Abstract
The phytochrome (phy) family of sensory photoreceptors transduce informational light signals to selected nuclear genes, inducing plant growth and developmental responses appropriate to the environment. Existing data suggest that one signaling pathway by which this occurs involves direct, intranuclear interaction of the photoactivated phy molecule with PIF3, a basic helix-loop-helix transcription factor. Here, we provide evidence from recently identified pif3 mutant alleles that PIF3 is necessary for early chloroplast greening and rapid phy-induced expression of nuclear genes encoding chloroplast components upon first exposure of seedlings to light. Therefore, these data indicate that PIF3 functions to transduce phy signals to genes involved in a critical facet of the early seedling deetiolation process, the generation of a functional photosynthetic apparatus. When transgenically expressed GUS:PIF3 fusion protein constructs were used, we found that PIF3 protein levels are rapidly and reversibly modulated by the photoreceptor over diurnal cycles in Arabidopsis seedlings. The PIF3 protein declines rapidly to a basal steady-state level upon initial light exposure, but reaccumulates to preirradiation levels in darkness during the subsequent night period. These data suggest that PIF3 may function in early phy signaling at the dark-to-light transition, not only during initial seedling deetiolation, but daily at dawn under diurnal light-dark cycles.
Plants constantly monitor the ambient environment for their most precious resource, photosynthetically active light, and modulate their growth and development to optimize solar energy capture and ensure reproduction. For this purpose, they have evolved a small network of informational photoreceptors that have differential, as well as partly overlapping, photosensory and/or physiological functions (1-4). Currently, nine such photoreceptors have been molecularly identified and functionally characterized in higher plants: two UVA/blue-light absorbing cryptochromes, cry1 and cry2 (2), two UVA/blue-light-absorbing phototropins, PHOT1 and PHOT2 (4), and five red light (R) and far-red light (FR)-absorbing phytochromes (phys), phyA-E (3, 5-7). In addition, there is long-standing evidence for an elusive UVB-light receptor(s), and more recent indications of additional UVA/blue-light receptors (8). Together, these photoreceptors perceive and integrate the fluctuating montage of incoming photosensory signals and regulate a progression of responses throughout the life cycle. The molecular mechanisms by which these signals are transduced through the relevant cellular response networks are an area of intense research interest (5-7, 9).
The phys are soluble, dimeric chromoproteins, with each subunit consisting of a linear tetrapyrrole chromophore, covalently linked to a polypeptide of ≈125 kDa (5, 10). The photosensory activity of the molecule resides in its capacity to switch reversibly between two conformers: the R-absorbing, biologically inactive Pr form, and the FR-absorbing, biologically active Pfr form. Studies with photoreceptor-null mutants have established that the five Arabidopsis phy family members have differential photosensory and/or physiological functions (albeit frequently partly redundant) (3, 5, 7, 11). This is most thoroughly documented for seedling deetiolation, where phyA is exclusively responsible for continuous FR (FRc) perception (12), and phyB is dominantly responsible for continuous R (Rc) perception in hypocotyl-growth regulation, although sharing this role with other phys in cotyledon-expansion and global gene-expression regulation (13).
It is well established that light-induced conversion of the phy molecule to its active Pfr form triggers an intracellular signaling process that culminates, within minutes, in the altered expression of target nuclear genes, initiating the transcriptional cascade responsible for the observed biological responses (3, 5, 7). Research efforts aimed at defining the molecular components and intracellular pathways involved in this signaling process have provided increasingly intriguing insights in recent years, punctuated by occasionally dramatic paradigm shifts. Two major developments in particular over the last 6 years dramatically altered prior concepts of phy signaling. The first was the accumulation of compelling evidence that the phy molecule, initially in the cytoplasm in its inactive Pr form, is triggered by light to translocate, within seconds to minutes, into the nucleus in its Pfr form (9, 14-16). This discovery provided an alternative to previously hypothesized second-messenger-based signaling pathways to nuclear genes. The second development was the identification of a constitutively nuclear, basic helix-loop-helix transcription factor as a phy-interacting protein, designated phytochrome-interacting factor 3 (PIF3) (17). Subsequent in vitro pull-down experiments showed that phy binding to PIF3 occurs specifically and reversibly only in the active Pfr form (18), gel-shift experiments showed that photoactivated phy can bind to PIF3 that is, in turn, bound to its DNA target motif (a G box sequence, CACGTG) (19), and fluorescent fusion protein studies have demonstrated intranuclear colocalization of phy and PIF3 molecules (16). These data led to the hypothesis that the phys may signal directly to target genes by physically binding to promoter-bound PIF3, potentially functioning as an integral, light-switchable component of a transcription-regulator complex (5, 19).
Initially, two additional sources of evidence provided what appeared to be compelling support for this hypothesis. First, and foremost, antisense PIF3-expressing transgenic seedlings generated in our initial study exhibited strikingly defective deetiolation in response to Rc, consistent with a critical functional role for PIF3 in phyB signaling (17). A subsequently observed reduction in the magnitude of the rapid, Rc-induced expression of the key photoresponsive circadian oscillator genes, CCA1 and LHY, in these same antisense lines was interpreted as further support for the central role of PIF3 in phyB signaling at the gene expression level (19). Furthermore, because the promoters of these genes contain G box motifs, and PIF3 was shown to bind to these promoter fragments, the data were considered to provide evidence that phyB could directly target these specific genes through the proposed phyB-PIF3-containing transcription-regulator complex at these promoters (19). Second, a microarray-based expression profile analysis of phyA-null mutants identified a small set of rapidly FRc-induced transcription factor-encoding genes that were completely dependent on phyA for light responsiveness, and found that the promoters of the majority of these genes contain G box motifs (12). These data led to the expanded hypothesis that these transcription factor genes may constitute a master set that are direct targets of phy-PIF3-containing transcription-regulator complexes, and that in turn regulate downstream targets in the transcriptional network.
