ABSTRACT
It has recently been suggested that oxygenic dismutation of NO into N2 and O2 may occur in the anaerobic methanotrophic “Candidatus Methylomirabilis oxyfera” and the alkane-oxidizing gammaproteobacterium HdN1. It may represent a new pathway in microbial nitrogen cycling catalyzed by a putative NO dismutase (Nod). The formed O2 enables microbes to employ aerobic catabolic pathways in anoxic habitats, suggesting an ecophysiological niche space of substantial appeal for bioremediation and water treatment. However, it is still unknown whether this physiology is limited to “Ca. Methylomirabilis oxyfera” and HdN1 and whether it can be coupled to the oxidation of electron donors other than alkanes. Here, we report insights into an unexpected diversity and remarkable abundance of nod genes in natural and engineered water systems. Phylogenetically diverse nod genes were recovered from a range of contaminated aquifers and N-removing wastewater treatment systems. Together with nod genes from “Ca. Methylomirabilis oxyfera” and HdN1, the novel environmental nod sequences formed no fewer than 6 well-supported phylogenetic clusters, clearly distinct from canonical NO reductase (quinol-dependent NO reductase [qNor] and cytochrome c-dependent NO reductase [cNor]) genes. The abundance of nod genes in the investigated samples ranged from 1.6 × 107 to 5.2 × 1010 copies · g−1 (wet weight) of sediment or sludge biomass, accounting for up to 10% of total bacterial 16S rRNA gene counts. In essence, NO dismutation could be a much more widespread physiology than currently perceived. Understanding the controls of this emergent microbial capacity could offer new routes for nitrogen elimination or pollutant remediation in natural and engineered water systems.
IMPORTANCE NO dismutation into N2 and O2 is a novel process catalyzed by putative NO dismutase (Nod). To date, only two bacteria, the anaerobic methane-oxidizing bacterium “Ca. Methylomirabilis oxyfera” and the alkane-oxidizing gammaproteobacterium HdN1, are known to harbor nod genes. In this study, we report efficient molecular tools that can detect and quantify a wide diversity of nod genes in environmental samples. A surprisingly high diversity and abundance of nod genes were found in contaminated aquifers as well as wastewater treatment systems. This evidence indicates that NO dismutation may be a much more widespread physiology in natural and man-made environments than currently perceived. The molecular tools presented here will facilitate further studies on these enigmatic microbes in the future.
KEYWORDS: nitric oxide (NO) dismutation, NO dismutase, oxygenic denitrification, contaminated aquifers, wastewater treatment, “Candidatus Methylomirabilis oxyfera”, HdN1
INTRODUCTION
Microbial nitrogen cycling has been intensively investigated for over a century and was thought to be rather well understood. Yet, recent discoveries of novel processes and microbes involved in the nitrogen cycle, e.g., methane-dependent nitrite and nitrate reduction (1–4), complete ammonia oxidation to nitrate by Nitrospira spp. (5, 6), and ammonia-oxidizing archaea (7), have demonstrated that our understanding of microbial nitrogen cycling may still be incomplete. Recently it was proposed that NO dismutation (NOD) to O2 and N2 (see equation below) may occur in the anaerobic methanotroph “Candidatus Methylomirabilis oxyfera” (NC10 phylum) and may also occur in the alkane-oxidizing gammaproteobacterium HdN1 (3, 8).
“Ca. Methylomirabilis oxyfera” oxidizes methane to CO2 and reduces nitrite via NO to N2 under strictly anoxic conditions. Interestingly, “Ca. Methylomirabilis oxyfera” possesses and highly expresses a complete aerobic methane oxidation pathway, including particulate methane monooxygenase (pMMO) (3). Metagenomic and physiological evidence suggests that the bacterium forms O2 to support the aerobic oxidation of methane under nitrite-reducing conditions (3). HdN1 grows on C6–C30 alkanes with oxygen, as well as nitrate and nitrite as electron acceptors. However, HdN1 does not harbor any fumarate-adding enzymes or other catalysts for anaerobic hydrocarbon activation, and it does not produce detectable alkyl-substituted succinates in anaerobically grown cultures (8). Instead, multiple copies of alkane monooxygenase genes were identified as the only means of alkane activation in HdN1. Therefore, HdN1 was suggested to utilize oxygen for its substrate activation when grown on alkanes with nitrate and nitrite as the electron acceptors (8).
In both “Ca. Methylomirabilis oxyfera” and HdN1, the O2 used for substrate activation is thought to be generated via NO dismutation, catalyzed by putative NO dismutases (Nod), which belong to the quinol-dependent NO reductase (qNor) family (9). NO dismutases exhibit amino acid substitutions at positions that are essential for electron transfer in canonical qNor, suggesting an electron-neutral reaction to be catalyzed by Nod (9, 10). In a “Ca. Methylomirabilis oxyfera” culture, 18O2 was indeed formed as an intermediate from 18O-labeled nitrite during nitrite-dependent methane oxidation (3). However, direct biochemical evidence for this activity of the enzyme is not yet available.
As a next step in addressing the occurrence and potential relevance of Nod-harboring microbes in natural systems, targeted detection assays for the gene or respective transcripts are required. However, the development of such assays is hampered by the extremely low number of reference sequences available for primer design. Two copies of putative nod genes have been identified in the genome of “Ca. Methylomirabilis oxyfera” and one in the HdN1 genome (3, 8). Recently, specific primer sets have been developed that are capable of detecting “Ca. Methylomirabilis oxyfera”-affiliated nod genes in a methane-oxidizing nitrite-reducing laboratory reactor inoculated with river sediments (11). Respective transcripts have also been found in water samples taken directly from marine oxygen minimum zones (12). However, evidence for the occurrence of a potentially wider diversity of putative nod genes in environmental systems is still lacking. Also, it is still unclear whether NOD can be coupled to the oxidation of electron donors other than alkanes and how important NOD might be in different systems with intensive N cycling. For example, such populations can be hypothesized to occur in contaminated aquifers or in wastewater treatment systems. Wastewater treatment systems especially offer a wealth of distinct niches for microbes involved in biological nitrogen removal (13).
Here, we provide a primary inquiry of the diversity and abundance of putative nod genes in such systems. A suite of primers capable of specifically detecting and quantifying a range of nod lineages was developed, and highly diverse and abundant environmental nod gene pools were recovered. Our results provide evidence for a widespread occurrence and high diversity of putative nod genes, suggesting that NOD could be an underestimated component of reductive nitrogen cycling in anthropogenically impacted and engineered water systems.
RESULTS
Primers targeting putative nod genes.
