Abstract
RNA interference was used to screen 3,314 Drosophila double-stranded RNAs, corresponding to ≈25% of Drosophila genes, for genes that affect the development of the embryonic nervous system. RNA-interference-mediated gene silencing in Drosophila embryos resulted in loss-of-function mutant phenotypes for 43 genes, which is 1.3% of the genes that were screened. We found 18 genes that were not known previously to affect the development of the nervous system. The functions of some of the genes are unknown. Other genes encode protein kinases, transcription factors, and signaling proteins, as well as proteins with other functions.
Keywords: embryonic nervous system, neural mutants, neural development
Injection of double-stranded RNA (dsRNA) in Caenorhabditis elegans or early Drosophila embryos results in destruction of the complementary species of mRNA by a process termed RNA interference (RNAi) (1, 2). An endonuclease, termed Dicer, catalyzes the cleavage of dsRNA into short segments of duplex RNA, each segment with 19 base pairs and two unpaired nucleotides at each end. The short dsRNA fragments bind to a multiprotein complex termed RISC, the RNA strands are unwound, and the antisense strand of RNA hybridizes to a complementary sequence in mRNA, thereby targeting that molecule of mRNA for destruction (3-5). Therefore, RNAi is a powerful tool that can be used to silence specific genes. In vertebrates, RNAi also is mediated by short dsRNA fragments of 12 nt in length; however, exposure of cells to long-chain dsRNA results in cell death (6, 7).
RNAi has been used successfully to screen large numbers of genes in C. elegans (8), Drosophila embryos (9), and Drosophila cell cultures (10, 11) to identify genes that are involved in biological phenomena, such as the cell cycle (8), cell growth and survival (10), cytoskeletal rearrangements (11), embryonic heart development (9), and the hedgehog (12) and wingless (13) signaling pathways. In this article, we describe the screening of a library of dsRNAs corresponding to ≈25% of the Drosophila genome for their effects on the development of the embryonic nervous system.
Materials and Methods
dsRNA Synthesis. We synthesized 5,849 cDNAs from the Drosophila embryo, head, larvae, and ovary (cDNA Set 1 collection, Berkeley Drosophila Genome Project consortium, available at www.bdgp.org) from the corresponding recombinant plasmids by PCR amplification. We used primers based on vector sequences surrounding the DNA inserts and a T3, T7, or Sp6 RNA polymerase initiation site, and cDNAs were purified by filtration using the QIAquick system (Qiagen, Valencia, CA) in a 96-well format, followed by ethanol precipitation. For cDNAs cloned in the pOT2 vector, primer sequences were as follows: 5′-ATTTAGGTGACACTATAGAACTCG-3′ and 5′-AAATTAATACGACTCACTATAGGG-3′. For cDNAs in the pBlueScript SK(-) vector, primers were as follows: 5′-GCGCAATTAACCCTCACTAAAGGG-3′ and 5′-TAATACGACTCACTATAGGGCGAA-3′. Purified PCR products were used for sense and antisense RNA synthesis by using T7, T3, or Sp6 MegaScript kits (Ambion, Austin, TX) according to the manufacturer's instructions and adapted to a 96-well format. Synthesized complementary RNA transcripts were then pooled, deproteinized with phenol saturated with an aqueous solution of sodium acetate, extracted with chloroform, and precipitated by ethanol at -20°C, yielding a pellet that was suspended in 400 μl of injection buffer (5 mM KCl/0.1 mM NaH2PO4, pH 7.8). Samples to be annealed were heated for 5 min at 90°C, allowed to cool to room temperature (4 h), and then precipitated with one volume of isopropanol. The resulting pellet was dissolved in 40 μl of injection buffer. Aliquots (1 μl) from each sample were used at a 1:100 to 1:200 dilution for agarose gel electrophoresis analysis of dsRNA, and sample absorbances were read at 260 and 280 nm. Pictures of dsRNA separated on agarose gels were compared for consistency of size with similar pictures of the corresponding PCR-amplified cDNAs.
Additional dsRNAs corresponding to predicted Drosophila mRNAs for protein kinases, protein phosphatases, some phosphatases and cyclases, and some dsRNA that failed to be synthesized starting from cDNAs in the Berkeley Drosophila Genome Project consortium cDNA collection were gifts from Philip Beachy and Lawrence Lum (The Johns Hopkins University School of Medicine, Baltimore), and they were synthesized as described (12). The resulting library of dsRNAs corresponded to 4,923 genes; 3,314 genes were screened in this study.