However, recent studies indicate that important elements of this overall concept are not supported by subsequent evidence, and a different potential mechanism of phy signaling has been uncovered. First, two laboratories have reported that, in contrast to the Rc hyposensitivity of the growth phenotype initially reported for the antisense PIF3 lines (17), bona fide loss-of-function pif3 mutants are hypersensitive to prolonged Rc administered from germination onward (16, 20). These data have been interpreted to indicate that PIF3 acts negatively rather than positively in phy signaling, as initially concluded for this response. Second, Schaefer and coworkers (16) have made the exciting discovery that light-induced phy entry into the nucleus and apparent interaction with PIF3 in nuclear speckles trigger rapid disappearance of the PIF3 protein (in <1 h) to below detectable levels, presumably by proteolytic degradation. Because PIF3 falls so rapidly below detectable levels, the data suggest that it may have only a limited transient role in seedling deetiolation. The data also suggest a mechanism of phy action involving light-induced proteolysis of a target transcription factor, a biochemical mechanism clearly distinct from the previously proposed direct action of the photoreceptor in transcriptional activator activity (5).
Because our central research interest is to define the primary events in phy signaling and transcriptional regulation, we have focused on the potential role of PIF3 in this process during the initial period of seedling deetiolation after exposure to light. Here, by using previously unreported pif3 mutant alleles, we report that, contrary to the proposed negative role of PIF3 that is apparent late in seedling development under prolonged Rc (16, 20), PIF3 is necessary for normal greening and chloroplast development during the critical early hours of deetiolation, indicative of a positive role in this process. When we used genome-wide microarray analysis of rapidly light-responsive genes, we identified expression-compromised, chloroplast component-encoding genes in the pif3 mutants that may be responsible for the defective greening phenotype, suggesting that PIF3 has a primary function in regulating phy-induced expression of a key subset of nuclear-encoded chloroplast proteins. Finally, we provide evidence that, rather than declining to negligible levels in light, PIF3 protein levels reach a lower steady state, and rapidly return to preirradiation levels in darkness, indicative of a potentially continuing, rather than a transient, function after deetiolation.
Materials and Methods
Isolation of pif3 Mutant Alleles. pif3-2 and pif3-3 were isolated in PCR screens of DNA pools from T-DNA (http://signal.salk.edu) and fast neutron (21) mutagenized Arabidopsis populations for disruption of the PIF3 gene, by using the general procedures described (11, 21). pif3-1 (SALK30753) was obtained from the public Ecker/Alonso sequenced collection of Arabidopsis insertional mutants. Specific primers used for genotyping are listed in Table 1, which is published as supporting information on the PNAS web site. Homozygous lines were selected for further analysis after outcrossing once (pif3-1 and pif3-2) or twice (pif3-3) to Col-0. A wild-type sibling (designated WT-s) of the selected homozygous pif3-3 line was retained and used as the genotypic control where possible, either alone or together with the standard Col-0 wild type (designated WT).
Seedling Growth and Irradiation. Seeds were sterilized and induced to germinate as described (11) and placed in darkness at 21°C. Dark-grown seedlings were kept in the dark for 4 days. For prolonged Rc and FRc treatments, seedlings were moved after 21 h in darkness to Rc or FRc at 21°C for 3 days. Measurements of light fluence rates, hypocotyl length, and cotyledon area were as described (11). Chlorophyll was extracted and quantified as described (22).
Microarray-Based Expression Profiling. Four-day-old dark-grown wild-type Col-0 and pif3-3 mutant seedlings were transferred to Rc (8 μmol·m-2·s-1) for 1 h or retained in darkness as controls. Three different biological replicates of each treatment were grown separately and extracted, processed, and analyzed independently. RNA was extracted and subjected to expression analysis by using the Affymetrix ATH1 microarray containing >22,000 genes as described in Supporting Text, which is published as supporting information on the PNAS web site.
RNA Blot Analysis. RNA analysis was performed as described (11) by using probes PCR-amplified from cDNA or genomic DNA with primer pairs listed in Table 1, and as described for LHY (19), 18S rRNA (23), and PRR9 (24).
Generation of β-Glucuronidase (GUS):PIF3 (GP3) Lines. A GUS:PIF3 fusion construct (17) was cloned into the pKF111 binary vector under the control of the 35S cauliflower mosaic virus promoter (17), and Arabidopsis (No-0) was transformed (25) and selected as described (17). Four-day-old dark-grown homozygous lines were assayed biochemically for GUS enzyme activity in seedling extracts by using a fluorometric assay as described (26), and three lines with high activity were chosen.
Results and Discussion
Reverse-Genetic Identification of pif3 Mutants. To verify and expand our initial antisense-based analysis of the functional role of PIF3 in phy signaling, we screened for loss-of-function mutants at the PIF3 locus induced by T-DNA insertion (27, 28) or fast-neutron bombardment (21). Three pif3 mutant alleles were identified: pif3-1 and pif3-2, bearing separate T-DNA insertions, and pif3-3 containing a 2.5-kbp deletion (Fig. 1A). pif3-1, now available in the public collections, is the same allele recently characterized by Kim et al. (20). Also shown is poc1, a mutant that we identified previously in a forward genetic screen for phy signaling mutants as carrying a T-DNA insertion in the promoter region of the PIF3 gene (29). This allele was used by Bauer et al. (16) in their recent report. The pif3-1 and pif3-2 alleles produced altered size transcripts that, if translated, are predicted to produce functionally null proteins lacking an intact basic helix-loop-helix domain, known to be necessary for dimerization and DNA binding (Fig. 1 A and B). The pif3-3 allele produced no detectable transcript and is likely null, whereas the poc1 mutant produced strongly reduced, normal-sized transcript levels in the dark (Fig. 1B), as previously reported (29). We were unable to reproduce by RNA blot analysis the previously reported Rc-induced increase in PIF3 transcript in the pif3 mutant measured by RT-PCR (29). Instead, the reduced transcript levels were retained unaltered in both Rc- and FRc-irradiated seedlings (Fig. 7, which is published as supporting information on the PNAS web site). These results indicate that, contrary to our previous interpretation (29), poc1 appears to represent a pif3 mutant allele of strongly reduced function, if not loss-of-function, rather than an aberrant light-induced overexpressor of PIF3.