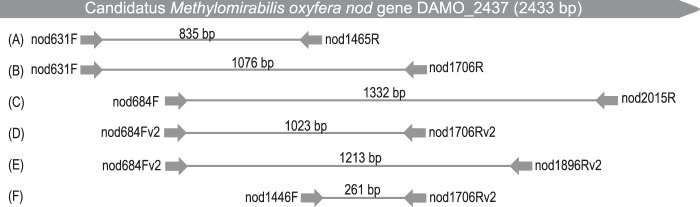
Nod belongs to the quinol-dependent nitric oxide reductase (qNor) family (9). Existing general qnor-targeting primers, such as qnorB2F and qnorB5R (14), have critical multiple mismatches to available nod sequences. Therefore, the development of suitable nod-specific primers was the first objective of this study. Initially, two forward primers (nod631F and nod684F) and four reverse primers (nod1465R, nod1706R, nod1896R, and nod2015R) designed to be selective for available nod sequences were developed (Fig. 1 and Table 1). PCR with DNA extracted from a “Ca. Methylomirabilis oxyfera” enrichment culture yielded the expected amplicon sizes, suggesting the primers to be functional. Based on the extended nod alignment, including sequences obtained from the two aquifers, adjusted forward (nod684Fv2) and reverse (nod1706Rv2 and nod1896Rv2) primers were iteratively developed and applied to the samples from water treatment systems.
FIG 1.
Scheme of the localization of nod-targeted primers developed in this study and the primer combinations used for clone library construction and qPCR. The positioning of nod primers refers to the “Ca. Methylomirabilis oxyfera” nod gene DAMO_2437. Primer combinations A to E were used for clone library construction, and primer set F was used for qPCR. The expected amplicon sizes are indicated; the scheme is not drawn to scale.
TABLE 1.
nod-targeted primers designed and applied in this study
| Designation | Sequence (5′ to 3′) | Positionsa |
|---|---|---|
| nod631F | TTCTTCTGGGGHGGYTGGG | 631–649 |
| nod684F | CTAYACHCACAACTGGCC | 684–701 |
| nod1465R | CGAAGAACAGGAACAGMACCATG | 1465–1443 |
| nod1706R | GGCTTGGCRATCCAGTAGAAG | 1706–1686 |
| nod1896R | GATGTTCCAGAAGTTRACGSC | 1896–1876 |
| nod2015R | ATGTTACCYTTKACACCGAAC | 2015–1995 |
| nod684Fv2 | STAYACHCAYAACTGGCC | 684–701 |
| nod1706Rv2 | GGCTTSGCRATCCAGTAGAAG | 1706–1686 |
| nod1896Rv2 | GATRTTCCAGAAGTTRACGSC | 1896–1876 |
| nod1446F | GGTGBYBTTCCTGTTCTTYRG | 1446–1466 |
Positions of target site according to the “Candidatus Methylomirabilis oxyfera” nod DAMO_2437 gene sequence.
High diversity of environmental nod genes.
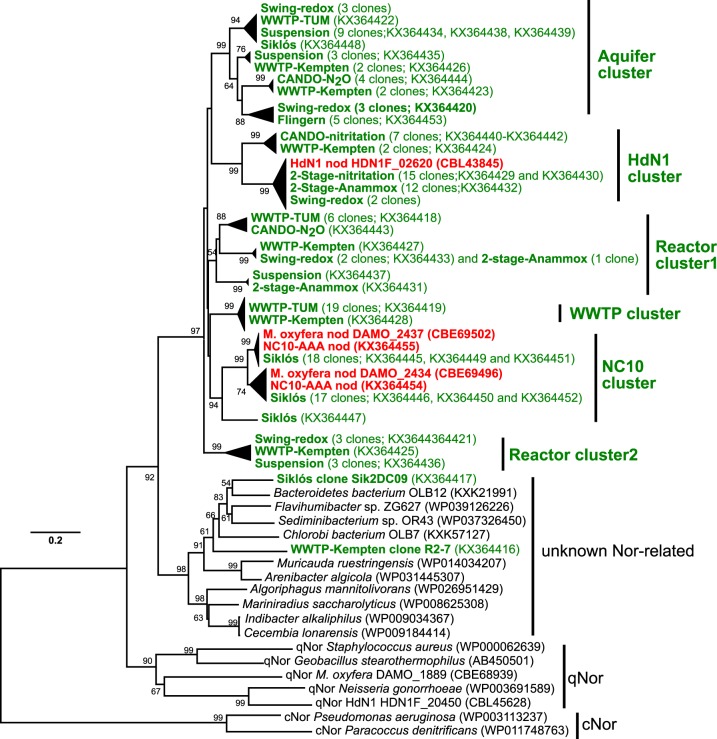
Amplicons of expected size were obtained from all analyzed samples by standard PCR, and amplicons were cloned and sequenced. Altogether, 149 sequences were obtained, of which 147 sequences resulted in “Ca. Methylomirabilis oxyfera” and HdN1 nod genes as top BLASTN matches. Only two sequences (accession no. KX364416, WWTP-Kempten clone R2-7; and accession no. KX364417, Siklós clone Sik2DC09) were more similar to unknown Nor-related sequences of microbes within the Fibrobacteres-Chlorobi-Bacteroidetes (FCB) superphylum (Fig. 2). This indicated a high specificity of the developed nod primers. Together with Nod from NC10 bacteria and HdN1, the environmental Nods (deduced amino acid sequences) formed a phylogenetic cluster distinct from known qNor and cNor with robust bootstrapping support (97%, 1,000 replicates). Six subclusters of putative nod genes were subsequently classified, tentatively named after the organism or habitat where they were first discovered (Fig. 2).
FIG 2.
Bootstrapped neighbor-joining phylogeny of putative Nod and selected qNor and cNorB sequences. Nod clones generated in this study are shown in green, and available reference sequences are in red. The 6 subclusters of Nod identified in this study are indicated. The accession numbers of reference sequences used and of selected nod sequences generated in this study are shown in parentheses. Bootstrap support (1,000 replicates) greater than 50% is indicated at the nodes. The scale bar represents 20% amino acid sequence divergence.
Almost all nod sequences recovered from the Siklós aquifer (using initial primer combinations A, B, and C) showed high nucleotide similarity (84 to 99%) to one of the two nod genes of “Ca. Methylomirabilis oxyfera”. The total numbers of clones that were either more closely related to DAMO_2437 (18 clones) or to DAMO_2434 (17 clones) were nearly equal, suggesting that those nod sequences could originate from “Ca. Methylomirabilis oxyfera”-related microbes with two nod paralogs in their genomes. In contrast, all nod sequences retrieved from the Flingern aquifer (primer pair A; ∼835 bp) were nearly identical (>99%) and exhibited only low sequence similarity to nod of both “Ca. Methylomirabilis oxyfera” (70 to 71% and 60 to 61%) and HdN1 (69% and 64%) on nucleotide and amino acid levels, respectively. This novel cluster of unidentified environmental nod genes was tentatively named the “aquifer cluster” (Fig. 2).
