Abstract
The cancer cells can acquire migration and invasion capacities during the metastasis process through the developmental regulatory program epithelial–mesenchymal-transition (EMT), and through its reverse process mesenchymal–epithelial transition cancer cells can recolonize at distant metastatic sites. Among the multifaceted effects exerted by this program, reorganization of actin cytoskeleton is the key mechanical drive for the invasive properties gained by cancer cells. Collapsin response mediator protein-1 (CRMP1) is a cytosolic phosphoprotein and originally characterized as the mediator of semaphorin 3A signaling involved in axon differentiation during neural development. Here we report that CRMP1 can act as a suppressor of tumorigenicity and metastasis in prostate cancer cells. We demonstrated that CRMP1 exhibited a decreased expression pattern in high-grade prostate cancer tissues and many prostate cancer cell lines, and its downregulation in cancer cells was attributed to histone deacetylation and direct repression of its gene by the EMT regulator Snail. Functional analyses revealed that CRMP1 suppressed EMT in prostate cancer cells, as its knockdown could trigger EMT and enhance in vitro invasion capacity, whereas its overexpression could inhibit EMT and suppress both in vitro invasion and in vivo metastasis capacities of prostate cancer cells. Moreover, CRMP1 overexpression could significantly confer resistance to EMT induced by Snail or transforming growth factor-β1 in prostatic epithelial cells and prostate cancer cells. Finally, we demonstrated that CRMP1 could associate with actin and WAVE1, an activator of actin nucleation complex Arp2/3, and also its knockdown could stabilize F-actin and trigger the formation of stress fibers in prostate cancer cells. Together, our study shows that CRMP1 acts an EMT and metastasis suppressor in prostate cancer cells via its regulation of actin polymerization and also suggests that targeting the CRMP1-actin signaling in actin organization could be a potential strategy for management of prostate cancer metastasis.
Introduction
It is widely accepted that the developmental process epithelial–mesenchymal transition (EMT) plays a critical role in cancer progression and metastasis, via reprogramming of epithelial-to-mesenchymal phenotype to lose the cell adhesion and gain migration capacity.1, 2, 3 The loss of E-cadherin function and disruption of adherens junctions are recognized as the hallmarks of EMT. During this cellular process, cancer cells not only gain enhanced invasiveness but also acquire malignant and cancer stem cell traits.4, 5, 6 This process is dynamic and plastic as the metastatic cancer cells can undergo the reverse process, mesenchymal–epithelial transition, to recolonize and proliferate at distant metastatic sites via different mechanisms, including epigenetic modifications and altered stromal interactions.7, 8, 9 The reorganization of actin cytoskeleton and formation of migratory membrane protrusions are the key mechanical drive for the morphology change and gained invasive properties of the metastatic cancer cells.3
Collapsin response mediator proteins (CRMPs), which were originally identified in developing nervous tissue, are characterized as the intracellular mediators of semaphorin 3A signaling pathway involved in axon growth and branching during neural development.10, 11, 12, 13, 14, 15 They belong to a family of highly homologous cytosolic phosphoproteins (CRMP1–5; MW 60–66 kDa), which show no enzymatic activity but can be phosphorylated by various kinases involved in semaphorin 3A or other axon-guidance pathways.16, 17 This particular function is largely mediated by binding of CRMPs to F-actin and microtubule, which is dependent on their oligomeric or phosphorylation status and causes disassembly or reorganization of cytoskeleton and growth cone cell membrane.18 Besides expression in developing nervous system, CRMPs also express and function in variety of cellular processes in peripheral tissues or organs.16
Altered expressions of different CRMPs have been observed in some malignant tumors, including lung and prostate cancers. CRMP1 is identified and characterized as an invasion suppressor of lung cancer and its reduced expression is associated with advanced tumor stage, lymph node metastasis, early relapse and shorter survival of non-small-cell lung cancer (NSCLC) patients.19 However, increased CRMP1 expression is observed in prostate tumors developed in a transgenic mouse model of metastatic neuroendocrine prostate cancer, suggesting that CRMP1 may have a role in promoting neuroendocrine transdifferentiation in prostate cancer.20 A long CRMP1 isoform (LCRMP1, p80-LCRMP1) is identified in lung adenocarcinoma cell lines, and functional analyses reveal that LCRMP1 can act as an enhancer of lung cancer cell invasion, opposite to its short isoform CRMP1 and its increased expression is associated with lymph node metastasis and poor survival of NSCLC patients.21, 22 In addition, CRMP4 is identified by proteomics approach and characterized as a metastasis suppressor of prostate cancer via unclear mechanism, and its downregulation is associated with early relapse and lymph node metastasis in prostate cancer patients.23 On the other hand, upregulated expressions of other CRMPs (including CRMP2, CRMP4 and CRMP5) are reported in several cancers, including colorectal, pancreatic, NSCLC and lung neuroendocrine carcinoma with their increased expressions correlated with metastasis and poor survival.24, 25, 26, 27 These observations suggest that different CRMPs or heterotetramers formed by different CRMPs may perform different functions in the advanced progression of cancers.
In this study, we investigated the functional significance of CRMP1 in EMT and metastasis regulation in prostate cancer cells. Our findings showed that CRMP1 exhibited a significant downregulation in high-grade prostate cancer tissues, partly attributed to histone deacetylation, and knockdown of CRMP1 could trigger EMT and promote invasion whereas its overexpression could reverse the EMT in prostate cancer cells via a mechanism of reorganization of cytoskeleton actin, and also CRMP1 gene was a direct target of the EMT-inducing transcription factor Snail.
Results
CRMP1 expression is downregulated in prostate cancer
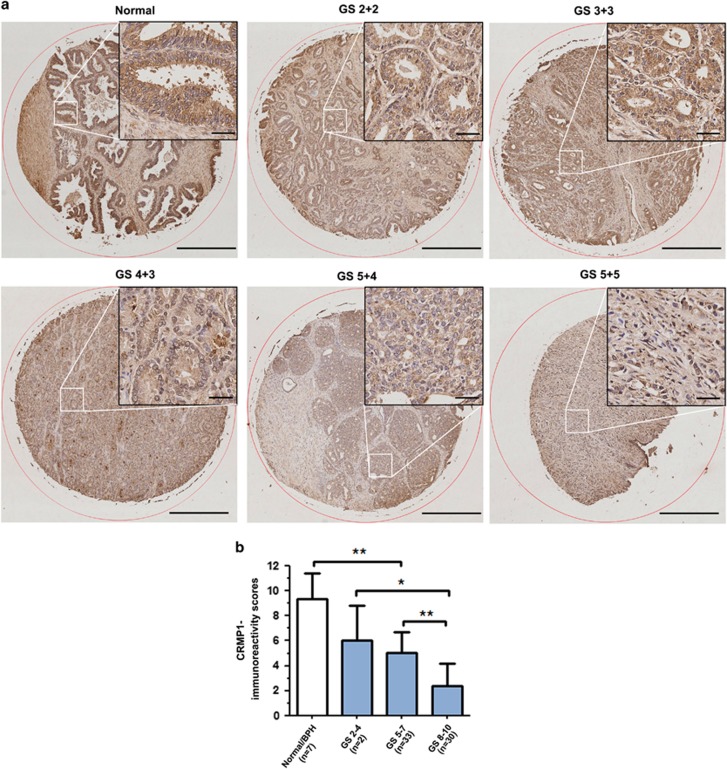
Immunohistochemistry of CRMP1 showed that normal prostatic epithelial cells exhibited moderate immunoreactivity in their cytoplasm, whereas the malignant cells in carcinoma lesions showed weak to negative immunoreactivity (Figure 1a). The specificity of CRMP1 antibody was validated before use (Supplementary Figures S1a–d). Immunoreactivity score analysis further demonstrated that higher Gleason scored lesions displayed significant lower CRMP1 immunoreactivity scores as compared with non-cancerous (normal or benign hyperplastic) tissues (Figure 1b). Reverse transcriptase–PCR analysis of CRMP1 expression performed in a panel of immortalized normal prostatic epithelial and prostate cancer cell lines also showed that only the short splice variant (CRMP1) transcripts but not the long variant (LCRMP1) were detected in all examined prostatic cell lines, with many prostate cancer cell lines expressing weaker levels as compared with the immortalized normal cell lines (Supplementary Figure S1e). Real-time PCR analysis further showed that the expression levels of CRMP1 were markedly reduced in two highly invasive and metastatic PC-3 sublines (PC-3M and PC-3M-LN4) as compared with their parental PC-3 cells (Supplementary Figure S1f). These findings indicated that CRMP1 exhibited a downregulated expression pattern in more advanced prostate cancer tissues and cells, suggesting that it might play a negative role in growth regulation of prostate cancer development and progression.
Figure 1.
