for transmembrane helices using TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM). Both subunits were predicted to have one transmembrane helix that spans amino acid residues 5 to 24, indicating that the UBC is likely membrane associated at the N terminus and that the bulk of the protein is exposed to the extracellular medium.
During the purification, it was possible that the U(IV) associated with the UBC became oxidized to U(VI). While our yields of protein after the column chromatography steps did not provide sufficient material for the bromo-PADAP colorimetric assay that measures U(VI) specifically, we conducted the bromo-PADAP colorimetric assay on the same strain UFO1 cells grown in the presence of 50 μM uranium after stirring the cells aerobically for 2 h. A similarly small amount of U(VI) was detected in the bromo-PADAP colorimetric assay on aerobically exposed cells as was measured from the anaerobic cells, 6.8 ± 4.4 and 5.2 ± 3.6 μM U(VI), respectively. The association of U(IV) to the UBC on intact strain UFO1 cells may protect the uranium from being oxidized by oxygen. Regardless of the oxidation state of the uranium on the UBC after purification, it was evident that, under physiological growth conditions, the UBC was binding U(IV).
UBC is present in the extracellular growth medium.
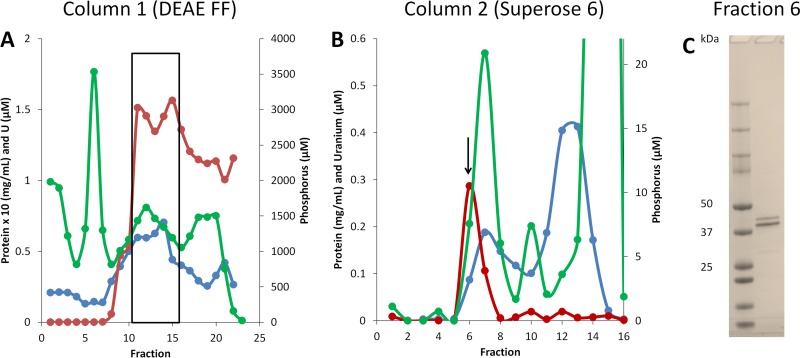
The UBC was also purified from the extracellular spent medium at the end of the growth phase (Fig. 3). This was purified using the same procedure used to obtain the UBC from the membrane and cytoplasmic fractions. However, in contrast to the membrane-associated UBC, the extracellular UBC bound to the DEAE FF column and eluted at NaCl concentrations from 260 to 360 mM (Fig. 3A). Purified extracellular UBC also contained only two major proteins that eluted off the Superose 6 column (Fig. 3B) at the same molecular masses, and MALDI mass spectrometry showed that these were the same two S-layer-type proteins (UFO1_4203 and UFO1_4202) identified in the membrane UBC (Fig. 2). However, the extracellular UBC contained only 0.3 uranium atoms per heterodimer (95 kDa), about 10-fold less than the amount of uranium in the membrane-associated UBC (see Table S3). The membrane and extracellular forms of the UBC were analyzed for phosphorus to determine if the uranium is sequestered by complexing with phosphate. However, no phosphorus was detected in the purified membrane UBC (Fig. 2), and, although uranium and phosphorous appeared to copurify with the extracellular UBC, the two elements did not overlap and were separated in the final purification step (Fig. 3B). Hence, uranium was not associated with phosphate in either the intracellular or the extracellular UBC. This also suggests that the UBC accumulated U(IV), since the insoluble U(VI) that was previously accumulated by cell suspensions of strain UFO1 was associated with phosphate in a 1:1 ratio (12).
FIG 3.
Purification of the uranium-binding complex from the extracellular medium. The uranium-binding complex (UBC) was purified from extracellular medium by two column chromatography steps, anion exchange (DEAE FF) (A) and size exclusion (Superose 6) (B). Fractions were then analyzed for protein (blue lines), uranium (red lines), and phosphorus (green lines). The fractions from column 1 used for the second purification step are denoted by a box in panel A. (C) Denaturing gel image for the purified UBC from fraction 6 of the Superose 6 column (arrow in panel B).
A total of 14 other metals were also examined by ICP-MS on the extracellular UBC fractions, including titanium, vanadium, manganese, iron, cobalt, nickel, copper, zinc, molybdenum, and tungsten. Of these metals, titanium, vanadium, cobalt, and tungsten were associated with the UBC in the peak fraction, although at much lower levels than uranium (see Table S4). Significant amounts of iron, nickel, and zinc were also present in the extracellular UBC peak fraction. However, as with phosphorus, the peaks of these metals did not overlap the uranium peak (see Table S4).
