Summary
In Asia, enthesitis‐related arthritis (ERA) is the most frequent category of juvenile idiopathic arthritis. ERA has a strong association with human leucocyte antigen (HLA)‐B27 and subclinical gut inflammation. In an HLA‐B27 transgenic rat model, the presence of Bacteroides bacteria in the gut appears to cause spondyloarthropathy (SpA). Thus, we studied gut microbiota in children with ERA. Stool specimens from 33 patients with ERA and 14 age‐matched healthy controls were studied; none had any gastrointestinal symptom, or had received a drug known to affect gut motility or microbiota in the preceding 6 weeks. From each specimen, a cDNA library for the V3 region of bacterial 16S rRNA was subjected to high‐throughput, massively parallel sequencing. Relationship of the specimens was studied using principal co‐ordinate analysis (PCoA), and abundances of various bacterial taxa and alpha diversity were compared between groups. In eight patients, a repeat faecal specimen was studied after 12 weeks of probiotic therapy. The 55 specimens yielded a median (range) of 397 315 (102 093–1 502 380) high‐quality reads each. In PCoA, gut microbiota from ERA showed a wider dispersion than those from controls. In patients, families Bacteroidaceae and Enterobacteriaceae were more abundant and Prevotellaceae were less abundant than in controls. Also, genera Bacteroides, Entercoccus and Klebsiella were over‐represented and genus Prevotella was under‐represented in ERA patients. Probiotic therapy led to a non‐significant increase in Prevotellaceae. Patients with ERA have a dysbiosis in the gut, with increased abundance of Bacteroides and reduction of Prevotella. Probiotic supplementation in a subset of patients did not reverse these changes significantly.
Keywords: gut, juvenile arthritis, microbiome
Introduction
Commensal micribiota of the gut, comprised of anaerobic and aerobic bacteria, and acquired at or shortly after birth, play a crucial role in body homeostasis. These microbiota harvest energy by fermenting the substances that reach the colon undigested, suppress the growth of pathogenic organisms and help in the development of intestinal microvasculature 1. In addition, they help in the development and maturation of gut‐mucosal and systemic immune systems, including the development and organization of tertiary lymphoid structures, such as Peyer's patches 2, 3.
A change in gut micribiota appears to alter host immune responses, thereby influencing the course of systemic immune‐inflammatory diseases 3, 4. In the K/B×N mouse model of arthritis, the disease is attenuated if the animals are raised in a germ‐free environment 5. Similar attenuation of disease has also been observed in germ‐free mice in the experimental autoimmune encephalomyelitis (EAE) model of multiple sclerosis 6. In both these models, mono‐colonization of the intestine with segmented filamentous bacteria belonging to genus Clostridium has been shown to enhance the T helper type 17 (Th17) immune response and to result in flare of autoimmune features 6, 7.
Interestingly, a particular group of bacteria can have varying effects in different diseases. For instance, in a human leucocyte antigen (HLA)‐B27 transgenic rat model of spondyloarthropathy (SpA), in which arthritis develops in rats raised under conventional conditions but not in those raised in germ‐free conditions 8, experimental colonization with Bacteroides spp. in the latter led to the induction of arthritis 8. By contrast, in the EAE mouse model, introduction of polysaccharide‐A positive B. fragilis attenuates the manifestations through induction of tolerogenic dendritic cells and interleukin (IL)−10 producing a regulatory T cell response 9.
Data on the role of gut microbiota in human arthritis have begun to emerge in recent years. Enthesitis‐related arthritis (ERA), a category of juvenile idiopathic arthritis (JIA), is a disease of children and young adults, which is characterized by arthritis and enthesitis and resembles the ankylosing spondylitis (AS) of adults. Several clinical and laboratory findings suggest that joint inflammation in ERA is related to perturbations in the intestinal tract. We have shown previously that children with ERA have (i) a demonstrable lymphoproliferative response to gut pathogens; (ii) an increased expression on peripheral blood and synovial fluid mononuclear cells of Toll‐like receptors which can sense bacterial products; and (iii) a Th1/Th17 predominant immune response 10, 11, 12, 13. In addition, these patients have a subclinical inflammation in the gut 14. ERA is also associated with the presence of the HLA‐B27 allele, similar to other spondyloarthropathies which are related to gut inflammation and gut micribiota. Further, this subset of JIA is most prevalent in geographical regions where gastrointestinal infections are common, comprising 36% of JIA in India in contrast to 10% in North America 15, 16. All these findings strongly suggest a pathogenic role for gut micribiota in ERA.
Some recent studies on stool specimens from patients with AS and ERA using bacterial 16S ribosomal‐DNA sequencing have revealed differences in the composition of gut micribiota compared to healthy controls 17, 18, 19. However, most of these studies have been performed in the developed world. Because ERA is particularly common in developing countries, where diet and intestinal micribiota are different than in the developed world, we compared gut microbiota in children with ERA and healthy controls in India.
Probiotics are preparations that contain live bacteria that have a beneficial effect on the host by multiple mechanisms, such as preventing growth of pathogens by altering gut micribiota or by altering the metabolites in the gut lumen. In addition, they also modulate the immune response. They have shown efficacy in animal models of immune‐inflammatory disorders such as rheumatoid arthritis, inflammatory bowel disease and also in small human studies 20. Thus, in order to determine if probiotics affect faecal microbiota in ERA, we also analysed stool samples from a small number of patients enrolled into a clinical trial of probiotics 21.