CRMP1 is downregulated in prostate cancer tissues. (a) CRMP1 immunohistochemistry. Representative micrographs show the CRMP1-immunostained tissue microarray spots of normal and malignant prostatic tissues. Magnification, × 40; bars, 500 μm. Insets show the indicated areas at higher magnification. Magnification, × 400; bars, 50 μm. The normal prostatic epithelial cells exhibited moderate granular immunostaining of CRMP1 in their cytoplasm. The malignant cells in low and moderately differentiated adenocarcinoma lesions (Gleason scores 2–7) showed weak cytoplasmic immunoreactivity, and showed barely detected or negative immunoreactivity in high-grade poorly differentiated lesions (Gleason scores 8–10). (b) CRMP1-IRS analysis performed on malignant and non-malignant (normal or benign prostatic hyperplasia) prostatic tissues. IRS scores were acquired by multiplying the level of staining intensity (negative=0, weak=1, moderate=2, strong=3) and percentage of positively stained cells (0%=0, <10%=1, 11–50%=2, 51–80%=3, >80%=4). Results showed that adenocarcinoma lesions of higher Gleason scores (Gleason score⩾5) showed significantly lower CRMP1 expression than normal or BPH tissues. *P<0.05; **P<0.01. IRS, immunoreactivity score.
Downregulation of CRMP1 in prostate cancer cells involves histone deacetylation
We next investigated whether epigenetic modifications would be involved in the downregulation of CRMP1 in prostate cancer cells. We found that in vitro treatments with two histone deacetylase inhibitors, trichostatin A and sodium butyrate, could significantly increase CRMP1 expression in one androgen receptor positive (LNCaP) and two androgen receptor-negative prostate cancer cell lines (DU145 and PC-3M), with trichostatin A consistently showing greater effect on increasing CRMP1 expression in all treated cell lines. Treatment with a DNA methylation inhibitor 5-aza-dC could also elevate CRMP1 expression but with less extent in all treated cancer cell lines (Supplementary Figure S2). These results indicated that histone deacetylation or endogenous histone deacetylase activities and DNA methylation could contribute to the epigenetic downregulation of CRMP1 expression in prostate cancer cells.
CRMP1 knockdown enhances cell growth and promotes invasion capacity of prostate cancer cells
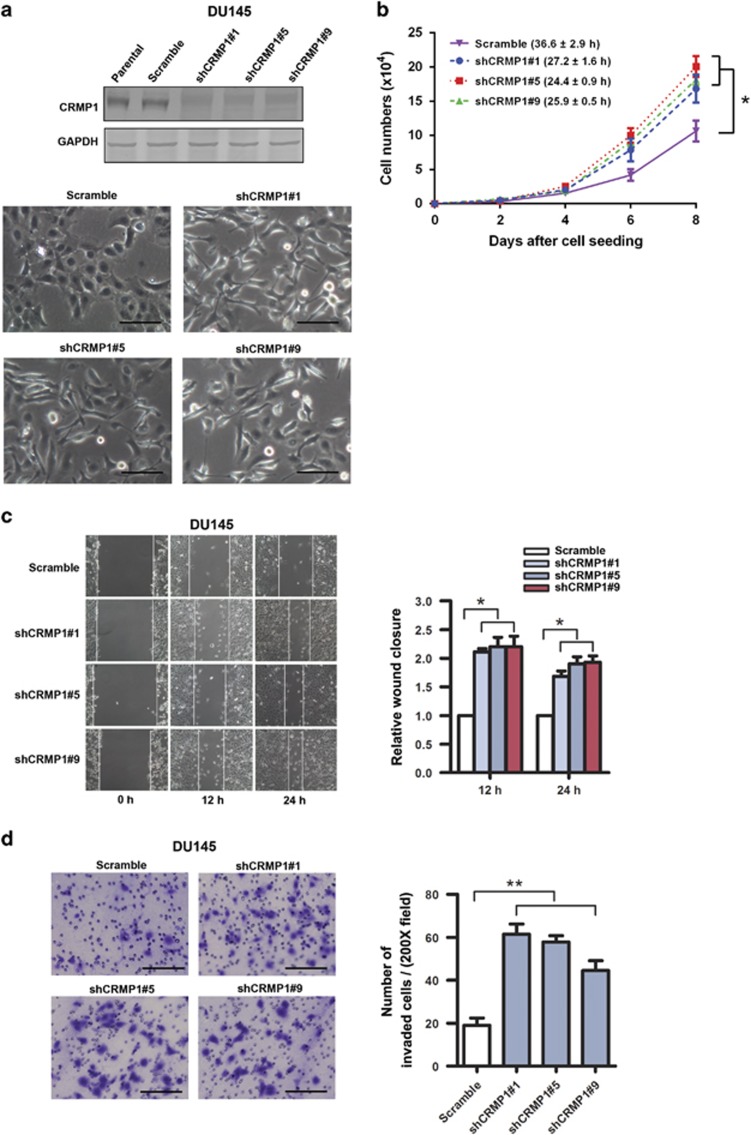
Since CRMP1 exhibited a downregulation pattern in high-grade prostate cancer, we first examined the functional significance of its knockdown in prostate cancer cell growth. We generated stable CRMP1-knockdown-transduced clones in DU145 prostate cancer cell line by lentiviral shCRMP1 infection for in vitro growth studies. Results showed that stable CRMP1-knockdown enhanced cell proliferation and induced distinct mesenchymal-like morphological features (including characteristic spindle-shaped morphology and loose intercellular contacts) in DU145-shCRMP1 infectants (Figures 2a and b). Results of wound healing and invasion assays also demonstrated that all examined shCRMP1 infectants showed enhanced migration and invasion capacities as compared with Scramble-shRNA infectants (Figures 2c and d). Together, these results indicated that knockdown of endogenous CRMP1 could enhance cell growth and promote in vitro migration and invasion capacities of prostate cancer cells.
Figure 2.
CRMP1 knockdown promotes cell proliferation, migration and invasion capacities of prostate cancer cells. (a) Top: Representative immunoblots validated that DU145-shCRMP1 infectants showed very weak or barely detectable CRMP1 expression, whereas the parental and DU145-shScramble infectants expressed a moderate level of endogenous CRMP1. Bottom: Representative phase-contrast images of DU145-shCRMP1 and DU145-shScramble infectants. DU145-shCRMP1 infectants appeared more spindle-shaped and less adhered to one another at subconfluence. Magnification, × 20; bars, 1 μm. (b) Proliferation assay by cell counting. DU145-shCRMP1 infectants proliferated at faster rates than the shScramble infectants. Brackets show the doubling time (hours). (c) Wound healing assay. Left: Representative images of scratched and recovering of wounded areas (marked by solid lines) on confluence monolayers of shCRMP1- and shScramble infectants taken at different time points. Right: Semiquantitative analysis of wound closure. Relative wound closure was determined by measuring the width of the wounds. Data are presented as fold changes relative to the width of the wounds made by control shScramble infectants. DU145-shCRMP1 infectants showed significantly higher migration capacity than the DU145-shScramble infectants. (d) Matrigel invasion assay. Left: representative micrographs of the crystal violet-stained migrated shCRMP1- and shScramble infectants. DU145-shCRMP1 infectants showed significantly higher invasion capacity than the DU145-shScramble infectants. Magnification, × 20; bars, 1 μm. *P<0.05; **P<0.01.
