ABSTRACT
Saccharomyces cerevisiae killer strains secrete a protein toxin active on nonkiller strains of the same (or other) yeast species. Different killer toxins, K1, K2, K28, and Klus, have been described. Each toxin is encoded by a medium-size (1.5- to 2.3-kb) M double-stranded RNA (dsRNA) located in the cytoplasm. M dsRNAs require L-A helper virus for maintenance. L-A belongs to the Totiviridae family, and its dsRNA genome of 4.6 kb codes for the major capsid protein Gag and a minor Gag-Pol protein, which form the virions that separately encapsidate L-A or the M satellites. Different L-A variants exist in nature; on average, 24% of their nucleotides are different. Previously, we reported that L-A-lus was specifically associated with Mlus, suggesting coevolution, and proposed a role of the toxin-encoding M dsRNAs in the appearance of new L-A variants. Here we confirm this by analyzing the helper virus in K2 killer wine strains, which we named L-A-2. L-A-2 is required for M2 maintenance, and neither L-A nor L-A-lus shows helper activity for M2 in the same genetic background. This requirement is overcome when coat proteins are provided in large amounts by a vector or in ski mutants. The genome of another totivirus, L-BC, frequently accompanying L-A in the same cells shows a lower degree of variation than does L-A (about 10% of nucleotides are different). Although L-BC has no helper activity for M dsRNAs, distinct L-BC variants are associated with a particular killer strain. The so-called L-BC-lus (in Klus strains) and L-BC-2 (in K2 strains) are analyzed.
IMPORTANCE Killer strains of S. cerevisiae secrete protein toxins that kill nonkiller yeasts. The “killer phenomenon” depends on two dsRNA viruses: L-A and M. M encodes the toxin, and L-A, the helper virus, provides the capsids for both viruses. Different killer toxins exist: K1, K2, K28, and Klus, encoded on different M viruses. Our data indicate that each M dsRNA depends on a specific helper virus; these helper viruses have nucleotide sequences that may be as much as 26% different, suggesting coevolution. In wine environments, K2 and Klus strains frequently coexist. We have previously characterized the association of Mlus and L-A-lus. Here we sequence and characterize L-A-2, the helper virus of M2, establishing the helper virus requirements of M2, which had not been completely elucidated. We also report the existence of two specific L-BC totiviruses in Klus and K2 strains with about 10% of their nucleotides different, suggesting different evolutionary histories from those of L-A viruses.
KEYWORDS: L-A helper virus, yeast killer toxins, double-stranded RNA virus, yeast virus, yeast wine strains
INTRODUCTION
Saccharomyces cerevisiae killer strains secrete protein toxins, which are lethal to sensitive strains of the same species (and, in some cases, to related yeast species) but not to themselves. Various killer toxins—K1, K2, K28, and Klus—have been described, with different modes of action: pore forming for K1 (and probably also K2) and blocking of DNA synthesis for K28 (1). Each toxin is encoded by a medium-size (1.5- to 2.3-kb) double-stranded RNA (dsRNA) virus: M1, M2, M28, and Mlus, respectively. The M dsRNA viruses show no sequence homology to each other, but their genome organizations are similar; the positive strand contains an open reading frame (ORF) in the 5′-end region that codes for the toxin precursor, or preprotoxin, which also provides immunity to the toxin (but not to different killer toxins). After the preprotoxin ORF, there is a unique internal AU-rich region, followed by a 3′-end region of variable length with no coding capacity (2, 3). M viruses depend on a dsRNA helper virus, L-A in the case of M1. L-A (4.6 kb) encodes the virion proteins: the major structural protein Gag (76 kDa) and the minor protein Gag-Pol (170 kDa), which is translated as a fusion of Gag and Pol ORFs by −1 ribosomal frameshifting. The Pol domain of Gag-Pol has motifs characteristic of viral RNA-dependent RNA polymerases (RdRp) (4–6). L-A and M dsRNAs are separately encapsidated in the same type of capsids, made by 60 asymmetric Gag dimers and 1 or 2 Gag-Pol molecules per capsid (7–9). The replication cycle of L-A (or its satellite M1 dsRNA) in yeast cytoplasm has been thoroughly investigated (10). Figure 1 summarizes the L-A and M1 replication cycles and the genomic organization of L-A, with cis signals on L-A required for frameshifting, encapsidation, and replication.
FIG 1.
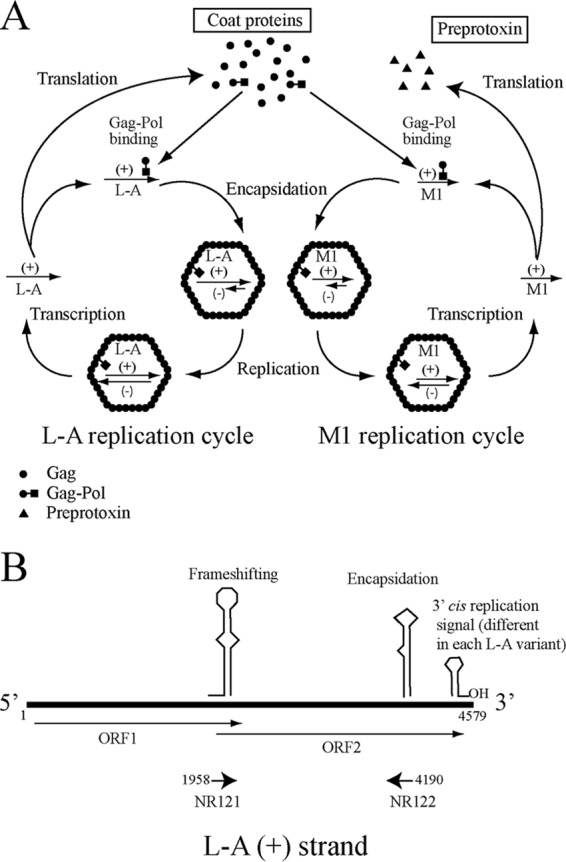
(A) Replication cycles of L-A virus and the killer toxin-encoding virus M1. L-A is transcribed by the transcriptase activity of Gag-Pol, yielding L-A positive strands (indicated by plus signs in parentheses) that are released from the virions into the cytoplasm and are either translated or encapsidated. Translation by ribosomes gives rise to Gag, the structural capsid protein, and Gag-Pol, the RNA polymerase. Gag-Pol interacts with L-A positive strands, triggering viral capsid assembly and encapsidation. The replicase activity of Gag-Pol inside the virions synthesizes the negative strand (indicated by a minus sign in parentheses) on the positive-strand template, forming the dsRNA genome. The replication cycle of M1 is similar to that of L-A and depends on L-A-encoded viral proteins Gag and Gag-Pol. Thus, M1 is a satellite virus of L-A. M1 positive strands are translated, giving rise to the preprotoxin, which is processed and secreted. (B) Genomic organization of L-A. The L-A positive strand and the two overlapping reading frames (ORF1 and ORF2) are indicated. Schematic representations of the secondary structures corresponding to the frameshifting region and to the cis signals required for encapsidation and replication in the 3′-end region of the L-A positive strand are shown. The sequences where oligonucleotides NR121 and NR122 anneal (indicated by arrows) are conserved in all L-A variants.
L-BC is another dsRNA virus that often coexists with L-A without being excluded, even though, in general, its copy number is about 10 to 20% that of L-A. L-BC has no helper activity to maintain M dsRNAs (11, 12). L-BC (4.6 kb) and L-A share the same genomic organization and mode of expression (13). S. cerevisiae viruses (ScVs) L-A and L-BC belong to the Totiviridae family and are present in the yeast cytoplasm. Like all mycoviruses, they are not infectious and are transmitted from mother cells to daughter cells through cell division or during mating (14). S. cerevisiae dsRNA viruses and killer toxins have been extensively reviewed (1, 15). In addition to the viruses of the Totiviridae family, some strains of S. cerevisiae harbor positive-strand RNA viruses of the Narnaviridae family, called 20S RNA and 23S RNA viruses (15).
Many host nuclear genes affect the maintenance of L-A and M viruses (16). MAK (for maintenance of killer) genes are required for killer activity. mak mutants cannot maintain M1 (or M2) and therefore are nonkillers; mak3, mak10, and mak31 mutants also lose L-A, due to defects in the N-acetyltransferase that acetylates the N terminus of Gag, a modification required for capsid assembly (17, 18). SKI genes have a negative effect on the viruses, and ski mutants show a superkiller phenotype, with increased levels of toxin and M1 (19). Most SKI gene products are components of the exosome, a complex involved in 3′-to-5′ RNA degradation (20), or modulators of its activity. In contrast, SKI1 (XRN1) codes for the main exonuclease involved in 5′-to-3′ mRNA degradation (21). Recently, it was reported for a yeast strain with the Sigma genome that the presence (or absence) of the K1 killer virus and the type of mitochondria can affect the phenotype of some chromosomal mutations, revealing a cytoplasmic contribution to the heritability of certain genes (22).
There are a number of variants of L-A, with as many as 26% of nucleotides different (23). The L-A virus in Klus strains (designated L-A-lus) is different from that in K1 strains or in most nonkiller laboratory strains. The same is true of K2 wine strains; a different L-A variant, named L-A-2, was found, confirming previous reports of differences in the L-A viruses in K1 or K2 laboratory killer strains based on T1 fingerprinting analysis and partial sequencing (11). The helper virus in K28 strains has been shown by Northern hybridization to be different from L-A (24). During the preparation of this report, the complete sequence of L-A-28 was deposited in GenBank (accession number KU845301) (25). The nucleotide sequence of L-A-28 is about 24% different from that of L-A, L-A-lus, or L-A-2. Different L-A variants show individual phenotypic properties. L-A is more sensitive to the growth of the host at elevated temperatures (37 to 39°C) than L-A-lus or L-A-2 and can easily be eliminated from the cells (26). Also, L-A is cured with high frequency by overexpression of the SKI1 5′ exonuclease (27), but L-A-lus and L-A-2 are not. Studies in the early 1980s reported a certain heterogeneity in L-A itself, with functional phenotypic variants that exhibited different abilities to maintain the K1 and K2 phenotypes (28, 29). Each killer strain carries only one type of M. When K1 and K2 haploid cells mate, the resulting diploids are only K1, a phenomenon known as exclusion (28). Exclusion is also observed between M1 and Mlus: cytoplasmic mixing experiments between K1 and Klus strains produce cells with either M1 or Mlus, but not both satellites together (23).
