Abstract
NifL is an antiactivator that tightly regulates transcription of genes required for nitrogen fixation in Azotobacter vinelandii by controlling the activity of its partner protein NifA, a member of the family of σ54-dependent transcriptional activators. Although the C-terminal region of A. vinelandii NifL shows homology to the transmitter domains of histidine protein kinases, signal transduction between NifL and NifA is conveyed by means of protein-protein interactions rather than by phosphorylation. Binding of the ligand 2-oxoglutarate to NifA plays a crucial role in preventing inhibition by NifL under conditions appropriate for nitrogen fixation. We have used a suppressor screen to identify a critical arginine residue (R306) in NifL that is required to release NifA from inhibition under appropriate environmental conditions. Amino acid substitutions at position 306 result in constitutive inhibition of NifA activity by NifL, thus preventing nitrogen fixation. Biochemical studies with one of the mutant proteins demonstrate that the substitution alters the conformation of NifL significantly and prevents the response of NifA to 2-oxoglutarate. We propose that arginine 306 is critical for the propagation of signals perceived by A. vinelandii NifL in response to the redox and fixed-nitrogen status and is required for a conformational switch that inactivates the inhibitory function of NifL under conditions appropriate for nitrogen fixation.
Keywords: 2-oxoglutarate, signal transduction, antiactivator, redox control, nitrogen regulation
Structural rearrangements of sensor proteins in response to environmental cues provide a fundamental mechanism for signal propagation within cells. However, although conformational changes have been well characterized in isolated signaling domains, mechanisms for signal transmission by means of interdomain interactions are frequently less well understood. For example, structural studies have identified ligand-induced conformational changes in the sensor domains of histidine protein kinases (HPKs), but it is not known how these changes are communicated to the kinase domain to control phosphoryl transfer.
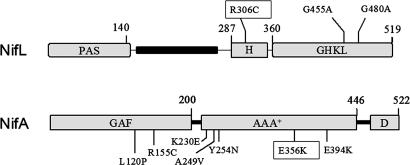
The Azotobacter vinelandii NifL regulatory protein is a histidine kinase-like protein that controls the expression of the genes required for nitrogen fixation in response to the redox, nitrogen, and carbon status. NifL is an antiactivator that tightly regulates the activity of its partner protein NifA, a member of the family of σ54-transcriptional activators (1, 2), by means of the formation of an inhibitory complex (3-5). The domain architecture of NifL is similar to that of some HPKs, with an N-terminal Per-Arnt-Sim (PAS) domain (6, 7) containing a flavin adenine dinucleotide (FAD) cofactor that senses the redox status (8, 9) and a C-terminal domain containing conserved residues corresponding to the N, G1, F, and G2 boxes that constitute the ATP-binding domain of the GHKL superfamily of ATPases (10-14) (Fig. 1). Unlike the HPKs, the GHKL domain of NifL does not exhibit ATP hydrolysis or transphosphorylation activity, but the binding of ADP to this domain strongly stimulates the inhibitory activity of NifL and the stability of the NifL-NifA complex (8, 15, 16). Also, the GHKL domain is involved in sensing the nitrogen status because it is the site of interaction with the signal transduction protein GlnK (17-19), the PII-like protein of A. vinelandii (20-23).
Fig. 1.
Domain structure of A. vinelandii NifL and NifA showing the mutations analyzed in this study.
Most HPKs contain an H-box motif located in a dimerization domain (also known as domain A) that contains the autophosphorylation site and interacts with the corresponding response regulator (24, 25). Although NifL contains an H-box motif (Fig. 1), the conserved histidine residue is not required for regulation of NifA activity (26) and neither autophosphorylation of NifL nor phosphotransfer to NifA has been detected (27, 28). These observations, together with the fact that NifA is not a member of the response-regulator family, suggest that NifL is not a bona fide HPK.
Whereas signals of the redox and fixed-nitrogen status are both perceived by NifL, the NifA protein is responsive to allosteric control by 2-oxoglutarate, a metabolic signal of the carbon status, which binds to the N-terminal GAF domain of this activator. Binding of 2-oxoglutarate to NifA antagonizes the influence of adenosine nucleotides on the NifL-NifA interaction, thus enabling NifA to escape from inhibition by NifL, under nitrogen-fixing conditions (29, 30). Our current model for activation of NifL implies that conformation changes caused either by changes in oxidation state of the PAS domain or the binding of GlnK to the GHKL domain are transmitted to the C-terminal region to enable NifL to inhibit NifA in the presence of 2-oxoglutarate. To identify residues that are involved in signal transmission, we have screened for NifL suppressor mutations that inhibit constitutive (NifL-resistant) forms of NifA. We have identified an arginine residue (R306) in the H motif of NifL that appears to be crucial for signal transmission because substitutions at this position prevent transcriptional activation by NifA under nitrogen-fixing conditions. We observe that the R306C mutation alters the conformation of NifL and that the mutant protein is competent to inhibit NifA in the presence of 2-oxoglutarate, thus preventing nitrogen fixation. The mutation also suppresses substitutions in conserved residues in the GHKL domain, suggesting that interactions between the nucleotide-binding domain and the H-motif region of NifL are important for signal transduction.