Methods
Patients and controls
We studied 33 patients fulfilling the International League of Associations for Rheumatology (ILAR) classification criteria for ERA 22, and 14 healthy children and young adults as controls. From each subject, a morning stool specimen was collected, transported immediately to the laboratory at 4°C and then frozen at −80°C until analysis. Children exposed to antibiotics, intra‐articular or systemic steroids, any other immunosuppressive drugs or symptomatic gastrointestinal infection during the last 6 weeks were excluded from the study. No patient had ever received biological therapy.
Repeat stool samples from eight patients who had received one capsule of a probiotic (VSL#3; Sun Pharmaceuticals, Mumbai, India) twice daily orally, with each capsule containing 112·5 billion bacterial cells belonging to eight species, namely Streptococcus thermophilus, Bifidobacterium breve, B. longum, B. infantis, Lactobacillus acidophilus, L. plantarum, L. paracasei and L. delbrueckii as part of a clinical trial 21, were collected after 12 weeks of probiotic administration.
Stool processing and 16S rRNA sequencing
One gram of stool sample was taken for DNA isolation. Cells were lysed using sodium dodecyl sulphate (SDS) lysis buffer and the phenol‐chloroform method was used for DNA extraction. After DNA extraction from stool, Illumina sequencing libraries were prepared using a one‐step polymerase chain reaction (PCR) in a 50‐μl reaction mixture that contained 200 ng input DNA, 6·25 pmol each of forward and reverse primers (Supporting information, Table S1) and KAPA Hi‐Fi PCR master mix (Kapa Biosystems, Boston, MA, USA). The PCR conditions were: an initial denaturation at 95ºC for 5 min, followed by 20 cycles of 95ºC, 65ºC and 72ºC for 1 min each and a final extension at 72ºC for 5 min. The amplification products were purified using 2% agarose gel electrophoresis, followed by recovery of amplicons of the desired length (GenElute Gel extraction kit; Sigma‐Aldrich, St Louis, MO, USA). The purified libraries were checked for size distribution, quantitated (Agilent Bioanalyser DNA1000; Agilent, Santa Clara, CA, USA) and normalized to 10 nM. The normalized libraries were pooled in sets of eight to 12 specimens each and sequenced in one lane of an IlluminaHiScan SQ sequencing flow cell using a standard 2 × 151‐cycle paired‐end multiplex sequencing format. The library pool was spiked with 30% Illumina PhiX control library to enhance sequence diversity for efficient base calling. Data were then demultiplexed using Illumina casava software.
Processing of sequence data
The raw paired‐end reads were trimmed to remove the primer sequences and merged using PANDAseq software 23. During this step, any sequences with an overlap of fewer than 20 nucleotides in opposing reads, merged length of < 100 nucleotides or containing any ambiguous nucleotide were purged. The merged reads were subjected to quality control using an NGSQC Toolkit (National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi, India) 24, and any reads with an average Phred score below 30 were excluded. Any chimeric sequences, identified using Usearch61 25, were also purged. The remaining high‐quality, non‐chimeric merged reads were assigned to operational taxonomic units (OTUs) using the UCLUST‐based subsampled open‐reference OTU picking protocol of qiime version 1.8 26. A representative sequence for each OTU was then aligned with the Greengenes core set alignment using the PyNAST tool 27 and a phylogenetic tree was constructed using the FastTree tool 28. Taxonomy was assigned to each OTU using the qiime's UCLUST Consensus Taxonomy Assigner against the Greengenes version 13.8 reference OTUs pre‐clustered at the 97% threshold (ftp://greengenes.microbio.me/greengenes_release/gg_13_5/gg_13_8_otus. tar. gz), using the software's default parameters. Any sequences that failed to align, and singleton OTUs (those with only one sequence in a specimen), unassigned OTUs and eukaryotic (chloroplast and mitochondrial) OTUs were removed. Further, to reduce noise, OTUs that were observed in fewer than 10% of stool specimens or accounted for fewer than 0·002% of reads in all the specimens taken together were purged. A specimen‐wise observation count of each OTU was tabulated as an OTU table in the ‘biom’ format (referred to hereafter as ‘filtered OTU table’).
Beta‐diversity analysis (comparison of specimens from controls versus ERA patients)
The filtered OTU table was assembled into a classic table format, where each row represented an OTU and each column represented a faecal specimen. The cells contained observation counts for a particular OTU in a particular specimen, normalized using a log‐frequency transformation, as follows:normalized value = log10 where OC represents the actual observed count of a particular OTU in a specimen, n is the sum of observed counts for all OTUs in a particular specimen (column total), Σx is the sum of n across all specimens (sum of column totals) and N is the total number of specimens in the table.
Beta diversity was then assessed using principal co‐ordinate analysis (PCoA) based on weighted UniFrac distance matrices.
Alpha‐diversity analysis
As species richness is affected by the depth of sequencing, the OTU table for each specimen was rarefied using PhyloSeq (version 1.12.2) to the same depth, i.e. the number of reads in the specimen with the fewest reads (specimen SE030 with 99 583 reads) 29. Measures of alpha diversity (observed, Chao 1 and abundance‐based coverage estimate (ACE) indices which measure species richness, and Shannon and Simpson indices which represent richness and evenness of taxa) were estimated 30 using PhyloSeq (version 1.12.2), and compared between groups using compare_alpha_diversity.py script of qiime 1.8, using a non‐parametric test with Bonferroni's correction for multiple comparisons.