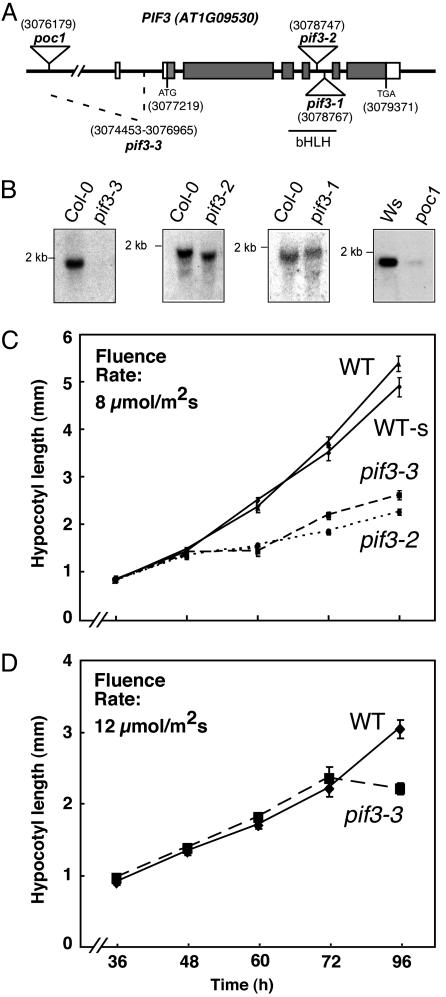
Fig. 1.
pif3 mutant identification and deetiolation in prolonged Rc. (A) Mutations identified in the Arabidopsis PIF3 gene (AT1G09530). T-DNA inserts in pif3-1, pif3-2, and poc1 are indicated at positions 3078747, 3078767, and 3076179, respectively, on chromosome I. pif3-3 carries a fast-neutron-induced deletion between positions 3074453 and 3076965. The location of the basic helix-loop-helix domain is indicated (bHLH). (B) RNA blots of 4-day-old, dark-grown wild-type and pif3 mutant seedlings. (C and D) Rc-induced hypersensitivity in pif3 mutants appears late in seedlings grown in Rc from germination onwards. Growth curves for hypocotyl length in WT, WT-s, pif3-2, and pif3-3 seedlings grown under Rc (8 μmol·m-2·s-1) (C) or Rc (12 μmol·m-2·s-1) (D) for 4 days.
pif3 Mutation Confers Seedling Hypersensitivity to Prolonged Rc.Contrary to expectation, the three more recently isolated pif3 mutants displayed shorter hypocotyls than wild-type seedlings when grown under Rc for 4 days (Fig. 8A, which is published as supporting information on the PNAS web site), confirming the recent observations of Kim et al. (20) and Bauer et al. (16). This gain of sensitivity to Rc in the hypocotyls was either not observed or marginal in the cotyledons (Fig. 8B). These results indicate that PIF3 plays a role in seedling deetiolation in prolonged Rc, with a function that might be more important in hypocotyl elongation than in cotyledon expansion. When grown under FRc, pif3 mutants were indistinguishable from wild-type seedlings (Fig. 8C), indicating that PIF3 does not participate in deetiolation under FRc. The essentially identical phenotypes of all four independent pif3 mutant alleles suggest that pif3-1, pif3-2, and poc1 (29) are likely to be loss-of-function null mutants for PIF3 like pif3-3.
These results are in direct contrast to the phenotype observed in PIF3 antisense lines previously described by this laboratory (17). Because the original interpretation of the biological function of PIF3 as a pivotally important positively acting transducer of phy signaling was based primarily on the strong hyposensitive deetiolation phenotype displayed by these PIF3 antisense lines, we attempted to determine the basis of this discrepancy. Further genetic analysis of the phenotypically strongest line, A22, used predominantly in our previous studies, indicated that the lesion responsible for the hyposensitive phenotype segregates independently of the antisense-encoding T-DNA insertion (data not shown), suggesting that this phenotype is not caused by suppression of PIF3 expression, but rather by a mutation at another locus. We have not yet molecularly identified this mutant locus. Regardless, the phenotype of the newly isolated, presumptively null pif3 mutants suggests that PIF3 acts negatively in phy-regulated seedling growth under prolonged Rc-irradiation, at least in suppression of hypocotyl elongation, as recently concluded (16, 20). These data are also retrospectively consistent with those of Halliday et al. (29), who reported that the poc1 mutant displays phyB-dependent hypersensitivity to Rc.
On the other hand, in detailed time course analyses, we found that the influence of PIF3 deficiency on hypocotyl extension rate appeared only late in the seedling growth response (48-72 h after initial Rc exposure) in a fluence rate-dependent manner (Fig. 1 C and D). This result raises the possibility that the hypocotyl hypersensitivity is an indirect consequence of PIF3-dependent events earlier in the light-induced seedling deetiolation response. Alternatively, this could be the result of the slightly higher levels (1.5- to 2-fold) of phyB that we observed in 4-day-old, Rc-grown pif3 seedlings compared to the wild type (Fig. 9, which is published as supporting information on the PNAS web site) (30).
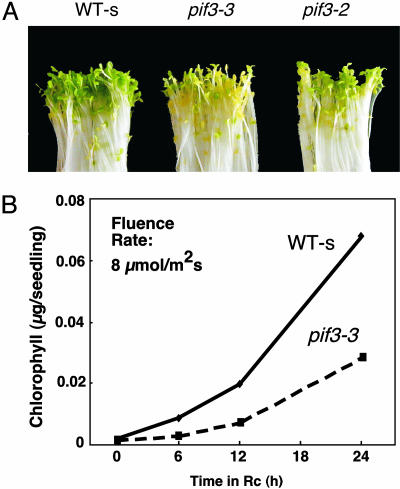
pif3 Mutant Seedlings Are Defective in Chlorophyll Accumulation. To determine whether PIF3 functions in early light-induced events in seedling deetiolation, we examined the phenotypic behavior of dark-grown pif3 seedlings over the first 24 h after transfer to Rc. Fig. 2 shows that these mutants display a substantial delay in visible greening and chlorophyll accumulation, indicative of defective chloroplast development (see also Figs. 10 and 11, which are published as supporting information on the PNAS web site).
Fig. 2.
pif3 mutants display delayed chlorophyll accumulation upon initial exposure to Rc. (A) Visual phenotype of WT-s and pif3 mutant seedlings grown in the dark for 4 days and transferred to Rc (10 μmol·m-2·s-1) for 24 h. (B) Chlorophyll accumulation in WT-s and pif3-3 mutant seedlings grown in the dark for 4 days and transferred to Rc (10 μmol·m-2·s-1) for 0, 6, 12, and 24 h.