Nowadays, many wine yeast strain collections are available as a result of screenings to select yeast starter cultures for winemaking, and they constitute good material with which to study the presence of dsRNA viruses in strains that may have evolved together in the same ecological niche. In our previous work, we reported the presence of two types of killer strains, K2 and Klus strains, in such a collection of wine yeast strains from different European regions and showed that there was a strong association between each L-A helper virus and its satellite virus (L-A-lus and Mlus; L-A-2 and M2), suggesting coevolution (23). Although the K2 killer toxin is active at the low pH of grape must and is thus technologically important for winemaking, since inoculated K2 killer strains may predominate over sensitive indigenous strains present in the must, the nucleotide sequence of the K2 helper virus, L-A-2, was not known, and the relationship of M2 with its helper virus was not clear. Therefore, in this work, we pursued several aims: (i) to complete the sequencing of the L-A-2 variant, (ii) to introduce L-A-2 and M2 from a K2 wine strain into standard laboratory strains, (iii) to analyze the possible maintenance of M2 by L-A and L-A-lus, and (iv) to study the L-BC variants present in Klus and K2 wild yeast populations. We also discuss the different degrees of variability for L-A and L-BC variants in S. cerevisiae strains. Finally, we speculate on the origin of the multicopy, virally encoded killer toxins from host genes.
RESULTS
L-A-2, the L-A variant in K2 killer strains.
In a previous study, we characterized L-A-lus, a variant of the L-A totivirus, which is the helper virus of the Klus-encoding dsRNA virus Mlus (3, 23). L-A-lus is frequently present in wine yeast strains. In the same screening, we found that most K2 strains carried a different L-A virus, which we named L-A-2. A region spanning nucleotides (nt) 210 to 1454 showed the same nucleotide sequence, which was different from those of L-A or L-A-lus, in various K2 wine strains. Following a strategy similar to that described previously (23), we decided to characterize this new L-A variant in more detail. First, we completed the sequencing of L-A-2 (4,580 nt) as described in Materials and Methods. Figure S1B in the supplemental material shows a ClustalW comparison of selected sequences from L-A, L-A-2, L-A-lus, and the recently reported L-A-28 (25). The overall identity ranges from 74 to 78%; L-A-2 and L-A-lus are most similar to each other (78% identity). However, two regions, the frameshifting region (nt 1958 to nt 2004) and the encapsidation signal (nt 4169 to nt 4203), show 100% identity, indicating their importance for the life cycle of L-A totiviruses. In the positive-strand 3′-end sequences, where the cis signals for replication reside, the last 11 nt are identical, except for two extra C's reported in the case of L-A-28. Predicted secondary structures for the 3′ ends of these four L-A variants are depicted in Fig. S1C. The effects of these structures on L-A replication have been analyzed in detail elsewhere (10). It is likely that the secondary structures at the 3′ ends play similar roles in the replication of the other variants.
There are two open reading frames on the positive strand of L-A-2: ORF1 (nt 30 to nt 2069) and ORF2 (nt 1958 to nt 4543). According to what is known for L-A (Fig. 1B), ORF1 encodes the L-A-2 major coat protein (Gag), while Gag-Pol is translated as a fusion product of ORF1 and ORF2 by a −1 ribosomal frameshift. In a Western blot, polyclonal antibodies against L-A Gag recognized the major coat protein of L-A-2 (Fig. S1D in the supplemental material) and a faint high-molecular-mass band, suggesting that the latter is the 171-kDa fused Gag-Pol protein. This cross-reactivity is not surprising, since both Gag proteins show 90% conservation.
Construction of K2 laboratory strains or strains carrying L-A-2 alone.
To better study this L-A-2 variant, we transferred the K2 killer trait from the K2 wine strain Ca7 to laboratory strains, as outlined in Fig. 2A. We followed the same experimental approach that we had used for Klus killer strains in our previous work (23). In this way, we generated strain 1137, which is our reference laboratory strain with a stable K2 killer phenotype (Fig. 2A; Table 1). In this strain, M2 was not lost by growth at high temperatures. Curing of M2 was achieved only by cycloheximide treatment and generated strain 1163 (L-A-2 M2-o) (Table 1). In this strain, as expected, the L-A-2 copy number increases as a result of M2 loss. Because strain 1137 and K2 strains from other laboratories (which we also analyzed in this work [see Fig. S2 in the supplemental material]) have different origins, we confirmed by reverse transcription-PCR (RT-PCR) that all of them carry M2 dsRNAs with the same coding sequence. The noncoding 3′-end sequences of M2 are unknown so far. However, since they all have similar sizes (about 1.5 kb), we consider them to be equivalent.
FIG 2.
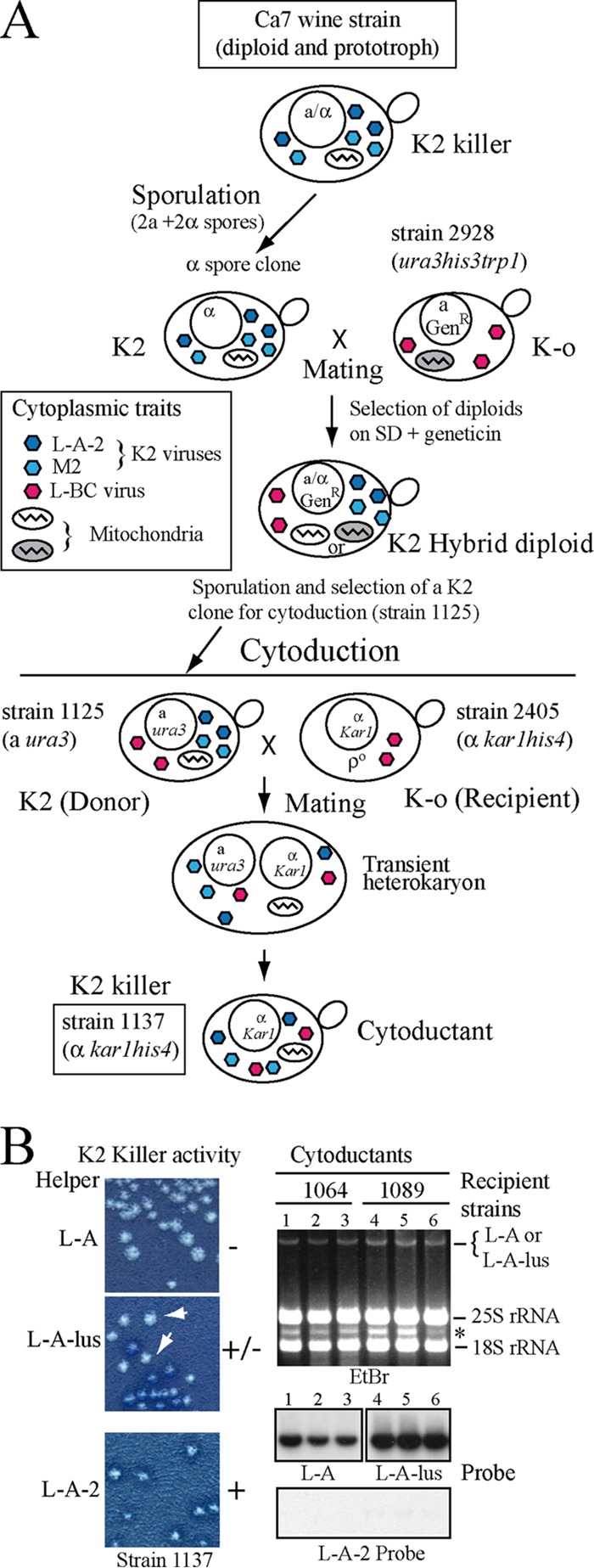
Construction of K2 strain 1137 and exclusion of K2 by L-A or L-A-lus. (A) Diagram of the experimental approach followed to introduce the K2 killer trait from wine strain Ca7 into laboratory strain 2405. The K2 strain Ca7 (diploid and prototroph) was induced to sporulate, and the spore clones were mated to laboratory strain 2928 (expressing Geneticin resistance [GenR] from a vector [23]). Hybrid diploids were selected on minimal medium (SD) with Geneticin. After sporulation, strain 1125 was selected (a ura3; K2). The cytoplasm of this K2 strain was then transferred by cytoduction into recipient strain 2405 (kar1 mutant defective in nuclear fusion and [rho0]). A transient heterokaryon was initially formed. After mitotic division, cytoductants carried the nucleus of the recipient strain and the mixed cytoplasms from the donor and recipient strains, and thus, they are K2 (strain 1137). Strain 1137 also carries L-BC virus (from strain 2928, since strain Ca7 is L-BC-o), and its mitochondria were inherited from the wine strain (information obtained from Fig. S5). The various cytoplasmic traits present in the cells are indicated as follows. K2 viruses L-A-2 and M2 are shown as dark-blue and light-blue hexagons, respectively; L-BC viruses are represented by red hexagons. Mitochondria from different parental strains are shown on a white or gray background. (B) K2 viruses are excluded by L-A or L-A-lus. L-A-2 and M2 viruses were transferred by cytoduction from strain 1125 into two strains isogenic with 2405 but containing L-A (strain 1064) or L-A-lus (strain 1089). (Left) (Top and center) K2 killer activity of isolated colonies from each strain, with the respective helper virus indicated. White arrows indicate colonies that have lost killer activity. (Bottom) K2 killer activity of strain 1137 carrying L-A-2 and M2, shown as a control. (Right) Total nucleic acids from three cytoductants from each recipient strain were separated in an agarose gel and were analyzed by Northern hybridization with probes specific for L-A (lanes 1 to 3) or L-A-lus (lanes 4 to 6). (Top) Ethidium bromide (EtBr)-stained gel. The band indicated by the asterisk corresponds to the 23S RNA narnavirus originally present in donor strain 1125. (Center) The two autoradiograms. (Bottom) The same samples (from lanes 1 to 6) annealed to the L-A-2-specific probe.
TABLE 1.
Strains used in this study
Unless otherwise indicated, laboratory strains were created in this work or previous work from our laboratory.
In descriptions of cytoductants, the first strain mentioned is the donor strain, and the second is the recipient strain.
From R. B. Wickner's laboratory (National Institutes of Health, USA).
From the EUROFAN collection (kindly supplied by J. L. Revuelta, University of Salamanca, Salamanca, Spain).