Methods
Mutagenesis and Isolation of Suppressor Mutations. PCR mutagenesis was carried out with TaqDNA polymerase by using standard reaction conditions. Reaction mixtures contained 10 ng of template plasmid pPMA (31) (encoding NifL and NifA-E356K), 40 pmol of each primer, 0.2 mM each deoxynucleoside triphosphate, 2 mM MgCl2, and 5 units of enzyme. Primers NifL1 (5′-CCGCCGCAAGGACAAGACC-3′) and P240 (5′-CCTTGCCGGTACCGGACTC-3′) were used to mutate the central and C-terminal domains of NifL. PCR products were purified, digested with MluI and MunI, and recloned into plasmid pPMA digested with the same enzymes. To identify nifL mutants that are able to suppress the effect of the nifA-E356K mutation (which renders NifA insensitive to NifL; ref. 31), ligation mixtures were electroporated into strain DH5α. The transformation mixture was incubated for 16 h, and plasmid DNA was then extracted and used to transform strain ET8000 containing the reporter plasmid pRT22, which carries a nifH-lacZ fusion (32). Transformants were selected on NFDM medium containing 1 mg/ml (NH4)2SO4 as nitrogen source, 20 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside, 30 μg/ml chloramphenicol, and 50 μg/ml ampicillin, and incubated aerobically. Mutants that restored inhibition by NifL were identified as white or light-blue colonies able to suppress the resistant phenotype of nifA-E356K. Plasmid DNA was recovered from selected colonies and reintroduced into the host strain to recheck phenotypes. DNA from selected mutants was isolated and sequenced to identify the mutations.
Site-directed mutagenesis of nifL was done by two-step PCR. The first step consisted of two PCRs, one of which was carried out with primer MS1 (5′-GGGAAAACCTCGCCGCCCCC-3′) and a reverse primer containing the desired mutation, and the other of which was carried out with a forward primer containing the desired mutation and primer L2 (5′-GTCGCTGTTCAGGTGGAGG-3′). PCR products were purified and used as a template for the second-step PCR, using the primers MS1 and L2 described above. The resulting fragment was cut with NotI and ApaI and cloned into plasmid pPR34 encoding wild-type NifL and NifA (32).
A. vinelandii Transformation and Analysis of Transformants. Mutations were introduced into the A. vinelandii genome by using plasmid pIM32, which contains a 1-kb region upstream of nifL, a spectinomycin-resistance cassette in the SmaI site upstream of the nifL promoter (33), and wild-type nifL and nifA sequences. This plasmid also contains an engineered EcoRI site close to the SmaI site within nifL to facilitate identification of recombinants by PCR. nifL and nifA mutations were cloned into pIM32, and the resultant plasmids were linearized and then transformed into competent A. vinelandii, as described (34).
The β-Galactosidase Assays. In vivo activity of NifL and NifA was measured in Escherichia coli strain ET8000 by using the reporter plasmid pRT22, which carries a nifH-lacZ translational fusion. NifL, NifA, and their mutant forms were expressed on a second plasmid from a constitutive promoter (32). The β-galactosidase assays were carried out as described (29, 35)
Protein Expression and Purification. Plasmids pTJ45, pDB737, and pIM15 were used for overexpression of Nhis6NifL, NifA, and NifA-E356K (28-30). For overexpression of Nhis6NifL-R306C, an NdeI-BamHI fragment encoding the corresponding mutation was cloned into plasmid pET28, resulting in plasmid pIM22. Plasmids pIM75 and pIM74, which overexpresses Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519), were made by PCR amplification using primers NifL-147 (5′-GGGAATTCCATATGAACAACCAGCGCTGATGATCG-3′) and NifL2 (5′-CGAAGGATCCTCAGGTGGAGGCCGAGAAGGG-3′). After digestion with NdeI and BamHI, the fragments were cloned into plasmid pET28. In all cases, protein overexpression was carried out in E. coli strain BL21(DE3) pLysS. Cultures were grown aerobically in Luria-Bertani broth, and expression from the T7 promoter was induced by addition of isopropyl β-d-thiogalactopyranoside to 1 mM. Proteins were purified as described (30, 31).
Open-Promoter Complex Assays. NifA-promoted catalysis of open-promoter complexes by σ54-RNA polymerase was used to assay NifA activity and its inhibition by NifL as described (16, 18).
Limited Trypsin Proteolysis. Trypsin proteolysis was performed in TA buffer (50 mM Tris·acetate, pH 7.9/100 mM potassium acetate/8 mM magnesium acetate/1 mM DTT) at room temperature. A trypsin/NifL weight ratio of 1:100 was used. The proteins were incubated with nucleotides for 5 min before digestion was started. Samples of 12 μl were removed at indicated time intervals to tubes on ice containing 12 μl of Laemmli loading buffer, and samples were heated at 100°C for 5 min before electrophoresis in 4-12% gradient gels using MES buffer (50 mM MES/50 mM Tris/0.1% SDS/1 mM EDTA).