Comparison of composition of faecal microbiota between groups
For identification of differentially abundant bacterial OTUs between controls and patients, we first undertook linear discriminant analysis effect size (LefSe) (linear discriminant analysis (LDA) coupled with effect size measurement) analysis 30; OTUs with log10 LDA score ≥ 2 (in either direction) and P‐values below 0·05 were considered as discriminating markers. As this analysis does not include correction for multiple hypothesis testing, we also compared the abundances of various bacterial taxa at different taxonomic levels in patients and controls using the Mann–Whitney U‐test, followed by the Benjamini–Hochberg false discovery rate (FDR) correction; FDR values below 0·10 were considered significant.
Analysis of paired data (before and after probiotic)
Data processing for this comparison was similar, except that filtering for low‐abundance OTUs removed OTUs with sequences that accounted for less than 0·001% of all the reads. For comparison of data before and after probiotic, abundances of individual taxa were compared using Wilcoxon's signed‐rank test with FDR correction. P‐ and FDR‐value cut‐offs used were similar to those for unpaired data.
Ethical considerations
The study protocol was approved by our institution's Ethics Committee and written informed consent was taken from either the patient or a parent.
Results
Study subjects
The median age of 33 children (32 male) with ERA was 15 (range = 5–20) years and the median duration of disease was 24 (1·5–120) months (Table 1). All the patients had active arthritis and were receiving non‐steroidal anti‐inflammatory drug (NSAID) therapy. No patients were receiving any immunosuppressive drugs, including disease modifying anti‐rheumatic drugs or biologicals. Median erythrocyte sedimentation rate was 80 (20–130) mm. The median age of 14 healthy controls (13 male) was similar [13 (5–22) years].
Table 1.
Clinical details of patients with enthesitis‐related arthritis (ERA) studied (n = 33)
| Characteristic | Value |
|---|---|
| Age (years), median (range) | 15 (5–10) |
| Duration of disease (months), median (range) | 24 (1·5–120) |
| Active arthritis, n (%) | 33 (100%) |
| Enthesitis, n (%) | 22 (66%) |
| Sacroiliitis, n (%) | 15 (45%) |
| Uveitis, n (%) | 0 |
| Inflammatory back pain, n (%) | 7 (21%) |
| HLA B27 positivity, n (%) | 31 (93%) |
| Erythrocyte sedimentation rate (Westergren method) (mm), median (range) | 80 (20–130) |
HLA = human leucocyte antigen.
Gut microbiota in patients with ERA versus controls
Median (range) number of high‐quality reads in specimens from 14 control subjects and 33 ERA were 272 327(136 563–761 951; total reads 4 728 631) and 397 315 (102 093–1 502 380; total reads 15 871 719), respectively. There was a total of 20 600 350 high‐quality reads (ERA: 15 871 719, healthy controls: 4 728 631), of which 586 852 reads were removed by filtering rare OTUs. These reads belonged to 17 539 non‐singleton OTUs. Of these, 533 OTUs, distributed in 10 phyla, were identified in at least five specimens each and accounted for > 0·002% of the total reads, and were analysed further.
On PCoA of weighted UniFrac OTU profile distances, the distributions of data points for faecal microbiota from patients with ERA overlapped with those from healthy controls (Fig. 1a). However, visual inspection of the graph reveals that data points for faecal microbiota from patients showed a wider dispersion than those from controls. There was no significant difference in measures of alpha diversity between healthy controls and patients with ERA (Fig. 1b).
Figure 1.
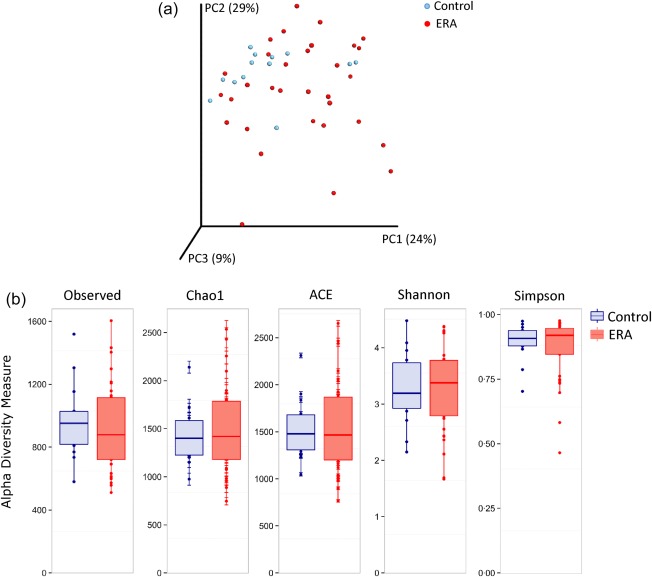
(a) Principal co‐ordinate analysis (beta diversity) of weighted UniFrac distances between specimens collected from patients with enthesitis‐related arthritis (ERA; in red) and healthy controls (in blue). (b) Comparison of measures of alpha diversity between specimens from patients with ERA (red) and healthy controls (blue) showed no significant difference. Median and interquartile range are shown. [Colour figure can be viewed at wileyonlinelibrary.com]
The abundance of various bacterial phyla, classes, orders, families and genera in the specimens in the two groups are shown in Supporting information, Table S2. On LefSe analysis, several phyla, classes, orders, families and genera showed high LDA scores (values > 2·0 and with uncorrected P‐values < 0·05; Supporting information, Table S3; however, the corrected P‐values, after applying FDR correction for multiple comparisons, were not significant for any taxon.