L-A-2 is excluded by L-A or by L-A-lus, and it is the helper virus of M2.
The presence of L-A-2 in most K2 wine strains suggested that L-A-2 showed a specific helper activity for M2 virus. To determine whether M2 could also be maintained by other L-A viruses in laboratory strains, we introduced L-A-2 and M2 from strain 1125 into strains isogenic with the L-A-o strain 2405 but containing either L-A (strain 1064) or L-A-lus (strain 1089). All cytoductants in the L-A-carrying strain were nonkillers (Fig. 2B, left, top). Northern hybridization showed that they harbored L-A and not L-A-2 (Fig. 2B, right, lanes 1 to 3), suggesting, first, that L-A excludes L-A-2 and, second, that L-A is not able to maintain M2. On the other hand, cytoductants in the L-A-lus-containing strain were initially K2 killers, showing killing halos of various sizes; successive single-colony isolations from K2 colonies gave rise to nonkiller colonies at a high frequency (Fig. 2B, left, center). Again, all nonkiller colonies carried L-A-lus and not L-A-2 (Fig. 2B, right, lanes 4 to 6). Thus, L-A-2 is also excluded by L-A-lus. L-A-lus seems to be able to function as a helper virus of M2 to a certain extent, but with time, M2 is lost, too.
Cytoduction of K2 viruses into recipient strains includes a transient heterokaryon stage with three types of virions together: L-A-2 and M2 from one parent and L-A or L-A-lus from the other. In order to eliminate the possibility that the presence of the toxin-encoding M2 virions might affect the final fate of the helper virus, similar mating experiments were performed using the M2-cured strain 1163 as the L-A-2 donor. In this case, two different approaches were used: either (i) analysis of the helper viruses in diploids after the mating of strain 1163 with two different strains containing either L-A or L-A-lus or (ii) analysis of haploids after the introduction of L-A-2 virions alone by cytoduction into the same strains made [rho0] in advance. Strain 1163 carries the kar1 mutation and is an α mating type strain. The a mating type partners were derivatives of strain 2928 (Table 1) carrying either L-A (strain 1127) or L-A-lus (strain 1098). In the first approach, analysis of 10 independent diploid colonies from each cross revealed that L-A-2 had been excluded by either L-A or L-A-lus. All these colonies were L-A-2-o (Fig. 3). The same observation was made for 10 cytoductants from each cytoduction experiment. This confirmed that the presence or absence of M2 did not affect the exclusion of L-A-2 by L-A or L-A-lus. The absence of L-A-2 (and the presence of the other L-A variant) was confirmed by RT-PCR analysis with specific oligonucleotides as mentioned in Materials and Methods.
FIG 3.
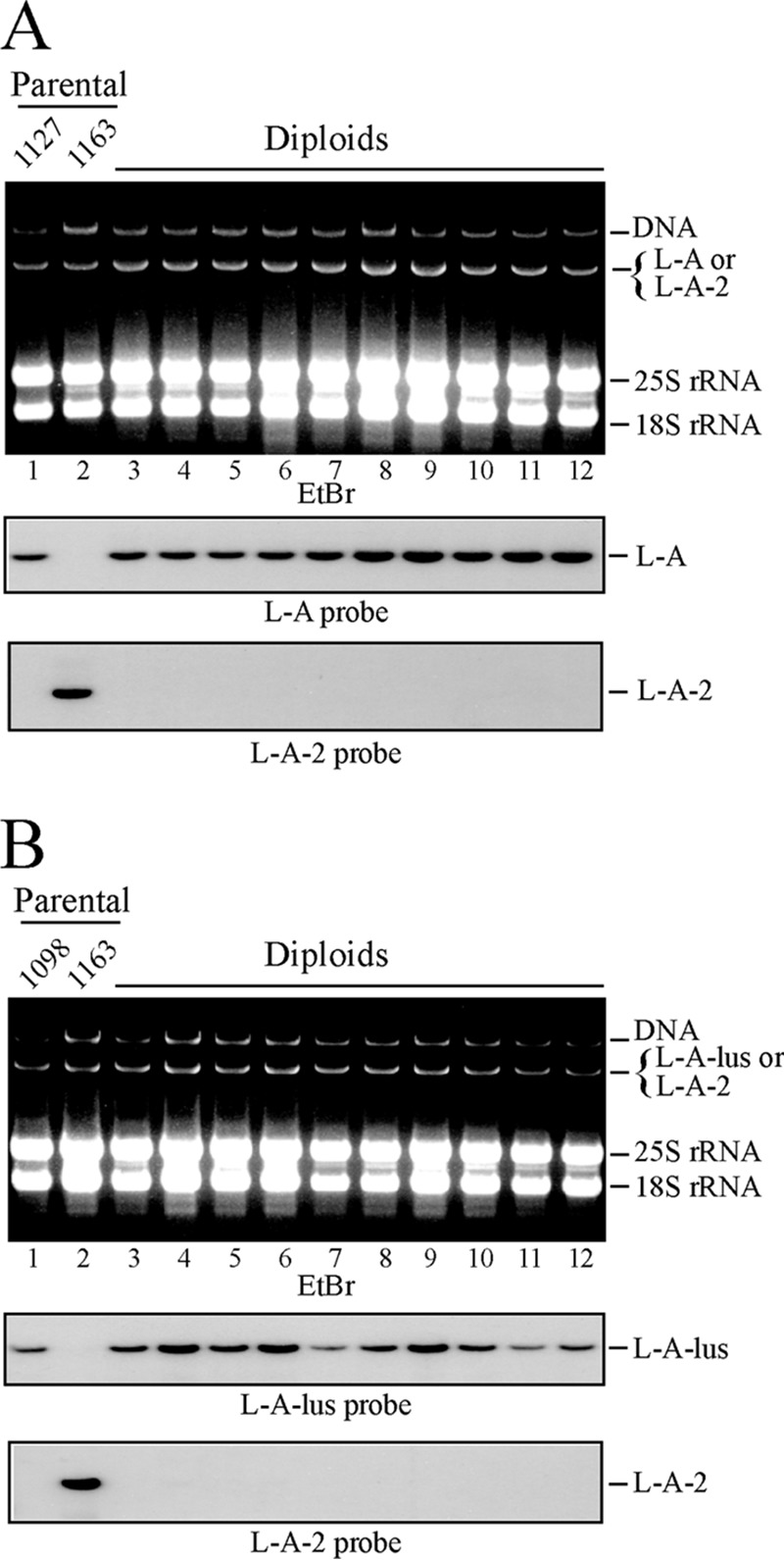
Exclusion of L-A-2 by L-A or L-A-lus. (A) Exclusion of L-A-2 by L-A. Strain 1163 (L-A-2) was mated with strain 1127 (L-A), and diploid colonies were selected. Total nucleic acids from the parental haploid strains (lanes 1 and 2) and from 10 independent diploid colonies (lanes 3 to 12) were separated in an agarose gel and were analyzed by Northern hybridization with specific probes. (Top) Ethidium bromide (EtBr)-stained gel. (Center and bottom) Autoradiograms of RNAs on blots detected with an L-A- or L-A-2-specific probe, respectively. (B) Exclusion of L-A-2 by L-A-lus. This experiment is similar to that for which results are shown in panel A, except that here the L-A-2-containing strain 1163 was mated with strain 1098, carrying L-A-lus. Ten independent diploid colonies were analyzed as described above, using probes against L-A-lus (center) or L-A-2 (bottom).
M2 maintenance by L-A or L-A-lus noncanonical virus depends on elevated helper virus copy numbers.
In the early 1980s, R. B. Wickner's laboratory used genetic methods to identify three phenotypic variants of L-A that affected the maintenance of the K1 and K2 toxins (28). Using specific probes now available for L-A and L-A-2, we checked which L-A species were present in three K2 strains from that laboratory. We found L-A-2 in strain 1384 and L-A in strains 1385 and 2360 (Fig. S2A in the supplemental material). The presence of L-A in K2 strains apparently contradicted our results, leading us to investigate the M2 requirements further. Strain 2360 is a ski7 mutant harboring enormous amounts of L-A, suggesting that large amounts of L-A viral proteins might overcome the dependency of M2 on L-A-2. Thus, we asked whether M2 could be maintained by increased amounts of noncanonical helper coat proteins overexpressed from a vector. Strain 2928 was transformed with plasmid p1290, which expresses Gag (from L-A) and a hybrid Gag-Pol whose Pol domain is from L-A-lus (23); then the cytoplasm of strain 1137 (L-A-2 and M2) was transferred to the transformed strain (Fig. 4A). Northern hybridization showed that in the cytoductants, L-A-2 was eliminated by coat protein overexpression; nevertheless, all the cytoductants were K2 killers, indicating that M2 could be maintained stably by the hybrid virions provided by the plasmid (Fig. 4B). This strain (strain 1149) (Table 1), which maintains M2 in the absence of L-A-2, was a good tool with which to analyze the complex relationship between a killer toxin-producing M virion and its helper virus or between different helper viruses. As seen above in cytoplasmic mixing experiments, L-A-2 was excluded by L-A or L-A-lus, neither of which had helper activity for M2. To confirm this, we now reintroduced M2 virions alone from strain 1149 into three isogenic strains with the noncanonical helper viruses: (i) strain 1064, carrying L-A in small amounts, (ii) strain 2404, carrying L-A in large amounts (at least 5-fold higher), and (iii) strain 1089, carrying L-A-lus (Fig. 4C). For the strain with L-A at a low copy number (strain 1064), all cytoductants were nonkillers; for the strains with L-A at a high copy number or with L-A-lus (all strains with L-A-lus have rather small amounts of the virus), a mixture of nonkiller and killer colonies was seen initially, but in successive colony isolations, the killer cells finally lost M2 (Fig. 4D). These results confirmed that M2 could be maintained only by L-A-2, not by L-A or L-A-lus, in these strains.
FIG 4.