Gene Expression Profiling. To examine the molecular basis of the defective greening phenotype and to define genes most likely to be early, if not direct, targets of phy signaling through PIF3, we performed a microarray analysis of light-induced changes in expression in wild-type and pif3-3 seedlings after initial transfer to Rc for 1 h. The primary data and all statistical parameters generated in this analysis are presented for all 22,000 genes present on the ATH1 chip in Table 2, which is published as supporting information on the PNAS web site. This analysis identified a total of 568 genes (Table 3, which is published as supporting information on the PNAS web site) that were induced (364 genes; Table 4, which is published as supporting information on the PNAS web site) or repressed (204 genes; Table 5, which is published as supporting information on the PNAS web site) in the wild type by 1-h Rc, in a statistically significant fashion compared to the dark control, and that exhibited statistically significant dependence on PIF3 (either positively or negatively) for this Rc responsiveness. The magnitude of the contribution of PIF3 to the Rc responsiveness of each gene was defined as the mean fold-induction ratio (MFIR) for induced genes, or mean fold-repression ratio (MFRR) for repressed genes (13) (see Supporting Text). This parameter quantitatively measures the magnitude of the change in expression induced by the Rc treatment in the wild type compared to the pif3 mutant, expressed as a ratio (wild type/pif3). A ratio of 1.0 signifies no detectable PIF3 contribution to the light-induced response, independently of whether the underlying response to light is intrinsically large or small for that gene. Deviations from 1.0 provide a quantitative measure of the robustness of the PIF3 contribution to the light response.
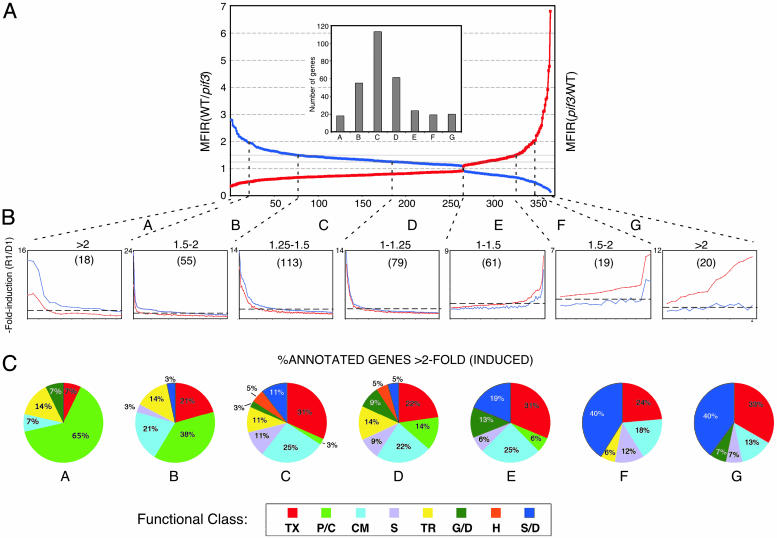
The 364 induced genes were arrayed in rank order of MFIR value, and further divided into bins according to the range of those values as depicted in Fig. 3. Genes at either extreme of this array are those exhibiting the strongest dependence on PIF3, either positively (left) or negatively (right), for Rc-induced expression, whereas those in the center exhibit the least PIF3 dependence. We define those genes with MFIR values ≥1.5 as being moderately to robustly dependent on PIF3 for Rc-responsiveness, either positively (bins A and B) or negatively (bins F and G), and those with MFIR values ≤1.5 as being marginally to minimally dependent on PIF3 for light responsiveness (bins C, D, and E) (Fig. 3A). In addition, to identify those genes responding quantitatively most robustly to the light signal, we arrayed the genes within each bin according to mean fold induction (MFI) in response to light (Fig. 3B), in wild type (bins A-D) or pif3 (bins E-G). We defined those genes with MFI values ≥2.0 as responding moderately to robustly to the Rc signal, whereas those with MFI values ≤2.0 were defined as being marginally to minimally Rc-responsive within the 1-h treatment period (dashed horizontal line in Fig. 3B). Although intrinsically arbitrary, these thresholds were chosen based on experience that differences in expression smaller than these appear less likely to be correlated with detectable changes in overt photomorphogenic phenotypes than greater differences. The genes with MFI values ≥2.0 were assigned to functional categories within each bin, and the percent of the annotated genes in each bin falling into each category are displayed in Fig. 3C. The expression levels detected by the microarray analysis were validated for selected genes by RNA blot analysis (Fig. 4).
Fig. 3.
PIF3 functions in early Rc-induced gene expression. (A) Induced genes arrayed in rank order of relative responsiveness to 1 h of Rc in wild-type (WT) compared to pif3-3 mutant seedlings. The curves depict the distribution of the 364 genes defined statistically both as significantly induced by 1 h of Rc in WT, and as significantly dependent on PIF3 for that Rc-responsiveness, arrayed in order of descending MFIR (see Supporting Text) for WT/pif3 [MFIR(WT/pif3)] (blue curve), and the reciprocal for pif3/WT [MFIR(pif3/WT)] (red curve). Vertical dashed lines divide the array into bins (A-G) according to MFIR value. (Inset) Histogram showing total number of genes in each bin. (B) Genes in each bin arrayed by mean fold induction (MFI) (see Supporting Text) of expression stimulated by 1 h Rc (R1) over the level in unirradiated, dark-control seedlings (D1), in WT seedlings for bins A-D, and in pif3 seedlings for bins E-G. Curves depict distribution of fold-induction values for WT (blue) and pif3 (red) seedlings. Note that scale varies between panels. Horizontal dashed line indicates 2-fold induction (R1/D1) in each bin. Value above bins indicates range of MFIR(WT/pif3) values for bins A-D, and the converse MFIR(pif3/WT) values for bins E-G. Numbers in parentheses are the numbers of genes in each bin. (C) Distribution of genes induced 2-fold or more by 1 h of Rc among functional categories, expressed as a percentage of the total annotated genes within each bin. TX, transcription; P/C, photosynthesis/chloroplast; CM, cellular metabolism; S, signaling; TR, transport; G/D, growth/development; H, hormones; S/D, stress/defense.
