ABSTRACT
Manipulation of biofilm formation in Shewanella is beneficial for application to industrial and environmental biotechnology. BpfA is an adhesin largely responsible for biofilm formation in many Shewanella species. However, the mechanism underlying BpfA production and the resulting biofilm remains vaguely understood. We previously described the finding that BpfA expression is enhanced by DosD, an oxygen-stimulated diguanylate cyclase, under aerobic growth. In the present work, we identify FlrA as a critical transcription regulator of the bpfA operon in Shewanella putrefaciens CN32 by transposon mutagenesis. FlrA acted as a repressor of the operon promoter by binding to two boxes overlapping the −10 and −35 sites recognized by σ70. DosD regulation of the expression of the bpfA operon was mediated by FlrA, and cyclic diguanylic acid (c-di-GMP) abolished FlrA binding to the operon promoter. We also demonstrate that FlhG, an accessory protein for flagellum synthesis, antagonized FlrA repression of the expression of the bpfA operon. Collectively, this work demonstrates that FlrA acts as a central mediator in the signaling pathway from c-di-GMP to BpfA-associated biofilm formation in S. putrefaciens CN32.
IMPORTANCE Motility and biofilm are mutually exclusive lifestyles, shifts between which are under the strict regulation of bacteria attempting to adapt to the fluctuation of diverse environmental conditions. The FlrA protein in many bacteria is known to control motility as a master regulator of flagellum synthesis. This work elucidates its effect on biofilm formation by controlling the expression of the adhesin BpfA in S. putrefaciens CN32 in response to c-di-GMP. Therefore, FlrA plays a dual role in controlling motility and biofilm formation in S. putrefaciens CN32. The cooccurrence of flrA, bpfA, and the FlrA box in the promoter region of the bpfA operon in diverse Shewanella strains suggests that bpfA is a common mechanism that controls biofilm formation in this bacterial species.
KEYWORDS: FlrA, Shewanella, biofilm, cyclic di-GMP, transcriptional factor
INTRODUCTION
Biofilm formed by bacteria is a multicell architecture for adaptation to diverse niches (1). Shewanella can form biofilm on a variety of surfaces, such as ferric oxides, electrodes, stainless steel, and glass (2–4). This genus is also renowned for an extracellular respiration ability to reduce iron oxide, electrodes, and other extracellular electron acceptors, including heavy metal ions. Such an ability has diverse potential applications in bioengineering and bioremediation (5). Biofilm formation in Shewanella benefits close contact with solid electron acceptors, thereby accelerating iron oxide reduction and improving current output, as well as inducing spatially stratified metabolic responses during contaminant exposure (4, 6, 7). Biofilm formed by Shewanella prevents microbially induced corrosion of steel and cast iron pipes (8, 9). On the other hand, biofilm formation of Shewanella also causes problems in some circumstances. For example, Shewanella forms biofilm on the surfaces of food processing equipment, causing contamination and spoilage of seafood, which can be rescued by inhibiting biofilm formation (10, 11). Unfortunately, the understanding of the mechanism underlying biofilm formation in Shewanella has been very limited, restricting the biological control and biotechnological manipulation of its biofilm.
Biofilm-promoting factor A (BpfA) is a key protein contributing to biofilm formation in some Shewanella species. BpfA belongs to a subfamily of the RTX (repeats in toxins) proteins typically containing 80- to 300-amino-acid repeats ordered in tandem (12). RTX proteins are extracellular proteins whose secretion depends on the type I secretion system (TISS) and that function as adhesins or toxins. The role of RTX proteins in biofilm formation has been investigated previously in several bacteria, such as Pseudomonas fluorescens and Pseudomonas putida (13, 14), Enterococcus faecalis (15), and Psychrobacter arcticus (16). P. putida encodes two RTX proteins, LapA and LapF, which play sequential roles in the initial stage and a later stage of biofilm development, respectively (17). In P. fluorescens Pfo-1, a low level of phosphate triggers the cleavage of LapA through the cyclic diguanylic acid (c-di-GMP) signaling pathway (18). In Shewanella oneidensis MR-1, BpfA contributes greatly to biofilm formation (19). In Shewanella sp. strain HRCR-1, BpfA exists predominantly in loosely associated extracellular polymeric substances of biofilm (20). The bpfA operon in Shewanella species such as S. oneidensis MR-1 and S. putrefaciens CN32 commonly contains seven genes, including a TISS for BpfA translocation. Disruption of the TISS blocks formation of pellicle, a type of biofilm formed at the air-liquid interface which is probably caused by interruption of BpfA translocation (21). A hyper-aggregating mutant of S. oneidensis MR-1 with enhanced biofilm formation exhibits increased transcription of a TISS component up to about 5-fold, and inactivation of the TISS results in the loss of the hyper-aggregation phenotype (22). High O2 tension induces autoaggregation of S. oneidensis MR-1 in a chemostat, and the transcription of TISS is also upregulated (23). Although these reports emphasize the importance of BpfA for biofilm formation, it is unclear how bpfA expression is regulated in response to environmental stimuli in these Shewanella species. We previously demonstrated that transcription of the bpfA operon in S. putrefaciens CN32 is increased with an increase in c-di-GMP level by a diguanylate cyclase (DosD) upon exposure to oxygen, thus promoting biofilm formation under conditions of aerobic growth (24). However, the responsive regulator that mediates the c-di-GMP regulation of the expression of the bpfA operon is still unidentified.
In the present work, we screened a transposon mutant library for potential regulators of the bpfA operon. FlrA was identified as a transcriptional repressor by binding to two boxes in the promoter of the bpfA operon. c-di-GMP weakened FlrA's binding to the promoter, therefore derepressing the expression of the bpfA operon. Transcriptional repression of bpfA by FlrA was also antagonized by an accessory protein of flagella, FlhG. Our work clarifies a central role of FlrA in the signaling pathway from DosD-derived c-di-GMP to BpfA-associated biofilm formation in S. putrefaciens CN32. Elucidation of the regulatory pathway of the bpfA operon by the bacterial second messenger c-di-GMP will deepen understanding of the physiology of these species under aerobic conditions.
RESULTS
FlrA represses the expression of the bpfA operon.
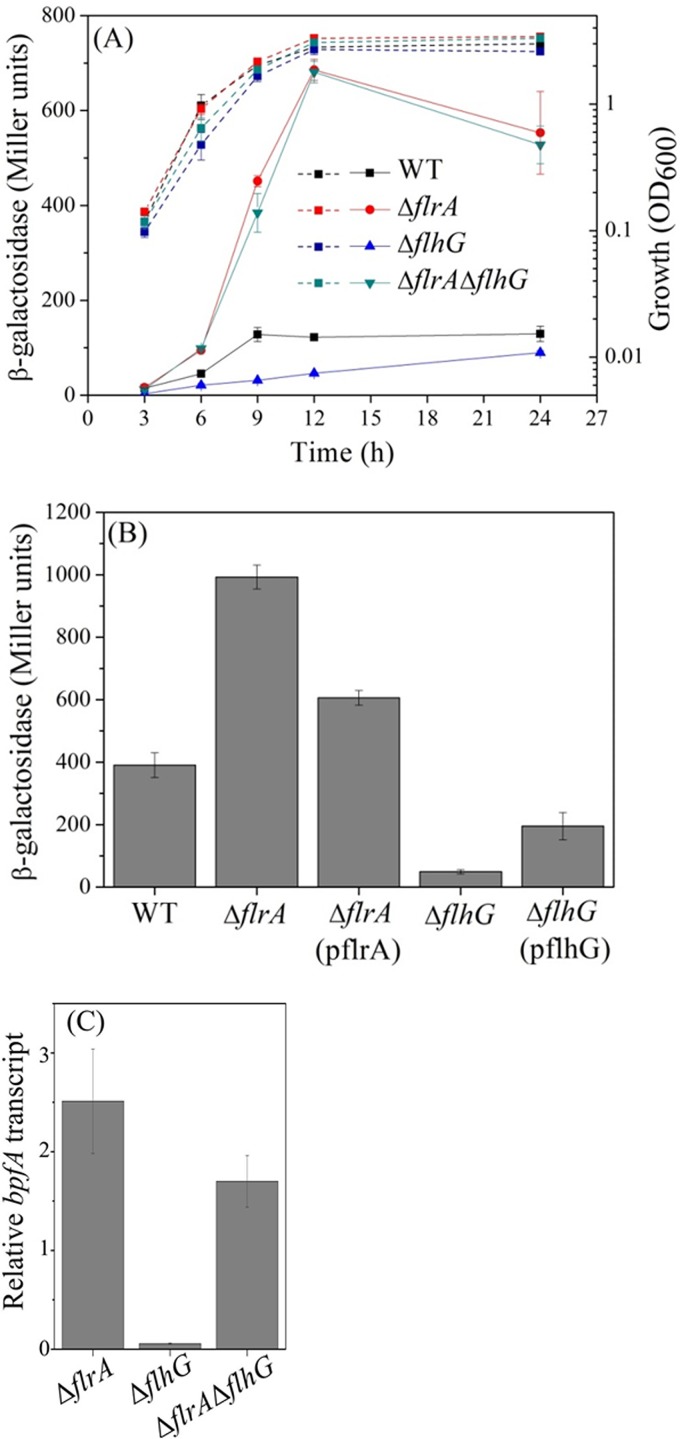
Our previous work identified the adhesin BpfA as an important factor contributing to biofilm formation in S. putrefaciens CN32 (24). To further disclose the regulation of bpfA transcription, we constructed a reporter strain, 3591Z, with a transcriptional fusion of lacZ into the bpfA operon in the genome of S. putrefaciens CN32 and performed transposon mutagenesis to screen mutants with altered β-galactosidase activity. A screening of approximately 9,000 mutants identified four white colonies and three dark-blue ones (data not shown), which suggested decreased and increased bpfA transcription, respectively. The β-galactosidase activity in these mutants was further verified during batch cultivation. Mutants that showed stable and dramatically changed activities were selected to determine the site of transposon insertion. One of the insertion sites was mapped to Sputcn32_2580, which encodes FlrA, a master transcription regulator of flagellar biosynthesis in S. oneidensis MR-1 (25). To detect the effect of FlrA on bpfA transcription, we deleted flrA in 3591Z (3591Z ΔflrA). The β-galactosidase activity in the parental 3591Z strain reached maximum at late-log phase (Fig. 1A), indicating the maximum expression of bpfA. Deletion of flrA increased the activity by 4- to 7-fold, while no obvious growth defect was observed compared with the growth of the parental strain (Fig. 1A). These results suggested that FlrA acts as a repressor of the bpfA operon.
