Abstract
To further resolve the genetic architecture of the inflammatory bowel diseases, ulcerative colitis and Crohn’s disease, we sequenced the whole genomes of 4,280 patients at low coverage, and compared them to 3,652 previously sequenced population controls across 73.5 million variants. We then imputed from these sequences into new and existing GWAS cohorts, and tested for association at ~12 million variants in a total of 16,432 cases and 18,843 controls. We discovered a 0.6% frequency missense variant in ADCY7 that doubles risk of ulcerative colitis. Despite good statistical power, we did not identify any other new low-frequency risk variants, and found that such variants explained little heritability. We detected a burden of very rare, damaging missense variants in known Crohn’s disease risk genes, suggesting that more comprehensive sequencing studies will continue to improve our understanding of the biology of complex diseases.
Introduction
Crohn’s disease and ulcerative colitis, the two common forms of inflammatory bowel disease (IBD), are chronic and debilitating diseases of the gastrointestinal tract that result from the interaction of environmental factors, including the intestinal microbiota, with the host immune system in genetically susceptible individuals. Genome-wide association studies (GWAS) have identified 215 IBD associated loci that have substantially expanded our understanding of the biology underlying these diseases1–8. The correlation between nearby common variants in human populations underpins the success of the GWAS approach, but this also makes it difficult to infer precisely which variant is causal, the molecular consequence of that variant, and often even which gene is perturbed. Rare variants, which plausibly have larger effect sizes, can be more straightforward to interpret mechanistically because they are correlated with fewer nearby variants. However, it remains to be seen how much of the heritability9 of complex diseases is explained by rare variants. Well powered studies of rare variation in IBD thus offer an opportunity to better understand both the biological and genetic architecture of an exemplar complex disease.
The marked drop in the cost of DNA sequencing has enabled rare variants to be captured at scale, but there remains a fundamental design question regarding how to most effectively distribute short sequence reads in two dimensions: across the genome, and across individuals. The most important determinant of GWAS success has been the ability to analyze tens of thousands of individuals, and detecting rare variant associations will require even larger sample sizes10. Early IBD sequencing studies concentrated on the protein coding sequence in GWAS-implicated loci11–14 , which can be naturally extended to the entire exome15–17. However, coding variation explains at most 20% of the common variant associations in IBD GWAS loci18, and others have more generally observed19 that the substantial majority of complex disease associated variants lie in non-coding, presumed regulatory, regions of the genome. Low coverage whole genome sequencing has been proposed20 as an alternative approach that captures this important non-coding variation, while being cheap enough to enable thousands of individuals to be sequenced. As expected, this approach has proven valuable in exploring rarer variants than those accessible in GWAS21,22, but is not ideally suited to the analysis of extremely rare variants.
Our aim was to determine whether low coverage whole genome sequencing provides an efficient means of interrogating these low frequency variants, and how much they contribute to IBD susceptibility. We present an analysis of the whole genome sequences of 4,280 IBD patients, and 3,652 population controls sequenced as part of the UK10K project23, both via direct comparison of sequenced individuals and as the basis for an imputation panel in an expanded UK IBD GWAS cohort. This study allows us to examine, on a genome-wide scale, the role of low-frequency (0.1%≤ MAF < 5%) and rare (MAF < 0.1%) variants in IBD risk.
Results
Whole genome sequencing of 7,932 individuals
Following quality control (Supplementary Note and Supplementary Table 1-2), whole genome sequences of 2,513 Crohn’s disease patients (median coverage 4x) and 1,767 ulcerative colitis patients (2x) were jointly analyzed with 3,652 population controls (7x) sequenced as part of the UK10K project23 (Figure 1). We discovered 87 million autosomal single nucleotide variants (SNVs) and 7 million short indels (Supplementary Note and Supplementary Table 3). We then applied support vector machines for SNVs and GATK VQSR24 for indels to distinguish true sites of genetic variation from sequencing artifacts (Figure 1, Supplementary Note). We called genotypes jointly across all samples at the remaining sites, followed by genotype refinement using the BEAGLE imputation software25. This procedure leverages information across multiple individuals and uses the correlation between nearby variants to produce high quality data from relatively low sequencing depth. We noted that genotype refinement was locally affected by poor quality sites that failed further quality control analyses, so we ran BEAGLE a second time after these exclusions, yielding a set of 73.5 million high quality sites (Supplementary Note, Supplementary Figure 1-3 and Supplementary Table 4). Over 99% of common SNVs (MAF ≥ 5%) were also found in 1000 Genomes Project Phase 3 Europeans, indicating high specificity. Among rarer variants, 54.6 million were not seen in 1000 Genomes, demonstrating the value of directly sequencing the IBD cases and UK population controls (Supplementary Table 5, Supplementary Figure 3).
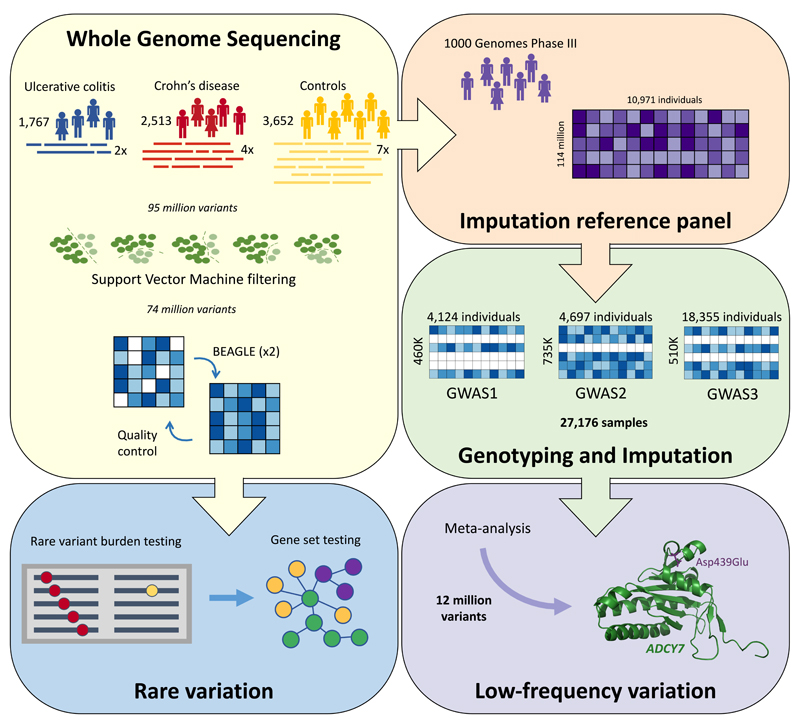
Figure 1. Overview of our study.