Figure 2a,b shows the results of comparison of abundances of various families and genera between patients with ERA and controls. In this analysis, bacteria belonging to families Bacteroidaceae, Enterobacteriaceae and Enterococcaceae had significantly higher abundances and those for family Prevotellaceae had a lower abundance in faeces from patients with ERA than in those from controls, even after correction for multiple comparisons (Fig. 2a). Similarly, bacteria belonging to genera Bacteroides, Enterococcus and Klebisella were more abundant, and those belonging to genus Prevotella were less abundant in the faeces of patients with ERA than in those from controls (Fig. 2b).
Figure 2.
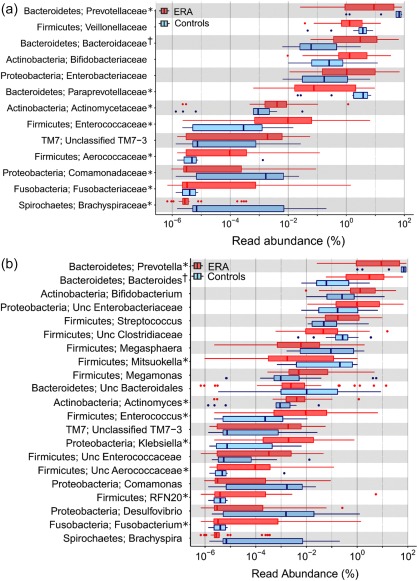
Differences in abundances of various bacterial families (a) and genera (b) in fecal microbiota from patients with enthesitis‐related arthritis (ERA; red) and healthy controls (blue). The data are shown using box‐plots and percentage values on a log10 scale. The ends of the boxes represent 25th–75th percentiles, and dots to their left or right indicate outliers. Data are shown for taxa where there was a significant intergroup difference using the Mann–Whitney U‐test. *Indicates taxa with corrected P‐values (following correction for multiple testing using the Benjamini–Hotchberg false discovery rate procedure) of < 0.10; †indicates those with corrected P‐values of < 0·05. [Colour figure can be viewed at wileyonlinelibrary.com]
Table 2 shows a comparison of relative abundance of some selected species belonging to the genus Bacteroides and Prevotella. This showed that some species belonging to genus Bacteroides, namely B. fragilis, B. plebeius and B. eggerthii, had higher abundance and some species belonging to genus Prevotella, i.e. P. copri and P. stercorea, had reduced abundance in the faecal microbiota of patients with ERA than in controls.
Table 2.
Comparison of relative abundance (%) of bacterial species of phylum bacteroidetes in fecal specimens from healthy controls and patients with enthesitis‐related arthritis (ERA)
| Species | Control | Patients | P | FDR P | ||
|---|---|---|---|---|---|---|
| Median | (Range) | Median | (Range) | |||
| Bacteroides plebeius | 2.6E‐04 | (0–0·99) | 0·01 | (0–52·88) | 0·0001 | 0·01 |
| Prevotella copri | 58.17457 | (0·79–77·16) | 8·92 | (0·02–67·53) | 0·0016 | 0·07 |
| B. fragilis | 1.2E‐03 | (0–0·07) | 0·01 | (0–17·96) | 0·0036 | 0·09 |
| Bacteroides eggerthii | 0 | (0–1·0E‐03) | 2·3E‐04 | (0–2·52) | 0·01 | 0·09 |
| P. stercorea | 3.855227 | (0·03‐15·70) | 0·08 | (1·2E‐03‐17·93) | 0·01 | 0·09 |
| B. caccae | 3.1E‐03 | (0–0·45) | 3·1E‐03 | (0–0·45) | 0·16 | 0·35 |
P‐values were calculated using the Mann–Whitney U‐test, and were corrected for multiple comparisons using the Benjamini–Hochberg procedure for false discovery rate (FDR).
Gut microbiota before and after probiotic administration
There were 5 834 497 high‐quality reads from 16 specimens from eight patients with paired data; 85 289 reads were removed because of filtering rare OTUs. Data for 875 OTUs across 11 phyla that accounted for at least 0·001% of all reads were analysed further. On PCoA analysis, no separation was noticed between the specimens collected before and after probiotic intake (Supporting information, Fig. S1a); also, the two types of specimens showed no difference in measures of alpha diversity (Supporting information, Fig. S1b).
Abundances of various bacterial taxa in the faeces collected at baseline and after probiotic administration (Supporting information, Table S4) showed no differences at phylum, class, order, family (Supporting information, Fig. S2a) and genus (Supporting information, Fig. S2b) levels.
Discussion
The intestinal tract plays a major role in the pathogenesis of HLA‐B27‐related diseases, and recent data on gut microbiome in animal models of these diseases and in patients with SpA suggest the presence of an intestinal dysbiosis. In our study, children with ERA in India showed an increased abundance in their faecal microbiota of bacterial families Bacteroideaceae, Enterobacteriaceae and Enterococcaceae, and genera Bacteroides and Enterococcus. Further, these children also had a larger proportion of Bacteroides fragilis and B. plebius and B. eggerthii in their faeces. By contrast, family Prevetollaceae and genus Prevotella were less abundant in the stool specimens from these patients. In a subset of our patients who received a probiotic, there was no significant change in the faecal microbiota during a 12‐week period.