To determine if the UBC could be purified from cells that were grown in the absence of uranium, the membrane and cytoplasmic fractions were applied to a DEAE FF column and the flowthrough and eluted fractions were analyzed by SDS-gel electrophoresis. In this case, the UBC was identified and bound tightly to the column and eluted at a high NaCl concentration (>850 mM). This concentration is higher than that needed to elute the extracellular UBC when purified from uranium-grown cells. These data suggest that the uranium-free UBC is negatively charged and that it is neutralized by uranium such that the extracellular UBC (containing 0.3 U atoms/heterodimer) is less charged, while the membrane-associated UBC (from uranium-grown cells), which contains 3.3 U atoms/heterodimer, does not bind to the anionic column. Surprisingly, the calculated pI of UFO1_4202 was 7.7 and that of UFO1_4203 was 9.2 according to ExPASy (http://web.expasy.org/compute_pi/), in which case, the heterodimer is predicted to have an overall positive charge at pH 8.0. However, the uranium-free protein clearly behaved as an acidic protein. Nonetheless, a significant amount of the membrane-associated UBC was produced by strain UFO1 even in the absence of uranium. An analysis of the spent growth medium from strain UFO1 grown in the absence of uranium by SDS-PAGE revealed that the extracellular UBC was also present in the absence of uranium (see Fig. S2), and the molecular mass of the UBC without uranium was determined by size exclusion chromatography to be the same as that of the uranium-bound form (15 MDa).
Production of the UBC is independent of U.
Genome-wide transcript profiling was performed using transcriptome sequencing (RNA-Seq) of RNA isolated from strain UFO1 cells grown to late log phase on R2 media with and without 50 μM uranium. Both UFO1_4202 and 4203 were among the most highly expressed genes under both conditions (see Table S5). However, uranium did not change their expression levels as seen by reverse transcription (RT)-PCR and RNA-Seq (see Fig. S3 and Table S2). UFO1_4202 and UFO1_4203 were expressed at levels that were about 100-fold greater than the expression levels of the other six genes in the UFO1 genome that encode proteins with S-layer domains. Hence, UFO1_4202 and UFO1_4203 appear to encode the main S-layer proteins in strain UFO1. The abundance of the transcripts from the other 6 genes was also not significantly affected by the presence of uranyl acetate in the growth medium (see Table S2). On the other hand, a total of 7 of the 4,793 genes in the UFO1 genome were upregulated 8-fold or more during growth in the presence of uranium. Unfortunately, all seven of these genes are annotated as encoding hypothetical proteins (see Table S6), and their involvement in the uranium response is unknown.
DISCUSSION
It was previously shown that cell suspensions of strain UFO1 sequester uranium in two different forms, with about 20% as precipitated uranyl phosphate (U[VI]), as seen in the mineral autunite (12). Other organisms have also been shown to immobilize uranium as insoluble U(VI). For example, cell suspensions of Bacillus sphaericus JG-A12 were shown to bind U(VI) coordinated to carboxyl and phosphate groups on the S-layer protein (17). B. sphaericus JG-7B and Sphingomonas sp. S15-S1 were also shown to immobilize U(VI) as uranium phosphate minerals similar to autunite, but only under acidic conditions, in a process relying on indigenous acid phosphatases (4).
The majority (80%) of the cell-associated uranium sequestered by cell suspensions of strain UFO1 was previously shown to be U(IV) that was likely bound to carboxyl and phosphoryl functional groups (12). Therefore, it seems that this is the same form of uranium (U[IV]) that we have characterized herein, which is proposed to be taken up by the UBC on the cell surface. Similar cell-surface U(IV) deposits were observed by Fourier transform infrared spectroscopy (FTIR) when multiple anaerobic bacterial communities were grown in the presence of U(VI). However, the identity of the proteins involved in binding the U(IV) were not identified from the bacterial communities (18). Some bacterial species, including Shewanella oneidensis MR-1 and Desulfovibrio vulgaris, have also been shown to reduce U(VI) and accumulate U(IV) as nanocrystalline uraninite (UO2) particles (19–21). However, this is not the case with strain UFO1 (12).