Variants were called from raw sequence reads in three groups of samples, and jointly filtered using support vector machines. The resulting genotypes were refined using BEAGLE and incorporated into the reference panel for a GWAS-imputation based meta-analysis, which discovered a low frequency association in ADCY7. A separate gene-based analysis identified a burden of rare damaging variants in certain known Crohn’s disease genes.
We also discovered 180,000 deletions, duplications and multiallelic copy number variants (CNVs) using GenomeStrip 2.026, but noted large differences in sensitivity between the three different sample sets (Supplementary Figure 4). Following quality control (Supplementary Note), including removal of CNVs with length < 60 kilobases, we observed an approximately equal number of variants in cases and controls, but retained only 1,475 CNVs. However, we still note a genome-wide excess of rare CNVs in controls (P=0.002), indicating that even after stringent filtering the data remains too noisy for meaningful conclusions to be drawn. We suggest that high coverage whole genome sequencing balanced in cases and controls will be required to evaluate the contribution of rare CNVs to IBD risk.
We individually tested 13 million SNVs and small indels with MAF ≥0.1% for association, and observed that we had successfully eliminated systematic differences due to sequence depth (λ1000_UC = 1.05, λ1000_CD = 1.04, λ1000_IBD =1.06, Supplementary Figure 5), while still retaining power to detect known associations. While we estimate that this stringent quality control produced well calibrated association test statistics for more than 99% of sites, this analysis yielded many extremely significant p-values at SNPs outside of known loci (e.g. ~7,000 with p < 10-15), 95% of which had an allele frequency below 5%. In contrast to GWAS, where routine procedures almost completely eliminate false positive associations, the heterogeneity of our sequencing depths makes it challenging to discern true associations from these data alone.
Imputation into GWAS
As noted by a previous study of type 2 diabetes27 with a similar design, our WGS dataset alone is not well powered to identify new associations, even if all samples were sequenced at the same depth. We therefore built a phased reference panel of 10,971 individuals from our low coverage whole genome sequences and 1000 Genomes Phase 3 haplotypes (Supplementary Note), in order to use imputation to leverage IBD GWAS to increase our power. Previous data have shown that such expanded reference panels significantly improve imputation accuracy of low-frequency variants28. We next generated a new UK IBD GWAS dataset by genotyping 8,860 IBD patients without previous GWAS data and combining them with 9,495 UK controls from the Understanding Society project (www.understandingsociety.ac.uk), all genotyped using the Illumina HumanCoreExome v12 chip. We then added previous UK IBD GWAS samples that did not overlap with those in our sequencing dataset29,30. Finally, we imputed all of these samples using the PBWT31 software and the reference panel described above, and combined these imputed genomes with our sequenced genomes to create a final dataset of 16,267 IBD cases and 18,843 UK population controls (Supplementary Table 6).
This imputation produced high quality genotypes at 12 million variants that passed typical GWAS quality control (Supplementary Note), and represented more than 90% of sites with MAF >0.1% that we could directly test in our sequences. Compared to the most recent meta-analysis by the International IBD Genetics Consortium32, which used a reference panel almost ten times smaller than ours, we tested an additional 2.5 million variants for association to IBD Because our GWAS cases and controls were genotyped using the same arrays, they should be not be differentially affected by the variation in sequencing depths in the reference panel, and thus not susceptible to the artifacts observed in the sequence-only analysis. Indeed, compared to the thousands of false-positive associations present in the sequence-only analysis, the imputation based meta-analysis revealed only four previously undescribed genome-wide significant IBD associations. Three of these had MAF > 10%, so we carried them forward to a meta-analysis of our data and published IBD GWAS summary statistics33.
Asp439Glu in ADCY7 doubles risk of ulcerative colitis
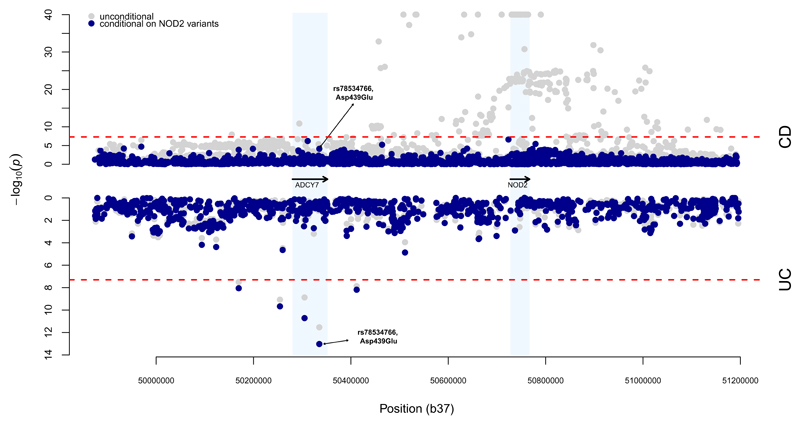
The fourth new association (P = 9x10-12) was a 0.6% missense variant (p.Asp439Glu, rs78534766) in ADCY7 that doubles risk of ulcerative colitis (OR=2.19, 95% CI =1.75-2.74), and is strongly predicted to alter protein function (SIFT = 0, PolyPhen = 1, MutationTaster = 1). This variant was associated (p=1x10-6) in a subset of directly genotyped individuals, suggesting the signal was unlikely to be driven by imputation errors. To further validate it we genotyped (Online Methods) an additional 450 ulcerative colitis cases and 3,905 controls (p=0.0009) and looked it up in 982 ulcerative colitis cases and 136,464 controls from the UK Biobank (p=0.0189). A meta-analysis of all three directly genotyped datasets showed genome-wide significant association (p=1.6x10-9), no evidence for heterogeneity (p=0.19) and clean cluster plots (Supplementary Table 7, Supplementary Figure 6). A previous report described an association between an intronic variant in this gene and Crohn’s disease34, but our signal at this variant (P = 2.9x10-7) vanishes after conditioning on the nearby associations at NOD2, (conditional P = 0.82). By contrast, we observed that p.Asp439Glu shows nominal association with Crohn’s disease after conditioning on NOD2 (P = 7.5x10-5, OR=1.40), while the significant signal remains for ulcerative colitis (Figure 2). Thus, one of the largest effect single alleles associated with ulcerative colitis lies, apparently coincidentally, only 300 kilobases away from a region of the genome that contains multiple large effect Crohn’s disease risk alleles (Figure 2).
