Abstract
Aim:
Differences in children's development and susceptibility to diseases and exposures have been observed by sex, yet human studies of sex differences in miRNAs are limited.
Materials & methods:
The genome-wide miRNA expression was characterized by sequencing-based EdgeSeq assay in cord blood buffy coats from 89 newborns, and 564 miRNAs were further analyzed.
Results:
Differential expression of most miRNAs was higher in boys. Neurodevelopment, RNA metabolism and metabolic ontology terms were enriched among miRNA targets. The majority of upregulated miRNAs (86%) validated by nCounter maintained positive-fold change values; however, only 21% reached statistical significance by false discovery rate.
Conclusion:
Accounting for host factors like sex may improve the sensitivity of epigenetic analyses for epidemiological studies in early childhood.
Keywords: : cord blood, early life, epigenetics, filtering, miRNA, miRNAome, newborns, next-generation sequencing, normalization procedures, sex differences
Epigenetic mechanisms influence gene expression without changes in DNA sequences. Unlike genetic mutations, which lead to permanent changes of gene structure, epigenetic modifications are reversible and responsive to different environmental factors including lifestyle, diet and exposure to chemicals. The most widely studied epigenetic marks are DNA methylation and histone modifications. Less is known about noncoding RNAs, often considered the third type of epigenetic marks [1–3]. Increasing evidence has shown that epigenetic modifications, including noncoding RNA, alter or control DNA expression and the degree in which DNA is transcribed as an adaptive response [4–6].
There is a growing interest in analyzing the role of miRNAs that represent an important group of noncoding RNAs. miRNAs are about 18–22 nucleotides in length and play an active role in the epigenetic regulation of gene expression in all living organisms with eukaryotic nuclear DNA [6–8]. Biogenesis of miRNAs begins in the nucleus, and they are transcribed mainly by RNA polymerase II [7]. Transcription of intergenic miRNAs occurs via independent promoters, whereas miRNAs in the introns of protein-encoding genes may use the same promoter as the proximal gene. Accumulated evidence has demonstrated that most of the known miRNAs participate in normal development, as well as disease pathology, and that miRNAs may be potential biomarkers for classifying tumors and identifying tissue injury [8–13]. miRNA profiles may also change in response to chemical exposures [14]. Recent studies have reported associations of miRNA expression with exposure to cigarette smoke, arsenic and air pollution [15–17]. miRNA regulation represents a mechanism through which genes involved in different biological processes can be regulated simultaneously [18]. Furthermore, miRNAs regulate genes that participate in several pathways ranging from the regulation of biosynthetic process to nervous system development [19–21].
Growing evidence has shown that host factors such as sex are associated with interindividual differences in gene expression in human and animal models. Mele et al. identified 92 protein-coding genes differentially expressed in males and females across multiple human tissues [22]. In agreement with human studies, Yang et al. identified 2871 genes from the mouse genome with differential expression by sex in multiple somatic tissues [23,24], while another study identified 25 of 376 miRNAs in fetal mouse lung that were also differentially expressed by sex [25]. Sex-specific differences were also shown for DNA methylation and histone acetylation. For example, Tsai et al. reported sex differences in histone acetylation in neonatal mouse brain [26]. Similarly, Xu et al. have demonstrated sex bias in the methylome and transcriptome of the human prefrontal cortex [27]. We recently reported on sex differences in genome-wide DNA methylation in newborns [28].
miRNA expression may also differ by sex but data are limited. In animal models, the majority of the reported studies have used brain samples [2,29]. For example, 149 miRNAs were described as differentially expressed in neonatal mouse brain, with almost one third regulated by sex-chromosome-mediated mechanisms, and approximately 72 identified miRNAs were potentially under estrogen regulation [29–31]. Another study has shown that sex-specific miRNA expression was induced in mouse brain after radiation, suggesting inherent sex-biased control of miRNA expression [32]. In that study, miR-29a and miR-29c were detected as significantly downregulated in the female brain when compared with male mice. As a consequence, DNA methyltransferases DNMT3a and DNMT3b, that are miR-29 family target genes, were reported as significantly upregulated in the female mice [32]. In addition, two studies conducted using rodent brain have reported estrogen regulation in DNA methylation and predicted regulation in miR-29a cluster [33,34]. Murphy et al. detected that miR-200 family was differentially expressed by sex in the brain of rat pups. They discovered that the miR-200 gene targets are linked to gonadotropin-releasing hormone receptor pathway suggesting that there is a relationship between gonadal hormone release-function and miRNAs [35]. To date, limited data are available on sex difference in miRNA expression in humans. One small study (n = 18) reported 40 miRNAs that were differentially expressed by sex in the human prefrontal cortex [20]. A few studies have examined sex-related differences in profiles of circulating plasma miRNAs in adults but were limited either by small sample size or low number of miRNAs assessed [36–39]. For example, one study found no differences by sex in miRNA expression of 108 miRNAs characterized in 20 adults [39]. In another human study, (n = 18) four miRNAs (out of 534 miRNAs) were significantly upregulated in women [38], while yet another study reported seven out of 179 candidate miRNAs to be differentially expressed by sex [36]. To our knowledge, no data are available on genome-wide miRNA expression (miRNAome) differences by sex in children. The aim of this study was to use targeted next-generation sequencing to investigate miRNA expression in cord blood and characterize the differences among newborn boys and girls.
Materials & methods
Samples
A random subset of term singletons (45 boys and 44 girls) with sufficient blood specimens available for miRNA analysis was selected from the CHAMACOS study, a well-characterized birth cohort followed by the Center for Environmental Research on Children's Health [40,41]. Study protocols were approved by the University of California, Berkeley Committee for Protection of Human Subjects. Written informed consent was obtained from all mothers.
RNA was purified from 20 μl of buffy coat separated by centrifugation from the cord blood and banked at -80°C the School of Public Health Biorepository. Cells were lysed in a guanidinium-based solution, followed by an acid–phenol:chloroform extraction and addition of the miRNA homogenate following manufacturer's instructions. Initial pilot studies performed in our laboratory indicated that the buffy coat fraction of blood that contains all white blood cells provides the highest RNA yields compared with clot and serum (data not shown). This data, together with a number of published studies demonstrating the utility of miRNA expression data from buffy coat fraction in human population studies [42–46] informed our decision to use RNA from buffy coats as starting material for the analysis of miRNA expression in cord blood of CHAMACOS newborns. Isolated RNA was purified by solid-phase extraction on glass-fiber filter columns provided in the mirVana™ miRNA isolation kit. Concentration and quality of RNA was measured using the NanoDrop 2000 Spectrophotometer (Thermo Scientific, MA, USA). Only samples with 260/280 ratio greater than 1.9 were retained for the analyses. RNA quality was assessed using the 2100 Bioanalyzer (Agilent, CA, USA). Purified RNA was stored at -80°C until analysis.
Analysis of miRNA expression by targeted sequencing
Isolated RNA from the 89 newborns was used for miRNA profiling experiments. After addition of three randomly selected replicates, we assayed a total of 92 samples. The EdgeSeq miRNA Whole Transcriptome Assay from HTG Molecular Diagnostics, Inc. (AZ, USA) was used to measure miRNA expression. This novel platform provides a hybrid system combining a nuclease-free library preparation for specific transcripts followed by next-generation sequencing (NGS) for quantitation of the transcripts [47]. It exhibits several key advantages including relatively simple sample preparation that does not require reverse transcription, adenylation or ligation, all steps that can potentially introduce bias. It also has a broad dynamic range with relatively high reproducibility as well as good sensitivity and specificity [48,49]. The assay version used in this study included a total of 2280 transcripts made up of 2256 mature miRNAs (referenced from miRBase v20), 13 housekeeping (HK) mRNAs, six positive controls (synthetic spike-ins and their complements), and five negative controls (plant genes) [50]. 100 ng of RNA was used per reaction to measure miRNA expression. Sample processing and library preparation of the samples were conducted following the protocol of the HTG EdgeSeq miRNA Sample Prep Pack and the Sequencing Tag Pack (HTG Molecular Diagnostics, Inc.). The positive control PhiX from Illumina was used in this experiment (Illumina, CA, USA). Concentration of the prepared dual-indexed libraries was measured on Qubit 3.0 and Bioanalyzer was used to assess the purity and size of the fragments of interest. Prior to pooling, Kapa qPCR (Kapa Biosystems, Inc., MA, USA) was used to quantify the NGS libraries. Libraries from the 92 samples were then pooled in equal amounts and clustered with a concentration of 5 pmol in one lane each of a single-read flow cell using the cBot (Illumina). 5% PhiX was also included, as per the loading guidelines for HTG EdgeSeq libraries. 50 cycles were sequenced on a HiSeq 2000 (Illumina) using high-output mode, with FASTQ-only output. Sequence analysis was carried out according to manufacturer instructions using EdgeSeq Parser software (HTG Molecular Diagnostics, Inc.).