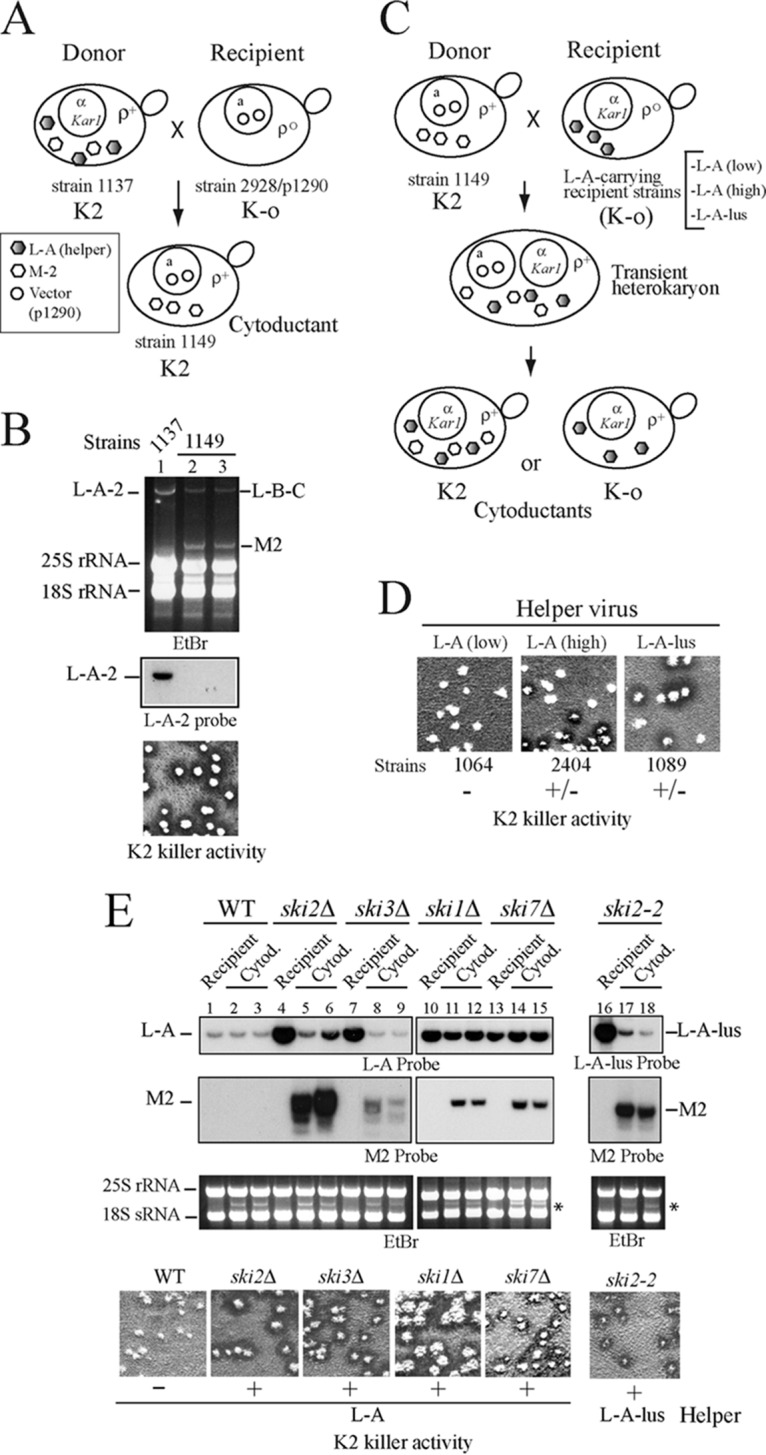
Helper activity of L-A variants. (A) Diagram of a cytoduction experiment to produce strain 1149, which maintains M2 virus by L-A proteins expressed from a vector. L-A-2 and M2 viruses (shaded and open hexagons, respectively) from strain 1137 were transferred to strain 2928 carrying plasmid p1290 (indicated by open circles in the nucleus). Although the cytoductants originally contained L-A-2 and M2, L-A-2 was eliminated by overexpression of viral proteins from the plasmid. (B) (Top) RNAs from two cytoductants (lanes 2 and 3) were analyzed in an agarose gel. RNAs from donor strain 1137 were used as a control (lane 1). Note that both cytoductants have lost L-A-2 but maintain large amounts of M2 dsRNA, visible as discrete bands in the ethidium bromide (EtBr)-stained gel (lanes 2 and 3). (Center) L-A-2 is detected by hybridization with an L-A-2-specific probe. (Bottom) K2 killer activity of isolated cytoductants. (C) Diagram of cytoduction experiments to introduce M2 virus from donor strain 1149 into three K-o recipient strains carrying L-A-lus or L-A helper viruses. The helper viruses (of any type) are indicated by shaded hexagons and M2 virus by open hexagons. (D) Killer assays of isolated cytoductants in each case. The recipient strain name is given below each image, and the respective helper virus is identified at the top. Only if an L-A variant has helper activity for M2 are the cytoductants K2 killers (+); otherwise they are nonkillers (−). (E) M2 maintenance by L-A or L-A-lus in ski mutants. M2 virions from strain 1161, which are maintained by viral proteins expressed from vector pI2L2, were introduced by cytoplasmic mixing into five strains carrying L-A: strain BY4741 (wild type [WT]) (lanes 1 to 3) or mutant derivative strains with deletions of SKI2 (strain Y05307) (lanes 4 to 6), SKI3 (strain Y05604) (lanes 7 to 9), SKI1 (strain Y04540) (lanes 10 to 12), or SKI7 (strain Y01852) (lanes 13 to 15). M2 virions from strain 1161 were also introduced into the L-A-lus-containing strain 1094, which carries the ski2-2 mutation (lanes 16 to 18). In each case, RNAs from the recipient strain and two independent cytoductants (Cytod.) were separated on an agarose gel and were analyzed by Northern hybridization with probes specific for L-A (or L-A-lus) and M2. All cytoductants carried 23S RNA (indicated by the asterisk), present in the donor strain but absent in the recipients. At the bottom, isolated colonies of independent cytoductants from each strain are analyzed for K2 killer activity.
Next, we checked what happened if the recipient strains carried ski mutations, which produce derepressed copy numbers of L-A or L-A-lus. For this purpose, we introduced M2 virions alone from strain 1161 (carrying M2 supported by coat proteins expressed from plasmid pI2L2) (Table 1) into recipient strains derived from the L-A-containing strain BY4741 with the SKI1, SKI2, SKI3, or SKI7 gene deleted. As seen in Fig. 4E, all cytoductants in the SKI deletion strains were stable killers, while L-A in the wild-type strain could not maintain M2. A similar result was obtained when the recipient strain with a ski2-2 mutation but carrying L-A-lus was used. Again, in this genetic background, M2 could be maintained by L-A-lus. These data thus indicate that the lack of specificity of M2 for its helper virus in these strains is due to the high copy numbers of the helper viruses they carry (or the increased expression of their coat proteins). We wondered whether strain 1385 (L-A, M2) (Fig. S2A in the supplemental material) might carry some mutation responsible for M2 maintenance by L-A, which would suggest that what we were observing was strain specific. When we first introduced the cytoplasm of strain 1385 into the L-A-o strain 37-4C (Table 1), there was a mixture of killer and nonkiller colonies, but in successive colony isolations, the K2 character was lost (Fig. S2B). The ability of strain 37-4C to maintain M2 was not impaired, since this strain was a stable K2 killer when M2 and L-A-2 were introduced from strain 1137 (not shown). These results suggest that the combination of M2 and L-A in strain 1385 is an exception to the general rule and a strain-specific phenomenon.
Strain 2405 (or its derivatives) does not carry mutant alleles that impair M2 maintenance by L-A.
The existence of mutations in the MKT1 gene has been correlated with the exclusion of M2 by L-A in certain strains (28). Because we used laboratory strain 2405 as the recipient strain for introducing the K2 killer character from wine yeast cells in the first instance (Fig. 2), we wanted to rule out the possibility that the strain might carry some mutant allele (in MKT1 or other gene) that impaired M2 maintenance by L-A helper viruses other than L-A-2. Thus, we carried out an additional experiment, mating directly germinating spores from the K2 wine strain Ca7 either with strain 2405 (L-A-o) or with its L-A-containing derivative strain 1064. Diploids were selected and were immediately sporulated. K2 killer activity and RNAs were analyzed both in the diploids and in selected meiotic segregants. As seen in Fig. S3 in the supplemental material, while the diploids derived from the parental L-A-o strain 2405 and subsequent meiotic segregants carried L-A-2 and M2 and were all stable K2 killers, the mating of spores of Ca7 (L-A-2, M2) to strain 1064 (L-A) resulted in nonkiller hybrids, and concomitantly, the meiotic segregants showed 4K− segregation. Northern hybridization with specific probes showed that in the latter diploids, L-A-2 had been excluded by L-A, and M2 was lost (Fig. S3). This confirms the data shown in Fig. 2 and indicates that M2 cannot be maintained by L-A, even in diploids resulting from mating between the L-A-containing strain 1064 and the K2 spores from Ca7, where any recessive allele needed for M2 maintenance by L-A should be complemented by the wild-type spore.
Different populations of L-BC virus.
During our recent work on the distribution of Klus or K2 viruses in a collection of 31 wine strains (23), we found by Northern hybridization with a 440-nt L-BC probe that about 42% of the strains tested (13 strains) carried L-BC. The signals on the blots, however, differed among the strains, suggesting either that the amounts of L-BC dsRNA were different or that the sequences could be somewhat different from that of the probe (Fig. 5A). Because different variants of L-A exist in wild strains, we wondered if the same was true for L-BC. Different types of L-BC, named L-B or L-C, had already been noticed, and in fact, the name L-BC was used because of a certain heterogeneity in L-BC virus populations (12, 30). When we synthesized random cDNAs from purified L-A-lus dsRNA from Klus strain EX229 (23), we also found a few L-BC clones (see Materials and Methods). One of these (clone 20) contained a 2.3-kb insert (Fig. 5B). Interestingly, the sequence of clone 20 showed about 12% of nucleotides different from those in the L-BC sequence deposited in GenBank (accession number U01060.1), suggesting the existence of a different variant of L-BC in the Klus strain. We designed specific oligonucleotides to amplify the rest of this L-BC by RT-PCR, including the 5′ and 3′ ends, which were amplified by rapid amplification of 3′ cDNA ends (3′ RACE) (Fig. 5B). The sequence thus obtained has the same length (4,614 bp) as L-BC and has been deposited in GenBank (accession number KT784813). With two pairs of oligonucleotides, RE635 and RE637 or RE635 and RE636, we could use RT-PCR to amplify a 1.2-kb cDNA fragment specific for L-BC from laboratory strains or from the L-BC variant in the Klus strain (which we designated L-BC-lus), respectively (Fig. 5C). In this way, we confirmed that three laboratory strains, strain BY4741 and two strains from Wickner's laboratory (strains 2928 and RE458), all had the same type of L-BC virus, which is identical to that sequenced by Bruenn's group (13). We were interested in analyzing the L-BC viruses in Klus and K2 wine strains. The pair of oligonucleotides that amplified L-BC-lus could also amplify a cDNA fragment of the same size from K2 strain 8F-13 (Fig. 5C), while the pair of oligonucleotides specific for L-BC could not. Interestingly, when we sequenced the RT-PCR fragment from the K2 strain, we found a type of L-BC different not only from the L-BC in laboratory strains but also from that in the Klus strain. Based on that evidence, we designated this variant in K2 strains L-BC-2 and sequenced it completely (GenBank accession number KX906605) as described in Materials and Methods. With the oligonucleotide pair RE635–RE636, we extended the analysis of L-BC to more Klus or K2 wine strains and sequenced the RT-PCR fragments. In summary, our sequencing data from three Klus strains (EX229, EX198, and Ca4) and three K2 strains (Lalvin EC1118, 8F-13, and S3920) show a close association between a specific K2 or Klus killer toxin-containing strain and a particular L-BC variant (L-BC-lus in Klus strains and L-BC-2 in K2 strains), suggesting that the strains have been isolated for a long period of time, allowing the L-BC viruses inside to evolve independently.