Figure 2. Association analysis for the NOD2/ADCY7 region in chromosome 16.
Results from the single variant association analysis are presented in gray, and results after conditioning on seven known NOD2 risk variants in blue. Results for Crohn’s disease (CD) are shown in the top half, and ulcerative colitis (UC) in the bottom half. The dashed red lines indicate genome-wide significance, at α = 5x10-8.
The protein encoded by ADCY7, adenylate cyclase 7, is one of a family of ten enzymes that convert ATP to the ubiquitous second messenger cAMP. Each has distinct tissue-specific expression patterns, with ADCY7 being expressed in haemopoietic cells. Here, cAMP modulates innate and adaptive immune functions, including the inhibition of the pro-inflammatory cytokine TNFα, itself the target of the most potent current therapy in IBD35. Indeed, myeloid-specific Adcy7 knockout mice (constitutive knockouts die in utero) show higher stimulus-induced production of TNFα by macrophages, impairment in B cell function and T cell memory, an increased susceptibility to LPS-induced endotoxic shock, and a prolonged inflammatory response36,37. In human THP-1 (monocyte-like) cells, siRNA knockdown of ADCY7 also leads to increased TNFα production.38 p.Asp439Glu affects a highly conserved amino acid in a long cytoplasmic domain immediately downstream of the first of two active sites and may affect the assembly of the active enzyme through misalignment of the active sites39.
Low-frequency variation makes a minimal contribution to IBD susceptibility
The associated variant in ADCY7 represents precisely the class of variant that our study design was intended to probe: below 1% MAF, OR ~2, and difficult to impute (only 1 copy of the non-reference allele was observed in the Phase 1 1000 Genomes, and INFO=0.7 when imputing33 from Phase 3), making it notable as our single discovery of this type. We had 66% power to detect that association, and reasonable power even for more difficult scenarios (e.g. 29% for 0.2% MAF and OR=2, or 11% for 0.5% MAF and OR=1.5). As noted by others40, heritability estimates for low frequency variants as a class are exquisitely sensitive to potential bias from technical and population differences. We therefore analyzed only the imputed GWAS samples to eliminate the effect of differential sequencing depth, and applied a more stringent SNP and sample quality control (Supplementary Note and Supplementary Figure 7). We used the restricted maximum likelihood (REML) method implemented in GCTA41 and estimated that autosomal SNPs with MAF > 0.1% explain 28.4% (s.e. 0.016) and 21.1% (s.e. 0.012) of the variation in liability for Crohn’s and ulcerative colitis, respectively. Despite SNPs with MAF < 1% representing approximately 81% of the variants included in this analysis, they explained just 1.5% of the variation in liability. While these results are underestimates due to limitations of our data and the REML approach, it seems very unlikely that a large fraction of IBD risk is captured by variants like ADCY7 p.Asp439Glu. Thus, our discovery of ADCY7 actually serves as an illustrative exception to a series of broader observations42 that low-frequency, high-risk variants are unlikely to be important contributors to IBD risk.
The role of rare variation in IBD risk
Our low coverage sequencing approach does not perfectly capture very rare and private variants because the cross-sample genotype refinement adds little information at sites where nearly all individuals are homozygous for the major allele. Similarly, these variants are difficult to impute from GWAS data: even using a panel of more than 32,000 individuals offers little imputation accuracy below 0.1% MAF28. Thus, while our sequence dataset was not designed to study rare variants, it is the largest to date in IBD, and has sufficient specificity and sensitivity to warrant further investigation (Supplementary Figure 8). Because enormous sample sizes would be required to implicate any single variant, we used a standard approach from exome sequencing43, where variants of a particular functional class are aggregated into a gene-level test. We extended Derkach et al’s Robust Variance Score statistic44 to account for our sequencing depth heterogeneity, because existing rare variant burden methods gave systematically inflated test statistics.
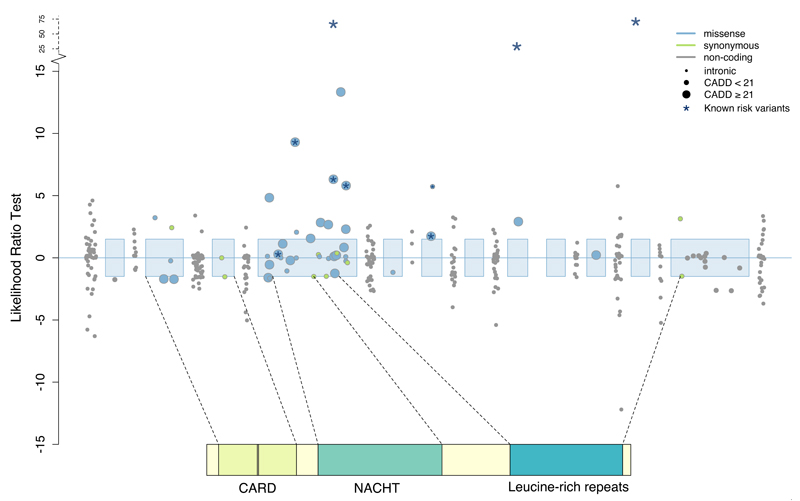
For each of 18,670 genes, we tested for a differential burden of rare (MAF ≤ 0.5% in controls, excluding singletons) functional or predicted damaging coding variation in our sequenced cases and controls (Online Methods, Supplementary Table 8-9). We detected a significant burden of damaging rare variants in the well-known Crohn’s disease risk gene NOD2 (Pfunctional = 1x10−7, Supplementary Figure 9), which was independent of the known low-frequency NOD2 risk variants (Online Methods). We noted that the additional variants (Figure 3) that contribute to this signal explain only 0.13% of the variance in disease liability, compared to 1.15% for the previously known variants11, underscoring the fact that very rare variants cannot account for much population variability in risk.