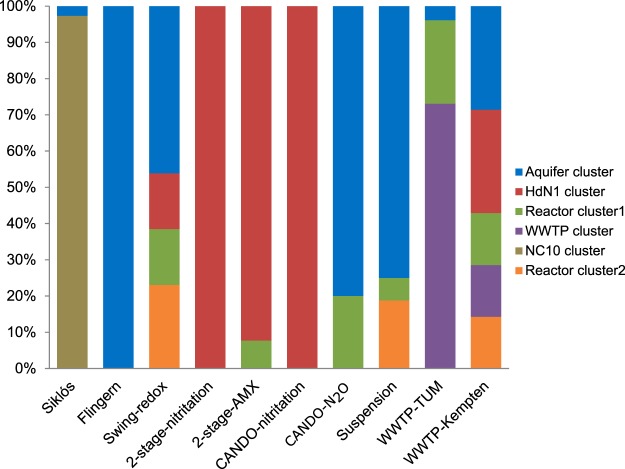
Most of the nod sequences obtained from the coupled two-stage cascade deammonification reactors (reactor performing partial nitrification [2-stage-nitritation] and reactor performing anammox [2-stage-AMX]), as well as 2 clones from the “swing-redox” reactor (i.e., the deammonification reactor that employed granule biomass) shared >98% nucleotide similarity with the nod gene of HdN1 (Fig. 2 and 3). The other 11 clones from the swing-redox reactor system were surprisingly diverse (Fig. 3), comprising not only the previously discovered aquifer cluster but also two novel “reactor clusters,” which were only distantly related to nod gene sequences of “Ca. Methylomirabilis oxyfera” (66% to 77%) and HdN1 (65 to 69%). nod gene pools within the coupled aerobic-anoxic nitrous decomposition operation (CANDO)-N2O and suspension reactors (i.e., deammonification reactor that employed suspended biomass) were again mostly affiliated with the aquifer cluster but also fell within the reactor clusters. Finally, a novel “WWTP cluster” was identified to dominate the nod gene pool in the WWTP-Technical University of Munich (TUM) plant, while samples from the WWTP in Kempten comprised the highest diversity of nod genes among all samples analyzed, with no fewer than 5 nod clusters detected (Fig. 3).
FIG 3.
Relative composition of putative nod gene clone libraries generated from the investigated samples. The affiliation of nod clones to the identified subclusters is given as shown in Fig. 2.
Nod gene abundance.
The abundance of nod genes in the investigated environments was notable (Fig. 4). In aquifer samples, nod abundance was ∼1.6 × 107 copies · g−1 for Flingern sediments and ∼5 × 108 copies for Siklós well sludge. At both sites, nod genes accounted for ∼2% of the total bacterial 16S rRNA gene counts. In the engineered water systems, absolute nod gene abundance was up to 3 orders of magnitude higher, with a maximum of ∼5 × 1010 copies · g−1 of sludge biomass in the CANDO reactor performing partial nitrification to nitrite (CANDO-nitritation) (Fig. 4). The relative abundance of nod versus bacterial 16S rRNA genes was still in a low range (<5%) for most samples. However, relative nod abundance was clearly elevated in the two-stage deammonification reactors (2-stage-nitritation and 2-stage-AMX), accounting for up to 10% of the bacterial 16S rRNA genes (Fig. 4).
FIG 4.
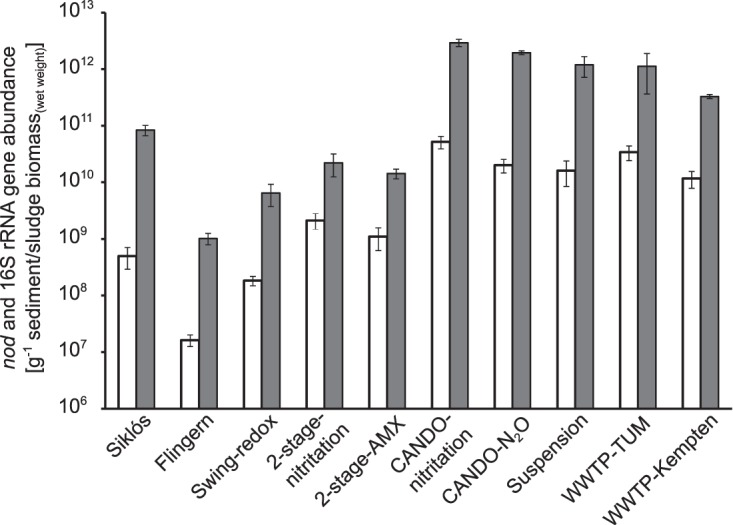
Absolute abundance of nod (open bars) and bacterial 16S rRNA (gray bars) in the investigated samples. Gene counts are shown as the average ± standard deviation (SD) from at least three technical qPCR replicates per sample, in which WWTP-TUM is from biologically replicated DNA extracts.
Characteristics of environmental Nod sequences.
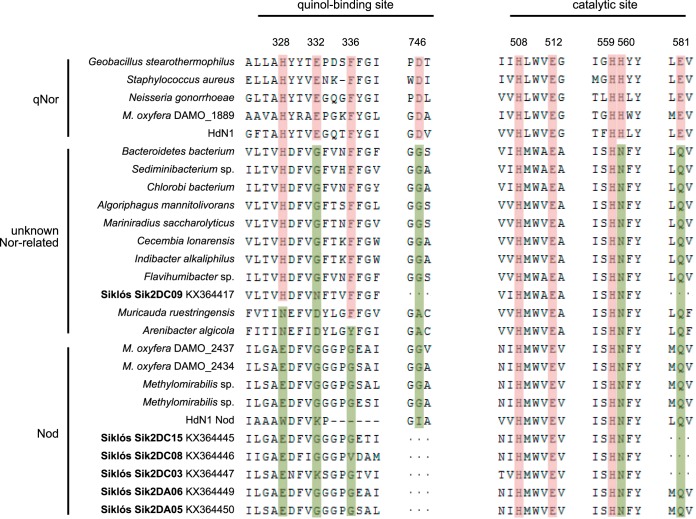
In canonical qNor, several functionally essential structures, such as the quinol-binding site, proton supply channel, and catalytic center, are constituted by highly conserved amino acids (15–17), which are often substituted for various amino acids in Nod. These amino acid deviations arguably disfavor a role of Nod as NO reductase and have been considered signatures for the function of NO disproportionation (9, 10). All these characteristic substitutions were also consistent in the environmental Nod sequences recovered in this study (Fig. 5, and the more extensive alignment of the environmental Nod sequences is shown in Fig. S1).
FIG 5.
Multiple-sequence alignment of selected putative Nod and qNor enzymes around the quinol-binding site and the catalytic site of qNor. Representative environmental Nod enzymes were deduced from the gene sequences generated in this study (in bold). Only five of the longest environmental Nod sequences obtained in this study are included here (see Fig. S1 for extended alignments, including further environmental Nod enzymes). The conserved residues for quinol-binding and catalytic functioning in qNor are highlighted in red, whereas substitutions at these sites in putative Nod and putative Nor are shown in green. The accession numbers are the same as in Fig. 2. The alignment was generated with ClustalW in MEGA6.
