Abstract
The global emergence of Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae poses a major public health threat requiring immediate and aggressive action. Some older generation antibiotics, such as trimethoprim, serve as alternatives for treatment of infections. Here, we determined the complete nucleotide sequence of plasmid pHS091147, which co-harbored the carbapenemase (blaKPC-2) and trimethoprim resistance genes (dfrA25) from a Klebsiella pneumoniae sequence type (ST) 11 clone recovered in Shanghai, China. pHS091147 had three replication genes, several plasmid-stability genes and an intact type IV secretion system gene cluster. Besides blaKPC-2 and dfrA25, pHS091147 carried several other resistance genes, including β-lactamase genes blaTEM-1 and blaCTX-M-14, sulphonamide resistance gene sul1, a quinolone resistance gene remnant (ΔqnrB2), and virulence associated gene iroN. Notably, the multidrug-resistance region was a chimeric structure composed of three subregions, which shared strong sequence homology with several plasmids previously assigned in Genbank. To our knowledge, this is the first report of the co-localization of blaKPC-2 and dfrA25 on a novel putative multi-replicon plasmid in a Klebsiella pneumoniae ST11 clone.
Introduction
A dramatic increase in the prevalence of Klebsiella pneumoniae carbapenemase (KPC)-producing K. pneumoniae is associated with a rise in morbidity and mortality, and poses an alarming clinical threat for hospitalized patients [1]. KPC-2 is the most common variant of the KPC enzymes. The large majority of KPC-2-producing K. pneumoniae in America and Europe belongs to sequence type (ST) 258 clone; in China, the majority belongs to the ST11 clone, which is a part of the same clonal complex as ST258 [2, 3].
Since the increase of carbapenem-resistant Enterobacteriaceae, which are frequently resistant to many different antibiotic substances, some older generation antibiotics (e.g., trimethoprim) have been used alternatively for treating infections [4, 5]. With a structure similar to that of folic acid, trimethoprim is a competitive inhibitor of dihydrofolate reductase [6, 7]. Bacterial resistance to trimethoprim can be inherited or acquired. The most common trimethoprim resistance mechanism involves acquisition of trimethoprim-resistant dihydrofolate reductase (dfr) gene [6, 7]. Until now, more than 25 different trimethoprim resistance dfrA genes have been identified; the majority are associated with mobile genetic elements such as plasmids, transposons or integrons [8, 9]. The gene dfrA25 was firstly detected as a gene cassette within a class 1 integron in Salmonella Agona [10]. In this study, we report the coexistence of blaKPC-2 and dfrA25 on a single putative multi-replicon plasmid obtained from an epidemic K. pneumoniae ST11 isolate recovered in China.
Materials and methods
Bacterial strain and plasmid
K. pneumoniae HS091147 used in this study was isolated in 2009 from a sputum sample at Huashan Hospital, Shanghai Medical College, Fudan University, China. Plasmid DNA was extracted from K. pneumoniae HS091147 (Qiagen plasmid mid kit; Qiagen, Germany) and transferred by electroporation (Micro-Pulser electroporator; Bio-Rad, USA) into E. coli DH5α. Transformants were selected on Luria-Bertani (LB) agar plates containing ampicillin (100 μg/ml) and imipenem (2μg/ml), then screened by a blaKPC-2 PCR assay. The primers targeting blaKPC-2 genes were described previously [11].
Antibiotic susceptibility testing
The minimal inhibitory concentrations (MICs) for K. pneumoniae HS091147, its transformant and E. coli DH5α were determined using the VITEK®2 COMPACT AST-GN13 (bioMérieux, France). E. coli ATCC 25922 was used as the quality control strain. All susceptibility tests were repeated three times and the results were interpreted according to the breakpoints suggested by the Clinical and Laboratory Standards Institute (CLSI) [12].
Multilocus Sequence Typing (MLST)
The sequencing types (STs) of K. pneumoniae strain HS091147 were determined by analyzing gapA, infB, mdh, pgi, phoE, rpoB, and tonB housekeeping genes. The results were compared with information provided in the multilocus sequence typing (MLST) databases (http://www.pasteur.fr/recherche/genopole/PF8/mlst/Kpneumoniae.html).
Sequencing and annotation of plasmid
Complete sequencing of the plasmid pHS091147 was performed with a shotgun approach using 454 GS Junior (Roche, Basel, Switzerland). The GenBank accession number is KX236178.
