Abstract
RecA- and RecBC-catalyzed repair in eubacteria assembles chromosomes fragmented by double-strand breaks. We propose that recA mutants, being unable to repair fragmented chromosomes, depend on various strategies designed to avoid chromosomal fragmentation. To identify chromosomal fragmentation-avoidance strategies, we screened for Escherichia coli mutants synthetically inhibited in combination with recA inactivation by identifying clones unable to lose a plasmid carrying the recA+ gene. Using this screen, we have isolated several RecA-dependent mutants and assigned them to three distinct areas of metabolism. The tdk and rdgB mutants affect synthesis of DNA precursors. The fur, ubiE, and ubiH mutants are likely to have increased levels of reactive oxygen species. The seqA, topA mutants and an insertion in smtA perturbing the downstream mukFEB genes affect nucleoid administration. All isolated mutants show varying degree of SOS induction, indicating elevated levels of chromosomal lesions. As predicted, mutants in rdgB, seqA, smtA, topA, and fur show increased levels of chromosomal fragmentation in recBC mutant conditions. Future characterization of these RecA-dependent mutants will define mechanisms of chromosomal fragmentation avoidance.
Chromosomal fragmentation due to double-strand DNA breaks and disintegrated replication forks is a major contributor to genome instability in all organisms (1, 2). The two major pathways used by eukaryotic cells to repair fragmented chromosomes are nonhomologous end joining and homologous recombination (3). Nonhomologous end joining is an imprecise repair, frequently involving loss of genetic information, but it is nevertheless an efficient way to repair double-strand breaks in cells of higher eukaryotes (4). However, disintegrated replication forks, having a single double-strand end, cannot be reassembled by nonhomologous end joining and require error-free recombinational repair (5, 6). The importance of recombinational repair in higher eukaryotes is illustrated by the inviability of recombinational repair mutants in mice (7) and by the rapid death of recombinational repair-defective vertebrate cells due to chromosomal fragmentation (8). In the model eubacterium Escherichia coli, recombinational repair of fragmented chromosomes is catalyzed by the RecA and RecBCD enzymes (9). RecBCD is an exonuclease/helicase that prepares the double-strand DNA ends of the break for RecA polymerization (10). RecA filament catalyzes homologous strand exchange of the broken DNA duplex with an intact sister duplex, thus creating an opportunity for double-strand break repair (11).
In recA or recBC mutants of E. coli, double-strand DNA breaks are not repaired (12) and cause chromosomal loss and cell death (13, 14). However, recA mutant E. coli strains are still 50% viable (15, 16), which indicates that the chromosomal fragmentation is not a frequent event in E. coli and suggests the existence of strategies designed to avoid chromosomal fragmentation. Inactivation of one of these hypothetical avoidance activities should make the corresponding mutant dependent on recombinational repair (”synthetically inhibited” in combination with recA inactivation), because both the avoidance of chromosomal fragmentation and the subsequent recombinational repair are not available in such a double mutant. Moreover, such RecA-dependent mutants are predicted to have elevated levels of chromosomal fragmentation.
There are three knockout mutants in E. coli that cause RecA dependence. The dam mutants are deficient in GATC site-specific DNA adenine methylase (17). Replication-caused transient hemimethylation of GATC sites is used by E. coli to identify the old DNA strand as the proper template for mismatch repair, whereas lack of GATC methylation in dam mutants results in double-strand DNA breaks due to a disoriented mismatch repair (18). The rdgB mutants depend on RecA and degrade their chromosomal DNA in recA- conditions (19), because they are deficient in ITP/XTPase, an enzyme believed to intercept the improper DNA precursor dITP, thus preventing hypoxanthine incorporation into DNA (20). In the absence of RdgB, hypoxanthine is proposed to occasionally incorporate into DNA in place of guanine, its subsequent excision by EndoV translating into chromosomal fragmentation (20). Last, fur mutants are deficient in the repression of iron assimilation and, as a result, experience iron overload (21), which is postulated to lead to increased oxidative DNA damage and RecA dependence in aerobic conditions (22). It is not known whether fur mutants suffer from chromosomal fragmentation.
The dam, rdgB, and fur mutants identify three potential strategies to avoid chromosomal fragmentation: (i) guidance of proteins that can damage DNA directly (the way Dam methylation guides mismatch-repair enzymes); (ii) quality control in the DNA precursor metabolism (rdgB); and (iii) reduction in the level of DNA-damaging species (fur). To find other strategies of chromosomal fragmentation avoidance or other activities that use the known strategies, we designed an approach to systematically search for RecA-dependent mutants in E. coli. We then tested the isolated mutants for elevated levels of chromosomal fragmentation.
Materials and Methods
Media, growth conditions, bacterial strains and plasmids, insertional mutagenesis, color screen, and mutant sequencing are described in Supporting Text, which is published as supporting information on the PNAS web site; see also Tables 1 and 2, which are published as supporting information on the PNAS web site.
