Abstract
A labile selenium donor compound monoselenophosphate is synthesized from selenide and ATP by selenophosphate synthetase (SPS). In the present study, Sps1 and Sps2 were cloned from a cDNA library prepared from human lung adenocarcinoma cells (NCIH441). The human lung Sps1 has been cloned as an ORF of 1,179 bp, identical in sequence to that of the recently revised human liver Sps1. The in-frame TGA codon of the lung Sps2 was genetically altered to TGT (Cys) to obtain the Sps2Cys gene. Expression of the recombinant plasmids containing Sps1 or Sps2Cys was highly toxic to Escherichia coli host cells grown aerobically. Accordingly, the human lung Sps homologs were characterized by an in vivo complementation assay using a selD mutant strain. An added selenium source and a low salt concentration (0.1-0.25% NaCl) in the medium were required for reproducible and sensitive in vivo complementation. Sps2Cys effectively complemented the selD mutant, and the resulting formate dehydrogenase H activity was as high as that of WT E. coli MC4100. In contrast, only a weak complementation of the selD mutant by the Sps1 gene was observed when cells were grown in selenite media. Better complementation with added l-selenocysteine suggested involvement of a selenocysteine lyase for mobilization of selenium. Based on this apparent substrate specificity of the Sps1 and Sps2 gene products we suggest that the Sps1-encoded enzyme depends on a selenium salvage system that recycles l-selenocysteine, whereas the Sps2 enzyme can function with a selenite assimilation system.
In many biological systems, the concentration of sulfur-containing compounds is on the order of 1,000 times greater than their selenium analogs (1). Thus a selenium-specific pathway for biosynthesis of proteins that contain selenocysteine (SeCys) residues inserted as directed by the UGA codon is required. In Escherichia coli the insertion of selenium into selenium-dependent enzymes and Se-containing tRNAs requires the participation of the selD gene product (2). This protein, later identified as selenophosphate synthetase (SPS), forms a highly reactive, reduced selenium compound, monoselenophosphate (3). Monoselenophosphate is the product of the reaction catalyzed by SPS in which the γ-phosphoryl group of ATP is transferred to selenide and inorganic phosphate and AMP are formed (4, 5). The mechanism of this reaction has yet to be determined. The identification of an essential cysteine residue, Cys-17, in the N-terminal glycine-rich region of the E. coli enzyme (SELD) has led to the assumption that this residue behaves as a nucleophile in the hydrolysis of ATP (6, 7). Positional isotope exchange experiments demonstrated that an enzyme phosphoryl-intermediate is formed during catalysis (8). However, repeated attempts to trap an enzyme thiophosphate intermediate have failed, suggesting that Cys-17 of E. coli SELD may have a different catalytic role.
The E. coli selD gene product and its homologs present in mammals and Drosophila can be divided into two major groups. One group of SPS enzymes, which have a cysteine or SeCys residue at the site corresponding to Cys-17 in E. coli SELD, can catalyze the selenide-dependent formation of monoselenophosphate in vitro (3, 4, 9, 10). Replacement of Cys-17 with serine results in the complete loss of activity with ATP and selenide as substrates (6). Analysis of the total genomic sequences of Methanococcus jannaschii (11) and Haemophilus influenzae (12) revealed that the selD gene in these organisms possesses a TGA codon at the position of the E. coli Cys-17, indicating the presence of a SeCys residue (9, 10). Moreover, SeCys-containing variants also were identified in mouse and human enzymes (9). Replacement of the SeCys residue of the mouse enzyme with cysteine decreased but did not abolish enzyme activity (13), whereas changing Cys-17 of SELD to Ser destroyed catalytic activity (6). The second group of SELD homologs present in human (14), mouse (9), and Drosophila (15) share high sequence similarity with the bacterial SPS but lack a cysteine or SeCys residue in the position that corresponds to Cys-17 of the E. coli enzyme. The human homolog has a threonine substitution (14), and a homolog from Drosophila melanogaster contains an arginine (15). The overproduced threonine-containing human enzyme weakly complemented a selD lesion in E. coli, and transfection of the gene into mammalian cells resulted in an increased 75Se labeling of mammalian selenium-dependent deiodinase (14).
Eukaryotic organisms including D. melanogaster, mouse, and human differ from bacteria and archaea in that they have a pair of Sps genes; one encodes a selenoenzyme capable of in vitro catalysis, and the other homolog encodes an enzyme exhibiting poor catalytic activity for selenide-dependent hydrolysis of ATP. Physiological roles of Sps1 and Sps2 remain to be elucidated, and the present study was initiated with an aim to clarify their roles in selenium homeostasis in mammals. It has been shown previously that Sps2 is specifically expressed at very early stages of development in vivo, namely in mouse embryonic sites of hematopoiesis (9). Based on the demonstration that Sps2 is a T cell activation gene, the up-regulation of Sps2 in T cells may have a role in directing the production of selenophosphate to the synthesis of selenoproteins involved in the immune response rather than in processes related to cell growth and division.