FIG 1.
FlrA and FlhG regulate the expression of bpfA. (A) The expression of bpfA was reported by lacZ transcriptionally fused into the bpfA operon in the genome. The wild type (WT) (3591Z) and the 3591Z ΔflrA, 3591Z ΔflhG, and 3591Z ΔflrA ΔflhG mutants were cultured in LB aerobically. Their growth and β-galactosidase activities were measured at the indicated time points. (B) Levels of expression of bpfA in 3591Z ΔflrA and 3591Z ΔflhG complemented with plasmid-borne flrA (pflrA) and flhG (pflhG), respectively. The wild type and ΔflrA and ΔflhG mutants were transformed with the empty vector. To all samples, we added 0.5% l-arabinose. (C) The levels of transcription of bpfA in the wild type and the ΔflrA, ΔflhG, and ΔflrA ΔflhG mutants were examined and compared by qRT-PCR. RNA was isolated from cultures grown to an OD of 1.0. Error bars indicate standard deviations from three samples.
FlrA as a transcription factor has orthologs in Vibrio and Pseudomonas species. It can interact with an ATPase and FleN and be antagonized by the latter in P. aeruginosa but not in V. cholerae (26, 27). Since S. putrefaciens CN32 encodes an FleN ortholog (FlhG), we tested whether FlhG interferes with the negative effect of FlrA on bpfA expression. Deletion of flhG (3591Z ΔflhG) decreased β-galactosidase activity compared with that of the parental strain. However, a deficiency of flrA along with an flhG mutation (3591Z ΔflrA ΔflhG) restored β-galactosidase activity to the high level seen with the ΔflrA single mutant (3591Z ΔflrA) (Fig. 1A). Complementation with plasmid-borne flhG and flrA largely restored the β-galactosidase activities in the corresponding mutants to the level in 3591Z (Fig. 1B). These results indicated that FlhG regulation of the expression of the bpfA operon was dependent on FlrA. Interestingly, β-galactosidase activity in 3591Z ΔflrA was fully recovered to the level in 3591Z by integrating a single copy of flrA into a neutral site in the genome of 3591Z ΔflrA (see Fig. S1 in the supplemental material). A possible explanation is that FlrA also regulates other genes but not the bpfA operon or flagellum cassette. Expression of FlrA in excess might alter the cross talk between the products of these genes, thereby affecting bpfA transcription. For example, FlrA positively regulates flhG transcription (Fig. S2), and hence overexpression of FlrA likely changes the ratio between FlrA and FlhG and the consequent expression of bpfA.
To obtain direct evidence for transcription, we used quantitative reverse transcription-PCR (qRT-PCR) to compare levels of bpfA transcripts in different strains. The flrA and flhG genes were deleted from the wild-type strain of S. putrefaciens CN32. Consistently with results based on β-galactosidase activity, the bpfA transcript level increased in the ΔflrA and the ΔflrA ΔflhG mutant but decreased in the ΔflhG mutant (Fig. 1C). With consideration of the above results together, FlrA and FlhG act as negative and positive regulators, respectively, for the transcription of bpfA in S. putrefaciens CN32.
FlrA and FlhG regulation of biofilm depends on BpfA.
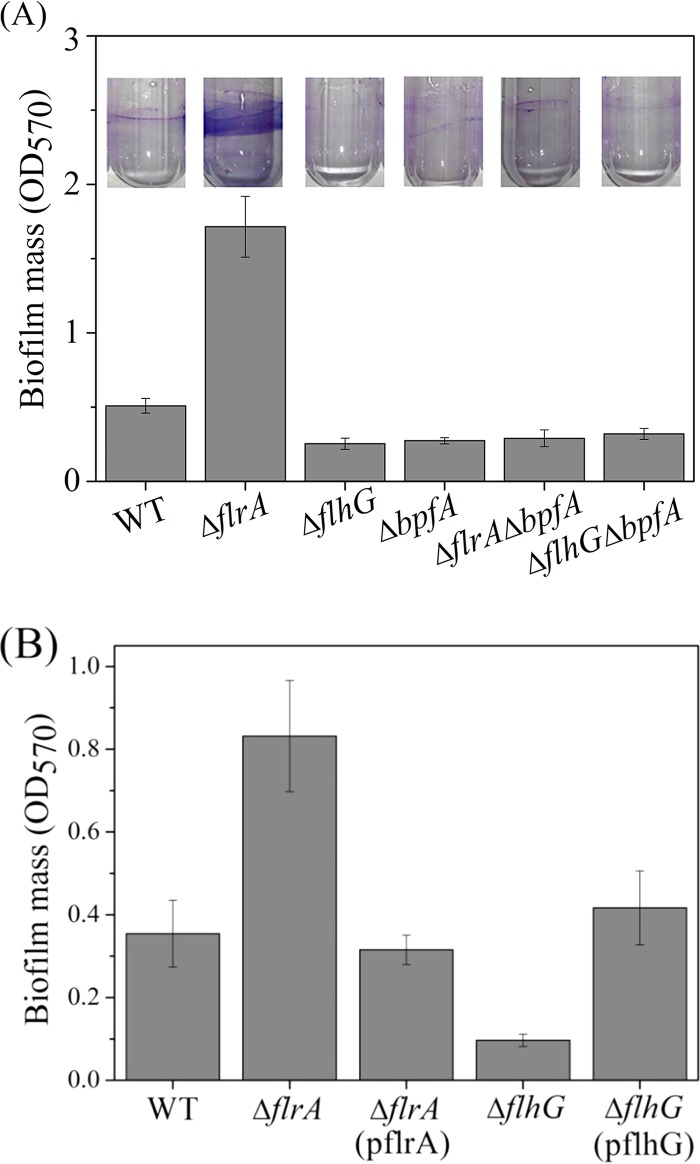
Since FlrA and FlhG are associated with bpfA transcription and since BpfA is required for biofilm development, we evaluated the role of FlrA and FlhG in biofilm formation. Compared to that of the wild-type strain, the biofilm formation of a strain with a deficiency of flrA increased and that of a strain with a deficiency of flhG was undermined (Fig. 2A), demonstrating the genes' opposite effects on biofilm formation. Deletion of bpfA along with an flrA mutation reduced the biofilm level to that of the ΔbpfA single mutant, indicating that FlrA regulation of biofilm was dependent on BpfA. Similar biofilm levels were observed for the ΔflhG, ΔbpfA, and ΔflhG ΔbpfA strains. Taken together, these results indicated that the effects of both FlrA and FlhG on biofilm formation were dependent on BpfA. The increased and reduced biofilm levels of the ΔflrA and ΔflhG single mutants, respectively, could be restored to the wild-type level by introducing plasmid-borne flrA or flhG (Fig. 2B).
FIG 2.
FlrA and FlhG regulate biofilm formation in a BpfA-dependent manner. (A) Biofilms formed by the wild type and mutants on the walls of glass tubes were quantified using a crystal violet assay after cells were grown in LB medium for 18 h. Images represent biofilms formed on glass walls. (B) Biofilms formed by the ΔflrA mutant and ΔflhG mutant complemented with flrA (pflrA) and flhG (pflhG), respectively. The wild type and ΔflrA and ΔflhG mutants were transformed with the empty vector. To all the samples, we added 0.5% l-arabinose. Error bars indicate standard deviations of results from three samples.