FIG 5.
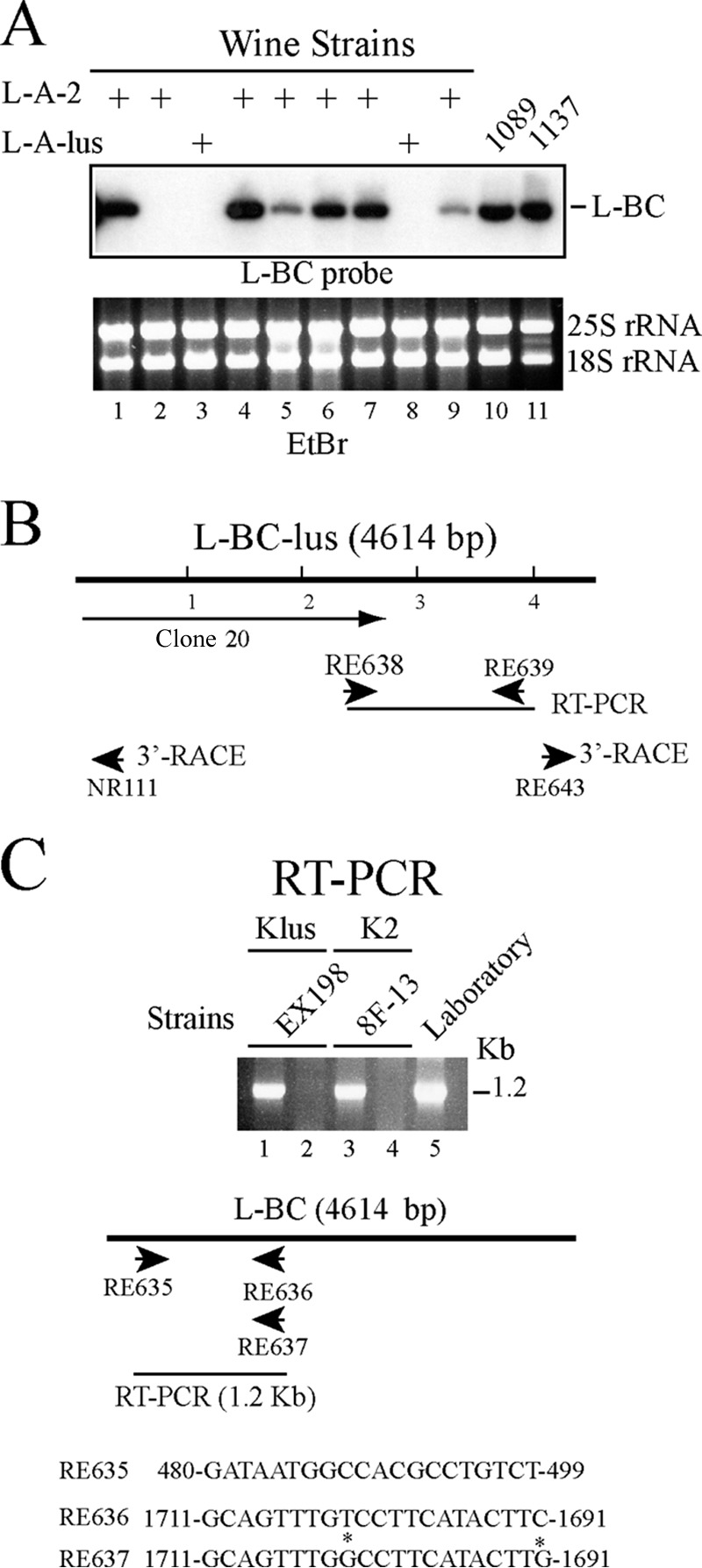
Different L-BC viruses in wine strains. (A) RNAs from a number of K2 or Klus wine strains (lanes 1 to 9) were hybridized with an L-BC probe made by runoff transcription from plasmid pRE442. Laboratory strains 1089 (Klus) and 1137 (K2) were also included (lanes 10 and 11). Ethidium bromide-stained rRNAs, used as loading controls, are shown below. The amounts of L-BC in lanes 5 and 9 are about 10 to 20% of the amounts of L-BC in other strains. (B) Strategy for sequencing L-BC-lus from strain EX198. Clone 20, obtained by random cDNA synthesis, is indicated. The rest of the sequence was obtained from RT-PCR clones or 3′ RACE. cDNA fragments were primed with the oligonucleotides indicated by the arrows. (C) RNAs from Klus wine strain EX198 (lanes 1 and 2), K2 strain 8F-13 (lanes 3 and 4), or laboratory strain BY4741 (lane 5) were used to amplify a fragment of 1.2 kb by RT-PCR with one of two pairs of oligonucleotides: RE635 plus RE636 (lanes 1 and 3) or RE635 plus RE637 (lanes 2, 4, and 5). The oligonucleotide sequences are given at the bottom. Note that the sequences of RE636 and RE637 differ only at two positions (marked by asterisks). RE636 was derived from L-BC in Klus strains, while RE637 comes from L-BC in laboratory strains.
Comparison of L-BC variants.
Most of the analysis discussed in this section was performed on the L-BC-lus variant, so we refer to L-BC-2 here only when comparing its nucleotide sequence (or the encoded proteins) to those of the other two variants (Fig. 6; see also Fig. S4 in the supplemental material). The genomic organization of L-BC-lus (and L-BC-2) is the same as that proposed for L-BC (13), with two overlapping frames, Gag and Pol (Fig. 6A), which are almost identical to those encoded by L-BC (about 96% identity). With respect to their nucleotide sequences, L-BC is closer to L-BC-2 (93% of nucleotides identical), while both L-BC and L-BC-2 showed 88% identity to L-BC-lus (Fig. S4). Certain features are worthy of note (Fig. 6; also Fig. S4). The 5′- and 3′-end sequences were basically identical. In the first 60 nt, only 2 nt were different, while in the last 60 nt, only 1 changed. Other parts differed 7 to 12%, with differences in certain stretches increasing up to 20%. The slippery site (1967-GGAUUUU-1973) and the adjacent stem-loop structure (nt 1973 to 1992) (Fig. 6B) in L-BC have been proposed as the region where the frameshift between Gag and Pol occurs. A minimum of 48 nt including this region, positioned upstream of a −1 out-of-frame β-galactosidase ORF, was sufficient to produce 2% expression of the reporter gene (13). Interestingly, nt 1987, which is C in L-BC, is U in L-BC-lus (or in L-BC-2). This change produces a less favorable interaction energy (ΔG = −5.24 kcal/mol) than in L-BC (ΔG = −7.72 kcal/mol) (Fig. 6B). Minimal changes in the stem-loop structure of the L-A virus frameshifting region produced major effects on frameshift efficiency (31, 32). The comparison of nucleotide sequences proximal to the 3′ ends also revealed a conserved sequence of 25 nt (nt 4359 to 4383), which folds into a stem-loop structure with a protruding A (Fig. S4). The structure, according to its free energy (ΔG = −11.53 kcal/mol), is quite stable. The surrounding nucleotide sequences, though quite conserved in the three variants, fold into completely different structures, suggesting a role of this conserved stem-loop in the L-BC virus cycle, perhaps in encapsidation. Oligonucleotides RE796 and RE797, which anneal in the frameshifting region or in this 25-nt interval, respectively, could amplify by RT-PCR a 2.4-kb fragment from all L-BC variants analyzed so far. With respect to putative cis signals for replication, a weak stem-loop structure proximal to the 3′ end is identical in the three variants (Fig. 6).
FIG 6.