Our primary finding was an increase in the abundance of genus Bacteroides in patients with ERA. Data on the abundance of genus Bacteriodes in such patients are also available from other studies. First, in a study from Alabama, USA, abundance of Bacteriodes in 25 patients with ERA was somewhat higher than in 13 controls, although the difference was not statistically significant. However, on further analysis, the authors found two distinct clusters among their patients – one of eight patients who had an increased abundance of Bacteroides and another of 17 patients who lacked this finding 18. This suggested that increased abundance of Bacteroides could be responsible for at least a subset of patients with ERA in that study. The more marked change in the abundance of these bacteria in our patients could suggest that these may be responsible for a larger proportion of cases of ERA in developing countries. Another recent study in children with other categories of JIA (mainly oligoarticular or polyarticular) has also reported an increase in Bacteroidetes and a reduction in Firmicutes 31. Thus, it appears that an increased abundance of Bacteroides, observed by us, is a common theme in patients with JIA.
In HLA‐B27 transgenic rats, intestinal colonization with Bacteroides spp. led to the development of arthritis and colitis, whereas the rats raised in a germ‐free environment did not develop the disease 8. A recent study comparing gut microbiota in HLA‐B27 transgenic rats to wild‐type rats showed an increased abundance of bacteria belonging to phylum Bacteroidetes in the caecal lumen and tissue. At species level, Bacteroides vulgatus and B. fragilis were found to be more abundant in transgenic rats 32. In a model of ankylosing enthesopathy, arthritis developed in germ‐free mice after colonization with a mixture of anaerobic bacteria; further, the gut microbiota of these mice showed an increased abundance of B. fragilis, B. ovatus and Enterococcus faecalis 33.
Studies on microbiota in human subjects with SpA are limited. In patients with AS, analysis of tissue from the terminal ileum revealed a higher abundance of bacteria belonging to the Bacteroidaceae family 34. Similarly, colonic biopsies of patients with Crohn's disease and ulcerative colitis also showed an increased abundance of Bacteroides 35. Faeces from patients with psoriatic arthritis have also shown an increased abundance of Bacteroidetes than those with psoriasis, but no arthritis 36.
Bacteroides in the gut are mainly commensal organisms. However, they contain virulence factors that can generate inflammatory responses. Agglutinins and histolytic enzymes of B. fragilis help these bacteria to adhere to the host mucosa and to cause tissue destruction, respectively 37. These bacteria are also known to evade the host immune response by decreasing the production of inducible nitric oxide synthase and inhibiting the phagocytosis by macrophages 38. In addition, the B. fragilis enterotoxin is known to disrupt the tight junctions in intestinal epithelium, leading to increased gut permeability in patients with inflammatory bowel disease (IBD) 39.
Another observation in our study was an increased abundance of members of the family Enterobacteriaceae, particularly those belonging to genus Klebsiella, in the stools of patients with ERA. This suggests a role for these bacteria in the causation of ERA. Previous studies on gut microbiome have not shown an increase in Enterobacteriaceae in patients with AS and JIA 18, 31, 34, and thus our finding is somewhat novel. However, it is supported by several additional pieces of evidence. First, members of Enterobacteriaceae, such as Salmonella, Yersenia and Shigella, have been identified as triggers for enetrically acquired reactive arthritis 40. Secondly, demonstrable lymphoproliferative T cell responses against enteric bacteria are observed more often in patients with ERA than in healthy people 10. Furthermore, using conventional stool culture techniques, faecal carriage of Klebsiella has been reported to be more common in patients with AS than in healthy people 41. In addition, in a collated data set from 1556 patients with AS from 16 countries, serum titres of antibodies to Klebsiella were higher than in healthy people 42. Also, the nitrogenase enzyme of Klebsiella pneumoniae has a sequence homology with HLA‐B27 and could play a role in the induction of joint inflammation via molecular mimicry 43. Another starch‐digesting enzyme, pullulanase, produced by Klebsiella, has sequence homology with collagens type I, III and IV, which are key components of spinal and synovial tissue, the primary site of involvement in AS 41. Interestingly, a high carbohydrate and low protein diet has been shown to be associated with increased faecal concentration of Klebsiella 44; whether or not this can increase the risk of ERA needs further study.
Patients with ERA in our study also had an increased abundance of bacteria belonging to the Enterococcaceae family as well as Enterococcus genus in their faecal microbiota. Several studies in HLA‐B27 transgenic and other animal models of IBD have shown a higher abundance of these bacteria, and linked it to the severity of colitis 33, 35, 45. However, data on abundances of these groups have not been reported in previous studies in AS or in JIA. Bacteria belonging to the Bifidobacteriaceae family were also more abundant in patients with ERA than in healthy controls – a finding similar to that reported previously in ERA and AS 17, 18. Members of Bifidobacterium genus are believed to influence the immune system through increased expression of IL‐12 in the gut lymphoid tissue and the generation of regulatory T cells 46.