Validation of differentially expressed miRNAs using nCounter miRNA expression assays
To validate our findings in another platform, we measured miRNA expression in a subset of miRNAs (n = 30) that were found to be upregulated (hsa-miR-1304-3p, hsa-miR-127-5p, hsa-miR-127-3p, hsa-miR-1469, hsa-miR-6724-5p, hsa-miR-4488, hsa-miR-4443, hsa-miR-4787-5p, hsa-miR-638, hsa-miR-3928-3p, hsa-miR-4448, hsa-miR-663a, hsa-miR-4516, hsa-miR-452-5p), downregulated (hsa-miR-374b-5p, hsa-miR-301a-3p, hsa-miR-30b-5p, hsa-let-7d-5p, hsa-miR-30e-3p, hsa-let-7e-5p, hsa-let-7f-5p, hsa-miR-331-3p, hsa-miR-30c-5p, hsa-let-7a-5p) or unchanged (hsa-miR-181a-5p, hsa-miR-148a-3p, hsa-miR-340-5p, hsa-miR-30e-5p, hsa-miR-30a-5p, hsa-miR-30d-5p) in boys compared with girls using a custom designed nCounter miRNA expression assay (NanoString Technologies, WA, USA). miRNAs chosen for validation were based on availability of the assay and fold change levels. Isolated RNAs from 142 CHAMACOS boys and girls (100 ng per sample) were used for the expression assay per the manufacturer's recommended protocol.
Statistical analysis
Development of miRNA-filtering steps
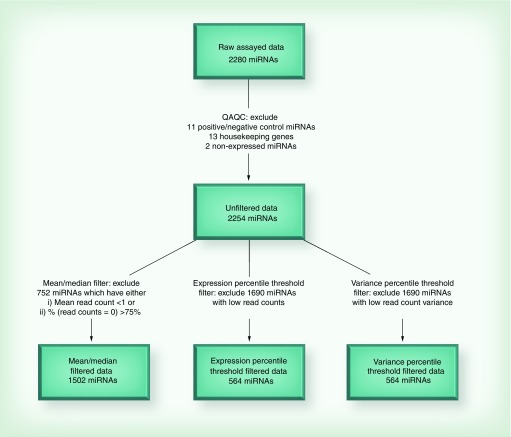
The count data from the 92 RNA samples (including three replicates) were generated and exported from the EdgeSeq assay for further filtering and processing steps. The data from three newborns were randomly selected and assayed as intra-assay technical replicates. After excluding probes for HK mRNAs, positive and negative controls and miRNAs with expression counts of 0 across all samples (n = 2), 2254 miRNAs remained in the analysis (Figure 1).
Figure 1. . miRNA filtering and diagnostics.
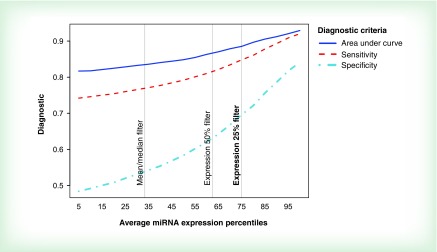
We tested three methods of miRNA filtering: 1) mean/median filters; 2) expression percentile threshold filtering and 3) variance percentile threshold filtering. The mean and median filtering method included only miRNAs that have: a mean read count over all 89 samples greater than one, and at least 25% of read counts across all samples that were non zero. The expression and variance percentile threshold filter included the 25% of miRNAs with the largest average read count and read count variance, respectively. To evaluate and compare the ability of each filter to exclude less informative miRNAs, we tested the ability of each filtered miRNA group to match intrasample replicates. We ran a multinomial logistic regression for each of the 2254 miRNAs, training on the first set of replicate samples. The outcome was the sample ID, and raw read counts were the explanatory variable. This classification algorithm was applied to the second set of replicate samples, rendering a set of predicted sample classifications for samples with known sample ID. We generated a confusion matrix for a multilevel variable and calculated three diagnostic criteria: area under the curve, sensitivity and specificity. The expression 25th percentile threshold filter was selected based on the high values observed using the diagnostic criteria. Figure 2 shows that diagnostic criteria tend to be higher for more aggressive filters 2 and 3. For example, for the mean/median filter, specificity was relatively low (0.54), and sensitivity was also less than desirable (0.74). Even at the 50th expression percentile, sensitivity was still only 0.79. This is why we applied the 25th percentile filter, and miRNAs with sensitivity above 0.84 were retained. In addition, the best observed average specificity was achieved in miRNA group with the highest expression. Furthermore, after the 25th expression percentile filter was applied, average area under the curve ranged from 0.89 to 0.93. Similar diagnostics were observed for the 25th percentile variance filter (data not shown). Since mean/median filtering was not quite adequate, we chose to proceed using more aggressive expression and variance threshold filtering at the 25th percentile yielding two groups of miRNAs: firstly, the highest expressing 25% of the miRNAs (n = 564) and secondly, the highest 25% of the miRNAs by variation in expression (n = 564).
Figure 2. . Classification diagnostics by increasing miRNA expression.
The best sensitivity and specificity were obtained when using the expression 25% filter. The mean/median filter was not adequate and achieved very low specificity.
We also compared methods of filtering by looking at overall counts per method and the overlap in retained miRNAs after each filtering procedure. There were 1128, 564 and 338 miRNAs after percentile threshold filtering at 50th, 25th and 15th expression thresholds, respectively. Firstly, the mean/median filter method retained a total of 1502 miRNAs. The expression and variance filters retained the same number of miRNAs (n = 564) because they both were set for the top 25th percentile. Overall, the most highly variable miRNAs also had the highest expression values. In fact, the overlap between the two groups was quite substantial. Of the two groups of top 25% highest expressors and top 25% of highest varying miRNAs, 544 miRNAs belong to both groups. Additionally, 20 miRNAs were unique to each of the two groups. Since the two groups were so similar, we chose to report all subsequent analyses using the results for the highest expressors group.
Normalization of HTG miRNA expression data
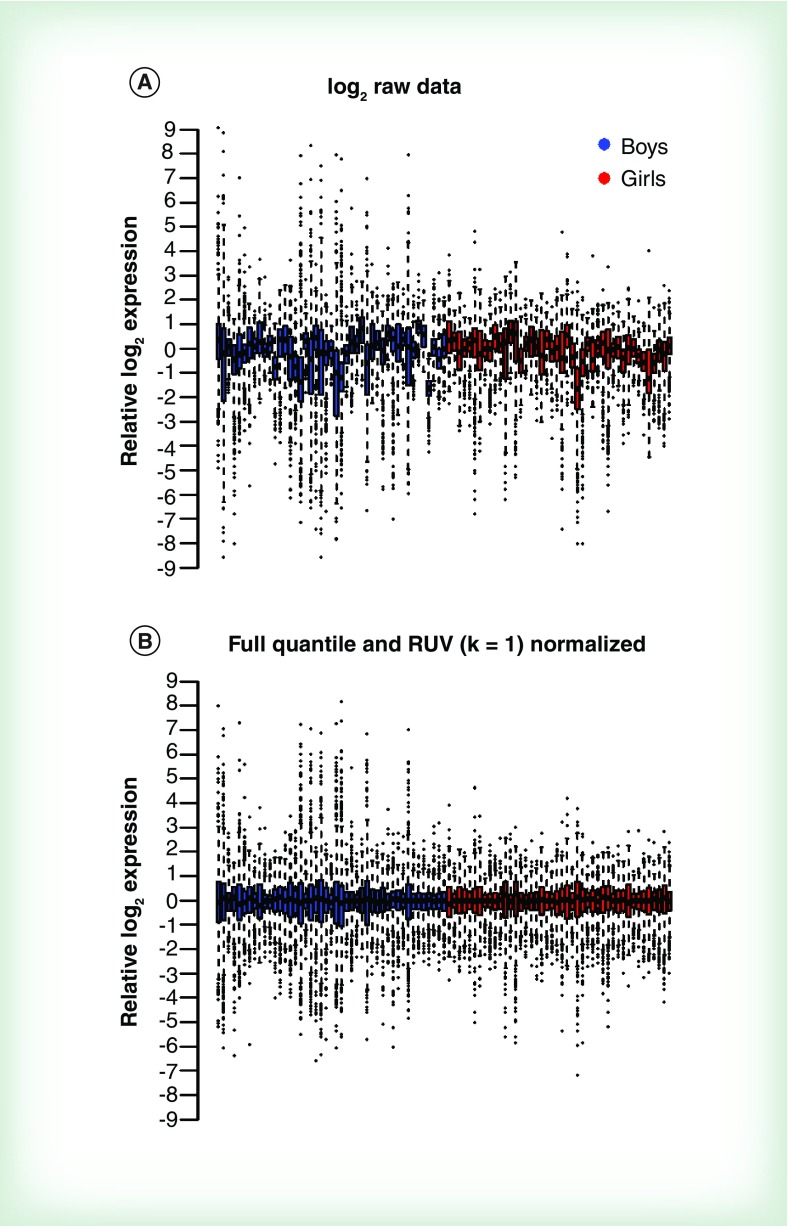
After applying the top 25% high expression filter, 564 miRNAs from 89 newborns were assessed for differential expression. We considered five normalization techniques: median house-keeping, upper quartile, trimmed mean, removed unwanted variance (RUV) and full quantile normalization [51,52]. We examined plots of the first and second principal components across samples and box plots of the normalized expression values to evaluate normalization procedures. Box plots revealed that full quantile normalization best centered and standardized expression distributions across samples. To further decrease technical variation, we applied RUV normalization after full quantile normalization. We found that full quantile with RUV best decreased overall variation among reads while maintaining separation in the first principal component by sex and this normalization procedure was used for subsequent analysis. Figure 3 shows box plots of relative log2 miRNA expression in boys and girls across 86 samples before and after normalization (full quantile and RUV). We calculated a mean SD of miRNA expression and found that overall variance was similar in boys and girls (average SD of log2 miRNA expression 2.879 and 2.878, respectively). However, using the Brown–Forsythe test for equality of variance, we found that 94 out of 564 miRNAs (top 25% highest expression) had unequal variances between boys and girls.