CRMP1 overexpression suppresses cell growth and malignant growth capacities of prostate cancer cells
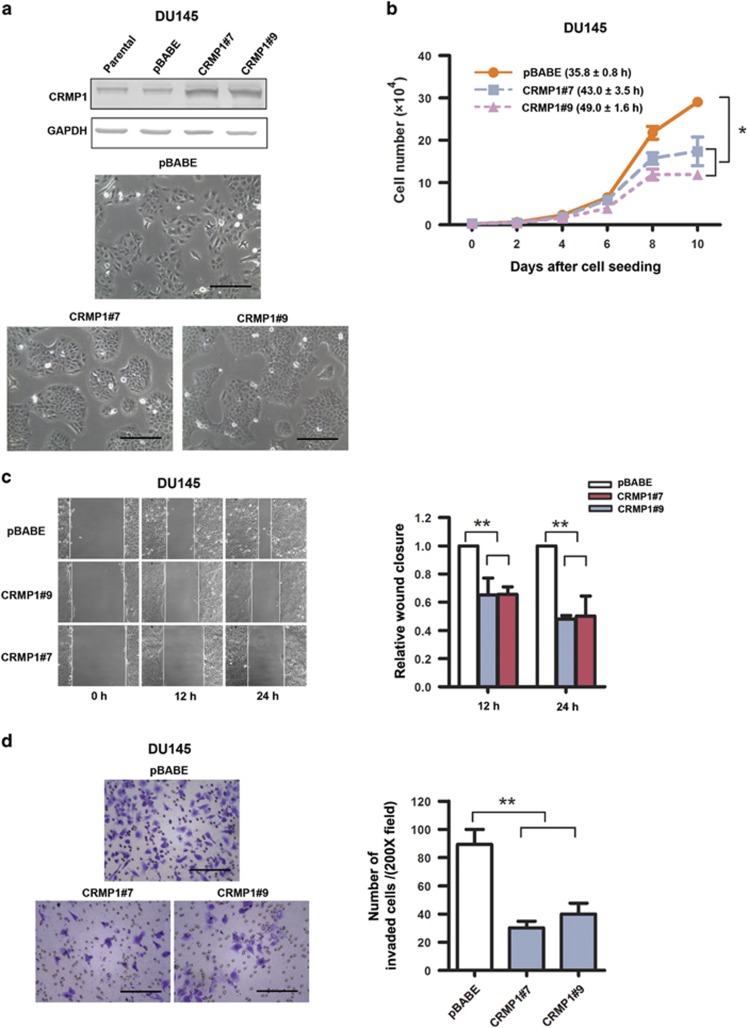
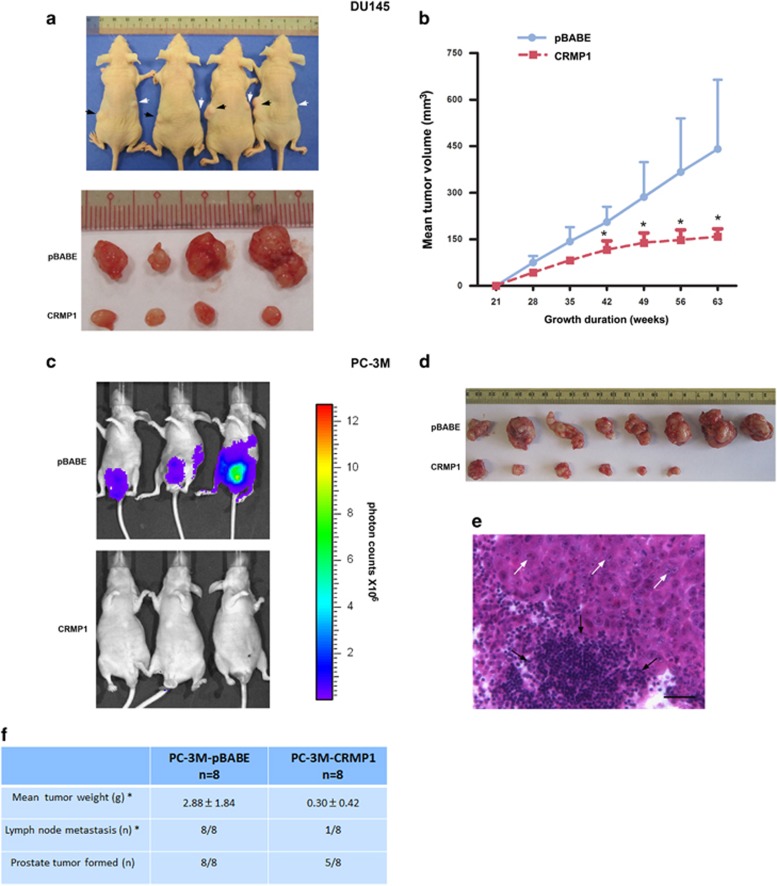
Since CRMP1 exhibited a decreased expression pattern in prostate cancer tissues and its knockdown could promote in vitro malignant growth of prostate cancer cells, we hypothesize that CRMP1 might play a negative role in prostate cancer growth. We then generated stable CRMP1-transduced infectants in two prostate cancer cell lines (DU145, PC-3 and its more invasive subline PC-3M) as in vitro models to further dissect its functional role in prostate cancer cell growth. All immunoblot-validated CRMP1 infectants displayed noticeable epithelial-like morphological features (apparent cobblestone shaped and grew as clusters in subconfluence) and proliferated at slower rates (Figures 3a and b and Supplementary Figures S3a–c, S4a and S4b). Moreover, all CRMP1 infectants exhibited significantly reduced in vitro migration and invasion capacities as compared with empty vector pBABE infectants (Figures 3c and d and Supplementary Figures S3d, S3e, S4c and S4d). In vivo tumorigenicity and metastasis studies also showed that CRMP1 overexpression could significantly suppress the tumor growth and lymph node metastasis capacities of CRMP1 infectants in host SCID mice (Figure 4). Together, these results indicated that CRMP1 could perform a tumor growth and metastasis suppressing function in prostate cancer cells.
Figure 3.
CRMP1 overexpression suppresses cell proliferation, migration and invasion capacities of DU145 prostate cancer cells. (a) Top: Immunoblot validation of CRMP1 expression in representative DU145-CRMP1 infectants. DU145-CRMP1 infectants expressed higher CRMP1 expression than the empty vector DU145-pBABE infectants and parental cells. Bottom: Representative images of DU145-CRMP1 and DU145-pBABE infectants. DU145-CRMP1 infectants appeared more cobblestone shaped and grew as clusters at subconfluence. Bars, 1 μm. (b) Cell counting assay. DU145-CRMP1 infectants proliferated at slower rates than the pBABE infectants. Brackets show the doubling time (hours). (c) Wound healing assay. DU145-CRMP1 infectants showed significantly slower migration capacity than the DU145-pBABE infectants. (d) Matrigel invasion assay. DU145-CRMP1 infectants exhibited significantly reduced invasion capacity than the DU145-pBABE infectants. Bars, 1 μm. *P<0.05; **P<0.01.
Figure 4.
CRMP1 overexpression suppresses tumorigenicity and metastasis in vivo of prostate cancer cells. (a) In vivo tumorigenicity assay. Top: Photograph shows the representative SCID mice bearing xenograft tumors formed by s.c. inoculation of DU145-CRMP1 (indicted by white arrows) and DU145-pBABE (black arrows) infectants in the same mice for comparison. Bottom: Photograph shows the dissected tumors formed by the inoculated infectants. (b) Growth curve of tumors formed by DU145 infectants in host mice for duration of 63 weeks. DU145-CRMP1 infectants formed significant smaller tumors than the DU145-pBABE infectants (n=8 for both). (c) Bioluminescence in vivo imaging. Representative bioluminescent images of mice at ninth week post-inoculation of PC-3M-CRMP1/Luc+ or PC-3M-pBABE/Luc+ cells. Significant tumor growth was detected in the prostate of mice, which received orthotopic inoculation of PC-3M-pBABE/Luc+ cells, as shown by intensive bioluminescent signals detected. However, no or barely detected weak bioluminescent signal was visualized in mice receiving inoculation of PC-3M-CRMP1/Luc+ cells. Scale bar shows the color intensity map for the bioluminescent signals expressed as photon counts. (d) Photograph shows the dissected prostate tumors formed by the inoculated CRMP1- or pBABE-transduced clones in host mice. Significant smaller tumors were formed by PC-3M-CRMP1/Luc+ clones. (e) Representative hematoxylin and eosin micrograph of aortic lymph node with prostate cancer metastasis in mouse receiving inoculation of PC-3M-pBABE/Luc+ cells. White arrows indicate the metastasizing prostate cancer cells and black arrows indicate the lymphocytes in lymph node. Magnification, × 400; bar, 50 μm. (f) Table summarizes the results of prostate tumors formed and lymph node metastasis detected in mice receiving orthotopic inoculation of PC-3M-CRMP1/Luc+ and PC-3M-pBABE/Luc+ cells. *P<0.05.
