Abstract
Mature Epstein-Barr virus (EBV) was purified from the culture medium of infected lymphocytes made functionally conditional for Zta activation of lytic replication by an in-frame fusion with a mutant estrogen receptor. Proteins in purified virus preparations were separated by gradient gel electrophoresis and trypsin-digested; peptides were then analyzed by tandem hydrophobic chromatography, tandem MS sequencing, and MS scans. Potential peptides were matched with EBV and human gene ORFs. Mature EBV was mostly composed of homologues of proteins previously found in a herpes virion. However, EBV homologues to herpes simplex virus capsid-associated or tegument components UL7 (BBRF2), UL14 (BGLF3), and EBV BFRF1 were not significantly detected. Instead, probable tegument components included the EBV and γ-herpesvirus-encoded BLRF2, BRRF2, BDLF2 and BKRF4 proteins. Actin was also a major tegument protein, and cofilin, tubulin, heat shock protein 90, and heat shock protein 70 were substantial components. EBV envelope glycoprotein gp350 was highly abundant, followed by glycoprotein gH, intact and furin-cleaved gB, gM, gp42, gL, gp78, gp150, and gN. BILF1 (gp64) and proteins associated with latent EBV infection were not detected in virions.
Keywords: lymphocyte, proteomics, virion, herpes virus
The experiments reported here determine the protein composition of mature enveloped Epstein-Barr (EB) virus (EBV). EBV protein composition has not been systematically studied since the proteins of purified enveloped and deenveloped EBV were initially displayed on polyacrylamide gels. Except for glycoproteins, EB virion protein annotations have usually been based on DNA sequence homology to a characterized herpesvirus ORF, with verification for EBV in specific instances.
EBV is a γ1-herpesvirus. The γ1- and γ2-herpesvirus genomes are mostly collinear and share smaller conserved gene clusters with α- and β-herpesvirus genomes. Herpesvirus family-conserved genes mostly encode proteins that are important for DNA replication, virus morphogenesis, or virion composition. Conserved herpesvirus genes are known to encode 5 capsid proteins, 5 envelope proteins, and 10 tegument proteins in at least one herpesvirus (1, 2). Accordingly, EBV BcLF1, BDLF1, BFRF3, BORF1, and BBRF1 ORFs are likely to encode the major, minor, and smallest capsid proteins (MCP, mCP, and sCP, respectively); the mCP-binding protein (mCPBP); and portal. Furthermore, the EBV BPLF1, BOLF1, BVRF1, BGLF1, BGLF4, BGLF2, BBRF2, BSRF1, BGLF3, and BBLF1 ORFs are likely to encode tegument proteins (3-5). EBV BNRF1 and BLRF2 probably encode γ-herpesvirus-unique tegument proteins (Table 1) (6-8). The EBV homologues of gB (BALF4), gH (BXLF2), and gL (BKRF2) have been detected by specific antibodies in EB virions. Moreover, EBV BLLF1, BZLF2, and BDLF3 ORF-specific antibodies have detected EBV-unique gp350, gp42, and gp150 in the virus (Table 1) (9). EBV BMRF2-encoded protein may also be in virus envelopes because it has an RGD (arginine-glycine-aspartic acid) motif that may be a ligand for an integrin coreceptor (10).
Table 1. Proteins detected in 10 μg of deglycosylated EBV.
| Peptides
|
||||||||
|---|---|---|---|---|---|---|---|---|
| Protein type | EBV ORF | HSV UL | Function | Coverage, % | Total, n | Unique, n | Area, units | MWobs, kDa |
| Capsid | BcLF1† | 19† | MCP | 67 | 112 | 62 | 23,628 | 155 |
| BDLF1 | 18† | mCP | 49 | 25 | 11 | 3,894 | 30 | |
| BFRF3 | 35† | sCP | 64 | 35 | 12 | 3,205 | 18 | |
| BORF1 | 38† | mCPBP | 40 | 17 | 9 | 1,576 | 40 | |
| BBRF1 | 6† | Portal | 34 | 14 | 14 | 300 | 68 | |
| (BdRF1) | 26.5 | Scaffold | 6 | 2 | 2 | 13 | ? | |
| Glycoprotein | BLLF1† | γ1 | gp350 | 29 | 31 | 15 | 4,067 | 350 |
| BXLF2† | 22† | gH | 41 | 30 | 18 | 1,344 | 88 | |
| BALF4 | * | gB-N | 54 | 22 | 14 | 1,014 | 78 | |
| BALF4 | * | gB-C | 50 | 25 | 21 | 966 | 58 | |
| BALF4† | 27† | gB-FL | 40 | 21 | 20 | 372 | 120 | |
| BZLF2† | γ | gp42 | 33 | 9 | 6 | 286 | 42 | |
| BBRF3 | 10† | gM | 17 | 4 | 4 | 74 | ? | |
| BILF2 | γ1 | gp78 | 10 | 2 | 2 | 54 | DG | |
| BLRF1 | 49.5† | gN | 8 | 1 | 1 | 40 | DG | |
| BDLF3† | γ | gp150 | 15 | 3 | 3 | 34 | 115 | |
| BKRF2† | 1† | gL | 18 | 2 | 1 | 21 | 25 | |
| BMRF2† | γ | epi ligand | 3 | 1 | 1 | 21 | DG | |
| Tegument | BPLF1 | 36† | LTP | 39 | 108 | 87 | 3,307 | 350 |
| BGLF2 | 16† | MyrPBP | 67 | 19 | 12 | 1,021 | 32 | |
| BOLF1 | 37† | LTPBP | 31 | 25 | 24 | 944 | 140 | |