Results
Isolation of a nifL Mutation That Suppresses the Resistant Allele nifA-E356K. The surfaces of A. vinelandii NifL that are required for the interaction with NifA are not well defined. Previous experiments have established that the N-terminal region of NifL is not essential for the interaction and, because the isolated C-terminal nucleotide-binding domain is not competent to bind NifA, a candidate region for the interaction is the central region located between residues 287 and 360 that contains the H-box motif (Fig. 1) (15, 32, 36). In an attempt to identify the NifL residues that are important for the NifA interaction, we used suppression mutagenesis, taking advantage of a well characterized mutant form of NifA that is resistant to inhibition by NifL (29, 31). We anticipated that the resistance phenotype of a NifA mutant unable to interact with NifL can be suppressed by a second-site suppressor mutation in nifL. The NifL-resistant mutant NifA-E356K (Fig. 1) was chosen to seek suppressor mutations because position 356 is conserved in NifA proteins but not in other members of the σ54-dependent activator family. To carry out the screening, we used a previously described two-plasmid system in E. coli, consisting of a nifHp-lacZ reporter and a second plasmid expressing nifL and nifA from a constitutive promoter (29, 31, 32, 35). After random PCR mutagenesis of the region encoding the central and C-terminal domains of NifL and introduction of the mutant library into the nifA-E356K background, we screened for suppressors on 20 μg/ml 5-bromo-4-chloro-3-indolyl β-d-galactopyranoside indicator plates containing excess fixed nitrogen under aerobic conditions. Colonies expressing nifL and the resistant allele nifA-E356K are blue under these conditions, and we anticipated that suppressors would give rise to white or light-blue colonies. Only one suppressor with the appropriate phenotype that expressed wild-type levels of NifL and NifA survived the screen. This mutation generated an arginine-to-cysteine substitution at position 306 in the central H-box motif of NifL (Fig. 1), adjacent to the conserved histidine residue at position 305 that is autophosphorylated in bona fide HPKs.
NifL-R306C Inhibits NifA Under Nitrogen-Fixing Conditions. We have previously demonstrated that the activity of the A. vinelandii NifL-NifA system is regulated in response to nitrogen and oxygen status in E. coli. However, the activity of NifA-E356K is constitutive and insensitive to NifL (Table 1, compare first and second rows). NifA-E356K exhibits considerably higher activity than wild-type NifA under anaerobic nitrogen-limiting conditions because NifL retains some inhibitory activity under these conditions (31, 35). The nifL-R306C mutation effectively suppressed the constitutive activity of nifA-E356K, although a low level of NifA activity was detectable under anaerobic, nitrogen-limiting conditions. (Table 1, row 3). To determine whether nifL-R306C is an allele-specific suppressor of nifA-E356K, the nifL-R306C mutation was combined with wild-type nifA. Surprisingly, NifA activity was also inhibited by NifL-R306C under conditions appropriate for nitrogen fixation (Table 1, row 4). Hence, nifL-R306C is not an allele-specific suppressor of nifAE356K, and it has the ability to inhibit transcriptional activation by both mutant and wild-type forms of NifA. When introduced into A. vinelandii, the nifL-R306C mutant had a Nif- phenotype and was unable to fix nitrogen on nitrogen-free medium containing molybdenum (see Fig. 6, which is published as supporting information on the PNAS web site). Therefore, the presence of the R306C substitution confers on NifL the ability to inhibit NifA under nitrogen-fixing conditions.
Table 1. Inhibition of NifA activity by NifL-R306C in vivo.
| β-Galactosidase activity, Miller units
|
|||||
|---|---|---|---|---|---|
| Anaerobic
|
Aerobic
|
||||
| Plasmid* | Proteins | −N† | +N‡ | −N | +N |
| pPR34 | NifL and NifA | 3,138 | 31 | 245 | 75 |
| pPMA | NifL and NifA-E356K | 28,918 | 30,130 | 35,538 | 30,957 |
| pIM17 | NifL-R306C and NifA-E356K | 859 | 52 | 290 | 199 |
| pIM18 | NifL-R306C and NifA | 113 | 25 | 122 | 81 |
| pPR54 | NifL (147-519) and NifA | 2305 | 49 | 2,522 | 471 |
| pIM27 | NifL-R306C (147-519) and NifA | 116 | 19 | 119 | 118 |
In all cases, strain ET8000 (rbs lacZ::IS1 gyrA hutCck) contained the nifH-lacZ reporter plasmid pRT22 and the indicated plasmid expressing the listed Nif regulatory proteins.
Cultures were grown in NFDM medium (see text) with casein hydrolysate (200 μg/ml) as nitrogen source (−N).
Cultures were grown in NFDM medium (see text) with (NH4)2 SO4 (1mg/ml) as nitrogen source (+N).
To determine whether the inhibitory effect of NifL-R306C requires the N-terminal PAS domain of NifL, we introduced the nifL-R306C mutation into a truncated version of NifL, NifL (147-519), that does not exhibit the redox response but is competent to inhibit NifA activity in response to the fixed-nitrogen status in E. coli (Table 1, row 5) (32). As in the case of NifL-R306C, the truncated mutant protein, NifL-R306C (147-519), was able to inhibit NifA activity under all of the conditions tested (Table 1, row 6). Hence, the N-terminal PAS domain of NifL is not required for constitutive inhibition of NifA by the NifL-R306C substitution. We also tested whether inhibition of NifA activity by the mutant NifL protein was influenced by the PII-like signal transduction proteins, by introducing appropriate plasmids into a glnB, ntrC background (E. coli strain RT8000) that does not express either PII or GlnK (35). Unlike wild-type NifL, NifL-R306C was competent to inhibit NifA activity under all conditions in this background (data not shown) implying that the PII-regulatory proteins are not required for inhibition by NifL-R306C.