SOS Induction. The level of SOS induction due to a particular mutation was measured in a derivative of AK43 (into which the mutation was introduced by P1 transduction to kanamycin resistance) according to the protocol of Miller (23) with the following modifications: (i) cells were grown in LB at 42°C without antibiotics; (ii) 200 μl of the culture with OD600 between 0.5 and 0.8 was combined with 2 ml of Z buffer (100 mM Na Phos, pH 7.0/10 mM KCl/1 mM MgSO4/50 mM 2-mercaptoethanol) containing 0.00125% SDS and 25 μl of chloroform; (iii) 60 μl of 4 mg/ml O-nitrophenyl β-d-galactoside in Z buffer was added to the opened cells; and (iv) color development was for 10 min with shaking at 28°C.
Quantification of Chromosomal Fragmentation by Pulsed-Field Gel Electrophoresis. Overnight 2-ml LB cultures were shaken at 22°C. In the morning, the cultures were diluted to OD600 0.02-0.06 (for further growth at 22°C) or to OD600 0.001-0.02 (for further growth at 37°C) into 2 ml of LB medium supplemented with 10 μCi/ml [32P]orthophosphoric acid (1 Ci = 37 GBq). Variations in dilutions accommodate slower growth of some strains. All strains were grown at 22°C for 1 h, and then 37°C samples were shifted to 37°C for 4 h. The 22°C samples were left at 22°C for another 5 h of growth. Growing cultures (OD600 for 37°C samples varied from 0.7 to 1.1; for 22°C samples, variation was from 0.4 to 0.6) were normalized to OD600 0.35, and 0.5 ml of the normalized cultures was pelleted and washed once with 1 ml of TE buffer (10 mM Tris/1 mM EDTA, pH 8.0) to remove unincorporated radioactivity. Plug preparation was as described (20). Half of the plug was inserted in 1% agarose gel on 0.5 × TBE buffer (89 mM Tris/89 mM boric acid/2.0 mM EDTA, pH 7.8) and run for 24 h at 6 V/cm with a switch time ramp 60-120 seconds, in CHEF-DRII Pulsed-Field Gel Electrophoresis (BioRad). The portion of the gel containing the Yeast Chromosome PFG Marker (New England Biolabs) was stained in 1 μg/ml ethidium bromide solution to estimate the size of fragmented chromosomes. The portion of the gel with radioactive samples was vacuum-dried for 2 h at 80°C to 3MM Whatman paper and directly scanned by PhosphorImager (FujiFilm FLA-3000, Fuji). The fraction of chromosomal DNA escaping from the well into the gel was calculated by dividing the counts found in the gel (starting at a position halfway between the center of the well and the center of the compression zone) by the total amount found in the whole lane, including the well.
Results
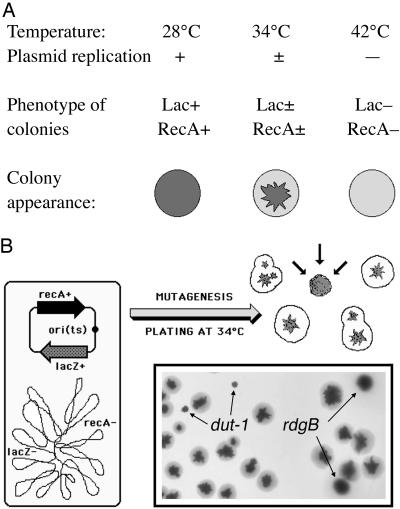
Color Screen. To search for chromosomal fragmentation avoidance activities, we set up a screen for recombination-dependent mutants in E. coli consisting of a lacZ recA mutant strain, which harbors a lacZ+ recA+ plasmid with a temperature-sensitive origin of replication. When plated on MacConkey-lactose agar, the resulting strain (S295) is RecA+ and forms purple colonies at 28°C, whereas at 42°C, it is RecA- and forms pale colonies (Fig. 1A). To screen for recA-dependent mutants with this system, we grew colonies at 34°C, the temperature that causes cells to lose the plasmid at a low rate, so that the colonies are sectored (Fig. 1). However, the colonies of a RecA-dependent mutant will be unable to form sectors; because such a mutant cannot grow without recA+ gene, growth of a mutant cell in the colony is inhibited once the cell loses the plasmid. Therefore, the colonies formed by RecA-dependent mutants are expected to be solidly purple and smaller (Fig. 1B).
Fig. 1.
The color screen for recA-dependent mutants. The screen looks for the inability to lose a temperature-sensitive plasmid carrying the recA+ gene. (A) The properties of the strain S295 [lacZ recA p(ori-Ts)-lacZ+-recA+] at three temperatures, plated on MacConkey-lactose agar. S295 is lacZ+ recA+ at 28°C, lacZ recA mutant at 42°C, and a mixture of lacZ+ recA+ and lacZ recA mutant cells (sectoring colonies) at 34°C. (B) The scheme of the experimental strain and the expected colony phenotype of a recA-dependent mutant (converging arrows). (Inset) Test plating of the original S295 strain and its two derivatives carrying mutations known to confer recA dependence, S296 (S295 rdgB3) and S297 (S295 dut-1), at 34°C. Sectoring colonies are those of the original strain, the small nonsectoring colonies are dut-1 mutants, and the midsize nonsectoring colonies are rdgB3 mutants.