Figure 3. . Comparative variability of miRNA expression in newborn boys and girls.
Box plots of (A) raw and (B) full quantile normalization with removed unwanted variance (k = 1) normalized to relative log2 expression. RUV: Removed unwanted variance.
Analysis of differential expression between boys & girls
With the EdgeR package [53], we used the Cox–Reid profile-adjusted likelihood to estimate the common and miRNA-specific dispersion using the full-quantile normalized expression values, taking into account sex and the factor of unwanted variation estimated by RUV [54]. We then performed a generalized linear model (GLM) likelihood ratio test to fit negative binomial GLMs with the Cox–Reid dispersion estimates using the EdgeR package. Results were very similar when we ran the analysis using the R package LIMMA. p-values were adjusted using the Bonferroni procedure.
Normalization & differential expression of nCounter miRNA expression data
miRNA counts were background corrected using the geometric mean of the negative control probes included in the nCounter assay. To account for sources of variation related to the nCounter platform, counts were first normalized relative to ligation controls. This was followed by a second normalization step using the geometric mean of three normalizers included in the custom panel that had low variance (hsa-miR-30d-5p, has-miR-181a-5p and hsa-miR-340-5p). Background correction and normalization were performed in nSolver 3.0 software (NanoString Technologies). Differential expression analysis was performed using the EdgeR package to fit a negative binomial GLM.
Coordinates of sex-associated miRNAs in the human genome & miRNA clusters
miRBase v.20 [55], the central publicly available repository for miRNA research data was used to retrieve information about miRNA genome coordinates and miRNA clusters [56]. Manhattan plots were used to show the distribution of the differentially expressed miRNAs by sex genome-wide. The R package qqman [57] was used to generate the plots. Chi-square tests were used to determine whether differentially expressed miRNAs were overrepresented in certain chromosomes compared with the overall distribution of all mature miRNAs across chromosomes in miRBase v20.
In silico miRNA target prediction & miRNA pathway analysis
The 94 miRNAs that were different by sex were used as input information for target predictions using ComiR [58] and Tarbase v6.0 [59]. ComiR [60] works as prediction tool that uses miRNA expression to improve and combine multiple miRNA targets for each of the four widely used prediction algorithms: MiRanda [61], TargetScan [62], PITA [63] and mirSVR [64]. The composite scores of the four algorithms are created using a support vector machine trained on Drosophila Ago1 immunoprecipitation data. The composite score threshold used in ComiR analysis was 0.75. Tarbase v.6.0 was used to identify experimentally validated targets from the 94 miRNAs hits using BiomaRt [65]. Two out of the 94 differentially expressed miRNAs (hsa-miR-15a-5p and hsa-miR-33a-5p) have experimentally validated mRNA targets in Tarbase v.6.0. Additionally, we found validated targets for all 94 miRNAs on a more recent version of miRTarBase [66], although the support for most of these was not very strong. However, 18 of the miRNAs had targets that had more than suggestive evidence. This provides additional support to the hits we report in this study.
A final list of miRNAs was generated by the union of predicted and validated mRNA targets and used as the input data for functional annotation analysis. It was conducted employing ConsensusPathDB [67,68], a pathway tool of the Max Planck Institute for Molecular Genetics. Using the Web interface of the database, an over-representation analysis was conducted. Raw p-values were calculated using the whole-human genome as a background list. It determines the size set of each gene ontology term (biological process gene ontology [GO] terms, level 4), and accounts for the number of genes present in our input list. Adjustment of p-values was performed using the default parameters together with the Benjamini–Hochberg false discovery rate (FDR), shown as q-values (q < 0.05). Visualization and categorization of GO terms by semantic similarity dimension reduction was performed by reduce + visualize gene ontologies (REVIGO) [69].
Results
Among the high-expression group of miRNAs, we identified 94 miRNAs that were differentially expressed in cord blood buffy coat samples between boys and girls after Bonferroni correction for multiple testing. Table 1 shows log2 expression from a total of 94 miRNAs that were differentially expressed in the EdgeSeq experiment. Furthermore, mean miRNA expression was higher in boys compared with girls for the majority of differentially expressed miRNAs (85 out of the 94 miRNAs).
Table 1. . Differentially expressed miRNAs by sex in newborns.
|
miRNA hsa-miR- |
MIMAT ID |
Boys |
Girls |
Overall |
LFC1 |
FC2 |
p-value |
|||
|---|---|---|---|---|---|---|---|---|---|---|
| log2 mean expression | Variance | log2 mean expression | Variance | log2 mean expression | Variance | |||||
| 6753-5p |
0027406 |
7.8 |
10.04 |
5.73 |
0.65 |
6.78 |
6.42 |
2.07 |
4.20 |
1.03 × 10-19 |
| 602 |
0003270 |
5.68 |
6.37 |
3.97 |
2.45 |
4.84 |
5.12 |
1.71 |
3.27 |
4.15 × 10-6 |
| 4417 |
0018929 |
6.58 |
5.75 |
4.99 |
2.21 |
5.79 |
4.6 |
1.59 |
3.01 |
3.85 × 10-6 |
| 452-5p |
0001635 |
6.08 |
4.37 |
4.52 |
1.08 |
5.31 |
3.33 |
1.56 |
2.95 |
2.42 × 10-11 |
| 4792 |
0019964 |
11.8 |
3.11 |
10.25 |
1.57 |
11.03 |
2.94 |
1.55 |
2.93 |
1.03 × 10-7 |
| 4739 |
0019868 |
7.96 |
6 |
6.41 |
0.44 |
7.2 |
3.82 |
1.55 |
2.93 |
1.18 × 10-15 |
| 4271 |
0016901 |
7.09 |
6.06 |
5.6 |
0.23 |
6.35 |
3.71 |
1.49 |
2.81 |
9.44 × 10-18 |
| 8071 |
0030998 |
6.55 |
1.67 |
5.11 |
0.63 |
5.84 |
1.67 |
1.44 |
2.71 |
2.30 × 10-16 |
| 3197 |
0015082 |
7.6 |
6.31 |
6.27 |
0.98 |
6.94 |
4.08 |
1.33 |
2.51 |
6.33 × 10-13 |
| 6799-5p |
0027498 |
8.09 |
3.03 |
6.76 |
0.79 |
7.43 |
2.35 |
1.33 |
2.51 |
7.08 × 10-11 |
| 6741-5p |
0027383 |
9.07 |
1.92 |
7.8 |
0.85 |
8.45 |
1.78 |
1.27 |
2.41 |
7.32 × 10-10 |
| 6875-5p |
0027650 |
5.18 |
3.14 |
3.91 |
0.54 |
4.55 |
2.24 |
1.27 |
2.41 |
3.17 × 10-11 |
| 8078 |
0031005 |
7.19 |
5.29 |
5.93 |
0.95 |
6.57 |
3.52 |
1.26 |
2.39 |
1.45 × 10-12 |
| 1292-3p |
0022948 |
8.24 |
2.34 |
7.03 |
1.91 |
7.64 |
2.47 |
1.21 |
2.31 |
2.11 × 10-4 |
| 6780b-5p |
0027572 |
7.19 |
5.08 |
6.04 |
0.32 |
6.62 |
3.03 |
1.15 |
2.22 |
1.74 × 10-12 |
| 3178 |
0015055 |
10.26 |
5.45 |
9.11 |
0.3 |
9.69 |
3.21 |
1.15 |
2.22 |
2.45 × 10-13 |
| 6126 |
0024599 |
10.72 |
3.44 |
9.6 |
0.25 |
10.16 |
2.16 |
1.12 |
2.17 |
6.15 × 10-12 |
| 6877-5p |
0027654 |
5.68 |
1.34 |
4.56 |
0.52 |
5.13 |
1.24 |
1.12 |
2.17 |
6.64 × 10-9 |
| 4449 |
0018968 |
6.16 |
2.09 |
5.06 |
0.52 |
5.62 |
1.6 |
1.1 |
2.14 |