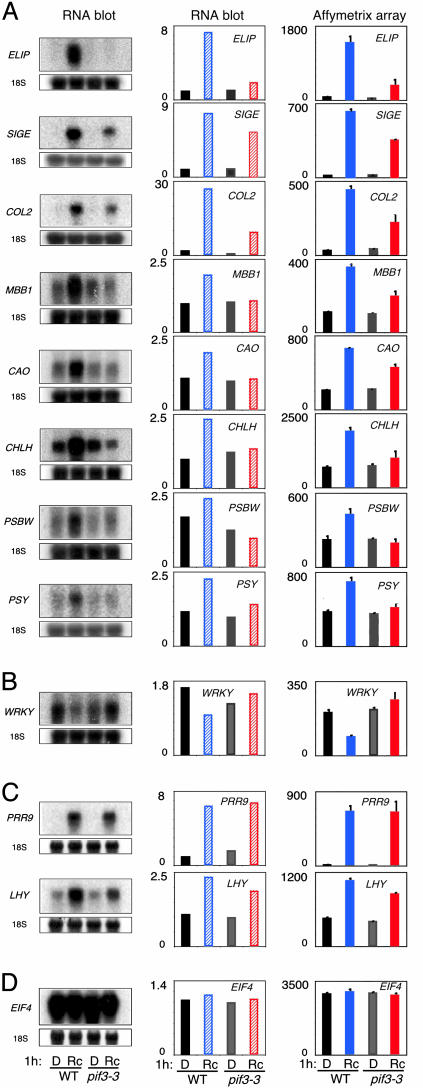
Fig. 4.
RNA blot validation of microarray data. Expression analysis of Rc- regulated PIF3-dependent genes. (Left) RNA blots of extracts from 4-day-old dark-grown wild-type (WT) and pif3-3 mutant seedlings irradiated with Rc (8 μmol·m-2·s-1) or maintained in the dark (D) for 1 h and hybridized with specific probes for Rc-induced PIF3-dependent genes ELIP (AT4G14690), SIGE (AT5G24120), COL2 (AT3G02389), MBB1 (AT3G17040), CAO (AT1G44446), CHLH (AT5G13630), PSBW (AT4G28660), and PSY (AT5G17230) (A); Rc-repressed PIF3-dependent gene WRKY (AT4G01250) (B); Rc- regulated PIF3-independent genes PRR9 (AT2G46790) and LHY (AT1G01060) (C); and constitutive control EIF4 (AT3G13920) (D). For normalization, the blots were reprobed for 18S RNA. (Center) Histograms of relative levels of expression of the genes shown in Left quantitated by using a phosphorimager. Levels are normalized to the 18S RNA and are expressed relative to the lowest expression value of all four samples set at 1. Expression levels are the average of two biological replicates, one shown in Left. (Right) Histograms of the expression levels obtained by microarray profiling for the corresponding genes.
The data provide intriguing insight into the role of PIF3 in rapid Rc-induced gene expression. Over 70% (10 of 14) of the genes most robustly dependent on PIF3 for ≥2-fold Rc-responsiveness are categorized as photosynthesis/chloroplast-related (Fig. 3C, bin A). When combined with bin B, this figure remains at 50% overall (21 of 42 annotated genes), indicating that PIF3 has a major role in transducing Rc-inductive signals to rapidly responsive genes involved in photosynthesis and chloroplast biogenesis. These genes (designated as gene set 1) are listed, together with the quantitative data used in their categorization, in Table 6, which is published as supporting information on the PNAS web site (see also Fig. 4). It is also striking that five of the six putative transcription factor genes in this gene set are zinc-finger proteins of various classes, whereas the other gene encodes the RNA polymerase σ subunit SigE involved in regulating chloroplast gene expression. The dependence of the light-induced expression of SIGE on PIF3 suggests that PIF3 may have a critical role in globally regulating the early, coordinate expression of chloroplast genome-encoded genes by means of control of the central chloroplast transcriptional machinery. Together, these data indicate an important role for PIF3 in regulating the early light-induced expression of nuclear, and, indirectly, plastid genes for chloroplast- and photosynthesis-related proteins, thereby providing an apparent explanation for the defective greening response observed in pif3 mutants.
It is notable that a subset of only six of the genes in gene set 1 (shown in bold type in Table 6) were identified as being phyB-regulated at 1 h of Rc in a previous study by using the 8,000-gene Affymetrix microarray (13), consistent with PIF3 involvement in phyB signaling to these genes. All six of these genes are functionally categorized as involved in photosynthesis (Table 6). Conversely, the remaining 40 genes in this set (Table 6) are potentially regulated by any of the five Arabidopsis phys (including phyB for those not on the 8,000-gene chip) in a PIF3-dependent manner. It is also notable that 26 genes previously identified (13) as phyB-regulated early response genes (designated gene set 6 here; see Table 7, which is published as supporting information on the PNAS web site) were not found to be PIF3-dependent for that response in the present study. Strikingly, this gene set includes four of the five putative transcription factor genes previously identified as rapidly phyB-induced in Rc (13), suggesting a phyB regulatory pathway not involving PIF3 that targets a limited subset of transcription factor genes.
By contrast to gene set 1, the genes with marginal or little dependence on PIF3 for Rc-responsiveness include substantially fewer chloroplast-related genes. This is apparent in Fig. 3C, bins C-E. It is also apparent in the corresponding expanded gene set (designated as gene set 2; 142 genes; Table 8, which is published as supporting information on the PNAS web site) formed by combining bins C-E of Fig. 3 (77 genes; minimal to marginal dependence on PIF3) with those genes defined as statistically Rc-induced, with MFI values of ≥2 in wild-type seedlings, but not statistically dependent on PIF3 for this Rc-responsiveness (an additional 65 genes; Table 8, bin X). Only 4% of gene set 2 (five genes) are functionally annotated as chloroplast-related. On the other hand, 36% of gene set 2 (43 genes) encode multiple classes of putative transcription factors that could function to regulate light-responsive genes downstream in the transcriptional network. Together, the genes in gene set 2 are interpreted as being Rc-induced by one or more members of the phy family, using factors other than PIF3 as intermediates in this process.