To verify this expectation, we have constructed two derivatives of the original strain, carrying known mutations that make E. coli dependent on RecA, rdgB3 (19) and dut-1 (24). We found that both rdgB3 (S296) and dut-1 (S297) derivatives of the original strain (i) form small flat colonies (rdgB) or do not form colonies (dut) at 42°C on LB agar (not shown) or (ii) form solidly colored colonies at 34°C on MacConkey-lactose agar, of either intermediate size (rdgB3) or small size (dut-1). We then mixed the wild-type, rdgB and dut-1 mutant cells at a 10:1:1 ratio and plated them on MacConkey-lactose plates, aiming to obtain 300 colonies per plate. After 48 h of incubation at 34°C, most of the colonies on the plates were sectored, but there were also a few solidly colored medium-size or small colonies (Fig. 1B Inset). Medium-size solidly colored colonies proved to be resistant to kanamycin, indicating the rdgB3::kan mutation, whereas small solidly colored colonies proved to be resistant to tetracycline [the dut-1 allele is linked to a nearby transposon 10 (Tn10)], demonstrating that the screen works.
To facilitate mutant identification and mutant phenotype interpretation, we used only insertional mutagenesis (see Supporting Text). After the initial mutant identification, the steps of the screen include (i) confirmation of the inability to sector on MacConkey-lactose agar; (ii) test for the ability to lose the plasmid and inhibition at 42°C; and (iii) transduction into AB1157 background to verify RecA dependence. We executed seven runs of the color screen with the following average statistics per run: >13,000 kanamycin-resistant colonies screened; 43 primary candidates checked for the inability to form sectors (Fig. 5 Left, which is published as supporting information on the PNAS web site); 38 secondary candidates tested for the ability to lose plasmid and temperature sensitivity; 12 mutants transduced into the AB1157 background; and two mutants confirmed as RecA-dependent in the AB1157 background (Fig. 5, the right four columns of spots). We kept the icdA mutant, which failed to confirm as RecA-dependent in the AB1157 background, as a negative control through our subsequent analyses. All of the original mutants in S295 were proven by blot hybridization to carry single inserts (data not shown).
A summary of the isolated mutants is given in Table 3, which is published as supporting information on the PNAS web site, whereas the position and orientation of the inserts relative to the chromosomal region are shown in Fig. 6, which is published as supporting information on the PNAS web site. We isolated seven independent insertions in tdk, two independent insertions in rdgB, and one insertion in each of the following genes: fur, seqA, smtA, topA, ubiE, and ubiH (Figs. 5 and 6). Our isolation of two independent rdgB inactivations showed that our screen for RecA-dependent mutants works, and that the number of loci, inactivation of which makes E. coli cells RecA-dependent, is limited. However, because no expected dam mutants turned up in the screen yet, the screen is not saturated.
SOS Induction in RecA-Dependent Mutants. Because recombinational repair in E. coli is the major repair system to mend chromosomal lesions, RecA-dependent mutants should have higher levels of chromosomal lesions. SOS response is a reaction of bacterial cells to chromosomal lesions and includes increased expression of ≈30 genes and decreased expression of 20 more genes (25, 26). The genes induced during the SOS response are repressed in the absence of chromosomal damage by the LexA repressor bound to their promoters (27). LexA autocleavage in the presence of RecA, activated by chromosomal lesions, leads to the derepression of the SOS regulon (28). A reporter gene under the control of a LexA-regulatable promoter identifies mutants elevated for SOS expression, presumably because they experience higher levels of chromosomal lesions.
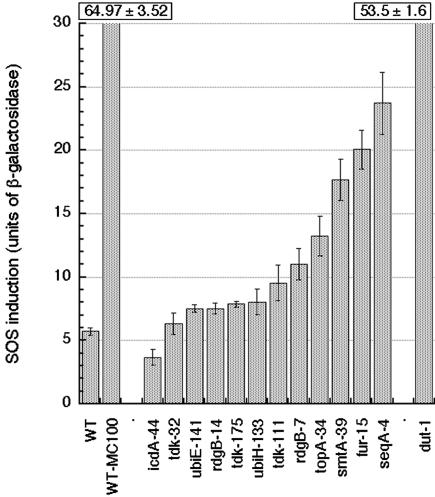
We introduced the isolated RecA-dependent mutants into a strain containing the lacZ+ gene under the control of an SOS-inducible sulA promoter (29). In a standard assay for β-galactosidase (23), the original strain shows six “units,” whereas the same strain incubated with 0.1 μg/ml Mitomycin C (a DNA crosslinking agent, which at this low dose induces a mild SOS response) shows 10 times higher levels of β-galactosidase (Fig. 2). A dut-1 mutant, known to be induced for SOS (24), serves as a positive control with >50 units. The icdA (negative control) and tdk-32 mutants show no SOS induction. Other RecA-dependent mutants show various levels of SOS induction (all statistically significant), from 1.3-times-induced ubiE-141 and rdgB-14 mutants to a 3.9-times-induced seqA mutant (Fig. 2). Thus, the SOS test indicates an increase in chromosomal lesions in RecA-dependent mutants.
Fig. 2.