Embryo Injection and Immunohistochemistry. Drosophila melanogaster, Oregon R, or yw strains were used. Freshly laid fertilized eggs were collected on agar-juice plates for 40-60 min, washed with water, and aligned on a microscope slide without dechorionization. Embryos were allowed to dry briefly to enable them to stick to the slide, and embryos were then covered with halocarbon oil 27 (Sigma). Capillaries for microinjection were prepared by using a PN-30 N micropipette puller (Narishige, Tokyo), and tips were opened by using a EG-400 microgrinder (Narishige). Embryos at the preblastoderm stage were injected with ≈100 pl of dsRNA, ≈1 μg/μl (0.1 ng) injected dsRNA, or pools of three dsRNA species (each at 1 μg/μl in 100 pl) by using a FemtoJet injection apparatus (Eppendorf) and incubated 24 h at 18°C to allow development to embryonic stage ≈15-16. Immunohistochemistry was performed by using a modification of the standard rapid protocol (14) that was adapted to a 96-multiwell format by using Durapore membrane filter plates (Multiscreen-DV, Millipore).
The mAbs (culture media) that recognize subsets of neurons in the embryonic CNS and peripheral nervous system (PNS) were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City) and were used as follows: mAb 22C10 (anti-futsch), 1:100 dilution, for both CNS and PNS neuron and axon subsets (15); mAb BP102, 1:100 dilution, to label some CNS axons (16); and mAb 1D4 (anti-FasII), 1:100 dilution, to stain some motor neurons and their axons (17). Ab-antigen complexes were detected by using biotinylated anti-mouse IgG (Vector Laboratories) and the Vectastain Elite ABC and DAB substrate kits (Vector Laboratories).
The morphology of injected embryos and patterns of Ab staining were inspected by using bright-field microscopy. Micro-photographs were taken by using an Axioplan microscope (Zeiss) equipped with a Photometrics Coolsnap HQ monochrome camera (Roper Scientific, Trenton, NJ; resolution, 1,392 × 1,040 pixels) and Micro*Color tunable RGB (red-green-blue) filters for digital imaging (Cambridge Research and Instrumentation, Woburn, MA). Digital images were acquired with IP LAB software (Scanalytics, Fairfax, VA) and further processed with photoshop (Adobe Systems, San Jose, CA).
Results and Discussion
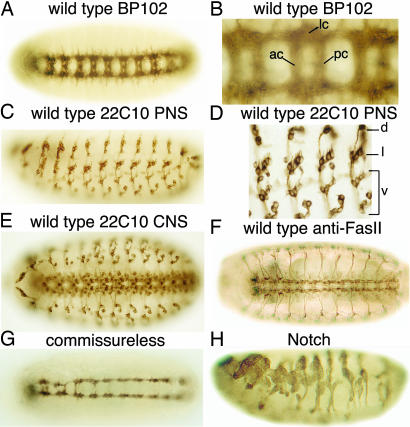
RNAi Screen for Genes Involved in Nervous System Development. RNAi-based gene silencing was used to identify genes affecting the embryonic development of the Drosophila nervous system. Early Drosophila embryos were injected with dsRNA, incubated to allow development to embryonic stage 15 or 16, fixed, incubated with mAbs directed against subsets of neurons in the nervous system, and then incubated with a secondary Ab directed against mouse IgG to which horseradish peroxidase had been covalently coupled to visualize the nervous system. The staining pattern of the ventral nerve cord (VNC) of a wild-type Drosophila embryo stained with mAb BP102, which stains the anterior and posterior commisures and longitudinal connectives of the VNC, is shown in Fig. 1 A and B. The pattern of cells in the PNS of a wild-type embryo stained with mAb 22C10 is shown in Fig. 1 C and D. The mAb 22C10 also stains a subset of neurons and neurites in the VNC. The patterns of cells stained with 22C10 both in the VNC and PNS of a wild-type embryo are shown in Fig. 1E. The staining pattern of a wild-type embryo stained with mAb 1D4 directed against fasciclin II (which labels a subset of neurons in the VNC, including motor neuron axons that innervate striated muscle cells in the periphery) is shown in Fig. 1F.
Fig. 1.
Wild-type patterns of the VNC or PNS of Drosophila embryos stained with mAb BP102 (A and B), 22C10 (C-E), or 1D4 (anti-FasII) (F). B and D show high magnifications of A and C, respectively. G shows the VNC of an embryo injected with commissureless dsRNA and stained with BP102. Most commissures are absent. H shows an embryo injected with Notch dsRNA stained with 22C10. Overproduction of PNS neurons and disorganization of the PNS can be seen. Ventral views of embryos are shown in A, B, and E--G; whereas, lateral views are shown in C, D, and H. lc, longitudinal connectives; ac, anterior commissures; pc, posterior commissures; d, dorsal cluster; l, lateral cluster; v, ventral cluster. In all images, anterior is to the left; in lateral views of embryos, dorsal is up.