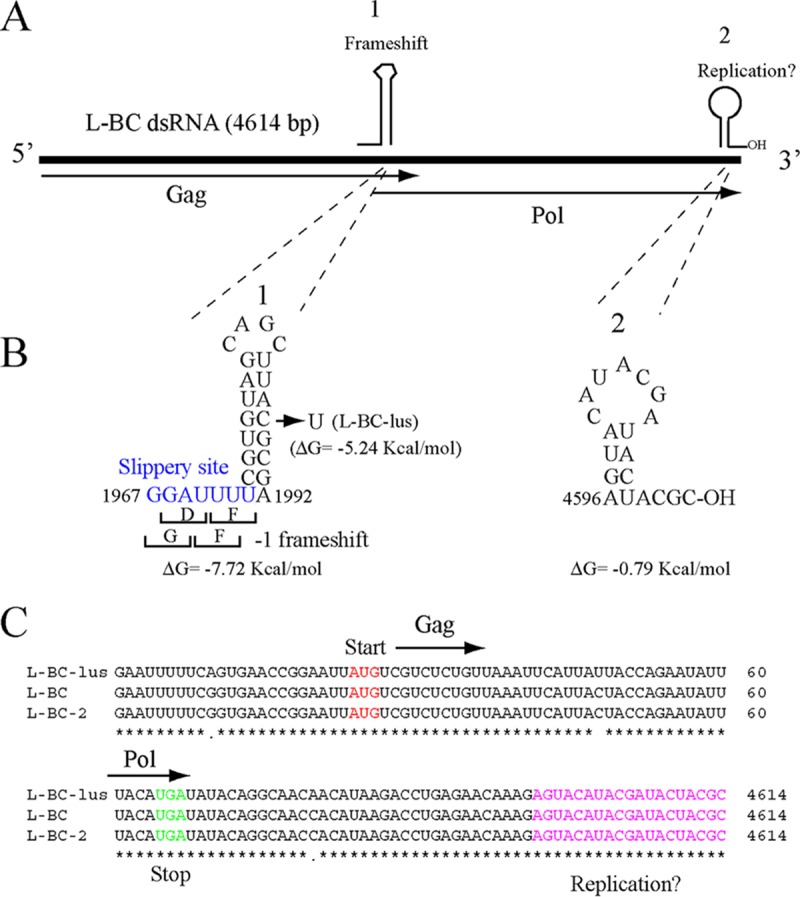
L-BC genomic organization and putative cis signals for frameshifting and replication. (A) Plus strand of L-BC dsRNA. Gag and Pol ORFs are represented by arrows. Pol is expressed as a Gag-Pol fusion protein by a −1 frameshift. Two cis signals, indicated as 1 and 2, are represented by schematic drawings above the plus strand. (B) Nucleotide sequences and predicted secondary structures of the two cis signals shown in panel A. For cis signal 1, the frameshifting region, with the slippery site in blue, is shown adjacent to a stem-loop structure. In L-BC-lus (and L-BC-2), the modification C1987U (indicated by the arrow) produces a change in the secondary-structure free energy. For cis signal 2, putative replication signals are identical in the three variants. (C) The nucleotide sequences of the 5′ and 3′ ends are shown with the start and stop codons for Gag and Pol, respectively. The putative replication signal is shown in pink.
Exclusion of different L-BC populations is independent of mitochondrial inheritance.
During the construction of the Klus laboratory strains 1083 and 1084 (23), spores from the Klus wine strain EX198 (L-A-lus, Mlus, L-BC-lus) were initially mated with laboratory strain 2928, which is L-A-o but carries L-BC. Thus, in the resulting diploids, we initially had two variants of L-BC. After sporulation, strains 1081 and 1082 (Table 1) were selected and were then used as donors of Klus viruses to produce strains 1083 and 1084. To find out what type of L-BC was present in this set of strains, we performed RT-PCR analysis with a pair of oligonucleotides specific for L-BC or for L-BC-lus. The results are shown in Fig. S5A in the supplemental material. Interestingly, the four haploid strains derived from the hybrid diploid carry only L-BC and not the L-BC-lus variant, indicating exclusion between these viruses. The virus present is that from the laboratory strain. Thus, Klus strains 1083 and 1084 carry the combination L-A-lus and L-BC, which is different from that found in nature (L-A-lus and L-BC-lus). Additionally, other strains generated in this work carry the combination L-A-2 and L-BC (e.g., strain 1137 [Table 1; Fig. 2A]), which is also different from the combination found in K2 wine strains (L-A-2 and L-BC-2). Thus, although each L-BC variant in wild strains is associated with a type of killer toxin-producing strain, there does not seem to be a dependency of a specific L-BC on a particular L-A.
We also wondered what happened with the other cytoplasmic trait, mitochondria, in the crosses mentioned above and if there was any particular association between mitochondria and L-BC inheritance. In yeast, when two strains carrying different mitochondria mate, only one type of mitochondria is found in the daughter cells after meiotic segregation (33). Thus, we analyzed the mitochondrial DNA (mtDNA) of strains EX198, 2928, and 1084 by restriction fragment length polymorphism (RFLP) analysis, a method frequently used to identify new wine strains. Klus wine strain EX198 and laboratory strain 2928 show completely different RFLP patterns, and strain 1084 carries the mitochondria from the Klus strain (Fig. S5B). A similar analysis of another wine strain, Ca4, that also carries L-BC-lus, and the laboratory-derived strain 1169, obtained by an experimental procedure similar to the one outlined in Fig. 2A, showed that strain 1169 inherited its mitochondria from strain 2928 and not from the wine strain Ca4 (Fig. S5B). Thus, in two independent crosses between laboratory strain 2928 and two wine strains, the offspring harbor the same L-BC variant from strain 2928, but the mitochondria come from a different parent, suggesting that they are independently inherited. Apart from the two strains mentioned above, we also analyzed the mtDNA patterns of four other laboratory strains generated from different Klus or K2 wine strains, including the K2 strains 1137 and Ca7, also shown in Fig. S5B. In four of the six strains analyzed, the mitochondria came from the wine strain, and in two, the mitochondria came from the laboratory strain (strain 2928). Thus, in general, the transmission of mitochondria and yeast totiviruses are not associated, as expected for cytoplasmic genetic elements that do not seem to interact physically.
DISCUSSION
L-A-2 is the helper virus of M2 and is excluded by L-A or L-A-lus.
In this work, we have characterized L-A-2, the L-A present in most K2 wine strains, and analyzed the M2 helper requirements in laboratory strains (either diploid or haploid). As shown here, the association of M2 and L-A-2 in most K2 wine strains is in accordance with the requirement of L-A-2 as the helper virus of M2 (Fig. 2 and 4D). During mating or cytoplasmic mixing, L-A-2 was excluded by L-A or L-A-lus, and since none of these viruses showed helper activity for M2, the K2 character was lost. We also demonstrated two exceptions to the L-A-2 requirement, both of which are related to elevated copy numbers of the noncanonical helper virus: (i) the provision of viral proteins without restriction by a vector and (ii) ski mutants, such as ski1, ski2, ski3, or ski7, with derepressed virus copy numbers. The effects of several ski mutations had already been reported by Wickner's group (34). In contrast to M2, M1 seems to be less strict about its helper requirements and can be maintained by L-A-H (L-A-2) or L-A-lus (23, 35). Also, maintenance of M28 by L-A has been reported (24). We do not know at present why different satellites may have different helper requirements, since what we observe in nature is the result of a fine-tuning between the helper and the satellite through a long period of coexistence. The dependency shown by the satellite on coat proteins provided by the helper for its own survival, while competing with the helper (but not displacing it, so as to avoid suicide), may have shaped this stable coexistence. This competition also explains why the curing of satellites by different means generally increases the copy number of the helper. The fine-tuning achieved during evolution in each case, however, does not rule out the possibility that some L-A helpers, which have evolved independently, could be used by more than one satellite. Only the encounter of one helper with a new satellite (probably rare in nature, but easily performed in laboratory strains) will provide information about the helper capacity of some L-A viruses for satellites other than those with which they have coevolved.
Three phenotypic variants of L-A have been described based on different K1 and K2 exclusion characteristics: L-A-E, L-A-HN, and L-A-H. L-A-E excluded M2 dsRNA (but not M1) in [NEX-o] strains but not in strains that carried [NEX] (28). L-A-HN was the L-A usually found in K1 killer strains, and L-A-H was the L-A present in the few original K2 strains examined (36). It was also reported that L-A-HN could maintain M2 in an MKT1 background but not in mkt1 mutants at temperatures above 20°C (28, 29). The differences of those L-A variants at the molecular level, however, were not known. As shown in Fig. S2A, our analysis of three K2 strains used in those studies revealed that one strain, which was NEX-o, carried L-A-2, and the other two (which were NEX) had L-A. Thus, the K2 exclusion observed when a strain carrying L-A-2 was mated with a nonkiller strain carrying L-A-E (28) agrees with what we know now: L-A-2 was excluded by a form of L-A, and this L-A was not able to maintain M2. Moreover, our data showed that exclusion of L-A-2 (and M2) by L-A was independent of mutations in the MKT1 gene (or in other genes) that might impair M2 maintenance by L-A (Fig. S3). Thus, the maintenance of M2 by L-A in certain laboratory strains was probably due to their genetic background; in fact, one strain shown in Fig. S2A carries the ski7-1 mutation, and the combination of L-A and M2 in strain 1385 is not stable after cytoduction into a different strain (37-4C) (Fig. S2B). A similar lack of specificity was found in our previous screening of 31 wine strains (Table 2 in reference 23), with a few cases in which L-A-lus and M2 were present together (exceptions to the general rule). M2 viruses from some of these strains, however, when transferred to a laboratory strain, were barely maintained by L-A-lus: their copy numbers were either greatly reduced or completely lost (N. Rodríguez-Cousiño and R. Esteban, unpublished data). This suggests, again, that the background of the strains was affecting M2 maintenance by L-A-lus. In those wild strains, we did not find the opposite (L-A-2 and Mlus), implying that if in the same ecological niche K1 and K2 strains ever mated, L-A-lus would have excluded L-A-2 and would have maintained M2 only in rare cases. We believe that the coexistence of K2 and Klus strains in wine environments is favored by the fact that mating is an infrequent event in nature (otherwise, as predicted by our data, K2 strains would tend to disappear).
L-A and L-BC totivirus variants in yeast strains.
The complete sequence of L-A-2 reported here complements the other three L-A variants so far identified (L-A, L-A-lus, and L-A-28). L-A-2 shares 74% identity with L-A and 76% or 78% identity with L-A-28 or L-A-lus, respectively. The association of each variant with different M dsRNAs encoding toxins suggests coevolution between the helpers and the toxin-producing viruses (23). In the viral genomes, the frameshifting and encapsidation regions are conserved except for nt 1991 in the frameshifting region, which can be U or C. These regions are also conserved on L-A viruses from sensu stricto yeasts other than S. cerevisiae, since primers based on these regions amplify cDNA fragments of the expected size from all of these viruses so far tested (Rodríguez-Cousiño and Esteban, unpublished). The degree of conservation of the encoded Gag and Pol ORFs ranges from 87 to 92%, while the central parts of Pol, where the conserved RdRp motifs reside, show >95% of their amino acids identical. Dendrograms of Gag and Pol from the different L-A variants (see Fig. S6 in the supplemental material) show that L-A-2 is closer to L-A-lus than to the other two L-A viruses. The fact that L-A-2 and L-A-lus are found in K2 and Klus killer yeasts that share the same habitat, wine environments (23, 37), suggests that they may have evolved from a common helper virus ancestor present in strains adapted to this ecological niche. In the same environments, K1 or K28 strains were not found (23). K1 strains seem to be confined mainly to laboratory collections, and K28 strains have been reported recently in Saccharomyces paradoxus (38, 39).