We have cloned Sps1 and Sps2 genes from the human lung adenocarcinoma cell line NCI-H441, in which the selenoprotein thioredoxin reductase is synthesized in much higher amounts than in normal cells (16). The human lung adenocarcinoma Sps1 was cloned as an ORF of 1,179 bp, and the nucleotide sequence was identical to that of the recently revised human liver Sps1 sequence. Sps2 had two point mutations, one of which was a Thr301Ala mutation. The in-frame opal codon directing SeCys in the lung Sps2 was genetically altered to a Cys-encoding codon, and the resulting gene was designated Sps2Cys.
Materials and Methods
Materials. E. coli strains MC4100 and WL400 (ΔselD) (2) were obtained from the E. coli Genetic Stock Center at Yale University, New Haven, CT.
DNA Manipulation. General DNA manipulations were performed as described by Sambrook and Russel (17). Restriction enzymes and DNA polymerase were purchased from Toyobo (Osaka) and Takara (Kyoto). Plasmid DNA was isolated by using a GFX Micro Plasmid Prep Kit from Amersham Pharmacia Bioscience. DNA ligation was performed by using DNA ligation kit version 1 (Takara). Ultrafree-DA purchased from Millipore was used to recover DNA fragments from SeaKem (Rockland, ME) GTG agarose gels. DNA sequencing was performed by using a DYEnamic ET terminator cycle sequencing kit (Amersham Pharmacia) and a model 310 capillary sequencer (Applied Biosystems). Each PCR was carried out in a 20-μl volume of 1× PCR buffer in the iCycler thermocycler system (BioRad).
Construction of Recombinant Plasmid Containing the Sps2Cys Gene. NCI-H441 cells were purchased from the American Type Culture Collection and maintained in RPMI medium 1640 with 10% heat-inactivated FBS (Sigma) at 37°C in humidified air with 5% CO2. When cells reached about 70% confluence mRNA was isolated by using the poly(A)pure mRNA isolation kit (Ambion, Austin, TX). cDNA synthesis was performed by using 3 μg of mRNA and an Omniscript RT kit (Qiagen, Valencia, CA). Human lung Sps2 gene was amplified by using a HotStarTaq PCR kit (Qiagen) and a set of primers (Sps2Up, 5′-CGGGATCCATGGCGGAAGCCTCGGCGA-3′; and Sps2Dw, 5′-CCCAAGCTTTCACGAGCTAGGCTCAGAGGA-3′. The underlined sequences indicate BamHI and HindIII sites, respectively. PCR products were ligated into the pCR2.1 vector by using a TOPO TA cloning kit (Invitrogen). Recombinant plasmids were subjected to DNA cycle sequencing in the sense and antisense directions. The in-frame TGA codon of human lung Sps2 was changed to the cysteine-encoding codon TGT with a QuikChange Site-Directed Mutagenesis Kit (Stratagene) using a set of primers (Sps2CysUp, 5′-ATGAAGGGCTGTGGCTGCAAG-3′ and Sps2CysDw, 5′-CTTGCAGCCACAGCCCTTCAT-3′) and pCR-Sps2 as a template. The resulting gene Sps2Cys was subcloned into the BamHI and HindIII sites of pQE30 (Qiagen), and the recombinant plasmid was designated pQE30Sps2Cys.
Construction of Recombinant Plasmid Containing Sps1 Genes. The Sps1 gene was amplified from the cDNA library of human lung adenocarcinoma cells by using a HotStarTaq PCR kit (Qiagen) and a pair of primers (Sps1UP, 5′-CGCGGATCCATGTCTACGCGGGAGTCCTTTA-3′ and Sps1Dw, 5′-ACGCGTCGACTCACGTTTTGAGTGGCCACTTG-3′. The underlined sequences indicate BamHI and SalI sites, respectively. The primers were designed based on the Sps1 sequence of 1,152 bp reported for human liver cDNA (14). The PCR product was subjected to DNA sequencing, and the Sps1 gene was found to have a frame-shift mutation caused by C insertion at 1130, resulting in extension of the ORF to 1,179 bp. To confirm the Sps1 sequence, another cDNA library was prepared by using a ReverTraAce RT-PCR kit (Toyobo), and the gene was amplified by using a KOD-plus DNA polymerase kit (Toyobo) with a set of primers (Sps1Up2, 5′-CGCGGATCCATGTCTACGCGGGAGTCCT T TA ACC-3′ and Sps1Dw2, 5′-ACGCGTCGACTTAAGAGGTGGCCCCGGGTGT-3′. The underlined sequences indicate BamHI and SalI sites, respectively. PCR products were incubated with 2 mM dATP and 10 units of Ex-TaqDNA polymerase (Takara) at 70°C for 3′ adenylation and cloned in the pCR2.1 vector. The DNA sequencing in the sense and antisense directions confirmed the C1130 insertion. The C1130 of human lung Sps1 was removed by QuikChange site-directed mutagenesis using a set of primers (ΔC1130Up, 5′-ATCGAGGTCGCACACAAGTGGCCACT-3′ and ΔC1130Dw, 5′-AGTGGCCACTTGTGTGCGACCTCGAT-3′) and pCR-Sps1 as a template. The entire subcloned gene was sequenced to confirm that no other mutation were present. Recombinant genes Sps1 and Sps1ΔC1130 were subcloned in BamHI and SalI sites of the expression vector pQE30, and the expression plasmids were designated pQE30Sps1 and pQE30Sps1ΔC1130.