To validate the screening strategy, we examined the ability of dsRNA injected into early embryos to reduce the levels of proteins that are known to be expressed in the embryonic nervous system and to reproduce phenotypes that are well described for genetic mutations. Injection of engrailed, embryonic lethal abnormal vision (elav), prospero, or FasII dsRNAs into embryos resulted in marked reductions in the respective protein levels, as shown by immunostaining with the corresponding Abs (see Fig. 5 A-H, which is published as supporting information on the PNAS web site). Injection of commissureless dsRNA into preblastoderm embryos resulted in embryos that lacked most commissures (Fig. 1G), and injection of Notch dsRNA resulted in hypertrophy of the PNS characteristic of the phenotype described for Notch mutants (Fig. 1H).
Injection of embryos with buffer alone usually resulted in ≈10 ± 5% of the stained embryos with defects in the structure of the nervous system. Each embryo was injected with ≈100 pl of solution containing dsRNA. However, injection of an embryo with ≥150 pl of buffer resulted in a much higher percentage of embryos with false-positive mutant nervous system phenotypes.
Very conservative criteria were used for evaluating neural mutant phenotypes, such as disruption of the structure of the CNS and/or PNS, or loss or gain of neurons. For each experiment, at least 50-100 embryos were injected with dsRNA, and only dsRNAs resulting in mutant phenotypes in >50% of the injected embryos were selected as candidates for further analysis. To eliminate false positives and determine whether the mutant phenotypes were reproducible, the candidate dsRNAs were injected into embryos in three to six or more experiments by two or more investigators. Only when ≥50% of injected embryos exhibited a mutant phenotype in reproducible experiments was a species of dsRNA stated to result in an RNAi-dependent mutant phenotype of the nervous system. Mutant phenotypes with penetrance of <50% were not considered further.
Genes Affecting the Embryonic Nervous System. A summary of the data is shown in Table 1. We screened 3,314 genes, which is ≈25% of the Drosophila genome, by RNAi and found 43 genes that affect the development of the embryonic nervous system (i.e., 1.3% of the tested dsRNAs). Loss-of-function mutant phenotypes that had not been reported previously were found for 18 genes. These mutant phenotypes are shown in Figs. 2, 3, 4. The mutant phenotypes of 22 of the 25 additional genes that were found that affect the development of the nervous system (except for lola, for which data are not shown, and Notch and commissureless, which are shown in Fig. 1) confirmed previously reported phenotypes found with genetic mutants, and they are shown in Figs. 6-8, which are published as supporting information on the PNAS web site.
Table 1. Summary of the no. of genes.
| Description | No. of genes |
|---|---|
| Genes screened by RNAi | 3,314 (≈25% of genome) |
| Genes that affect neural development | 43 (1.30%) |
| Novel phenotypes | 18 |
| Genes with unknown functions | 8 |
| Genes with known functions | 10 |
| Known phenotypes | 25 |
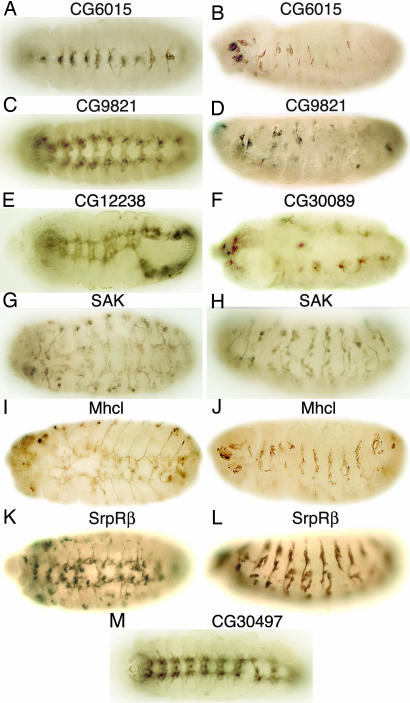
Fig. 2.
RNAi mutant phenotypes of genes with unknown functions. Stage 13 (A and B) or 15/16 (C-M) embryos were stained with BP102 (A, C, E, and M) or 22C10 (B, D, and F-L) mAb after injection of each indicated dsRNA. Ventral (A, C, E-G, I, K, and M) and lateral (B, D, H, J, and L) views of embryos are shown.
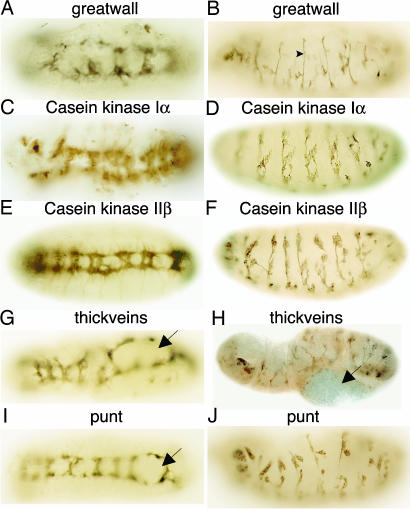
Fig. 3.