6.36 × 10-9 |
| 4281 |
0016907 |
12.32 |
2.52 |
11.22 |
1.22 |
11.77 |
2.16 |
1.1 |
2.14 |
1.90 × 10-4 |
| 4478 |
0019006 |
6.09 |
1.45 |
5.06 |
0.69 |
5.58 |
1.34 |
1.03 |
2.04 |
1.84 × 10-6 |
| 1914-5p |
0007889 |
4.36 |
1.41 |
3.33 |
0.53 |
3.85 |
1.24 |
1.03 |
2.04 |
1.96 × 10-7 |
| 4516 |
0019053 |
13.87 |
1.71 |
12.87 |
1.54 |
13.37 |
1.86 |
1 |
2.00 |
1.93 × 10-3 |
| 3142 |
0015011 |
7.22 |
1.62 |
6.22 |
0.31 |
6.73 |
1.21 |
1 |
2.00 |
2.82 × 10-9 |
| 663a |
0003326 |
9.84 |
2.52 |
8.85 |
0.54 |
9.35 |
1.77 |
0.99 |
1.99 |
1.30 × 10-7 |
| 663b |
0005867 |
5.95 |
2.87 |
4.97 |
0.76 |
5.46 |
2.05 |
0.98 |
1.97 |
2.13 × 10-6 |
| 8069 |
0030996 |
6.1 |
1.29 |
5.14 |
0.45 |
5.62 |
1.1 |
0.96 |
1.95 |
3.73 × 10-8 |
| 4505 |
0019041 |
6.68 |
0.87 |
5.73 |
0.79 |
6.21 |
1.05 |
0.95 |
1.93 |
9.84 × 10-5 |
| 6729-5p |
0027359 |
6.96 |
2.43 |
6.02 |
0.3 |
6.5 |
1.59 |
0.94 |
1.92 |
2.36 × 10-10 |
| 3940-5p |
0019229 |
6.09 |
2.49 |
5.16 |
0.41 |
5.63 |
1.67 |
0.93 |
1.91 |
2.84 × 10-8 |
| 6798-5p |
0027496 |
6.07 |
1.15 |
5.15 |
0.58 |
5.62 |
1.07 |
0.92 |
1.89 |
2.37 × 10-5 |
| 3652 |
0018072 |
5.13 |
1.88 |
4.24 |
0.29 |
4.69 |
1.28 |
0.89 |
1.85 |
1.13 × 10-8 |
| 6829-5p |
0027558 |
4.85 |
0.75 |
3.97 |
0.5 |
4.41 |
0.82 |
0.88 |
1.84 |
3.60 × 10-6 |
| 6786-5p |
0027472 |
9.05 |
2.86 |
8.18 |
0.26 |
8.62 |
1.75 |
0.87 |
1.83 |
4.08 × 10-9 |
| 4651 |
0019715 |
7.41 |
1.05 |
6.56 |
0.74 |
6.99 |
1.08 |
0.85 |
1.80 |
9.75 × 10-4 |
| 6124 |
0024597 |
6.31 |
1.05 |
5.46 |
0.45 |
5.89 |
0.92 |
0.85 |
1.80 |
7.96 × 10-6 |
| 4448 |
0018967 |
12.1 |
1.03 |
11.26 |
0.83 |
11.69 |
1.1 |
0.84 |
1.79 |
1.17 × 10-2 |
| 3928-3p |
0018205 |
4.08 |
2.18 |
3.25 |
1.08 |
3.67 |
1.79 |
0.83 |
1.78 |
1.51 × 10-2 |
| 149-3p |
0004609 |
8.91 |
1.41 |
8.08 |
0.35 |
8.5 |
1.05 |
0.83 |
1.78 |
1.85 × 10-7 |
| 638 |
0003308 |
8.77 |
2.42 |
7.94 |
0.44 |
8.36 |
1.6 |
0.83 |
1.78 |
1.10 × 10-8 |
| 4787-5p |
0019956 |
10.49 |
1.33 |
9.73 |
0.62 |
10.11 |
1.11 |
0.76 |
1.69 |
3.14 × 10-5 |
| 3656 |
0018076 |
11.38 |
2.84 |
10.64 |
0.75 |
11.02 |
1.93 |
0.74 |
1.67 |
1.58 × 10-5 |
| 3648 |
0018068 |
4.03 |
2.16 |
3.3 |
0.62 |
3.67 |
1.52 |
0.73 |
1.66 |
1.97 × 10-6 |
| 127-3p |
0000446 |
6.41 |
0.52 |
5.68 |
0.57 |
6.05 |
0.67 |
0.73 |
1.66 |
1.30 × 10-3 |
| 4443 |
0018961 |
9.61 |
1.46 |
8.9 |
0.35 |
9.26 |
1.03 |
0.71 |
1.64 |
6.48 × 10-6 |
| 6088 |
0023713 |
7.44 |
0.67 |
6.76 |
0.54 |
7.1 |
0.71 |
0.68 |
1.60 |
1.89 × 10-3 |
| 4488 |
0019022 |
13.06 |
1.97 |
12.39 |
0.86 |
12.73 |
1.52 |
0.67 |
1.59 |
1.91 × 10-3 |
| 4463 |
0018987 |
7.42 |
0.85 |
6.75 |
0.3 |
7.09 |
0.69 |
0.67 |
1.59 |
2.82 × 10-5 |
| 6724-5p |
0025856 |
5.96 |
1.5 |
5.3 |
0.25 |
5.63 |
0.98 |
0.66 |
1.58 |
6.06 × 10-6 |
| 616-5p |
0003284 |
6.35 |
0.7 |
5.69 |
0.48 |
6.02 |
0.69 |
0.66 |
1.58 |
1.26 × 10-3 |
| 4419b |
0019034 |
4.64 |
0.64 |
3.99 |
0.53 |
4.32 |
0.69 |
0.65 |
1.57 |
1.70 × 10-2 |
| 4534 |
0019073 |
5.03 |
0.49 |
4.39 |
0.49 |
4.71 |
0.59 |
0.64 |
1.56 |
1.26 × 10-3 |
| 6086 |
0023711 |
5.37 |
0.39 |
4.74 |
0.28 |
5.06 |
0.43 |
0.63 |
1.55 |
4.62 × 10-5 |
| 4442 |
0018960 |
4.8 |
0.75 |
4.18 |
0.6 |
4.49 |
0.76 |
0.62 |
1.54 |
4.67 × 10-2 |
| 2861 |
0013802 |
10.22 |
1.76 |
9.63 |
0.31 |
9.93 |
1.12 |
0.59 |
1.51 |
3.12 × 10-5 |
| 7845-5p |
0030420 |
4.46 |
1.26 |
3.88 |
0.48 |
4.17 |
0.95 |
0.58 |
1.49 |
3.72 × 10-3 |
| 1469 |
0007347 |
4.45 |
1.96 |
3.87 |
0.43 |
4.16 |
1.27 |
0.58 |
1.49 |
1.06 × 10-5 |
| 1207-5p |
0005871 |
6.51 |
0.59 |
5.93 |
0.38 |
6.22 |
0.57 |
0.58 |
1.49 |
1.70 × 10-3 |
| 6085 |
0023710 |
6.08 |
0.46 |
5.5 |
0.37 |
5.79 |
0.5 |
0.58 |
1.49 |
2.53 × 10-3 |
| 6851-5p |
0027602 |
4.29 |
0.74 |
3.71 |
0.35 |
4 |
0.62 |
0.58 |
1.49 |
1.40 × 10-3 |
| 6727-5p |
0027355 |
6.7 |
1.89 |
6.13 |
0.68 |
6.42 |
1.36 |
0.57 |
1.48 |
2.31 × 10-3 |
| 6894-5p |
0027688 |
5.59 |
0.48 |
5.02 |
0.43 |
5.31 |
0.53 |
0.57 |
1.48 |
1.67 × 10-2 |
| 762 |
0010313 |
9.03 |
1.53 |
8.47 |
0.13 |
8.75 |
0.91 |
0.56 |
1.47 |
1.06 × 10-6 |
| 4481 |
0019015 |
4.5 |
0.61 |
4 |
0.31 |
4.25 |
0.52 |
0.5 |
1.41 |
2.58 × 10-2 |
| 5787 |
0023252 |
10.29 |
1.76 |
9.8 |
0.54 |
10.05 |
1.2 |
0.49 |
1.40 |
2.80 × 10-2 |
| 4763-3p |
0019913 |
6.32 |
0.45 |
5.84 |
0.36 |
6.08 |
0.46 |
0.48 |
1.39 |
5.47 × 10-3 |
| 4674 |
0019756 |
4.66 |
2.61 |
4.19 |
0.65 |
4.43 |
1.68 |
0.47 |
1.39 |
2.23 × 10-4 |
| 6764-3p |
0027429 |
6.02 |
0.41 |
5.55 |
0.27 |
5.79 |
0.39 |
0.47 |
1.39 |
1.05 × 10-2 |
| 127-5p |
0004604 |
7.59 |
0.72 |
7.12 |
0.22 |
7.36 |
0.52 |
0.47 |
1.39 |
8.90 × 10-3 |
| 4497 |
0019032 |
14.53 |
0.84 |
14.06 |
0.39 |
14.3 |
0.67 |
0.47 |
1.39 |
1.52 × 10-2 |
| 566 |
0003230 |
5.93 |
0.42 |
5.47 |
0.36 |
5.7 |
0.44 |
0.46 |
1.38 |
2.23 × 10-2 |
| 6781-5p |
0027462 |
4.4 |
1.29 |
3.95 |
0.43 |
4.18 |
0.91 |
0.45 |
1.37 |
1.12 × 10-2 |
| 1268b |
0018925 |
8.86 |
0.49 |
8.41 |
0.4 |
8.64 |
0.49 |
0.45 |
1.37 |
1.44 × 10-2 |
| 6791-5p |
0027482 |
4.83 |
0.32 |
4.39 |
0.22 |
4.61 |
0.32 |
0.44 |
1.36 |
1.75 × 10-2 |
| 6090 |
0023715 |
10.55 |
1.77 |
10.12 |
0.32 |
10.34 |
1.09 |
0.43 |
1.35 |
9.37 × 10-5 |
| 6756-5p |
0027412 |
5.87 |
0.55 |
5.44 |
0.16 |
5.66 |
0.4 |
0.43 |
1.35 |
1.42 × 10-3 |
| 4690-5p |
0019779 |
4.45 |
0.55 |
4.03 |
0.22 |
4.24 |
0.42 |
0.42 |
1.34 |
1.12 × 10-2 |
| 1227-5p |
0022941 |
9.57 |
0.53 |
9.16 |
0.23 |
9.37 |
0.42 |
0.41 |
1.33 |
6.75 × 10-4 |
| 4758-5p |
0019903 |
4.4 |
0.39 |
4 |
0.29 |
4.2 |
0.38 |
0.4 |
1.32 |
5.54 × 10-3 |
| 6821-5p |
0027542 |
5.52 |
0.8 |
5.13 |
0.32 |
5.32 |
0.6 |
0.39 |
1.31 |
1.72 × 10-2 |
| 4466 |
0018993 |
11.64 |
1.27 |
11.26 |
0.26 |
11.45 |
0.8 |
0.38 |
1.30 |
4.46 × 10-4 |
| 6803-5p |
0027506 |
7.48 |
2.1 |
7.1 |
0.25 |
7.29 |
1.21 |
0.38 |
1.30 |
2.12 × 10-5 |
| 7109-3p |
0028116 |
5.84 |
0.48 |
5.48 |
0.13 |
5.66 |
0.34 |
0.36 |
1.28 |
1.61 × 10-2 |
| 708-5p |
0004926 |
4.79 |
0.25 |
5.16 |
0.31 |
4.97 |
0.31 |
-0.37 |
-1.29 |
9.31 × 10-3 |
| 590-5p |
0003258 |
7.05 |
0.47 |
7.46 |
0.35 |
7.26 |
0.45 |
-0.41 |
-1.33 |
1.76 × 10-2 |
| 1537-3p |
0007399 |
4.78 |
0.45 |
5.21 |
0.31 |
4.99 |
0.42 |
-0.43 |
-1.35 |
1.94 × 10-2 |
| 15a-5p |
0000068 |
13.13 |
0.63 |
13.65 |
0.43 |
13.39 |
0.59 |
-0.52 |
-1.43 |
7.45 × 10-3 |
| 424-3p |
0004749 |
3.99 |
0.43 |
4.54 |
0.35 |
4.26 |
0.46 |
-0.55 |
-1.46 |
9.19 × 10-4 |
| 33a-3p |
0004506 |
3.93 |
0.66 |
4.5 |
0.57 |
4.21 |
0.69 |
-0.57 |
-1.48 |
7.56 × 10-3 |
| 33b-5p |
0003301 |
5.2 |
0.76 |
5.78 |
0.85 |
5.49 |
0.88 |
-0.58 |
-1.49 |
2.29 × 10-2 |
| 33a-5p |
0000091 |
5.86 |
0.79 |
6.46 |
0.48 |
6.16 |
0.72 |
-0.6 |
-1.52 |
3.83 × 10-2 |
| 4454 |
0018976 |
11.37 |
1.14 |
12.44 |
0.71 |
11.9 |
1.21 |
-1.07 |
-2.10 |
1.08 × 10-2 |
| 20a-3p |
0004493 |
4.28 |
2.12 |
5.42 |
0.56 |
4.85 |
1.66 |
-1.14 |
-2.20 |
6.73 × 10-3 |
| 7975 | 0031178 | 8.63 | 4.19 | 10.46 | 1.65 | 9.53 | 3.76 | -1.83 | -3.56 | 1.88 × 10-2 |
FC2: Fold change; LFC1: Log2-fold change; MIMAT: Mature miRNA miRBase accession.