We have also found that the Klus and K2 wine strains carry L-BC variants (named L-BC-lus and L-BC-2) different from the L-BC in laboratory strains (either K1 or nonkillers). In contrast to L-A populations (in which as many as 26% of nucleotides are different), the L-BC nucleotide sequences are quite conserved, with only about 10% of their nucleotides different. This may reflect different evolutionary rates of these two types of totiviruses in the same host or, alternatively, distinct evolutionary histories in separate hosts before the introduction of one of the viruses into yeasts carrying the other to generate strains with both together. While killer totiviruses (and thus L-A) are found in S. cerevisiae and other yeasts from the sensu stricto cluster, such as S. paradoxus or Saccharomyces uvarum (38–40), as well as in nonrelated yeasts, L-BC seems to be less widespread. We have found it to be present only in S. cerevisiae strains and absent from several S. paradoxus and S. uvarum strains so far examined (Rodríguez-Cousiño and Esteban, unpublished). In genome databases, the totiviruses most closely related to L-BC are in Scheffersomyces segobiensis (41) and Delisea pulchra. This suggests that L-BC ancestors may have resided in other yeasts before being introduced into S. cerevisiae killer strains by horizontal transfer. After that event, L-A and L-BC have evolved together, resulting in strains with different L-A variants and different accompanying L-BC viruses. We have proposed that the toxin-encoding satellite RNAs may have exerted selective pressure to render the helpers better fit to support them. If that is the case, the absence of that pressure in L-BC viruses, which lack helper activity for any satellite RNA, may be correlated with a slower evolution rate (Fig. S6).
Since the L-A variants are associated with different M satellites, it would be interesting to know the origin and evolution of these satellites. We believe that they originated from host mRNAs transcribed from genes encoding preprotoxins that were encapsidated after acquiring signals for encapsidation and replication, most likely from the helper genome itself. This hypothesis is supported by the existence of ORFs with similarities to the preprotoxins in yeast chromosomes, either in the same host or in other ancestrally related yeasts. In the former case, the YFR020W ORF was found to be similar to Klus preprotoxin (3). According to a BLAST search against GenBank in October 2016, putative ancestors for K2 and K1 are present in other yeasts, such as Kazachstania africana or Kluyveromyces lactis. The KAR_0A00110 ORF from K. africana CBS 2517 shows a significant similarity to K1 preprotoxin (26% identity in an interval that covers 76% of the ORF). With respect to K2 preprotoxin, putative ancestors with higher conservation are found in K. africana (37% identity in an interval covering 79% of the ORF) and K. lactis (38% identity with 61% coverage of the ORF). A K28 preprotoxin-related degenerated ORF is also present in S. cerevisiae JM195.
MATERIALS AND METHODS
Yeast strains and media.
The laboratory strains used in this study are summarized in Table 1. The wild wine strains used (prototrophic and homothallic) were described in a previous report (23). The media and incubation conditions have been described previously (23). Curing of M2 was achieved by incubating K2 strains on YPAD (1% yeast extract, 2% peptone, 0.04% adenine, 2% dextrose) plates supplemented with 0.02% uracil in the presence of 0.1 μg/ml cycloheximide.
Cytoduction and killer assay.
Cytoduction is a type of mating in which cytoplasmic mixing occurs without nuclear fusion because one of the parental strains is a kar1 mutant, defective in nuclear fusion (42). This allows cytoplasmic traits, such as killer viruses or mitochondria, to pass from a donor strain to a recipient strain, which is [rho0] (respiration deficient). The cytoduction procedure is summarized in Fig. 2A and has been described in detail previously (23). Killer assays were performed by replica plating of isolated colonies on methylene blue (MB) plates seeded with a lawn of the sensitive strain 5x47. MB plates were incubated at 25°C for 2 to 3 days, unless otherwise noted. Around the killer colonies, a halo of growth inhibition of the sensitive strain is visible.
Preparation of total nucleic acids and Northern hybridization.
Total RNAs were obtained from 1-ml stationary-phase cultures, separated on agarose gels, and denatured in the gels before transfer to neutral nylon membranes (GE Healthcare). The details of nucleic acid preparation and Northern hybridization conditions have been provided elsewhere (43, 44). 32P-labeled-specific probes were made by T3 or T7 runoff transcription from plasmids that had been linearized with appropriate restriction enzymes. Plasmids used to prepare probes to detect L-A, L-A-lus, M1, M2, or L-BC dsRNAs have been described elsewhere (23). The L-A-2 probe was synthesized from plasmid pRE1311, which contains a HindIII-EcoRI fragment of 831 bp of L-A-2 cDNA (from nt 502 to nt 1332), inserted between the EcoRI and HindIII sites of the Bluescript KS(+) vector.
cDNA synthesis and sequencing of L-A-2 dsRNA.
As reported previously (23), oligonucleotides NR88 and NR80 (see Table S7 in the supplemental material) could amplify a 1.3-kb cDNA fragment from the L-A dsRNA present in seven different K2 wine strains by RT-PCR. The sequence was the same in all of the fragments. As described in Results, we introduced K2 killer viruses from wine strain Ca7 into a laboratory strain, producing strain 1137. L-A-2 from this strain was then used for subsequent cDNA synthesis by RT-PCR with appropriate oligonucleotides to complete its sequence. Figure S1A shows a diagram of the L-A-2 cDNA fragments sequenced. The conditions for annealing and first-strand synthesis were those described for the 3′ RACE protocol (see below). Depending on the experiments, either Go Taq DNA polymerase (Promega) or AccuPrime Taq DNA Polymerase, High Fidelity (Invitrogen) was used, and the annealing temperatures differed. Finally, the sequences from the ends of L-A-2 were obtained by 3′ RACE. For this purpose, L-A-2 dsRNA was purified from strain 1163 (strain 1137 cured of M2 by cycloheximide treatment) by CF-11 cellulose chromatography as described elsewhere (45). Then the 3′ ends of each strand were A-tailed using poly(A) polymerase and the conditions recommended by the supplier (Epicentre). The conditions for annealing with an oligo(dT) primer (oligonucleotide NR67) and for cDNA synthesis using SuperScript reverse transcriptase (Invitrogen) have been described previously (3). For PCR amplification, we used primer NR68 and either primer NR125, for the amplification of the 3′ end of the negative strand, or primer NR126, for the amplification of the 3′ end of the positive strand. In all cases, PCR products were purified and directly sequenced.
cDNA synthesis and sequencing of two L-BC variants from Klus and K2 wine strains.
In the course of the cDNA synthesis and cloning of L-A-lus (23), we obtained some random clones from L-BC, since this dsRNA was also present in strain EX229 and copurified with L-A-lus. Those random cDNAs were cloned in the Bluescript KS(+) vector and were sequenced. The sequence of a gap between nt 2544 and 4015 was obtained by RT-PCR, and the sequences of the ends of the molecule were obtained by 3′ RACE. Figure 5 includes a diagram of the sequencing strategy. This L-BC was named L-BC-lus, to differentiate it from the L-BC found in K2 strains, which was designated L-BC-2 (see Results). The full sequence of L-BC-2 from strain S3920 was also obtained from RT-PCR fragments amplified with specific primers designed according to previous sequences. The sequences and annealing positions of those primers are shown in Table S7 in the supplemental material.
Other procedures.
Enzyme digestions and cloning procedures were carried out by following standard methods (46). Plasmid DNA was obtained using the Wizard Plus SV Minipreps DNA purification system (Promega). DNA fragments for cloning were purified with a QIAquick gel extraction kit (Qiagen). Before the sequencing of PCR fragments, oligonucleotides present in the samples were removed by Sepharose chromatography using MicroSpin S-400 HR columns (GE Healthcare). Antibodies against the Gag protein of L-A virus have been described elsewhere (47). Yeast cells were transformed using lithium acetate to permeabilize the cells (48). RFLP analysis of mtDNA was performed according to reference 49. Plasmids pI2L2 and p1290 are 2μm-derived plasmids that express L-A or L-A-lus virion proteins under the control of the PGK1 promoter and carry the TRP1 gene as a selective marker (23, 50). The identities of the L-A variants in the exclusion experiments for which results are shown in Fig. 2 and 3 were confirmed by RT-PCR and sequencing using oligonucleotides NR88 and NR80 (for both L-A-2 and L-A-lus) or oligonucleotides RE548 and RE549, specific for L-A (23). The DNA primers used are listed in Table S7 in the supplemental material. RNA secondary-structure predictions were carried out using the MFOLD program (51).
Accession number(s).
The nucleotide sequences and the encoded Gag and Gag-Pol proteins appear in NCBI/GenBank under accession numbers KC677754.1 for L-A-2, KT784813 for L-BC-lus, and KX906605 for L-BC-2.
Supplementary Material
ACKNOWLEDGMENTS
This work has been supported by grants BFU2010-15768 and BFU2014-52418-P from the Spanish Ministry of Economy and Competitiveness (MINECO).
We thank Pilar Gómez for invaluable technical assistance.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02991-16.