Herein, we characterized the uranium-binding complex (UBC) and showed that it is composed of two subunits with S-layer domains, which are encoded by two of the most abundant proteins in the cell. Indeed, transcriptional analysis indicated that the UBC is the main protein component that makes up the S-layer of strain UFO1. In support of this, the Mep45 protein that makes up the major outer membrane S-layer protein of the firmicute Selenomonas ruminantium is a homolog of UFO1_4203 with 37% identity (22). Unlike the S-layer of B. sphaericus JG-A12 that binds U(VI) (17), the strain UFO1 S-layer appears to bind U(IV). In strain UFO1, the U(IV) is likely coordinated by carboxyl groups from glutamic and aspartic acids, as well as phosphate groups from phosphorylated serine and threonine residues (12). Analysis of the amino acid composition of the predicted extracellular exposed regions of the UBC heterodimer (UFO1_4202 and UFO1_4203) showed a large amount of these amino acid residues accounting for approximately 25% of the total amino acids.
S-layers provide a protective barrier for microorganisms, protecting them from a wide array of environmental threats, including high temperatures, low pHs, gamma radiation, and metals (23). The B. sphaericus strain JG-A12 S-layer has been shown to bind a wide range of metals cations, including U(VI). In the case of strain UFO1, several metals other than U, including Ti, V, Co, and W, were associated with the UBC, as potentially were also Fe, Ni, and Zn, which were present in the peak fraction, but whose chromatographs did not match the U chromatograph. No metals other than U were directly added to the growth medium, but were present as trace contaminants in the various medium components. It is possible that some of these metals accumulate on the UBC if they are present at significant concentrations in the growth medium. A potential mechanism was proposed for B. sphaericus strain JG-A12 in which U(VI) adsorption might damage the S-layer, resulting in S-layer repair (17, 24). In the case of strain UFO1, the S-layer protein was detected in the extracellular medium (see Fig. S2 in the supplemental material), a potential result of either a damaged S-layer sloughing off growing cells or from the remains of dead cells that lysed during growth of the culture. However, the presence of the UBC in the spent media did not depend on the presence of uranium in the growth medium. In addition, the UBC isolated from the spent medium of cells grown with uranium contained less uranium (0.3 U atoms/heterodimer) than the UBC isolated from the membrane (3.3 U atoms/heterodimer). This indicates that the loss and repair of the S-layer in strain UFO1 under the conditions used herein were not primarily driven by U binding, but were normal physiological processed that occurred in the absence of uranium. This is supported by the observation that uranium exposure during growth did not increase the expression levels of the UBC genes. Although S-layer proteins were shown previously to bind uranium (17), the uranium that was bound was in the U(VI) oxidation state, rather than the U(IV) state seen here with the UBC from strain UFO1. The binding of U(IV) to cell surfaces was also seen from bacterial communities grown in the presence of uranium (18). However, the proteins involved and the amount of uranium bound per protein were not determined.
The UBC of strain UFO1 bound U(IV), rather than the U(VI) that was added to the growth medium in the form of uranyl acetate, indicating a system for reducing uranium by growing strain UFO1 cells. The reduction of U(VI) by bacterial species does not appear to be catalyzed by dedicated or specialized reductases, but rather by systems that are primarily used for reducing other compounds (25). In D. vulgaris, tetraheme cytochrome c3 together with a periplasmic hydrogenase, its electron donor, were shown to be responsible for the majority, but not all, of the uranium reductase activity, while the alternate uranium reductases have yet to be identified (21, 25, 26). In Shewanella putrefaciens strain 200, a link between U(VI) reduction and nitrite reduction was discovered in the form of a mutant that simultaneously lost the ability to use both compounds as sole electron acceptors for growth (27). A transcription profiling experiment in S. oneidensis MR-1 followed by loss-of-function mutant analysis identified several proteins that were required for full U(VI) reductase activity, including cytochromes MtrA, MtrC, and CymA, as well as the outer membrane protein MtrB and a protein involved in menaquinone biosynthesis (MenC) (6). In strain UFO1, there are no identifiable homologs of the tetraheme cytochrome c3 from D. vulgaris, or homologs of MtrA, MtrB, MtrC, and MenC from S. oneidensis MR-1. However, there is a homolog with 25% identity to S. oneidensis MR-1 CymA (UFO1_4127), the tetraheme cytochrome subunit of a membrane-bound nitrite reductase. The expression of this protein was not significantly affected by the presence of uranium in the medium, but UFO1_4127 would make an intriguing starting point for investigating the U(VI) reduction capabilities of strain UFO1, since it has connections to nitrite reduction and shows homology to CymA.