| BVRF1 | 25† | Capsid-assoc. | 45 | 20 | 17 | 656 | 58 | |
| BBLF1 | 11† | MyrP | 87 | 16 | 4 | 430 | 15‡ | |
| BGLF1 | 17† | Packaging | 23 | 6 | 6 | 122 | 58 | |
| BSRF1 | 51† | PalmP | 19 | 5 | 4 | 120 | 27‡ | |
| BGLF4 | 13† | TS kinase | 25 | 7 | 7 | 109 | 47 | |
| BNRF1† | γ | MTP | 54 | 87 | 51 | 15,845 | 140 | |
| BLRF2† | γ | Unknown | 64 | 35 | 12 | 3,205 | 19 | |
| BRRF2 | γ | Unknown | 73 | 34 | 23 | 3,152 | 72‡ | |
| BDLF2 | γ | Unknown | 32 | 13 | 10 | 304 | 54 | |
| BKRF4 | γ | Unknown | 18 | 7 | 3 | 219 | 42‡ | |
| BORF2§ | 39 | RNR-L | 35 | 19 | 19 | 249 | 90 | |
| BALF2§ | 29 | ssDNABP | 25 | 16 | 16 | 227 | 130 | |
| BXLF1§ | 23 | TK | 14 | 10 | 7 | 145 | 72 | |
| BMRF1§ | 42 | dsDNABP | 25 | 7 | 7 | 118 | 49 | |
| Host | N/A | Actin | 70 | 63 | 22 | 10,022 | 45 | |
| Host | N/A | Hsp70 | 61 | 54 | 33 | 2,713 | 72 | |
| Host | N/A | Cofilin | 69 | 37 | 15 | 2,670 | 17 | |
| Host | N/A | β-Tubulin | 74 | 37 | 22 | 2,290 | 54 | |
| Host | N/A | Enolase | 54 | 30 | 22 | 1,402 | 49 | |
| Host | N/A | Hsp90 | 53 | 48 | 38 | 1,394 | 90 | |
Coverage as a percentage of ORF, total number of detected peptides, number of unique peptides, total peptide area for all peptides assigned to each protein, and observed molecular sizes are shown from the sequencing of 10 μg of partially deglycosylated EB virion proteins. The observed molecular mass (MWobs) for glycoproteins is from nondeglycosylated virion protein sequencing experiments. Corresponding HSV ORF (HSV UL) and putative function are also shown. γ and γ1 refer to proteins that have only γ- or γ1- herpesvirus homologues. ?, protein was detected over a range of sizes; DG, glycoprotein was only detected in deglycosylated state; N/A, not applicable.
HSV UL27 (gB) does not undergo cleavage into N- and C-terminal polypeptides.
Known virion protein.
Nonglycoprotein with observed molecular size deviating > 10% from that predicted.
Highly abundant infected cell proteins; significance discussed in text.
Herpesviruses also incorporate specific cell proteins. Purified cytomegalovirus and pseudorabies virus contain actin (11, 12). Furthermore, cryoelectron microscopic tomography of herpes simplex virus (HSV) has identified 7-nm fibrils in the tegument that are likely to be polymerized actin (13). Purified cytomegalovirus also contains β2 microglobulin, annexin II, CD55, and CD59 (14-16).
Materials and Methods
Liquid Chromatography Tandem MS (MS/MS) Sequencing of Purified EBV. Three purified mature extracellular EB virion (B95-8 strain) preparations were sequenced a total of four times: (i) 2 μg of purified EB virion proteins, (ii)10 μg of purified EB virion proteins after incubation with N- and O-glycanases (degly), (iii and iv) Mixtures of 5 μg of purified EB virion proteins modified by a 12C (light) isotope-coded affinity tag (ICAT) and 5 μg of 1% Nonidet P-40 (NP-40)-treated or 1% NP-40 and 0.5% deoxycholate (ND)-treated virus from the same preparation modified by a 13C (heavy) ICAT. Details of virus purification, gel electrophoresis, and liquid chromatography MS/MS analysis are in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results
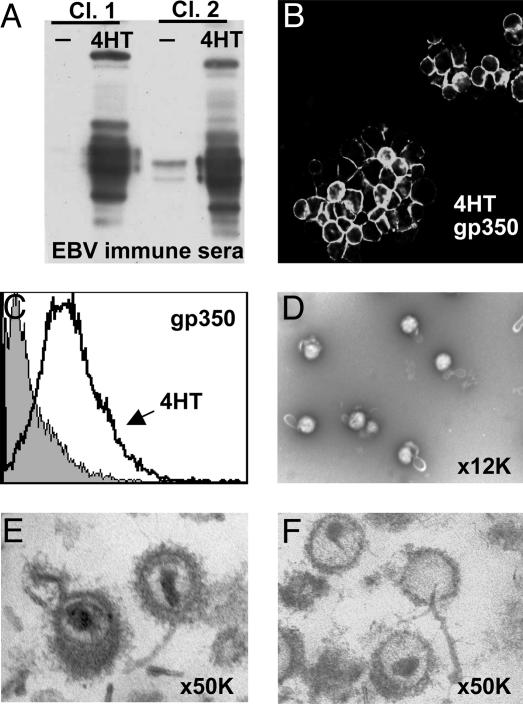
Induction of Lytic EBV Infection in Lymphoblasts by Using Conditional ZHT Fusion Protein. The EBV Zta activator of lytic gene expression (17) was fused to the 4-hydroxytamoxifen (4HT)-responsive mutated estrogen receptor hormone-binding domain (ZHT) to make a putative 4HT-dependent inducer of EBV replication. Stable clones of EBV-latently infected B95-8 lymphoblasts that conditionally express nuclear ZHT were derived by transfection with pcDNA3-ZHT and limit dilution cloning in selective media. Aliquots of these clones were tested for 4HT-dependent lytic EBV protein expression and gp350 surface expression (Fig. 1 A-C). Clones negative before and almost uniformly gp350-positive 2 days after 4HT addition were used in subsequent experiments.
Fig. 1.
Induction of EBV lytic replication in ZHT-converted lymphoblasts. (A) Western blot of 4HT-dependent induction of lytic proteins in B95-8/ZHT cells. (B and C) Gp350 surface staining of 4HT-induced B95-8/ZHT lymphoblasts by fluorescence microscopy (B) and FACS analysis (C). (D) Electron microscopy with negative uranyl acetate staining performed on purified EB virions at ×12,000 magnification. (E and F) Electron microscopy of fixed and stained thin sections of EB virions (E) and NP-40-treated EB virions (F)at ×50,000 magnification.