RNAi mutant phenotypes of protein kinases. Stage 15/16 embryos were stained with BP102 (A, C, E, G, and I), 22C10 (D, F, H, and J), or 1D4 (B) mAb after injection with the indicated dsRNA. Ventral (A, C, E, G, and I) or lateral (B, D, F, H and J) views of embryos are shown. The arrowhead in B indicates a segmental nerve, and arrows in G-I indicate hole-like structures.
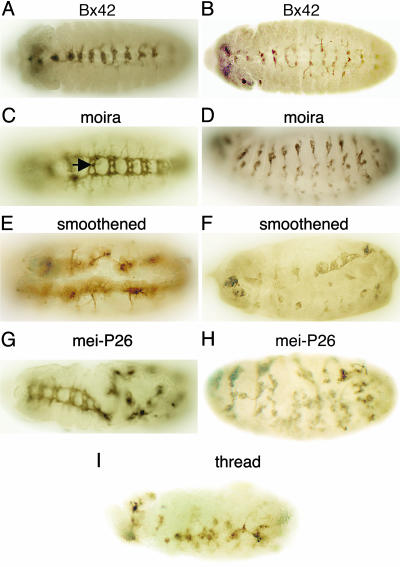
Fig. 4.
Additional RNAi mutant phenotypes. Stage 13 (A and B) or 15/16 (C-I) embryos were stained with BP102 (A, C, E, and G) or 22C10 (B, D, F, H, and I) mAb after injection of the dsRNA indicated at top of each image. Ventral (A-C, E, and G) and lateral (D, F, H, and I) views of embryos are shown. The arrow in C indicates abnormal commissures.
RNAi-induced mutant phenotypes for eight genes whose functions are unknown are shown in Fig. 2. However, general functions can be inferred for some of the proteins encoded by these genes that exhibit homology to proteins whose functions have been identified. CG6015 encodes a protein that exhibits homology to RNA-splicing factor PRP17, which was identified originally in yeast and is conserved in vertebrates (18). Although not essential for splicing in yeast, PRP17 (also known as CDC40) is required for cell-cycle progression (19). Silencing of CG6015 gene expression by RNAi resulted in hypoplasia of the VNC (Fig. 2 A) because of cessation of development at stages 14 and 15. Axons of midline neurons are misrouted (data not shown). In some mutants, the longitudinal connectives and commissures do not form, and severe hypoplasia of the PNS can be detected (Fig. 2B). The interesegmental nerve also is abnormal (data not shown). CG9821 is a gene of unknown function encoding a protein that exhibits limited homology to a transcription coactivator. The neural phenotype observed after injection of CG9821 dsRNA into early embryos includes underdeveloped commissures, resulting in widening of the distance between the longitudinal connectives of the VNC (Fig. 2C) and severe hypoplasia of the PNS (Fig. 2D). CG12238 is a gene of unknown function encoding a protein with a plant homeodomain-finger domain. DNA insertions in this gene are recessive lethal (20); however, to our knowledge, no further information about the mutant phenotype has been reported. Suppression of CG12238 gene expression by RNAi resulted in embryos that lacked some commissures (Fig. 2E). The CG30089 gene encodes a protein that contains an α-hydrolase domain; in an RNAi-based screen in Drosophila cell culture, CG30089 resulted in a reduced cell-viability phenotype (10). Suppression of the CG30089 gene by RNAi in embryos resulted in a mutant phenotype with abnormalities in the formation of ectodermal cells and severe loss of neurons, longitudinal connectives, and commissures in the VNC (Fig. 2F). SAK encodes a protein with homology to polo-like kinases, which are a family of kinases that are key regulators of mitosis in yeast, flies, and mammals (21). No mutants have been described for SAK in Drosophila. However, knockout mice lacking Sak undergo gastrulation but fail to develop beyond 7.5 days postcoitum, and they display an increased number of cells that stop in late mitosis and enter the apoptotic pathway (22). We observed mosaic phenotypes in SAK RNAi mutants. The most severe RNAi mutant phenotypes exhibit disruption of development at very early stages, suggesting a primary role of SAK in cell proliferation (data not shown). More moderate phenotypes exhibit a reduction in the number of neurons, absence of the longitudinal connectives, and disorganization of the VNC (Fig. 2G) and the PNS (Fig. 2H). Myosin heavy-chain-like (Mhcl) gene encodes a protein similar to a myosin heavy-chain protein, containing a myosin motor domain and a PDZ domain (23). No mutations of this gene have been reported in Drosophila. However, the role of nonmuscle myosins in the embryonic development of the nervous system has been described extensively in rodents (24). RNAi-dependent suppression of Mhcl gene function resulted in loss of neurons and disorganization of the VNC (Fig. 2I) and loss of neurons in the PNS (Fig. 2 J). Signal recognition particle receptor β (SrpRβ) is the Drosophila homolog of an endoplasmic reticulum membrane-associated GTP-binding protein that regulates vesicular trafficking of secretory and membrane proteins (25, 26). SrpRβ mutant alleles have been reported in Drosophila that are recessive lethal (27); however, no description of the embryo mutant phenotype has been reported. The RNAi-induced SrpRβ phenotype includes disorganization of some neuromeres in the VNC (Fig. 2K) and loss of neurons in the lateral and ventral clusters of the PNS (Fig. 2L). CG30497 is a gene that encodes a protein of unknown function that does not contain any obvious structural domain. A P-element insertion in this gene has been generated that interacts genetically with Pannier, a gene regulator essential for dorsal closure and development of the epidermis (28). In addition, CG30497 dsRNA reduced viability of Drosophila cell cultures (10). We find that embryos injected with CG30497 dsRNA have breaks in the longitudinal connectives of the VNC (Fig. 2M).