The nonbiological cleanup of uranium by physicochemical methods is expensive, and there has been great interest in biological methods to sequester uranium (4, 9). The in situ biostimulation of U(VI) reduction by amending groundwater with electron donors such as ethanol has been shown to decrease soluble uranium levels to less than the EPA drinking water standard (30 μg/liter) (4). However, the insoluble U(IV) is readily reoxidized to soluble U(VI) once the electron source amendments are discontinued (8, 28). Uraninite is particularly susceptible to oxidation by O2, nitrate, and ferric iron (6–8). Longer-term U immobilization has been achieved in situ through the use of long-lived electron donors, such as emulsified vegetable oil (EVO). A single injection of EVO has been shown to decrease soluble uranium concentrations to below the starting amount for more than 4 months (29). Precipitation of U(VI) with phosphate has also been explored. Due to mobility issues of phosphate in situ, organophosphates have been used as phosphate sources with some success (5). In addition, B. sphaericus JG-7B and Sphingomonas sp. S15-S1 have been shown to use internal phosphate sources to mineralize uranium (4). The S-layer of B. sphaericus JG-A12 has been shown to bind U(VI), and the use of S-layer proteins to immobilize uranium in contaminated environments has been proposed (17). The S-layer of strain UFO1, which contains the UBC and binds U(IV), is another potential tool for U sequestration in contaminated environments.
MATERIALS AND METHODS
Growth conditions.
Strain UFO1 was grown anaerobically under an argon atmosphere without shaking at 25°C in a modified version of R2 broth. The medium contained the following components per liter: 0.5 g peptone, 0.5 g casein hydrolysate, 0.5 g yeast extract, 0.3 g KH2PO4, 0.3 g sodium pyruvate, 0.25 g MgSO4·7H2O, 3.2 g sodium fumarate, and 0.5 g cysteine. Where indicated, uranium was added in the oxidized form as uranyl acetate (U[VI]).
Uranium partitioning experiment.
To determine the cellular localization of uranium in strain UFO1, cultures (300 ml) of strain UFO1 were grown to late log phase (20 h) in triplicate experiments on modified R2 broth containing 25 and 50 μM uranium. To determine the amount of unbound and extracellular high-molecular-weight (HMW) bound uranium, the supernatant was collected after 50 ml of culture was harvested at 7,000 rpm. The supernatant was concentrated from 50 ml to 1 ml using a 3-kDa Amicon ultra centrifugal filter (Merck Millipore Ltd., Billerica, MA). The flowthrough (extracellular low-molecular-weight [LMW]) and retained (extracellular HMW) fractions were then analyzed by ICP-MS for uranium. To determine the amount of cell-associated uranium, the remaining 250 ml of culture was harvested at 7,000 rpm, and the pellet was washed once with 50 ml of 50 mM Tris, pH 8.0, before suspending in 1 ml of distilled water (dH2O) in a glass test tube. The sample was then evaporated to dryness in a 115°C oven and suspended in 1 ml of concentrated nitric acid. The sample was again evaporated to dryness before suspending in 2% nitric acid prior to ICP-MS analysis. Results were calculated as the percent of total uranium in each of the fractions compared to the amount of uranium added to the original culture.
U(VI) quantitation.
Cultures (50 ml) of strain UFO1 were grown to late log phase (20 h) in triplicate experiments on modified R2 broth containing no added or 50 μM uranium. Cells were harvested in an anaerobic chamber (95% Ar, 5% H2) (Coy Laboratory Products, Grass Lake, MI) and were suspended in 5 ml anoxic double-distilled water (ddH2O), thereby concentrating the cells 10-fold. A portion of the cells was removed from the chamber and was stirred slowly in the presence of oxygen for 2 h. The amounts of U(VI) in the anaerobic and aerobically exposed samples were colorimetrically determined using 2-(5-bromo-2-pyridylazo)-5-diethylaminophenol (bromo-PADAP) under anaerobic conditions as previously described (15). Briefly, the following were added to 100 μl of concentrated cells with mixing between each addition: 200 μl complexing solution [68 mM (1,2-cyclohexylenedinitrilo)tetraacetic acid, 120 mM sodium fluoride, and 256 mM sulfosalicylic acid in ddH2O], 200 μl buffer (1 M triethanolamine, pH 7.85), 1 ml ethanol (200 proof), and 200 μl 0.05% bromo-PADAP in ethanol. The volume of the assay was then increased to 2.5 ml with ddH2O before incubation at room temperature for 40 min. The absorbance (578 nm) was measured on a SpectraMax 190 microplate reader (Molecular Devices, Sunnyvale, CA). The amounts of U(VI) in the samples were determined using a standard curve of U(VI) (0 to 100 μM) dissolved in the strain UFO1 cell suspension from the no-uranium-added culture to control for interfering compounds, such as phosphate. The total amounts of uranium in the samples were determined using ICP-MS as described below.