Figure 3. Associations between NOD2 and Crohn’s disease.
Each point represents the contribution of an individual variant to our NOD2 burden test. Three common variants (rs2066844, rs2066845, rs2066847) are shown for scale, and the six rare variants identified by targeted sequencing are starred. Exonic regions (not to scale) are marked in blue, with their corresponding protein domains highlighted.
Some genes implicated by IBD GWAS had suggestive p-values, but did not reach exome-wide significance (P=5x10-7, Supplementary Table 10), so we combined individual gene results into two sets: (i) 20 genes that had been confidently implicated in IBD risk by fine-mapping or functional data, and (ii) 63 additional genes highlighted by less precise GWAS annotations (Supplementary Note, Supplementary Table 11). We tested these two sets (after excluding NOD2, which otherwise dominates the test) using an enrichment procedure43 that allows for differing direction of effect between the constituent genes (Supplementary Note, Supplementary Table 12). We found a burden in the twelve confidently implicated Crohn’s disease genes that contained at least one damaging missense variant (Pdamaging = 0.0045). By contrast, we saw no signal in the second, more generic set of genes (P=0.94, Figure 4, Table 1).
Figure 4. Burden of rare damaging variants in Crohn’s disease.
Each point represents a gene in our confidently implicated (green) or generically implicated (blue) gene sets. Genes are ranked on the x-axis from most enriched in cases to most enriched in controls, and position on the y-axis represents significance. The purple shaded region indicates where 75% of all genes tested lie. Our burden signal is driven by a mixture of genes where rare variants are risk increasing (e.g. NOD2) and risk decreasing (IL23R).
Table 1. Burden of rare, predicted damaging (CADD ≥ 21) coding variation in IBD gene sets.
| Gene set | Constituents | Phenotype | P-value |
|---|---|---|---|
| NOD2 | NOD2 | CD | 4.00 x 10-07 |
| Other IBD genes implicated by causal coding or eQTL variants (genes in brackets had zero contributing rare variants) | CARD9, FCGR2A, IFIH1, IL23R, MST1, (SMAD3), TYK2, (IL10), IL18RAP, (ITGAL), NXPE1, TNFSF8 | UC | 0.46153 |
| ATG16L1, CARD9, CD6, FCGR2A, FUT2, IL23R, MST1, (NOD2), PTPN22, (SMAD3), TYK2, ERAP2, (IL10), IL18RAP, (IL2RA), (SP140), TNFSF8 | CD | 0.00448 | |
| CARD9, FCGR2A, IL23R, MST1, (SMAD3), TYK2, (IL10), IL18RAP, TNFSF8 | IBD | 0.00261 | |
| Other IBD GWAS genes | Genes implicated by two or more candidate gene approaches in Jostins et al (2012) | UC | 0.95123 |
| CD | 0.94382 | ||
| IBD | 0.93070 |
We extended this approach to evaluate rare regulatory variation, using enhancer regions described by the FANTOM5 project (Supplementary Table 13). Within each robustly defined enhancer45, we tested all observed rare variants, as well as the subset predicted to disrupt or create a transcription factor binding motif18. We combined groups of enhancers with cell- and/or tissue-type specific expression, in order to improve power in an analogous fashion to the gene set tests above. However, none of these tissue or cell specific enhancer sets had a significant burden of rare variation after correction for multiple testing (Supplementary Table 14).
Discussion
We investigated the role of low frequency variants of intermediate effect in IBD risk through a combination of low-coverage whole genome sequencing and imputation into GWAS data (Figure 5). We discovered an association to a low frequency missense variant in ADCY7, which represents one of the strongest ulcerative colitis risk alleles outside of the major histocompatibility complex. The most straightforward mechanistic interpretation of this association is that loss-of-function of ADCY7 reduces production of cAMP, leading to an excessive inflammatory response that predisposes to IBD. Previous evidence suggested that general cAMP-elevating agents that act on multiple adenylate cyclases might, in fact, worsen IBD46. While members of the adenylate cyclase family have been considered potential targets in other contexts 39, specific upregulation of ADCY7 has not yet been attempted, raising the intriguing possibility that altering cAMP signalling in a leukocyte-specific way might offer therapeutic benefit in IBD.