RNAi-induced mutant phenotypes for five dsRNAs corresponding to protein kinase genes are shown in Fig. 3. greatwall (gwl) encodes a protein containing a Ser-Thr protein kinase domain. Data from a screen for gain-of-function mutants suggested that gwl is involved in motor axon guidance and synaptogenesis in larvae (29). More recently, gwl loss-of-function mutations that result in mitotic delay in larval neuroblasts and defective condensation of chromosomes in Drosophila cell cultures have been described (30). Embryos injected with gwl dsRNA lack some commissures, have breaks in the longitudinal connectives, and lack some segmental nerves (Fig. 3 A and B), compared with wild-type embryos stained with anti-FasII mAb 1D4 (Fig. 1F). The overall disruption of the nervous system suggests an essential role for gwl in the development of the embryonic nervous system. Casein kinase Iα (CkIα) is an isoform of the casein kinase protein family that has various functions, including cell-cycle regulation, DNA repair (31), and hedgehog (12), and wingless (13, 32) signaling. RNAi-induced CkIα phenotypes exhibited disruption of both commissures and longitudinal connectives in the VNC (Fig. 3C) and hypoplasia of neurons in the PNS (Fig. 3D). The defects that we observed for embryos injected with CkIα dsRNA are consistent with the known requirement for CkIα in both wingless (32) and hedgehog (12) signaling and the requirement of these pathways for CNS development (33). Also, phosphorylation by CkIα results in inactivation of armadillo, a protein in the wingless pathway that is required early in development for neuroblast fate determination, and later, for the construction of the axonal scaffold (34). Casein kinase II (CkII) protein is a heterotetramer composed of two catalytic (α) and two regulatory (β) subunits. CkII catalyzes the phosphorylation of a number of proteins required for signal transduction, cell-cycle regulation, DNA metabolism, etc. (35). RNAi phenotypes of embryos injected with CkIIβ dsRNA exhibit defects in the longitudinal connectives and commissures of the VNC (Fig. 3E) and have disorganized PNS neurons (Fig. 3F). A hypomorphic allele of CkIIβ has been described that has a reduced number of neurons in the mushroom bodies of adult Drosophila (36), and a behavioral mutant, andante (37), with lengthened circadian rhythms has been reported. We find that CkIIβ also is required during the development of the embryonic nervous system. Two genes, thickveins and punt, were identified that encode receptors in the decapentaplegic (dpp)/transforming growth factor β (TGF-β) pathway (38). Both genes are necessary for embryonic dorsal-ventral polarity, cell proliferation, and differentiation of many tissues. RNAi-induced mutant phenotypes of thickveins (Fig. 3 G and H) and punt (Figs. 3 I and J) exhibit severe disruption of the VNC and the PNS, including defects in neuromere organization. The hole-like structures in Fig. 3 G-I may result from defects in ectoderm development.