Sex-associated miRNAs & their distribution by chromosomes & participation in clusters
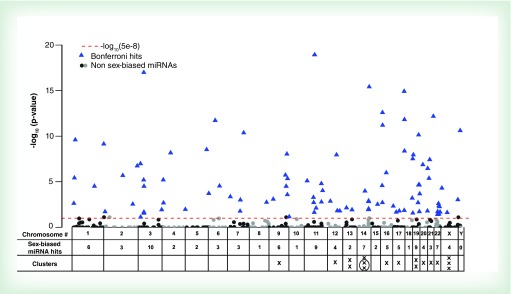
The majority of the miRNA hits were found on autosomes (96%), with only a small fraction (4%) identified on the X chromosome and none on the Y chromosome (Supplementary Table 1 & Figure 4). This does not differ significantly from the distribution of all-mature miRNAs located in autosomes and sex chromosomes (∼7%) according to miRBase v20. Similarly, many of the chromosomes with the most sex-associated hits were those that also have the most miRNAs in them (Chr 1, 9, 11, 14 and 19), representing 39% of the hits. Two chromosomes, Chr 3 and 22, were significantly enriched for miRNAs differentially expressed in boys and girls. For instance, we found that 10.5% of the sex-biased hits were in Chr 3, whereas only 4.8% of all-mature miRNAs (miRBase v20) are located in Chr 3 (Chi-square p = 0.008). While about 2.6% of all-mature miRNAs are located in Chr 22, we observed 7.4% of sex-associated hits in this chromosome (Chi-square p = 0.003).
Figure 4. . Manhattan plot of -log10 Bonferroni-adjusted p-values for the group of miRNAs filtered by their expression in different chromosomes.
The dashed red line indicates the Bonferroni cut-off set at 8 × 10-5 that was calculated by dividing an alpha = 0.05 by 564 tests. Blue triangles represent miRNAs that were differentially expressed between boys and girls after Bonferroni correction.
To evaluate the participation of the miRNAs differentially expressed by sex in our newborns in miRNA-clusters, we used the miRBase database, with the cluster criteria that included miRNA genes located within 10 Kb of distance on the same chromosome and sometimes functionally connected across different chromosomes [70,71]. Among 94 sex-associated miRNAs, 17 belong to 16 well-known miRBase clusters distributed across 11 different chromosomes (chr 9, 12, 13, 14, 16, 17, 19, 20, 21, 22; Supplementary Table 1 & Figure 4). Among those 11 chromosomes, the number of miRNA clusters linked to sex-biased miRNAs ranged from 1 to 3. On chromosome 14 and X, three sex-biased miRNAs belonged to miRBase miRNA clusters. However, at closer examination, two of the clusters on Chr14, per miRBase, in fact belong to the same large cluster that was previously described as C14MC [72,73]. There is currently no uniform nomenclature for miRNA clusters, for example, C14MC has also been referred to the miR-379/miR656 cluster [74], the Mirg cluster [75] or the miR 379/miR-410 cluster [76].
Pathway analysis of differentially expressed miRNAs
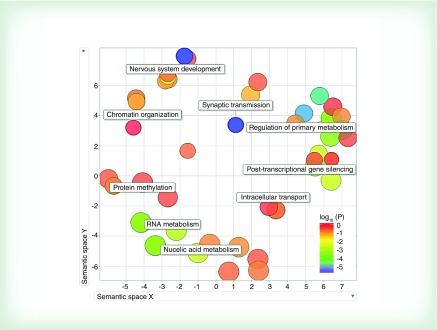
A total of 571 predicted mRNA targets was identified by in silico analysis for the 94 miRNAs differentially expressed by sex. According to Tarbase (v 6.0), only 2 of the 94 hits had experimentally validated targets (hsa-miR-15a-5p and hsa-miR-33a-5p had 188 and 98 mRNA targets, respectively). Examining the union of predicted and validated miRNA target genes (n = 771) for enrichment of particular GO terms, we identified 100 pathways that were significantly enriched after controlling for FDR (FDR p < 0.05; Supplementary Table 2). The miRNA-related GO terms showed that the most prominent sex associations in newborns fell into nine broad categories including: nervous system development, synaptic transmission, regulation of primary metabolism, post-transcriptional gene silencing, intracellular transport, nucleic acid metabolism, RNA metabolism, protein methylation, and chromatin organization (Figure 5).
Figure 5. . Visualization of enriched gene ontology terms.
Gene ontology terms significantly enriched (q ≤ 0.05) in miRNAs differentially expressed by sex.
Validation of differentially expressed miRNAs by nCounter miRNA expression assay
To validate our findings from the EdgeSeq miRNA Whole Transcriptome Assay, we selected 30 miRNAs that were either unchanged, upregulated or downregulated between boys and girls to run in the nCounter miRNA expression array (Supplementary Table 3). The unchanged miRNAs (determined by EdgeSeq) remained unchanged between boys and girls when assayed by nCounter. Of the miRNAs selected for validation that were upregulated in the EdgeSeq assay, 12 of the 14 (86%) also had positive log2-fold change values by nCounter. Of those, three miRNAs were significant (FDR-adjusted p < 0.05; hsa-miR-1469, hsa-miR-4488, hsa-miR-663a) and several (n = 5) were borderline significant (FDR-adjusted p < 0.10). The set of downregulated miRNAs did not validate in the nCounter assay, however, it is important to note that the downregulated miRNAs from the EdgeSeq assay were not as strongly associated with sex (larger p-values) compared with the upregulated miRNAs and that the majority of hits were upregulated.
Discussion
In the current study, we assessed miRNAome in cord blood buffy coat samples of 89 newborns, and identified 94 miRNAs differentially expressed between boys and girls. For most of these miRNAs, expression levels were higher in boys than in girls. We were able to validate some, but not all of the upregulated miRNAs using another assay platform (nCounter). Sex-associated hits were distributed unevenly among chromosomes and were linked with 16 miRNA clusters according to the miRBase registry. Among 100 identified pathways for predicted in silico gene targets, categories for nervous system development, RNA metabolic processes and post-transcriptional gene silencing were significantly enriched for sex-biased miRNAs. To our knowledge, there are no previous publications on sex-related differences in the miRNAome of newborn children.
Although some methods and pipelines for preprocessing of miRNA data have been published [52,77–78], there is no clear consensus on which filtering and normalization steps are the most effective for analysis of miRNAome datasets. To achieve detailed characterization of the miRNAome by next-generation sequencing, we first explored three different data filtering approaches to minimize technical variability. Using diagnostic criteria of specificity and sensitivity to select the most appropriate filtering methods, we found that the highest levels of sensitivity and specificity were best achieved when we applied a filter using the top 25% expression or variance threshold. Following filtering steps, the most commonly used normalization technique for miRNA sequencing data in other studies is quantile normalization [53,57]. We tested quantile normalization in addition to four other common normalization techniques and found that full quantile normalization followed by RUV normalization produced the most optimal results. RUV has been successfully applied in the analysis of large datasets derived from RNA-seq [79] and metabolomic [80] experiments [54]. Our filtering and normalization pipeline resulted in minimization of technical variability, and enabled us to identify biological variability of miRNA expression in boys and girls. This newly developed pipeline may be useful for analysis of miRNA sequencing data generated in future population studies.