Extracellular EBV Purification and Deenvelopment with NP-40 or ND. Five days after 4HT addition to ZHT-expressing cells, EBV was collected from the 2.5 liters of cell culture medium and purified by two cycles of velocity centrifugation on Dextran T10 gradients (18). Electron microscopy of negatively stained pellets revealed mostly intact virus with partially angular profiles that likely reflect the underlying capsid (Fig. 1D). Contaminating structures were not observed. Of the virions, 1-5% had stain within the envelope, which is indicative of partial disruption. After pelleting through 30% glycerol, virus preparations that were fixed, stained, embedded, and sectioned were also predominantly enveloped virus (Fig. 1E). The tegument was eccentrically distributed, as has been rigorously described for HSV (13). Some envelopes had extra membrane and some envelope fragments were evident.
To identify proteins that remain virus capsid-associated after partial or complete envelope dissolution, purified virus was treated with NP-40 or ND, and nucleocapsids with varying amounts of residual tegument were recovered as a pellet by centrifugation through 30% glycerol. Fixed, stained, and embedded NP-40-treated virus had very little residual membrane, variable amounts of tegument, and mostly intact nucleocapsids. Some nucleocapsids that lacked tegument were leaking DNA, usually through a capsid vertex (Fig. 1F).
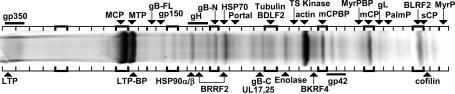
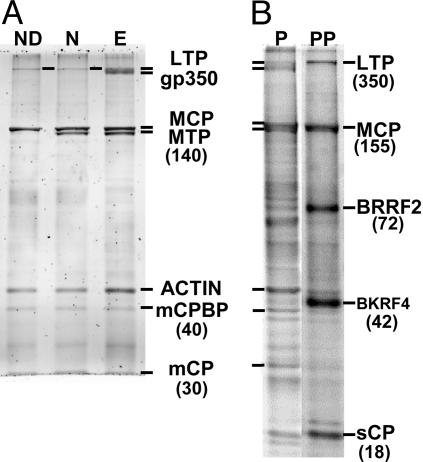
Liquid Chromatography MS/MS Analysis of Proteins Associated with Purified EBV. SYPRO Ruby- or Coomassie brilliant blue-stained, 18-cm SDS/PAGE (5-15%) gels with 2 or 10 μg of purified EBV had remarkably similar protein patterns compared with those reported for [35S]Met-labeled virus (Fig. 2) (18). The doublet of MCP and major tegument protein (MTP) was apparent at 155 and 140 kDa. Diffuse gp350 staining at the top of the gel was removed by NP-40 or ND treatment, exposing the putative large tegument protein (LTP) (Fig. 3A). A 45-kDa protein was equally as intense as MCP and decreased relative to MCP with NP-40 or ND treatment (Fig. 3A), indicative of an unknown major inner tegument protein; this protein was β-actin (see below).
Fig. 2.
Coomassie stain of SDS/PAGE 5-15% gel of purified EB virions. Arrows indicate gel slices from which the indicated proteins were found in peak abundance. Bands that were inconsistently observed in virion preparations and contained only low-abundance, non-EBV proteins are not labeled. Protein names and sizes are given in Table 1; abbreviations are explained in the text.
Fig. 3.
EB virion proteins before and after deenvelopment and virion phosphoproteins. (A) Purified EBV proteins were separated by SDS-polyacrylamide gels directly (lane E) or after treatment with NP-40 (lane N) or ND (lane ND) and stained with Sypro-Ruby. Molecular sizes are given in parenthesis. (B) Purified EB virion proteins were separated on SDS-polyacrylamide gels and stained with Sypro-Diamond to detect phosphoproteins (lane PP). The same gel was restained with Sypro-Ruby to visualize all virion proteins (lane P). BKRF4 is in small type because of its low abundance in virions and phosphopeptides from two host-encoded proteins (protein kinase A and neural-encoded death-determining protein 5) were also detected at 42 kDa (Table 6).
Stained proteins ranging in size from that of LTP and gp350 to 10 kDa and all intervening gel segments were divided into 3-mm slices. Proteins were in-gel digested with trypsin. Peptides were extracted and analyzed by high-pressure reversed-phase nanoflow column chromatography coupled to an on-line lcq deca xp plus proteome x workstation (ThermoFinnigan, Cambridge, MA) for nanospray MS/MS sequencing with intermittent MS scanning. sequest software was used to compare MS/MS data with the nonredundant protein database and the EBV B95-8 genome translated in all six potential reading frames.
To better detect glycoprotein peptides, 10 μg of EBV protein was incubated with deglycosylases before SDS/PAGE (EBV degly) and sequenced (Table 1). Stained gp350 was smaller but still diffuse and larger than the 140-kDa nascent protein. Partial shifts of other EBV glycoproteins were detected by Western blot by using EBV-immune human sera (Fig. 5 and Table 2, which are published as supporting information on the PNAS web site). Partial deglycosylation was probably due to uncleaved O-linked sugars with complex modifications (19).
Relative abundance was estimated for each protein based on gel staining intensity, the number of unique and total peptides sequenced by MS/MS, the overall percentage sequenced (coverage), and total area of peptides detected in MS scans in the peak slice for that protein (Table 1).
EBV Capsid Proteins. Sequencing of the 155-, 40-, and 30-kDa gel slices from 10 μg of degly or 2 μg of native enveloped virus revealed peptides encoded by EBV BcLF1, BDLF1, and BORF1, homologues of MCP, mCP, and mCPBP (Fig. 2 and Table 1). Indeed, MCP at 155 kDa was the most heavily stained protein in enveloped, NP-40-treated, and ND-treated virus (Fig. 3A). MCP peptides were 5-fold more abundant than peptides from the next most frequently detected protein in the 155-kDa slice, which was MTP. MTP is slightly smaller than MCP and was the dominant component of the next smaller gel slice (Fig. 2).
mCP and mCPBP peptides were the most frequently sequenced in the 30- and 40-kDa gel slices, respectively (Fig. 2 and Table 1). sCP peptides were split between the 17- and 19-kDa slices and were half as abundant as BLRF2 peptides (a probable tegument protein, see below) in the 19-kDa slice. Overall, 67% of MCP, 49% of mCP, 40% of mCPBP, and 64% of sCP amino acid residues were specifically identified in the 10-μg virus preparation (Table 1).