SOS induction in RecA-dependent mutants. Mutations were introduced into the strain containing a sulA::lacZ+ construct (AK43), and the level of β-galactosidase was determined in cells growing in LB at 42°C (conditions in which RecA dependence was originally observed). Values are averages of three to six independent measurements ± SE. WT, AK43; WT-MC100, AK43 grown in the presence of 100 ng/ml mitomycin C.
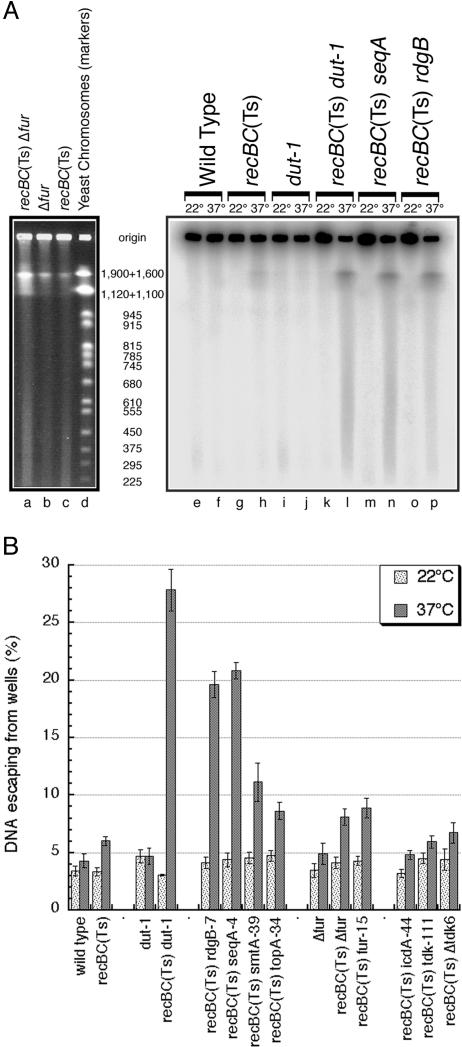
RecA-Dependent Mutants Suffer Chromosomal Fragmentation. To test our prediction that all RecA-dependent mutants suffer from increased chromosomal fragmentation, we introduced the isolated mutations into recBC(Ts) strain SK129, defective at 37°C in both the RecBCD-promoted linear DNA degradation and RecBC-catalyzed recombinational repair of double-strand breaks (30). The isolated RecA-dependent mutants proved to be inhibited at 42°C when introduced into the recBC(Ts) strain (data not shown), indicating their dependence on the RecBCD enzyme, as is generally the case with known RecA-dependent mutants (20, 24, 31). We measured the degree of chromosomal fragmentation in the double mutants using pulsed-field gel electrophoresis (Fig. 3). Intact chromosomal DNA stays in the wells during pulsed-field gels, whereas linear subchromosomal fragments migrate into the gel according to their size (32). The chromosomal DNA in all tested strains show fragmentation levels of 3-5% at 22°C (Fig. 3B). After 4 h at 37°C, the fragmentation is not significantly increased in wild-type cells or dut-1 and Δfur mutants but is increased to 6% in the recBC(Ts) mutant (Fig. 3 A, lanes f, h, and j, and B), setting up the background for the double mutants. The dut-1 recBC(Ts) double mutant offers a positive control for massive chromosomal fragmentation (24) with ≈30% chromosomal DNA migrating from wells after only 4 h at 37°C (Fig. 3 A, lane l, and B). We found that the rdgB, seqA, smtA, topA, and fur mutants, if combined with the recBC(Ts) defect, leach chromosomal DNA into the gel at 37°C, revealing levels of chromosomal fragmentation ranging from 9 to 21% (Fig. 3 A and B). On the other hand, the icdA mutant and two tdk mutants showed no increase in chromosomal fragmentation over the recBC(Ts) level (Fig. 3B). We conclude that the color screen for RecA dependence identifies mutants whose chromosomes suffer fragmentation during regular growth, but not all RecA-dependent mutants exhibit chromosomal fragmentation.
Fig. 3.
RecA-dependent mutants reveal chromosomal fragmentation when combined with a recBC(Ts) defect. (A) Pulsed-field gel electrophoresis detection of chromosomal DNA fragmentation. The relevant genotypes of strains are shown above the gels. DNA loading was equalized by OD600 of the original cultures. The length in kbp of the yeast chromosomes (markers) is indicated between Left and Right. The bands in the chromosomal smears with an apparent molecular weight of 1.6-1.9 mbp correspond to the “compression zone,” typical of pulsed-field gels. (Left) Ethidium bromide-stained portion of a representative gel, showing migration of the chromosomal smear relative to the molecular weight markers. Strains were grown for 2 h at 42°C. (Right) Direct radioactivity scanning of 32P-labeled chromosomal DNA in another representative gel. The cells were grown either at 22°C for 6 hours (total) or at 37°C for 4 hours. Δfur, AK108; recBC(Ts) Δfur, AK110. (B) A plot of the values (averages of 5-10 independent measurements ± SE) for chromosomal fragmentation in various mutant and control strains, at 22°C versus 37°C. Chromosomal fragmentation is measured as DNA that escaped from the well, expressed as percent of total DNA.