It is notable that a subset of 35 genes in gene set 2 (designated as gene set 4; Table 9, which is published as supporting information on the PNAS web site) was identified previously (13), by using the 8,000-gene chip, as rapidly Rc-induced, but not dependent on phyB for the light responsiveness. By implication, these genes are regulated by phyA, -C, -D, and/or -E in a PIF3-independent fashion. Interestingly, 11 of the 16 putative transcription factor genes previously identified as Rc-induced in a phyB-independent manner (13) are identified here as also being induced independently of PIF3 (gene set 4; Table 9). These data suggest a major phyA, -C, -D, and/or -E-mediated Rc signaling pathway that utilizes factors other than PIF3 to target a subset of putative transcription factor genes.
The genes in bins F and G of Fig. 3 exhibit an “inverse” dependence on PIF3 for Rc-induced expression, whereby the absence of PIF3 results in moderately to robustly enhanced light induction relative to wild type (see gene set 3; Table 10, which is published as supporting information on the PNAS web site). No chloroplast-related genes are present in this gene set. Strikingly, however, 40% of the functionally classified genes (13 genes) in this set are involved in stress/defense responses. These results suggest that the pif3 mutant may be displaying a rapid light-induced stress response.
An analysis parallel to that described above for the 364 Rc-induced genes was conducted for the 204 Rc-repressed genes (gene set 5; Table 11 and Fig. 12, which are published as supporting information on the PNAS web site). No clear general pattern of PIF3 involvement in the regulation of this gene set was immediately apparent.
Previously, we postulated that PIF3 may mediate phy regulation of the circadian clock by means of control of the expression of the central oscillator genes, CCA1 and LHY (12, 19). However, only a minimal reduction in light-induced expression of these genes was detected in the pif3 mutant compared to the wild type at 1 h of Rc (see gene set 2; Table 8 and Fig. 4). To determine whether these small changes might impact circadian oscillator function under longer-term diurnal conditions, we examined the free-running expression of LHY in light-dark cycle-entrained seedlings shifted to continuous light. No significant difference in oscillatory pattern was detected between pif3 and the wild type (Fig. 13, which is published as supporting information on the PNAS web site), indicating that PIF3 does not have a major role in regulating clock function.
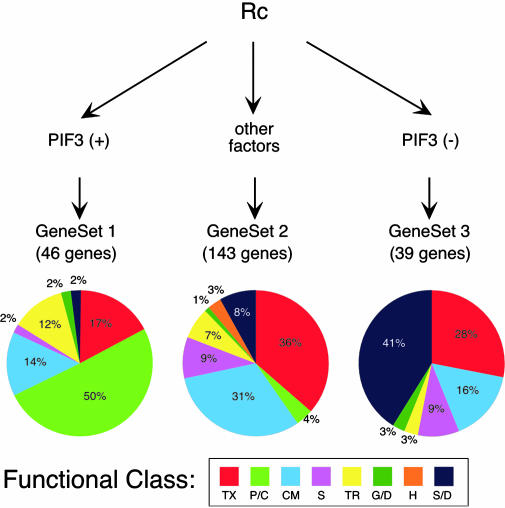
Signal Channeling by the phy Network. Taken together, the above data suggest that PIF3 has at least a partially specialized role focused on transducing light signals from photoactivated phy molecules to inducible early response genes involved in functional chloroplast biogenesis (Fig. 5; gene set 1). Conversely, the larger subset of induced genes (gene set 2), which responds rapidly and robustly to phy-mediated Rc signals via a transduction pathway(s) that appears to be quantitatively largely independent of PIF3, contains very few chloroplast-related genes. Instead, this gene set has a strong representation of putative transcription factor genes, many of which have been previously identified as potential primary targets of phy signaling (13). This finding suggests, in simplified terms, a potential bifurcation in phy signaling pathways whereby PIF3 plays an important role in transducing phy signals to chloroplast-related genes, but other factors dominate in transducing such signals to a subset of transcriptional master regulators, perhaps predominantly involved in other aspects of phy-regulated seedling deetiolation (Fig. 5).
Fig. 5.
PIF3 selectively channels phy signals to rapidly induced, chloroplast-related nuclear genes. The simplified scheme depicts the major sets of rapidly, Rc-induced genes identified here, and the postulated action of PIF3 and other unidentified signaling intermediates in differentially transducing phy signals to these different gene sets, which in turn appear to be predominantly involved in different facets of the cellular response program. The number of genes in each gene set and the distribution of the annotated genes within each set among functional categories are indicated (see Tables 6, 8, and 10 for gene lists). PIF3(+) indicates that PIF3 is necessary for Rc-induced expression. PIF3(-) indicates that the absence of PIF3 results in Rc-enhanced expression of these genes in the pif3 mutant relative to the wild type. Functional categories as in Fig. 3.
The function of PIF3 in the regulation of gene set 3, many members of which appear to be stress-related, is not clear. One possibility is that PIF3 acts more or less directly in response to phy signals to suppress otherwise light-induced expression of these genes, as depicted schematically in Fig. 5. Alternatively, the absence of PIF3 in the pif3 mutant and the consequent delay in production of a subset of potentially key chloroplast components might lead to photooxidative stress caused by the absence or misassembly of chlorophyll-protein complexes. In this case, the observed light-induced expression may be an indirect consequence of the absence of PIF3, independent of normal phy signaling pathways.