Bacteria belonging to the Prevotella group were less abundant in faeces of patients with ERA, both at family and genus levels. A similar low abundance of Prevotellaceae has been reported previously in ileal biopsies from patients with AS 34. However, this finding has not been reported previously in patients with JIA. Expansion of P. copri has been reported in rheumatoid arthritis 46, although P. histicola has been shown to attenuate arthritis in animal models of RA 47. However, RA has very different pathogenesis to spondyloarthropathies, including ERA. In our population, healthy controls showed a high abundance of Prevotella; similar data have been shown in another study from India 48. In this context, it may be interesting to know that low fibre and high protein diets are associated with a reduction of Prevotella in the gut 49. In India, the diet has high fibre and low protein content, and this may account for high Prevotella in healthy subjects.
Another interesting component of our study was the comparison of faecal microbiota before and after administration of a probiotic preparation. This treatment led to a slight increase in the abundance of Prevotella and a decrease in the abundance of Bifidobacterium. A recent study using genus/species‐specific real time PCR had shown an increase in Lactobacillus and Bifidobacterium after consumption of probiotic yoghurt in patients with inflammatory bowel disease as well as healthy people 50. However, a recent systematic review 51 concluded that probiotic supplementation did not alter faecal microbiota in healthy adults, except in one study in which the probiotic group had increased beta diversity compared to placebo 52.
We believe that our results are fairly robust, as we studied a fairly large number of patients as well as controls. Furthermore, our study was performed in a developing world setting, in which ERA is the predominant category of JIA. The main limitation of our study was that our patients were receiving treatment for arthritis and this, by itself, could have had an effect on the gut microbiota. However, it is difficult to obviate this limitation in this disease, particularly in tertiary‐care settings in developing countries, as patients often present late and have marked symptoms. However, by contrast, our study was also helped by the fact that none of our patients were receiving biologicals or disease‐modifying anti‐rheumatic drugs – thus obviating a limitation with studies from developed countries. Also, the effect of probiotics was studied in only a small number of subjects.
In conclusion, our data suggests the existence of a dysbiosis in the guts in children with ERA with increased abundance of bacterial groups that may promote a proinflammatory state. This, together with previous data from various experimental models of arthritides, suggests that this alteration of gut microbiota can change the gut and the systemic immune response, and thus play a role in the causation of this type of JIA. Hence, restoration of gut microbiota towards normal may be helpful in ameliorating this form of arthritis. In this respect, our data on gut microbiota following administration of a probiotic in the usual dose, albeit limited by a small sample size, were not encouraging, and suggest that this intervention may not be adequate for achieving either a major change in gut microbiota or a clinical response 21. However, it may be worthwhile trying a higher than usual dose of a probiotic, different formulation of a probiotic or faecal microbial transplantation in the treatment of this condition.
Disclosure
The authors have no disclosutres to declare.
Supporting information
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Results of beta and alpha diversity analysis for patients with enthesitis‐related arthritis (ERA) before and after probiotic administration. (a) Principal co‐ordinate analysis (beta diversity) of weighted UniFrac distances between specimens collected from patients with ERA at baseline (red) and after administration of a probiotic preparation for 12 weeks (blue). (b) Comparison of measures of alpha diversity between specimens from patients with ERA at baseline (red) and after probiotic (blue).
Supporting Figure 1b
Fig. S2. Abundances of bacterial families (a) and genera (b) in faecal microbiota from patients with enthesitis‐related arthritis (ERA) at baseline (red) and after probiotic administration (blue). Data are shown using box‐plots and percentage values on a log10 scale. The ends of the boxes represent 25th to 75th centiles, and any dots to the left or right of the boxes indicate outliers.
Supporting Figure 2b
Table S1. Sequences of primers used for 16S rRNA sequencing
Table S2. Relative percentage abundance of each taxonomic group in faecal specimens from healthy controls and patients with enthesitis‐related arthritis (ERA). The data at each taxonomic level are shown in the ascending order of P‐values. Column ‘P’ shows uncorrected P‐values using the Mann–Whitney U‐test for comparison of controls versus patients, and column ‘FDR_P’ shows values for false discovery rates using the Benjamini–Hochberg procedure to control for multiple comparisons. Cells with FDR values < 0.10 are highlighted in grey
Table S3. Linear discriminant analysis effect size (LefSe) linear discriminant analysis (LDA) scores for abundances of different taxonomic groups in patients with enthesitis‐related arthritis (ERA) versus healthy controls
Table S4. Relative percentage abundance of each taxonomic group in faecal specimens from patients with enthesitis‐related arthritis (ERA) before and after 12 weeks of probiotic treatment. The data at each taxonomic level are shown in the ascending order of P‐values. Column ‘P’ shows uncorrected P‐values using Wilcoxon's signed‐rank test for comparison of controls versus patients and column ‘FDR_P’ shows values for false discovery rates using the Benjamini–Hochberg procedure to control for multiple comparisons
Acknowledgements
This work was funded by a grant from Department of Biotechnology, Government of India, New Delhi to A. A.