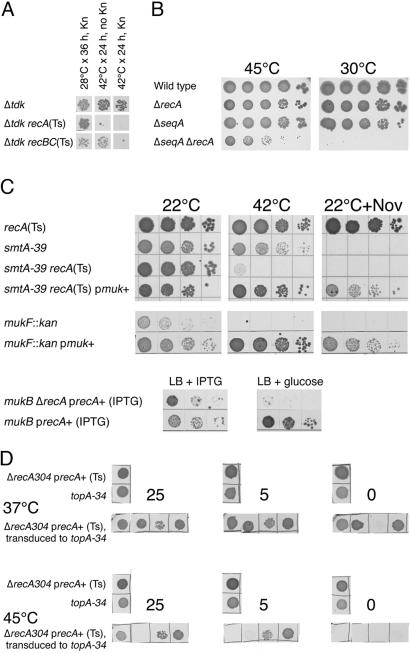
Further Characterization of tdk, seqA, smtA, and topA Mutants. To further confirm and characterize some of our RecA-dependent mutants, we constructed precise deletions of the corresponding genes or used available knockout alleles. A precise deletion of tdk made cells dependent on RecA (Fig. 4A). Curiously, this Δtdk::kan mutant depends on RecBC only in the presence of kanamycin (Fig. 4A). We have also constructed a precise seqA deletion, which rendered the strain harboring it somewhat cold-sensitive. Although we were able to construct a ΔseqA ΔrecA mutant at 45°C, which is close to the temperature limit for E. coli, the strain was barely viable at this temperature and unable to form colonies below 34°C (Fig. 4B).
Fig. 4.
Characterization of RecA dependence of tdk, seqA, smtA, and topA mutants. Mid-log cultures of various mutant combinations grown in appropriate conditions were serially diluted and spot-tested in conditions nonper-missive for either conditional mutations or plasmid replicons. The strains, plasmids, and conditions are as follows: (A) Δtdk, AK141; Δtdk recA(Ts), AK142; Δtdk recBC(Ts), AK143; and Kn, 50 μg/ml kanamycin. (B) Wild-type, AB1157; ΔrecA, JC10287; ΔseqA, ER20; and ΔseqA ΔrecA, ER23. Plates were developed for 72 h. (C) recA(Ts), JC9941; smtA-39 recA(Ts), L-143; pmuk+, pEAK25; mukF::kan, L-138; mukB ΔrecA, L-139; mukB, L-137; precA+ IPTG, pCY579; Nov, 20 μg/ml novobiocin; IPTG, 1 mM IPTG; glucose, 0.2% glucose. Plates were developed for 24 h at 42°C or for 48 h at 22°C (D) ΔrecA304, JC10287; precA+ (Ts), pEAK2; topA-34, AK144. Four independent transductants of the ΔrecA304 topA-34 pEAK2 genotype were spotted at 37°C and 45°C on LB supplemented with three concentrations of NaCl (indicated above the spotting panels): 25, 5 (regular LB), or 0 g/liter.
The RecA dependence of our smtA mutant (Fig. 4C Top) could have been similar to the RecA dependence of the ubiHE mutants (Table 3), because distant homologs of the smtA gene (blast search E value 1e-09 to 1e-05) include ubiE genes [Smith-Waterman report against Nrprot on Colibri database for smtA (http://genolist.pasteur.fr/Colibri/genome.cgi)]. On the other hand, smtA is the first gene in a four-gene operon, and our smtA insertion could interfere with the expression of the downstream mukFEB genes, implicated in nucleoid condensation and/or partitioning (33). The mukFEB mutants in AB1157 are inhibited by the DNA gyrase inhibitor novobiocin at concentrations of 2 μg/ml (AB1157 itself shows no inhibition by 200 μg/ml novobiocin), whereas our smtA mutant is inhibited by 20 μg/ml of the drug (Fig. 4C Top), suggesting a partial mukFEB defect in our smtA mutant. Complete inactivation of mukFEB by our smtA insertion is unlikely, though, because mukFEB-null mutants are temperature-sensitive (34) (Fig. 4C Center), whereas our smtA mutant is not. In fact, because of the Ts phenotype, we would not be able to isolate complete mukFEB inactivation in our screen. To demonstrate that the RecA dependence of the smtA-39 mutant is due to the mukFEB defect, we have cloned the chromosomal region comprising the inactivated smtA-39 allele, as well as the downstream muk genes, on a plasmid with a copy number of 20 per chromosome. The resulting plasmid (pEAK25) conferred resistance to both high temperature and novobiocin to a bona fide mukF::kan mutant (Fig. 4C Center), indicating that it complements the mukFEB defect. The same muk+ plasmid also complemented the temperature and novobiocin sensitivity of the smtA-39 recA(Ts) strain (Fig. 4C Top), indicating that the RecA dependence of smtA-39 mutation is due to muk defect. To see whether muk recA mutants are indeed inviable, we have constructed a mukB::kan ΔrecA mutant, in which the recA defect was complemented by an isopropyl β-d-thiogalactoside (IPTG)-dependent plasmid carrying the functional recA+ gene (Fig. 4C Bottom). Although the plasmid showed a high degree of instability in the mukB mutant, it was nevertheless evident that the strain cannot grow if IPTG in the medium is replaced with glucose, conditions that inhibit the plasmid replication (Fig. 4C Bottom). Thus, we conclude that muk mutants depend on RecA for viability.