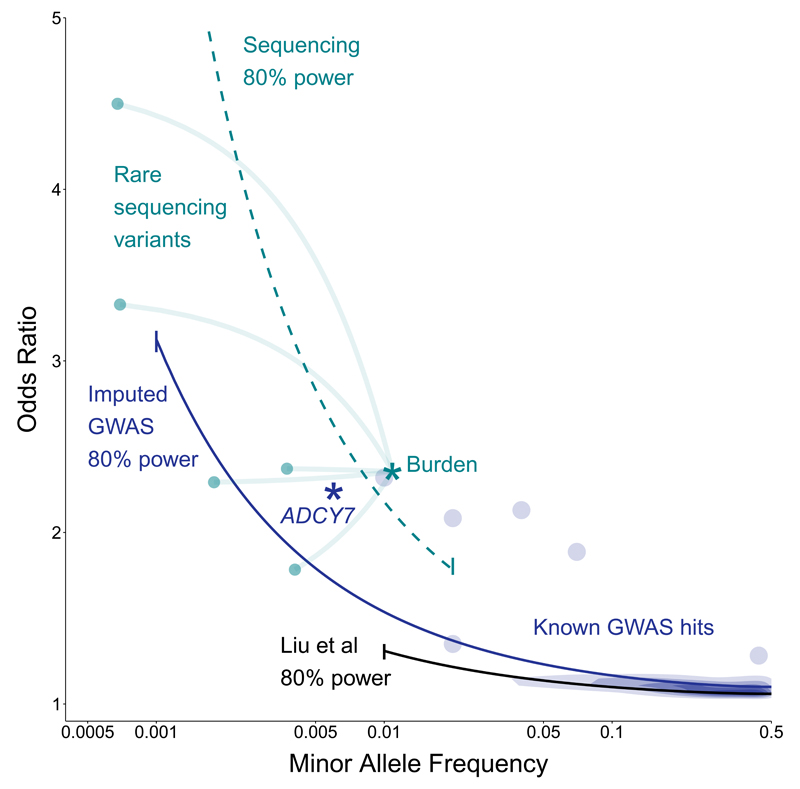
Figure 5. Relative power of this study compared to previous GWAS.
The black line shows the path through frequency-odds ratio space where the latest IIBDGC meta-analysis had 80% power. The purple line (imputed GWAS) and green line (sequencing) shows the same for this study. The earlier study had more samples but restricted their analysis to MAF > 1%. Purple density and points show known GWAS loci, with our novel ADCY7 association (p.Asp439Glu) highlighted as a star. Green points show a subset of our sequenced NOD2 rare variants, and the green star shows their equivalent position when tested by gene burden, rather than individually.
In order to maximize the number of IBD patients we could sequence, and thus our power to detect association, we sequenced our cases at lower depth than the controls available to us via managed access. While joint and careful analysis largely overcame the bias this introduces, this is just one example of the complexities associated with combining sequencing data from different studies. Such challenges are not just restricted to low coverage whole-genome sequencing designs; variable pulldown technology and sequencing depth in the 60,000 exomes in the Exome Aggregation Consortium47 necessitated a simultaneous analysis of such analytical complexity and computational intensity that it would be prohibitive at all but a handful of research centers. Therefore, if rare variant association studies are to be as successful as those for common variants, computationally efficient methods and accepted standards for combining sequence datasets need to be developed.
We have participated in one such joint analysis by contributing to the Haplotype Reference Consortium28 (HRC), which has collected WGS data from more than 32,000 individuals into a reference panel that allows accurate imputation of low-frequency and common variants. Indeed, imputation into GWAS from the HRC is as accurate as low-coverage sequencing at allele frequencies as low as 0.05%28, so by far the most effective way to discover complex disease associations to variants in this range is to re-analyze the huge quantities of existing GWAS data with improved imputation. While projects like ours have provided wider public benefit through the HRC, there is little need for future low-coverage whole genome sequencing projects in complex disease.
Despite our study being specifically designed to interrogate both coding and non-coding variation, our sole new association was a missense variant. This is perhaps unsurprising, as the only previously identified IBD risk variants with similar frequencies and odds ratios are protein-altering changes to NOD2, IL23R and CARD9. More generally, the alleles with largest effect sizes at any given frequency tend to be coding18, and are therefore the first to be discovered when new technologies expand the frequency spectrum of genetic association studies. This pattern is further reinforced by the contrast between the tantalizing evidence we found for a burden of very rare coding variants in previously implicated IBD genes and the absence of any signal across the enhancer regions we tested. This distinction emphasizes how dramatically better we can distinguish likely functional from neutral variants in coding compared to non-coding sequence. For example, if we include all rare coding variants (MAF ≤ 0.5% in controls, N=136) in IBD genes the P-value is 0.2291, compared to P=0.0045 when using the subset of 54 coding variants with CADD ≥ 21. Therefore, the identification of rare variant burdens in the non-coding genome will require not only tens of thousands of samples to be sequenced, but also much better discrimination between functional and neutral variants in regulatory regions.
Nonetheless, it is likely that rare variants play an important role in IBD risk, and that many such alleles are regulatory, as is the case for common risk variants. The ADCY7 association offers a direct window on a new IBD mechanism, but would probably eventually have been discovered through HRC imputation in existing GWAS samples, and is a relatively meager return compared to the number of loci discovered more simply by increasing GWAS sample size33. Making real progress on rare variant association studies will require much larger numbers of deep exomes or whole genomes, especially if “ultra-rare” variants are as important in IBD as they are in, for example, schizophrenia48, . Extrapolating10 for IL23R, the IBD gene with the most significant coding burden (p=0.0005) after NOD2, we would require roughly 20,000 cases to reach genome-wide significance; as we noted above the challenge is even greater for non-coding regions where functional variants cannot currently be distinguished from neutral. Together, our discoveries suggest that a combination of continued GWAS coupled to new imputation reference panels, and large scale deep sequencing studies will be needed to complete our understanding of the genetic basis of complex diseases.
Data availability
Whole genome sequence data that supports this study has been deposited in the European Genome-phenome Archive (EGA) under the accession codes EGAD00001000409 and EGAD00001000401. Genotype data is available under accession code EGAS00001000924.
Online Methods
Preparation of genome-wide genetic data
Sample ascertainment and sequencing
British IBD cases, diagnosed using accepted endoscopic, histopathological and radiological criteria, were sequenced to low depth (2-4x) using Illumina HiSeq paired-end sequencing. Population controls, also sequenced to low depth (7x) using the same protocol, were obtained from the UK10K project. Supplementary Table 2 provides details on sample numbers and quality control filters. Case sequence data was aligned to the human reference used in Phase II of the 1000 Genomes project49. Control data was aligned to an earlier human reference (1000 Genomes Phase I)50, and then updated to the same reference as the cases using BridgeBuilder, a tool we developed (Supplementary Note).