Results and discussion
HS091147 and its E. coli DH5α transformant (T-pHS091147)
K. pneumoniae HS091147 was characterized according to the species-specific MLST schemes of the Pasteur database and found to belong to the K. pneumoniae ST11 clone. ST11 was previously determined as the prevalent clone associated with the spread of KPC in Asia (particularly in China) [2, 13], while other β-lactamases (e.g., CTX-M-type ESBLs) did not show similar epidemiological characteristics [14–16].
The MICs of K. pneumoniae HS091147 and its transformant are shown in Table 1. Notably, the original HS091147 strain was resistant to all 17 antibiotics tested (ampicillin, ampicillin/sulbactam, cefazolin, cefotetan, ceftazidime, ceftriaoxone, cefepime, aztreonam, ertapenem, imipenem, amikcin, tobramycin, ciprofloxacin, levofloxacin, nitrofurantoin, piperacillin/tazobactam and trimethoprim/sulfamethoxazole), and 7 of these (ampicillin, ampicillin/sulbactam, cefazolin, ceftriaoxone, aztreonam, imipenem, and trimethoprim/sulfamethoxazole) were fully transferable to the recipient, E. coli DH5α strain via the plasmid pHS091147. Furthermore, the transformant T-pHS091147 was intermediately resistant to ertapenem and piperacillin/tazobactam when compared to the DH5α background.
Table 1. Antibiotic resistance profiles for K. pneumoniae HS091147 and its transformant (T-pHS091147) in E. coli DH5α.
| Antibiotics | Strain | ||
|---|---|---|---|
| HS091147 | DH5α | T-pHS091147 | |
| Ampicillin | ≥32 (R) | ≤2 (S) | ≥32 (R) |
| Ampicillin/Sulbactam | ≥32 (R) | ≤2 (S) | ≥32 (R) |
| Cefazolin | ≥64 (R) | ≤4 (S) | ≥64 (R) |
| Cefotetan | ≥64 (R) | ≤4 (S) | ≤4 (S) |
| Ceftazidime | ≥64 (R) | ≤1 (S) | 4 (S) |
| Ceftriaoxone | ≥64 (R) | ≤1 (S) | ≥64 (R) |
| Cefepime | ≥64 (R) | ≤1 (S) | 2 (S) |
| Aztreonam | ≥64 (R) | ≤1 (S) | 16 (R) |
| Ertapenem | ≥8 (R) | ≤0.5 (S) | 1 (I) |
| Imipenem | ≥16 (R) | ≤1 (S) | 8 (R) |
| Amikcin | ≥64 (R) | ≤2 (S) | ≤2 (S) |
| Tobramycin | ≥16 (R) | ≤1 (S) | ≤1 (S) |
| Ciprofloxacin | ≥4 (R) | ≤0.25 (S) | ≤0.25 (S) |
| Levofloxacin | ≥8 (R) | ≤0.25 (S) | ≤0.25 (S) |
| Nitrofurantoin | ≥512 (R) | ≤16 (S) | ≤16 (S) |
| Piperacillin/Tazobactam | ≥128 (R) | ≤4 (S) | 64 (I) |
| Trimethoprim/Sulfamethoxazole | ≥320 (R) | ≤20 (S) | ≥320 (R) |
R, resistant; I, intermediate resistant; S, susceptible
General features of plasmid pHS091147
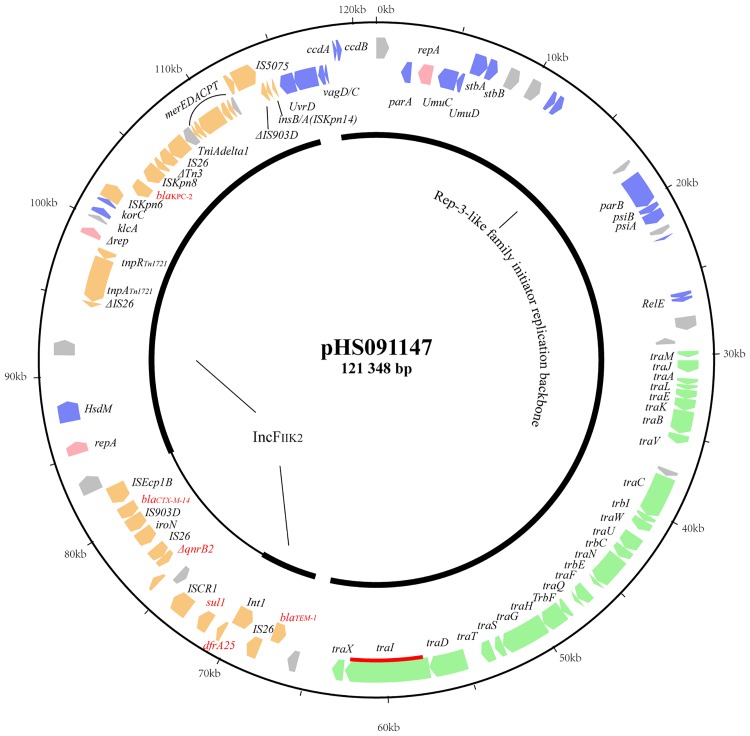
Plasmid pHS091147 was determined to be a circular molecule, 121,348 bp in size with an average G+C content of 52.8%. Annotation revealed 107 predicted open reading frames. One hundred and one of these frames encoded proteins homologous to proteins with known functions and assigned to other sequenced plasmids in GenBank. Using the approach by Norman et al. [17], pHS091147 was determined to carry genes involved in replication, stability, propagation and adaptation (Fig 1).