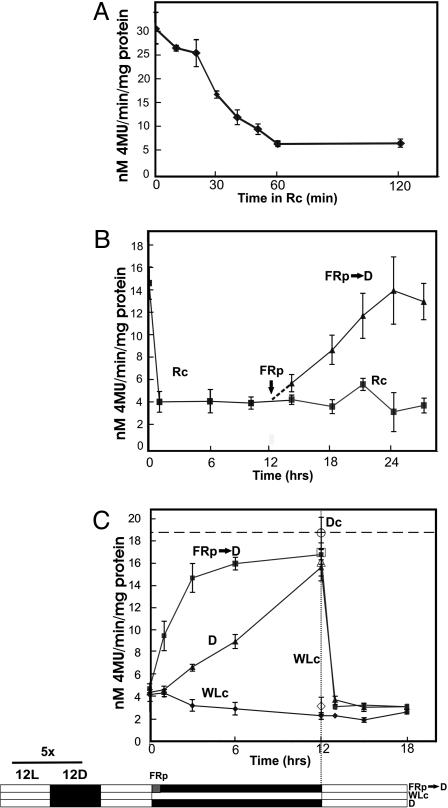
Light-Regulated PIF3 Protein Dynamics. Bauer et al. (16) used PIF3:GFP protein fusions to show that the PIF3 protein levels present in dark-grown seedlings decline rapidly below the level of detection, histochemically and immunochemically, upon transfer to light. We have confirmed these histochemical observations by using transgenic Arabidopsis expressing a GUS:PIF3 fusion protein (Fig. 14, which is published as supporting information on the PNAS web site), and have quantitated the changes biochemically by measuring GUS enzymatic activity in seedling extracts (Fig. 6). In general agreement with Bauer et al. (16), we found that GUS:PIF3 levels decline with a half-time of ≈30 min in Rc (Fig. 6A and Fig. 15, which is published as supporting information on the PNAS web site). However, rather than declining to undetectable levels, the GUS:PIF3 protein established a new steady-state level after ≈1 h of Rc, at ≈20% of the original dark level (Fig. 6B). This apparent discrepancy with the histochemical and immunochemical observations may reflect higher limits of detection for these latter assays.
Fig. 6.
Light regulation of PIF3 protein levels. GUS activity was measured fluorometrically in extracts of transgenic Arabidopsis seedlings expressing GUS:PIF3 fusion protein driven by the constitutive 35S CaMV promoter. (A) Rapid Rc-induced degradation of GUS:PIF3 in 4-day, dark-grown seedlings transferred to Rc (10 μmol·m-2·s-1) for 2 h. (B) Rc-induced GUS:PIF3 degradation and reaccumulation in darkness after a FRp (FRp → D) in seedlings grown 4 days in the dark before transfer to Rc. (C) phy-regulated PIF3 protein levels in green seedlings grown for 5 days in light/dark (L/D) diurnal cycles (12 h/12 h) before transfer to 12 h darkness with (FRp → D) or without (D) a preceding FRp, or continued maintenance in WLc, and subsequent exposure to WLc again at 12 h for all treatments. Open symbols at the 12-h time point are from an identical parallel experiment that included control seedlings maintained in continuous darkness throughout (Dc; open circle).
Interestingly, we found that PIF3 levels increased rapidly again to preirradiation levels in seedlings transferred to darkness at 12 h after a short FR pulse (FRp) (Fig. 6B). This result indicates that the lower steady-state levels of PIF3 induced by Rc require the continued presence of the active Pfr form of phytochrome to sustain these levels, at least during early deetiolation. To determine whether this behavior is retained in fully deetiolated green seedlings, we examined GUS:PIF3 levels in seedlings grown for 5 days under light-dark diurnal cycles. The data show that the PIF3 levels in these seedlings are maintained at 10-20% of the dark control during the light cycle and remain at that level under an extended continuous white light (WLc) treatment that replaces the normal dark period (subjective night period) (Fig. 6C). By contrast, these levels increase to dark control levels during the normal 12-h diurnal dark period, and the rate of this increase is accelerated dramatically by a FRp at the beginning of the dark period (Fig. 6C). Reexposure of the seedlings to white light after the diurnal dark period results in a reduction in PIF3 levels to the steady-state WLc levels within 1 h. These data indicate that PIF3 protein levels undergo significant and rapid diurnal fluctuations in response to light signals, and that this process is induced and sustained by the Pfr form of the phy molecule in a dynamically reversible manner. No evidence of circadian regulation of this process is apparent (Fig. 6C).
Conclusions
The evidence presented here and elsewhere (16, 20) indicates that our previous conclusion that PIF3 functions centrally and pleiotropically to mediate the overall phy-induced seedling deetiolation process (5, 17, 19, 29) is incorrect. This previous conclusion was based largely on the characterization of a PIF3-antisense expressing line, A22, which exhibited a robust, pleiotropic reduction in Rc-induced seedling deetiolation. This phenotype in the A22 line now appears to be the result of an inadvertently induced mutation at a separate locus unrelated to PIF3.
On the other hand, characterization of the bona fide pif3 loss-of-function mutants presented here indicates that PIF3 does, nevertheless, have an important function in a crucial facet of the deetiolation process. The data show that PIF3 functions in mediating the initial phases of light-induced chloroplast development after first exposure of seedlings to light, through regulation of a subset of rapidly photoresponsive nuclear genes encoding plastid and photosynthesis-related components. Because rapid establishment of a fully functional photosynthetic apparatus upon emergence of germinating seedlings from subterranean darkness into sunlight is central to successful seedling establishment and competitive early growth, PIF3 may play an important biological role in this process.
The expression profiles of rapidly Rc-induced genes suggest an intriguing potential bifurcation in the phy signaling pathways involved in seedling deetiolation. The data suggest that the phys may signal selectively through PIF3 to induce a subset of genes dominated by chloroplast-related components, as mentioned, whereas they may signal predominantly through other as yet unidentified factors to induce a subset of genes enriched in putative transcription-factor genes that may function to control the transcriptional network regulating other facets of the deetiolation process, such as hypocotyl and cotyledon cell expansion.
The exciting discovery by Bauer et al. (16) of rapid, phy-induced, intranuclear degradation of the PIF3 protein suggests a previously undescribed mechanism of phy signaling through PIF3 interaction. This mechanism is fundamentally different to that of phy acting as an integral functional component of transcription regulator complexes at target promoters that we previously hypothesized on the basis of in vitro DNA-binding gel shift experiments (5, 19). Although these two potential mechanisms of phy action are not necessarily mutually exclusive, there is currently no direct evidence for phy-modulated PIF3 transcriptional regulator activity. Therefore, it is possible that the molecular mechanism of phy action is to induce the proteolytic degradation of PIF3, and perhaps other related transcription factors, potentially via the ubiquitin-proteosome pathway, an increasingly prominent theme in a wide variety of plant signaling systems (31).