CRMP1 knockdown induces EMT but its overexpression reverses it in prostate cancer cells
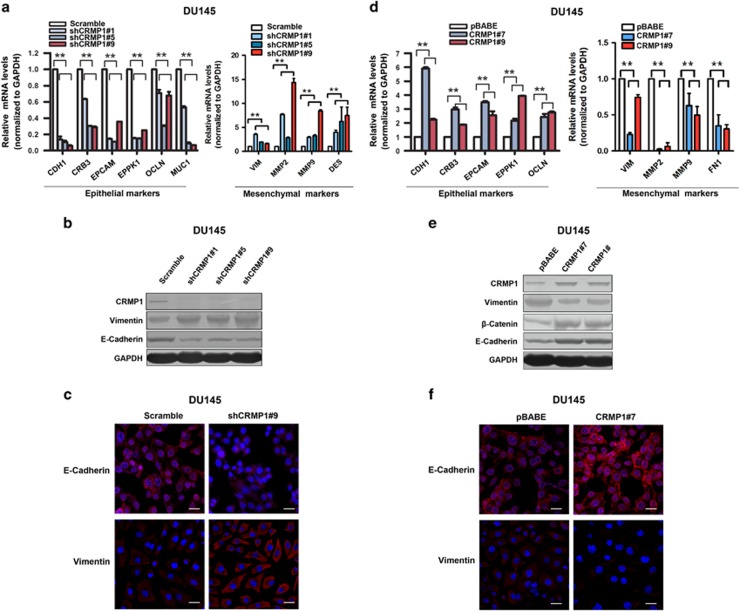
To further elucidate the tumor suppressing function of CRMP1 in prostate cancer growth, we next analyzed the expression profile of some EMT-associated markers in prostate cancer cells with either stable knockdown or overexpression of CRMP1. Real-time PCR analysis showed that DU145-shCRMP1 infectants showed significant downregulation of several epithelial cell markers, including CDH1, CRB3, EPCAM, EPPK1, OCLN and MUC1, but accompanied with upregulation of some mesenchymal cell markers, including VIM, MMP2, MMP9 and DES (Figure 5a). Immunoblot analysis validated the decreased protein expression of E-cadherin but increased expression of vimentin in DU145-shCRMP1 infectants (Figure 5b). Immunofluorescence study also revealed that knockdown of CRMP1 led to decreased E-cadherin but increased vimentin immunoreactivity in DU145-shCRMP1 infectants as compared with DU145-shScramble infectants (Figure 5c). These results suggested that CRMP1 knockdown triggered an EMT phenotype in DU145 prostate cancer cells. In contrast, overexpression of CRMP1 reversed the EMT process (or induced the mesenchymal–epithelial transition) in DU145-CRMP1 infectants, as evidenced by increased expression of epithelial markers but decreased expression of mesenchymal markers at both mRNA and protein levels (Figures 5d and e). Immunofluorescence also confirmed an increased E-cadherin but decreased vimentin immunoreactivity in DU145-CRMP1 infectants (Figure 5f). Similar results were also validated in another prostate cancer cell line PC-3 (Supplementary Figure S5). Together, these results suggested that CRMP1 could play a negative role in EMT regulation in prostate cancer cells.
Figure 5.
CRMP1 knockdown induces EMT but its overexpression induces mesenchymal–epithelial transition in DU145 prostate cancer cells. Real-time PCR, immunoblot and immunofluorescence analyses of EMT markers in (a–c) DU145-shCRMP1 and (d–f) DU145-CRMP1 infectants. (a, d) Real-time PCR analysis. Results showed that (a) DU145-shCRMP1 infectants expressed significantly lower levels of epithelial cell markers, including CDH1, CRB3, EPCAM, EPPK1, OCLN and MUC1, but higher levels of mesenchymal cell markers, including VIM, MMP2, MMP9 and DES, whereas (d) DU145-CRMP1 infectants expressed higher levels of epithelial cell markers but lower levels of mesenchymal cell markers (VIM, MMP2, MMP9 and FN1). Expression levels were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). Data were presented as the mean±s.d. obtained from three independent experiments. **P<0.01 DU145-shCRMP1 or DU145-CRMP1 infectants versus shScramble- or pBABE infectants. (b, e) Immunoblot analysis. A significant reduced E-cadherin expression but an increased vimentin expression was shown in (b) DU145-shCRMP1 infectants as compared with shScramble infectants, whereas a decreased vimentin expression but increased expressions of E-cadherin and β-catenin were revealed in (e) DU145-CRMP1 infectants as compared with pBABE infectants. (c, f) Immunofluorescence. A significant reduction of E-cadherin but increase of vimentin immunosignals were detected in (c) DU145-shCRMP1 infectants, whereas an increase of E-cadherin but a reduction of vimentin immunosignals were seen in (f) DU145-CRMP1 infectants. Nuclei were counterstained by 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI). Magnification, × 400; bars, 50 μm.
CRMP1 confers resistance to EMT induced by Snail or TGF-β1 in prostatic cells
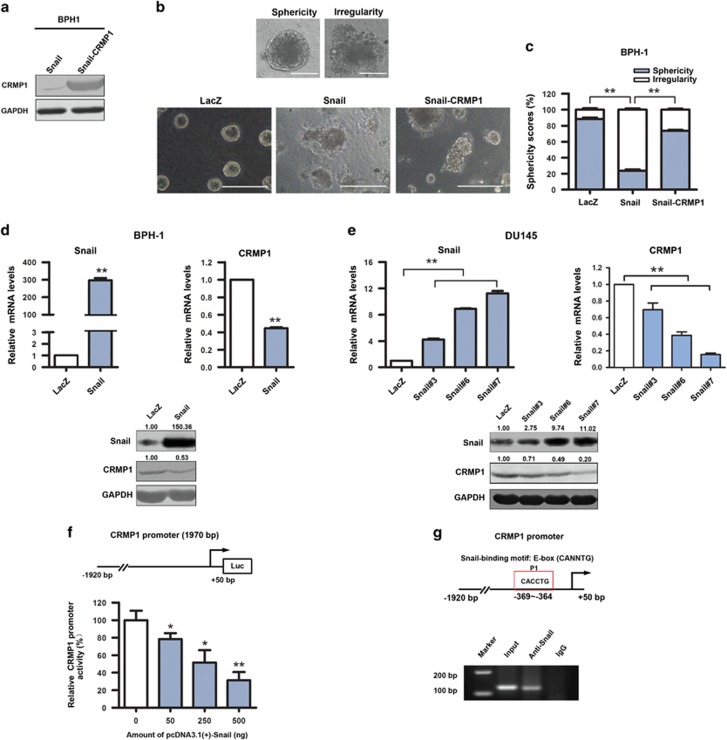
It is characterized that Snail, an EMT-activating transcription factor, plays a significant role in invasion and metastasis of prostate cancer.28 Our previous report shows that Snail is significantly upregulated in several androgen-insensitive and metastatic prostate cancer cell lines, and also its overexpression can trigger EMT phenotype in BPH-1 immortalized prostatic epithelial cells.29 We then evaluated the functional significance of CRMP1 in Snail-induced EMT in BPH-1 cells. Three-dimensional culture assay showed that the BPH-1-Snail infectants formed irregular aggregates in Matrigel (characterized as a mesenchymal morphological feature), whereas the control BPH-1-LacZ infectants formed mainly regular spheres (characterized as an epithelial morphological feature) (Figures 6a–c). However, upon CRMP1 transduction, majority of colonies formed by BPH-1-Snail-CRMP1 infectants appeared as spherical morphology (Figure 6c), suggesting that CRMP1 could significantly counteract the action of Snail on EMT induction in prostatic epithelial cells. Recent advances show that the secreted cytokine TGF-β is a potent driver of EMT in cancer progression, mainly mediated through its transactivation of EMT-inducing transcription factors including Snail.30 Upon TGF-β1 treatment, cultured DU145-pBABE infectants underwent distinct mesenchymal morphological change (prominently elongated), whereas the DU145-CRMP1 infectants remained unchanged and showed typical epithelial morphology (cobblestone shaped) same as the untreated cells (Supplementary Figure S6a). Immunoblot analysis also revealed that TGF-β1 treatment could time dependently induce EMT in DU145-pBABE infectants as shown by a significant decrease of E-cadherin and an increase of vimentin expression. However, similar treatment induced no significant change in protein levels of E-cadherin and vimentin in DU145-CRMP1 infectants, suggesting CRMP1 overexpression could resist the TGF-β1-induced EMT transdifferentiation (Supplementary Figure S6b). Taken together, these findings indicated that CRMP1 could counteract the actions of Snail and TGF-β1 in EMT induction in prostatic cells.
Figure 6.
(a–c) CRMP1 confers resistance to Snail-induced EMT in BPH-1 prostatic epithelial cells. (a) Immunoblot of CRMP1 expression in BPH-1-Snail and BPH-1-Snail-CRMP1 infectants. (b, c) Three-dimensional (3D) culture assay and scoring analysis of spherical 3D spheres (sphericity) or irregular aggregates (irregularity) formed by the BPH-1 infectants. (b) Top: Representative phase-contrast images of a regular sphere with smooth contour (sphericity) and an irregular aggregate (irregularity) formed by the BPH-1 infectants. Bars, 1 μm. Bottom: Representative images of spheres and irregular aggregates formed by LacZ (control), Snail and Snail-CRMP1 infectants. Bars, 5 μm. (c) Scoring analysis showed that both the BPH-1-LacZ and BPH-1-Snail-CRMP1 infectants formed mostly regular spheres (with sphericity scores of 88.4 and 73.6%, respectively), as compared with BPH-1-Snail infectants that formed considerably less regular spheres (sphericity score of 23.6%). Data are presented as mean±s.d. from three independent experiments performed in triplicate. **P<0.01. (d–g) Snail acts as the direct repressor of CRMP1 gene. (d, e) Reverse transcriptase–PCR and immunoblot analyses of CRMP1 expression in BPH-1-Snail and DU145-Snail infectants. Results showed that both BPH-1-Snail and DU145-Snail infectants showed significant reduction of CRMP1 expression, as compared with control LacZ vector-transduced cells. Figures shown above the immunoblots indicate the relative levels of Snail and CRMP1 expressed in infectants. **P<0.01 versus LacZ vector-transduced cells. (f) Luciferase reporter assay of CRMP1 gene promoter performed in HEK293FT cells. CRMP1-Luc reporter, driven by the proximal CRMP1 promoter, was significantly repressed by Snail in a dose-dependent manner. *P<0.05, **P<0.01 versus empty vector-transfected cells. (g) Chromatin immunoprecipitation assay performed on CRMP1 gene promoter in HEK293FT cells transfected with pcDNA3.1-Snail. Top: Diagram shows the location of Snail-binding site or E-box identified in the proximal CRMP1 promoter. A 146-bp fragment could be PCR-amplified in anti-Snail-immunoprecipitated DNA but not IgG-immunoprecipitated DNA in Snail-transfected HEK293FT cells.
