Summary
Killer‐cell immunoglobulin‐like receptors (KIRs) are components of two fundamental biological systems essential for human health and survival. First, they contribute to host immune responses, both innate and adaptive, through their expression by natural killer cells and T cells. Second, KIR play a key role in regulating placentation, and hence reproductive success. Analogous to the diversity of their human leucocyte antigen class I ligands, KIR are extremely polymorphic. In this review, we describe recent developments, fuelled by methodological advances, that are helping to decipher the KIR system in terms of haplotypes, polymorphisms, expression patterns and their ligand interactions. These developments are delivering deeper insight into the relevance of KIR in immune system function, evolution and disease.
Keywords: expression, haplotypes, killer‐cell immunoglobulin‐like receptors, ligands, natural killer cell, polymorphism
Abbreviations
- HCMV
human cytomegalovirus
- HIV
human immunodeficiency virus
- HSV
herpes simplex virus
- KIR
killer‐cell immunoglobulin‐like receptors
- NK cell
natural killer cell
Introduction
Killer‐cell immunoglobulin‐like receptors (KIRs) are type I transmembrane glycoproteins belonging to the immunoglobulin superfamily. They are primarily expressed on natural killer (NK) cells but they are also expressed on subsets of CD4, CD8 and γδ T cells.1, 2, 3, 4, 5, 6, 7, 8 Comprising both activating and inhibitory forms they represent an archetypal paired receptor system.9 The best characterized ligands for KIR are HLA class I molecules that express either the Bw4, C1 or C2 motif (Fig. 1).
Figure 1.
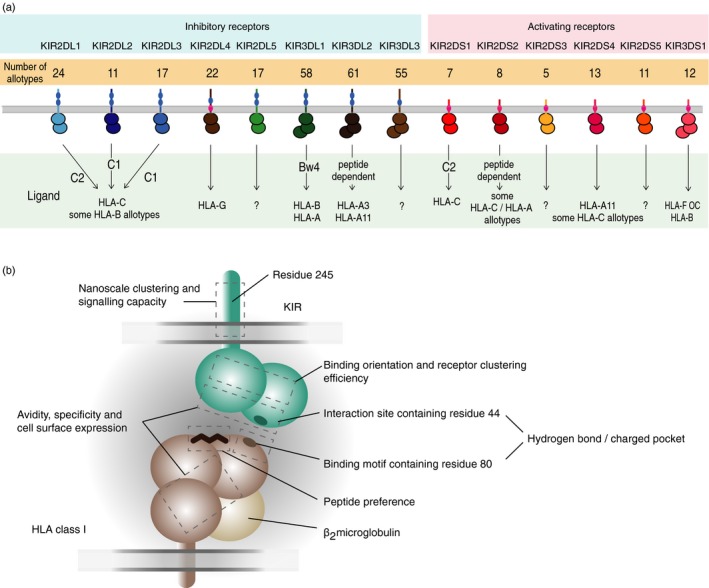
Killer‐cell immunoglobulin‐like receptors (KIR) proteins and their ligand interactions. (a) KIR have either two or three immunoglobulin‐like extracellular domains, KIR2D or KIR3D, respectively. They are either activating or inhibitory depending on the structure of their intracellular domain. Inhibitory KIR have long cytoplasmic tails (KIR**L*) that contain immunoreceptor tyrosine‐based inhibitory motifs (ITIM) that transduce inhibitory signals to the natural killer (NK) cell. Activating KIR have short cytoplasmic tails (KIR**S*) with a positively charged amino acid residue in their transmembrane region. The charged residue allows KIR proteins to associate with the TYROBP (DAP12) transmembrane signalling polypeptide, which acts as an activating signal transduction element because it contains an immunoreceptor tyrosine‐based activation motif (ITAM) in its cytoplasmic domain. KIR3DL1 and KIR3DS1, which are encoded by alleles of the same gene, KIR3DL1/S1, thus have opposing functions. KIR differentially bind HLA‐A, ‐B or ‐C allotypes and dimorphisms in the HLA class I α domains are the major determinants for this interaction. The binding motifs are referred to as C1 and C2 in HLA‐C and Bw4 in HLA‐B and HLA‐A. The precise KIR binding motif of HLA‐A*11, which can be recognized by KIR2DS2, KIR2DS4 and KIR3DL2, has not been determined.10, 11 Interactions may also be sensitive to polymorphism outside the HLA and KIR binding motifs and to the presented peptide sequence. The ligands for activating KIR and some inhibitory KIR are presently not well‐defined. OC, open conformers (b) Schematic to show how polymorphism in different parts of the KIR and HLA class I molecules diversifies their interactions. Key residues are KIR position 44 and HLA position 80, which control specificity and KIR position 245 that influences inhibitory signal strength, as discussed in the text.