The RecA dependence of our topA-34 allele was elusive: some of the ΔrecA precA+(Ts) → topA-34 transductants were unable to grow at 42°C and higher, in conditions where the recA+ plasmid is lost, indicating RecA dependence, but some others would grow (Fig. 4D Center), suggesting rapid suppression of the topA defect. Because topA codes for Topo I, the enzyme responsible for maintaining proper supercoiling of the bacterial chromosome by relaxing DNA underwound by DNA gyrase, to reveal the phenotype of our topA mutant, we modified growth conditions to change cellular DNA superhelicity. Low osmolarity of the medium (35) and growth temperatures >40°C (36) cause a decrease in cellular DNA superhelicity. We found that changes in both the osmolarity and growth temperature differentially affect the viability of individual topA-34 recA clones; however, in low osmolarity LB at 45°C, topA-34 recA mutants are reliably inviable (Fig. 4D Right).
Discussion
We designed a screen for synthetic lethals in E. coli, based on the appearance of nonsectoring colonies indicating the inability to lose a plasmid carrying a “query” gene of choice. The idea of this screen has been adopted from the work of Holm and colleagues (37-39), who successfully used a similar screen to look for synthetic lethals in the yeast Saccharomyces cerevisiae. We worked in the DH5α strain of E. coli, because it forms compact and almost colorless colonies on MacConkey plates. Other E. coli strains tested, derivatives of MG1655, AB1157, and BW13711, form widespread pinkish colonies on MacConkey, making it difficult to distinguish between sectoring and nonsectoring phenotypes. In our hands, sectoring on LB agar plus 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) was obscured by the heavy background color; however, others used this combination with more success in a similar screen for synthetic lethals (40).
Using the color screen, we looked for E. coli mutants that depended on RecA, the central gene of recombinational repair in eubacteria. We have isolated the following RecA-dependent mutants: fur, rdgB, seqA, smtA, tdk, topA, ubiE, and ubiH. All isolated mutants (except one tdk mutant) were induced for SOS to varying degrees. RecA dependence and SOS induction indicate chromosomal damage, and we indeed detected various levels of chromosomal fragmentation in rdgB, fur, seqA, smtA, and topA mutants. Although RecA-dependent mutants are unified by these phenotypes, specific mechanisms leading to chromosomal fragmentation in the isolated mutants are likely to be different.
Isolation of fur as a RecA-dependent mutant confirms the reported fur recA synthetic lethality ascribed to increased oxidative DNA damage (22). We now report elevated chromosomal fragmentation in fur mutants, providing the reason for RecA dependence. RecA dependence of ubiH and ubiE mutations, which block two consecutive ring decoration stages in the biosynthesis of ubiquinone (41), the electron carrier in electron-transfer reaction, may also be due to the elevated level of oxidative DNA damage. Unfortunately, in combination with the recBC(Ts) mutation, ubiE and ubiH mutants showed extremely slow growth even at 22°C, so we could not determine the level of chromosomal fragmentation in these two mutants. The ubiH and ubiE mutants also proved to be different from other isolated RecA-dependent mutants by being RecA-dependent only in the presence of kanamycin (all our mutants are resistant to kanamycin because of insertion of a kanamycin-resistance cassette) and, in this respect, were “conditionally” RecA-dependent.
Isolation of tdk inactivation as synthetically inhibited with recA was not completely unexpected in light of the recently found dut recA synthetic lethality (24), because both dUTPase (gpdut) and thymidine kinase (gptdk) contribute to the formation of dUMP, an intermediate in dTTP biosynthesis (42), and because dut rec lethality is suppressed by amplification of tdk+ (E.A.K. and A.K., unpublished work). The metabolic defect of tdk mutants is unclear, though, because it is unlikely to be due to significant uracil incorporation into DNA or thymineless death-like phenomena (tdk mutants do not require thymine). The reason we have isolated several independent insertions in tdk, which is a relatively small gene (618 bp), is also unclear. Our findings of no or low SOS induction in tdk mutants and no detectable increase in chromosomal fragmentation were surprising.
The smtA gene forms an operon with the mukFEB genes, involved in nucleoid administration. Our data indicate that inadequate expression of mukFEB due to the smtA insertion causes RecA dependence, perhaps via interference of improper nucleoid condensation and/or partitioning with replication fork progress. This interpretation is supported by our isolation of an insertion in seqA, another gene implicated in nucleoid administration (33). SeqA tracts associated with the cell envelope are hypothesized to guide the newly synthesized daughter DNAs toward opposite poles of the cell (43). Therefore, the synthetic inhibition of seqA recA double mutants could be due to the same interference of improper nucleoid partitioning with replication fork progress. On the other hand, SeqA binds hemimethylated GATC sites at the chromosomal origin and thus hides the origin from the DnaA initiator protein to prevent premature initiation (44). seqA mutants show initiation asynchrony and overinitiation (44), which may be another reason for chromosomal fragmentation. Besides dam and seqA, the only other known null mutations that cause asynchronous initiation of chromosomal DNA replication are in topA, encoding Topoisomerase I, the main DNA relaxation activity of eubacterial cell (45). Our isolation of a topA insertion as a RecA-dependent mutant again points to replication asynchrony as a potential cause of chromosomal fragmentation, However, we found that, in contrast to the known strong topA alleles, our topA-34 mutation increases the level of supercoiling only slightly (not shown). Interestingly, although the growth defect of topA-null mutants is relieved in media with low osmolarity, because of the relaxation of cellular DNA under these conditions (35), our topA-34 mutant becomes reliably RecA-dependent only at high temperatures and in conditions of low osmolarity. Thus, the initial characterization of topA-34 mutant suggests that its RecA dependence is unlikely to be due to the associated change in supercoiling.