Specifically, His328 and Asp746 (Geobacillus stearothermophilus qNor numbering) form hydrogen bonds with NO and OH groups of quinol, respectively, coordinated by Glu332 and Phe336 (16). In Nod, however, these highly conserved residues are replaced by various amino acids that are unlikely to provide proper quinol-binding capability (9, 10). Consistently, all environmental Nod sequences covering the quinol-binding region had deviations at sites His328, Glu332, and Phe336 (Fig. 5). Information for site Asp746 is unfortunately not covered by the length of our environmental Nod sequences. Furthermore, one of the three nonheme metal (often FeB) coordinating His residues in qNor, His560, was consistently replaced by asparagine in Nod (Fig. 5), possibly leading to an altered active-site configuration. One of the two highly conserved glutamate residues (Glu581) suggested to be a potential terminal proton donor in qNor (16) was exchanged for a glutamine in Nod, which was also observed for all environmental Nod sequences covering that region (Fig. 5 and S1). Intriguingly, the two more deeply branching unknown Nor-related sequences retrieved in this study, as well as the genomic sequences discovered via BLAST (all of them were annotated as Nor, including genes of Muricauda ruestringensis and Arenibacter algicola) in the same phylogenetic cluster (Fig. 2) possessed the same substitutions as Nod at the catalytic site and partially also at the quinol-binding site (Fig. 5).
DISCUSSION
Detecting environmental nod genes.
Nitric oxide dismutation (NOD) to O2 and N2 (Eq. 1) is an emergent process and a potential ecophysiology not well documented for biological systems to date. Genes of the putative NO dismutase (Nod) were first reported for “Ca. Methylomirabilis oxyfera” and HdN1 (3, 8). Although several other related laboratory enrichments have been described (e.g., in references 18–21), information on environmental nod gene occurrence remains scarce. Relatively short (329- to 426-bp) nod gene sequences closely related to that of “Ca. Methylomirabilis oxyfera” were recently reported from a nitrite-reducing methane-oxidizing laboratory reactor inoculated with river sediments (11). Closely related nod gene transcripts have also been found directly in total transcriptome libraries from marine oxygen minimum zones (12). Still, a comparative survey of nod gene pools in different natural habitats has not been conducted to date.
In this study, we provide primary evidence for the existence of an extensive diversity of nod genes in a range of natural and engineered water systems (Table 1 and Fig. 3). Although the employed nod primers were initially developed from a very limited number of reference sequences, environmental nod genes clearly distinct from those of “Ca. Methylomirabilis oxyfera” and HdN1 were successfully detected from the contaminated aquifers, which were investigated with the first generation of primers. Only the Siklós site was queried with the three initial primer pairs (Table 2). However, differences in the affiliation of resulting nod libraries were not observed, suggesting a similar performance of the assays. The improved primer pair D (nod684Fv2/nod1706Rv2) was then developed iteratively, based on actual environmental nod sequence data obtained with the first assays. It covers an ∼1,000-bp region and several distinctive sites of the “Ca. Methylomirabilis oxyfera” nod gene, and it was capable of recovering a surprising diversity of putative nod gene lineages from the engineered water systems (Fig. 3). Based on these results, the use of primer pair D for recovering nod gene diversities from environmental samples is recommended.
TABLE 2.
Environmental samples investigated for nod genes in this study
| Environment | Designation | Main process(es)a | Reference(s) | Primer pairb | No. of clonesd | Nod clusters |
|---|---|---|---|---|---|---|
| BTEX-impacted aquifer in Siklósc | Siklós | 26, 27 | A | 16 | 2 | |
| C | 9 | |||||
| B | 13 | |||||
| BTEX-impacted aquifer in Flingern | Flingern | 28, 37 | A | 5 | 1 | |
| Swing-redox deammonification reactor | Swing-redox | Nitritation and anammox | 23 | D | 13 | 4 |
| Two-stage sequencing batch deammonification reactor cascade | 2-stage-nitritation | Nitritation | D | 16 | 1 | |
| 2-stage-AMX | Anammox | D | 14 | 2 | ||
| CANDO reactor system | CANDO-nitritation | Nitritation | 39 | D | 7 | 1 |
| CANDO-N2O | Nitrous denitritation to N2O | 39 | D | 5 | 2 | |
| Suspension deammonification reactor | Suspension | Nitritation and anammox | 23 | D | 16 | 3 |
| Wastewater treatment plant, Garching | WWTP-TUM | Nitritation and anammox | 38 | D | 25 | 3 |
| Wastewater treatment plant, Kempten | WWTP-Kempten | Nitritation and anammox | 22 | D | 6 | 5 |
| E | 4 |
Diversity and abundance of environmental nod gene lineages.
This study provides a proof of concept of the general detectability and diversity of putative nod gene pools in terrestrial water systems. A “classical” cloning-and-sequencing approach was chosen, as it allows for more direct and less cost-intensive rounds in iterative primer development than with next-generation sequencing. The read length of Sanger sequencing was clearly also beneficial to retrieve full sequence information from some of the rather long amplicons (>1,000 bp) generated. Still, we are aware that the small size of some of our clone libraries prohibits more elaborate statistical comparison of the nod gene pools recovered. Future studies should incorporate a next-generation sequencing (NGS) strategy also for this gene marker.
Still, of all samples analyzed, the WWTP in Kempten and the swing-redox reactor samples appeared most diverse, spanning 5 and 4 of the proposed 6 nod gene clusters, respectively (Fig. 3). Both are biological nutrient removal systems removing nitrogen by partial nitritation, followed by anammox (22–24). The biomass samples were from thick biofilm carriers for Kempten and large biomass granules from the swing-redox reactor. Thus, nod gene diversity potentially reflected the typically high structural and physicochemical heterogeneity of such habitats (25).
The nod gene pools recovered from the two benzene, toluene, ethylbenzene, and xylene (BTEX)-impacted aquifers were strikingly different, possibly reflecting distinct contamination and redox scenarios at the sites (26–28). While the absolute abundance of the nod gene was one order of magnitude higher in Siklós than that in Flingern (Fig. 4), relative nod gene abundances were comparable in the two aquifers. This can likely be explained by the distinct nature of the samples, with more organic well sump material being taken in Siklós, while highly mineral sediments were taken in Flingern. The absolute abundance of bacterial 16S rRNA gene counts at the Flingern site was consistent with previous studies (28, 29), but the relative abundance of nod genes in both aquifers and most of the wastewater samples was only a few percent. This is within the typical range of the relative abundances of other nitrogen cycling genes that have been found in activated sludge (30). In contrast, relative nod gene abundance was clearly elevated in the coupled 2-stage-nitritation and 2-stage-AMX reactors, where nod genes accounted for up to 10% of the bacterial 16S rRNA gene counts. This was intriguing given that these reactors were designed for partial nitrification and anammox, and a high abundance of anammox organisms can be assumed. The affiliation of the nod genes detected in these systems should be a subject of further investigation.