The functional activity and development of KIR‐expressing lymphocytes are modulated by interactions between these receptors and their ligands.12, 13, 14 A major function of circulating cytotoxic NK cells is to recognize and eliminate cells that fail to express self HLA class I molecules in the surveillance for virus‐infected or transformed cells.15, 16 By contrast, a major function of non‐cytotoxic NK cells in the uterus is to secrete cytokines to regulate placentation during pregnancy. This occurs through a mechanism of maternal allogeneic recognition involving interaction between KIR on maternally derived uterine NK cells with HLA on fetally derived cells.17 The KIR system acts to diversify NK cell stimulation potential through specificity of interaction and strength of signalling. In this regard, weakly inhibitory KIR/HLA combinations permit a lower threshold for cell activation than do strongly inhibitory KIR/HLA combinations.
KIR genes are located in the leucocyte receptor complex on human chromosome 19q13.4. The genes are variably present in the germline between individuals, forming haplotypes with diverse gene content (Fig. 2), and numerous alleles exist for many of the genes. Despite the major implications of KIR variation for human health it is known that genome‐wide studies have poorly captured the diversity at the KIR locus. Through focused analyses, constituent polymorphism has been described at the basic levels – gene content of haplotypes, copy number, alleles and their frequencies. Resulting information has supported genetic, functional and disease investigation. In this review we discuss the outstanding challenges in KIR analysis and the recent methodological developments that are facilitating new discoveries.
Figure 2.
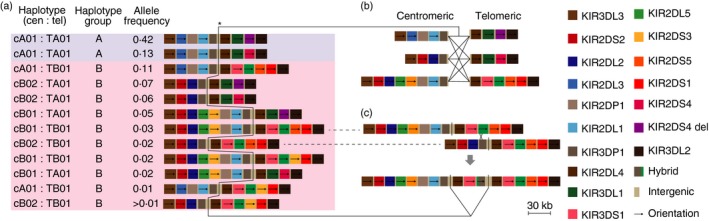
Structural haplotypes of the KIR gene cluster and recombination mechanisms. Numerous killer‐cell immunoglobulin‐like receptor (KIR) haplotypes with different gene content have been described. These haplotypes have been generated through serial duplications and deletions of chromosomal segments containing KIR genes. The distinction between alleles and genes is, therefore, sometimes blurred; for example KIR2DS3 can be located in two different positions within the KIR locus. (a) The arrangements of genes in 12 common European haplotypes18 are shown. Typically, a person inherits between 14 and 24 KIR genes (between 7 and 12 KIR genes per haplotype). KIR2DP1 and KIR3DP1 are pseudogenes. Two broad haplotypes exist – A (light blue background) and B (pink background), resulting in genotypes that are an ‘AA’, ‘AB’ or ‘BB’. A haplotypes have a single arrangement of seven expressed genes that encode mostly inhibitory KIR, which are diversified by allelic variation. B haplotypes have varied gene arrangements and tend to comprise more activating genes and less allelic diversity. The A haplotype can be divided into two types depending on whether the KIR2DS4 gene is full‐length (KIR2DS4) or carries a frameshift deletion (KIR2DS4 del). (b) Diversity has been generated by homologous recombination, particularly at a recombination hotspot (*) centrally sited within the gene cluster,19 which has shuffled the centromeric (cen) and telomeric (tel) parts of the locus encompassing allelic and gene‐content motifs. (c) Further diversity has been generated through continuing cycles of unequal crossing‐over (non‐allelic homologous recombination), which result in re‐assortment and addition or subtraction of genes in a ‘cut & paste’‐like manner.20, 21 So called fusion genes composed of parts of different KIR genes have been generated by unequal crossover events when the recombination has occurred within genes.21, 22
KIR genetics
Functional consequences of KIR polymorphism
Four influential discoveries cultivated the fundamental principle that genetic variation of KIR has a direct impact on NK cell function, and stimulated ongoing research into the impact of this variation on human health; (i) the KIR genomic region has variable gene content,23 (ii) KIR allelic variation affects KIR allotype function,24, 25 (iii) there are multiple alleles for each KIR gene26 and (iv) this genetic variation correlates with ability to control disease or reproduce27, 28, 29, 30 (Fig. 3). Even a single nucleotide mutation can dramatically change receptor expression,25, 31, 32, 33 ligand specificity34, 35, 36 or signalling strength. KIR gene copy number variation influences NK cell education, shaping the NK cell repertoire.37 Lastly, combinatorial diversity of KIR and HLA class I alleles also impacts NK cell function, because any given KIR allotype has differential reactivity to the allotypes of its cognate HLA class I ligand.38, 39, 40, 41, 42, 43 In this scenario one may expect specific alleles or combinations to be beneficial for resisting specific infections, or stimulating immune‐mediated disease.44 This combinatorial diversity is amplified by hosting the KIR and HLA genes on separate chromosomes, and the result is many millions of different cognate KIR/HLA class I genotypes, tending to individuality.45, 46 Hence, through evolving multiple genetic mutations in the KIR locus, the human population probably generates and maintains considerable diversity in immunity to evolutionarily nimble and diverse pathogens.17 That there is little in common between human, chimpanzee and orang‐utan KIR loci,47, 48 and that distributions of KIR genes track with ethnicity and geography49, 50, 51 are testament to this hypothesis of rapid evolution.