Protein purification and column chromatography.
Strain UFO1 was grown in a 20-liter pH-adjusted fermenter in modified R2 broth containing 50 μM uranium. The fermenter was stirred at 300 rpm, with an overlay of 20% CO2 balanced with N2 gas at 25°C. Cells were grown to late log phase (16 h) before harvesting using continuous centrifugation. Cells were frozen in liquid N2 and stored at −80°C until use. A sample (3 liters) of the culture supernatant was retained for extracellular protein purification. The spent medium was buffered with 50 mM Tris and the pH was adjusted with NaOH to 8.0 before storing at 4°C. A cell extract containing both membrane and cytoplasmic components of strain UFO1 was prepared by lysing 20 g of strain UFO1 in 60 ml of 50 mM Tris, pH 8.0, by sonicating in the presence of 50 mg/liter lysozyme and 50 mg/liter DNase. The extract was then centrifuged at 8,000 rpm for 30 min and filtered through Whatman 1 filter paper and through a 0.22-μM Stericup vacuum-driven filter.
Anion exchange chromatography was carried with a 150-ml DEAE FF column (GE Healthcare Life Sciences, Pittsburgh, PA) using an equilibration buffer of 50 mM Tris, pH 8.0. An NaCl gradient from 0 to 1 M was used to elute bound proteins at a flow rate of 10 ml/min. Cation exchange chromatography utilized a 5 ml HiTrap SP HP column (GE Healthcare Life Sciences) with 20 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 7.0, as the equilibration buffer. Proteins were eluted with an NaCl gradient from 0 to 1 M at a flow rate of 10 ml/min. Size exclusion chromatography was carried out with a Superose 6 column (GE Healthcare Life Sciences) using an equilibration buffer of 50 mM Tris, pH 8.0, containing 150 mM NaCl at a flow rate of 0.5 ml/min. In each case, fractions were collected and analyzed for uranium and phosphate by ICP-MS and for protein using the Bradford method (30).
ICP-MS analysis.
Samples for ICP-MS analysis were diluted 1:15 with 2% (vol/vol) nitric acid. All samples were centrifuged at 2,000 × g for 10 min in a Beckman Allegra 6R centrifuge at 25°C. Metal analyses were performed in triplicate experiments using an Agilent 7500ce octopole ICP-MS in FullQuant mode using an internal standard with inline addition and a multielement external standard curve as previously described (31). Samples were loaded via a Cetac ASX-520 autosampler. Sample introduction, data acquisition, and processing were performed using Agilent Mass Hunter version B.01.01.
Identification of proteins by MALDI-MS.
Proteins identified by MALDI-MS were separated using denaturing SDS-PAGE (4 to 20% Criterion gels; Bio-Rad). Gel bands of interest were cut out and digested for 16 h at 37°C with recombinant porcine trypsin (Roche Applied Science, Indianapolis, IN). Digested peptides were dissolved in 30% acetonitrile (vol/vol), 0.1% trifluoroacetic acid (TFA) (vol/vol) and were spotted onto a MTP 384 Massive MADLI target (Bruker Daltonics, Billerica, MA). Peptides were analyzed using a Bruker Daltonics Autoflex MALDI–time of flight mass spectrometer in reflection mode using positive ion detection. The mass list was analyzed using Mascot's peptide mass fingerprint tool (version 2.1, Matrix Science) against the published genome sequence of strain UFO1 (11).
Transcript profiling using RNA sequencing.