Portal (BBRF1) is estimated to be at 12 molecules per nuclear HSV capsid and has been detected in mature HSV virions by Western blot (20, 21). Sequencing of the 68-kDa slice from 2 or 10 μg of enveloped virus revealed peptides spaced throughout the EBV BBRF1 ORF (Table 1). Portal peptides were similar in detection frequency as five other proteins in this weakly stained slice. Nevertheless, the reproducible finding of significant numbers of portal peptides at the appropriate size is strong evidence that portal is in mature EBV.
Two peptides of capsid scaffold protein and two peptides of protease were detected in the 10-μg degly analysis at low peptide areas, consistent (see below) with scaffold or protease not being significant virion components.
The known abundance of herpesvirus capsid proteins, with 955 MCP, 900 sCP, 640 mCP, 320 mCPBP, and 12 portal molecules per virion, provides an internal reference for relative protein abundance. The total masses for MCP, mCP, sCP, mCPBP, and portal capsid protein, calculated from the predicted molecule number per virion times molecular weight, are 147, 22, 16, 13, and 0.8, respectively. Thus, the relative masses of MCP, mCP, sCP, mCPBP, and portal (normalized to MCP) in virions are 100, 15, 12, 9, and 0.6, respectively. This correlated best with relative total MS-detected peptide areas of 100, 16, 14, 7, and 1.3 and less well with the relative total peptides identified by MS/MS of 100, 22, 31, 15, and 13 (Table 1).
EBV Envelope Glycoproteins. Although glycosylated peptides cannot be detected by MS/MS sequencing and EBV glycoproteins have many predicted N- and O-glycosylation sites (Table 2), most EBV glycoproteins were detected near their reported size during sequencing of 2 μg of native virion proteins (Table 1, MWobs). To minimize the effect of glycosylation on peptide detection, analyses of the 10-μg degly preparation provided the primary data set for detection and abundance estimations.
Despite extensive glycosylation, gp350 peptides corresponding to 8% of BLLF1 were abundant from 280 to 350 kDa in the 2-μg EBV native protein experiment. In the 10-μg degly experiment, coverage was 40% and gp350 peptides were the most abundantly sequenced peptides in the lightly stained gel slices from 205 to 310 kDa. Peptides reported in Table 1 reflect only the one slice containing the most gp350. LTP peptides were the most abundantly sequenced in the 350-kDa slice of the 10-μg gel (Fig. 2).
Seven peptides comprising 12% of EBV gB (BALF4) were detected at 125 kDa in 2 μg of enveloped virus, which increased to 20 peptides and 40% coverage at 97 kDa in the 10 μg of degly (Table 1). These findings are consistent with previous reports that mature EB virions have a small amount of full-length gB (9). In the 2-μg nondeglycosylated gel, a further five gB N-terminal peptides were found at 78 kDa, and 13 C-terminal peptides were found in the 58-kDa slice (Fig. 6, which is published as supporting information on the PNAS web site). EBV gB contains a consensus furin cleavage site, and these results suggest that enveloped mature EBV contains both full-length and likely furin-cleaved gB, similar to the γ2-herpesvirus KSHV (22).
EBV gH, gL, and gp42 can form a complex, which would be depleted for gp42 if gp42 associates with MHC class II proteins in B95-8 marmoset lymphoblasts, as in human lymphoblasts (23). EBV gH (BXLF2) peptides were among the most abundant peptides detected at 88 and 85 kDa in the 2- and 10-μg degly gels. One EBV gL (BKRF2) peptide was detected at 27 kDa in the 2-μg gel, and another peptide corresponding to 18% of gL was detected at 13 kDa in 10-μg degly gel. Furthermore, gp42 (BZLF2) was detected at 42 kDa in 2 μg of virus protein, and six peptides corresponding to 33% coverage of gp42 were detected at 27 kDa in the 10 μg of degly.
EBV gp150 (BDLF3) is highly glycosylated and migrates substantially above its predicted size of 26 kDa. Only a single gp150 peptide was detected several times in the 2-μg gel at 115 kDa, and three unique peptides from 15% of gp150 were detected at 80 kDa in the 10-μg degly gel.
EBV gM (BBRF3) and gN (BLRF1) are conserved herpesvirus glycoprotein complexes that have not been detected in EB virions. Peptides of gM corresponding to 12-17% coverage were in multiple slices from 40 to 180 kDa in the 2-μg or 10-μg gels, which is consistent with prior descriptions of gM as gp48/84/113. Herpesvirus gMs have eight predicted membrane helices, which likely cause anomalous mobility. EBV gN was not detected in any slice from the 2-μg gel. Only a single C-terminal peptide corresponding to 8% of gN was detected at 10 kDa in the 10-μg degly gel, close to the expected gN size. A nontryptic peptide search found the peptide that would result from cleavage after the putative gN signal peptide (S33-Y53), increasing total coverage to 28%.
Two gp78 (BILF2) peptides corresponding to 10% of 248 amino acids were detected multiple times from 32 to 36 kDa, corresponding to partially or fully deglycosylated gp78. The unique peptide that would arise as a consequence of cleavage of the predicted signal peptide (FFSDLVKFENVTAHAGAR) was also detected in a nontryptic search, confirming this cleavage.
Only a single peptide corresponding to 3% of gp55 (BMRF2) was in four slices between 225 and 350 kDa in the 10-μg degly EBV protein preparation. BMRF2 multiple hydrophobic domains could account for anomalous migration but not the low-level detection of only a single peptide of 357-aa BMFR2.