Functionality of environmental Nod.
Quinol-dependent NO reductase (qNor) reduces NO to N2O, with electrons accepted from quinol (16). Although Nod belongs to the qNor family, it lacks proper quinol-binding sites and has altered catalytic center configuration. This compromises the activity of Nod as a conventional NO reductase and has been discussed as a possible signature of a role in NO disproportionation (9, 10). All environmental Nod gene sequences recovered in this study possessed substitutions similar to those found in the genes of “Ca. Methylomirabilis oxyfera” and HdN1 (Fig. 4 and S1), supporting that putative environmental nod genes were actually recovered, although we cannot provide direct evidence for NOD activity or actual O2 formation in this study. Nevertheless, the primary detection of these genes in the investigated water systems is an important prerequisite for follow-up studies on their potential expression and biochemical activity in the future.
The two more deeply branching nod-like sequences (KX364416 and KX364417) recovered were in a phylogenetic cluster between known qNor and Nod genes, tentatively named “unknown Nor-related genes” in this study (Fig. 2). This cluster also included genomic sequences of members of the Fibrobacteres-Chlorobi-Bacteroidetes (FCB) superphylum, recovered via BLAST, all of them annotated as Nor. Intriguingly, these sequences carry the same residual substitutions as Nod at the catalytic site and partially at the quinol-binding site (Fig. 5). Even more surprisingly, the Nor-related gene of Arenibacter algicola, an aerobic degrader of polycyclic aromatic hydrocarbons (31), retained all the residual substitutions characteristic of Nod (Fig. 5). To the best of our knowledge, direct biochemical evidence for the physiological function of any member of this gene cluster is not available. Studying the expression and functioning of these previously unknown Nor-related genes in members of the FCB superphylum can provide valuable further cues on the potential role of the putative nod genes detected in this study. Still, we cannot exclude at this stage that these could also just be atypical qNors using different electron-supplying mechanisms for NO reduction. Without direct biochemical evidence, clear functional connotations cannot be ascertained for the novel gene clusters detected in the environment. More detailed studies on NO respiration by pure cultures, such as HdN1 and A. algicola, can shed further light on this enigmatic process.
Potential environmental relevance of nod-harboring populations.
Nitrate/nitrite reduction via NO dismutation can be referred to as oxygenic denitrification to facilitate discussion. It can also be referred to as a potential new oxygenic route in nitrous denitritation, in line with the nomenclature proposed for biological nitrogen removal systems (13). It is interesting to consider whether the putative nod genes detected in the different systems could be affiliated with microbes other than “Ca. Methylomirabilis oxyfera” and HdN1, and the physiologies to which they could be connected.
Although “Ca. Methylomirabilis oxyfera” possesses multiple NO reductases in addition to Nod, the N2O reductase is missing (3, 10). Therefore, the reduction of NO2− to N2 seems to proceed essentially via NO dismutation in this bacterium. In contrast, HdN1 contains a full canonical denitrification pathway in addition to Nod (8), indicating that conventional and oxygenic denitrification pathways could coexist in a single microbe. In the environment, it can be anticipated that microorganisms performing oxygenic denitrification will compete for nitrate/nitrite with conventional denitrifiers, as well as microbes mediating dissimilatory nitrate reduction to ammonia (DNRA). The niche partitioning between conventional denitrifiers and microbes mediating DNRA is driven by various environmental controls, such as the availability of organic carbon or the ratio of available nitrite to nitrate (32). However, the environmental parameters defining the ecophysiological niche of putative oxygenic denitrifiers are still far from clear.
Both “Ca. Methylomirabilis oxyfera” and HdN1 lack anaerobic catabolic pathways for their alkane substrates. Thus, they are suggested to rely on O2 formed via NO dismutation to activate and oxidize hydrocarbons when growing under nitrate- and nitrite-reducing conditions. Methane and alkanes are among the most stable compounds that require high energy for activating the first C-H bond (33). Therefore, it can be speculated that O2 formed via NO dismutation could provide a competitive advantage for oxygenic denitrifiers thriving on recalcitrant compounds in anoxic environments.
It is also tempting to hypothesize that oxygenic denitrifiers theoretically couple aerobic catabolic processes other than alkane oxidation to oxygenesis. With an ability to rely on aerobic catabolism under both aerobic and nitrate/nitrite reduction, oxygenic denitrifiers could capitalize on ecological advantages under hypoxic or fluctuating redox conditions with transient availability of nitrate/nitrite. While “Ca. Methylomirabilis oxyfera” is a strict anaerobe and can be inhibited by short exposure to low levels of O2 (34), HdN1 is much more versatile, capable of using nitrate, nitrite, and O2 as electron acceptors when growing on alkanes (8). Our finding of a high abundance of nod genes nearly identical to HdN1 in several wastewater treatment systems (Fig. 2 to 4), as well as the fact that HdN1 was initially isolated from activated sludge (35), could suggest a potential relevance of HdN1 relatives in such systems. In the Flingern aquifer, a previous study has revealed a surprising peak abundance of aerobic toluene monooxygenase (tmoA) genes in the highly reduced core of the anoxic BTEX plume (28). The nod genes recovered from the Flingern aquifer in the present study potentially explain this unexpected high abundance of tmoA genes in highly reduced sediments with an at least transient supply of nitrate.
Conclusions.
Our results reveal a hitherto unrecognized ubiquity and abundance of putative nod genes in terrestrial water systems. The wide phylogenetic diversity detected suggests that NOD capacity may exist in microbes other than “Ca. Methylomirabilis oxyfera” and HdN1. Although no direct evidence for an actual activity of the detected Nod-harboring populations is provided here, this primary study provides important molecular cues to follow up on this. Attempts to enrich and isolate putative oxygenic denitrifiers with a range of electron donors are currently ongoing and essential to further substantiate our hypothesis. The fostering of microbes with a capacity for oxygenic denitrification, which may bypass N2O as an intermediate of canonical NO reduction, could also be an attractive strategy in minimizing problematic N2O emissions in wastewater treatment (36). A more detailed understanding of populations potentially carrying a capacity for NOD could be vital for a more comprehensive understanding of microbial N cycling as well as for the development of novel bioremediation strategies and engineering solutions for biological nitrogen removal.
MATERIALS AND METHODS
Primer design.