FlrA functions as a repressor of the bpfA operon.
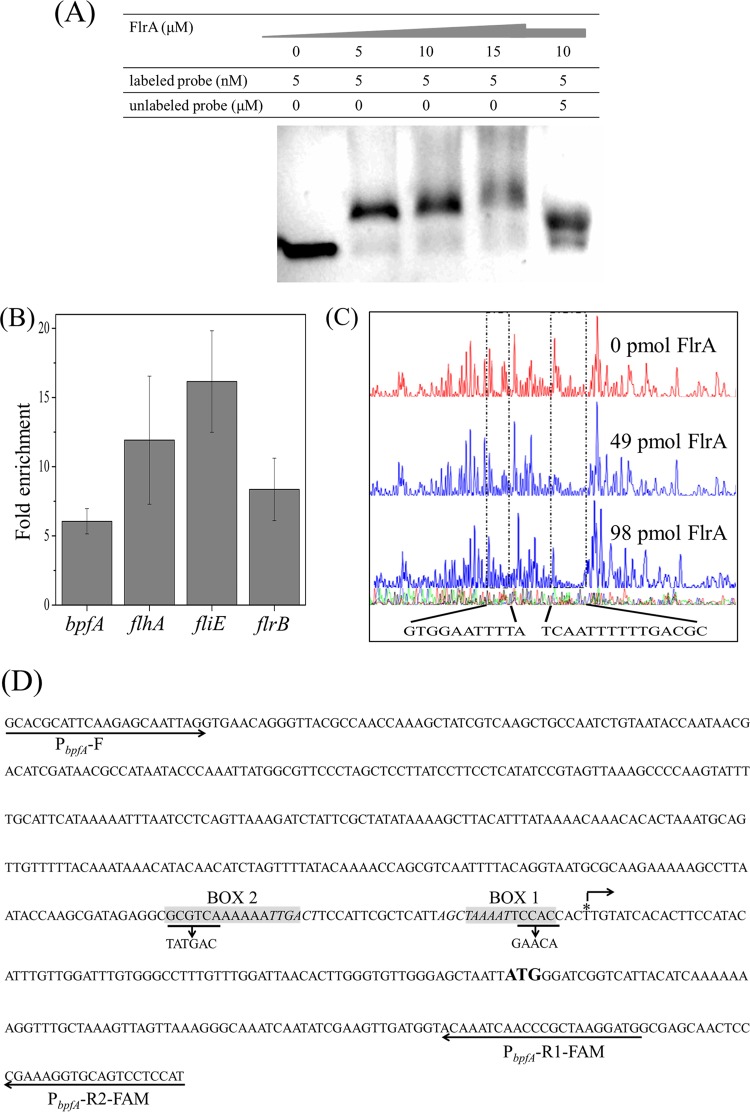
After confirming FlrA as a regulator of biofilm formation via BpfA, we asked whether FlrA could function as a direct regulator to control the transcription of the bpfA operon. In enzyme-linked immunosorbent assays (EMSAs), purified recombinant FlrA (Fig. S3) was incubated with a 6-carboxyfluorescein (FAM)-labeled DNA (577 bp) spanning from bp −480 to +97, relative to the translational start site of the bpfA operon. Binding of FlrA to the promoter of the bpfA operon was evidenced by the shifts of DNA bands (Fig. 3A). The binding was outcompeted by DNA with the same sequence but without FAM labeling (Fig. 3A). The in vitro assay supported the idea that FlrA can directly bind to the promoter of the bpfA operon. Further, we tested the in vivo binding of FlrA to the promoter of the bpfA operon by chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) analysis. In a strain of CN32FlrA-FLAG expressing FlrA with an N-terminal FLAG epitope from its native site in the genome, FlrA demonstrated binding to the promoter of the bpfA operon (Fig. 3B). The FlrA also showed binding to the promoters of flrA, fliE, and flrB (Fig. 3B), all of which belong to the flagellum cassette and are under the direct regulation of FlrA. The in vitro and in vivo assays supported the finding that FlrA can directly bind to the promoter of the bpfA operon, confirming its simultaneous regulation of the bpfA operon and flagellum cassette through direct binding.
FIG 3.
FlrA binds to two boxes in the promoter region of the bpfA operon. (A) FlrA binds to DNA fragments of the operon promoter in EMSAs. The 5′-FAM-labeled DNA was incubated with recombinant FlrA in increasing concentrations. Unlabeled DNA with the same sequence was used for competition. (B) ChIP-qPCR analysis of FlrA binding to different promoters. DNA fragments of promoters of bpfA, flhA, and fliE pulled down by FlrA were quantified using qRT-PCR. Fold enrichment was calculated as the CT of the ChIP samples relative to the CT of nonantibody samples using the formula 2−(CTIP − CTNegative). Error bars indicate standard deviations from three samples. (C) Determination of FlrA binding sites in the DNase I footprinting assay. FAM-labeled DNA that was the same as that used in EMSAs was incubated with FlrA in various amounts. Electropherograms indicate the protection pattern of DNA in the absence and presence of FlrA in the indicated concentrations. The protected DNA determined by sequencing is also shown. (D) Nucleotide sequence of the promoter region of the bpfA operon. The transcriptional start site, indicated by an asterisk, was determined by the primer extension assay using two primers (PE-bpfA-R1, PE-bpfA-R2) in independent assays. The sequences of two FlrA boxes are shown shaded. Sequences of the putative −10 and −35 regions are shown in italics. The translation start site, ATG, is in bold. Mutations introduced into FlrA boxes are underscored, and nucleotides after the substitutions are also indicated by arrows.
We next determined the precise site of FlrA binding to the promoter of the bpfA operon by performing both DNase I footprinting and primer extension analysis. When the same DNA probe as that used in an EMSA was preincubated with FlrA and then digested by DNase I, two DNA regions (box 1, GTGGAATTTTA; box 2, TCAATTTTTTGACGC) were protected by FlrA in a concentration-dependent manner (Fig. 3C). The transcription start site (TSS) of the bpfA operon was determined by the primer extension assay to be bp −76 upstream of the start codon (ATG), and this was supported by two independent assays using two primers (PbpfA-R1-FAM and PbpfA-R2-FAM) (Fig. 3D and S4). Therefore, FlrA-binding box 1 and box 2 were located at bp −79 and −109 upstream of the TSS, respectively. Furthermore, the same region recognized by σ70 was independently predicted by two online software programs, BPROM and Virtual Footprint (28, 29). Predicted −35 and −10 regions (TTGAT/AGCTAAAAT) were located at bp −84 and −107, respectively, upstream of the start codon, ATG. Thus, box 1 and box 2 overlapped the −10 and −35 regions, respectively (Fig. 3D). These results supported the hypothesis that FlrA acts as a transcriptional repressor of the bpfA operon, presumably by competing binding sites with σ70.
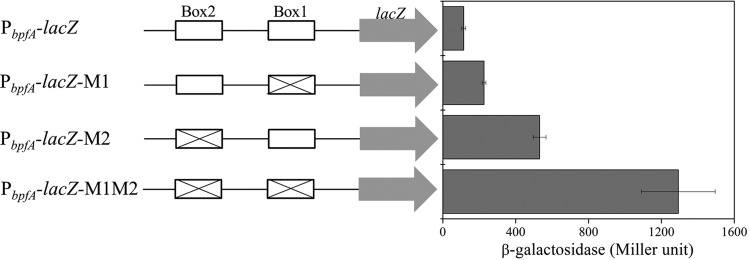
To evaluate in vivo the role of the two FlrA-binding boxes in the transcription of the bpfA operon, we constructed several reporter constructs carrying the lacZ gene, which is transcriptionally fused to the promoter with or without a mutation in the two boxes. The mutation sites are outside the −10 and −35 regions but still inside box 1 and box 2, avoiding interference with the DNA binding of σ70 for transcription initiation (Fig. 3D). The activity of β-galactosidase was measured to evaluate the strengths of promoters with a mutation in these boxes. Mutations in both box 1 and box 2 increased the transcription activity of the promoter compared to that of the wild-type promoter (Fig. 4). Thus, a full activity of FlrA to repress the transcription of bpfA involves both of the binding sites of box 1 and box 2.
FIG 4.
The two FlrA boxes are essential for FlrA repression of bpfA expression. Plasmids containing the promoter of the wild type (PbpfA-lacZ) or promoters with a mutation in FlrA box 1 (PbpfA-lacZ-M1), box 2 (PbpfA-lacZ-M2), or both (PbpfA-lacZ-M1M2) were introduced into the wild-type strain. The β-galactosidase activities of overnight cultures were measured to evaluate promoter strength. Error bars indicate standard deviations of results from three samples.
c-di-GMP interferes with the binding of FlrA to the promoter of bpfA.