Fig 1. Circular map of pHS091147.
Genes are color-coded dependent upon functional annotations as follows: pink, replication; blue, stability; green, propagation; orange, adaptation (the MDR region); grey, other functions and hypothetical proteins. The relaxase gene (traI) is indicated by the red bar. Red text highlights the resistance genes: blaKPC-2, blaTEM-1, blaCTX-M-14, dfrA25, sul1 and ΔqnrB2.
Replication, stability and propagation of pHS091147
Plasmid pHS091147 contains three replication genes, one of which was truncated with loss of function (Δrep in Fig 1, position 98903 to 99463). The other two genes, repFIIK2 (position 85132 to 85887) and an unassigned gene (position 2795 to 3745), were located between the MDR region and the plasmid backbone, respectively. The repFIIK2 gene is frequently reported in KPC-encoding plasmids and may be the primary vehicle for the dissemination of blaKPC [18, 19]. The unassigned gene, encoding a Rep-3-like family initiator replication protein, was only found in 28 plasmid sequences deposited in GenBank. Among these, the presence of two single-replicon plasmids, pNJST258C3 and pNJST258N3 (GenBank accession numbers: CP006925 and CP006921, respectively), suggested the likelihood that the unassigned replicons function in initiation of replication.
Genes that encode replication (repA), stability (e.g., ParA, stbA/stbB and UnuC/UmuD surrounding the repA gene) and propagation were located within the unassigned Rep-3-like family backbone of pHS091147. In addition, we found a full complement of conjugation machinery including a type IV secretion system (T4SS) and relaxase, which enabled mobilization [17].
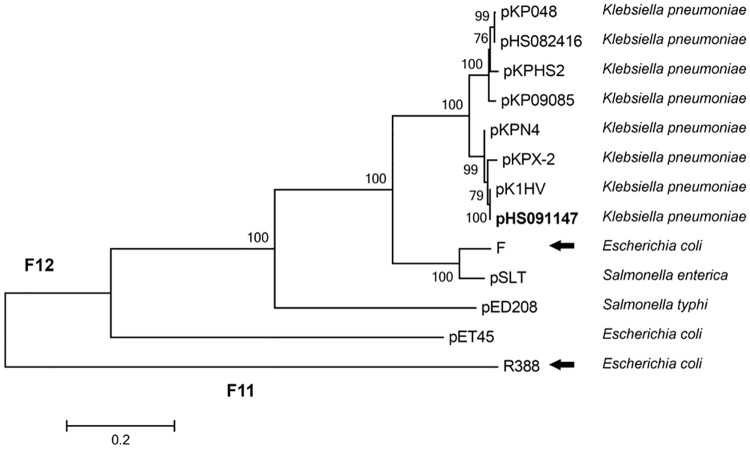
Relaxase, the only common component in all transmissible plasmids, can be used to classify plasmids and infer phylogenetic relationships [20–22]. Plasmids can be classified in six mobility (MOB) families: MOBF, MOBP, MOBQ, MOBH, MOBC, and MOBV, according to the relaxase sequence. MOBF, a well-characterized family, contains several subfamilies [20–22]. Phylogenetic analysis showed that the relaxase encoded by pHS091147 was grouped in the F12 subfamily (Fig 2). Comparison of the pHS091147 relaxase protein sequence revealed 79% identity with the sequence found in F (GenBank accession number: AP001918), the prototype plasmid for the F12 subfamily (IncF12), and 100% identity with that of pK1HV (GenBank accession number: HF545434), a K. pneumoniae multidrug resistance plasmid harboring qnrS1 isolated in Vietnam.