Genotype calling and quality control
Variants were joint called across 8,424 samples, using samtools and bcftools for SNVs and INDELs, and GenomeSTRiP for copy number variants. Copy number variants were filtered using standard GenomeSTRiP quality metrics as described in the Supplementary Note. SNVs were filtered using support vector machines (SVMs) trained on variant quality statistics output from samtools. Each variant was required to pass with a minimum score of 0.01 from at least two out of five independent SVM models. Indels were filtered using GATK VQSR, with a truth sensitivity threshold of 97% (VQSLOD score of 1.0659).
Genotype refinement and further quality control
Following initial SNV and INDEL quality control, genotypes at all passing sites were refined via BEAGLE25. Variants were then filtered again to remove those showing significant evidence of deviation from Hardy-Weinberg equilibrium (HWE) in controls (PHWE<1x10-7), a significant frequency difference (P < 1x10-3) in samples sequenced at the Wellcome Trust Sanger Institute versus the Beijing Genomics Institute, >10% missing genotypes following refinement (posterior probability < 0.9), SNPs within three base pairs of an INDEL, and allow only one INDEL to pass when clusters of INDELs were separated by two or fewer base pairs. Following these exclusions, a second round of genotype refinement was performed. Sample quality control was then applied to remove samples with an excessive heterozygosity rate (µ ± 3.5σ), duplicated or related individuals, and individuals of non-European ancestry (Supplementary Note and Supplementary Figure 10).
Novel GWAS samples
A further 11,768 British IBD cases and 10,484 population control samples were genotyped on the Human Core Exome v12 chip. Detailed information on ascertainment, genotyping and quality control are described elsewhere33.
Existing GWAS cohorts
1,748 Crohn’s disease cases and 2,936 population controls genotyped on the Affymetrix 500K chip, together with 2,361 ulcerative colitis cases and 5,417 population controls genotyped on the Affymetrix 6.0 array, were obtained from the Wellcome Trust Case Control Consortium (WTCCC)29,30. Both datasets were converted to build 37 using liftOver51.
Imputation
The whole genome sequences described above were combined with 2504 samples from the Phase 3 v5 release of the 1000 Genomes project (2013-05-02 sequence freeze) to create a phased imputation reference panel enriched in IBD-associated variants. We used PBWT52 to impute from this reference panel (114.2 million total variants) into the three GWAS panels described above, after removing overlapping samples. This results in imputed whole genome sequences for 11,987 cases and 15,189 controls (Supplementary Table 6).
Common and low-frequency variation association testing
Association testing and meta-analysis
We tested for association to ulcerative colitis, Crohn’s disease and IBD separately within the sequenced samples and three imputed GWAS panels using SNPTEST v2.5, performing an additive frequentist association test conditioned on the first ten principal components for each cohort (calculated after exclusion of the MHC region). We filtered out variants with MAF < 0.1%, INFO < 0.4, or strong evidence for deviations from HWE in controls (pHWE<1x10-7), and then used METAL (release 2011-03-05)53 to perform a standard error weighted meta-analysis of all four cohorts. Only sites for which all cohorts passed our quality control filters were included in our meta-analysis.
Quality control
The output of the fixed-effects meta-analysis was further filtered, and sites with high evidence for heterogeneity (I2>0.90) were discarded. In addition, we discarded all genome-wide significant variants for which the meta-analysis p-value was not lower than all of the cohort-specific p-values. Finally, and in order to minimise the false positive associations due to mis-imputation, sites which did not have an info score ≥ 0.8 in at least three of the four datasets (two of the three for Crohn’s disease and ulcerative colitis) were removed.
Locus definition
A linkage disequilibrium (LD) window was calculated for every genome-wide significant variant in any of the three traits (Crohn’s disease, ulcerative colitis, IBD), defined by the left-most and right-most variants that are correlated with the main variant with an r2 of 0.6 or more. The LD was calculated in the GBR and CEU samples from the 1000 Genomes Phase 3, release v5 (based on 20130502 sequence freeze and alignments). Loci with overlapping LD windows, as well as loci whose lead variants were separated by 500kb or less, were subsequently merged, and the variant with the strongest evidence of being associated was kept as the lead variant for each merged locus. This process was conducted separately for each trait. A locus was annotated as known when there was at least one variant in it that was previously reported (Supplementary Table 15) to be of genome-wide significance (irrespective of the LD between that variant and the most associated variants in the locus), and as novel otherwise.
Conditional analysis
Conditional analyses were conducted using SNPTEST 2.5 54, as for the single variant association analysis. P-values were derived using the score test (default in SNPTEST v2.5). In order to fully capture the NOD2 signal when investigating the remaining signal in the region, we conditioned on seven variants which are known to be associated: rs2066844, rs2066845, rs2066847, rs72796367, rs2357623, rs184788345, and rs104895444.
Replication of the ADCY7 association
Following quality control33, an additional 450 UK ulcerative colitis cases and 3905 population controls (Dupuytren’s contracture cases), genotyped using the Illumina Human Core Exome array v12, were available for replication. An additional 982 ulcerative colitis cases and 136,464 controls from the UK Biobank, genotyped on either the UK Biobank Axiom or UK BiLEVE array, formed a second replication cohort . Quality control of the UK biobank data was performed as previously described (http://biobank.ctsu.ox.ac.uk/crystal/docs/genotyping_qc.pdf), and non-British or Irish individuals were excluded from further analysis. Cases were defined as those with self-reported ulcerative colitis or an ICD10 code of K51 in their Hospital Episode Statistics (HES) record. Controls were defined as those individuals without a self-diagnosis or hospital record of ulcerative colitis or Crohn’s disease (HES = K50). Logistic regression conditional on 10 principal components test was carried out in both replication cohorts. We used METAL (release 2011-03-05)53 to perform a standard error weighted meta-analysis of all three directly genotyped cohorts.