Figure 3.
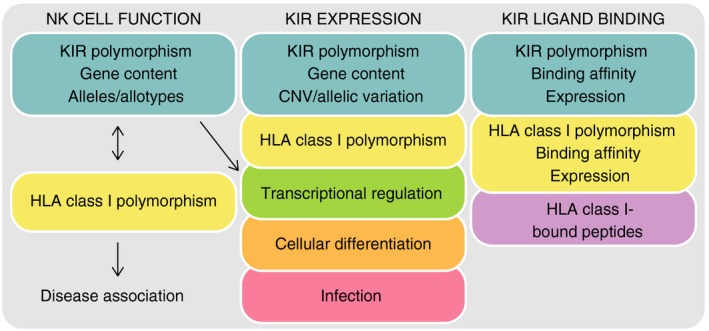
Major factors that influence natural killer (NK) cell function, expression and killer‐cell immunoglobulin‐like receptors (KIR) ligand binding.
KIR diversity and balancing selection
The interest in KIR genetic variation was piqued from early studies involving relatively small numbers and simple methods. The reason is that multiple diverse genotypes were detected in few individuals23, 26, 52 and the only plausible explanation was a highly heterogeneous genetic system with multiple common variants. These early studies therefore provided the first evidence that KIR may be subject to natural selection that maintains high diversity, just like it does for HLA.19, 53 The Yucpa population from Venezuela has low genomic diversity as a consequence of serial founder effects.54 There are only two common KIR haplotypes in the Yucpa (one A and one B; see Fig. 2 for nomenclature), but between them they carry all of the expressed KIR genes.55 This situation is extremely unlikely without the impact of a form of natural selection called balancing selection, which maintains genetic variation of specific loci in the population.55 Indeed, all human populations studied to date have a representation of KIR A and B haplotypes51, 56 and the prevailing hypothesis is that the A haplotypes are good for fighting infection57, 58 whereas B haplotypes are more beneficial for reproduction.17, 59
The impact of balancing selection is also seen clearly in the DNA sequences of KIR genes.19, 38 Namely, a greater number of common sequences, and greater divergence between them is present in the KIR locus than would be expected if there were no selection, and when compared with other parts of the genome. A good example is KIR3DL1/S1, which has over 100 alleles characterized (Fig. 4)60 and has three divergent allele lineages that have been maintained by balancing selection for millions of years.61 A synergy of population genetics, phylogenetic analysis and comparison of nucleotide substitution rates among codons showed that this diversity is focused towards the parts of the KIR molecule that bind the HLA class I and peptide.61 The prediction that these major lineages of KIR3DL1/S1 have distinct ligand HLA/peptide‐binding properties has been borne out with crystallographic and functional studies.35, 39, 41, 42, 62, 63, 64, 65 Expansion of the phylogenetic analyses to include other KIR molecules revealed natural selection has consistently been focused towards residues that affect interaction with the HLA class I ligand,66 as well as those that affect the signalling properties of the receptor.67
Figure 4.