In our study, the majority of miRNA hits differentially expressed between boys and girls (96%) were in autosomes. This preliminary finding points to possible regional differences in miRNA regulation and impact. Based on chromosomal distribution of the sex-biased miRNAs, we detected that 54% of our hits were located on seven chromosomes. Those with the highest number of locations ranging between six and ten included chromosomes of three main size groups of human karyotype including large Chr3 and X, medium-sized Chr 9, 11 and 14, and the small Chr 19 and 22. Some other chromosomes have fewer hits (1–5). In addition, we identified two miRNAs that belong to C14MC cluster (hsa-miR-127-3p and hsa-127-5p) that may act in concert since they also belong to a common precursor (mature miRNA start 101349372 and 101349338, respectively). Our data show similar expression levels for these two miRNAs. In our study, we did not find a significant enrichment of differentially expressed miRNAs in sex chromosomes. There are at least 113 miRNAs, representing approximately 7% of total human miRNAs, located on the X chromosome, and only two miRNAs on the Y chromosome [81]. In our study, miR-424-3p was differentially expressed in the initial EdgeSeq experiment. This miRNA and several other X chromosome-linked miRNAs (e.g., miR-221/222 cluster, miR-98, miR-106a, miR-424 and miR-18b) have been implicated in immune regulation [82]. It is possible that the number of differentially expressed miRNAs in boys and girls may change as they get older since previous studies have shown that sex hormones can affect regulation of miRNAs [83,84].
Previous miRNA studies [36,85], as well as our findings reported here, indicate the importance of accounting for sex differences when analyzing miRNA data. Similar observations have been reported for other epigenetic marks including DNA methylation and histone acetylation [28,86–87]. Recent publications have provided strong evidence that sex influences genome-wide DNA methylation in human biofluids including blood and saliva [88,89]. Furthermore, Cordero et al. identified 117 miRNAs present in buffy coat samples, which were differentially methylated between men and women [90], some of which overlap with the cord blood miRNAs identified in our study. It is possible that differences in oxidative stress, inflammation, growth and birth outcomes observed between newborn boys and girls [91–93] could be controlled, at least in part, by differentially expressed miRNAs. Age is another host factor that can bias epigenetic markers [94,95]. However, miRNAome has not yet been analyzed by age in infants or children, an important area for future research.
Additionally, the statistically significant miRNAs confirmed in the validation analysis (hsa-miR-4488, hsa-miR-663a, hsa-miR-1469) are biologically relevant. For example, miR-4488 expression levels in white blood cells were lower in Behcet patients who experience systemic inflammation and high levels of IL-6, demonstrating a potential role in inflammation [96]. Recent studies have demonstrated differences in IL-6 levels between newborn boys and girls, with lower levels reported in girls [91], consistent with our miRNA findings. Serum expression of miR-663a has been associated with autism spectrum disorder [97], which corroborates with our pathway analysis identifying many neurodevelopmental ontology terms that were strongly enriched among miRNA targets observed in CHAMACOS newborns. Neurodevelopmental ontology terms have also been identified in epigenetic studies of sex differences in animals [2]. Our data also corroborate some of the other sex-biased GO terms found in animal studies. For example, Murphy et al. reported synapsis-related ontologies in rat cortex tissue, and Shao et al. identified developmental processes, growth, cellular, RNA processes and metabolic processes as sex-biased ontology terms in a nonhuman model [35,98]. In addition, our findings are in agreement with a study in children that reported sex differences in brain development and maturation rates assessed by magnetic resonance imaging in older children (6–17 years of age) [99]. This suggests that our miRNA expression data may contribute to better understanding of sex differences in cognitive mechanisms in early life. miRNA expression analysis in animal models and human surrogate tissues such as blood may also add a new layer in the complex and still not well-understood epigenetic processes that take place in human brain neurodevelopment.
Although there was a substantial overlap between the sex-associated pathways reported in animals and our results in human cord blood, some of the sex-biased ontology terms may be newborn specific or tissue specific. Many human population studies of epigenetic marks use blood as a surrogate for other target tissues because it is more readily available and does not require invasive procedures like biopsies. Many well-characterized birth cohorts and other longitudinal studies with banked blood samples rely on them for analysis of DNA methylation and gene expression. Epigenetic changes in blood are relevant even if they do not capture all changes observed in organ-specific tissue [100]. An increasing number of studies have demonstrated that levels of circulating miRNAs released by cells into the bloodstream are correlated with tissue miRNA levels [101,102]. Another limitation with use of blood specimens is that cell-type heterogeneity can act as a source of bias if cell-type proportions are associated both with miRNA expression levels and sex. Other studies have demonstrated that miRNA expression can be affected by cell composition [103]. However, in our study, blood cell proportions were not different in cord blood samples of CHAMACOS boys and girls [104]. While use of cord blood is a potential limitation in our study, the growing consensus is that blood can serve as an appropriate and informative model for studies of epigenetic mechanisms in birth cohorts. The effects of host factors in blood need further investigation and this data may contribute to identifying sensitive windows [105] that can be used as opportunities for therapy [106].
Our study also focused solely on newborns, which may not be generalizable to other ages. Furthermore, although only three of the upregulated miRNAs were validated with high significance, additional miRNAs were also more highly expressed in boys than girls by nCounter consistent with EdgeSeq – that adds up to more than 78% of tested upregulated miRNAs. Additionally, all six miRNAs that were not different in boys and girls by EdgeSeq had the same result by nCounter. The discordance between different miRNA expression platforms is well known. In fact, the miRNA quality control study compared 12 different platforms and reported the average validation rate of any two platforms was 55% [107], indicating that miRNAs that were not significant may still be biologically relevant. Additional replication on miRNAome sex differences in a larger number of samples, various age groups, as well as other populations is warranted. Finally, the main focus of our analysis was limited to the top 25% highly expressed miRNAs that produced the most reliable results. With the increase in the number of publicly available and better characterized miRNA data from population and mechanistic studies, the role of sex on miRNA expression and clustering can be explored in more depth.
Conclusion
Our study described the differential expression of the miRNAome in newborns boys and girls, which provides novel resources for better understanding of epigenetic regulation in early life stages. Using pathway analysis, we found that neurodevelopment, RNA metabolism and metabolic ontology terms were enriched among the targets from sex-associated miRNA. To our knowledge, this is one of the first studies to characterize the miRNAome in children using next-generation sequencing with a focus on differences by sex.
Future perspective
Understanding of molecular mechanisms that can explain the basis for sex differences in susceptibility to diseases and environmental exposures is of critical importance, and miRNA expression has an excellent potential to produce critical insights. Given that very limited data so far are available for miRNAome in human tissues, more studies in different age groups and populations, in healthy subjects and patients with various health conditions are warranted. Additional interesting angle of future research will be on longitudinal studies of sex differences for epigenetic marks, and whether sex can modify the relationship of environmental exposures with these modifications.
Executive summary.
Among more than 2200 miRNAs characterized by next-generation sequencing in cord blood from 89 newborn children, we identified 94 miRNAs differentially expressed by sex. In our validation experiments, most upregulated miRNAs remained more highly expressed in boys but only a few were statistically significant by FDR.
A majority (96%) of these miRNAs were located on autosomes, and their expression was higher in boys than in girls.
The sex-associated miRNA gene targets were mainly involved in nervous system development, nucleic acid metabolism and transcription control.
Accounting for host factors like sex in miRNA expression and other epigenomic analyses is essential for human studies and may increase sensitivity of an assessment of their relation with health and environmental exposures.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge laboratory and field CHAMACOS staff and especially, the CHAMACOS participants for their contributions. The authors thank Dr Yousefi for his helpful comments on the bioinformatics analysis.
Footnotes
Disclaimer
The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of EPA or NIEHS.