REFERENCES
- 1.Schmitt MJ, Breinig F. 2006. Yeast viral killer toxins: lethality and self-protection. Nat Rev Microbiol 4:212–221. doi: 10.1038/nrmicro1347. [DOI] [PubMed] [Google Scholar]
- 2.Schmitt MJ, Tipper DJ. 1995. Sequence of the M28 dsRNA: preprotoxin is processed to an α/β heterodimeric protein. Virology 213:341–351. doi: 10.1006/viro.1995.0007. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-Cousiño N, Maqueda M, Ambrona J, Zamora E, Esteban R, Ramírez M. 2011. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl Environ Microbiol 77:1822–1832. doi: 10.1128/AEM.02501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Icho T, Wickner RB. 1989. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J Biol Chem 264:6716–6723. [PubMed] [Google Scholar]
- 5.Dinman JD, Icho T, Wickner RB. 1991. A −1 ribosomal frame shift in a double-stranded RNA virus of yeast forms a Gag–Pol fusion protein. Proc Natl Acad Sci U S A 88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruenn JA. 2003. A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases. Nucleic Acids Res 31:1821–1829. doi: 10.1093/nar/gkg277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng RH, Caston JR, Wang GJ, Gu F, Smith TJ, Baker TS, Bozarth RF, Trus BL, Cheng N, Wickner RB, Steven AC. 1994. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J Mol Biol 244:255–258. doi: 10.1006/jmbi.1994.1726. [DOI] [PubMed] [Google Scholar]
- 8.Ribas JC, Wickner RB. 1998. The Gag domain of the Gag-Pol fusion protein directs incorporation into the L-A double-stranded RNA viral particles in Saccharomyces cerevisiae. J Biol Chem 273:9306–9311. doi: 10.1074/jbc.273.15.9306. [DOI] [PubMed] [Google Scholar]
- 9.Fujimura T, Esteban R. 2000. Recognition of RNA encapsidation signal by the yeast L-A double-stranded RNA virus. J Biol Chem 275:37118–37126. doi: 10.1074/jbc.M005245200. [DOI] [PubMed] [Google Scholar]
- 10.Esteban R, Fujimura T, Wickner RB. 1989. Internal and terminal cis-acting sites are necessary for in vitro replication of the L-A double-stranded RNA virus of yeast. EMBO J 8:947–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Field LJ, Bobek LA, Brennan VE, Reilly JD, Bruenn JA. 1982. There are at least two yeast viral double-stranded RNAs of the same size: an explanation for viral exclusion. Cell 31:193–200. doi: 10.1016/0092-8674(82)90419-6. [DOI] [PubMed] [Google Scholar]
- 12.Sommer SS, Wickner RB. 1982. Yeast L dsRNA consists of at least three distinct RNA's; evidence that the non-Mendelian genes [HOK], [NEX] and [EXL] are on one of these dsRNA's. Cell 31:429–441. doi: 10.1016/0092-8674(82)90136-2. [DOI] [PubMed] [Google Scholar]
- 13.Park CM, Lopinski JD, Masuda J, Tzeng TH, Bruenn JA. 1996. A second double-stranded RNA virus from yeast. Virology 216:451–454. doi: 10.1006/viro.1996.0083. [DOI] [PubMed] [Google Scholar]
- 14.Wickner RB. 1991. Yeast RNA virology: the killer systems, p 263–296. In Broach JR, Pringle J, Jones E (ed), The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis, and energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 15.Wickner RB, Fujimura T, Esteban R. 2013. Viruses and prions of Saccharomyces cerevisiae. Adv Virus Res 86:1–36. doi: 10.1016/B978-0-12-394315-6.00001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickner RB, Bussey H, Fujimura T, Esteban R. 1995. Viral RNA and the killer phenomenon of Saccharomyces, p 221–226. In Kück U. (ed), The Mycota, vol II. Genetics and biotechnology. Springer Verlag, Berlin, Germany. [Google Scholar]
- 17.Tercero JC, Wickner RB. 1992. MAK3 encodes an N acetyltransferase whose modification of the L-A Gag NH2 terminus is necessary for virus particle assembly. J Biol Chem 267:20277–20281. [PubMed] [Google Scholar]
- 18.Polevoda B, Sherman F. 2001. NatC N-terminal acetyltransferase of yeast contains three subunits, Mak3p, Mak10p and Mak31p. J Biol Chem 276:20154–20159. doi: 10.1074/jbc.M011440200. [DOI] [PubMed] [Google Scholar]
- 19.Toh-E A, Guerry P, Wickner RB. 1978. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J Bacteriol 136:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomecki R, Drazkowska K, Dziembowski A. 2010. Mechanisms of RNA degradation by the eukaryotic exosome. Chembiochem 11:938–945. doi: 10.1002/cbic.201000025. [DOI] [PubMed] [Google Scholar]
- 21.Larimer FW, Hsu CL, Maupin MK, Stevens A. 1992. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120:51–57. doi: 10.1016/0378-1119(92)90008-D. [DOI] [PubMed] [Google Scholar]
- 22.Edwards MD, Symbor-Nagrabska A, Dollard L, Gifford DK, Fink GR. 2014. Interactions between chromosomal and nonchromosomal elements reveal missing heritability. Proc Natl Acad Sci U S A 111:7719–7722. doi: 10.1073/pnas.1407126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodríguez-Cousiño N, Gómez P, Esteban R. 2013. L-A-lus, a new variant of the L-A totivirus found in wine yeasts with Klus killer toxin-encoding Mlus double-stranded RNA: possible role of killer toxin-encoding satellite RNAs in the evolution of their helper viruses. Appl Environ Microbiol 79:4661–4674. doi: 10.1128/AEM.00500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitt MJ, Tipper DJ. 1990. K28, a unique double-stranded RNA killer virus of Saccharomyces cerevisiae. Mol Cell Biol 10:4807–4815. doi: 10.1128/MCB.10.9.4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konovalovas A, Serviené E, Serva S. 2016. Genome sequence of Saccharomyces cerevisiae double-stranded RNA virus L-A-28. Genome Announc 4:e00549-16. doi: 10.1128/genomeA.00549-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sommer SS, Wickner RB. 1982. Co-curing of plasmids affecting killer double-stranded RNAs of Saccharomyces cerevisiae: [HOK], [NEX], and the abundance of L are related and further evidence that M1 requires L. J Bacteriol 150:545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Esteban R, Vega L, Fujimura T. 2008. 20S RNA narnavirus defies the antiviral activity of SKI1/XRN1 in Saccharomyces cerevisiae. J Biol Chem 283:25812–25820. doi: 10.1074/jbc.M804400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wickner RB. 1980. Plasmids controlling exclusion of the K2 killer double-stranded RNA plasmid of yeast. Cell 21:217–226. doi: 10.1016/0092-8674(80)90129-4. [DOI] [PubMed] [Google Scholar]
- 29.Wickner RB. 1983. Killer systems in Saccharomyces cerevisiae: three distinct modes of exclusion of M2 double-stranded RNA by three species of double-stranded RNA, M1, L-A-E, and L-A-HN. Mol Cell Biol 3:654–661. doi: 10.1128/MCB.3.4.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.El-Sherbeini M, Tipper DJ, Mitchell DJ, Bostian KA. 1984. Virus-like particle capsid proteins encoded by different L double-stranded RNAs of Saccharomyces cerevisiae: their roles in maintenance of M double-stranded killer plasmids. Mol Cell Biol 4:2818–2827. doi: 10.1128/MCB.4.12.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tzeng T-H, Tu C-L, Bruenn JA. 1992. Ribosomal frameshifting requires a pseudoknot in the Saccharomyces cerevisiae double-stranded RNA virus. J Virol 66:999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dinman JD, Wickner RB. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol 66:3669–3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger KH, Yaffe MP. 2000. Mitochondrial DNA inheritance in Saccharomyces cerevisiae. Trends Microbiol 8:508–513. doi: 10.1016/S0966-842X(00)01862-X. [DOI] [PubMed] [Google Scholar]
- 34.Ridley SP, Sommer SS, Wickner RB. 1984. Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol Cell Biol 4:761–770. doi: 10.1128/MCB.4.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannig EM, Leibowitz MJ, Wickner RB. 1985. On the mechanism of exclusion of M2 double-stranded RNA by L-A-E double-stranded RNA in Saccharomyces cerevisiae. Yeast 1:57–65. doi: 10.1002/yea.320010107. [DOI] [PubMed] [Google Scholar]
- 36.Wickner RB. 1986. Double-stranded RNA replication in yeast: the killer system. Annu Rev Biochem 55:373–395. doi: 10.1146/annurev.bi.55.070186.002105. [DOI] [PubMed] [Google Scholar]
- 37.Maqueda M, Zamora E, Alvarez ML, Ramírez M. 2012. Characterization, ecological distribution and population dynamics of Saccharomyces sensu stricto killer yeasts in the spontaneous grape must fermentations of southwestern Spain. Appl Environ Microbiol 78:735–743. doi: 10.1128/AEM.06518-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pieczynska M, de Visser JAGM, Korona R. 2013. Incidence of symbiotic dsRNA ‘killer’ viruses in wild and domesticated yeast. FEMS Yeast Res 13:856–859. doi: 10.1111/1567-1364.12086. [DOI] [PubMed] [Google Scholar]
- 39.Chang S-L, Leu J-Y, Chang T-H. 2015. A population study of killer viruses reveals different evolutionary histories of two closely related Saccharomyces sensu stricto yeasts. Mol Ecol 24:4312–4322. doi: 10.1111/mec.13310. [DOI] [PubMed] [Google Scholar]
- 40.Ivannikova YV, Naumova ES, Naumov GI. 2007. Viral dsRNA in the wine yeast Saccharomyces bayanus var. uvarum. Res Microbiol 158:638–643. doi: 10.1016/j.resmic.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Taylor DJ, Ballinger MJ, Bowman SM, Bruenn JA. 2013. Virus-host co-evolution under a modified nuclear genetic code. PeerJ 1:e50. doi: 10.7717/peerj.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conde J, Fink GR. 1976. A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc Natl Acad Sci U S A 73:3651–3655. doi: 10.1073/pnas.73.10.3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramírez-Garrastacho M, Esteban R. 2011. Yeast RNA viruses as indicators of exosome activity: human exosome hCsl4p participates in RNA degradation in Saccharomyces cerevisiae. Yeast 28:821–832. doi: 10.1002/yea.1909. [DOI] [PubMed] [Google Scholar]
- 44.Fujimura T, Esteban R, Esteban LM, Wickner RB. 1990. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell 62:819–828. doi: 10.1016/0092-8674(90)90125-X. [DOI] [PubMed] [Google Scholar]
- 45.Franklin RM. 1966. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc Natl Acad Sci U S A 55:1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 47.Fujimura T, Ribas JC, Makhov AM, Wickner RB. 1992. Pol of Gag-Pol fusion protein required for encapsidation of viral RNA of yeast L-A virus. Nature 359:746–749. doi: 10.1038/359746a0. [DOI] [PubMed] [Google Scholar]
- 48.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 49.Querol A, Barrio E, Ramón D. 1992. A comparative study of different methods of yeast strain characterization. Syst Appl Microbiol 15:439–446. doi: 10.1016/S0723-2020(11)80219-5. [DOI] [Google Scholar]
- 50.Wickner RB, Icho T, Fujimura T, Widner WR. 1991. Expression of yeast L-A double-stranded RNA virus proteins produces derepressed replication: a ski− phenocopy. J Virol 65:155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuker M, Mathews DH, Turner DH. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p 11–43. In Barciszewski J, Clark BFC (ed), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Dordrecht, Netherlands. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