Fig 2. Phylogenetic analysis of plasmid-encoded relaxase homologs.
Plasmid pHS091147 (in bold) and twelve other protein sequences were aligned, and the tree was generated with MEGA5 using the maximum-likelihood method. Solid black arrows point to the prototype plasmids for the MOBF12 and MOBF11 subfamilies. Other relaxase sequences of plasmids pKP048 (GenBank accession number FJ628167), pHS082416 (KF724507), pKHS2 (CP003224), pKP09085 (KF719970), pKPN4 (CP000649), pKPX-2 (AP012056), pK1HV (HF545434), F (AP001918), pSLT (AE006471), pED208 (AF411480), pET45 (CU468132) and R388 (BR000038) were obtained from GenBank.
The adaptation region is highly mosaic-like
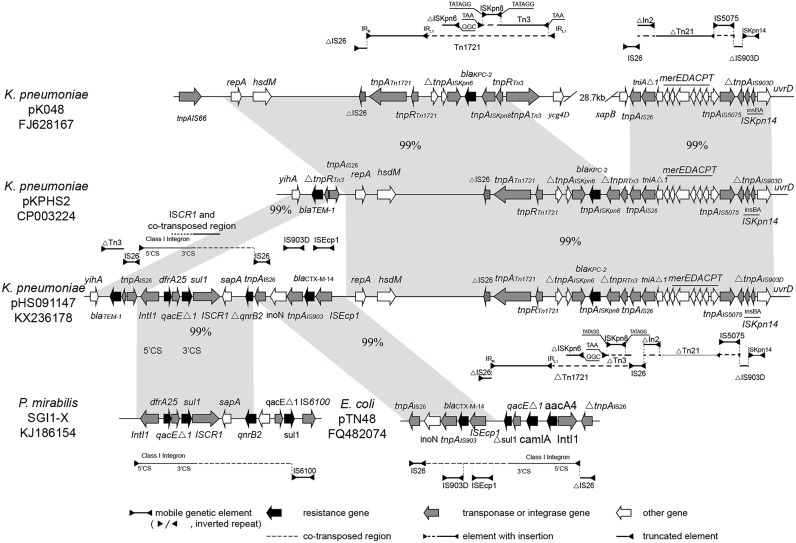
The adaptation region, a continuous 49,218 bp IncFIIK2 fragment, harbored five resistance genes (including blaKPC-2 and dfrA25), one truncated ΔqnrB2 gene and some genetic elements like insertion sequences (ISs), transposons and integrons. It was highly mosaic-like and can be divided into three subregions that share strong homology with several different plasmids (Fig 3).
Fig 3. Comparative analysis of the multidrug-resistance region of plasmid pHS091147.
The relevant parts of pKP048, pKPHS2, SGI1-X and pTN48 are shown to highlight the syntenic regions. The horizontal line shown above or below the schematics (with one or both ends demarcated by solid triangles to indicated inverted repeats) represents intact, interrupted or truncated ISs, transposons and integrons as appropriate. Interruptions in the structures above are indicated as dashed lines. Regions with similar sequences are indicated in gray with corresponding percentages between the plasmids.