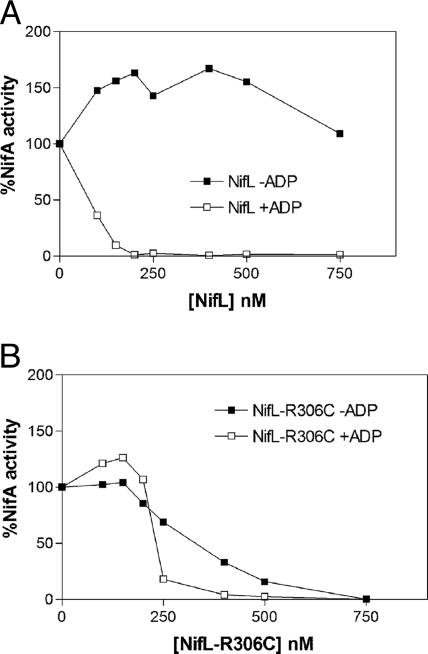
NifL-R306C Inhibits NifA Activity in the Absence of ADP. To investigate the characteristics of the nifL-R306C mutation further, we purified the mutant protein as an N-terminal hexahistidine fusion and analyzed its in vitro properties. The ability of NifL to inhibit NifA was determined by measuring the formation of open-promoter complexes by NifA at the nifH promoter, in the presence of σ54-RNA polymerase and integration host factor (IHF), with GTP as substrate for nucleotide hydrolysis by NifA. Gel-retardation assays were used to quantitate the formation of heparin-stable nucleoprotein complexes (16). The presence of adenosine nucleotides, particularly ADP, stabilizes complexes between NifL and NifA and increases the inhibitory activity of NifL (15, 16). Hence, wild-type Nhis6NifL inhibits NifA in the presence of ADP, whereas in the absence of this nucleotide, NifA is resistant to Nhis6NifL (Fig. 2A). By contrast, Nhis6NifL-R306C was able to inhibit NifA in the absence of ADP, although its inhibitory influence was stimulated by ADP (Fig. 2B).
Fig. 2.
Response of NifA to oxidized Nhis6NifL and Nhis6NifL-R306C. NifA activity was measured by the formation of open-promoter complexes, as described in Methods, and plotted relative to the extent of NifA activity in the absence of NifL. Each data point is the mean of at least two independent experiments. All assays contained 4 mM GTP as hydrolyzable nucleotide and 250 nM NifA (calculated as a dimer). (A) Response of NifA to the indicated concentration of Nhis6NifL (calculated as a tetramer) in the absence (▪) or presence (□) of 0.05 mM ADP. (B) Response of NifA to the indicated concentration of Nhis6NifL-R306C (calculated as a tetramer) in the absence (▪) or presence (□) of 0.05 mM ADP.
To determine whether the integrity of the nucleotide-binding (GHKL) domain is essential for NifL-R306C to inhibit NifA activity under nitrogen-fixing conditions, we used two substitutions of conserved residues in the GHKL domain of NifL (NifL-G455A and NifL-G480A) that reduce the binding of ADP substantially and, consequently, severely impair inhibition of NifA activity by NifL in vivo (S. Perry, N. Shearer, R. L., and R. D., unpublished data). Interestingly, the nifL-R306C mutation suppressed the regulatory defects exhibited by the mutations in the nucleotide-binding domain and, in particular, restored strong inhibition of NifA activity in response to the fixed-nitrogen status in vivo (Table 2). However, the double mutants apparently lost the ability conferred by NifLR306C to inhibit NifA fully under nitrogen-fixing conditions (compare Tables 1 and 2). These results support the observation that ADP is not essential for NifL-R306C to inhibit NifA in vitro. They also suggest that the integrity of the NifL nucleotide-binding domain is more important for inhibition of NifA by NifL-R306C under nitrogen-limiting conditions.
Table 2. Suppression of mutations in the nucleotide-binding domain of NifL by NifL-R306C.
| β-Galactosidase activity, Miller units
|
|||||
|---|---|---|---|---|---|
| Anaerobic
|
Aerobic
|
||||
| Plasmid | Proteins | −N | +N | −N | +N |
| pNLG455 | NifL-G455A and NifA | 19,623 | 11,441 | 31,054 | 14,491 |
| pIM23 | NifL-R306C, G455A, and NifA | 1,619 | 17 | 486 | 106 |
| pNLG480 | NifL-G480A and NifA | 25,538 | 20,165 | 31,335 | 12,977 |
| pIM25 | NifL-R306C, G480A, and NifA | 6,315 | 18 | 1,417 | 28 |
Assay conditions were identical to those shown in Table 1.