In summary, after N- and partial O-deglycosylation, complete analysis of 10 μg of enveloped virus detected very abundant peptides from gp350/220, moderately abundant peptides from gH, less abundant peptides from furin-cleaved and intact gB, and even less abundant peptides from gM, gp42, gL, gp78, gp150, and gN. A single peptide of gp55 (BMRF2) was detected at levels substantially less than portal peptides. Peptides of gp64 (BILF1), LMP-1, and LMP-2 were not detected in any virus preparation.
EBV Tegument or Capsid-Associated Proteins. As anticipated, the EBV LTP is encoded by BPLF1 and the MTP is encoded by BNRF1. From 10 μg of virus protein, 87 LTP peptides encoding 39% of BPLF1 and 51 MTP peptides encoding 54% of BNRF1 were the dominant peptides detected at 350 and 140 kDa, respectively (Fig. 2 and Table 1). Also, 24 peptides of the LTP-binding protein encoded by BOLF1 were moderately abundant at 140 kDa.
Furthermore, peptides from 87% of the 8-kDa, BBLF1-encoded myristoylated phosphoprotein (MyrP) and from 67% of the BGLF2-encoded, MyrP-binding protein were at 15 and 32 kDa, respectively. Moreover, peptides from 45% of the EBV BVRF1 homologue of HSV UL25, which is thought to be the “cork” that prevents genomic DNA from escaping through portal (24), were the second most abundant peptides at 58 kDa.
Peptides from 23% of the EBV capsid-associated UL17 protein homologue (BGLF1), 19% of the EBV palmitoylated protein homologue (BSRF1), and 25% of the EBV protein kinase (BGLF4) were detected at levels similar to portal, at sizes of 58, 27, and 47 kDa, respectively.
Surprisingly, only one peptide of the 32 kDa, BBRF2-encoded putative homologue of HSV UL7 was detected at levels substantially less than portal in the 2-μg or 10-μg degly experiments (Table 3, which is published as supporting information on the PNAS web site). Furthermore, no BGLF3 peptides were detected, even though the HSV homologue, UL14, is a tegument protein (25). BBRF2 and BGLF3 were the only predicted tegument proteins not significantly detected in virus.
In addition to MTP, four γ-herpesvirus-unique proteins were detected. BLRF2 peptides were predominant at 19 kDa and accounted for 64% of its 162 amino acids. Peptides of 73% and 61% of 537-aa BRRF2 were abundantly detected at 72 and 85 kDa. Because 72 kDa is larger than expected, BRRF2 likely undergoes posttranslational modification, including phosphorylation (see below). Peptides from 32% of EBV BDLF2 were frequently detected at 54 kDa, and peptides from 18% of BKRF4 were at 42 kDa, which is larger than the anticipated size of 24 kDa. BLRF2, BRRF2, BDLF2, and BKRF4 are weakly conserved among γ-herpesviruses and are more likely to be tegument than nucleocapsid components.
Peptides of four other EBV proteins were small components of lightly stained slices but were detected close to their anticipated size at a level similar to portal: the single-stranded DNA-binding protein (BALF2) at 130 kDa, ribonucleotide reductase large subunit (BORF2) at 95 kDa, thymidine kinase (BXLF1) at 72 kDa, and processivity factor (BMRF1) at 49 kDa (Table 1). Nine other EBV-encoded proteins were detected in amounts much less than portal (Table 3) and are thus unlikely to be substantial components of the EB virion.
Most surprisingly, β-actin peptides were dominantly detected in the intensely stained 45-kDa band, which decreased somewhat with NP-40 and ND deenvelopment (Fig. 3A). Actin peptides totaling 70% coverage were detected at least 20-fold more frequently in this gel slice than peptides of other proteins, with similar results in four analyses of three different virus preparations.
Peptides of several other cell proteins were detected at levels greater than portal. Peptides representative of 74% and 69% of 444-aa β-tubulin and the 166-aa, actin-binding protein, cofilin, were the most abundant peptides detected at 54 and 17 kDa, close to their expected sizes. Furthermore, peptides representative of 53% and 61% of 724-aa heat shock protein (Hsp)-90β and 646-aa Hsp70 were abundant peptides in the lightly stained 90- and 72-kDa slices. Enolase was detected at 49 kDa. Thus, β-tubulin, cofilin, Hsp70 and possibly Hsp90β, and enolase are likely EB-virion-associated. Other cell proteins were detected at levels 1- to 3-fold that of portal and are included in a more extensive list of possible EBV components (Table 4, which is published as supporting information on the PNAS web site).
Localization of Proteins Within Enveloped EBV. ICAT was used to more accurately compare protein associations with envelope or tegument as estimated by their stability to NP-40 or ND treatment. Purified enveloped EBV proteins modified by a 12C (light) ICAT were mixed with equivalent amounts of NP-40- or ND-treated virions from the same preparation that had been subsequently modified by a 13C (heavy) ICAT. MS/MS sequencing of trypsin-treated gel slices indicated that nearly all assigned cysteine-containing peptides were modified by heavy or light ICAT reagent.
ICAT ratios for proteins with more than one cysteine-containing peptide detected in each experiment are summarized, including 95% confidence intervals, in Table 5, which is published as supporting information on the PNAS web site. To account for differences in matching the amount of untreated virions to NP-40- or ND-treated virions, ratios from both experiments were normalized to give a MCP ratio of 1.00. Perhaps because of partial extraction of MCP by ND treatment, this normalization appeared to slightly inflate the ND ratios such that mCP increased (144%) relative to MCP after ND extraction. In contrast, 18-19% residual gH was seen after NP-40 or ND treatment, and the gB C terminus was reduced to 27% or 43% levels by NP-40 or ND extraction, confirming efficient deenvelopment by both treatments.
MTP was unique among tegument components by being 82% stable to NP-40 but 87% extracted by ND. In contrast, EBV BRRF2, BLRF2, and MyrP-binding protein (BGLF2) remained 92%, ≈100%, and 55% capsid-associated with NP-40 and were not further extracted by ND. Similarly, actin, α-tubulin, and β-tubulin remained 78%, 74%, and 90% capsid-associated after NP-40 treatment and 106%, 99%, and 87% capsid-associated after ND treatment. Moreover, Hsp70 remained 51% and 94% capsid-associated after NP-40 or ND extraction. These data indicate that BRRF2, BLRF2, α-tubulin, β-tubulin, and actin are strongly capsid-associated, consistent with a possible role in the intracytoplasmic fate of EBV capsids after infection as well as in morphogenesis.