The first subregion, which co-harbored blaKPC-2 and blaTEM-1, showed >99% homology with K. pneumoniae plasmid pKPHS2 (GenBank accession number CP003224), which was also found in Huashan Hospital in 2011. Carbapenemase gene blaKPC-2 was located on a Tn1721 transposon variant that was truncated by IS26, thus forming Tn1721-blaKPC-2-ΔTn3-IS26. It was the dominant blaKPC-2 genetic structure in forty-two non-duplicated, blaKPC-2-postive K. pneumoniae strains isolated in Huashan Hospital between August 2006 (when the first blaKPC-2-positive clinical K. pneumoniae isolate was detected) and October 2010 (when the blaKPC-2-positive isolates were detected continuously and steadily) [23]. It was fused, in turn, with the left end of ΔTn21-IS5075-ΔIS903D-ISKpn14. Both the Tn1721-derived blaKPC-2-bearing transposon and the ΔTn21-IS5075-ΔIS903D-ISKpn14 contiguous region were highly similar to pKP048 (GenBank accession number FJ628167), a K. pneumoniae multidrug resistance plasmid carring blaKPC-2, blaDHA-1, qnrB4, and armA isolated in Zhejiang province of China. Given that the first subregion was highly related to pKP048, we inferred that this long segment derived from a pKP048-like plasmid following insertion of IS26 into the Tn1721 variant. Subsequently, recombination mediated by the inserted IS26 and the IS26 adjacent to ΔTn21 occurred and the intervening 35.3 kb fragment was deleted as a consequence [24]. The genetic environment of blaTEM-1 was consistent with the IS26-blaTEM configuration reported in Australia in 2011 and located in a distance of 14kb from the blaKPC-2 fragment [25]. The other two subregions were located in-between.
The second subregion, IntI1-dfrA25-sul1-ISCR1-ΔqnrB2, shared >99% identity with the MDR region of the Salmonella genomic island, SGI1-X in P. mirabilis PmC162 (GenBank accession number KJ186154). The In207 class 1 integron carried 5’-CS, which contained the int1, dfrA25-attC gene cassettes array (confers trimethoprim resistance) and the conserved segment, 3’-CS (qacΔ1 and sul1Δ). The 3’-CS segment was followed by ISCR1-ΔqnrB2; the qnrB2 gene was truncated by IS26 flanking the third subregion.
The third subregion exhibited >99% homology to an E. coli CTX-M-14-encoding plasmid, pTN48 (GenBank accession number FQ482074), which included blaCTX-M-14 and its environment (IS903 and ISEcp1B), IS26 and iroN. The blaCTX-M genes were usually located within adjacent ISEcp1 areas, which provide a promoter for resistance gene expression [26, 27]. Moreover, CTX-M-14 was a highly malleable β-lactamase with broad opportunity to evolve. Many novel CTX-M-type ESBLs variants derived from CTX-M-14-like β-lactamase genes, culminating in higher MIC values [28, 29]. The high prevalence of CTX-M-14-producing Enterobacteriaceae has recently been reported in different provinces of China, with incidences ranging from 28.2% to 48.4% [14, 15, 30, 31]. In addition, the third subregion included iroN, the outer membrane siderophore receptor gene identified in Salmonella spp. iroN, which mediated utilization of structurally-related catecholate siderophores, was critical for virulence of the iroBCDEN gene cluster [32, 33]. It cannot function alone, and must operate in association with other virulence genes [33].
In conclusion, we describe here the complete sequence of a novel putative multi-replicon plasmid (pHS091147) obtained from a multidrug-resistant K. pneumoniae ST11 isolate, which carried five resistance genes (including blaKPC-2, blaCTX-M-14, blaTEM-1, sul1 and dfrA25). Structural analysis showed that pHS091147 was highly mosaic and composed of parts previously identified in other plasmids of Enterobacteriaceae origin, suggesting that homologous recombination and horizontal gene transmission mediated by mobilizable elements played critical roles in evolution of the plasmid. The identified genes mediate resistance to last-line antimicrobial agents (carbapenems) and other, older generation antibiotics (i.e., trimethoprim). The location of these genes together on a single plasmid poses a serious epidemiological, clinical and public health threat.
Acknowledgments
We are grateful to Dr. Stephen H. Gregory (Providence, RI, USA) for his contributions to writing and editing this manuscript. This study was supported by the National Natural Science Foundation of China (Grants 81071396 and 81572031) and Shanghai Natural Science Fund (No. 14ZR1404500).
Data Availability
All relevant data are within the paper.
Funding Statement
Support was provided by the National Natural Science Foundation of China (Grants 81071396 and 81572031), Shanghai Natural Science Fund (No. 14ZR1404500).