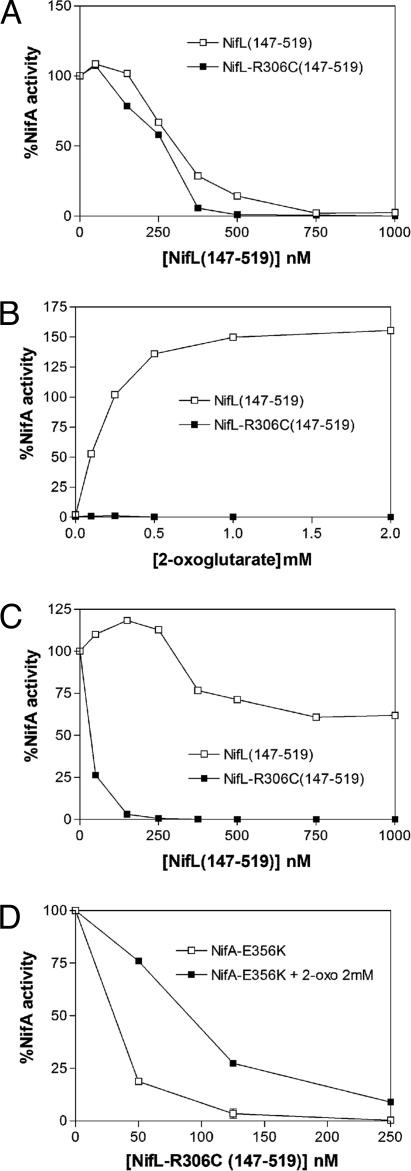
NifL R306C Overrides the Effect of 2-Oxoglutarate on NifA Activity. Binding of 2-oxoglutarate to the GAF domain of NifA relieves inhibition by the reduced ADP-bound form of NifL, enabling NifA to escape from inhibition by NifL under conditions appropriate for nitrogen fixation (29, 30). However, the allosteric effect of 2-oxoglutarate on NifA activity is overridden under oxidizing or excess fixed-nitrogen conditions, ensuring that NifA is inhibited under adverse environmental conditions. We considered the possibility that the nifL-R306C mutation locks NifL in a form that prevents the response of NifA to 2-oxoglutarate under conditions appropriate for nitrogen fixation. Because oxidized NifL overrides the effect of 2-oxoglutarate on NifA, we made use of the truncated version of NifL, NifL (147-519), which lacks the PAS domain and does not exhibit a redox response (32). Recall that nifL-R306C is competent to inhibit NifA under all conditions in the absence of the PAS domain in vivo (Table 1). In the presence of ADP, NifL (147-519) inhibits NifA, but this inhibition is relieved by the binding of 2-oxoglutarate to the GAF domain of NifA (19, 29, 30). As anticipated, Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519) were both effective in inhibiting open-promoter complex formation by NifA in the presence of ADP (Fig. 3A). However, whereas inhibition of NifA activity by Nhis6NifL (147-519) was alleviated in response to 2-oxoglutarate as demonstrated previously, NifA activity was not responsive to 2-oxoglutarate in the presence of Nhis6NifL-R306C (147-519), and inhibition was observed irrespective of the concentration of this ligand (Fig. 3B). Therefore, it seems likely that the ability of NifL-R306C to inhibit NifA in vivo under nitrogen-fixing conditions is due to its ability to override the influence of 2-oxoglutarate on NifA activity.
Fig. 3.
Response of NifA proteins to Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519). NifA activity was measured by the formation of open-promoter complexes and plotted relative to the extent of NifA activity in the absence of NifL. Each data point is the mean of at least two independent experiments. All assays contained 4 mM GTP as hydrolyzable nucleotide, 0.05 mM ADP, and 250 nM NifA (calculated as a dimer). (A) Response of NifA to the indicated concentration of Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519) (calculated as a dimer). (B) Response of NifA to 2-oxoglutarate. NifA activity is plotted relative to the extent of NifA activity in the absence of NifL and 2-oxoglutarate. Reactions contained 750 nM Nhis6NifL (147-519) (□) or 500 nM Nhis6NifL-R306C (147-519) (▪), and the concentration of 2-oxoglutarate is indicated on the x axis. (C) Response of NifA-E356K to the indicated concentration of Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519) (calculated as a dimer). (D) Response of NifA-E356K to the indicated Nhis6NifL-R306C (147-519) concentration in the absence (□) or presence (▪) of 2 mM 2-oxoglutarate.
As anticipated from the in vivo suppression data, Nhis6NifL-R306C (147-519) was competent to inhibit NifA-E356K activity in vitro in the presence of ADP, in contrast to Nhis6NifL (147-519), which was ineffective (Fig. 3C). NifA-E356K was apparently more sensitive to Nhis6NifL-R306C (147-519) than wild-type NifA at equivalent protein concentrations (compare Fig. 3 A and C). We were also interested in determining whether the NifA-E356K mutant was able to respond to 2-oxoglutarate in the presence of NifL-R306C. We observed that inhibition of NifA-E356K activity was responsive to 2-oxoglutarate at low Nhis6NifL-R306C (147-519) concentrations (Fig. 3D), in contrast to wild-type NifA (Fig. 3B), which exhibited no response to this ligand, irrespective of the Nhis6NifL-R306C (147-519) concentration (data not shown). These differences may account for the in vivo properties of the nifLR306C, nifA-E356K double mutant, which retains some NifA activity under anaerobic nitrogen-limiting conditions in E. coli (Table 1, second and third rows). This property is apparent also in A. vinelandii because, in contrast to the single nifL-R306C mutant, which is unable to fix nitrogen, the nifL-R306C, nifA-E356K double mutant is Nif+ (Fig. 6).