Cultures of strain UFO1 (50 ml) were grown to late log phase (20 h) in modified R2 broth without (in triplicate experiments) and with (in duplicate experiments) 50 μM uranium added to the growth medium. RNA was isolated from the cultures and reverse transcription-PCR (RT-PCR) was performed as previously described (32). Primers for RT-PCR are listed in Table 1. Two micrograms of each RNA sample was subjected to rRNA depletion with the ScriptSeq Ribo-Zero (bacteria) kit (Epicentre, Madison, WI) according to the manufacturer's instructions, followed by ethanol precipitation. The samples were loaded on an RNA 6000 Pico chip (Agilent Technologies, Santa Clara, CA) and analyzed on a Bioanalyzer 2100 instrument (Agilent Technologies) to ensure the removal of rRNA. Samples that showed residual DNA contamination were subjected to additional DNase treatment with the Turbo DNA-free DNase kit (Life Technologies, Carlsbad, CA), followed by ethanol precipitation and a Bioanalyzer quality check to ensure no DNA contamination remained. A 4.4-ng sample of RNA was used to generate di-tagged cDNA libraries with the ScriptSeq v2 RNA-seq library preparation kit (Epicentre). The cDNA was purified with AMPure XP beads (Beckman Coulter, Indianapolis, IN), and the library was amplified with FailSafe PCR enzyme (Epicentre) with an index added to each sample (ScriptSeq Index Primer set 1; Epicentre). The amplified library was purified with AMPure XP beads. The samples were analyzed on the Bioanalyzer 2100 with a high sensitivity DNA chip (Agilent Technologies). The libraries were quantified with the Quant-iT PicoGreen DNA assay kit (Life Technologies, Carlsbad, CA). The libraries were pooled and sequenced on a MiSeq platform (150 cycles, single read) following the manufacturer's protocol (Illumina, Inc., San Diego, CA).
TABLE 1.
Reverse transcription-PCR primers
| Primer | Sequence (5′–3′) |
|---|---|
| UFO1_4202 F | GGAAAGTTTGTCGATGGTA |
| UFO1_4202 R | GGTCCTATGATGCTGTAA |
| UFO1_4203 F | CCGTAAGTAATTGTACCC |
| UFO1_4203 R | CCCTAAAGCTTATACAACTC |
The resulting reads in FASTQ format were mapped onto the genome of strain UFO1 (GenBank accession no. GCA_000725345.1) using EDGE-Pro version 1.3.1 (33). The GenBank annotations in generic feature format (GFF) were converted to a PTT format for use with EDGE-Pro using a custom perl script. Differential expressions of genes in response to the presence of uranium in the growth medium were assessed using the DESeq package available in Bioconductor version 2.13 (34). The P values that we reported were adjusted using the Benjamini-Hochberg procedure for multiple hypothesis testing.
Supplementary Material
ACKNOWLEDGMENTS
This material by the Ecosystems and Networks Integrated with Genes and Molecular Assemblies (ENIGMA) (http://enigma.lbl.gov), a scientific focus area program at Lawrence Berkeley National Laboratory is based upon work supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research under contract number DE-AC02-05CH11231.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.03044-16.
REFERENCES
- 1.Markich SJ. 2002. Uranium speciation and bioavailability in aquatic systems: an overview. ScientificWorldJournal 2:707–729. doi: 10.1100/tsw.2002.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beneš P. 1999. The environmental impacts of uranium mining and milling and the methods of their reduction, p. 225–246. In Choppin GR, Khankhasayev MK (ed), Chemical separation technologies and related methods of nuclear waste management. Springer, Dordrecht, Netherlands. [Google Scholar]
- 3.Riley RG, Zachara JM. 1992. Chemical contaminants on DOE lands and selection of contaminant mixtures for subsurface science research. US Department of Energy, Office of Scientific and Technical Information, Washington, DC. [Google Scholar]
- 4.Merroun ML, Nedelkova M, Ojeda JJ, Reitz T, Fernández ML, Arias JM, Romero-González M, Selenska-Pobell S. 2011. Bio-precipitation of uranium by two bacterial isolates recovered from extreme environments as estimated by potentiometric titration, TEM and X-ray absorption spectroscopic analyses. J Hazard Mater 197:1–10. doi: 10.1016/j.jhazmat.2011.09.049. [DOI] [PubMed] [Google Scholar]