References
- 1.Sbrana F, Malacarne P, Viaggi B, Costanzo S, Leonetti P, Leonildi A, et al. Carbapenem-sparing antibiotic regimens for infections caused by Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in intensive care unit. Clin Infect Dis. 2013;56(5):697–700. 10.1093/cid/cis969 [DOI] [PubMed] [Google Scholar]
- 2.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–12. 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 3.Chen YT, Lin JC, Fung CP, Lu PL, Chuang YC, Wu TL, et al. KPC-2-encoding plasmids from Escherichia coli and Klebsiella pneumoniae in Taiwan. J Antimicrob Chemother. 2014;69(3):628–31. 10.1093/jac/dkt409 [DOI] [PubMed] [Google Scholar]
- 4.Falagas ME, Vardakas KZ, Roussos NS. Trimethoprim/sulfamethoxazole for Acinetobacter spp.: A review of current microbiological and clinical evidence. Int J Antimicrob Agents. 2015;46(3):231–41. 10.1016/j.ijantimicag.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 5.Wilkowski P, Ciszek M, Dobrzaniecka K, Sanko-Resmer J, Labus A, Grygiel K, et al. Successful treatment of urinary tract infection in kidney transplant recipients caused by multiresistant Klebsiella pneumoniae producing New Delhi Metallo-beta-lactamase (NDM-1) with strains genotyping. Transplant Proc. 2016;48(5):1576–9. 10.1016/j.transproceed.2016.01.060 [DOI] [PubMed] [Google Scholar]
- 6.Huovinen P, Sundstrom L, Swedberg G, Skold O. Trimethoprim and sulfonamide resistance. Antimicrob Agents Chemother. 1995;39(2):279–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skold O. Resistance to trimethoprim and sulfonamides. Vet Res. 2001;32(3–4):261–73. 10.1051/vetres:2001123 [DOI] [PubMed] [Google Scholar]
- 8.Lee JC, Oh JY, Cho JW, Park JC, Kim JM, Seol SY, et al. The prevalence of trimethoprim-resistance-conferring dihydrofolate reductase genes in urinary isolates of Escherichia coli in Korea. J Antimicrob Chemother. 2001;47(5):599–604. [DOI] [PubMed] [Google Scholar]
- 9.Peirano G, Agerso Y, Aarestrup FM, dos Reis EM, dos Prazeres Rodrigues D. Occurrence of integrons and antimicrobial resistance genes among Salmonella enterica from Brazil. J Antimicrob Chemother. 2006;58(2):305–9. 10.1093/jac/dkl248 [DOI] [PubMed] [Google Scholar]
- 10.Agerso Y, Peirano G, Aarestrup FM. dfrA25, a novel trimethoprim resistance gene from Salmonella Agona isolated from a human urine sample in Brazil. J Antimicrob Chemother. 2006;58(5):1044–7. 10.1093/jac/dkl366 [DOI] [PubMed] [Google Scholar]
- 11.Woodford N, Tierno PM Jr., Young K, Tysall L, Palepou MF, Ward E, et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A beta-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother. 2004;48(12):4793–9. 10.1128/AAC.48.12.4793-4799.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CLSI. Performance standards for antimicrobial susceptibility testing; Twenty-fourth informational supplement. Wayne, PA: Clinical and Laboratory Standards Institute. 2014;CLSI document M100-S24.