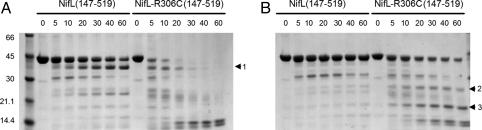
NifL-R306C Exhibits a Conformational Change. Because NifL-R306C is not an allele-specific suppressor of NifA-E356K and is a potent inhibitor of wild-type NifA, we rationalized that this NifL mutation may induce a conformational change that locks NifL in a form that is competent to inhibit wild-type NifA even in the presence of 2-oxoglutarate. Limited trypsin proteolysis was used to probe the conformations of Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519). Binding of ADP to NifL protects the C-terminal domain from proteolytic digestion (32). In the absence of ADP, Nhis6NifL (147-519) exhibited a major cleavage product (marked 1 in Fig. 4A) that results from trypsin cleavage in the C terminus of NifL (32). This band is not apparent in the presence of ADP, which protects the C-terminal domain from proteolysis and increases the stability of Nhis6NifL (147-519) significantly (Fig. 4B). The proteolysis pattern of Nhis6NifL-R306C (147-519) was surprisingly different from that of Nhis6NifL (147-519) and was considerably more sensitive to trypsin digestion in the absence of ADP (Fig. 4A). Although ADP decreased the rate of digestion of Nhis6NifL-R306C (147-519), enhanced sensitivity to trypsin was evident also under these conditions in comparison with Nhis6NifL (147-519) (Fig. 4B). Two bands (marked 2 and 3) corresponding to C-terminal fragments (32) were more evident after cleavage of Nhis6NifL-R306C (147-519) in the presence of ADP. These results suggest that the R306C protein remains responsive to the binding of ADP, and the mutation significantly alters the conformation of NifL.
Fig. 4.
Limited trypsin proteolysis of Nhis6NifL (147-519) and Nhis6NifL-R306C (147-519) in the absence (A) or presence (B) of 2 mM ADP. We incubated 2 μM protein with trypsin (weight ratio, 100:1) for the times (in min) indicated above each lane. Arrowheads indicate the proteolysis products described in the text.
Influence of nifL-R306C on Other nifA Mutations. Although nifLR306C was isolated originally as a suppressor of the nifL- resistant nifA allele nifA-E356K, it seems likely from our experiments that the mutation influences the inhibitory properties of NifL rather than the specificity of the NifL-NifA interaction. Therefore, it was of interest to check whether nifL-R306C is able to suppress other mutations that render NifA resistant to NifL. Such mutations have been isolated in both the N-terminal GAF and AAA+ domains of NifA (Fig. 1) (31). NifL-R306C inhibited the AAA+ domain substitutions NifA-A249V and NifA-Y254N, and it also partially suppressed the NifL-resistant phenotype of NifA-E394K in the in vivo reporter assay in E. coli. As in the case of wild-type NifA, NifL-R306C inhibited the activity of these mutant NifA proteins under conditions appropriate for nitrogen fixation. However, the GAF domain substitutions NifA-L120P and NifA-R155A and the AAA+ domain substitution NifAE230K were not suppressed by nifL-R306C in vivo (see Table 3, which is published as supporting information on the PNAS web site). Clearly, only some of the NifL-resistant NifA mutants are susceptible to inhibition as a consequence of the altered conformation of NifL-R306C.
R306 Is Essential for the Normal Function of NifL. To examine whether the phenotype of nifL-R306C is specific to the arginine-to-cysteine substitution, we constructed a series of substitutions at position 306 by site-directed mutagenesis. Strikingly, all tested substitutions had very similar in vivo phenotypes to that of the cysteine substitution, resulting in constitutive inhibition of NifA activity in E. coli irrespective of whether polar, hydrophobic, or charged side chains were introduced (see Fig. 7, which is published as supporting information on the PNAS web site). Western blotting indicated that the mutant proteins accumulated to the same extent as wild-type NifL under the conditions used for the in vivo assays. These results clearly demonstrate that the arginine residue at position 306 is essential for appropriate regulation of NifA activity by NifL.
Discussion
The regulatory dialogue between the NifL and NifA proteins involves reciprocal conformational changes in which the binding of 2-oxoglutarate to the GAF domain of the transcriptional activator NifA plays a major role in modulating the response to the NifL antiactivator (29, 30) (Fig. 5). In this study, we have characterized a substitution in NifL that constitutively inhibits NifA, giving rise to a Nif- phenotype. The mutant protein is apparently locked in an antiactivation conformation so that NifL inhibits NifA under conditions that are appropriate for nitrogen fixation, even when 2-oxoglutarate is available. This substitution may enable NifL to interact with the altered conformation of NifA that is induced by the binding of 2-oxoglutarate, or alternatively, the mutant protein may lock NifA in an inactive form that is unable to undergo the conformational change normally induced by binding of this ligand.
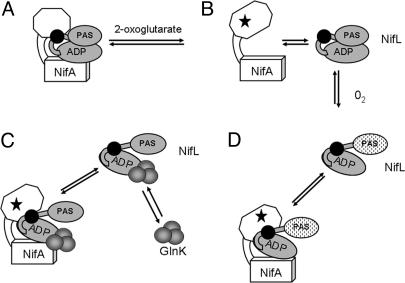
Fig. 5.