CRMP1 gene is directly repressed by Snail
As ectopic expression of CRMP1 could confer resistance to Snail/TGF-β1-induced EMT in prostatic cells, we could infer that CRMP1 could be a downstream component of Snail/TGF-β signaling pathway. To verify this, we then examined the CRMP1 expression in Snail-transduced prostatic cells. Expression analyses showed that CRMP1 was significantly downregulated at both mRNA and protein levels in both BPH-1-Snail and DU145-Snail infectants, suggesting an inverse correlation between Snail and CRMP1 expression (Figures 6d and e). We next determined whether the suppression of CRMP1 expression could be a direct transcriptional repression of its gene promoter by Snail or indirectly via other signaling pathways. Luciferase reporter assay showed that the ectopic transfected Snail could dose dependently repress the transactivation of CRMP1-Luc reporter, driven by a 1.9 kb-fragment of CRMP1 5′-flanking region (Figure 6f). Furthermore, chromatin immunoprecipitation assay identified a Snail-enriched site located at the proximal P1 region of the CRMP1 promoter, which contained a Snail-binding E-box sequence CACCTG (located at −369 to −364 bp) (Figure 6g). Together, these results indicated that Snail could directly repress the CRMP1 gene, leading to EMT induction in prostate cancer cells.
CRMP1 can interact with actin and its knockdown promotes cytoskeletal actin filament formation in prostate cancer cells
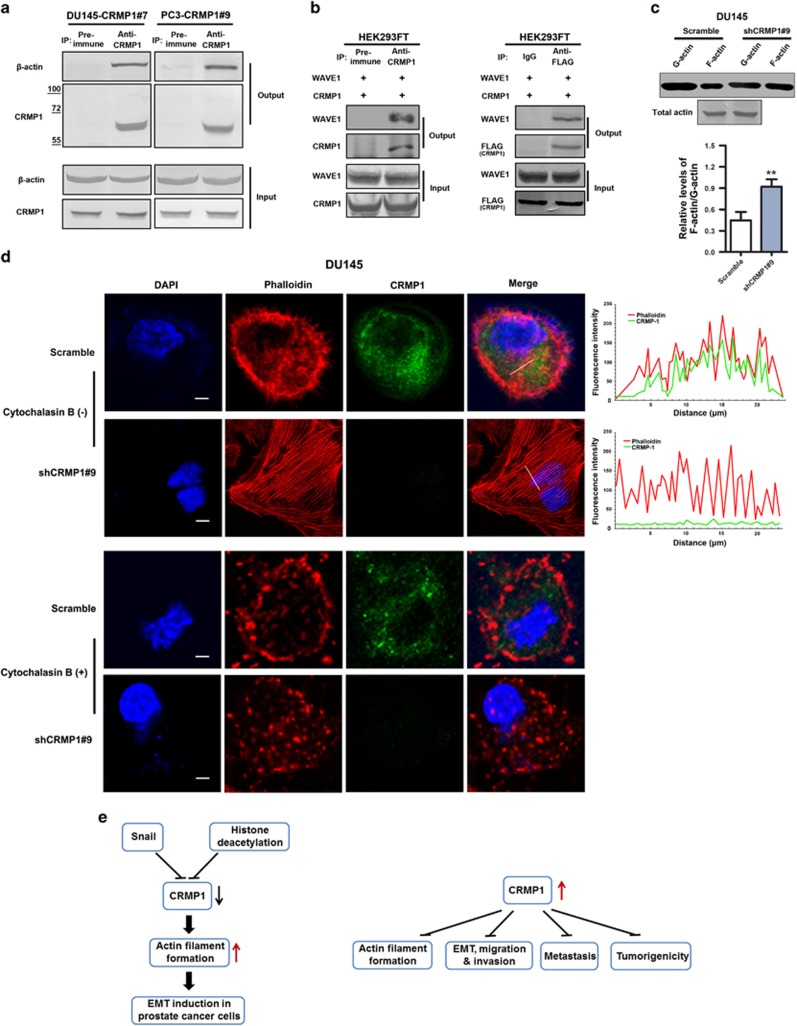
We hypothesize that the regulation of EMT by CRMP1 in prostate cancer cells could be mediated through its action on the remodeling of F-actin organization. To verify this, we explored its interaction with actin and also significance on F-actin polymerization in prostate cancer cells with either CRMP1 knockdown or overexpression. Immunoprecipitation performed in DU145-CRMP1 and PC-3-CRMP1 infectants showed that CRMP1 was associated with β-actin (Figure 7a). Moreover, immunoprecipitation performed in HEK293FT cells co-transfected with CRMP1 and WAVE1 (Wishott-Aldrich syndrome protein (WASP) family verprolin-homologous protein-1, an activator of actin nucleation core complex Arp2/3 in actin polymerization) showed that CRMP1 was associated with WAVE1 (Figure 7b), but not with other activation factors (including WAVE2, WAVE3, WASP and N-WASP) (data not shown), suggesting that CRMP1 could play a role in the activation of actin nucleation via the WAVE1 nucleation pathway. Since actin polymerization plays an important role in cell migration, we next examined the effect of CRMP1 knockdown on the F-actin organization. F-actin/G-actin in vivo assay showed that knockdown of endogenous CRMP1 could enhance the relative F-actin/G-actin ratio or favor the F-actin polymerization in DU145-shCRMP1 infectants (Figure 7c), and also suggested that CRMP1 could inhibit F-actin stabilization. Immunofluorescence further revealed that CRMP1 shared partial co-localization with F-actin in the cytoplasm of prostate cancer cells. Results also showed that knockdown of endogenous CRMP1 could cause increased formation of stress fibers or F-actin filaments (but reduction of cortical actin bundles) and enhance focal adhesion in DU145-shCRMP1 infectants. However, treatment with cytochalasin B, a potent inhibitor of F-actin polymerization, could abolish the formation of stress fibers in DU145-shCRMP1 infectants (Figure 7d). On the contrary, the assembly of stress fibers was inhibited by overexpression of CRMP1 in the invasive PC-3M cells (Supplementary Figure S7). These results suggested that CRMP1 could abrogate the formation of stress fibers in prostate cancer cells, likely via its association with WAVE1 and suppressing its activity in the activation of actin nucleation complex Arp2/3. Together, these results indicate that CRMP1 plays a negative role in the assembly of F-actin filaments via its association with actin and WAVE1 in the activation of actin nucleation core complex Arp2/3, and also EMT regulation in prostate cancer cells.
Figure 7.
CRMP1 is associated with actin and WAVE1 and its knockdown promotes cytoskeletal actin filament formation in prostate cancer cells. (a) Immunoprecipitation (IP) performed in CRMP1 infectants of DU145 and PC-3 cells. Results showed that CRMP1 antibody but not pre-immune rabbit serum could immunoprecipitate β-actin in both DU145-CRMP1 and PC-3-CRMP1 infectants. (b) IP performed in HEK293 cells co-transfected with CRMP1 and WAVE1. Results showed that both anti-CRMP (left) and anti-FLAG (right) antibodies could immunoprecipitate WAVE1 in transfected HEK293FT cells. (c) F-actin/G-actin in vivo assay. Upper: representative immunblots show the monomeric G-actin and polymeric F-actin expressed in DU145-shCRMP1 and DU145-Scramble infectants. The immunosignal intensities of F-actin relative to that of G-action were quantified (n=3 for shCRMP1-/Scramble infectants). Immunoblots of total actin from DU145-shCRMP1 and DU145-Scramble infectants used in F-actin/G-actin assay are shown. Lower: semiquantitative analysis of F-actin/G-actin ratio in DU145 infectants. Results showed that CRMP1 knockdown could enhance the F-actin/G-actin ratio in DU145-shCRMP1 infectants as compared with DU145-Scramble infectants. **P<0.01 versus Scramble infectants. (d) Combined rhodamine-phalloidin staining for F-actin and CRMP1 immunofluorescence in DU145-shCRMP1 and DU145-Scramble infectants with or without cytochalasin B treatment. Upper two panels: In the absence of cytochalasin B, phalloidin-stained subcortical actin bundles, which outlined the cell boundaries, were visualized in DU145-Scramble infectants. CRMP1 and phalloidin-stained F-actin shared partial co-localization to the cytoplasm of the DU145-Scramble infectants. Prominent phalloidin-stained F-actin filaments or stress fibers were clearly visualized in DU145-shCRMP1 but not DU145-Scramble infectants. Fluorescent images were analyzed for intensity profiles using software FV10-ASW (Olympus, Tokyo, Japan). Pearson's correlation coefficient was measured for co-localization correlation of intensity distributions between two channels, with 10 cells totally analyzed in each experiment. The white lines in the merged images indicate the portions of cells where fluorescence intensities were measured. Pearson's coefficient: DU145-shCRMP1, 0.018±0.005; DU145-Scramble, 0.486±0.135. Lower two panels: In the presence of cytochalasin B, phalloidin-stained F-actin filaments were not seen in DU145-shCRMP1 infectants. Magnification, × 600; bars, 10 μm. (e) Schematic diagrams illustrating the transcriptional repression of CRMP1 by Snail and histone deacetylation and also its inhibitory effects in malignant and metastatic growth of prostate cancer cells via its regulation of cytoskeletal actin organization and suppression of EMT.