Multiple Alignment and Phylogenetic Tree. The multiple protein sequence alignment was carried out with clustal w (18) using the sequences of Methanopyrus kandleri selD (GenBank accession no. AE010430), M. jannaschii selD (GenBank accession no. F64498), Geobacter sulfurreducens selD (GenBank accession no. AAR33938), D. melanogaster Sps1 (GenBank accession nos. O18373, O18597, and Q9V700) and Sps2 (GenBank accession no. Q8IPC0), E. coli selD (GenBank accession nos. P16456 and P78172), H. influenzae selD (GenBank accession no. P43911), Yersinia pestis selD (GenBank accession no. Q8ZEK1), Mus musculus (mouse) Sps1 (GenBank accession no. Q8BH69) and Sps2 (GenBank accession no. P97364), Caenorhabditis elegans probable SPS (GenBank accession no. O62461), Caenorhabditis briggsae hypothetical SPS (GenBank accession no. CAE58592), Campylobacter jejuni selD (GenBank accession no. Q9PMF9), Thermoanaerobacter tengcongensis selD (GenBank accession no. Q8R8W3), Salmonella typhimurium selD (GenBank accession no. NP460263), Aquifex aeolicus selD (GenBank accession no. O67139), Rhizobium meliloti selD (GenBank accession no. Q931D0), and Eubacterium acidaminophilum selD1 (GenBank accession no. AJ245960) and selD2 (GenBank accession no. AJ249161). The phylogenetic tree was calculated by the neighbor-joining method applied to the distance of difference derived from the multiple alignments (18).
Plasmid Stability Test. A colony from the transformation plate was inoculated into 2 ml of LB medium containing 50 μg/ml carbenicillin and incubated until the culture became slightly turbid. Cells were streaked on agar plates containing carbenicillin, and single colonies were used to inoculate 2 ml of LB-1% glucose containing either 0 or 50 μg/ml carbenicillin. After growth at 37°C slightly turbid cultures were diluted to 10-5, and cells were streaked on LB agar containing 1% isopropyl β-d-thiogalactoside (IPTG) with or without 50 μg/ml added ampicillin; 10-6 dilutions of cells were plated on LB agar with or without added ampicillin.
Complementation of an E. coli selD Mutation by Human Lung Sps1 and Sps2Cys. In this study, E. coli, strain WL400 (ΔselD) cells (2) transformed with the Sps1 or Sps2Cys gene, were tested for formate dehydrogenase H (FDHH) activity in an in vitro assay (19). Fresh transformant cells were streaked on LB-glucose agar plates and grown for 2 days at 30°C in Anaerocult A mini filled with CO2 (Merck). The plates then were overlaid with soft agar containing 1 mg/ml benzyl viologen, 0.25 M sodium formate, and 25 mM KH2PO4 (19-21). Development of a purple color upon reduction of the viologen dye indicated that the bacterial cells contained catalytically active FDHH.
Results
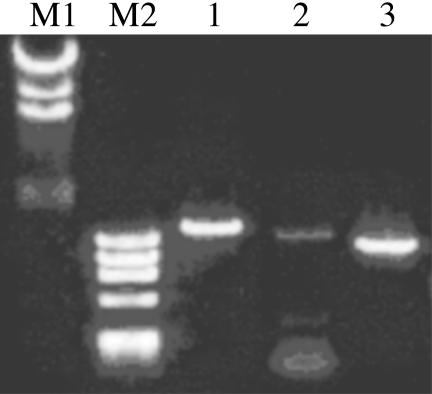
RT-PCR of Sps1 and Sps2 from Human Lung Adenocarcinoma Cells. Human Sps1 and Sps2 genes were amplified from the cDNA library synthesized from human lung adenocarcinoma cells (NCI-H441) (Fig. 1). The PCR product of Sps1 gave an intense signal on agarose gel electrophoresis. In contrast, a very weak amplification signal was obtained for Sps2, whose specific amplification required the presence of Q solution (Qiagen); no signals were obtained without the optimizer solution even when Mg concentration and thermal programs were varied.
Fig. 1.
RT-PCR of human lung Sps genes from mRNA extracted from NCIH441. Lane M1, λ/HindIII molecular markers; lane M2, ΦX174/HaeIII molecular markers; lane 1, cytosolic thioredoxin reductase (TrxR1) amplified as a positive control; lane 2, human lung Sps2 gene amplified in the presence of Q solution; lane 3, Sps1 gene amplified in the presence of 1.5 mM MgCl2 without Q solution.