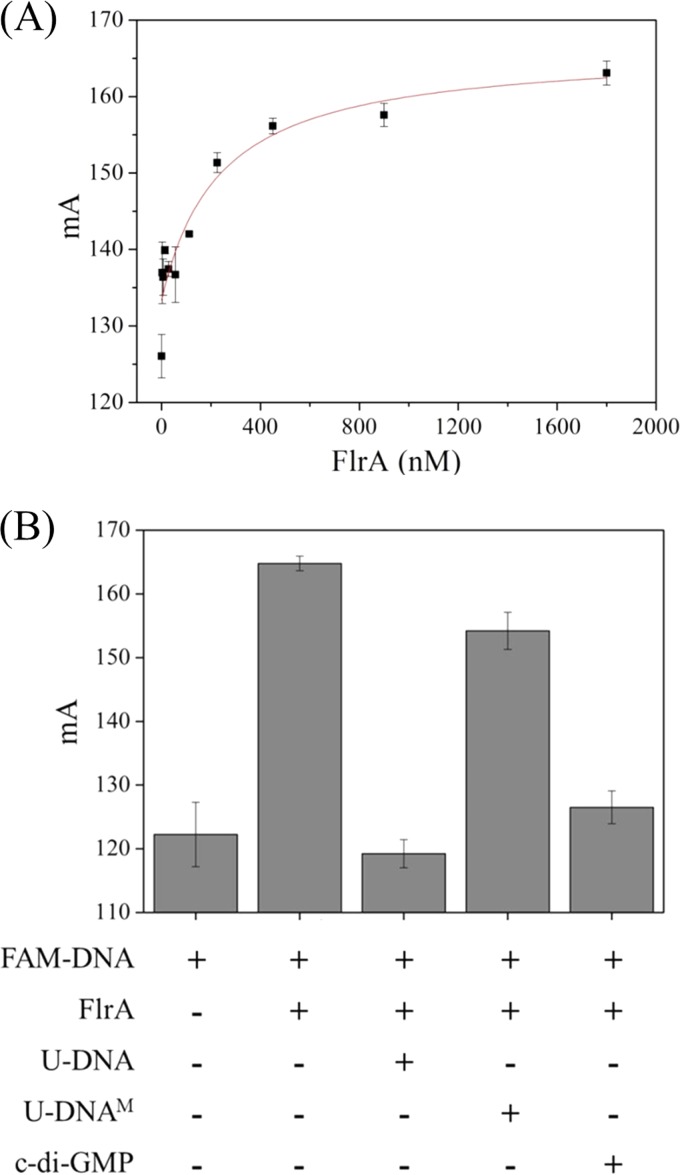
FleQ, an FlrA ortholog in Pseudomonas aeruginosa, functions as a c-di-GMP-responsive regulator, and its residues involved in the biding of c-di-GMP have been characterized (30). FlrA also possesses these conserved residues (Fig. S5) (30), which suggests that the activity of FlrA may be tuned by c-di-GMP, a second messenger found in many bacteria (31). We first tested the effect of c-di-GMP on the binding of FlrA to the FlrA box ib bpfA promoter region in vitro via fluorescence polarization (FP). When the FAM-labeled DNA of the promoter containing box 2 was incubated with FlrA, fluorescence anisotropy of the DNA increased with the increase of FlrA's concentration (Fig. 5A), indicating the binding of FlrA to the labeled DNA. The binding was outcompeted by an unlabeled DNA with the same sequence but was almost not affected by a DNA with a mutation in box 2, as observed before (Fig. 5B). Moreover, binding of FlrA to the labeled DNA was almost completely abolished in the presence of c-di-GMP (Fig. 5B). This in vitro data supported the hypothesis that a small molecule of c-di-GMP inactivated the binding of FlrA to the promoter of the bpfA operon.
FIG 5.
c-di-GMP represses the binding of FlrA to the promoter of the bpfA operon as determined by fluorescence polarization. (A) The binding of FlrA to FAM-labeled DNA covering box 2 of the promoter region was determined by fluorescence polarization. FAM-labeled DNA (15 nM) was incubated with FlrA at various concentrations. mA, mili-anisotropy value. (B) The binding of FlrA (450 nM) to this FAM-labeled DNA (FAM-DNA; 15 nM) was abolished by 25 μM c-di-GMP. The binding was outcompeted by unlabeled DNA with the same sequence (U-DNA) but not by unlabeled DNA with a mutation in box 2 (U-DNAM). Error bars indicate standard deviations from three samples.
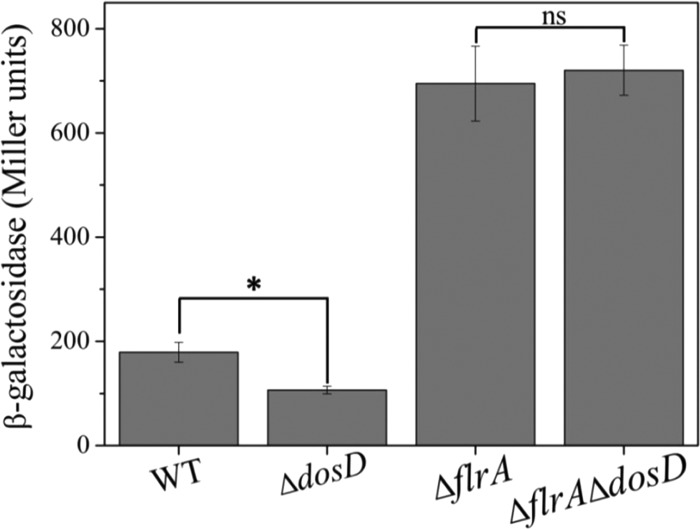
We have identified DosD as a diguanylate cyclase for c-di-GMP synthesis in S. putrefaciens CN32. Deletion of DosD results in a level of intracellular c-di-GMP that is decreased by approximately 50% under aerobic growth conditions (24). A decreased level of intracellular c-di-GMP is expected to increase the transcriptional repression of the bpfA operon by FlrA, thus reducing the expression of the bpfA operon. Indeed, deletion of dosD to a lower intracellular c-di-GMP level significantly reduced the transcriptional activity of the bpfA promoter, as indicated by expression of lacZ fused to the promoter (Fig. 6). Deletion of flrA along with deletion of dosD increased the promoter activity to a level comparable to that in the ΔflrA mutant (Fig. 6). These results indicated that the effect of c-di-GMP derived from DosD on bpfA transcription depends on FlrA. Taking these results together, we concluded that the regulation of FlrA on the transcription of the bpfA operon is modulated by c-di-GMP.
FIG 6.
The regulation of DosD on the expression of the bpfA operon was dependent on FlrA. The β-galactosidase activities in overnight cultures of the wild type (3591Z), 3591Z ΔdosD, 3591Z ΔflrA, and 3591Z ΔflrA ΔdosD were measured to evaluate the expression of the bpfA operon. Error bars indicate standard deviations from three samples. *, significantly different [P < 0.05]; ns, not significantly different.
DISCUSSION
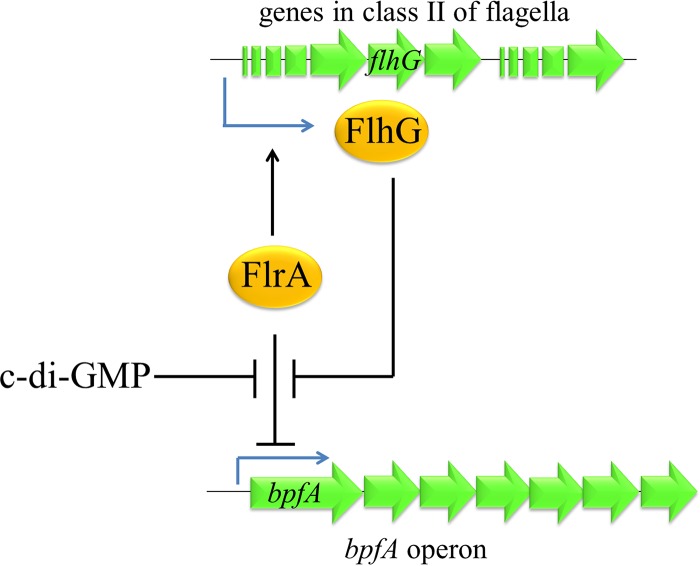
The present work reveals FlrA as a c-di-GMP-responsive regulator of S. putrefaciens CN32 that controls the expression of an operon encoding the adhesin BpfA and its accessory system, thereby regulating biofilm formation (Fig. 7). FlrA acts as a repressor of the bpfA operon by binding to two boxes in the promoter region of the bpfA operon, and the binding is abolished by c-di-GMP. c-di-GMP is a key messenger controlling biofilm formation in many bacteria, including Shewanella (32). Increasing the c-di-GMP level in S. oneidensis MR-1 by deletion of pdeB, encoding a c-di-GMP phosphodiesterase, decreases cell motility and increases biofilm formation (33). Many Shewanella species, such as S. oneidensis MR-1, encode a large number of putative proteins with conserved GGEDF and/or EAL domains, presumably involved in c-di-GMP synthesis, hydrolysis, or binding as effectors (34), suggesting that c-di-GMP might play a role in the physiology and niche adaptation of these species. However, the activities of most of these putative proteins have not been examined. We previously revealed that BpfA plays an essential role in the aerobic biofilm formation of S. putrefaciens CN32, which is regulated by the diguanylate cyclase DosD, whose activity is controlled by oxygen (24). However, there had been a missing component in this c-di-GMP signaling pathway which regulates BpfA-dependent biofilm formation, that is, the factor that mediates c-di-GMP's regulation of the transcription of the bpfA operon. In the present work, we identify FlrA as a responsive regulator connecting DosD with the bpfA operon.