References
- 1. Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system. Science 2010; 330:1768–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci USA 2002; 99:15451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014; 157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal alpha‐defensins by intestinal Paneth cells in response to bacteria. Nat Immunol 2000; 1:113–8. [DOI] [PubMed] [Google Scholar]
- 5. Berer K, Mues M, Koutrolos M et al Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479:538–41. [DOI] [PubMed] [Google Scholar]
- 6. Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Pro‐inflammatory T‐cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108(Suppl. 1):4615–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu HJ, Ivanov II, Darce J et al Gut‐residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 2010; 32:815–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Taurog JD, Richardson JA, Croft JT et al The germfree state prevents development of gut and joint inflammatory disease in HLA‐B27 transgenic rats. J Exp Med 1994; 180:2359–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ochoa‐Repáraz J, Mielcarz DW, Wang Y et al A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol 2010; 3:487–95. [DOI] [PubMed] [Google Scholar]
- 10. Saxena N, Misra R, Aggarwal A. Is enthesitis related arthritis type of juvenile idiopathic arthritis a form of chronic reactive arthritis? Rheumatology (Oxf) 2006; 45:1129–32. [DOI] [PubMed] [Google Scholar]
- 11. Myles A, Aggarwal A. Expression of Toll‐like receptors 2 and 4 is increased in peripheral blood and synovial fluid monocytes of patients with enthesitis related arthritis subtype of juvenile idiopathic arthritis. Rheumatology (Oxf) 2011; 50:481–8. [DOI] [PubMed] [Google Scholar]
- 12. Myles A, Rahman M, Aggarwal A. Membrane‐bound Toll‐like receptors are over‐expressed in peripheral blood and synovial fluid mononuclear cells of enthesitis related arthritis category of juvenile idiopathic arthritis (JIA‐ERA) patients and lead to secretion of inflammatory mediators. J Clin Immunol 2012; 32:488–96. [DOI] [PubMed] [Google Scholar]
- 13. Mahendra A, Misra R, Aggarwal A. Th1 and Th17 predominance in enthesitis related arthritis form of juvenile idiopathic arthritis. J Rheumatol 2009; 36:1730–06. [DOI] [PubMed] [Google Scholar]
- 14. Orlando A, Renna S, Perricone G, Cottone M. Gastrointestinal lesions associated with spondyloarthropathies. World J Gastroenterol 2009; 15:2443–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kunjir V, Venuopalan A, Chopra A. Profile of indian patients with juvenile onset chronic inflammatory joint disease using the ILAR classification criteria for JIA: a community‐based cohort study. J Rheumatol 2010; 37:1756–62. [DOI] [PubMed] [Google Scholar]
- 16. Beukelman T, Ringold S, Davis TE, DeWitt EM, Pelajo CF, Weiss PF. Disease‐modifying antirheumatic drug use in the treatment of juvenile idiopathic arthritis: a cross‐sectional analysis of the CARRA registry. J Rheumatol 2012; 39:1867–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stebbings S, Munro K, Simon MA et al Comparison of the faecal microflora of patients with ankylosing spondylitis and controls using molecular methods of analysis. Rheumatology (Oxf) 2002; 41:1395–401. [DOI] [PubMed] [Google Scholar]
- 18. Stoll ML, Kumar R, Morrow CD et al Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis‐related arthritis. Arthritis Res Ther 2014; 16:486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gill T, Asquith M, Rosenbaum JT, Colbert RA. The intestinal microbiome in spondyloarthritis. Curr Opin Rheumatol 2015; 27:319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bedaiwi MK, Inman RD. Microbiome and probiotics: link to arthritis. Curr Opin Rheumatol 2014; 26:410–5. [DOI] [PubMed] [Google Scholar]
- 21. Shukla A, Gaur P, Aggarwal A. Effect of probiotics on clinical and immune parameters in enthesitis‐related‐arthritis category of juvenile idiopathic arthritis. Clin Exp Immunol 2016; 195:301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petty RE, Southwood TR, Manners P et al International league of associations for rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton 2001. J Rheumatol 2004; 31:390–2. [PubMed] [Google Scholar]
- 23. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired‐end assembler for Illumina sequences. BMC Bioinformatics 2012; 13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patel RK, Jain M. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS ONE 2012; 7:e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010; 26:2460–1. [DOI] [PubMed] [Google Scholar]
- 26. Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol 2012; Chapter 1:Unit 1E.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 2010; 26:266–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009; 26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLOS ONE 2013; 8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gotelli NJ, Anne C. Measuring and estimating species richness, species diversity, and biotic similarity from sampling data In: Levin SA, ed. Encyclopedia of biodiversity, 2nd edn Waltham, MA: Academic Press, 2013:195–211. [Google Scholar]
- 31. Tejesvi MV, Arvonen M, Kangas SM et al Faecal microbiome in new‐onset juvenile idiopathic arthritis. Eur J Clin Microbiol Infect Dis 2016; 35:363–70. [DOI] [PubMed] [Google Scholar]
- 32. Lin P, Bach M, Asquith M et al HLA‐B27 and human β2‐microglobulin affect the gut microbiota of transgenic rats. PLoS One 2014; 9:e105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sinkorová Z, Capková J, Niederlová J, Stepánková R, Sinkora J. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H‐2(k)) male mice. Hum Immunol 2008; 69:845–50. [DOI] [PubMed] [Google Scholar]
- 34. Costello ME, Ciccia F, Willner D et al Intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol 2015; 67:686–91. [DOI] [PubMed] [Google Scholar]