Discussion
In the present study, we demonstrated that the semaphorin 3A signaling mediator CRMP1, which displayed a significant downregulation expression in high-grade prostate cancer and highly invasive and metastatic prostate cancer cell lines, could act as a tumor growth and metastasis suppressor in prostate cancer, as evidenced by its overexpression could suppress both in vitro invasion capacity and in vivo tumor growth and lymph node metastasis of prostate cancer cells. This downregulation pattern and also its invasion and metastasis suppressor function as characterized here in prostate cancer cells are similar to that as shown previously in NSCLC19 and also to another isoform CRMP4 shown in prostate cancer.23 The negative correlation between its expression and Gleason score also suggests that CRMP1 may be useful for prognosis and as being a negative predictor for relapse and metastasis of prostate cancer. We also characterized that this metastasis suppression function is dependent on its negative regulation on the EMT induction, as its overexpression could confer resistance to Snail/TGF-β1-induced EMT but in contrast its knockdown could induce EMT in prostate cancer cells.
Reorganization of cytoskeleton is believed to be the major mechanical drive for cell migration in the processes of EMT and also cancer metastasis.3 It is characterized that CRMPs play a role in the regulation of cytoskeleton dynamics in neuronal outgrowth mediated by semaphorin 3A or GTPase signaling via their interaction with the actin filaments and microtubules.18 Here, we demonstrated that CRMP1 could form a complex with actin and WAVE1, a key activator of actin nucleation core complex Arp2/3 in actin polymerization, and its knockdown could facilitate the F-actin or stress fiber assembly in prostate cancer cells. It is also reported that another CRMP member CRMP2 can interact with kinesin-1 and link kinesin-1 to the Sra1/WAVE1 complex in the CRMP2-induced axon differentiation in developing hippocampal neurons,31 suggesting that interaction with actin–WAVE1 complex could be a common mechanism through which CRMPs regulate the F-actin assembly in migrating cells. Based on these findings, we hypothesize that CRMP1 can form a complex with actin and WAVE1 to regulate the activity of actin nucleation core complex Arp2/3 in F-actin polymerization, and through this mechanism that CRMP1 functions to suppress EMT and thus invasion and metastasis in prostate cancer cells.
CRMP1 can be expressed in two alternative splice variants, the short (p62, CRMP1) and the long isoforms (p80, LCRMP1), with the latter possessing an extended N-terminus. The long isoform LCRMP1 is identified and expressed in rat brain tissues and also human lung adenocarcinoma cell lines.21, 32 Contrary to CRMP1 expression, high LCRMP1 expression is shown to be associated with lymph node metastasis and poor survival of NSCLC patients.21 Moreover, functional analyses show that opposite to CRMP1, LCRMP1 can function to promote both in vitro and in vivo invasion and metastasis abilities of lung cancer cells by enhancing filopodia formation via its interaction with the actin regulator Cdc42 (a GTPase of Rho family) in the GTPase signaling pathway and also WAVE1 in activation of F-actin nucleation complex.21, 22 As shown by truncation analysis, the LCRMP1-induced filopodia formation is dependent on its extended N-terminus, which is absent in CRMP1. It is also claimed that LCRMP1 but not CRMP1 can bind to WAVE1 via its conserved N-terminal region and also it can form LCRMP1–CRMP1 heterodimers to counteract the invasion suppression function mediated by CRMP1, by interrupting their interaction with WAVE1 and disrupting the actin nucleation in filopodia formation.22 However, in this study we did not detect the mRNA transcripts of LCRMP1 but only the CRMP1 in all examined prostatic cell lines, indicating that LCRMP1 or the putative LCRMP1–CRMP1 heterodimers as detected in lung cancer cells might not be expressed in the prostate cancer cells. It is speculated that the absence of LCRMP1 in prostate cancer cells may help to explain the discrepancy between CRMP1 and LCRMP1 on their association with WAVE1, as demonstrated here in prostate cancer and previously in lung cancer cells. However, it still needs further investigation to support this possibility.
The regulation of CRMP1 expression in cancer cells is still not well characterized. It is previously shown that CRMP1 expression can be negatively modulated by the tumor necrosis factor-α and inflammation-associated cyclooxygenase-2 activities, via the binding of induced or activated transcription factors NF-κB (p50), Sp1 and C/EBPα to their respective regulatory elements present at the promoter region of CRMP1 gene.25, 33 Here we provided evidences showing that downregulation of CRMP1 in prostate cancer cells was partially attributed to epigenetic modifications, largely by histone deacetylation and less by DNA methylation, and also direct repression by the EMT-regulatory transcription factor Snail, which plays critical roles in prostate cancer progression and metastasis.28, 29 On the other hand, our unpublished findings also showed that the CRMP1 gene could be directly transactivated by an orphan nuclear receptor estrogen-related receptor β (not shown), which is characterized to function as a tumor suppressor in prostate cancer.34 Together, these results suggest that CRMP1 expression can be regulated by multiple genetic and epigenetic factors, and their differential expression may also indirectly reflect their different roles played in the early and advanced development of prostate cancer.
In conclusion, the current study shows that CRMP1 can perform an invasion and metastasis suppressor function in prostate cancer cells, via its association between actin and WAVE1 to inhibit actin filament formation and then suppress cell migration and invasion. Our study also demonstrates the functional significance of its downregulation in advanced growth of prostate cancer, with a resultant effect of promotion of EMT and metastasis potential of cancer cells (Figure 7e).