On the other hand, our data indicate that the phy-induced degradation of the PIF3 protein is a rapidly reversible and highly dynamic process, rather than representing an apparent irreversible depletion of PIF3 within 1 h of light exposure as reported (16). These data suggest that, rather than acting only briefly and transiently during the initial phases of seedling deetiolation, PIF3 remains potentially functionally important in fully green seedlings, where the observed robust phy-regulated fluctuations in PIF3 levels under diurnal light-dark cycles may have signaling significance.
Supplementary Material
Acknowledgments
We thank Barbara Simpson for technical assistance, David Hantz and greenhouse staff for plant care, Ben Bolstad and Matt Hudson for statistical analysis assistance, Andrew Likuski for help retrieving data from the Munich Information Center for Protein Sequences, and all members of the laboratory for support and discussion. The research was supported by a postdoctoral fellowship from the Spanish Ministry of Science and Technology (to E.M.), and by National Institutes of Health Grant GM47475, Department of Energy Grant DE-FG03-87ER13742, and U.S. Department of Agriculture Grant 5335-21000-010-00D (to P.H.Q.).
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 20, 2004.
Abbreviations: R, red light; FR, far-red light; FRc, continuous FR; Rc, continuous R; phy, phytochrome; MFI, mean fold induction; MFIR, MFI ratio; FRp, FR pulse; WLc, continuous white light.
See accompanying Biography on page 16088.
References
- 1.Kendrick, R. E. & Kronenberg, G. H. M. (1994) Photomorphogenesis in Plants (Kluwer Academic, Dordrecht, The Netherlands), 2nd Ed.
- 2.Cashmore, A. R., Jarillo, J. A., Wu, Y. J. & Liu, D. (1999) Science 284, 760-765. [DOI] [PubMed] [Google Scholar]
- 3.Smith, H. (2000) Nature 407, 585-591. [DOI] [PubMed] [Google Scholar]
- 4.Briggs, W. R. & Olney, M. A. (2001) Plant Physiol. 125, 85-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quail, P. H. (2002) Nat. Rev. Mol. Cell. Biol. 3, 85-93. [DOI] [PubMed] [Google Scholar]
- 6.Quail, P. H. (2002) Curr. Op. Cell Biol. 14, 180-188. [DOI] [PubMed] [Google Scholar]
- 7.Wang, H. & Deng, X.-W. (2004) in The Arabidopsis Book, eds. Somerville, C. R. & Meyerowitz, E. M. (Am. Soc. Plant Biologists, Rockville, MD).
- 8.Somers, D. E., Kim, W.-Y. A. & Geng, R. A. (2004) Plant Cell 16, 769-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy, F. & Schafer, E. (2002) Annu. Rev. Plant Biol. 53, 329-355. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser, C. (2001) J. Biol. Chem. 276, 11453-11456. [DOI] [PubMed] [Google Scholar]
- 11.Monte, E., Alonso, J. M., Ecker, J. R., Zhang, Y., Li, X., Young, J., Austin-Phillips, S. & Quail, P. H. (2003) Plant Cell 15, 1962-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tepperman, J. M., Zhu, T., Chang, H.-S., Wang, X. & Quail, P. H. (2001) Proc. Natl. Acad. Sci. USA 98, 9437-9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tepperman, J. M., Hudson, M. E., Khanna, R., Zhu, T., Chang, H.-S., Wang, X. & Quail, P. H. (2004) Plant J. 38, 725-739. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto, K. & Nagatani, A. (1996) Plant J. 10, 859-868. [DOI] [PubMed] [Google Scholar]
- 15.Nagy, F. & Schäfer, E. (2000) Curr. Opin. Plant Biol. 3, 450-454. [DOI] [PubMed] [Google Scholar]
- 16.Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K. C. S., Adam, E., Fejes, E., Schaefer, E. & Nagy, F. (2004) Plant Cell 16, 1433-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni, M., Tepperman, J. M. & Quail, P. H. (1998) Cell 95, 657-667. [DOI] [PubMed] [Google Scholar]
- 18.Ni, M., Tepperman, J. M. & Quail, P. H. (1999) Nature 400, 781-784. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-García, J. F., Huq, E. & Quail, P. H. (2000) Science 288, 859-863. [DOI] [PubMed] [Google Scholar]
- 20.Kim, J., Yi, H., Choi, G., Shin, B., Song, P.-S. & Choi, G. (2003) Plant Cell 15, 2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, X., Song, Y. J., Century, K., Straight, S., Ronald, P., Dong, X. N., Lassner, M. & Zhang, Y. L. (2001) Plant J. 27, 235-242. [DOI] [PubMed] [Google Scholar]
- 22.Chory, J. (1992) Development (Cambridge, U.K.) 115, 337-354. [Google Scholar]
- 23.Cantón, F. R. & Quail, P. H. (1999) Plant Physiol. 121, 1207-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Makino, S., Matsushika, A., Kojima, M., Yamashino, T. & Mizuno, T. (2002) Plant Cell Physiol. 43, 58-69. [DOI] [PubMed] [Google Scholar]
- 25.Bechtold, N., Ellis, J. & Pelletier, G. (1993) C. R. Acad. Sci. 316, 1194-1199. [Google Scholar]
- 26.Somers, D. E. & Quail, P. H. (1995) Plant J. 7, 413-427. [DOI] [PubMed] [Google Scholar]
- 27.Alonso, J. M. A., Stepanova, A. N. A., Leisse, T. J. A., Kim, C. J. A., Chen, H. A., Shinn, P. A., Stevenson, D. K. A., Zimmerman, J. A., Barajas, P. A., Cheuk, R. A., et al. (2003) Science 301, 653-657. [DOI] [PubMed] [Google Scholar]
- 28.Young, J. C. A., Krysan, P. J. A. & Sussman, M. R. (2001) Plant Physiol. 125, 513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halliday, K. J., Hudson, M., Ni, M., Qin, M. & Quail, P. H. (1999) Proc. Natl. Acad. Sci. USA 96, 5832-5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagner, D., Koloszvari, M. & Quail, P. H. (1996) Plant Cell 8, 859-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan, J. A., Shirasu, K. & Deng, X. W. (2003) Nat. Rev. Genet. 4, 948-958. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.