Model for regulation of NifA activity by NifL. The PAS-and ADP-binding (GHKL) domains of NifL are shown in gray, and the H-box region is represented by a black circle. The GAF and AAA+ domains of NifA are indicated by an open octagon and cube, respectively. For simplicity, the interacting partners are shown as monomers. (A) In the absence of 2-oxoglutarate, both the reduced and oxidized forms of NifL inhibit the activity of NifA, provided that adenosine nucleotide is bound to NifL (8, 15, 19). (B) Binding of 2-oxoglutarate to the GAF domain of NifA (indicated by stars) induces a conformational change that releases inhibition by the reduced form of NifL, enabling NifA to activate transcription (29, 30). (C) Oxidation of the flavin in the PAS domain of NifL (indicated by stippled ovals) causes a conformational change that enables NifL to inhibit NifA in the presence of 2-oxoglutarate. (D) Binding of the signal transduction protein GlnK (indicated by gray circles) to the C-terminal domain of NifL, enables the reduced form of NifL to interact with NifA, in the presence of 2-oxoglutarate. It is possible that the GlnK-NifL interaction promotes a conformational change similar to that induced by the oxidation of NifL. The GlnK-NifL-NifA ternary complex is formed under nitrogen-excess conditions when GlnK is primarily in the nonuridylylated form. Uridylylation of GlnK under nitrogen-limiting conditions prevents this interaction (17-19). We infer that the R306C substitution locks NifL in a conformation that is analogous to that shown in C and D, so that it is competent to inhibit NifA irrespective of other signals.
The R306C substitution is located in the H-box region of NifL adjacent to a histidine residue that corresponds to the autophosphorylation site of the HPKs. However, this histidine residue is functionally redundant, and NifL does not seem to exhibit autokinase activity (26). Nevertheless, this region of NifL may have structural similarity to the dimerization and phosphotransfer domain (domain A) of the HPKs (24). Although, it is conceivable that the R306C substitution could influence dimerization of NifL, both NifL (147-519) and NifL-R306C (147-519) chromatographed as dimers on gel filtration (data not shown), and therefore, we favor the hypothesis that this substitution influences protein conformation rather than oligomerization. Clearly, the arginine at position 306 is essential for the normal function of A. vinelandii NifL because other substitutions at this position give identical phenotypes to that of R306C. An arginine residue at this position is also observed in Pseudomonas stutzeri NifL but not in other NifL proteins, implying that the function of this residue is context-dependent. We infer that arginine 306 is critical for the propagation of signals perceived by A. vinelandii NifL in response to the redox and fixed-nitrogen status and also that it is required for a conformational switch that normally inactivates the inhibitory function of NifL under conditions that are appropriate for nitrogen fixation (Fig. 5).
The altered behavior of R306C with respect to the ADP requirement for inhibition of NifA activity provides a potential clue to the nature of the conformational change elicited by this mutation. Unlike wild-type NifL, the mutant protein is able to inhibit NifA in the absence of ADP. This property implies that the NifL-R306C-NifA complex is more stable in the absence of adenosine nucleotides than the wild-type complex, congruent with the finding that the NifL-R306C mutation is competent to suppress substitutions in the nucleotide-binding GHKL domain in vivo. However, the presence of the GHKL domain is required for NifL-R306C to inhibit NifA, and we observe that ADP is necessary for inhibition of NifA activity by the oxidized form of NifL-R306C, when 2-oxoglutarate is present (data not shown). These results are consistent with the in vivo suppression data (Table 2), which demonstrates that NifLR306C is less effective in suppressing nucleotide-binding mutants under nitrogen-limiting conditions when the intracellular concentration of 2-oxoglutarate increases. Overall these results suggest that the binding of ADP to the GHKL domain contributes to the conformational changes that are required to inhibit NifA in the presence of 2-oxoglutarate.
In the HPKs, interactions between the dimerization and phosphotransfer domain (domain A) and the catalytic GHKL domain (domain B) modulate kinase and phosphatase activity (10, 13). For example, suppressors of mutations in the G2 region of the nucleotide-binding domain of the histidine kinase EnvZ are located in domain A, suggesting a model in which residues in the phosphotransfer domain interact functionally with the ATP-binding face of the GHKL domain (domain B) to regulate enzymatic function (37, 38). By analogy, the R306C mutation may alter the topological relationship between the H-box region and the GHKL domain of NifL, thus enabling inhibition of NifA, even under conditions that are appropriate for nitrogen fixation, when 2-oxoglutarate levels are elevated. Thus, although signal transmission by NifL does not involve phosphoryl transfer reactions, the interdomain movements that are necessary to modulate the inhibitory function of NifL may be similar to those required to control the catalytic function of the HPKs.
Supplementary Material
Acknowledgments
I.M-A. was supported by a fellowship from the Spanish Ministerio de Educación y Ciencia. R.L and R.D. were supported by a grant from the U.K. Biotechnology and Biological Sciences Research Council.
Author Contributions: R.D. designed research; I.M.-A. and R.L. performed research; I.M.-A., R.L., and R.D. analyzed data; and I.M.-A. and R.D. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HPK, histidine protein kinase; PAS, Per-Arnt-Sim.