Materials and methods
Human prostatic tissues and immunohistochemistry
Sets of prostate cancer tissue microarray slides containing 80 validated cases (PR803a; US Biomax, Inc., Rockville, MD, USA) were used for CRMP1 immunoreactivity score analysis. Immunohistochemistry was performed on antigen-retrieved sections using a rabbit polyclonal CRMP1 antibody and a streptavidin-biotin-peroxidase amplification method.35 The polyclonal CRMP1 antibody was raised in rabbits against the bacterially expressed His-tagged human recombinant protein and immunoblot-validated before use (Supplementary Figures S1a–d). The CRMP1 immunosignal intensity detected in prostatic sections was evaluated by a method of semiquantitative immunoreactivity score analysis as described previously.36
Cell lines and cell culture
One immortalized non-tumorigenic human prostatic epithelial cell line (BPH-1), five prostate cancer cell lines (LNCaP, PC-3, PC-3M, PC-3-M-LN4 and DU145) were used in this study. Besides, human lung carcinoma cell line A549 (cultured in F-12 K medium with 10% fetal bovine serum) and rodent neuroblastoma-glioma hybrid cell line NG108-15 (cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum) were used as positive expression controls of both short and long variants of CRMP. LNCaP, PC-3, DU145, A549 and NG109-15 cells were obtained from ATCC (Manassas, VA, USA); BPH-1 was provided by Dr Hayward SW (Vanderbilt University, Nashville, TN, USA); metastatic PC-3M and PC-3M-LN4 by Dr Fidler I (MD Anderson Cancer Center, Houston, TX, USA). PC-3M and PC-3M-LN4 cells were cultured in RPMI-1640 medium with 10% fetal bovine serum, 1% penicillin and streptomycin; and other cells were cultured in different media with supplements as described previously.35, 37
Plasmid construction and retroviral transduction
(1) Full-length cDNAs of human CRMP1 and CRMP4 were PCR-amplified using the pDNR-Dual-CRMP1 and pLX304-CRMP4 plasmids (GenBank accession numbers: NM_001313.3, NM_001387.2; Harvard PlasmID Database), FLAG-tagged and subcloned into pBABE-puro retroviral vector as pBABE-CRMP1 for retroviral transduction and pcDNA3.1(+) as pcDNA3.1-CRMP1 and pcDNA3.1-CRMP4 for transfection. His-tagged CRMP1 cDNA was subcloned into pET507a plasmid for recombinant protein production in Rosetta competent cells. pcDNA3.1-Snail and pLenti6/V5-Snail were constructed as described previously.29 pGL3-CRMP1-Luc, containing an insert of human CRMP1 5′ flanking region (−1920 to +50), was provided by Dr Wu CC (Academia Sinica, Taipei City, Taiwan). pcDNA3.1-His-WASF1 (WAVE1) was kindly provided by Dr Staruschenko A (Wisconsin Medical College, Milwaukee, WI, USA). All plasmid constructs were confirmed by DNA sequence before use. (2) Stable CRMP1-transduced clones were generated from prostatic cells (including DU145, PC-3, PC-3-M, BPH-1-Snail) using pBABE-CRMP1 following procedures on retroviral transduction as described previously.38 BPH-1-Snail and DU145-Snail-transduced cells were established as described previously.29
RNA interference
Sets of lentiviral plasmids pLKO.1-shCRMP1 (EG1400; Open Biosystems, Lafayette, CO, USA; Thermo Scientific, Waltham, MA, USA) with sequences targeting to CRMP1 mRNA and pLKO.1-Scramble with scramble sequence as non-targeting control were used for gene knockdown studies. Infectious shRNA-lentiviruses were packaged in 293FT cells, purified and used to infect prostatic cells for generation of stable transduced clones as described previously.34
In vitro treatments and cell growth analyses
(1) In vitro treatments. For epigenetic modification studies, cultured cells were treated with a DNA methyltransferase inhibitor 5-aza-2′deoxycytidine (5-aza-dC; 0.1 μm) or two histone deacetylase inhibitors, trichostatin A (0.2 μg/ml) and sodium butyrate (1 mm) for 24–72 h before RNA analysis. Besides, cells were also treated with recombinant human TGF-β1 (0.625 ng/ml; PeproTech, Rocky Hill, NJ, USA) for 12–24 h for EMT induction. (2) Cell viability of CRMP1/shCRMP1-transduced cells was assessed by trypan blue-exclusion assay as described previously.34 (3) Cell migration capacity of CRMP1/shCRMP1-tranduced cells was evaluated by wound healing assay as described previously,39 with the confluent cells starved for 24 h before making scratches. Relative wound closure was calculated as scratch width at specific time minus that at 0 h. (4) Matrigel transwell invasion assay was performed as described previously,29 with cells starved for 24 h before cell seeding for the assay. (5) Three-dimensional culture was performed on Snail-/Snail-CRMP1-transduced BPH-1 cells using a Matrigel-based overlay three-dimensional culture method as described previously.29 Results of all assays were obtained from three independent experiments and repeated in triplicate.
RNA and protein analyses
(1) SYBR-based real-time PCR analysis was performed as described previously.36 The relative gene expression levels were determined by the comparative CT method (2−ΔΔCT) after normalization to glyceraldehyde 3-phosphate dehydrogenase.40 Oligonucleotide sequences of primer pairs are listed in the Supplementary Table S1. (2) Immunoblot analysis on cellular proteins extracted from different transduced cells was performed using a colorimetric detection method as described previously.38 Primary antibodies used include CRMP1 (a rabbit polyclonal antibody generated as described above and also one obtained from Abcam, Cambridge, UK), E-cadherin (rabbit monoclonal; Cell Signaling, Danvers, MA, USA), β-catenin (rabbit monoclonal; Cell Signaling), Snail (rabbit monoclonal; Cell Signaling), vimentin (mouse monoclonal; Sigma-Aldrich, St Louis, MO, USA), WAVE1 (goat polyclonal; Santa Cruz Biotechnology, Dallas, TX, USA), pan-actin (mouse polyclonal; Santa Cruz Biotechnology), β-actin (mouse polyclonal; Santa Cruz Biotechnology), FLAG (mouse monoclonal; Wako, Osaka, Japan) and glyceraldehyde 3-phosphate dehydrogenase (rabbit monoclonal; Cell Signaling).
Fluorescence microscopy
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline, permeabilized with 0.1% Triton X-100 in phosphate-buffered saline, blocked with 5% normal goat serum and stained with primary antibodies at 4 °C overnight, followed by TRITC- or Cy3-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) in dark. For combined F-actin staining, cells were also incubated with rhodamine-phalloidin (50 μg/ml; Sigma-Aldrich). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI). Confocal immunofluorescence was performed as described previously.36
Immunoprecipitation and chromatin immunoprecipitation
(1) Immunoprecipitation assay was performed on cell lysates of CRMP1-transduced prostatic cells using CRMP1- or FLAG-antibody, or pre-immune rabbit serum or IgG as control, following procedures as described previously.36 (2) Chromatin immunoprecipitation assay of CRMP1 promoter was performed on HEK293 cells transfected with pcDNA3.1-Snail following procedures as described previously.34 Sonicated chromatin extracts were immunoprecipitated with a Snail-antibody. Immunoprecipitated DNA was analyzed by PCR with primers specific for CRMP1 promoter (forward: ACACCCCCGCAGATGTTC; reverse: AAAGGAGCCCTGAAAACCAT).
F-actin/G-actin in vivo assay
The actin polymerization state in shCRMP1-transduced cells was determined by an F-actin (filamentous form)/G-actin (globular form) in vivo assay (Cytoskeleton, Inc., Denver, CO, USA). Briefly, cells were lysed in an F-actin stabilizing buffer (50 mm PIPES, pH 6.9, 50 mm NaCl, 5 mm MgCl2, 5 mm EGTA, 5% glycerol, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.1% Tween-20, 0.1% 2-mercaptoethanol, 0.001% Antifoam C and protease inhibitors). The cell lysates were centrifuged at 106 g to separate the F-actin (pellets) from G-actin (supernatants). F-actin in pellet fraction was then depolymerized in water containing 8 m urea. Actin in each fraction was resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and quantitated by immunoblotting with an anti-actin antibody. The immunosignal intensities were quantified using gel densitometry software (ImageJ, NIH, Bethesda, MD, USA) and the ratio of F-actin/G-actin was determined.
Luciferase reporter assay
Luciferase reporter assay was performed on HEK293 cells co-transfected with pcDNA3.1-Snail, pGL3-CRMP1-Luc and pRL-SV40 using a Renilla firefly dual-luciferase reporter system (Promega, Madison, WI, USA) as described previously,37 with the firefly luciferase activity normalized to Renilla luciferase by pRL-SV40 and expressed as fold relative to pcDNA3.1 vector control. Results were obtained from three independent experiments performed in triplicate.
In vivo tumorigenicity and metastasis assay
(1) In vivo tumorigenicity assay was performed on CRMP1- and vector-transduced prostatic cells and sizes of xenograft tumors were measured following procedures as described previously.38 (2) For metastasis study, CRMP1- or pBABE-transduced and firefly luciferase-labeled PC-3M-Luc cells (3 × 105 cells per 10 μl sterile phosphate-buffered saline) were inoculated orthotopically into the dorsal prostate of anesthetized 7-week-old intact nude mice (n=8 per group) using Hamilton syringes, followed by growth for 9 weeks. The orthotopic tumor growth during 4–8 weeks was monitored bi-weekly by palpation. At eighth and ninth weeks, animals bearing prostate tumors were anesthetized with isoflurane and injected intraperitoneally with D-luciferin (0.15 mg/g body weight), followed by bioluminescence in vivo imaging (FMT 1000 Fluorescent Tomography System; PerkinElmer, Waltham, MA, USA) for detection of orthotopic tumors and metastasis. Luminescent images were acquired and analyzed for tumor growth or metastasis using TrueQuant software (PerkinElmer).
Statistical analysis
All data are presented as mean±s.d. from at least three independent experiments. Statistically analyses of data were performed using two-tail Student's t-test or Mann–Whitney U-test (only on in vivo metastasis study). P<0.05 was considered as statistically significant.