Since only limited nod sequences were available from public databases at the beginning of this study, an iterative improvement was applied during primer development. “Ca. Methylomirabilis oxyfera” nod DAMO_2434 (CBE69496), DAMO_2437 (CBE69502), HdN1 nod HDN1F_02620 (CBL43845), and two nod sequences (KX364454 and KX364455) assembled from the metagenome of a NC10-AAA enrichment culture (4) were aligned with selected qnor and cnorB sequences in MEGA6 by the ClustalW algorithm. Based on this initial nod alignment, forward and reverse primers that covered all five nod sequences were developed (Table 1). These first primer pairs (combinations A, B, and C) were tested using environmental DNA extracted from contaminated aquifer sediments (Table 2). The resulting aquifer nod-like sequences were then included in the nod alignment. Sequence information of the amplicons generated with the first primers was then used to further degenerate and optimize the specificity of a second generation of primers (combinations D and E). Of these, primer set D performed well and was used to recover putative nod sequences from different wastewater treatment plants and laboratory-scale reactor systems. The divergent nod sequences attained from engineered water systems further extended the nod alignment, based on which a final forward primer (nod1446F) was designed to allow quantitative PCR (qPCR) analysis in combination with the reverse primer nod1706Rv2 (primer combination F).
Sampling and sites.
The samples used in this study were collected from various sites, as listed in Table 2. Siklós sediment samples were collected in April 2015 from the bottom of monitoring well ST2 in the center of a xylene plume in Siklós, Hungary (26, 27), while Flingern sediments were taken by push-coring from the upper fringe of a toluene plume (6.4 m below ground) in Flingern, Düsseldorf, Germany, in September 2013 (28, 37). Sediments were transported to the lab in cooling boxes and were then frozen at −20°C before DNA extraction.
Samples from wastewater treatment plants (WWTP) and laboratory-scale reactors were collected in September 2015. Activated sludge samples from a WWTP in Garching, Germany, next to the Technical University of Munich (TUM), were collected from the denitrifying basin, which receives clarified nitrate-rich effluent from an upstream trickling filter (38). Biofilm carriers from the WWTP in Kempten, Germany, were collected from the deammonification basin (22).
Biomass samples were also collected from two single-stage deammonification bioreactors, operating under alternating oxic/anoxic conditions (23). Here, the deammonification reactor that employed suspended biomass was designated suspension, and the other, which employed granule biomass, was designated swing-redox (Table 2). Biomass samples from a two-stage sequencing batch cascade deammonification system were also analyzed. The system comprises a reactor performing partial nitrification and a reactor performing anammox, which were designated 2-stage-nitritation and 2-stage-AMX, respectively (Table 2). Biomass samples were collected from a system implementing the coupled aerobic-anoxic nitrous decomposition operation (CANDO) process, aiming for simultaneous nitrogen removal, greenhouse gas mitigation, and energy recovery (39). The two reactors of the CANDO system, one performing partial nitrification to nitrite and one nitrous denitritation to N2O, were designated CANDO-nitritation and CANDO-N2O, respectively (Table 2). A comprehensive redefinition of the nomenclature for biological processes contributing to nitrogen removal in such engineered water systems can be found elsewhere (13).
DNA isolation.
DNA was isolated from aquifer sediments as previously described (14), with a minor modification of the final precipitation of DNA being done at 20,000 × g and 4°C for 30 min, instead of at 20°C. For DNA isolation from WWTP and reactor samples, 0.5 to 1.0 ml of homogenized biomass or sludge was pipetted into 1.5-ml Eppendorf tubes, which were spun at 13,000 rpm for 1 min. The supernatant was then removed, and the remaining biomass was weighed. For samples from WWTP Kempten, biofilm from carriers was put into a 1.5-ml Eppendorf tube and weighed. DNA isolation was done as described above (14), omitting the second bead-beating step. DNA was extracted in triplicate from WWTP-TUM, and other samples were nonreplicated. DNA concentration and quality were checked by UV spectrophotometry (NanoDrop ND-1000; Isogen Life Science, The Netherlands) and standard agarose gel electrophoresis.
PCR and qPCR.
DNA samples diluted 10- or 100-fold were used as the template for nod gene PCR analysis. The primer pairs used are listed in Table 1, and their positioning on the “Ca. Methylomirabilis oxyfera” nod gene and the expected amplicon size are given in Fig. 1. To recover potentially increased nod diversity, gradient PCR with the following cycling conditions was performed: a 3-min initial dissociation at 96°C, followed by 35 cycles of amplification (45 s at 95°C, 60 s at 52 to 62°C, and 90 s at 72°C), and a final 5-min extension at 72°C. All PCRs were performed in 25-μl reaction mixtures containing nuclease-free H2O, 1× PCR buffer, 1.5 mM MgCl2, 0.1 mM dinucleotide triphosphates (dNTPs), 0.5 U Taq polymerase (all Fermentas GmBH, Basel, Switzerland), 5 μg of bovine serum albumin (BSA; Roche Diagnostics GmbH, Basel, Switzerland), 0.5 μM each primer, and 1 μl of template DNA. PCR products were checked by standard agarose gel electrophoresis.
To quantify potential nod-harboring microorganisms and their relative abundance in each sample, qPCR targeting nod as well as the bacterial 16S rRNA gene was performed, using primer pairs nod1446F/nod1706Rv2 and Ba519F/Ba907R (29), respectively. The specificity of the primer pair nod1446F/nod1706Rv2 was verified by cloning and sequencing its amplicons using Siklós DNA. All 8 sequenced clones were nod. Synthetic nod gene (440 bp, “Ca. Methylomirabilis oxyfera” DAMO_2437) and Escherichia coli 16S rRNA gene (980-bp) fragments, covering the respective primer sites with a >60-bp flanking region at each end, were used as respective standards for nod and 16S rRNA (gBlocks; Integrated DNA Technologies, Leuven, Belgium). Sample DNA in 10- and 100-fold dilutions for nod and in 100- and 1,000-fold dilutions for 16S rRNA was quantified. Standard and samples were quantified in triplicate, repeated in at least two independent qPCR runs for each assay. qPCR experiments (25-μl reaction volume) were carried out with the MX3000p cycler (Agilent, Santa Clara, CA, USA). 2× GoTaq SYBR green master mix (Promega, Madison, WI, USA) with Rox as the reference dye was used. The qPCR annealing temperatures used for nod and 16S rRNA were 57°C and 52°C, respectively. qPCR analysis with an efficiency of 100% ± 10% was used for calculation. The absolute nod and 16S rRNA gene counts of each sample were calculated per gram (wet weight) of sediment or biomass used in DNA extraction.
Cloning, sequencing, and phylogenetic analysis.
PCR products of different annealing temperatures from each sample were pooled and purified with PCRextract spin columns (5 Prime, Hamburg, Germany), according to the manufacturer's protocol. Purified PCR products were cloned and sequenced as previously described (29). The high-quality sequences obtained were translated to amino acids in MEGA6 and then aligned with selected qNor and cNor sequences with the ClustalW algorithm, with default settings. A phylogenetic tree was constructed based on an amino acid alignment with MEGA6 using the neighbor-joining method. The robustness of the tree topology was tested by bootstrap analysis (1,000 replicates).