Construction of an Expression Plasmid Containing the Sps2Cys Gene. The PCR product for Sps2 was cloned into the TA cloning vector pCR2.1, and the resulting plasmid was designated pCRSps2. DNA sequence analysis revealed two point mutations in the ORF of 1,347 bp; A901 → G resulted in a Thr301Ala change but G1221A was a silent mutation of Val-407. A mutant cDNA containing a TGT (Cys) codon in place of TGA (SeCys) was obtained by oligonucleotide mutagenesis, and the resulting plasmid was designated pCR-Sps2Cys. pCR-Sps2Cys was digested with BamHI and HindIII, and the 1.2-kb fragment was inserted into the corresponding site of pQE30 to yield pQE30Sps2Cys.
Construction of Expression Plasmids Containing Sps1. The Sps1 gene was cloned from a human lung cDNA library prepared by using a Omniscript RT-PCR kit and a HotStarTaq PCR kit. The PCR primer was designed on the sequence of human liver Sps1 (14), and nucleotide sequencing identified a frame-shift mutation caused by the C1130 insertion. Another cDNA library was made with a ReverTraAce RT-PCR kit, and the Sps1 gene was amplified by using KOD-plus DNA polymerase. The frame shift caused by the C1130 insertion was reproduced in the second round of RT-PCR, and the human lung Sps1 gene encoding an ORF of 1,179 bp was designated Sps1. The human liver Sps1 sequence, which recently was revised as an ORF encoding 1,179 bp, now was completely consistent with the sequence of the human lung Sps1.
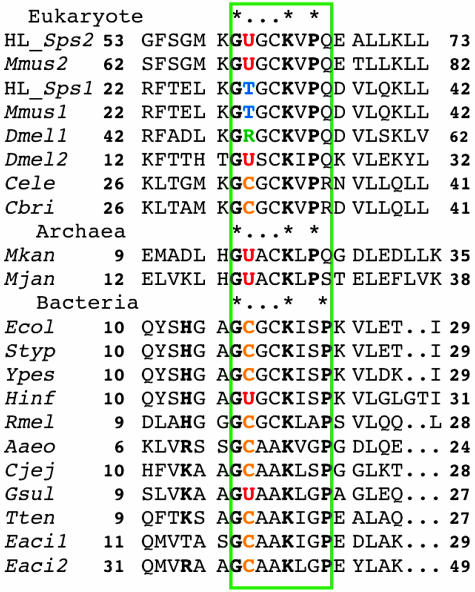
Multiple Alignment and Phylogenic Tree. The Cys-17 in E. coli SPS (SELD) occurs in a glycine-rich sequence, -Gly-16-Cys-17-Gly-18-Cys-19-Lys-20-Ile-21-Ser-22-Pro-23. Multiple alignments of amino acid sequences of bacterial, archaea, and eukaryote SPS variants identified conserved sequences corresponding to the E. coli glycine-rich sequence (Figs. 2 and 3). The residue corresponding to Gly-16 in E. coli SPS is conserved in all of the sequences. Residues corresponding to Gly-18 and Cys-19 also are found in many SPS homologs, including human lung SPS1 and SPS2, but they are replaced with alanine in bacterial and archea SPS variants. Lys-20 is a catalytically essential residue in E. coli SELD (22), and it is conserved in all of the SPS homologs. One of three hydrophobic residues, valine, isoleucine, or leucine, follows the Lys-20, and a helix-breaking l-proline occurs either in position 22 or 23. If the highly conserved region from Gly-16 to Ile/Val/Leu-21 is in either an α-helix or a loop conformation, Lys-20 could be close enough to interact with Cys-17, thus effectively lowering the pKa value for the thiol ionization.
Fig. 2.
Partial amino acid sequence alignment of SPS homologs from the human lung cancer cell (HuLung), mouse (Mmus), D. melanogaster (Dmel), C. elegans (Cele), C. briggsae (Cbri), Methanopyrus kandleri (Mkan), M. jannaschii (Mjan), E. coli (Ecol), S. typhimurium (Styp), Y. pestis (Ypes), H. influenzae (Hinf), R. meliloti (Rmel), A. aeolicus (Aaeo), C. jejuni (Cjej), G. sulfurreducens (Gsul), T. tengcongensis (Tten), and E. acidaminophilum (Eaci1 and Eaci2).
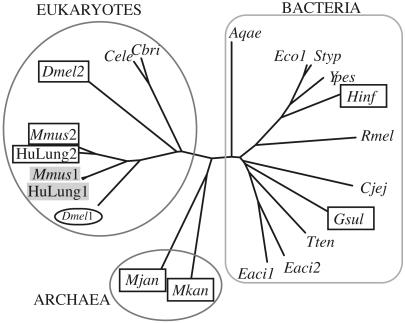
Fig. 3.