- 35. Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J Med Res 2015; 142:23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scher JU, Ubeda C, Artacho A et al Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol 2015; 67:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wexler HM. Bacteroides: the good, the bad, and the nitty‐gritty. Clin Microbiol Rev 2007; 20:593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Vieira JM, Vallim DC, Ferreira EO et al Bacteroides fragilis interferes with iNOS activity and leads to pore formation in macrophage surface. Biochem Biophys Res Commun 2005; 326:607–13. [DOI] [PubMed] [Google Scholar]
- 39. Berkes J, Viswanathan VK, Savkovic SD, Hecht G. Intestinal epithelial responses to enteric pathogens: effects on the tight junction barrier, ion transport, and inflammation. Gut 2003; 52:439–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yu D, Kuipers JG. Role of bacteria and HLA‐B27 in the pathogenesis of reactive arthritis. Rheum Dis Clin North Am 2003; 29:21–36. [DOI] [PubMed] [Google Scholar]
- 41. Eastmond CJ, Calguner M, Shinebaum R, Cooke EM, Wright V. A sequential study of the relationship between faecal Klebsiella aerogenes and the common clinical manifestations of ankylosing spondylitis. Ann Rheum Dis 1982; 41:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rashid T, Ebringer A, Wilson C. The link between Klebsiella and ankylosing spondylitis in worldwide geographical locations. Curr Rheumatol Rev 2016. [Epub ahead of print]. [PubMed] [Google Scholar]
- 43. Rashid T, Ebringer A. Ankylosing spondylitis is linked to Klebsiella – the evidence. Clin Rheumatol 2007; 26:858–64. [DOI] [PubMed] [Google Scholar]
- 44. Rashid T, Ebringer A, Tiwana H, Fielder M. Role of Klebsiella and collagens in Crohn's disease: a new prospect in the use of low‐starch diet. Eur J Gastroenterol Hepatol 2009; 21:843–9. [DOI] [PubMed] [Google Scholar]
- 45. Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel disease in interleukin‐10 knockout mice. Am J Pathol 2002; 160:2253–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Scher JU, Sczesnak A, Longman RS et al Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2013; 2:e01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marietta EV, Murray JA, Luckey DH et al Human gut‐derived prevotella histicola suppresses inflammatory arthritis in humanized mice. Arthritis Rheumatol 2016. doi: 10.1002/art.39785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bhute S, Pande P, Shetty SA et al Molecular characterization and meta‐analysis of gut microbial communities illustrate enrichment of Prevotella and Megasphaera in Indian subjects. Front Microbiol 2016; 7:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simpson HL, Campbell BJ. Review article: Dietary fibre‐microbiota interactions. Aliment Pharmacol Ther 2015; 42:158–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shadnoush M, Hosseini RS, Khalilnezhad A, Navai L, Goudarzi H, Vaezjalali M. Effects of probiotics on gut microbiota in patients with inflammatory bowel disease: a double‐blind, placebo‐controlled clinical trial. Korean J Gastroenterol 2015; 65:215–21. [DOI] [PubMed] [Google Scholar]
- 51. Kristensen NB, Bryrup T, Allin KH, Nielsen T, Hansen TH, Pedersen O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med 2016; 8:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ferrario C, Taverniti V, Milani C et al Modulation of fecal Clostridiales bacteria and butyrate by probiotic intervention with Lactobacillus paracasei DG varies among healthy adults. J Nutr 2014; 144:1787–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article at the publisher's website:
Fig. S1. Results of beta and alpha diversity analysis for patients with enthesitis‐related arthritis (ERA) before and after probiotic administration. (a) Principal co‐ordinate analysis (beta diversity) of weighted UniFrac distances between specimens collected from patients with ERA at baseline (red) and after administration of a probiotic preparation for 12 weeks (blue). (b) Comparison of measures of alpha diversity between specimens from patients with ERA at baseline (red) and after probiotic (blue).
Supporting Figure 1b
Fig. S2. Abundances of bacterial families (a) and genera (b) in faecal microbiota from patients with enthesitis‐related arthritis (ERA) at baseline (red) and after probiotic administration (blue). Data are shown using box‐plots and percentage values on a log10 scale. The ends of the boxes represent 25th to 75th centiles, and any dots to the left or right of the boxes indicate outliers.
Supporting Figure 2b
Table S1. Sequences of primers used for 16S rRNA sequencing
Table S2. Relative percentage abundance of each taxonomic group in faecal specimens from healthy controls and patients with enthesitis‐related arthritis (ERA). The data at each taxonomic level are shown in the ascending order of P‐values. Column ‘P’ shows uncorrected P‐values using the Mann–Whitney U‐test for comparison of controls versus patients, and column ‘FDR_P’ shows values for false discovery rates using the Benjamini–Hochberg procedure to control for multiple comparisons. Cells with FDR values < 0.10 are highlighted in grey
Table S3. Linear discriminant analysis effect size (LefSe) linear discriminant analysis (LDA) scores for abundances of different taxonomic groups in patients with enthesitis‐related arthritis (ERA) versus healthy controls
Table S4. Relative percentage abundance of each taxonomic group in faecal specimens from patients with enthesitis‐related arthritis (ERA) before and after 12 weeks of probiotic treatment. The data at each taxonomic level are shown in the ascending order of P‐values. Column ‘P’ shows uncorrected P‐values using Wilcoxon's signed‐rank test for comparison of controls versus patients and column ‘FDR_P’ shows values for false discovery rates using the Benjamini–Hochberg procedure to control for multiple comparisons