Heritability explained
The SNP heritability analysis was performed on the dichotomous case-control phenotype using constrained REML in GCTA41 with a prevalence of 0.005 and 0.0025 for Crohn’s disease and ulcerative colitis respectively. Hence, all reported values of h2g are on the underlying liability scale. To further eliminate spurious associations we computed genetic relationship matrices (GRMs) restricted to all variants with MAF ≥ 0.1%, imputation r2 ≥ 0.6, missing rate ≤ 1% and Hardy-Weinberg equilibrium P-value ≤ 1x10-7 in controls for each GWAS cohort. We further checked the reliability and robustness of our estimates by performing a joint analysis across all autosomes, a joint analysis between common (MAF≥1%) and rare variants (0.1%≤MAF<1%), and LD-adjusted analysis using LDAK55 (Supplementary Note, Supplementary Table 16, Supplementary Figure 7).
Rare variation association testing
Additional variant quality control
Additional site filtering was undertaken, as rare variant association studies are more susceptible to differences in read depth between cases and controls (Supplementary Figure 11). This included removing singletons, as well as sites with: missingness rate > 0.9 when calculated using genotype probabilities estimated from the samtools genotype quality (GQ) field; low confidence observations comprising ≥ 1% of non-missing data, or; INFO < 0.6 in the appropriate cohorts.
Association testing
Individual gene and enhancer burden tests were performed using an extension of the Robust Variance Score statistic44 (Supplementary Note), to adjust for the systematic coverage bias between cases and controls. This required the estimation of genotype probabilities directly from samtools (using the genotype quality score), as genotype refinement using imputation results in poorly calibrated probabilities at rare sites. Burden tests were performed across sites with a MAF ≤ 0.5% in controls and within genes defined by Ensembl, or enhancers as based on its inclusion in the FANTOM5 ‘robustly-defined’ enhancer set45. For each gene, two sets of burden tests were performed: all functional coding variants and all predicted damaging (CADD ≥ 21) functional coding variants (Supplementary Table 8). For each enhancer, burden tests were repeated to include all variants falling within the region, and just the subset predicted to disrupt or create a transcription factor binding motif (Supplementary Note).
NOD2 independence testing
We evaluated the independence of the rare NOD2 signal from the known common coding variants in this gene (rs2066844, rs2066845, and rs2066847). Individuals with a minor allele at any of these sites were assigned to one group, and those with reference genotypes to another. Burden testing was performed for this new phenotype in both variant sets that contained a significant signal in Crohn’s disease vs controls.
Set definition
The individual burden test statistic was extended to test across sets of genes and enhancers using an approach based on the SMP method43, whereby the test statistic for a given set is evaluated against the statistics from the complete set (e.g. all genes), to account for residual case-control coverage bias. The sets of genes confidently associated with IBD risk were defined based on implication of specific genes in ulcerative colitis, Crohn’s disease or IBD risk through fine-mapping, eQTL and targeted sequencing studies (Supplementary Table 11). The broader set of IBD genes was defined as any remaining genes implicated by two or more candidate gene approaches in Jostins et al (2012)56. Enhancer sets were defined as those showing positive differential expression in each of 69 cell types and 41 tissues, according to Andersson et al45 (Supplementary Table 17).
Supplementary Material
Acknowledgements
We would like to thank all individuals who contributed samples to the study. This work was co-funded by the Wellcome Trust [098051] and the Medical Research Council, UK [MR/J00314X/1]. Case collections were supported by Crohn’s and Colitis UK. KMdL, LM, YL, CAL, CAA and JCB are supported by the Wellcome Trust [098051; 093885/Z/10/Z]. KMdL is supported by a Woolf Fisher Trust scholarship. CAL is a clinical lecturer funded by the NIHR. HU is supported by the Crohn’s & Colitis Foundation of America (CCFA), and the Leona M. and Harry B. Helmsley Charitable Trust. We acknowledge support from the Department of Health via the NIHR comprehensive Biomedical Research Centre awards to Guy’s and St Thomas’ NHS Foundation Trust in partnership with King’s College London and to Addenbrooke’s Hospital, Cambridge in partnership with the University of Cambridge, and the BRC to the Oxford IBD cohort study, University of Oxford. This research was also supported by the NIHR Newcastle Biomedical Research Centre. The UK Household Longitudinal Study is led by the Institute for Social and Economic Research at the University of Essex and funded by the Economic and Social Research Council. The survey was conducted by NatCen and the genome-wide scan data were analysed and deposited by the Wellcome Trust Sanger Institute. Information on how to access the data can be found on the Understanding Society website https://www.understandingsociety.ac.uk/. We are grateful for genotyping data from the British Society for Surgery of the Hand Genetics of Dupuytren's Disease consortium, and Lorraine Southam for assistance with genotype intensities. This research has been conducted using the UK Biobank Resource.