- 5.Martinez RJ, Wu CH, Beazley MJ, Andersen GL, Conrad ME, Hazen TC, Taillefert M, Sobecky PA. 2014. Microbial community responses to organophosphate substrate additions in contaminated subsurface sediments. PLoS One 9:e100383. doi: 10.1371/journal.pone.0100383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bencheikh-Latmani R, Williams SM, Haucke L, Criddle CS, Wu L, Zhou J, Tebo BM. 2005. Global transcriptional profiling of Shewanella oneidensis MR-1 during C (VI) and U(VI) reduction. Appl Environ Microbiol 71:7453–7460. doi: 10.1128/AEM.71.11.7453-7460.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holmes DE, Finneran KT, O'Neil RA, Lovley DR. 2002. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl Environ Microbiol 68:2300–2306. doi: 10.1128/AEM.68.5.2300-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu W-M, Carley J, Luo J, Ginder-Vogel MA, Cardenas E, Leigh MB, Hwang C, Kelly SD, Ruan C, Wu L, Van Nostrand J, Gentry T, Lowe K, Mehlhorn T, Carroll S, Luo W, Fields MW, Gu B, Watson D, Kemner KM, Marsh T, Tiedje J, Zhou J, Fendorf S, Kitanidis PK, Jardine PM, Criddle CS. 2007. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ Sci Technol 41:5716–5723. doi: 10.1021/es062657b. [DOI] [PubMed] [Google Scholar]
- 9.Brim H, McFarlan SC, Fredrickson JK, Minton KW, Zhai M, Wackett LP, Daly MJ. 2000. Engineering Deinococcus radiodurans for metal remediation in radioactive mixed waste environments. Nat Biotechnol 18:85–90. doi: 10.1038/71986. [DOI] [PubMed] [Google Scholar]
- 10.Ray AE. 2007. Discovery and characterization of a novel anaerobe with a potential role in bioremediation of metal-contaminated subsurface environments. PhD dissertation Idaho State University, Pocatello, ID. [Google Scholar]
- 11.Brown SD, Utturkar SM, Magnuson TS, Ray AE, Poole FL, Lancaster WA, Thorgersen MP, Adams MW, Elias DA. 2014. Complete genome sequence of Pelosinus sp. strain UFO1 assembled using single-molecule real-time DNA sequencing technology. Genome Announc 2:e00881-14. doi: 10.1128/genomeA.00881-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray AE, Bargar JR, Sivaswamy V, Dohnalkova AC, Fujita Y, Peyton BM, Magnuson TS. 2011. Evidence for multiple modes of uranium immobilization by an anaerobic bacterium. Geochim Cosmochim Acta 75:2684–2695. doi: 10.1016/j.gca.2011.02.040. [DOI] [Google Scholar]
- 13.Maithreepala R, Doong RA. 2009. Transformation of carbon tetrachloride by biogenic iron species in the presence of Geobacter sulfurreducens and electron shuttles. J Hazard Mater 164:337–344. doi: 10.1016/j.jhazmat.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 14.Liu C, Zachara JM, Foster NS, Strickland J. 2007. Kinetics of reductive dissolution of hematite by bioreduced anthraquinone-2,6-disulfonate. Environ Sci Technol 41:7730–7735. doi: 10.1021/es070768k. [DOI] [PubMed] [Google Scholar]
- 15.Johnson D, Florence T. 1971. Spectrophotometric determination of uranium(VI) with 2-(5-bromo-2-pyridylazo)-5-diethylaminophenol. Anal Chim Acta 53:73–79. doi: 10.1016/S0003-2670(01)80072-6. [DOI] [Google Scholar]
- 16.Renshaw JC, Butchins LJ, Livens FR, May I, Charnock JM, Lloyd JR. 2005. Bioreduction of uranium: environmental implications of a pentavalent intermediate. Environ Sci Technol 39:5657–5660. doi: 10.1021/es048232b. [DOI] [PubMed] [Google Scholar]
- 17.Merroun ML, Raff J, Rossberg A, Hennig C, Reich T, Selenska-Pobell S. 2005. Complexation of uranium by cells and S-layer sheets of Bacillus sphaericus JG-A12. Appl Environ Microbiol 71:5532–5543. doi: 10.1128/AEM.71.9.5532-5543.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martins M, Faleiro ML, da Costa AMR, Chaves S, Tenreiro R, Matos AP, Costa MC. 2010. Mechanism of uranium (VI) removal by two anaerobic bacterial communities. J Hazard Mater 184:89–96. doi: 10.1016/j.jhazmat.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 19.Dohnalkova A, Marshall MJ, Kennedy DW, Gorby YA, Shi L, Beliaev A, Apkarian R, Fredrickson JK. 2005. The role of bacterial exopolymers in metal sorption and reduction. Microsc Microanal 11(Suppl 2):116–117. doi: 10.1017/S1431927605506688.15817140 [DOI] [Google Scholar]