Acknowledgments
We thank Dr Isaiah Fidler for providing the PC-3M and PC-3M-LN4 cell lines, Dr Andrew ML Chan for the A549 cell line, Dr Cheng-Chung Wu for the pGL3-CRMP1-Luc and Dr Alexander Staruschenko for the pcDNA3.1-His-WASF1. This work was supported by an earmarked research grant from the Research Grants Council of Hong Kong (project no. CUHK4411/06M) and a research grant from the National Natural Science Foundation of China.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer 2002; 2: 442–454. [DOI] [PubMed] [Google Scholar]
- Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell 2008; 14: 818–829. [DOI] [PubMed] [Google Scholar]
- Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev 2009; 28: 15–33. [DOI] [PubMed] [Google Scholar]
- Polyak K, Weinberg RA. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat Rev Cancer 2009; 9: 265–273. [DOI] [PubMed] [Google Scholar]
- Scheel C, Weinberg RA. Cancer stem cells and epithelial-mesenchymal transition: concepts and molecular links. Semin Cancer Biol 2012; 22: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. The ins and outs of the epithelial to mesenchymal transition in health and disease. Annu Rev Cell Dev Biol 2011; 27: 347–376. [DOI] [PubMed] [Google Scholar]
- Hugo H, Ackland ML, Blick T, Lawrence MG, Clements JA, Williams ED et al. Epithelial–mesenchymal and mesenchymal–epithelial transitions in carcinoma progression. J Cell Physiol 2007; 213: 374–383. [DOI] [PubMed] [Google Scholar]
- Gunasinghe NP, Wells A, Thompson EW, Hugo HJ. Mesenchymal-epithelial transition (MET) as a mechanism for metastatic colonisation in breast cancer. Cancer Metastasis Rev 2012; 31: 469–478. [DOI] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 2013; 19: 1438–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature 1995; 376: 509–514. [DOI] [PubMed] [Google Scholar]
- Minturn JE, Fryer HJL, Geschwind DH, Hockfield S. Toad-64, a gene expressed early in neuronal differentiation in the rat, is related to Unc-33, a C. elegans gene involved in axon outgrowth. J Neurosci 1995; 15: 6757–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byk T, Dobransky T, Cifuentes-Diaz C, Sobel A. Identification and molecular characterization of Unc-33-like phosphoprotein (Ulip), a putative mammalian homolog of the axonal guidance-associated unc-33 gene product. J Neurosci 1996; 16: 688–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene 1996; 180: 157–163. [DOI] [PubMed] [Google Scholar]
- Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem 1997; 69: 2261–2269. [DOI] [PubMed] [Google Scholar]
- Fukada M, Watakabe I, Yuasa-Kawada J, Kawachi H, Kuroiwa A, Matsuda Y et al. Molecular characterization of CRMP5, a novel member of the collapsin response mediator protein family. J Biol Chem 2000; 275: 37957–37965. [DOI] [PubMed] [Google Scholar]
- Charrier E, Reibel S, Rogemond V, Aguera M, Thomasset N, Honnorat J. Collapsin response mediator proteins (CRMPs): involvement in nervous system development and adult neurodegenerative disorders. Mol Neurobiol 2003; 28: 51–64. [DOI] [PubMed] [Google Scholar]
- Yamashita N, Goshima Y. Collapsin response mediator proteins regulate neuronal development and plasticity by switching their phosphorylation status. Mol Neurobiol 2012; 45: 234–246. [DOI] [PubMed] [Google Scholar]
- Schmidt EF, Strittmatter SM. The CRMP family of proteins and their role in Sema3A signaling. Adv Exp Med Biol 2007; 600: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih JY, Yang SC, Hong TM, Yuan A, Chen JJ, Yu CJ et al. Collapsin response mediator protein-1 and the invasion and metastasis of cancer cells. J Natl Cancer Inst 2001; 93: 1392–1400. [DOI] [PubMed] [Google Scholar]
- Hu Y, Ippolito JE, Garabedian EM, Humphrey PA, Gordon JI. Molecular characterization of a metastatic neuroendocrine cell cancer arising in the prostates of transgenic mice. J Biol Chem 2002; 277: 44462–44474. [DOI] [PubMed] [Google Scholar]
- Pan SH, Chao YC, Chen HY, Hung PF, Lin PY, Lin CW et al. Long form collapsin response mediator protein-1 (LCRMP-1) expression is associated with clinical outcome and lymph node metastasis in non-small cell lung cancer patients. Lung Cancer 2010; 67: 93–100. [DOI] [PubMed] [Google Scholar]
- Pan SH, Chao YC, Hung PF, Chen HY, Yang SC, Chang YL et al. The ability of LCRMP-1 to promote cancer invasion by enhancing filopodia formation is antagonized by CRMP-1. J Clin Invest 2011; 121: 3189–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Pang J, Li LY, Liu WP, Di JM, Sun QP et al. Expression profiling identifies new function of collapsin response mediator protein 4 as a metastasis-suppressor in prostate cancer. Oncogene 2010; 29: 4555–4566. [DOI] [PubMed] [Google Scholar]
- Meyronet D, Massoma P, Thivolet F, Chalabreysse L, Rogemond V, Schlama A et al. Extensive expression of collapsin response mediator protein 5 (CRMP5) is a specific marker of high-grade lung neuroendocrine carcinoma. Am J Surg Pathol 2008; 32: 1699–1708. [DOI] [PubMed] [Google Scholar]
- Wu CC, Lin JC, Yang SC, Lin CW, Chen JJ, Shih JY et al. Modulation of the expression of the invasion-suppressor CRMP-1 by cyclooxygenase-2 inhibition via reciprocal regulation of Sp1 and C/EBPalpha. Mol Cancer Ther 2008; 7: 1365–1375. [DOI] [PubMed] [Google Scholar]
- Hiroshima Y, Nakamura F, Miyamoto H, Mori R, Taniguchi K, Matsuyama R et al. Collapsin response mediator protein 4 expression is associated with liver metastasis and poor survival in pancreatic cancer. Ann Surg Oncol 2013; 20: S369–S378. [DOI] [PubMed] [Google Scholar]
- Oliemuller E, Peláez R, Garasa S, Pajares MJ, Agorreta J, Pio R et al. Phosphorylated tubulin adaptor protein CRMP-2 as prognostic marker and candidate therapeutic target for NSCLC. Int J Cancer 2013; 132: 1986–1995. [DOI] [PubMed] [Google Scholar]
- Smith BN, Odero-Marah VA. The role of Snail in prostate cancer. Cell Adh Migr 2012; 6: 433–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu JH, Yu S, Hayward SW, Chan FL. Development of a three-dimensional culture model of prostatic epithelial cells and its use for the study of epithelial-mesenchymal transition and inhibition of PI3K pathway in prostate cancer. Prostate 2009; 69: 428–442. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 2013; 25: 76–84. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Yoshimura T, Tsuboi D, Kawabata S, Kaneko-Kawano T, Shirataki H et al. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol 2005; 25: 9920–9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung T, Ng Y, Cheong A, Ng CH, Tan I, Hall C et al. p80 ROKα binding protein is a novel splice variant of CRMP-1 which associates with CRMP-2 and modulates RhoA-induced neuronal morphology. FEBS Lett 2002; 532: 445–449. [DOI] [PubMed] [Google Scholar]
- Gao M, Yeh PY, Lu YS, Chang WC, Kuo ML, Cheng AL. NF-κB p50 promotes tumor cell invasion through negative regulation of invasion suppressor gene CRMP-1 in human lung adenocarcinoma cells. Biochem Biophys Res Commun 2008; 376: 283–287. [DOI] [PubMed] [Google Scholar]
- Yu S, Wong YC, Wang XH, Ling MT, Ng CF, Chen S et al. Orphan nuclear receptor estrogen-related receptor-β suppresses in vitro and in vivo growth of prostate cancer cells via p21WAF1/CIP1 induction and as a potential therapeutic target in prostate cancer. Oncogene 2008; 27: 3313–3328. [DOI] [PubMed] [Google Scholar]
- Cheung CP, Yu S, Wong KB, Chan LW, Lai FMM, Wang XH et al. Expression and functional study of estrogen receptor-related receptors in human prostatic cells and tissues. J Clin Endocr Metab 2005; 90: 1830–1844. [DOI] [PubMed] [Google Scholar]
- Yu S, Xu Z, Zou C, Wu D, Wang Y, Yao X et al. Ion channel TRPM8 promotes hypoxic growth of prostate cancer cells via an O2-independent and RACK1-mediated mechanism of HIF-1α stabilization. J Pathol 2014; 234: 514–525. [DOI] [PubMed] [Google Scholar]
- Yu S, Wang MW, Yao XQ, Chan FL. Establishment of a novel immortalized human prostatic epithelial cell line stably expressing androgen receptor and its application for the functional screening of androgen receptor modulators. Biochem Biophys Res Commun 2009; 382: 756–761. [DOI] [PubMed] [Google Scholar]
- Yu S, Wang X, Ng CF, Chen S, Chan FL. ERRγ suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res 2007; 67: 4904–4914. [DOI] [PubMed] [Google Scholar]
- Wu D, Yu S, Jia L, Zou C, Xu Z, Xiao L et al. Orphan nuclear receptor TLX functions as a potent suppressor of oncogene-induced senescence in prostate cancer via its transcriptional co-regulation of the CDKN1A (p21WAF1/CIP1 and SIRT1 genes. J Pathol 2015; 236: 103–115. [DOI] [PubMed] [Google Scholar]
- Lui K, Huang Y, Choi HL, Yu S, Wong KB, Chen S et al. Molecular cloning and functional study of rat estrogen receptor-related receptor gamma in rat prostatic cells. Prostate 2006; 66: 1600–1619. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.