Dendrogram showing the divergence of SPS homologues. Boxes around human lung Sps2 (HuLung2), mouse Sps2 (Mmus2), D. melanogaster Sps2 (Dmel2), H. influenzae selD (Hinf), G. sulfurreducens Sps (Gsul), M. jannaschii Sps (Mjan), and Methanopyrus kandleri Sps (Mkan) denote their special character as selenoenzymes. Mouse Sps1 (Mmus1) and human lung Sps1 (HuLung1) designated in shaded boxes appeared to have Thr, whereas the D. melanogaster Sps1 (Dmel1) has Arg residue at the corresponding position, suggested by the multiple sequence alignment of all of the available SPS homologs.
Expression of Recombinant Sps Genes in E. coli. Despite extensive efforts, expression of recombinant human lung Sps genes in E. coli cells has not been successful. We have subcloned Sps genes in a number of vectors, including pUC18, pKK233-3, pET32, and pQE30, and used host strains JM109, DH5α, BL21(DE3), and M15. E. coli ABLE-K strain (Toyobo) that suppresses the plasmid copy number by 1/10th that of normal E. coli hosts also was tested with the recombinant plasmids, pQE30Sps1 and pQE30Sps2Cys. However, no Coomassie blue-stained bands corresponding to SPS proteins were detected on SDS/PAGE. Expression of Sps1 and Sps2Cys genes in E. coli WL400 cells under anaerobic conditions gave similar results.
Plasmid Stability Test. Recombinant plasmids normally are stable and are retained by a very high fraction of host cells even after growth for many generations in the absence of antibiotics. However, problems of plasmid instability can arise when a gene whose product is toxic to the host cell is present in the plasmid. To determine the fraction of cells carrying the recombinant Sps gene, freshly transformed cells grown in the absence or presence of 50 μg/ml carbenicillin were plated on four agar media of differing composition (Table 1). When E. coli WL400 cells harboring the pQE30 vector were grown under aeration, most of the cells retained the plasmid and grew on the ampicillin-containing agar. When the host cell transformed with the Sps2Cys gene was grown in the absence of carbenicillin, the recombinant plasmid was very unstable and none of the cells retained the recombinant plasmid. When the pQE30Sps2Cys transformants were grown in the presence of carbenicillin, there were fewer viable cells and the plasmid was barely retained in the surviving cells. The β-lactamase enzyme, secreted in the medium in the early stages of growth, may have degraded the selective antibiotic, thus allowing growth of cells lacking the plasmid. The human lung Sps1 gene appeared to be stably retained in the E. coli host cells, but growth was much slower than the cells with pQE30 vector alone. Normally, addition of IPTG to the agar will prevent colony formation by cells containing both the inducible gene and a functional target plasmid, but not the growth of cells lacking the plasmid or mutants that have lost the ability to express target DNA. The large fraction of cells able to grow on IPTG-containing LB agar suggested that a majority of the recombinant cells were unable to express the recombinant gene. When the growth test was carried out in the presence of carbenicillin, most of the surviving cells were found to be insensitive to IPTG induction. Sps1Δ1130C was made from the Sps1 gene by removing the C1130. The behavior of recombinant Sps1Δ1130C gene resembled that of Sps2, indicating the recombinant plasmid was unstable in the host cell. The cells transformed with Sps1Δ1130C gene could not retain the recombinant plasmid either in the absence or presence of carbenicillin.
Table I. Plasmid stability test.
| Without carbenicillin*
|
With carbenicillin*
|
||||||
|---|---|---|---|---|---|---|---|
| Media | pQE30 | Sps2Cys | Sps1 | Sps1Δ1130C | Sps2Cys | Sps1 | Sps1Δ1130C |
| LB agar | 2,600 | 1,390 | 220 | 450 | 70 | 70 | 860 |
| + Ampicillin | 2,300 | 0 | 220 | 0 | 0 | 170 | 0 |
| + IPTG | 290 | 745 | 68 | 202 | 35 | 165 | 45 |
| + Ampicillin, IPTG | 300 | 0 | 6 | 12 | 8 | 79 | 0 |
All viable cells can grow on the plate with no additives; cells that retain plasmid will grow on the plates containing ampicillin; cells that have lost plasmid or lost the ability to express target DNA will grow in the presence of IPTG; and mutants that retain plasmid but have lost the ability to express target DNA will grow in the presence of both ampicillin and IPTG.
E. coli WL400 was transformed with pQE30Sps2Cys, pQE30Sps1, and Sps1Δ1130C genes, respectively, and grown in 2 ml of LB containing 1% glucose with 50 g/ml carbenicillin or without carbenicillin.
The Medium Optimization for in Vivo Complementation Assay. In previous studies, the in vivo selD complementation tests have been carried out by monitoring FDHH activity of E. coli cells grown in media such as LB agar containing d-glucose (20, 21). We found that lower NaCl concentrations (0.1-0.25%) in the media and supplementation with selenium markedly improved the reproducibility and sensitivity of the complementation assay. Accordingly, the agar medium used consisted of 1.6% polypepton, 1% yeast extract, 1% agar, and 0.1% NaCl (pH 7), 10 μM Na2MoO4, and 1 μM Na2SeO3 or 1 μM l-SeCys. Fresh transformant cells were streaked on agar plates and incubated at 30°C in a plastic bag filled with CO2. Addition of IPTG to the agar medium interfered with color development and was not included. Sodium selenite, up to 10 μM or l-SeCys, 0.1 or 1 μM, served as a satisfactory selenium source. However, 10 μM l-SeCys inhibited the growth of E. coli MC4100 (WT), WL400 (ΔSelD), and E. coli WL400 cells transformed with Sps2Cys or Sps1. Selenite, >10 μM, also is toxic for many bacterial species (23).