- 20.Marshall MJ, Beliaev AS, Dohnalkova AC, Kennedy DW, Shi L, Wang Z, Boyanov MI, Lai B, Kemner KM, McLean JS, Reed SB, Culley DE, Bailey VL, Simonson CJ, Saffarini DA, Romine MF, Zachara JM, Fredrickson JK. 2006. c-type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis. PLoS Biol 4:e268. doi: 10.1371/journal.pbio.0040268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lovley DR, Widman PK, Woodward JC, Phillips E. 1993. Reduction of uranium by cytochrome c3 of Desulfovibrio vulgaris. Appl Environ Microbiol 59:3572–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kojima S, Kaneko J, Abe N, Takatsuka Y, Kamio Y. 2011. Cadaverine covalently linked to the peptidoglycan serves as the correct constituent for the anchoring mechanism between the outer membrane and peptidoglycan in Selenomonas ruminantium. J Bacteriol 193:2347–2350. doi: 10.1128/JB.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerbino E, Carasi P, Mobili P, Serradell M, Gómez-Zavaglia A. 2015. Role of S-layer proteins in bacteria. World J Microbiol Biotechnol 31:1877–1887. doi: 10.1007/s11274-015-1952-9. [DOI] [PubMed] [Google Scholar]
- 24.Jankowski U, Merroun ML, Selenska-Pobell S, Fahmy K. 2010. S-Layer protein from Lysinibacillus sphaericus JG-A12 as matrix for AuIII sorption and Au-nanoparticle formation. J Spectrosc 24:177–181. doi: 10.1155/2010/319249. [DOI] [Google Scholar]
- 25.Wall JD, Krumholz LR. 2006. Uranium reduction. Annu Rev Microbiol 60:149–166. doi: 10.1146/annurev.micro.59.030804.121357. [DOI] [PubMed] [Google Scholar]
- 26.Payne RB, Gentry DM, Rapp-Giles BJ, Casalot L, Wall JD. 2002. Uranium reduction by Desulfovibrio desulfuricans strain G20 and a cytochrome c3 mutant. Appl Environ Microbiol 68:3129–3132. doi: 10.1128/AEM.68.6.3129-3132.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade R, DiChristina TJ. 2000. Isolation of U(VI) reduction-deficient mutants of Shewanella putrefaciens. FEMS Microbiol Lett 184:143–148. doi: 10.1111/j.1574-6968.2000.tb09005.x. [DOI] [PubMed] [Google Scholar]
- 28.Wu W-M, Carley J, Green SJ, Luo J, Kelly SD, Nostrand JV, Lowe K, Mehlhorn T, Carroll S, Boonchayanant B, Löfller FE, Watson D, Kemner KM, Zhou J, Kitanidis PK, Kostka JE, Jardine PM, Criddle CS. 2010. Effects of nitrate on the stability of uranium in a bioreduced region of the subsurface. Environ Sci Technol 44:5104–5111. doi: 10.1021/es1000837. [DOI] [PubMed] [Google Scholar]
- 29.Gihring TM, Zhang G, Brandt CC, Brooks SC, Campbell JH, Carroll S, Criddle CS, Green SJ, Jardine P, Kostka JE, Lowe K, Mehlhorn TL, Overholt W, Watson DB, Yang Z, Wu WM, Schadt CW. 2011. A limited microbial consortium is responsible for extended bioreduction of uranium in a contaminated aquifer. Appl Environ Microbiol 77:5955–5965. doi: 10.1128/AEM.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Cvetkovic A, Menon AL, Thorgersen MP, Scott JW, Poole FL II, Jenney FE Jr, Lancaster WA, Praissman JL, Shanmukh S, Vaccaro BJ, Trauger SA, Kalisiak E, Apon JV, Siuzdak G, Yannone SM, Tainer JA, Adams MW. 2010. Microbial metalloproteomes are largely uncharacterized. Nature 466:779–782. doi: 10.1038/nature09265. [DOI] [PubMed] [Google Scholar]
- 32.Scott IM, Rubinstein GM, Lipscomb GL, Basen M, Schut GJ, Rhaesa AM, Lancaster WA, Poole FL, Kelly RM, Adams MW. 2015. A new class of tungsten-containing oxidoreductase in Caldicellulosiruptor, a genus of plant biomass-degrading thermophilic bacteria. Appl Environ Microbiol 81:7339–7347. doi: 10.1128/AEM.01634-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Magoc T, Wood D, Salzberg SL. 2013. EDGE-pro: estimated degree of gene expression in prokaryotic genomes. Evol Bioinform Online 9:127–136. doi: 10.4137/EBO.S11250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.