References
- 1.Morett, E. & Segovia, L. (1993) J. Bacteriol. 175, 6067-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Studholme, D. J. & Dixon, R. (2003) J. Bacteriol. 185, 1757-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Argudo, I., Little, R., Shearer, N., Johnson, P. & Dixon, R. (2004) J. Bacteriol. 186, 601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, D. K., Narberhaus, F. & Kustu, S. (1994) Proc. Natl. Acad. Sci. USA 91, 103-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz, R. A., Klopprogge, K. & Grabbe, R. (2002) J. Mol. Microbiol. Biotechnol. 4, 235-242. [PubMed] [Google Scholar]
- 6.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhulin, I. B., Taylor, B. L. & Dixon, R. (1997) Trends Biochem. Sci. 22, 331-333. [DOI] [PubMed] [Google Scholar]
- 8.Hill, S., Austin, S., Eydmann, T., Jones, T. & Dixon, R. (1996) Proc. Natl. Acad. Sci. USA 93, 2143-2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macheroux, P., Hill, S., Austin, S., Eydmann, T., Jones, T., Kim, S.-O., Poole, R. & Dixon, R. (1998) Biochem. J. 332, 413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolanin, P. M., Thomason, P. A. & Stock, J. B. (2002) Genome Biol. 3, 3013.1-3013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanco, G., Drummond, M., Woodley, P. & Kennedy, C. (1993) Mol. Microbiol. 9, 869-880. [DOI] [PubMed] [Google Scholar]
- 12.Dutta, R. & Inouye, M. (2000) Trends Biochem. Sci. 25, 24-28. [DOI] [PubMed] [Google Scholar]
- 13.West, A. H. & Stock, A. M. (2001) Trends Biochem. Sci. 26, 369-376. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka, T., Saha, S. K., Tomomori, C., Ishima, R., Liu, D., Tong, K. I., Park, H., Dutta, R., Qin, L., Swindells, M. B., et al. (1998) Nature 396, 88-92. [DOI] [PubMed] [Google Scholar]
- 15.Money, T., Jones, T., Dixon, R. & Austin, S. (1999) J. Bacteriol. 181, 4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eydmann, T., Söderbäck, E., Jones, T., Hill, S., Austin, S. & Dixon, R. (1995) J. Bacteriol. 177, 1186-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rudnick, P., Kunz, C., Gunatilaka, M. K., Hines, E. R. & Kennedy, C. (2002) J. Bacteriol. 184, 812-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Little, R., Colombo, V., Leech, A. & Dixon, R. (2002) J. Biol. Chem. 277, 15472-15481. [DOI] [PubMed] [Google Scholar]
- 19.Little, R., Reyes-Ramirez, F., Zhang, Y., van Heeswijk, W. C. & Dixon, R. (2000) EMBO J. 19, 6041-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arcondéguy, T., Jack, R. & Merrick, M. (2001) Microbiol. Mol. Biol. Rev. 65, 80-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ninfa, A. & Atkinson, M. (2000) Trends Microbiol. 8, 172-179. [DOI] [PubMed] [Google Scholar]
- 22.Meletzus, D., Rudnick, P., Doetsch, N., Green, A. & Kennedy, C. (1998) J. Bacteriol. 180, 3260-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Colnaghi, R., Rudnick, P., He, L., Green, A., Yan, D., Larson, E. & Kennedy, C. (2001) Microbiology 147, 1267-1276. [DOI] [PubMed] [Google Scholar]
- 24.Tomomori, C., Tanaka, T., Dutta, R., Park, H., Saha, S. K., Zhu, Y., Ishima, R., Liu, D., Tong, K. I., Kurokawa, H., et al. (1999) Nat. Struct. Biol 6, 729-734. [DOI] [PubMed] [Google Scholar]
- 25.Castelli, M. E., Cauerhff, A., Amongero, M., Soncini, F. C. & Vescovi, E. G. (2003) J. Biol. Chem. 278, 23579-23585. [DOI] [PubMed] [Google Scholar]
- 26.Woodley, P. & Drummond, M. (1994) Mol. Microbiol. 13, 619-626. [DOI] [PubMed] [Google Scholar]
- 27.Lee, H.-S., Narberhaus, F. & Kustu, S. (1993) J. Bacteriol. 175, 7683-7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Austin, S., Buck, M., Cannon, W., Eydmann, T. & Dixon, R. (1994) J. Bacteriol. 176, 3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Argudo, I., Little, R. & Dixon, R. (2004) Mol. Microbiol. 52, 1731-1744. [DOI] [PubMed] [Google Scholar]
- 30.Little, R. & Dixon, R. (2003) J. Biol. Chem. 278, 28711-28718. [DOI] [PubMed] [Google Scholar]
- 31.Reyes-Ramirez, F., Little, R. & Dixon, R. (2002) J. Bacteriol. 184, 6777-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Söderbäck, E., Reyes-Ramirez, F., Eydmann T, Austin, S., Hill, S. & Dixon, R. (1998) Mol. Microbiol. 28, 179-192. [DOI] [PubMed] [Google Scholar]
- 33.Brewin, B., Woodley, P. & Drummond, M. (1999) J. Bacteriol. 181, 7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouncey, N. J., Mitchenall, L. A. & Pau, R. N. (1995) J. Bacteriol. 177, 5294-5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reyes-Ramirez, F., Little, R. & Dixon, R. (2001) J. Bacteriol. 183, 3076-3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lei, S., Pulakat, L. & Gavini, N. (1999) J. Bacteriol. 181, 6535-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, Y. & Inouye, M. (2002) Mol. Microbiol. 45, 653-663. [DOI] [PubMed] [Google Scholar]
- 38.Cai, S. J., Khorchid, A., Ikura, M. & Inouye, M. (2003) J. Mol. Biol. 328, 409-418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.