Identification of Phosphorylation Sites in EB Virion Proteins. Sypro-Diamond, which binds phosphorylated S, T, or Y, stained EB virion proteins at the sizes of LTP, MCP, BRRF2, BKRF4, and sCP (Fig. 3B, lane PP). Searches for S, T, and Y residues with additional masses of 80 Da because of phosphate modification identified 28 potential phosphopeptides, corresponding to 10 EBV and 5 cell proteins (Table 6, which is published as supporting information on the PNAS web site). These data indicate that LTP (BPLF1), MCP (BcLF1), BRRF2, and sCP (BFRF3) are the 350-, 155-, 72-, and 20-kDa phosphoproteins, respectively. HSV sCP phosphorylation was described in ref. 26. Two β-actin phosphopeptides likely account for the weak phosphoprotein stain at 45 kDa. Phosphopeptides from gB, gM, BDLF2, BBLF1, BKRF4, and Hsp90 were also detected. Interestingly, phosphopeptides RLPLSSTTD(Tp)EDDQLPR from BPLF1 and IEDVG(Sp)DEEDDGKDKK from Hsp90 have consensus virion kinase, varicella-zoster virus ORF47, sites (S/T-X-D/E-D/E) (27), consistent with phosphorylation by BGLF4, the EBV protein kinase homologue, which was also detected in virions.
Discussion
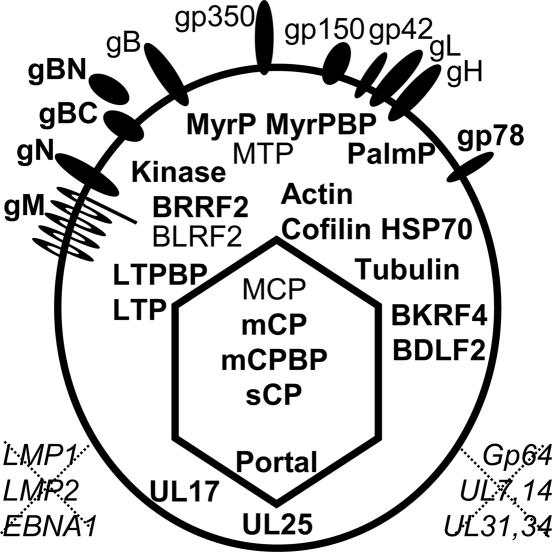
The data presented here, in general, confirm the supposition that the EBV genome is packaged in a capsid, tegument, and envelope composed principally of EBV-encoded homologues of conserved herpesvirus virion proteins (Fig. 4) (4, 28). Mature EBV was composed of EBV MCP, mCP, mCPBP, sCP, and portal capsid protein homologues. Based on analogy with HSV, six MCP and two triplexes of two mCP and one mCPBP condense around the EBV protease (BVRF2)-processed scaffold protein (BdRF1) into 150 hexons, each of which has six sCP molecules and 12 pentons. After capsid assembly, scaffold protein is degraded by protease. Persistence of scaffold or protease polypeptides was uncertain, and only trace amounts were detected. Protease and scaffold are therefore unlikely to have a role in mature virions.
Fig. 4.
EB virion proteins. Proteins not previously detected in EBV are in bold. Proteins that were not detected are shown in italics outside the virion. Unique long (UL) designations refer to the EBV homologues of HSV tegument proteins as in Table 1.
Genome-length herpes DNA, with much of the charged phosphate neutralized by polyamines, enters the capsid through a portal (BBRF1) dodecamer. Portal was consistently detected in mature EBV in relative amounts that are compatible with a dodecamer per virion. Portal may orient the capsid for the application of specific tegument proteins, movement through cellular compartments, envelopment, and DNA release near nuclear pores. The electron microscopy images of DNA being extruded from deenveloped capsids (Fig. 1) likely demarcate the portal vertex (out of plane of section).
EBV encodes several sequence-specific DNA-binding proteins that might increase infection efficiency if they were encapsidated with DNA. For example, persistent latent EBV infection of B lymphocytes depends on EB nuclear antigen 1 (EBNA-1)-mediated genome replication and maintenance. If EBNA-1 bound to cognate DNA sites were encapsidated with EBV DNA, it would increase DNA entry into infected cell nuclei and enhance transcription. However, EBNA-1 was not detected in any of the mature virion preparations, and other EBV-encoded transcription or replication factors were rarely detected.
Other EBV-infected cell nucleotide- or polynucleotide-binding proteins were detected in mature EBV, below portal levels, including the single-stranded DNA-binding protein, ribonucleotide reductase, thymidine kinase, processivity factor, and cellular histones. In contrast to purified simian virus 40, wherein histones are as abundant as capsid protein (29), histones were substantially less abundant than portal. Detection of these highly abundant nuclear proteins likely reflects a low level of virion contamination with nuclear debris.
HSV capsids acquire tegument proteins in the nucleus before budding through the innernuclear membrane. The EBV homologue (BVRF1) to the capsid-associated UL25 protein was readily detected, whereas the EBV homologue (BGLF1) of HSV UL17 packaging protein was similar to portal in abundance in mature EBV. However, peptides of BBRF2, the homologue of HSV UL7, which has been associated with extracellular virus, were rarely detected in mature EBV. Although BFRF1 has been detected in enveloped EBV prepared from infected cells (30), we did not detect significant amounts of peptides from BFRF1 or from the BFRF1-binding protein, BFLF2, in mature EBV (31).