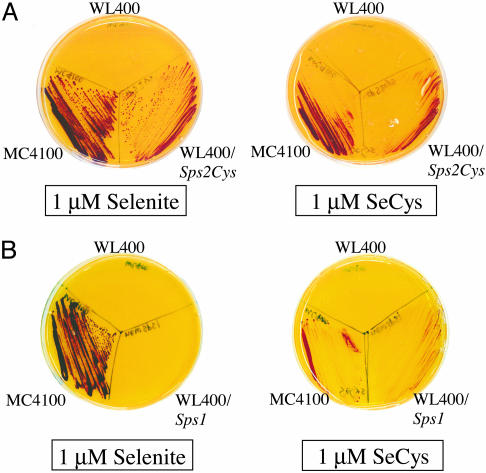
Complementation of selD Lesion by Human Lung Sps1 and Sps2Cys. The human lung Sps1 or Sps2Cys genes cloned in pQE30 complemented the bacterial selD deficiency to different extents. Although the active site SeCys-60 was genetically replaced with a Cys residue, human lung Sps2Cys allowed E. coli WL400 cells to produce FDHH activity on the agar media supplemented with 1 μM sodium selenite. The FDHH activity detected for WL400/pQE30Sps2Cys was as high as that of WT MC4100 in which intact selD was functional. When selenium was supplied as 1 μM l-SeCys, the detected FDHH activity was less than with sodium selenite, suggesting that selenite was a better selenium source for Sps2Cys than l-SeCys (Fig. 4A). In contrast, E. coli cells transformed with the Sps1 gene showed low FDHH activity when sodium selenite was added as a selenium source. Interestingly, the Sps1 gene allowed E. coli host cells to produce detectable FDHH activity when l-SeCys was added as the selenium source (Fig. 4B). Although the method of detection of E. coli FDHH activity is indirect and thus a nonquantitative determination of in vivo SPS activity, the higher FDHH activity observed suggests that the human lung SPS1 generated monoselenophosphate from selenium that was mobilized from free l-SeCys. This finding is consistent with previous reports on the mobilization of selenium from l-SeCys, providing a better selenium source for SelD in vivo.
Fig. 4.
SelD complementation by human lung Sps genes. E. coli MC4100 (WT), WL400 (selD), and WL400 transformed with pQE30Sps2Cys (A) or pQE30Sps1 (B) were grown anaerobically at 30°C on a plate containing 10 μM sodium molybdate and 1 μM sodium selenite or 1 μM l-SeCys. Soft agar containing 0.75% agar, 1 mg/ml benzyl viologen, 0.25 M sodium formate, and 25 mM KH2PO4 was laid over the culture. Colonies with active FDHH reduced benzyl viologen, which develops a deep blue color.
Discussion
The present study has demonstrated that human lung adenocarcinoma cells (NCI-H441) possess mature mRNA molecules encoding Sps1 and Sps2. Recombinant genes, Sps1 and Sps2Cys, were highly toxic to E. coli host cells when grown under aerobic conditions. However, the two gene products could be characterized in an in vivo complementation assay using the selD-deficient E. coli WL400 strain grown under anaerobic conditions. For optimal expression of the Sps genes it was necessary to decrease the NaCl content of the usual rich medium and add Se and Mo supplements. The mutant Sps2Cys complemented the selD mutation as effectively as the intact selD does in WT strain MC4100. The presence of Sps2Cys allowed the E. coli host to produce the SeCys-containing FDHH when the medium was supplemented with sodium selenite or l-SeCys. In contrast, complementation with Sps1 was relatively ineffective, particularly when selenite was used as a selenium source for FDHH synthesis.
The specific gene amplification of Sps2 from the cDNA library of lung adenocarcinoma cells required the addition of Q solution; yet the yield was still smaller than the amplified Sps1 gene when compared on agarose gel electrophoresis. Because the two homologs share high sequence homology, it is likely that the in vivo transcription of Sps2 was much lower than Sps1 in the adenocarcinoma cells.