Financial & competing interests disclosure
This work was supported by grants 1R01ES023067 and PO1 ES009605 from the National Institute of Environmental Health Science (NIEH) and RD83451301 from the US Environmental Protection Agency (EPA). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
- 1.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr. Res. 2007;61(5 Pt 2):R24–R29. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MM, Nugent BM. At the frontier of epigenetics of brain sex differences. Front. Behav. Neurosci. 2015;9:221. doi: 10.3389/fnbeh.2015.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maccani MA, Marsit CJ. Epigenetics in the placenta. Am. J. Reprod. Immunol. 2009;62(2):78–89. doi: 10.1111/j.1600-0897.2009.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Javierre BM, Hernando H, Ballestar E. Environmental triggers and epigenetic deregulation in autoimmune disease. Discov. Med. 2011;12(67):535–545. [PubMed] [Google Scholar]
- 5.Galea S, Uddin M, Koenen K. The urban environment and mental disorders: epigenetic links. Epigenetics. 2011;6(4):400–404. doi: 10.4161/epi.6.4.14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2011;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Al-Kafaji G, Al-Mahroos G, Alsayed NA, Hasan ZA, Nawaz S, Bakhiet M. Peripheral blood microRNA-15a is a potential biomarker for type 2 diabetes mellitus and pre-diabetes. Mol. Med. Rep. 2015;12(5):7485–7490. doi: 10.3892/mmr.2015.4416. [DOI] [PubMed] [Google Scholar]
- 9.Dhayat SA, Husing A, Senninger N, et al. Circulating microRNA-200 family as diagnostic marker in hepatocellular carcinoma. PLoS ONE. 2015;10(10):e0140066. doi: 10.1371/journal.pone.0140066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bottoni A, Calin GA. MicroRNAs as main players in the pathogenesis of chronic lymphocytic leukemia. Microrna. 2014;2(3):158–164. doi: 10.2174/2211536602666131126002337. [DOI] [PubMed] [Google Scholar]
- 11.Seifoleslami M, Khameneie MK, Mashayekhi F, et al. Identification of microRNAs (miR-203/miR-7) as potential markers for the early detection of lymph node metastases in patients with cervical cancer. Tumour Biol. 2015 doi: 10.1007/s13277-015-4265-6. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 12.Tomaszewski D. Biomarkers of brain damage and postoperative cognitive disorders in orthopedic patients: an update. Biomed. Res. Int. 2015;2015:402959. doi: 10.1155/2015/402959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang L, Xu Y, Jin X, et al. A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res. Treat. 2015;154(2):423–434. doi: 10.1007/s10549-015-3591-0. [DOI] [PubMed] [Google Scholar]
- 14.Taylor EL, Gant TW. Emerging fundamental roles for non-coding RNA species in toxicology. Toxicology. 2008;246(1):34–39. doi: 10.1016/j.tox.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Hou L, Barupal J, Zhang W, et al. Particulate air pollution exposure and expression of viral and human microRNAs in blood: the Beijing truck driver air pollution study. Environ. Health Perspect. 2016;124(3):344–350. doi: 10.1289/ehp.1408519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rager JE, Bailey KA, Smeester L, et al. Prenatal arsenic exposure and the epigenome: altered microRNAs associated with innate and adaptive immune signaling in newborn cord blood. Environ. Mol. Mutagen. 2014;55(3):196–208. doi: 10.1002/em.21842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, Wang R, Strulovici-Barel Y, et al. Persistence of smoking-induced dysregulation of miRNA expression in the small airway epithelium despite smoking cessation. PLoS ONE. 2015;10(4):e0120824. doi: 10.1371/journal.pone.0120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309(5740):1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 19.Hatziapostolou M, Polytarchou C, Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol. Metab. 2013;24(7):361–373. doi: 10.1016/j.tem.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Ziats MN, Rennert OM. Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry. 2014;19(7):848–852. doi: 10.1038/mp.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun E, Shi Y. MicroRNAs: small molecules with big roles in neurodevelopment and diseases. Exp. Neurol. 2015;268:46–53. doi: 10.1016/j.expneurol.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Mele M, Ferreira PG, Reverter F, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Coster S, van Leeuwen DM, Jennen DG, et al. Gender-specific transcriptomic response to environmental exposure in Flemish adults. Environ. Mol. Mutagen. 2013;54(7):574–588. doi: 10.1002/em.21774. [DOI] [PubMed] [Google Scholar]
- 24.Yang X, Schadt EE, Wang S, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mujahid S, Logvinenko T, Volpe MV, Nielsen HC. miRNA regulated pathways in late stage murine lung development. BMC Dev. Biol. 2013;13:13. doi: 10.1186/1471-213X-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai HW, Grant PA, Rissman EF. Sex differences in histone modifications in the neonatal mouse brain. Epigenetics. 2009;4(1):47–53. doi: 10.4161/epi.4.1.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Wang F, Liu Y, Yu Y, Gelernter J, Zhang H. Sex-biased methylome and transcriptome in human prefrontal cortex. Hum. Mol. Genet. 2014;23(5):1260–1270. doi: 10.1093/hmg/ddt516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yousefi P, Huen K, Dave V, Barcellos L, Eskenazi B, Holland N. Sex differences in DNA methylation assessed by 450 K BeadChip in newborns. BMC Genomics. 2015;16:911. doi: 10.1186/s12864-015-2034-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pak TR, Rao YS, Prins SA, Mott NN. An emerging role for microRNAs in sexually dimorphic neurobiological systems. Pflugers Arch. 2013;465(5):655–667. doi: 10.1007/s00424-013-1227-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morgan CP, Bale TL. Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol. Sex Differ. 2012;3(1):22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morgan CP, Bale TL. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J. Neurosci. 2011;31(33):11748–11755. doi: 10.1523/JNEUROSCI.1887-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koturbash I, Zemp F, Kolb B, Kovalchuk O. Sex-specific radiation-induced microRNAome responses in the hippocampus, cerebellum and frontal cortex in a mouse model. Mutat. Res. 2011;722(2):114–118. doi: 10.1016/j.mrgentox.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Raiche J, Rodriguez-Juarez R, Pogribny I, Kovalchuk O. Sex- and tissue-specific expression of maintenance and de novo DNA methyltransferases upon low dose X-irradiation in mice. Biochem. Biophys. Res. Commun. 2004;325(1):39–47. doi: 10.1016/j.bbrc.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Mani ST, Thakur MK. In the cerebral cortex of female and male mice, amyloid precursor protein (APP) promoter methylation is higher in females and differentially regulated by sex steroids. Brain Res. 2006;1067(1):43–47. doi: 10.1016/j.brainres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Murphy SJ, Lusardi TA, Phillips JI, Saugstad JA. Sex differences in microRNA expression during development in rat cortex. Neurochem. Int. 2014;77:24–32. doi: 10.1016/j.neuint.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ameling S, Kacprowski T, Chilukoti RK, et al. Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study. BMC Med. Genomics. 2015;8(1):61. doi: 10.1186/s12920-015-0136-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 38.Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating miRNA biomarker signatures. PLoS ONE. 2011;6(6):e20769. doi: 10.1371/journal.pone.0020769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mooney C, Raoof R, El-Naggar H, Sanz-Rodriguez A, Jimenez-Mateos EM, Henshall DC. High throughput qPCR expression profiling of circulating microRNAs reveals minimal sex- and sample timing-related variation in plasma of healthy volunteers. PLoS ONE. 2015;10(12):e0145316. doi: 10.1371/journal.pone.0145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eskenazi B, Gladstone EA, Berkowitz GS, et al. Methodologic and logistic issues in conducting longitudinal birth cohort studies: lessons learned from the Centers for Children's Environmental Health and Disease Prevention Research. Environ. Health Perspect. 2005;113(10):1419–1429. doi: 10.1289/ehp.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eskenazi B, Harley K, Bradman A, et al. Association of in utero organophosphate pesticide exposure and fetal growth and length of gestation in an agricultural population. Environ. Health Perspect. 2004;112(10):1116–1124. doi: 10.1289/ehp.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health Perspect. 2010;118(6):763–768. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donzelli S, Blandino G, Muti P. Use of buffy coat miRNA profiling for breast cancer prediction in healthy women. Methods Mol. Biol. 2016;1379:13–19. doi: 10.1007/978-1-4939-3191-0_2. [DOI] [PubMed] [Google Scholar]
- 44.Georgiadis P, Hebels DG, Valavanis I, et al. Omics for prediction of environmental health effects: blood leukocyte-based cross-omic profiling reliably predicts diseases associated with tobacco smoking. Sci. Rep. 2016;6:20544. doi: 10.1038/srep20544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebels DG, Georgiadis P, Keun HC, et al. Performance in omics analyses of blood samples in long-term storage: opportunities for the exploitation of existing biobanks in environmental health research. Environ. Health Perspect. 2013;121(4):480–487. doi: 10.1289/ehp.1205657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voellenkle C, van Rooij J, Cappuzzello C, et al. MicroRNA signatures in peripheral blood mononuclear cells of chronic heart failure patients. Physiol. Genomics. 2010;42(3):420–426. doi: 10.1152/physiolgenomics.00211.2009. [DOI] [PubMed] [Google Scholar]
- 47.Girard L, Rodriguez-Canales J, Behrens C, et al. An expression signature as an aid to the histologic classification of non-small cell lung cancer. Clin. Cancer Res. 2016;22(19):4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reinholz M, Liu Q, Schwartz M, Luecke J, Roberts C, LaFleur B. European Society of Human Genetics. Barcelona, Spain: 21–24 May 2016. Verification of miRNA expression using nuclease protection and targeted next-generation sequencing (NGS) Presented at. [Google Scholar]
- 49.Thompson D, Botros I, Harrison H, Modur V. Merck Technology Symposium. Long Branch, NJ: 6 May 2014. Automated high fidelity miRNA expression profiling using nuclease protection coupled with next generation sequencing. Presented at. [Google Scholar]