Accession number(s).
Representative nod sequences as well as two unknown nor-related sequences obtained in this study were deposited at NCBI under the accession numbers KX364418 to KX364453, KX364416, and KX364417. The two nod paralogs assembled from the metagenome of NC10-AAA enrichment (4) can be found under the accession numbers KX364454 and KX364455.
Supplementary Material
ACKNOWLEDGMENTS
We thank Claus Lindenblatt (Chair of Urban Water Systems Engineering, Technical University of Munich) for his assistance in WWTP sampling and providing lab-scale reactor samples. We also thank Katharina Ettwig (Radboud University Nijmegen, The Netherlands) for providing NC10 enrichments DNA for initial primer testing.
This research has received funding from the European Research Council (ERC) under the European Union's Seventh Framework Programme (FP7/2007-2013), grant agreement 616644 (POLLOX) to T.L. We also acknowledge funding by the Helmholtz Society and by a bilateral interaction project (Revisiting DeHu) funded by the German Ministry of Education and Research (BMBF, grant 01DS14037 to T.L.) and the Hungarian National Research, Development and Innovation Office (NKFIH, grant TéT_12_DE-1-2013-0007 to A.T.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02750-16.
REFERENCES
- 1.Haroon MF, Hu S, Shi Y, Imelfort M, Keller J, Hugenholtz P, Yuan Z, Tyson GW. 2013. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature 500:567–570. doi: 10.1038/nature12375. [DOI] [PubMed] [Google Scholar]
- 2.Raghoebarsing AA, Pol A, van de Pas-Schoonen KT, Smolders AJ, Ettwig KF, Rijpstra WI, Schouten S, Sinninghe Damste JS, Op den Camp HJ, Jetten MS, Strous M. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918–921. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 3.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MM, Schreiber F, Dutilh BE, Zedelius J, de Beer D, Gloerich J, Wessels HJ, van Alen T, Luesken F, Wu ML, van de Pas-Schoonen KT, Op den Camp HJ, Janssen-Megens EM, Francoijs KJ, Stunnenberg H, Weissenbach J, Jetten MS, Strous M. 2010. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 4.Ettwig KF, Zhu B, Speth DR, Keltjens JT, Jetten MSM, Kartal B. 2016. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci U S A 113:12792–12796. doi: 10.1073/pnas.1609534113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Kessel MAHJ, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, Kartal B, Jetten MSM, Lucker S. 2015. Complete nitrification by a single microorganism. Nature 528:555–559. doi: 10.1038/nature16459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daims H, Lebedeva EV, Pjevac P, Han P, Herbold C, Albertsen M, Jehmlich N, Palatinszky M, Vierheilig J, Bulaev A, Kirkegaard RH, von Bergen M, Rattei T, Bendinger B, Nielsen PH, Wagner M. 2015. Complete nitrification by Nitrospira bacteria. Nature 528:504–509. doi: 10.1038/nature16461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Francis CA, Beman JM, Kuypers MMM. 2007. New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19–27. doi: 10.1038/ismej.2007.8. [DOI] [PubMed] [Google Scholar]
- 8.Zedelius J, Rabus R, Grundmann O, Werner I, Brodkorb D, Schreiber F, Ehrenreich P, Behrends A, Wilkes H, Kube M, Reinhardt R, Widdel F. 2011. Alkane degradation under anoxic conditions by a nitrate-reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environ Microbiol Rep 3:125–135. doi: 10.1111/j.1758-2229.2010.00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ettwig KF, Speth DR, Reimann J, Wu ML, Jetten MS, Keltjens JT. 2012. Bacterial oxygen production in the dark. Front Microbiol 3:273. doi: 10.3389/fmicb.2012.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reimann J, Jetten MM, Keltjens J. 2015. Metal enzymes in “impossible” microorganisms catalyzing the anaerobic oxidation of ammonium and methane, p 257–313. In Kroneck PMH, Sosa Torres ME (ed), Sustaining life on planet Earth: metalloenzymes mastering dioxygen and other chewy gases, vol 15 Springer International Publishing, Basel, Switzerland. doi: 10.1007/978-3-319-12415-5_7. [DOI] [Google Scholar]
- 11.Bhattacharjee AS, Motlagh AM, Jetten MS, Goel R. 2016. Methane dependent denitrification–from ecosystem to laboratory-scale enrichment for engineering applications. Water Res 99:244–252. doi: 10.1016/j.watres.2016.04.070. [DOI] [PubMed] [Google Scholar]
- 12.Padilla CC, Bristow LA, Sarode N, Garcia-Robledo E, Gomez Ramirez E, Benson CR, Bourbonnais A, Altabet MA, Girguis PR, Thamdrup B, Stewart FJ. 2016. NC10 bacteria in marine oxygen minimum zones. ISME J 10:2067–2071. doi: 10.1038/ismej.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weissbach M, Criddle CS, Drewes JE, Koch K. 24 November 2016. A proposed nomenclature for biological processes that remove nitrogen. Environ Sci Water Res Technol. doi: 10.1039/C6EW00216A. [DOI] [Google Scholar]
- 14.Pilloni G, Granitsiotis MS, Engel M, Lueders T. 2012. Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes. PLoS One 7:e40467. doi: 10.1371/journal.pone.0040467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiro Y. 2012. Structure and function of bacterial nitric oxide reductases: nitric oxide reductase, anaerobic enzymes. Biochim Biophys Acta 1817:1907–1913. doi: 10.1016/j.bbabio.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto Y, Tosha T, Pisliakov AV, Hino T, Sugimoto H, Nagano S, Sugita Y, Shiro Y. 2012. Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus. Nat Struct Mol Biol 19:238–245. doi: 10.1038/nsmb.2213. [DOI] [PubMed] [Google Scholar]
- 17.Terasaka E, Okada N, Sato N, Sako Y, Shiro Y, Tosha T. 2014. Characterization of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus: enzymatic activity and active site structure. Biochim Biophys Acta 1837:1019–1026. doi: 10.1016/j.bbabio.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, van Dijk G, Fritz C, Smolders AJP, Pol A, Jetten MSM, Ettwig KF. 2012. Anaerobic oxidization of methane in a minerotrophic peatland: enrichment of nitrite-dependent methane-oxidizing bacteria. Appl Environ Microbiol 78:8657–8665. doi: 10.1128/AEM.02102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu S, Zeng RJ, Burow LC, Lant P, Keller J, Yuan Z. 2009. Enrichment of denitrifying anaerobic methane oxidizing microorganisms. Environ Microbiol Rep 1:377–384. doi: 10.1111/j.1758-2229.2009.00083.x. [DOI] [PubMed] [Google Scholar]