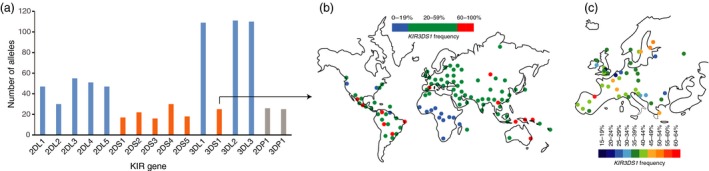
KIR allele counts and KIR3DS1 frequency worldwide plots. (a) Number of alleles reported in the December 2015 release of the IPD KIR database.60 The advent of killer‐cell immunoglobulin‐like receptor (KIR) analysis by next‐generation sequencing is rapidly increasing the number of recognized KIR alleles.68 Blue are genes encoding inhibitory KIR, orange activating KIR and grey are pseudogenes. There are 753 alleles in total. (b) As an example of how KIR gene frequencies can vary significantly across populations, KIR3DS1 gene frequency across the world and (c) Europe are shown (data from The Allele Frequency Net Database56).
KIR and HLA co‐evolution
Specific combinations of cognate KIR and HLA class I ligand correlate in frequency across the world, indicating that co‐evolution between them continues in modern humans.69 Again, high‐resolution analysis of the Yucpa population yields further insight into this phenomenon. The population has extremely high frequency of HLA‐C7 and corresponding high frequency of a KIR2DL3 allotype, unique to the Yucpa, having reduced C7 binding.55 By comparison, the KhoeSan population from Southern Africa has one of the highest aggregate frequencies of HLA‐C allotypes expressing the C2 motif. Here, a KIR allotype that switched binding specificity entirely from C2 to C1 has evolved.34 Both of these binding changes are due to substitutions at residues subject to balancing selection (Fig. 5).66 Hence, in both populations it appears that KIR have been able to respond rapidly and specifically to HLA class I frequency changes (of unknown aetiology). In contrast, in the Māori of New Zealand, a very low frequency of HLA‐B allotypes expressing the Bw4 motif (again of unknown aetiology) has been countered by an increase in frequency of HLA‐A allotypes that express KIR ligands.46 Such studies of human populations at high resolution reveal functionally important changes occurring on a fine scale, the sum of which throughout human evolution has resulted in the strong signature of balancing selection throughout the KIR locus.
Figure 5.
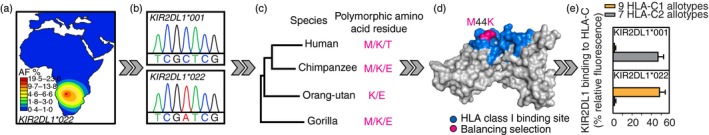
From discovery to function. Amino acid residues that have been subject to balancing selection can have dramatic effects on HLA class I recognition. Multiple novel KIR alleles may be discovered during population studies, and molecular analysis identifies the most functionally important. An example is KIR2DL1*022, which was discovered in the southern African KhoeSan population (a) and which differs from its parental allele KIR2DL1*001 by a single nucleotide substitution in codon 44 (b).34 Phylogenetic analyses that included the most closely related KIR from other hominoid species identified that residue 44 has been subject to balancing selection (c).66 Residue 44 occurs at the HLA binding site in the D1 protein domain of the killer‐cell immunoglobulin‐like receptors (KIR) molecule (d) (PDB: 1IM9).70 Substitution of methionine 44 (KIR2DL1*001) for lysine 44 (KIR2DL1*022) switches the specificity of the receptor from HLA‐C2 to HLA‐C1 (e). The methodological pipeline described above links population‐based analyses to functional mapping through sequence/phylogeny analysis and structural biology.