FIG 7.
Schematic of the proposed regulation of FlrA on the expression of the bpfA operon. The repression of FlrA on the transcription of the bpfA operon is inhibited by c-di-GMP and FlhG. FlrA is also the master regulator of flagellum synthesis.
Shewanellae are phylogenetically closer to Vibrio than to Pseudomonas. However, the regulatory mode of FlrA in S. putrefaciens CN32 is similar to that of its ortholog in P. aeruginosa, FleQ, but different from that of its ortholog in V. cholerae, FlrA. In V. cholerae, the FlrA regulon has been limited in flagellum-coding genes to date, and the FlrA regulation of flagellum synthesis seems unaffected by FlhG (26). The deficiency of FlrA in Vibrio results in the altered expression of some virulence genes and binding to epithelial cells, while it is still unclear whether FlrA directly regulates the transcription of these genes (35, 36). In contrast, FleQ in P. aeruginosa regulates many biofilm-related genes, such as pel and psl for exopolysaccharide synthesis and cdrAB, coding for an adhesin. Moreover, FleQ's regulation of the pel operon's transcription is antagonized by FleN, an FlhG ortholog (37). As with these characteristics of FleQ, FlrA in S. putrefaciens CN32 controls biofilm formation through regulating transcription of the bpfA operon, which is also antagonized by FlhG (Fig. 7). Our present results suggest a potential interaction between FlrA and FlhG to control the transcription of the bpfA operon; however, we still cannot exclude the possibility that other mechanisms may be involved. It has been reported that FlhG affects the number and location of flagella not as a result of FlrA but through direct interaction with flagellar C-ring proteins (38).
Despite its similarity to FleQ in Pseudomonas, FlrA of S. putrefaciens has special features in its regulatory mode. FleQ functions both as a repressor and as an activator in the expression of several biofilm-related genes in P. aeruginosa in response to the c-di-GMP level. It represses the transcription of the pel and cdr operons by binding to two or three boxes around −35/−10 regions under conditions of low c-di-GMP levels, while activating their transcription with an increase of the intracellular c-di-GMP level (39). In contrast, FlrA of S. putrefaciens CN32 binds to two boxes overlapping the −10 and −35 regions in the promoter of bpfA, presumably by repressing the binding of σ70 and subsequent initiation of the transcription of the bpfA operon. Therefore, the binding site of FlrA to the bpfA promoter indicates that FlrA functions as a repressor only of the bpfA operon. Moreover, the activity of FlrA as a transcriptional repressor can be tuned by c-di-GMP, since c-di-GMP abolished the binding of FlrA to the promoter of the bpfA operon.
Orthologs of FlrA in Pseudomonas and Vibrio species are master regulators of flagellum synthesis (35, 40). Thus, FlrA is supposed to be involved in flagellum synthesis in S. putrefaciens CN32. Indeed, the transcription of several flagellar genes and the phenotype of cell motility decreased in the flrA-deficient mutant (see Fig. S2 in the supplemental material). Biofilm and motility, representing sessile and planktonic states, respectively, are mutually exclusive lifestyles of bacteria in response to the fluctuation of diverse environmental conditions, such as nutrients and stress (1). This work reveals a coordination of biofilm and motility by FlrA in S. putrefaciens CN32. Interestingly, flrA and bpfA cooccur in the genomes of many Shewanella species (Fig. S6A and B). Moreover, two FlrA boxes are also found in the promoter regions of the genes coding for BpfA orthologs. Sequences of one box are highly conserved, and those of another box are relatively varied (Fig. S6C). These results suggest that FlrA regulation of bpfA-dependent biofilms is a common mechanism in these bacteria. However, the distribution of dosD is not as wide as those of FlrA and BpfA (24). In Shewanella species containing FlrA and BpfA but not DosD, other genes coding enzymes for c-di-GMP synthesis or hydrolysis are expected to be involved in tuning FlrA activity and consequent bpfA transcription.
The bpfA operon is the first target of FlrA identified to date in Shewanella species, except for genes involved in flagellum synthesis. Further investigation into the FlrA regulon is under way to comprehensively understand its regulatory mechanism and consequent physiological role in this bacterial species.
MATERIALS AND METHODS
Strains and growth conditions.
S. putrefaciens CN32 and Escherichia coli strains (Table 1) were cultured in Luria broth (LB) medium at 30°C and 37°C, respectively. Chemicals were added when needed at the following concentrations: 100 μg ml−1 diaminopimelic acid, 50 μg ml−1 kanamycin, 34 μg ml−1 chloramphenicol, and 20 μg ml−1 gentamicin for E. coli strains and 10 μg ml−1 chloramphenicol and gentamicin for S. putrefaciens strains.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. putrefaciens | ||
| CN32 | Wild type | ATCC |
| 3591Z | CN32 derivative with lacZ transcriptionally fused into the bpfA operon | This study |
| CN32FLAG-FlrA | CN32 derivative with FLAG translationally fused into the 5′ end of flrA | This study |
| 3591Z ΔflrA | 3591Z derivative with deletion of Sputcn32_2580 | This study |
| 3591Z ΔflhG | 3591Z derivative with deletion of Sputcn32_2560 and flhG | This study |
| 3591Z ΔflrA ΔflhG | 3591Z derivative with double deletion of flrA and flhG | This study |
| CN32 ΔflrA | CN32 derivative with deletion of flrA | This study |
| CN32 ΔflhG | CN32 derivative with deletion of flhG | This study |
| CN32 ΔflrA ΔflhG | CN32 derivative with double deletion of flrA and flhG | This study |
| CN32 ΔbpfA | CN32 derivative with deletion of Sputcn32_3591 | 24 |
| CN32 ΔflrA ΔbpfA | CN32 derivative with double deletion of flrA and bpfA | This study |
| CN32 ΔflhG ΔbpfA | CN32 derivative with double deletion of flhG and bpfA | This study |
| E. coli | ||
| JM109 | General cloning host for plasmid manipulation | New England Biolabs |
| MG1655 | Derivative of E. coli K-12 | ATCC |
| WM3064 | Auxotrophic to DAP; used for conjugation | 48 |
| BL21(DE3) | Expression host carrying the T7 RNA polymerase | Novagen |
| Plasmids | ||
| pRL27 | Tn5 delivery vector, Kmr | 41 |
| pRE112 | Suicide vector, Cmr | 49 |
| pRE-flrA | pRE112 derivative to delete flrA | This study |
| pRE-flhG | pRE112 derivative to delete flhG | This study |
| pRE-PbpfA-lacZ | pRE112 derivative to construct 3591Z | This study |
| pRE-flag-flrA | pRE112 derivative to construct CN32FLAG-FlrA | This study |
| pET-28a(+) | Bacterial expression vector | Novagen |
| pET-FlrA | pET-28a(+) derivative to express His-tagged FlrA | This study |
| pJN105 | Broad-host-range vector carrying the araBAD promoter | 50 |
| pflrA | pJN105 derivative to express FlrA | This study |
| pflhG | pJN105 derivative to express FlhG | This study |
| pBBR1MCS-5 | Broad-host-range plasmid | 51 |
| PbpfA-lacZ | pBBR1MCS5 derivative bearing the wild-type bpfA promoter fused with lacZ | This study |
| PbpfA-lacZ-M1 | PbpfA-lacZ derivative with a mutation in FlrA box 1 | This study |
| PbpfA-lacZ-M2 | PbpfA-lacZ derivative with a mutation in FlrA box 2 | This study |
| PbpfA-lacZ-M1M2 | PbpfA-lacZ derivative with a mutation in FlrA box 1 and box 2 | This study |
DAP, diaminopimelic acid.
Strain construction.
Mutants with an in-frame deletion of the desired genes were constructed as described previously (24). Briefly, two flanking regions of a target gene were amplified, ligated, and inserted into pRE112. The generated plasmids were introduced into strains of S. putrefaciens CN32 by conjugation with E. coli WM3064. After a two-step selection, mutants with a deletion of a target gene were obtained, with the DNA sequence at mutation sites being verified by sequencing.