It was reported by Kim et al. (13) that replacement of SeCys by cysteine in fetal mouse Sps2 reduced but did not eliminate enzyme activity. Accordingly, the human lung adenocarcinoma Sps2 was genetically altered in this study to the cysteine-containing homolog. The selD-defective host harboring recombinant Sps2Cys gene exhibited FDHH activity equal to that of the WT E. coli strain, indicating a high catalytic potency of Sps2Cys gene product for selenophosphate synthesis. In fact, human lung Sps2Cys was able to use both selenite-derived and l-SeCys-derived selenium as substrate. In contrast, Sps1, the human homolog containing threonine, only weakly complemented a selD mutation in E. coli when sodium selenite was the selenium source. The human lung Sps1 specifically required SeCys-derived selenium for monoselenophosphate production, suggesting the involvement of l-SeCys β-lyase enzymes. A similar requirement was observed with a mutant form of SPS from E. coli that contained serine in place of Cys-17. This mutant was inactive in vitro with selenide as selenium source, but when selenium mobilized from SeCys by a lyase protein was supplied a low, but detectable, level of selenophosphate was produced (20). Based on these observations it seems that human Sps1 and the E. coli Cys-17/Ser SPS mutant are functional analogs in terms of selenium substrate requirement.
The apparent participation of l-SeCys β-lyase enzymes in selenophosphate biosynthesis in bacteria was first observed for selenoprotein A of the glycine reductase complex in Clostridium sticklandii. 75Se derived from [75Se]-SeCys was incorporated into the selenoprotein more efficiently than 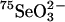 (24). It also was demonstrated that the NifS protein from Azobactor vinelandii (25) and the NifS-like proteins from E. coli (20) effectively mobilized selenium from free SeCys and the selenium was used for monoselenophosphate production by SPS. Furthermore, Lacourciere (23) demonstrated that selenium mobilized from l-SeCys by NifS-like protein was specifically incorporated into E. coli FDHH.
(24). It also was demonstrated that the NifS protein from Azobactor vinelandii (25) and the NifS-like proteins from E. coli (20) effectively mobilized selenium from free SeCys and the selenium was used for monoselenophosphate production by SPS. Furthermore, Lacourciere (23) demonstrated that selenium mobilized from l-SeCys by NifS-like protein was specifically incorporated into E. coli FDHH.
NifS-like proteins have a conserved cysteine residue near the active site, and the residue is proposed to bind selenium and deliver it as a substrate for selenium metabolizing enzymes such as SPS (26). It seems likely that the human lung Sps1 also used the E. coli system that provides protein-bound selenium as the substrate.
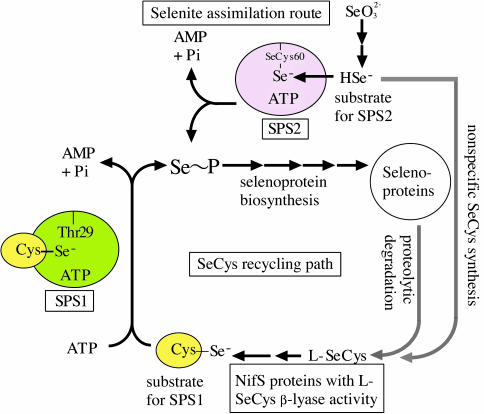
The substrate specificity of human lung Sps gene products characterized in the present study has allowed us to hypothesize that Sps2 functions in the pathway of de novo synthesis of selenophosphate from selenite after reduction of the latter by intracellular thiols. The reduced selenium is bound to the SeCys-60 residue of Sps2 to form an enzyme-substrate complex (Fig. 5). Alternatively, for Sps1 catalysis, NifS-like proteins in mammalian cells would supply an elemental form of selenium derived via a SeCys-salvage pathway. In bacteria selenide can be converted to a trace amount of free l-SeCys through nonspecific incorporation via the cysteine biosynthetic pathway. Alternatively, SeCys can be formed from selenoproteins through proteolysis during protein turnover. These pathways could provide a source of l-SeCys for the NifS-like proteins. The lethal effect of recombinant Sps genes on the E. coli host under aerobic conditions presumably could be caused by the high levels of this reactive selenium compound produced in the cells. If the enzyme reaction product is equally toxic for mammalian cells, the cellular damage may be controlled by limiting the supply of required substrate for Sps1 product formation or by extremely low expression of Sps2, respectively.
Fig. 5.
Hypothetical selenium assimilation routes in the lung adenocarcinoma cell NCI-H441. Up-regulation of SPS2, capable of using selenide derived from selenite, provides a bypass route, which directly converts selenide into monoselenophosphate, leading to an increased cellular selenium pool. The SeCys-60 residue in Sps2 is proposed to provide a selenide binding site for enzyme-substrate complex formation. SPS1 that lacks a SeCys or Cys residue in the corresponding glycine-rich sequence would require a selenium-delivery system in which activated selenium is supplied as a perselenide (-S-SeH) derivative.
Further study is required to identify the mammalian selenium-delivery protein that can serve as a substrate for the Thr-containing SPS1 protein and also to experimentally prove that a SeCys/Cys residue in SPS2 protein/E. coli SELD actually functions as a substrate-binding residue for the reduced form of selenite.
Acknowledgments
This work was supported in part by Grant-in-Aid for Encouragement of Young Scientist 11780499 from the Ministry of Education, Science, and Culture of Japan, the Mishima Kaiun Memorial Foundation, the Japan Science Society, and the Kurozumi Medical Foundation.