- 50.HTG Molecular. HTG EdgeSeq miRNA whole transcriptome assay (2016) www.htgmolecular.com/sites/default/files/HTG%20EdgeSeq%20miRNA%20WTA%20sales%20sheet.pdf
- 51.Garmire LX, Subramaniam S. Evaluation of normalization methods in mammalian microRNA-Seq data. RNA. 2012;18(6):1279–1288. doi: 10.1261/rna.030916.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tam S, Tsao MS, McPherson JD. Optimization of miRNA-seq data preprocessing. Brief. Bioinform. 2015;16(6):950–963. doi: 10.1093/bib/bbv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jacob L, Gagnon-Bartsch JA, Speed TP. Correcting gene expression data when neither the unwanted variation nor the factor of interest are observed. Biostatistics. 2016;17(1):16–28. doi: 10.1093/biostatistics/kxv026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.miRBase. www.mirbase.org/
- 57.Turner SD. qqman: an R package for visualizing GWAS results using Q-Q and Manhattan plots. biorXiv. 2014 http://dx.doi.org/10.1101/005165 [Google Scholar]
- 58.Coronnello C, Benos PV. ComiR: combinatorial microRNA target prediction tool. Nucleic Acids Res. 2013;41:W159–W164. doi: 10.1093/nar/gkt379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vergoulis T, Vlachos IS, Alexiou P, et al. TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support. Nucleic Acids Res. 2012;40:D222–D229. doi: 10.1093/nar/gkr1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.ComiR: Combinatorial miRNA target prediction tool. www.benoslab.pitt.edu/comir/ [DOI] [PMC free article] [PubMed]
- 61.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4 doi: 10.7554/eLife.05005. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39(10):1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 64.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11(8):R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4(8):1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.miRTarBase. http://mirtarbase.mbc.nctu.edu.tw/
- 67.ConsensusPathDB. http://consensuspathdb.org/
- 68.Kamburov A, Stelzl U, Lehrach H, Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE. 2011;6(7):e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo L, Lu Z. Global expression analysis of miRNA gene cluster and family based on isomiRs from deep sequencing data. Comput. Biol. Chem. 2010;34(3):165–171. doi: 10.1016/j.compbiolchem.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 71.Guo L, Yang S, Zhao Y, Zhang H, Wu Q, Chen F. Global analysis of miRNA gene clusters and gene families reveals dynamic and coordinated expression. Biomed. Res. Int. 2014;2014:782490. doi: 10.1155/2014/782490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaille J. A large imprinted microRNA gene cluster at the mouse Dlk1–Gtl2 domain. Genome Res. 2004;14(9):1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morales-Prieto DM, Chaiwangyen W, Ospina-Prieto S, et al. MicroRNA expression profiles of trophoblastic cells. Placenta. 2012;33(9):725–734. doi: 10.1016/j.placenta.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Glazov EA, McWilliam S, Barris WC, Dalrymple BP. Origin, evolution, and biological role of miRNA cluster in DLK–DIO3 genomic region in placental mammals. Mol. Biol. Evol. 2008;25(5):939–948. doi: 10.1093/molbev/msn045. [DOI] [PubMed] [Google Scholar]
- 75.Bortolin-Cavaille ML, Dance M, Weber M, Cavaille J. C19MC microRNAs are processed from introns of large Pol-II, non-protein-coding transcripts. Nucleic Acids Res. 2009;37(10):3464–3473. doi: 10.1093/nar/gkp205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Noguer-Dance M, Abu-Amero S, Al-Khtib M, et al. The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum. Mol. Genet. 2010;19(18):3566–3582. doi: 10.1093/hmg/ddq272. [DOI] [PubMed] [Google Scholar]
- 77.Sarver AL. Toward understanding the informatics and statistical aspects of micro-RNA profiling. J. Cardiovasc. Transl. Res. 2010;3(3):204–211. doi: 10.1007/s12265-010-9180-z. [DOI] [PubMed] [Google Scholar]
- 78.Meyer SU, Pfaffl MW, Ulbrich SE. Normalization strategies for microRNA profiling experiments: a ‘normal’ way to a hidden layer of complexity? Biotechnol. Lett. 2010;32(12):1777–1788. doi: 10.1007/s10529-010-0380-z. [DOI] [PubMed] [Google Scholar]
- 79.Bernard E, Jacob L, Mairal J, Vert JP. Efficient RNA isoform identification and quantification from RNA-Seq data with network flows. Bioinformatics. 2014;30(17):2447–2455. doi: 10.1093/bioinformatics/btu317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Livera AM, Sysi-Aho M, Jacob L, et al. Statistical methods for handling unwanted variation in metabolomics data. Anal. Chem. 2015;87(7):3606–3615. doi: 10.1021/ac502439y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pinheiro I, Dejager L, Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33(11):791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- 82.Dai R, Ahmed SA. Sexual dimorphism of miRNA expression: a new perspective in understanding the sex bias of autoimmune diseases. Ther. Clin. Risk. Manag. 2014;10:151–163. doi: 10.2147/TCRM.S33517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morgan CP, Bale TL. Sex differences in microRNA regulation of gene expression: no smoke, just miRs. Biol. Sex Differ. 2012;3(1):22. doi: 10.1186/2042-6410-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang Z, Wang L. Regulation of microRNA expression and function by nuclear receptor signaling. Cell Biosci. 2011;1(1):31. doi: 10.1186/2045-3701-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sharma S, Eghbali M. Influence of sex differences on microRNA gene regulation in disease. Biol. Sex Differ. 2014;5(1):3. doi: 10.1186/2042-6410-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Huen K, Yousefi P, Bradman A, et al. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ. Mol. Mutagen. 2014;55(3):209–222. doi: 10.1002/em.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCarthy MM, Auger AP, Bale TL, et al. The epigenetics of sex differences in the brain. J. Neurosci. 2009;29(41):12815–12823. doi: 10.1523/JNEUROSCI.3331-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.El-Maarri O, Becker T, Junen J, et al. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122(5):505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- 89.Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PLoS ONE. 2010;5(4):e10028. doi: 10.1371/journal.pone.0010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cordero F, Ferrero G, Polidoro S, et al. Differentially methylated microRNAs in prediagnostic samples of subjects who developed breast cancer in the European Prospective Investigation into Nutrition and Cancer (EPIC-Italy) cohort. Carcinogenesis. 2015;36(10):1144–1153. doi: 10.1093/carcin/bgv102. [DOI] [PubMed] [Google Scholar]
- 91.Diaz-Castro J, Pulido-Moran M, Moreno-Fernandez J, et al. Gender specific differences in oxidative stress and inflammatory signaling in healthy term neonates and their mothers. Pediatr. Res. 2016;80(4):595–601. doi: 10.1038/pr.2016.112. [DOI] [PubMed] [Google Scholar]
- 92.Nagy E, Loveland KA, Orvos H, Molnar P. Gender-related physiologic differences in human neonates and the greater vulnerability of males to developmental brain disorders. J. Gend. Specif. Med. 2001;4(1):41–49. [PubMed] [Google Scholar]
- 93.Verburg PE, Tucker G, Scheil W, Erwich JJ, Dekker GA, Roberts CT. Sexual dimorphism in adverse pregnancy outcomes – a retrospective Australian Population Study 1981–2011. PLoS ONE. 2016;11(7):e0158807. doi: 10.1371/journal.pone.0158807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Almen MS, Nilsson EK, Jacobsson JA, et al. Genome-wide analysis reveals DNA methylation markers that vary with both age and obesity. Gene. 2014;548(1):61–67. doi: 10.1016/j.gene.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 95.Simons RL, Lei MK, Beach SR, et al. Economic hardship and biological weathering: the epigenetics of aging in a U.S. sample of black women. Soc. Sci. Med. 2016;150:192–200. doi: 10.1016/j.socscimed.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Woo MY, Yun SJ, Cho O, Kim K, Lee ES, Park S. MicroRNAs differentially expressed in Behcet disease are involved in interleukin-6 production. J. Inflamm. (Lond.) 2016;13:22. doi: 10.1186/s12950-016-0130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mundalil Vasu M, Anitha A, Thanseem I, et al. Serum microRNA profiles in children with autism. Mol. Autism. 2014;5:40. doi: 10.1186/2040-2392-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shao CC, Xu MJ, Chen YZ, Tao JP, Zhu XQ. Comparative profiling of microRNAs in male and female Rhipicephalus sanguineus . Appl. Biochem. Biotechnol. 2015;176(7):1928–1936. doi: 10.1007/s12010-015-1688-x. [DOI] [PubMed] [Google Scholar]
- 99.De Bellis MD, Keshavan MS, Beers SR, et al. Sex differences in brain maturation during childhood and adolescence. Cereb. Cortex. 2001;11(6):552–557. doi: 10.1093/cercor/11.6.552. [DOI] [PubMed] [Google Scholar]
- 100.Houseman EA, Kim S, Kelsey KT, Wiencke JK. DNA methylation in whole blood: uses and challenges. Curr. Environ. Health Rep. 2015;2(2):145–154. doi: 10.1007/s40572-015-0050-3. [DOI] [PubMed] [Google Scholar]
- 101.Grasso M, Piscopo P, Confaloni A, Denti MA. Circulating miRNAs as biomarkers for neurodegenerative disorders. Molecules. 2014;19(5):6891–6910. doi: 10.3390/molecules19056891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med. Res. Rev. 2012;32(2):326–348. doi: 10.1002/med.20215. [DOI] [PubMed] [Google Scholar]
- 103.Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012;5(3):492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yousefi P, Huen K, Quach H, et al. Estimation of blood cellular heterogeneity in newborns and children for epigenome-wide association studies. Environ. Mol. Mutagen. 2015;56(9):751–758. doi: 10.1002/em.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chiu YH, Hsu HH, Coull BA, et al. Prenatal particulate air pollution and neurodevelopment in urban children: examining sensitive windows and sex-specific associations. Environ. Int. 2016;87:56–65. doi: 10.1016/j.envint.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bardin J. Neurodevelopment: unlocking the brain. Nature. 2012;487(7405):24–26. doi: 10.1038/487024a. [DOI] [PubMed] [Google Scholar]
- 107.Mestdagh P, Hartmann N, Baeriswyl L, et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods. 2014;11(8):809–815. doi: 10.1038/nmeth.3014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.