The S. putrefaciens CN32 derivative of 3591Z containing lacZ transcriptionally fused to the bpfA operon in the genome (PbpfA-lacZ fusion) was constructed to report the transcription from the bpfA operon promoter in vivo. The open reading frame of lacZ from E. coli MG1655 was cloned into the pRE112 vector, flanked by the promoter and the coding region of bpfA. S. putrefaciens was transformed with the generated plasmid. After a two-step selection, we obtained the 3591Z strain with a lacZ insertion into the genome, and it was under the control of the bpfA operon promoter. The flrA and flhG genes were deleted from 3591Z, generating 3591Z ΔflrA, 3591Z ΔflhG, and 3591Z ΔflrA ΔflhG.
To construct a strain of CN32FLAG-FlrA expressing FlrA with an N-terminal tag of FLAG, the suicide vector pRE-flag-flrA was introduced into the wild-type strain S. putrefaciens CN32 by conjugation. After a two-step selection and sequencing procedure, we obtained CN32FLAG-FlrA expressing FLAG-FlrA from its native transcription and translation initiation signals. All strains and vectors used in this study are listed in Table 1.
Transposon mutagenesis and screening.
To identify factors that regulate bpfA expression, transposon mutagenesis was performed using 3591Z as a parental strain. The suicide plasmid pRL27, carrying a transposase, mini-Tn5 transposon, and an RK6 origin (41), was introduced into 3591Z by conjugation. The resultant cells were then plated on LB agar plates containing kanamycin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to select mutants containing the mini-Tn5 transposon. After incubation at 30°C for 24 to 48 h, white or dark-blue colonies were inoculated into LB for analysis of bacterial growth and β-galactosidase activity according to the method developed by Miller (42).
Mutants with increased β-galactosidase activity, compared with that of the parental strain 3591Z, were selected to map the insertion site of a transposon according to a modified nested semiarbitrary PCR method (43). Briefly, during the first round of PCR, genomic DNA was amplified using two primers, with one (pRL27Etdx or pRL27Extsx) hybridizing to one end of mini-Tn5 and the other (a mixture of five degenerate primers, arb1, arb2, arb3, arb4, and arb5) hybridizing to an arbitrary sequence with a 5′-GC clamp. One microliter of the product was subjected to a second round of PCR using the pRL27Intdx/pRL27Intsx primer pair and arb6, with the same sequence as the 5′-GC clamp. The second-round product was purified and sequenced to determine the insertion site of a transposon. Primers used are listed in Table 2.
TABLE 2.
Primers used in this study
| Application(s) and primer | DNA sequence |
|---|---|
| Transposon mutagenesis | |
| pRL27Etdx | CCAGAAAGTGAGGGAGCCA |
| pRL27Extsx | GACAACAAGCCAGGGATG |
| pRL27Intdx | GAGTCGACCTGCAGGCATGC |
| pRL27Intsx | CGCACTGAGAAGCCCTTAGAGC |
| arb1 | GGCCACGCGTCGACTAGTACNNNNNNNNNNAGAG |
| arb2 | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC |
| arb3 | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT |
| arb4 | GGCCACGCGTCGACTAGTACNNNNNNNNNNAACGC |
| arb5 | GGCCACGCGTCGACTAGTACNNNNNNNNNNTCGAC |
| arb6 | GGCCACGCGTCGACTAGTAC |
| EMSA, footprinting, and primer extension | |
| PbpfA-F | GCACGCATTCAAGAGCAATTAG |
| PbpfA-R1-FAM | FAM-ACAAATCAACCCGCTAAGGATG |
| PbpfA-R2-FAM | FAM-ATGGAGGACTGCACCTTTCG |
RNA isolation and qRT-PCR.
Total RNA was isolated from S. putrefaciens cultures grown to an optical density at 600 nm (OD600) of 1.0 using RNAiso Plus (TaKaRa Co., China) according to the manufacturer's instructions. Total RNA was treated with DNase I to digest genomic DNA and subjected to PCR analysis to ensure the complete digestion of the DNA. DNA-free RNA was then used for the reverse transcription to synthesize cDNA using the PrimeScript RT reagent kit (TaKaRa Co., China). Quantitative reverse transcription-PCR (qRT-PCR) was performed with the StepOne real-time PCR system (Applied Biosystems, USA) using synthesized cDNA and the SYBR Premix Ex Taq kit (TaKaRa Co., China). Relative expression values of target genes were obtained from three determinations, with normalization against 16S rRNA using the method of 2–ΔΔCT, where CT is the threshold cycle.
Biofilm formation and quantification.
The biomass of biofilm was quantified as described before (24). Overnight cultures were inoculated into fresh LB to an OD600 of 0.01 in glass tubes and incubated at 30°C for 18 h with shaking at 100 rpm. Biofilm formed on the tube walls was stained using 0.5% crystal violet for 15 min at room temperature, rinsed with 10 mM phosphate-buffered saline (PBS), pH 7.2, and air dried. Absorbed crystal violet was dissolved using 33% (vol/vol) glacial acetic acid. The absorption of dissolved crystal violet at 570 nm was determined for quantification of biofilm mass.
FlrA production and purification.
FlrA was cloned into the pET-28a(+) vector (Novagen, USA) between NcoI and XhoI sites, and the resulting plasmid, pET-flrA, was sequenced to verify the flrA sequence. FlrA with an N-terminal 6×His tag was expressed in E. coli BL21(DE3) and purified. In detail, E. coli BL21(DE3) carrying pET-flrA was subcultured from an overnight culture into fresh LB and grown at 37°C with shaking at 200 rpm. After growth to an OD600 of 0.6 to 0.8, the culture was transferred to 16°C for 30 min before addition of 0.1 mM isopropyl-β-d-thiogalactoside (IPTG) to induce the expression of FlrA. After induction for 12 h, cells were harvested, resuspended in a lysis buffer (50 mM HEPES [pH 8.0], 250 mM NaCl, 0.1% Triton X-100, and 10% glycerol), and lysed by sonication. The supernatant after centrifugation was loaded onto a 1.0-ml preequilibrated nitrilotriacetic acid (Ni-NTA; Qiagen, USA) and sequentially washed with wash buffer (50 mM Tris-HCl [pH 9.0], 300 mM KCl, and 10% glycerol) containing imidazole at concentrations of 10, 20, 50, and 100 mM. The FlrA was eluted with elution buffer (50 mM HEPES [pH 8.0], 250 mM NaCl, 0.1% Triton X-100, 250 mM imidazole, and 10% glycerol), dialyzed against storage buffer (50 mM HEPES [pH 8.0], 250 mM NaCl, 0.1% Triton X-100, 50% glycerol) at 4°C overnight, and stored at −70°C until use.
EMSA.
Electrophoretic mobility shift assay (EMSA) was performed to test the binding of FlrA to DNA fragments of the bpfA operon promoter in vitro. The promoter probe of the bpfA operon was amplified using the PbpfA-F and PbpfA-R1-FAM primer pair with 6-carboxyfluorescein (FAM) labeling at its 5′ end (Table 2). This probe covered the intergenic region between bpfA and its upstream gene Sputcn_3590 from bp −97 to +480, relative to the bpfA translational start site. In detail, the probe of 0.1 pmol (40 ng) was incubated with various amounts of FlrA in a reaction buffer [50 mM Tris-HCl (pH 8.0), 100 mM KCl, 2.5 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.01 mM ATP, 25 ng poly(dI-dC), and 10% glycerol] in a total volume of 20 μl. Besides poly(dI-dC), sheared salmon sperm DNA (100 ng μl−1) was added as a nonspecific competitor. After incubation at 25°C for 30 min, the mixture was loaded into a 2% agarose gel for electrophoresis in a 1× TBE buffer (89 mM Tris, 89 mM boric acid, 2 mM EDTA). Gels were scanned with an ImageQuant LAS 4000 mini (GE Healthcare, USA).
DNase I footprinting assay.
To determine the binding sites of FlrA in the promoter of the bpfA operon, DNase I footprinting was performed as described previously, with modification (44, 45). In detail, 1.5 pmol (600 ng) of the same FAM-labeled probe as used in the EMSA was incubated with various amounts of FlrA in a total volume of 40 μl. The mixture was preincubated at 25°C for 30 min for the binding between DNA and FlrA. Then, 0.015 unit DNase I (Promega, USA) was added and the mixture was incubated for 1 min for DNA digestion; then the reaction was immediately stopped by addition of 140 μl of a DNase I stop solution (200 mM unbuffered sodium acetate, 30 mM EDTA, and 0.15% SDS). The DNA in the mixture was extracted with phenol-chloroform, precipitated with ethanol, and finally dissolved in 30 μl of MilliQ water. Preparation of a DNA ladder, electrophoresis, and data analysis were the same as described before, except that the GeneScan-LIZ500 size standard (Applied Biosystems, USA) was used.
Primer extension analysis.