Injection of Bx42 dsRNA in early embryos resulted in severe hypoplasia and disorganization of the CNS and PNS in stage-13 embryos (Fig. 4 A and B, respectively). Injected embryos did not develop beyond stage 14, similar to embryos injected with CG6015 dsRNA. Bx42 is a nuclear transcription factor required for the formation of different tissues during development. Phenotypic analysis of wing imaginal disks suggests that Bx42 is involved in the Notch signaling pathway (39). Treatment of Drosophila cell cultures with Bx42 dsRNA resulted in cell death (10). However, no genetic mutants of Bx42 have been described. The moira gene encodes an RNA polymerase II transcription factor that is essential for the transcriptional regulation and activity of multiple homeotic and segmentation genes, including antennapedia (40). moira is ubiquitously expressed in early development and is highly concentrated in the CNS at later stages of embryonic development (41). Embryos injected with moira dsRNA exhibited abnormal commissures (Fig. 4C) and loss of neurons in the PNS (Fig. 4D). smoothened encodes a transmembrane protein required for hedgehog signaling (42, 43). Injection of smoothened dsRNA resulted in embryos that lack commissures, have broken longitudinal connectives in the VNC (Fig. 4E), and also exhibit loss of cells and disorganization of the PNS (Fig. 4F). mei-P26 plays a role in germ cell development and cell proliferation (44). mei-P26 encodes a protein that exhibits homology to the brain tumor (brat) protein (45) and contains two B-box zinc finger domains near the N terminus of the protein and six NHL repeats, which form a β-propeller domain near the C terminus of the protein. mei-P26 exhibits amino acid sequence similarity to a heterogeneous family of proteins, the tripartite motif (TRIM) proteins, which may be expressed during embryonic development in vertebrates and include genes that are mutated in inherited human diseases, such as Opitz syndrome (46). We observed severe RNAi-dependent phenotypic abnormalities of the nervous system in embryos injected with mei-P26 dsRNA, such as breaks in the VNC, disorganization of the VNC (Fig. 4G), and disorganization of the PNS (Fig. 4H). The zygotic expression of the mei-P26 gene in the embryo is restricted to germ cells (47); whether mei-P26 affects neural development directly or indirectly is not known. thread is an antiapoptotic protein and is a Drosophila homolog of mouse and human inhibitor of apoptosis-1 (48). In thread mutants, embryonic development is arrested and cells undergo apoptosis (48). thread negatively regulates apoptosis by binding to a caspase, i.e., a proapoptotic protease (48). Treatment of cultured Drosophila cells with thread dsRNA also resulted in massive cell death (10). Injection of thread dsRNA into early embryos resulted in a marked loss of neurons and disorganization of both the VNC and the PNS (Fig. 4I), possibly because of the lack of regulation of apoptosis.
A summary of the 18 genes found by RNAi that exhibit novel mutant phenotypes that affect the development of the nervous system is given in Table 2. Suppression of 25 additional genes by RNAi resulted in “known” mutant phenotypes of the nervous system. The commissureless and Notch RNAi mutant phenotypes are shown in Fig. 1 G and H, respectively. RNAi-dependent mutant phenotypes of the nervous system for Delta, big brain, echinoid, robo, slit, frazzled, plexin A, Egf receptor, and patched are shown in Fig. 6. RNAi-dependent mutant phenotypes for eight genes that encode proteins that regulate gene expression are shown in Fig. 7. Injection of dsRNA corresponding to Su(H), mastermind, engrailed, sequoia, tramtrack, daughterless, twist, and SoxNeuro resulted in mutant phenotypes of the nervous system. The mutant phenotype for Su(H) differed from the phenotype reported for genetic mutants (49), perhaps because Su(H) dsRNA contains nucleotide sequences that are similar to sequences in some species of mRNA from different genes that encode basic helix-loop-helix proteins. Other RNAi mutant phenotypes resembled phenotypes reported for genetic mutants.
Table 2. Genes with novel RNAi-dependent mutant phenotypes that affect the nervous system.
| Gene | Number | Function |
|---|---|---|
| Function unknown | ||
| CG6015 | CG6015 | |
| CG9821 | CG9821 | |
| CG12238 | CG12238 | |
| CG30089 | CG30089 | |
| CG30497 | CG30497 | |
| SAK | CG7186 | |
| Myosin heavy chain like | CG10218 | |
| Signal recognition | CG33162 | |
| particle receptor β | ||
| Protein kinases | ||
| Casein kinase Iα | CG2028 | Ser-Thr protein kinase |
| Casein kinase IIβ | CG15524 | Ser-Thr protein kinase regulatory subunit |
| thick veins | CG14026 | Type I TGF-β receptor |
| punt | CG7904 | Type II TGF-β receptor |
| greatwall | CG7719 | Ser-Thr protein kinase |
| Other | ||
| Bx42 | CG8264 | Transcription factor |
| moira | CG18740 | Transcription factor |
| smoothened | CG11561 | Hedgehog pathway |
| mei-P26 | CG12218 | Germ cell development |
| thread | CG12284 | Apoptosis inhibitor |
TGF, transforming growth factor.
RNAi mutant phenotypes for dsRNAs corresponding to Bazooka, shotgun, three rows, Cyclin A, and cut up are shown in Fig. 8.
A summary of the 43 genes found by RNAi that affect the development of the nervous system is given in Table 3, which is published as supporting information on the PNAS web site. Additional photomicrographs of many mutant RNAi embryos and periodic updates on the RNAi screening are available at http://flyembryo.nhlbi.nih.gov/flyembryo.
Similar RNAi-dependent mutant phenotypes were found with some genes. For example, similar mutant phenotypes were found with patched, a hedgehog receptor (50), smoothened, and casein kinase Iα, which function in the hedgehog signal transduction pathway (43). Similarly, genes in the Notch pathway (i.e., Notch, Delta, bigbrain, echinoid, and mastermind), with the exception of Su(H), resulted in the expected hypertrophic PNS.