Abbreviations: SPS, selenophosphate synthetase; SeCys, selenocysteine; FDHH, formate dehydrogenase H; IPTG, β-d-thiogalactoside.
References
- 1.Sliwkowski, M. X. & Stadtman, T. C. (1985) J. Biol. Chem. 260, 3140-3144. [PubMed] [Google Scholar]
- 2.Leinfelder W., Forchhammer, K., Veprek, B., Zehelein, E. & Boeck, A. (1990) Proc. Natl. Acad. Sci. USA 87, 543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glass, R. S., Singh, W. P., Jung, W., Veres, Z., Scholz, T. D. & Stadtman, T. C. (1993) Biochemistry 32, 12555-12559. [DOI] [PubMed] [Google Scholar]
- 4.Ehrenreich, A., Forchhammer, K., Tormay, P., Veprek, B. & Boeck, A. (1992) Eur. J. Biochem. 206, 767-773. [DOI] [PubMed] [Google Scholar]
- 5.Veres, Z., Tsai, L., Scholz, T. D., Politino, M., Balaban, R. S. & Stadtman, T. C. (1992) Proc. Natl. Acad. Sci. USA 89, 2975-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim, I., Veres, Z. & Stadtman, T. C. (1992) J. Biol. Chem. 267, 19650-19654. [PubMed] [Google Scholar]
- 7.Veres, Z., Kim, I. Y., Scholz, T. D. & Stadtman, T. C. (1994) J. Biol. Chem. 269, 10597-10603. [PubMed] [Google Scholar]
- 8.Mullins, L. S., Hong, S. B., Gibson, G. E., Walker, H., Stadtman, T. C. & Rauschel, F. M. (1997) J. Am. Chem. Soc. 119, 6684-6685. [Google Scholar]
- 9.Guimaraes, M. J., Peterson, D., Vicari, A., Cocks, B. G., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., Ferrick, D. A., Kastelein, R. A., Bazan, J. F. & Zlotnick, A. (1996) Proc. Natl. Acad. Sci. USA 93, 15086-15091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilting, R., Schorling, S., Persson, B. C. & Boeck, A. (1997) J. Mol. Biol. 266, 637-641. [DOI] [PubMed] [Google Scholar]
- 11.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischman, R. D., Sutton, G. G., Black, J. A., Fitzgerald, L. M., Clayton, R. A., Gocayne, J. D., et al. (1996) Science 273, 1058-1073. [DOI] [PubMed] [Google Scholar]
- 12.Fleishmann, R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A. & Merrick, J. M. (1995) Science 269, 496-512. [DOI] [PubMed] [Google Scholar]
- 13.Kim, I. Y., Guimaraes, M. J., Zlotnik, A., Bazan, J. F. & Stadtman, T.C. (1997) Proc. Natl. Acad. Sci. USA 94, 418-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low, S. C., Harney, J. W. & Berry M. J. (1995) J. Biol. Chem. 270, 21659-21664. [DOI] [PubMed] [Google Scholar]
- 15.Persson, B.C., Boeck, A., Jackle, H. & Vorbruggen, G. (1997) J. Mol. Biol. 274, 174-180. [DOI] [PubMed] [Google Scholar]
- 16.Tamura, T. & Stadtman, T. C. (1996) Proc. Natl. Acad. Sci. USA 93, 1006-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sambrook, J. & Russel, D. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 18.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1995) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mandrand-Berthelot, M. A., Wee, M. Y. K. & Haddock, B. A. (1978) FEMS Mirobiol. Lett. 4, 37-40. [Google Scholar]
- 20.Lacourciere, G. M., Mihara, H., Kurihara, T., Esaki, N. & Stadtman T. C. (2000) J. Biol. Chem. 275, 23769-23773. [DOI] [PubMed] [Google Scholar]
- 21.Lacourciere, G. M. & Stadtman T. C. (1998) J. Biol. Chem. 273, 30921-30926. [DOI] [PubMed] [Google Scholar]
- 22.Kim, I. Y., Veres, Z. & Stadtman, T. C. (1993) J. Biol. Chem. 268, 27020-27025. [PubMed] [Google Scholar]
- 23.Lacourciere, G. M. (2002) J. Bacteriol. 184, 1940-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stadtman, T. C., Dilworth, G. L. & Chen, C. S. (1979) in Proceedings of the Third International Symposium on Organic Selenium and Tellurium Compounds, eds. Cagniant, D. & Kirsh, G. (Universite de Metz, Metz, France), pp. 115-130.
- 25.Zheng, L., White, R. H., Cash, V. L. & Dean, D. R. (1994) Biochemistry 33, 4714-4720. [DOI] [PubMed] [Google Scholar]
- 26.Bordo, D., Forlani, F., Spallarossa, A., Colnaghi, R., Carpen, A., Bologensi, M. & Pagani, S. (2001) Biol. Chem. 382, 1245-1252. [DOI] [PubMed] [Google Scholar]