KIR allelic variation
There is little doubt that the most basic level of presence/absence of KIR impacts NK cell activity, and that the number of those genes present can influence NK cell development37 or control of disease.71 However, the prevailing theme of the genetic studies to date is that analysis of gene content identifies the fundamental tenets, which are then refined following higher‐resolution analyses of the alleles. This stems from the realization that KIR alleles have different magnitudes of effect, creating hierarchical series of phenotypes. From this, testable hypotheses can be formed about the extent to which each gene or allele, in conjunction with ligand variation, can predispose to a phenotype. For example, the cell surface expression of KIR3DL1 and the ability to recognize HLA‐Bw4 ligand vary according to allele, and hence KIR3DL1 alleles should be analysed accordingly in the disease context.25, 38, 58, 72 Similarly, polymorphism in the extracellular domain of KIR2DL1 influences the binding affinity to HLA‐C2.34, 40 Over 200 associations of KIR genes with disease resistance or susceptibility are published,56, 73 and the handful studied at high resolution are proving informative.58, 74, 75, 76 The field is therefore ripe for harvesting new information through in‐depth analyses.68, 77
KIR imputation
A statistical model that infers KIR genotypes from whole‐genome single nucleotide polymorphism data has been developed with about 97% accuracy for the majority of KIR genes, and can distinguish the broad A/B haplotypes.77 This method overcomes many of the obstacles caused by the complexity of gene content diversity to enable efficient KIR disease association analyses in large cohorts, and access to the wealth of previously generated single nucleotide polymorphism array data.78
KIR disease association studies
Combinations of KIR and HLA class I variants influence resistance to infections, susceptibility to autoimmune diseases and pregnancy syndromes, as well as outcome after haematopoietic stem cell transplantation (see The KIR and Diseases Database; Table 1).79 The influence of HLA class I and KIR gene variation on human immunodeficiency virus (HIV) disease outcome has been particularly well studied (Fig. 6).80 The variation at these genes has been described as a double‐edged sword because a particular genotype that confers protection from one disease (e.g. infection) could bestow increased risk to another disease type (e.g. autoimmunity or cancer).67, 69, 81 Population stratification is an important consideration in KIR association studies. It becomes problematic to analyse genes under strong selection and rapid evolution, such as the KIR, in admixed (mixed ancestry) populations where gene frequencies can vary significantly between subpopulations (Fig. 4). Approaches for KIR genetic analysis are improving in resolution and throughput (Fig. 7). Statistical developments offer additional strategies, such as principle component analysis of ancestry‐informative markers in case–control analysis to address admixture issues.82
Table 1.
KIR and HLA databases and resources
| Immuno Polymorphism Database | www.ebi.ac.uk/ipd/ |
| Allele Frequency Net Database | www.allelefrequencies.net/ |
| dbMHC and dbLRC | www.ncbi.nlm.nih.gov/gv/mhc/ |
| www.ncbi.nlm.nih.gov/gv/lrc/ | |
| LRC Haplotype Project | vega.sanger.ac.uk/info/data/LRC_Homo_sapiens.html |
The Immuno Polymorphism Database (IPD) provides a repository for killer‐cell immunoglobulin‐like receptors (KIR) sequences and includes, allele alignments, fully sequenced KIR haplotypes, donor KIR B‐content group calculator, allele ethnicity tool and primer/probe search tool.83 The Allele Frequency Net Database (AFND) provides a repository for KIR allele frequencies and listings of KIR disease associations.56 dbMHC and dbLRC databases provides sequences and frequency distributions for alleles of the MHC and leucocyte receptor complex (LRC) as well as an alignment viewer, primer/probe search tool and typing kit interface.84 LRC Haplotype Project provides DNA sequence data from several human LRC haplotypes including contig maps, annotation and a catalogue of all single nucleotide polymorphisms and insertions/deletions.85
Figure 6.
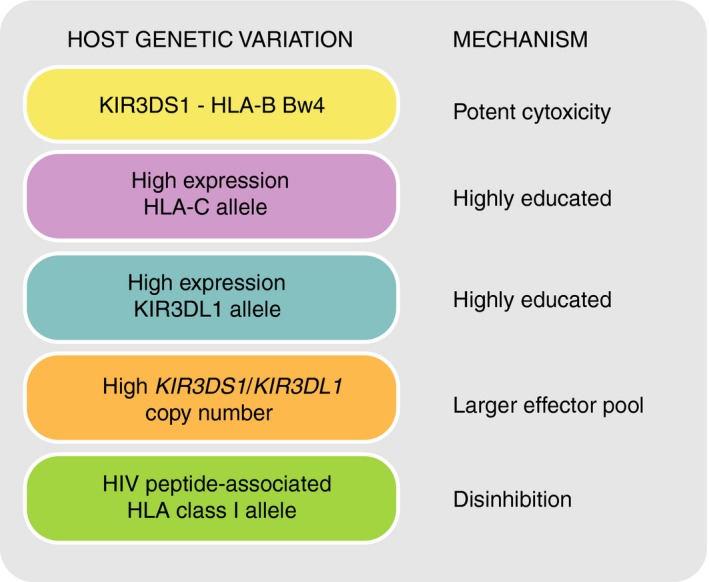