Footnotes
Author contributions
YL, KMdL, LJ, LM, JCB and CAA performed statistical analysis. YL, KMdL, LJ, LM, JCL, CAL, EGS, JR, MaP, SN, and SMC processed the data. TA, CE, NAK, AH, CH, JCM, JCL, CM, WGN, JS, AS, MT, HU, DCW, NJP, CWL, CGW, MP, and CGM contributed samples/materials. YL, KMdL, LM, JCL, MP, CAL, NAK, JCB and CAA wrote the paper. All authors read and approved the final version of the manuscript. JCM, MP, CWL, TA, NJP, JCB and CAA conceived & designed experiments.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–989. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parkes M, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–832. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki K, et al. A Genome-Wide Association Study Identifies 2 Susceptibility Loci for Crohn’s Disease in a Japanese Population. Gastroenterology. 2013;144:781–788. doi: 10.1053/j.gastro.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kenny EE, et al. A genome-wide scan of Ashkenazi Jewish Crohn’s disease suggests novel susceptibility loci. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julià A, et al. A genome-wide association study identifies a novel locus at 6q22.1 associated with ulcerative colitis. Hum Mol Genet. 2014;23:6927–6934. doi: 10.1093/hmg/ddu398. [DOI] [PubMed] [Google Scholar]
- 7.Yang S-K, et al. Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut. 2014;63:80–87. doi: 10.1136/gutjnl-2013-305193. [DOI] [PubMed] [Google Scholar]
- 8.Ellinghaus D, et al. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuk O, et al. Searching for missing heritability: designing rare variant association studies. Proc Natl Acad Sci U S A. 2014;111:E455–64. doi: 10.1073/pnas.1322563111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaudoin M, et al. Deep Resequencing of GWAS Loci Identifies Rare Variants in CARD9, IL23R and RNF186 That Are Associated with Ulcerative Colitis. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt KA, et al. Negligible impact of rare autoimmune-locus coding-region variants on missing heritability. Nature. 2013;498:232–235. doi: 10.1038/nature12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prescott NJ, et al. Pooled sequencing of 531 genes in inflammatory bowel disease identifies an associated rare variant in BTNL2 and implicates other immune related genes. PLoS Genet. 2015;11:e1004955. doi: 10.1371/journal.pgen.1004955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Do R, et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh T, et al. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016;19:571–577. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang H, Fang M, Jostins L, Mirkov MU, Boucher G. Association mapping of inflammatory bowel disease loci to single variant resolution. bioRxiv. 2015 doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farh KK-H, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2014 doi: 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.CONVERGE consortium. Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature. 2015;523:588–591. doi: 10.1038/nature14659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danjou F, et al. Genome-wide association analyses based on whole-genome sequencing in Sardinia provide insights into regulation of hemoglobin levels. Nat Genet. 2015;47:1264–1271. doi: 10.1038/ng.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.UK10K Consortium et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526:82–90. doi: 10.1038/nature14962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Browning BL, Browning SR. Improving the accuracy and efficiency of identity-by-descent detection in population data. Genetics. 2013;194:459–471. doi: 10.1534/genetics.113.150029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Handsaker RE, et al. Large multiallelic copy number variations in humans. Nat Genet. 2015;47:296–303. doi: 10.1038/ng.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuchsberger C, et al. The genetic architecture of type 2 diabetes. Nature. 2016;536:41–47. doi: 10.1038/nature18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCarthy S, Das S, Kretzschmar W, Durbin R. A reference panel of 64,976 haplotypes for genotype imputation. bioRxiv. 2015 doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.UK IBD Genetics Consortium et al. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334. doi: 10.1038/ng.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durbin R. Efficient haplotype matching and storage using the positional Burrows–Wheeler transform (PBWT) Bioinformatics. 2014;30:1266–1272. doi: 10.1093/bioinformatics/btu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. doi: 10.1038/ng.3760. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li YR, et al. Meta-analysis of shared genetic architecture across ten pediatric autoimmune diseases. Nat Med. 2015;21:1018–1027. doi: 10.1038/nm.3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahle MK, Myhre AE, Aasen AO, Wang JE. Effects of forskolin on Kupffer cell production of interleukin-10 and tumor necrosis factor alpha differ from those of endogenous adenylyl cyclase activators: possible role for adenylyl cyclase 9. Infect Immun. 2005;73:7290–7296. doi: 10.1128/IAI.73.11.7290-7296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duan B, et al. Distinct roles of adenylyl cyclase VII in regulating the immune responses in mice. J Immunol. 2010;185:335–344. doi: 10.4049/jimmunol.0903474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang LI, Sternweis PC, Wang JE. Zymosan activates protein kinase A via adenylyl cyclase VII to modulate innate immune responses during inflammation. Mol Immunol. 2013;54:14–22. doi: 10.1016/j.molimm.2012.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Risøe PK, et al. Higher TNFα responses in young males compared to females are associated with attenuation of monocyte adenylyl cyclase expression. Hum Immunol. 2015;76:427–430. doi: 10.1016/j.humimm.2015.03.018. [DOI] [PubMed] [Google Scholar]
- 39.Pierre S, Eschenhagen T, Geisslinger G, Scholich K. Capturing adenylyl cyclases as potential drug targets. Nat Rev Drug Discov. 2009;8:321–335. doi: 10.1038/nrd2827. [DOI] [PubMed] [Google Scholar]
- 40.Bhatia G, et al. Subtle stratification confounds estimates of heritability from rare variants. bioRxiv. 2016:048181. doi: 10.1101/048181. [DOI] [Google Scholar]
- 41.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen G-B, et al. Estimation and partitioning of (co)heritability of inflammatory bowel disease from GWAS and immunochip data. Hum Mol Genet. 2014;23:4710–4720. doi: 10.1093/hmg/ddu174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell SM, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derkach A, et al. Association analysis using next-generation sequence data from publicly available control groups: The robust variance score statistic. Bioinformatics. 2014;30:2179–2188. doi: 10.1093/bioinformatics/btu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmerman NP, Kumar SN, Turner JR, Dwinell MB. Cyclic AMP dysregulates intestinal epithelial cell restitution through PKA and RhoA. Inflamm Bowel Dis. 2012;18:1081–1091. doi: 10.1002/ibd.21898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Exome Aggregation Consortium et al. Analysis of protein-coding genetic variation in 60,706 humans. bioRxiv. 2016:030338. doi: 10.1101/030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genovese G, et al. Increased burden of ultra-rare protein-altering variants among 4,877 individuals with schizophrenia. Nat Neurosci. 2016;19:1433–1441. doi: 10.1038/nn.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The 1000 Genomes Project Consortium. The 1000 Genomes Project Phase II. 2011 Available at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/phase2_reference_assembly_sequence/hs37d5.fa.gz.
- 50.The 1000 Genomes Project Consortium. The 1000 Genomes Project Phase I. 2010 Available at ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/human_g1k_v37.fasta.gz.
- 51.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–8. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Durbin R. Efficient haplotype matching and storage using the positional Burrows-Wheeler transform (PBWT) Bioinformatics. 2014;30:1266–1272. doi: 10.1093/bioinformatics/btu014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 55.Speed D, Balding DJ. MultiBLUP: improved SNP-based prediction for complex traits. Genome Res. 2014;24:1550–1557. doi: 10.1101/gr.169375.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole genome sequence data that supports this study has been deposited in the European Genome-phenome Archive (EGA) under the accession codes EGAD00001000409 and EGAD00001000401. Genotype data is available under accession code EGAS00001000924.