- 13.Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895 10.3389/fmicb.2016.00895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Zhou K, Zheng B, Zhao L, Shen P, Ji J, et al. High prevalence of ESBL-producing Klebsiella pneumoniae causing community-onset infections in China. Front Microbiol. 2016;7:1830 10.3389/fmicb.2016.01830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quan J, Zhao D, Liu L, Chen Y, Zhou J, Jiang Y, et al. High prevalence of ESBL-producing Escherichia coli and Klebsiella pneumoniae in community-onset bloodstream infections in China. J Antimicrob Chemother. 2017;72(1):273–80. 10.1093/jac/dkw372 [DOI] [PubMed] [Google Scholar]
- 16.Yano H, Uemura M, Endo S, Kanamori H, Inomata S, Kakuta R, et al. Molecular characteristics of extended-spectrum beta-lactamases in clinical isolates from Escherichia coli at a Japanese tertiary hospital. PLoS One. 2013;8(5):e64359 10.1371/journal.pone.0064359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Norman A, Hansen LH, Sorensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364(1527):2275–89. 10.1098/rstb.2009.0037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang Y, Yu D, Wei Z, Shen P, Zhou Z, Yu Y. Complete nucleotide sequence of Klebsiella pneumoniae multidrug resistance plasmid pKP048, carrying blaKPC-2, blaDHA-1, qnrB4, and armA. Antimicrob Agents Chemother. 2010;54(9):3967–9. 10.1128/AAC.00137-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Chavda KD, Melano RG, Jacobs MR, Levi MH, Bonomo RA, et al. Complete sequence of a bla(KPC-2)-harboring IncFII(K1) plasmid from a Klebsiella pneumoniae sequence type 258 strain. Antimicrob Agents Chemother. 2013;57(3):1542–5. 10.1128/AAC.02332-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiang DR, Li JJ, Sheng ZK, Yu HY, Deng M, Bi S, et al. Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 Strain in China. Antimicrob Agents Chemother. 2015;60(3):1343–8. 10.1128/AAC.01488-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcillán-Barcia MP, Francia MV, de La Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiology Reviews. 2009;33(3):657–87. [DOI] [PubMed] [Google Scholar]
- 22.Alvarado A, Garcillan-Barcia MP, de la Cruz F. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One. 2012;7(7):e40438 10.1371/journal.pone.0040438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen P, Zhang Y, Li G, Jiang X. Characterization of the genetic environment of the blaKPC-2 gene among Klebsiella pneumoniae isolates from a Chinese hospital. Braz J Infect Dis. 2016;20(4):384–8. 10.1016/j.bjid.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bi D, Jiang X, Sheng ZK, Ngmenterebo D, Tai C, Wang M, et al. Mapping the resistance-associated mobilome of a carbapenem-resistant Klebsiella pneumoniae strain reveals insights into factors shaping these regions and facilitates generation of a 'resistance-disarmed' model organism. J Antimicrob Chemother. 2015;70(10):2770–4. 10.1093/jac/dkv204 [DOI] [PubMed] [Google Scholar]
- 25.Bailey JK, Pinyon JL, Anantham S, Hall RM. Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J Antimicrob Chemother. 2011;66(4):745–51. 10.1093/jac/dkq529 [DOI] [PubMed] [Google Scholar]
- 26.Zong Z, Ginn AN, Dobiasova H, Iredell JR, Partridge SR. Different IncI1 plasmids from Escherichia coli carry ISEcp1-blaCTX-M-15 associated with different Tn2-derived elements. Plasmid. 2015;80:118–26. 10.1016/j.plasmid.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 27.Poirel L, Decousser JW, Nordmann P. Insertion sequence ISEcp1B is involved in expression and mobilization of a blaCTX-M-lactamase gene. Antimicrobial Agents and Chemotherapy. 2003;47(9):2938–45. 10.1128/AAC.47.9.2938-2945.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He D, Chiou J, Zeng Z, Liu L, Chen X, Zeng L, et al. Residues distal to the active site contribute to enhanced catalytic activity of variant and hybrid beta-lactamases derived from CTX-M-14 and CTX-M-15. Antimicrob Agents Chemother. 2015;59(10):5976–83. 10.1128/AAC.04920-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D'Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type beta-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303(6–7):305–17. 10.1016/j.ijmm.2013.02.008 [DOI] [PubMed] [Google Scholar]
- 30.Zhong YM, Liu WE, Liang XH, Li YM, Jian ZJ, Hawkey PM. Emergence and spread of O16-ST131 and O25b-ST131 clones among faecal CTX-M-producing Escherichia coli in healthy individuals in Hunan Province, China. J Antimicrob Chemother. 2015;70(8):2223–7. 10.1093/jac/dkv114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Zhou Y, Guo S, Chang W. High prevalence and risk factors of fecal carriage of CTX-M type extended-spectrum beta-lactamase-producing Enterobacteriaceae from healthy rural residents of Taian, China. Front Microbiol. 2015;6:239 10.3389/fmicb.2015.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sorsa LJ, Dufke S, Heesemann J, Schubert S. Characterization of an iroBCDEN gene cluster on a transmissible plasmid of uropathogenic Escherichia coli: evidence for horizontal transfer of a chromosomal virulence factor. Infect Immun. 2003;71(6):3285–93. 10.1128/IAI.71.6.3285-3293.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caza M, Lepine F, Milot S, Dozois CM. Specific roles of the iroBCDEN genes in virulence of an avian pathogenic Escherichia coli O78 strain and in production of salmochelins. Infect Immun. 2008;76(8):3539–49. 10.1128/IAI.00455-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.