As EBV capsids bud through the inner nuclear membrane, they acquire an envelope highly enriched for gB (BALF4) and depleted for gp350 (BLLF1), whereas mature extracellular enveloped EBV is enriched for gp350 relative to gB, consistent with a model in which EBV capsids are initially enveloped at the innernuclear membrane, deenveloped in the endoplasmic reticulum, and reenveloped at the transgolgi or plasma membrane (32), as is now recognized for other herpesviruses. Overall, despite more extensive gp350 glycosylation, gp350 peptides were much more frequently detected in extracellular EBV than gB peptides. These data support the notion that gp350 is the dominant EBV glycoprotein of the extracellular virus envelope.
EBV gB is unusual in having a critical, EBV-specific, role in initial nuclear egress as well as a herpesvirus family role in envelope fusion (33). Our data provide new evidence that much of mature EB virion gB is furin-cleaved. Full-length and furin-cleaved gB are moderately abundant potential fusogens in extracellular EBV envelopes (33).
Overall, the abundance of gH/gL and gM/gN are consistent with their putative critical roles in entry fusion and envelopment, respectively. However, the failure to detect significant amounts of BMRF2 in enveloped extracellular EBV raises a question as to whether the BMRF2 RGD motif mediates cell-cell virus spread rather than mature virion infection.
By analogy to HSV UL36, the EBV LTP homologue BPLF1 is expected to associate with perinuclear deenveloped cytoplasmic capsids and recruit other tegument proteins, including BOLF1, the EBV LTP-binding protein homologue of HSV UL37. BPLF1, BOLF1, BBLF1, the homologue to the UL11 MyrP, BGLF2, the homologue to the UL16 MyrP-binding protein, and BSRF1, the homologue to the UL51 palmitoylated protein, are likely essential for reenvelopment in the cytoplasm or plasma membrane and were detected in mature EB virions.
EBV BGLF4, the TS protein kinase homologue, was also a component of mature virions. Virion protein phosphorylations included one in LTP that matched the consensus substrate for the varicella-zoster virus BGLF4 homologue (27). After adsorption, penetration, and uncoating, the capsid and tegument must traverse the cytoplasm in the opposite direction. The HSV protein kinase homologue, UL13, phosphorylates multiple virion proteins in assembly and disassembly. These phosphorylations could modify virion protein activities so as to foster ingress toward the newly infected cell nucleus rather than egress.
Interestingly, EBV had at least five γ-herpesvirus-unique tegument proteins, MTP (BNRF1), BLRF2, BRRF2, BDLF2, and BKRF4. Some homologues have been detected in other γ-herpesviruses (34). These proteins may have functions similar to those of the missing HSV homologues and allow γ-herpesviruses to accomplish related functions in differentiated cells critical for γ-herpesvirus infection.
Cell-encoded proteins are surprisingly major components of the mature EB virion tegument and are likely important for virion morphogenesis and infection. β-Actin was one of the most abundant tegument proteins and cofilin, tubulin, Hsp90, and Hsp70 were also readily detected. Capsids likely accumulate these proteins as mediators of morphogenesis. High-level presence in mature virions may also indicate a role in virion stability or ingress. Tubulin is most likely related to capsid movement from the perinuclear cytoplasm to definitive envelopment in the golgi or plasma membrane, whereas MyrP, MyrP-binding protein, BSRF1, actin, and cofilin are likely incorporated during the envelopment process. The usurping of host cytoskeletal machinery by viruses during morphogenesis has been well documented and is likely to be a common theme in enveloped virus egress (35). Other actin-associated proteins (profilin, ezrin-radixin-moesin proteins, ARP3, ARP2/3 subunit 4, and α-actinin) were also detected at portal levels or higher. Hsp90, Hsp70, and CCT chaperone complexes, including cyclophilin A, may regulate actin filament formation. The persistence of cell cytoskeleton protein associations with capsids through NP-40 and ND extraction may be due to their polymerized state.
These investigations not only confirmed suppositions but substantially altered knowledge of mature EBV protein composition. Significant amounts of peptides from BBRF2 (HSV UL7 virion protein homologue); BFRF1 (UL34 homologue), which had been detected in cell-associated enveloped EBV (30); or BFLF2 (BFRF1-binding protein) were not detected in mature virions. Instead, EBV BLRF2, BRRF2, and BDLF2 encoded unanticipated EB virion proteins, likely tegument components. Furthermore, β-actin was among the most abundant tegument proteins; α- and β-tubulin, cofilin, Hsp90, and Hsp70 were also readily detected. Furin-cleaved gB was a significant component of mature virion envelopes; gp78, gM, and gN were also detected. Proteins associated with latent infection were not detected in virions.
Supplementary Material
Acknowledgments
We thank Wade Gibson, Patricia Spear, Lynn Enquist, Thomas Shenk, and Bernard Roizman for helpful advice; Maria Ericsson and Elizabeth Benecchi for performing the electron microscopy; and Lisa Kattenhorn for unpublished data. E.J. was supported by National Institutes of Health Grant 1K-08 AI49943-03. E.C.-M. was supported by Leukemia and Lymphoma Society Special Fellowship LLS-3499. This work was supported by National Cancer Institute Grants CA47006, CA87661, and CA85180.
Author contributions: E.J., M.L., and E.K. designed research; E.J., M.L., S.W., E.C.-M., D.I., and D.S. performed research; E.C.-M. contributed new reagents/analytic tools; E.J., M.L., M.R.C., and D.S. analyzed data; and E.J., M.L., M.R.C., E.C.-M., and E.K. wrote the paper.
Abbreviations: NP-40, 1% Nonidet P-40; ND, NP-40/0.5% deoxycholate; MCP, major capsid protein; mCP, minor capsid protein; sCP, smallest capsid protein; HSV, herpes simplex virus; EB, Epstein-Barr; EBV, EB virus; 4HT, 4-hydroxytamoxifen; MS/MS, tandem MS; ICAT, isotope-coded affinity tag; degly, deglycosylated purified EB virion proteins; MTP, major tegument protein; LTP, large tegument protein; MyrP, myristoylated phosphoprotein; Hsp, heat shock protein.