The transcriptional start site of the bpfA operon was determined using the fluorescence-based primer extension method described previously, with modification (46). Briefly, total RNA was isolated from cultures of the S. putrefaciens CN32 wild-type strain grown to an OD600 of 1.0. The cDNA was synthesized using the Primer Extension System (Promega, USA) according to the manufacturer's instructions. Total RNA of 10 μg was annealed with 5′-FAM-labeled primers (PbpfA-R1-FAM or PbpfA-R2-FAM) (Table 2). The resulting cDNA was precipitated with 2.5 volumes of 95% ethanol and 0.1 volume of 3 M sodium acetate, washed with 75% ethanol, air dried, and dissolved in 15 μl of MilliQ water. The size of the cDNA was determined by capillary electrophoresis using an Applied Biosystems 3730 DNA analyzer, with a GeneScan-LIZ500 size standard (Applied Biosystems, USA) as a molecular weight ladder. The transcription start site was then analyzed based on the sequences of the first-strand synthesis primers and the cDNA size mapping to the bpfA promoter.
ChIP and qRT-PCR analysis.
Mid-exponential-phase culture (OD600 of 0.6) of 20 ml was treated with 1% formaldehyde in PBS, pH 7.4, at room temperature for 30 min. Cross-linking was quenched with 250 mM glycine at room temperature for 10 min. Cells were washed three times with PBS, resuspended in 500 μl of a lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 0.5% Triton X-100, 0.5 mM DTT, and protease inhibitor cocktail from Sigma), and lysed by sonication with the SCIENTZ08 ultrasonicator (Scientz, China). The lysate was centrifuged at 12,000 × g for 20 min at 4°C. The supernatant was diluted with chromatin immunoprecipitation (ChIP) dilution buffer (Tris-HCl [pH 8.0], 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, and protease inhibitor cocktail from Sigma). Diluted lysate was incubated with a 1:300 dilution of anti-FLAG antibody (Cell Signaling Technology [CST], USA) as the ChIP sample or of IgG (CST, USA) as the negative control. After incubation at 4°C overnight with rotation, protein G magnetic beads (CST, USA) were added, and the mixture was incubated at 4°C for 4 h with rotation. Beads were washed with low-salt buffer (50 mM HEPES [pH 7.5], 0.1% deoxycholate, 1% Triton X-100, 1 mM EDTA, 150 mM NaCl) and high-salt buffer (50 mM HEPES [pH 7.5], 0.1% deoxycholate, 1% Triton X-100, 1 mM EDTA, 500 mM NaCl). Cross-linked complex was eluted by incubation with 150 μl of a freshly prepared elution buffer (1% SDS, 0.1 M NaHCO3) at 65°C with gentle vortexing for 30 min. Cross-links were reversed by the addition of 6 μl of 5 M NaCl and 2 μl protease K (20 mg/ml; TaKaRa, Japan) and incubation at 65°C for 4 h. DNA was purified using a PCR purification kit (Qiagen, USA).
The qPCR was performed as described above to quantitatively determine the amount by which a ChIP sample was enriched for individual loci. Fold enrichment was calculated using the formula 2−(CTIP − CTNegative), where CT is the fractional threshold cycle of the immunoprecipitation (IP) samples and negative controls.
Site-directed mutagenesis of the FlrA-binding DNA boxes.
The same promoter region of the bpfA operon as tested in the EMSA was used to examine the functions of two FlrA boxes. Mutations in the FlrA-binding boxes were introduced by overlapping PCR using two primer pairs with expected substitutions at the 5′ ends of a joint primer. The resulting DNA fragments with expected mutations were ligated with lacZ, inserted into pBBR1MCS5, and then introduced into the wild-type strain of S. putrefaciens CN32. The activity of β-galactosidase was measured to evaluate the transcription from mutated promoters.
FP.
The binding activity of FlrA to the promoter DNA of the bpfA operon was assayed by fluorescence polarization (47). The DNA of 30 bp (AGAGGCGCGTCAAAAAATTGACTTCCATTC) covering FlrA box 2 in the promoter region was chosen. For the competition assay, a site mutation of the same FlrA box 2 as used for the above-described experiment was introduced into the 30-bp DNA (AGAGGCTATGACAAAAATTGACTTCCATTC).The FAM-labeled and unlabeled synthetic oligonucleotides of 30 bases were synthesized by General Biosystems (China). The two strands of complementary oligonucleotides were annealed in annealing buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl) by heating them to 95°C for 10 min and slowly cooling them (30 min) to room temperature. FlrA was serially diluted in the same storage buffer as used for FlrA purification. To analyze the binding affinity, 15 nM DNA with 5′-labeled FAM was incubated with various concentrations of FlrA in a fluorescence polarization (FP) buffer (50 mM Tris-HCl [pH 8.0], 100 mM KCl, 2.5 mM MgCl2, 0.5 mM DTT, 0.01 mM ATP) at 25°C for 30 min before FP measurement. For competition assay, 15 nM FAM-labeled DNA and 450 nM FlrA were incubated with 1.5 μM unlabeled DNA containing wild-type or mutated FlrA box 2. In addition, 15 nM FAM-labeled DNA and 450 nM FlrA were incubated with 25 μM c-di-GMP to analyze c-di-GMP's effect on FlrA's binding to DNA. The fluorescence anisotropy was measured using SpectraMax M5 (Molecular Devices, USA), and the results are presented in mili-anisotropy values. The change in mili-anisotropy value as a function of FlrA concentration was used to determine the equilibrium dissociation constant (Kd) for the interaction between FlrA and FAM-labeled DNA. The data were fitted to the equation mA = mAf + [(mAb − mAf) · x/(Kd + x)], where mA indicates the measured mili-anisotropy value, mAf the mili-anisotropy value of the free FAM-labeled DNA, mAb the mili-anisotropy value of the bound FAM-labeled DNA, and x the FlrA concentration.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuzhi Hong from Rutgers University and Albert Remus Romero Rosana from the University of Alberta for valuable suggestions and critical comments.
This work was supported by the National Natural Science of Foundation of China (grant 31571286 to Yuan-Yuan Cheng), the Natural Science Foundation of Anhui Province (grant 1608085QC47 to Yuan-Yuan Cheng), the Natural Science Foundation of the Department of Education of Anhui Province (grant KJ2015A010 to Yuan-Yuan Cheng), and the Scientific Research Foundation for Talented Scholars from Anhui University (grant J01006042 to Yuan-Yuan Cheng).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02410-16.
REFERENCES
- 1.Stoodley P, Hall-Stoodley L, Costerton JW. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Doghri I, Rodrigues S, Bazire A, Dufour A, Akbar D, Sopena V, Sable S, Lanneluc I. 2015. Marine bacteria from the French Atlantic coast displaying high forming-biofilm abilities and different biofilm 3D architectures. BMC Microbiol 15:231. doi: 10.1186/s12866-015-0568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu JL, Zhao AF, Feng LF, Gao HC. 2016. Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica. Int J Food Microbiol 217:146–155. doi: 10.1016/j.ijfoodmicro.2015.10.020. [DOI] [PubMed] [Google Scholar]
- 4.Renslow RS, Babauta JT, Majors PD, Beyenal H. 2013. Diffusion in biofilms respiring on electrodes. Energy Environ Sci 6:595–607. doi: 10.1039/C2EE23394K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hau HH, Gralnick JA. 2007. Ecology and biotechnology of the genus Shewanella. Annu Rev Microbiol 61:237–258. doi: 10.1146/annurev.micro.61.080706.093257. [DOI] [PubMed] [Google Scholar]
- 6.Cao B, Majors PD, Ahmed B, Renslow RS, Silvia CP, Shi L, Kjelleberg S, Fredrickson JK, Beyenal H. 2012. Biofilm shows spatially stratified metabolic responses to contaminant exposure. Environ Microbiol 14:2901–2910. doi: 10.1111/j.1462-2920.2012.02850.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lies DP, Hernandez ME, Kappler A, Mielke RE, Gralnick JA, Newman DK. 2005. Shewanella oneidensis MR-1 uses overlapping pathways for iron reduction at a distance and by direct contact under conditions relevant for biofilms. Appl Environ Microbiol 71:4414–4426. doi: 10.1128/AEM.71.8.4414-4426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubiel M, Hsu CH, Chien CC, Mansfeld F, Newman DK. 2002. Microbial iron respiration can protect steel from corrosion. Appl Environ Microbiol 68:1440–1445. doi: 10.1128/AEM.68.3.1440-1445.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HB, Hu C, Hu XX, Yang M, Qu JH. 2012. Effects of disinfectant and biofilm on the corrosion of cast iron pipes in a reclaimed water distribution system. Water Res 46:1070–1078. doi: 10.1016/j.watres.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Bagge D, Hjelm M, Johansen C, Huber I, Grami L. 2001. Shewanella putrefaciens adhesion and biofilm formation on food processing surfaces. Appl Environ Microbiol 67:2319–2325. doi: 10.1128/AEM.67.5.2319-2325.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao A, Zhu J, Ye X, Ge Y, Li J. 2016. Inhibition of biofilm development and spoilage potential of Shewanella baltica by quorum sensing signal in cell-free supernatant from Pseudomonas fluorescens. Int J Food Microbiol 230:73–80. doi: 10.1016/j.ijfoodmicro.2016.04.015. [DOI] [PubMed] [Google Scholar]