- 20.He Z, Geng S, Cai C, Liu S, Liu Y, Pan Y, Lou L, Zheng P, Xu X, Hu B. 2015. Anaerobic oxidation of methane coupled to nitrite reduction by halophilic marine NC10 bacteria. Appl Environ Microbiol 81:5538–5545. doi: 10.1128/AEM.00984-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luesken FA, van Alen TA, van der Biezen E, Frijters C, Toonen G, Kampman C, Hendrickx TL, Zeeman G, Temmink H, Strous M, Op den Camp HJ, Jetten MS. 2011. Diversity and enrichment of nitrite-dependent anaerobic methane oxidizing bacteria from wastewater sludge. Appl Microbiol Biotechnol 92:845–854. doi: 10.1007/s00253-011-3361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leix C, Drewes JE, Koch K. 2016. The role of residual quantities of suspended sludge on nitrogen removal efficiency in a deammonifying moving bed biofilm reactor. Bioresour Technol 219:212–218. doi: 10.1016/j.biortech.2016.07.134. [DOI] [PubMed] [Google Scholar]
- 23.Lackner S, Lindenblatt C, Horn H. 2012. ‘Swinging ORP’ as operation strategy for stable reject water treatment by nitritation-anammox in sequencing batch reactors. Chem Eng J 180:190–196. doi: 10.1016/j.cej.2011.11.043. [DOI] [Google Scholar]
- 24.Leix C, Hartl R, Zeh C, Beer F, Drewes JE, Koch K. 2016. Performance and N2O formation of the deammonification process by suspended sludge and biofilm systems—a pilot-scale study. Water 8:578. doi: 10.3390/w8120578. [DOI] [Google Scholar]
- 25.Vlaeminck SE, Terada A, Smets BF, De Clippeleir H, Schaubroeck T, Bolca S, Demeestere L, Mast J, Boon N, Carballa M, Verstraete W. 2010. Aggregate size and architecture determine microbial activity balance for one-stage partial nitritation and anammox. Appl Environ Microbiol 76:900–909. doi: 10.1128/AEM.02337-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Táncsics A, Szoboszlay S, Szabo I, Farkas M, Kovacs B, Kukolya J, Mayer Z, Kriszt B. 2012. Quantification of subfamily I.2.C catechol 2,3-dioxygenase mRNA transcripts in groundwater samples of an oxygen-limited BTEX-contaminated site. Environ Sci Technol 46:232–240. doi: 10.1021/es201842h. [DOI] [PubMed] [Google Scholar]
- 27.Táncsics A, Farkas M, Szoboszlay S, Szabo I, Kukolya J, Vajna B, Kovacs B, Benedek T, Kriszt B. 2013. One-year monitoring of meta-cleavage dioxygenase gene expression and microbial community dynamics reveals the relevance of subfamily I.2.C extradiol dioxygenases in hypoxic, BTEX-contaminated groundwater. Syst Appl Microbiol 36:339–350. doi: 10.1016/j.syapm.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Larentis M, Hoermann K, Lueders T. 2013. Fine-scale degrader community profiling over an aerobic/anaerobic redox gradient in a toluene-contaminated aquifer. Environ Microbiol Rep 5:225–234. doi: 10.1111/1758-2229.12004. [DOI] [PubMed] [Google Scholar]
- 29.Winderl C, Anneser B, Griebler C, Meckenstock RU, Lueders T. 2008. Depth-resolved quantification of anaerobic toluene degraders and aquifer microbial community patterns in distinct redox zones of a tar oil contaminant plume. Appl Environ Microbiol 74:792–801. doi: 10.1128/AEM.01951-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geets J, de Cooman M, Wittebolle L, Heylen K, Vanparys B, De Vos P, Verstraete W, Boon N. 2007. Real-time PCR assay for the simultaneous quantification of nitrifying and denitrifying bacteria in activated sludge. Appl Microbiol Biotechnol 75:211–221. doi: 10.1007/s00253-006-0805-8. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez T, Rhodes G, Mishamandani S, Berry D, Whitman WB, Nichols PD, Semple KT, Aitken MD. 2014. Polycyclic aromatic hydrocarbon degradation of phytoplankton-associated Arenibacter spp. and description of Arenibacter algicola sp. nov., an aromatic hydrocarbon-degrading bacterium. Appl Environ Microbiol 80:618–628. doi: 10.1128/AEM.03104-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft B, Tegetmeyer HE, Sharma R, Klotz MG, Ferdelman TG, Hettich RL, Geelhoed JS, Strous M. 2014. The environmental controls that govern the end product of bacterial nitrate respiration. Science 345:676–679. doi: 10.1126/science.1254070. [DOI] [PubMed] [Google Scholar]
- 33.Rabus R, Boll M, Heider J, Meckenstock RU, Buckel W, Einsle O, Ermler U, Golding BT, Gunsalus RP, Kroneck PMH, Kruger M, Lueders T, Martins BM, Musat F, Richnow HH, Schink B, Seifert J, Szaleniec M, Treude T, Ullmann GM, Vogt C, von Bergen M, Wilkes H. 2016. Anaerobic microbial degradation of hydrocarbons: from enzymatic reactions to the environment. J Mol Microbiol Biotechnol 26:5–28. doi: 10.1159/000443997. [DOI] [PubMed] [Google Scholar]
- 34.Luesken FA, Wu ML, Op den Camp HJ, Keltjens JT, Stunnenberg H, Francoijs KJ, Strous M, Jetten MS. 2012. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera': kinetic and transcriptional analysis. Environ Microbiol 14:1024–1034. doi: 10.1111/j.1462-2920.2011.02682.x. [DOI] [PubMed] [Google Scholar]
- 35.Ehrenreich P, Behrends A, Harder J, Widdel F. 2000. Anaerobic oxidation of alkanes by newly isolated denitrifying bacteria. Arch Microbiol 173:58–64. doi: 10.1007/s002030050008. [DOI] [PubMed] [Google Scholar]
- 36.Law YY, Ye L, Pan YT, Yuan ZG. 2012. Nitrous oxide emissions from wastewater treatment processes. Philos Trans R Soc Lond B Biol Sci 367:1265–1277. doi: 10.1098/rstb.2011.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller H, Bosch J, Griebler C, Damgaard LR, Nielsen LP, Lueders T, Meckenstock RU. 2016. Long-distance electron transfer by cable bacteria in aquifer sediments. ISME J 10:2010–2019. doi: 10.1038/ismej.2015.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koch K, Helmreich B, Drewes JE. 2015. Co-digestion of food waste in municipal wastewater treatment plants: effect of different mixtures on methane yield and hydrolysis rate constant. Appl Energ 137:250–255. doi: 10.1016/j.apenergy.2014.10.025. [DOI] [Google Scholar]
- 39.Scherson YD, Wells GF, Woo SG, Lee J, Park J, Cantwell BJ, Criddle CS. 2013. Nitrogen removal with energy recovery through N2O decomposition. Energ Environ Sci 6:241–248. doi: 10.1039/C2EE22487A. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.