References
- 1.Gompels, U. A., Craxton, M. A. & Honess, R. W. (1988) J. Virol. 62, 757-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davison, A. J. & Taylor, P. (1987) J. Gen. Virol. 68, 1067-1079. [DOI] [PubMed] [Google Scholar]
- 3.de Jesus, O., Smith, P. R., Spender, L. C., Elgueta Karstegl, C., Niller, H. H., Huang, D. & Farrell, P. J. (2003) J. Gen. Virol. 84, 1443-1450. [DOI] [PubMed] [Google Scholar]
- 4.Davison, A. J., Dargan, D. J. & Stow, N. D. (2002) Antiviral Res. 56, 1-11. [DOI] [PubMed] [Google Scholar]
- 5.Russo, J. J., Bohenzky, R. A., Chien, M. C., Chen, J., Yan, M., Maddalena, D., Parry, J. P., Peruzzi, D., Edelman, I. S., Chang, Y. & Moore, P. S. (1996) Proc. Natl. Acad. Sci. USA 93, 14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baer, R., Bankier, A. T., Biggin, M. D., Deininger, P. L., Farrell, P. J., Gibson, T. J., Hatfull, G., Hudson, G. S., Satchwell, S. C., Seguin, C., et al. (1984) Nature 310, 207-211. [DOI] [PubMed] [Google Scholar]
- 7.Cameron, K. R., Stamminger, T., Craxton, M., Bodemer, W., Honess, R. W. & Fleckenstein, B. (1987) J. Virol. 61, 2063-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serio, T. R., Angeloni, A., Kolman, J. L., Gradoville, L., Sun, R., Katz, D. A., Van Grunsven, W., Middeldorp, J. & Miller, G. (1996) J. Virol. 70, 8047-8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutt-Fletcher, L. M. & Lake, C. M. (2001) Curr. Top. Microbiol. Immunol. 258, 51-64. [DOI] [PubMed] [Google Scholar]
- 10.Tugizov, S. M., Berline, J. W. & Palefsky, J. M. (2003) Nat. Med. 9, 307-314. [DOI] [PubMed] [Google Scholar]
- 11.Baldick, C. J., Jr., & Shenk, T. (1996) J. Virol. 70, 6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong, M. L. & Chen, C. H. (1998) Virus Res. 56, 191-197. [DOI] [PubMed] [Google Scholar]
- 13.Grunewald, K., Desai, P., Winkler, D. C., Heymann, J. B., Belnap, D. M., Baumeister, W. & Steven, A. C. (2003) Science 302, 1396-1398. [DOI] [PubMed] [Google Scholar]
- 14.Wright, J. F., Kurosky, A., Pryzdial, E. L. & Wasi, S. (1995) J. Virol. 69, 4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spear, G. T., Lurain, N. S., Parker, C. J., Ghassemi, M., Payne, G. H. & Saifuddin, M. (1995) J. Immunol. 155, 4376-4381. [PubMed] [Google Scholar]
- 16.Rose, J. S. & Grundy, J. E. (1992) J. Gen. Virol. 73, 507-512. [DOI] [PubMed] [Google Scholar]
- 17.Grogan, E., Jenson, H., Countryman, J., Heston, L., Gradoville, L. & Miller, G. (1987) Proc. Natl. Acad. Sci. USA 84, 1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolyniuk, M., Wolff, E. & Kieff, E. (1976) J. Virol. 18, 289-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwase, H. & Hotta, K. (1993) Methods Mol. Biol. 14, 151-159. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., Juhas, R. M., Thomsen, D. R., Homa, F. L., Burch, A. D., Weller, S. K. & Brown, J. C. (2001) J. Virol. 75, 10923-10932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel, A. H. & MacLean, J. B. (1995) Virology 206, 465-478. [DOI] [PubMed] [Google Scholar]
- 22.Baghian, A., Luftig, M., Black, J. B., Meng, Y. X., Pau, C. P., Voss, T., Pellett, P. E. & Kousoulas, K. G. (2000) Virology 269, 18-25. [DOI] [PubMed] [Google Scholar]
- 23.Li, Q., Spriggs, M. K., Kovats, S., Turk, S. M., Comeau, M. R., Nepom, B. & Hutt-Fletcher, L. M. (1997) J. Virol. 71, 4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheaffer, A. K., Newcomb, W. W., Gao, M., Yu, D., Weller, S. K., Brown, J. C. & Tenney, D. J. (2001) J. Virol. 75, 687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada, K., Goshima, F., Takakuwa, H., Yamada, H., Daikoku, T. & Nishiyama, Y. (1999) J. Gen. Virol. 80, 2423-2431. [DOI] [PubMed] [Google Scholar]
- 26.McNabb, D. S. & Courtney, R. J. (1992) J. Virol. 66, 4839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kenyon, T. K., Homan, E., Storlie, J., Ikoma, M. & Grose, C. (2003) J. Med. Virol. 70, Suppl. 1, S95-S102. [DOI] [PubMed] [Google Scholar]
- 28.Davison, A. J. (2002) Vet. Microbiol. 86, 69-88. [DOI] [PubMed] [Google Scholar]
- 29.Moyne, G., Harper, F., Saragosti, S. & Yaniv, M. (1982) Cell 30, 123-130. [DOI] [PubMed] [Google Scholar]
- 30.Farina, A., Santarelli, R., Gonnella, R., Bei, R., Muraro, R., Cardinali, G., Uccini, S., Ragona, G., Frati, L., Faggioni, A. & Angeloni, A. (2000) J. Virol. 74, 3235-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lake, C. M. & Hutt-Fletcher, L. M. (2004) Virology 320, 99-106. [DOI] [PubMed] [Google Scholar]
- 32.Gong, M. & Kieff, E. (1990) J. Virol. 64, 1507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haan, K. M., Lee, S. K. & Longnecker, R. (2001) Virology 290, 106-114. [DOI] [PubMed] [Google Scholar]
- 34.Bortz, E., Whitelegge, J. P., Jia, Q., Zhou, Z. H., Stewart, J. P., Wu, T. T. & Sun, R. (2003) J. Virol. 77, 13425-13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luftig, R. B. & Lupo, L. D. (1994) Trends Microbiol. 2, 178-182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.