- 12.Satchell KJ. 2011. Structure and function of MARTX toxins and the large repetitive RTX proteins. Annu Rev Microbiol 65:71–90. doi: 10.1146/annurev-micro-090110-102943. [DOI] [PubMed] [Google Scholar]
- 13.Martinez-Gil M, Yousef-Coronado F, Espinosa-Urgel M. 2010. LapF, the second largest Pseudomonas putida protein, contributes to plant root colonization and determines biofilm architecture. Mol Microbiol 77:549–561. doi: 10.1111/j.1365-2958.2010.07249.x. [DOI] [PubMed] [Google Scholar]
- 14.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 15.Paganelli FL, de Been M, Braat JC, Hoogenboezem T, Vink C, Bayjanov J, Rogers MRC, Huebner J, Bonten MJM, Willems RJL, Leavis HL. 2015. Distinct SagA from hospital-associated clade A1 Enterococcus faecium strains contributes to biofilm formation. Appl Environ Microbiol 81:6873–6882. doi: 10.1128/AEM.01716-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinsa-Leasure SM, Koid C, Tiedje JM, Schultzhaus JN. 2013. Biofilm formation by Psychrobacter arcticus and the role of a large adhesin in attachment to surfaces. Appl Environ Microbiol 79:3967–3973. doi: 10.1128/AEM.00867-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Gil M, Ramos-Gonazlez MI, Espinosa-Urgel M. 2014. Roles of cyclic di-GMP and the Gac system in transcriptional control of the genes coding for the Pseudomonas putida adhesins LapA and LapF. J Bacteriol 196:1484–1495. doi: 10.1128/JB.01287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol 9:e1000587. doi: 10.1371/journal.pbio.1000587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theunissen S, De Smet L, Dansercoer A, Motte B, Coenye T, Van Beeumen JJ, Devreese B, Savvides SN, Vergauwen B. 2010. The 285 kDa Bap/RTX hybrid cell surface protein (SO4317) of Shewanella oneidensis MR-1 is a key mediator of biofilm formation. Res Microbiol 161:144–152. doi: 10.1016/j.resmic.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 20.Cao B, Shi LA, Brown RN, Xiong YJ, Fredrickson JK, Romine MF, Marshall MJ, Lipton MS, Beyenal H. 2011. Extracellular polymeric substances from Shewanella sp. HRCR-1 biofilms: characterization by infrared spectroscopy and proteomics. Environ Microbiol 13:1018–1031. doi: 10.1111/j.1462-2920.2010.02407.x. [DOI] [PubMed] [Google Scholar]
- 21.Liang YL, Gao HC, Chen JR, Dong YY, Wu L, He ZL, Liu XD, Qiu GZ, Zhou JZ. 2010. Pellicle formation in Shewanella oneidensis. BMC Microbiol 10:291. doi: 10.1186/1471-2180-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Windt W, Gao HC, Kromer W, Van Damme P, Dick J, Mast J, Boon N, Zhou JZ, Verstraete W. 2006. AggA is required for aggregation and increased biofilm formation of a hyper-aggregating mutant of Shewanella oneidensis MR-1. Microbiology 152:721–729. doi: 10.1099/mic.0.28204-0. [DOI] [PubMed] [Google Scholar]
- 23.McLean JS, Pinchuk GE, Geydebrekht OV, Bilskis CL, Zakrajsek BA, Hill EA, Saffarini DA, Romine MF, Gorby YA, Fredrickson JK, Beliaev AS. 2008. Oxygen-dependent autoaggregation in Shewanella oneidensis MR-1. Environ Microbiol 10:1861–1876. doi: 10.1111/j.1462-2920.2008.01608.x. [DOI] [PubMed] [Google Scholar]
- 24.Wu C, Cheng YY, Yin H, Song XN, Li WW, Zhou XX, Zhao LP, Tian LJ, Han JC, Yu HQ. 2013. Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep 3:1945. doi: 10.1038/srep01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao T, Shi MM, Ju LL, Gao HC. 2015. Investigation into FlhFG reveals distinct features of FlhF in regulating flagellum polarity in Shewanella oneidensis. Mol Microbiol 98:571–585. doi: 10.1111/mmi.13141. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava D, Hsieh ML, Khataokar A, Neiditch MB, Waters CM. 2013. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol Microbiol 90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dasgupta N, Ramphal R. 2001. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol 183:6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences. Nova Science Publishers, Hauppauge, NY. [Google Scholar]
- 29.Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189. doi: 10.1093/bioinformatics/bti635. [DOI] [PubMed] [Google Scholar]
- 30.Matsuyama BY, Krasteva PV, Baraquet C, Harwood CS, Sondermann H, Navarro M. 2016. Mechanistic insights into c-di-GMP-dependent control of the biofilm regulator FleQ from Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E209–E218. doi: 10.1073/pnas.1523148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romling U, Galperin MY, Gomelsky M. 2013. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol Mol Biol Rev 77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao L, Rakshe S, Leff M, Spormann AM. 2013. PdeB, a cyclic di-GMP specific phosphodiesterase that regulates Shewanella oneidensis MR-1 motility and biofilm Formation. J Bacteriol 195:3827–3833. doi: 10.1128/JB.00498-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredrickson JK, Romine MF, Beliaev AS, Auchtung JM, Driscoll ME, Gardner TS, Nealson KH, Osterman AL, Pinchuk G, Reed JL, Rodionov DA, Rodrigues JLM, Saffarini DA, Serres MH, Spormann AM, Zhulin IB, Tiedje JM. 2008. Towards environmental systems biology of Shewanella. Nat Rev Microbiol 6:592–603. doi: 10.1038/nrmicro1947. [DOI] [PubMed] [Google Scholar]
- 35.Syed KA, Beyhan S, Correa N, Queen J, Liu J, Peng F, Satchell KJ, Yildiz F, Klose KE. 2009. The Vibrio cholerae flagellar regulatory hierarchy controls expression of virulence factors. J Bacteriol 191:6555–6570. doi: 10.1128/JB.00949-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo G, Huang L, Su Y, Qin Y, Xu X, Zhao L, Yan Q. 2016. flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg Microbes Infect 5:e85. doi: 10.1038/emi.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol 69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schuhmacher JS, Rossmann F, Dempwolff F, Knauer C, Altegoer F, Steinchen W, Dorrich AK, Klingl A, Stephan M, Linne U, Thormann KM, Bange G. 2015. MinD-like ATPase FlhG effects location and number of bacterial flagella during C-ring assembly. Proc Natl Acad Sci U S A 112:3092–3097. doi: 10.1073/pnas.1419388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res 40:7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dasgupta N, Wolfgang MC, Goodman AL, Arora SK, Jyot J, Lory S, Ramphal R. 2003. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol 50:809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 41.Larsen RA, Wilson MM, Guss AM, Metcalf WW. 2002. Genetic analysis of pigment biosynthesis in Xanthobacter autotrophicus Py2 using a new, highly efficient transposon mutagenesis system that is functional in a wide variety of bacteria. Arch Microbiol 178:193–201. doi: 10.1007/s00203-002-0442-2. [DOI] [PubMed] [Google Scholar]
- 42.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 43.Lauro FM, Tran K, Vezzi A, Vitulo N, Valle G, Bartlett DH. 2008. Large-scale transposon mutagenesis of Photobacterium profundum SS9 reveals new genetic loci important for growth at low temperature and high pressure. J Bacteriol 190:1699–1709. doi: 10.1128/JB.01176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Cen XF, Zhao GP, Wang J. 2012. Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J Bacteriol 194:5237–5244. doi: 10.1128/JB.00989-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lloyd AL, Marshall BJ, Mee BJ. 2005. Identifying cloned Helicobacter pylori promoters by primer extension using a FAM-labelled primer and GeneScan analysis. J Microbiol Methods 60:291–298. doi: 10.1016/j.mimet.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 47.Moerke NJ. 2009. Fluorescence polarization (FP) assays for monitoring peptide-protein or nucleic acid-protein binding. Curr Protoc Chem Biol 1:1–15. doi: 10.1002/9780470559277.ch090102. [DOI] [PubMed] [Google Scholar]
- 48.Dehio C, Meyer M. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol 179:538–540. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Edwards RA, Keller LH, Schifferli DM. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149–157. doi: 10.1016/S0378-1119(97)00619-7. [DOI] [PubMed] [Google Scholar]
- 50.Newman JR, Fuqua C. 1999. Broad-host-range expression vectors that carry the l-arabinose-inducible Escherichia coli araBAD promoter and the araC regulator. Gene 227:197–203. doi: 10.1016/S0378-1119(98)00601-5. [DOI] [PubMed] [Google Scholar]
- 51.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.