In conclusion, genes were found in this study that, directly or indirectly, affect the development of the nervous system. Some of the most interesting genes that were found have unknown functions, whereas other genes encode proteins of known function that were not previously known to affect the development of the embryonic nervous system. Further studies are required to determine how these genes affect the development of the nervous system.
Supplementary Material
Acknowledgments
We thank Philip Beachy and Lawrence Lum for the gift of ≈600 dsRNAs and Corey Goodman (University of California, Berkeley) for the gift of mAb 1D4.
Author contributions: A.C.R. and M.N. designed research; A.I.I., A.C.R., P.P., S.Y., B.M., H.-P.L., S.-H.Y., H.H., and V.G. performed research; M.S. contributed new reagents/analytic tools; A.I.I., A.C.R., P.P., S.Y., B.M., H.-P.L., H.H., and M.N. analyzed data; and A.C.R., S.Y., and M.N. wrote the paper.
Abbreviations: dsRNA, double-stranded RNA; RNAi, RNA interference; VNC, ventral nerve cord; PNS, peripheral nervous system.
References
- 1.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- 2.Kennerdell, J. R. & Carthew, R. W. (1998) Cell 95, 1017-1026. [DOI] [PubMed] [Google Scholar]
- 3.Hannon, G. J. (2002) Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- 5.Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta, S., Schoer, R. A., Egan, J. E., Hannon, G. J. & Mittal, V. (2004) Proc. Natl. Acad. Sci. USA 101, 1927-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hannon, G. J. & Conklin, D. S. (2004) Methods Mol. Biol. 257, 255-266. [DOI] [PubMed] [Google Scholar]
- 8.Gonczy, P., Echeverri, C., Oegema, K., Coulson, A., Jones, S. J., Copley, R. R., Duperon, J., Oegema, J., Brehm, M., Cassin, E., et al. (2000) Nature 408, 331-336. [DOI] [PubMed] [Google Scholar]
- 9.Kim, Y. O., Park, S. J., Balaban, R. S., Nirenberg, M. & Kim, Y. (2004) Proc. Natl. Acad. Sci. USA 101, 159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutros, M., Kiger, A. A., Armknecht, S., Kerr, K., Hild, M., Koch, B., Haas, S. A., Consortium, H. F., Paro, R. & Perrimon, N. (2004) Science 303, 832-835. [DOI] [PubMed] [Google Scholar]
- 11.Kiger, A., Baum, B., Jones, S., Jones, M., Coulson, A., Echeverri, C. & Perrimon, N. (2003) J. Biol. 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lum, L., Yao, S., Mozer, B., Rovescalli, A., Von Kessler, D., Nirenberg, M. & Beachy, P. A. (2003) Science 299, 2039-2045. [DOI] [PubMed] [Google Scholar]
- 13.Matsubayashi, H., Sese, S., Lee, J. S., Shirakawa, T., Iwatsubo, T., Tomita, T. & Yanagawa, S. (2004) Mol. Cell. Biol. 24, 2012-2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel, N. H. (1994) Methods Cell Biol. 44, 445-487. [DOI] [PubMed] [Google Scholar]
- 15.Fujita, S. C., Zipursky, S. L., Benzer, S., Ferrus, A. & Shotwell, S. L. (1982) Proc. Natl. Acad. Sci. USA 79, 7929-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seeger, M., Tear, G., Ferres-Marco, D. & Goodman, C. S. (1993) Neuron 10, 409-426. [DOI] [PubMed] [Google Scholar]
- 17.Vactor, D. V., Sink, H., Fambrough, D., Tsoo, R. & Goodman, C. S. (1993) Cell 73, 1137-1153. [DOI] [PubMed] [Google Scholar]
- 18.Mount, S. M. & Salz, H. K. (2000) J. Cell Biol. 150, F37-F44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahan, O. & Kupiec, M. (2004) Nucleic Acids Res. 32, 2529-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bourbon, H. M., Gonzy-Treboul, G., Peronnet, F., Alin, M. F., Ardourel, C., Benassayag, C., Cribbs, D., Deutsch, J., Ferrer, P., Haenlin, M., et al. (2002) Mech. Dev. 110, 71-83. [DOI] [PubMed] [Google Scholar]
- 21.Barr, F. A., Sillje, H., H. & Nigg, E., A. (2004) Nat. Rev. Mol. Cell Biol. 5, 429-440. [DOI] [PubMed] [Google Scholar]
- 22.Hudson, J. W., Kozarova, A., Cheung, P., Macmillan, J. C., Swallow, C. J., Cross, J. C. & Dennis, J. W. (2001) Curr. Biol. 11, 441-446. [DOI] [PubMed] [Google Scholar]