Besides the isolated insertion mutants, there are missense mutations in genes coding for enzymes directly involved in DNA replication that also make E. coli cells dependent on RecA. The polA and lig mutants have defects in Okazaki fragment maturation, whereas dut mutants have shorter Okazaki fragments. The corresponding genes code for DNA polymerase I, DNA ligase, and dUTPase, all three proteins being either indispensable or required for growth in rich media. Mutants with partial defects in either one of the three activities are synthetic lethal with recA inactivation and show signs of chromosomal fragmentation (24, 46, 47), suggesting that problems with Okazaki fragments somehow translate into double-strand DNA breaks. Okazaki-fragment-maturation-defective mutants in yeast also depend on recombinational repair (reviewed in ref. 48). Certain point mutations in the DNA cleaving (GyrA) subunit of DNA gyrase in Salmonella typhimurium, again an indispensable enzyme involved in chromosomal replication, also lead to RecA dependence and signs of chromosomal fragmentation (49).
Conclusion
On the basis of our analysis, we suggest three major contributors to chromosomal fragmentation in E. coli under regular growth conditions: (i) unscheduled initiation of chromosomal replication (seqA, dam, and topA) and/or defects in nucleoid administration (seqA, mukFEB, and topA); (ii) defects in DNA precursor synthesis and quality control (rdgB and dut); and (iii) defects leading to an increased level of reactive oxygen species (fur and ubiEH). If the recombinational repair-dependent mutants in the DNA replication functions (polA, lig, and gyrA) are added to this list, there will be one more source of chromosomal fragmentation: defects in the elongation phase of DNA replication. For a few mutants from this list, specific mechanisms leading to recombinational repair dependence have been investigated (20, 24, 50); for the majority of the RecA-dependent mutants, the underlying mechanisms of chromosomal fragmentation and its avoidance remain to be determined.
Supplementary Material
Acknowledgments
We thank Frederick Blattner (University of Wisconsin, Madison), Alvin John Clark (University of Arizona, Tucson), John Cronan (University of Illinois at Urbana-Champaign), Richard P. Cunningham (State University of New York, Albany), Richard Kolodner (University of California at San Diego), Peter Kuempel (University of Colorado, Boulder), Sydney Kushner (University of Georgia, Athens), William Metcalf (University of Illinois at Urbana-Champaign), and James Sawitzke (National Cancer Institute) for providing strains and plasmids. This work was supported by Grant MCB-0196020 from the National Science Foundation.
Author contributions: E.A.K. and A.K. designed research; E.A.K., E.R., L.M., J.Z., and A.K. performed research; E.A.K., E.R., L.M., J.Z., and A.K. analyzed data; and A.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: IPTG, isopropyl β-d-thiogalactoside.
References
- 1.Myung, K. & Kolodner, R. (2003) DNA Repair 2, 243-258. [DOI] [PubMed] [Google Scholar]
- 2.Pierce, A. J., Stark, J. M., Araujo, F. D., Moynahan, M. E., Berwick, M. & Jasin, M. (2001) Trends Cell Biol. 11, S52-S59. [DOI] [PubMed] [Google Scholar]
- 3.Pfeiffer, P., Goedecke, W. & Obe, G. (2000) Mutagenesis 15, 289-302. [DOI] [PubMed] [Google Scholar]
- 4.Valerie, K. & Povirk, L. F. (2003) Oncogene 22, 5792-5812. [DOI] [PubMed] [Google Scholar]
- 5.Kuzminov, A. (1999) Microbiol. Mol. Biol. Rev. 63, 751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paques, F. & Haber, J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim, D.-S. & Hasty, P. (1996) Mol. Cell. Biol. 16, 7133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonoda, E., Sasaki, M. S., Buerstedde, J.-M., Bezzubova, O., Shinohara, A., Ogawa, H., Takata, M., Yamaguchi-Iwai, Y. & Takeda, S. (1998) EMBO J. 17, 598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sargentini, N. J. & Smith, K. C. (1986) Radiat. Res. 107, 58-72. [PubMed] [Google Scholar]
- 10.Kowalczykowski, S. C. (2000) Trends Biochem. Sci. 25, 156-165. [DOI] [PubMed] [Google Scholar]
- 11.Roca, A. I. & Cox, M. M. (1997) Prog. Nucleic Acid Res. Mol. Biol. 56, 129-223. [DOI] [PubMed] [Google Scholar]
- 12.Krasin, F. & Hutchinson, F. (1977) J. Mol. Biol. 116, 81-98. [DOI] [PubMed] [Google Scholar]