- 23.Tzolovsky, G., Millo, H., Pathirana, S., Wood, T. & Bownes, M. (2002) Mol. Biol. Evol. 19, 1041-1052. [DOI] [PubMed] [Google Scholar]
- 24.Brown, M. E. & Bridgman, P. C. (2004) J. Neurobiol. 58, 118-130. [DOI] [PubMed] [Google Scholar]
- 25.Jekely, G. (2003) BioEssays 25, 1129-1138. [DOI] [PubMed] [Google Scholar]
- 26.Legate, K. R. & Andrews, D. W. (2003) J. Biol. Chem. 278, 27712-27720. [DOI] [PubMed] [Google Scholar]
- 27.Spradling, A. C., Stern, D., Beaton, A., Rhem, E. J., Laverty, T., Mozden, N., Misra, S. & Rubin, G. M. (1999) Genetics 153, 135-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pena-Rangel, M. T., Rodriguez, I. & Riesgo-Escovar, J. R. (2002) Genetics 160, 1035-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kraut, R., Menon, K. & Zinn, K. (2001) Curr. Biol. 11, 417-430. [DOI] [PubMed] [Google Scholar]
- 30.Yu, J., Fleming, S. L., Williams, B., Williams, E. V., Li, Z., Somma, P., Rieder, C. L. & Goldberg, M. L. (2004) J. Cell Biol. 164, 487-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos, J. A., Logarinho, E., Tapia, C., Allende, C. C., Allende, J. E. & Sunkel, C. E. (1996) J. Cell Sci. 109, 1847-1856. [DOI] [PubMed] [Google Scholar]
- 32.Liu, C., Li, Y., Semenov, M., Han, C., Baeg, G. H., Tan, Y., Zhang, Z., Lin, X. & He, X. (2002) Cell 108, 837-847. [DOI] [PubMed] [Google Scholar]
- 33.Patel, N. H., Schafer, B., Goodman, C. S. & Holmgren, R. (1989) Genes Dev. 3, 890-904. [DOI] [PubMed] [Google Scholar]
- 34.Loureiro, J. & Peifer, M. (1998) Curr. Biol. 8, 622-632. [DOI] [PubMed] [Google Scholar]
- 35.Allende, J. E. & Allende, C. C. (1995) FASEB. J. 9, 313-323. [DOI] [PubMed] [Google Scholar]
- 36.Heisenberg, M., Heusipp, M. & Wanke, C. (1995) J. Neurosci. 15, 1951-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akten, B., Jauch, E., Genova, G. K., Kim, E. Y., Edery, I., Raabe, T. & Jackson, F. R. (2003) Nat. Neurosci. 6, 251-257. [DOI] [PubMed] [Google Scholar]
- 38.Ruberte, E., Marty, T., Nellen, D., Affolter, M. & Basler, K. (1995) Cell 80, 889-897. [DOI] [PubMed] [Google Scholar]
- 39.Negeri, D., Eggert, H., Gienapp, R. & Saumweber, H. (2002) Mech. Dev. 117, 151-162. [DOI] [PubMed] [Google Scholar]
- 40.Arnosti, D. N. (2002) Insect Biochem. Mol. Biol. 32, 1257-1273. [DOI] [PubMed] [Google Scholar]
- 41.Crosby, M. A., Miller, C., Alon, T., Watson, K. L., Verrijzer, C. P., Goldman-Levi, R. & Zak, N. B. (1999) Mol. Cell. Biol. 19, 1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcedo, J., Ayzenzon, M., Von Ohlen, T., Noll, M. & Hooper, J. E. (1996) Cell 86, 221-232. [DOI] [PubMed] [Google Scholar]
- 43.Lum, L. & Beachy, P. A. (2004) Science 304, 1755-1759. [DOI] [PubMed] [Google Scholar]
- 44.Page, S. L., McKim, K. S., Deneen, B., Van Hook, T. L. & Hawley, R. S. (2000) Genetics 155, 1757-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arama, E., Dickman, D., Kimchie, Z., Shearn, A. & Lev, Z. (2000) Oncogene 19, 3706-3716. [DOI] [PubMed] [Google Scholar]
- 46.Reymond, A., Meroni, G., Fantozzi, A., Merla, G., Cairo, S., Luzi, L., Riganelli, D., Zanaria, E., Messali, S., Cainarca, S., et al. (2001) EMBO J. 20, 2140-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomancak, P., Beaton, A., Weiszmann, R., Kwan, E., Shu, S., Lewis, S. E., Richards, S., Ashburner, M., Hartenstein, V., Celniker, S. E. & Rubin, G. M. (2002) Genome Biol. 3, RESEARCH0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lisi, S., Mazzon, I. & White, K. (2000) Genetics 154, 669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lecourtois, M. & Schweisguth, F. (1995) Genes Dev. 9, 2598-2608. [DOI] [PubMed] [Google Scholar]
- 50.Marigo, V., Davey, R. A., Zuo, Y., Cunningham, J. M. & Tabin, C. J. (1996) Nature 384, 176-179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.