- 13.Capaldo, F. N. & Barbour, S. D. (1975) J. Mol. Biol. 91, 53-66. [DOI] [PubMed] [Google Scholar]
- 14.Skarstad, K. & Boye, E. (1993) J. Bacteriol. 175, 5505-5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capaldo, F. N., Ramsey, G. & Barbour, S. D. (1974) J. Bacteriol. 118, 242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miranda, A. & Kuzminov, A. (2003) Genetics 163, 1255-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer, B. R. & Marinus, M. G. (1994) Gene 143, 1-12. [DOI] [PubMed] [Google Scholar]
- 18.Wang, T.-C. V. & Smith, K. C. (1986) J. Bacteriol. 165, 1023-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clyman, J. & Cunningham, R. P. (1987) J. Bacteriol. 169, 4203-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bradshaw, J. S. & Kuzminov, A. (2003) Mol. Microbiol. 48, 1711-1725. [DOI] [PubMed] [Google Scholar]
- 21.Keyer, K. & Imlay, J. (1996) Proc. Natl. Acad. Sci. USA 93, 13635-13640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Touati, D., Jacques, M., Tardat, B., Bouchard, L. & Despied, S. (1995) J. Bacteriol. 177, 2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 24.Kouzminova, E. A. & Kuzminov, A. (2004) Mol. Microbiol. 51, 1279-1295. [DOI] [PubMed] [Google Scholar]
- 25.Courcelle, J., Khodursky, A., Peter, B., Brown, P. O. & Hanawalt, P. C. (2001) Genetics 158, 41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernández de Henestrosa, A. R., Ogi, T., Aoyagi, S., Chafin, D., Hayes, J. J., Ohmori, H. & Woodgate, R. (2000) Mol. Microbiol. 35, 1560-1572. [DOI] [PubMed] [Google Scholar]
- 27.Friedberg, E. C., Walker, G. C. & Siede, W. (1995) DNA Repair and Mutagenesis (Am. Soc. Microbiol., Washington, DC).
- 28.Little, J. W. (1991) Biochimie 73, 411-421. [DOI] [PubMed] [Google Scholar]
- 29.Ossanna, N. & Mount, D. W. (1989) J. Bacteriol. 171, 303-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kushner, S. R. (1974) J. Bacteriol. 120, 1213-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuzminov, A. (1995) Mol. Microbiol. 16, 373-384. [DOI] [PubMed] [Google Scholar]
- 32.Michel, B., Ehrlich, S. D. & Uzest, M. (1997) EMBO J. 16, 430-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiraga, S. (2000) Annu. Rev. Genet. 34, 21-59. [DOI] [PubMed] [Google Scholar]
- 34.Yamanaka, K., Ogura, T., Niki, H. & Hiraga, S. (1996) Mol. Gen. Genet. 250, 241-251. [DOI] [PubMed] [Google Scholar]
- 35.Higgins, C. F., Dorman, C. J., Stirling, D. A., Waddell, L., Booth, I. R., May, G. & Bremer, E. (1988) Cell 52, 569-584. [DOI] [PubMed] [Google Scholar]
- 36.Adamcik, J., Viglasky, V., Valle, F., Antalik, M., Podhradsky, D. & Dietler, G. (2002) Electrophoresis 23, 3300-3309. [DOI] [PubMed] [Google Scholar]
- 37.Kranz, J. E. & Holm, C. (1990) Proc. Natl. Acad. Sci. USA 87, 6629-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merrill, B. J. & Holm, C. (1998) Genetics 148, 611-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merrill, B. J. & Holm, C. (1999) Genetics 153, 595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernhardt, T. G. & De Boer, P. A. (2004) Mol. Microbiol. 52, 1255-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meganathan, R. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 642-656. [Google Scholar]
- 42.Neuhard, J. & Kelln, R. A. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), Vol. 1, pp. 580-599. [Google Scholar]
- 43.Brendler, T., Sawitzke, J., Sergueev, K. & Austin, S. (2000) EMBO J. 19, 6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, M., Campbell, J. L., Boye, E. & Kleckner, N. (1994) Cell 77, 413-426. [DOI] [PubMed] [Google Scholar]
- 45.Olsson, J. A., Nordstrom, K., Hjort, K. & Dasgupta, S. (2003) J. Mol. Biol. 334, 919-931. [DOI] [PubMed] [Google Scholar]
- 46.Gottesman, M. M., Hicks, M. L. & Gellert, M. (1973) J. Mol. Biol. 77, 531-547. [DOI] [PubMed] [Google Scholar]
- 47.Monk, M. & Kinross, J. (1972) J. Bacteriol. 109, 971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuzminov, A. (2001) Proc. Natl. Acad. Sci. USA 98, 8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garí, E., Figueroa-Bossi, N., Blanc-Potard, A.-B., Spirito, F., Schmid, M. B. & Bossi, L. (1996) Mol. Microbiol. 21, 111-122. [DOI] [PubMed] [Google Scholar]
- 50.Garí, E., Bossi, L. & Figueroa-Bossi, N. (2001) Genetics 159, 1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.