Host genetic variation and functions of HLA class I and killer‐cell immunoglobulin‐like receptors (KIR) in human immunodeficiency virus type 1 (HIV‐1) resistance. HIV‐1 down‐regulates HLA class I expression on the surface of infected CD4+ T cells so as to evade CD8+ T‐cell lysis.86, 87 However, this action exposes the infected cells to recognition and lysis by natural killer (NK) cells through KIRs. The effectiveness of an NK cell response, and consequently the outcome of infection, are linked to the host's KIR genes, their copy number and the expression levels of their ligands. More specifically, resistance to HIV‐1 correlates with; (i) the compound genotype, KIR3DS1 ‐ HLA‐B Bw4 (with isoleucine at position 80); KIR3DS1 binds HLA‐F open conformers, which can be expressed on HIV‐infected activated CD4+ T cells.88, 89 In this case NK cells expressing KIR3DS1 could also degranulate more potently in response to HIV‐infected Bw4+ CD4+ T cells and suppress viral replication.90 (ii) HLA‐C alleles that confer high cell surface expression91, 92; this may occur because the higher‐expressing HLA alleles result in highly educated NK cells, in addition to more efficient presentation of HIV epitopes to cytotoxic T cells, (iii) high expression KIR3DL1 alleles in Bw4+ individuals; again this could be due to highly educated KIR3DL1+ NK cells with greater activation potential when the ligand is down‐regulated by HIV,58 (iv) Increased copy number of KIR3DS1 alone or KIR3DL1 in the presence of HLA‐B Bw4; probably related to the clonal distribution of KIR, whereby the frequency of NK cells expressing a given KIR correlates linearly with gene copy number, (v) HLA allotypes that form complexes with HIV peptides that bind weakly to inhibitory KIR or strongly to activating KIR. Through any of these mechanisms, NK cells can exert selection pressure on HIV through KIR.63, 93 Conversely, HIV peptides that complex with HLA allotypes and bind inhibitory KIR with high affinity represent NK cell ‘escape’ variants via inhibition of NK cell function.94, 95
Figure 7.
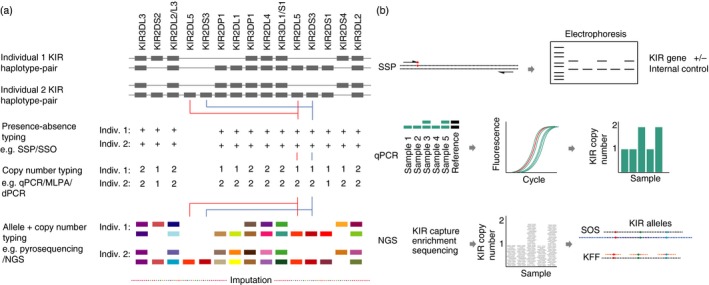
KIR genotyping methodology. (a) Representative haplotype‐pairs of two individuals are shown along with the expected results from different typing approaches; (i) presence/absence typing by PCR‐sequence‐specific primers (SSP) or sequence‐specific oligonucleotide (SSO) probes, (ii) copy number typing by quantitative PCR (qPCR),96 multiplex ligation‐dependent probe amplification (MLPA)97 and digital PCR (dPCR),98 (iii) allele and copy number typing by pyrosequencing and next‐generation sequencing (NGS), (iv) imputation infers KIR genotypes from single nucleotide polymorphism (SNP) data. (b) Schemes illustrating different typing approaches. SSP‐specificity relies on single nucleotide differences at the 3′ end of primers to distinguish different KIR genes. Real‐time qPCR combines SSP with fluorescently labelled probes to distinguish KIR genes and a reference gene (always two copies) amplifications in a multiplex reaction. KIR gene copy number is calculated by relative quantification. With NGS typing, oligonucleotide probes are used to capture the KIR genomic region. A bioinformatics pipeline converts sequence data into genotypes. Gene copy number is determined by relative read depth‐ratio of KIR genes compared with a reference gene (always two copies). Allele typing is achieved by filtering reads specific for genes based on alignment to all known reference alleles from KIR coding sequences (e.g. Son of Samtools (SOS)68). In parallel, sequence data can be probed with specific sequence search strings (‘in silico SSO’) to determine which alleles are present (e.g. KIR Filter Fish (KFF)68).
KIR ligand binding
KIR and HLA allotype interactions
Three key advancements have progressed understanding of the recognition of HLA class I by KIR; (i) the crystal structures of KIR in complex with their HLA class I ligands,10, 35, 70, 99 (ii) the development of soluble KIR36, 100, 101 and (iii) their use in a multiplex immunoassay against a broad panel of HLA class I allotypes102, 103 (Fig. 3). These tools have enabled fine‐scale analyses of KIR and HLA class I interaction diversity.
Two‐immunoglobulin domain inhibitory KIRs (KIR2DL1‐3) bind HLA‐C (Fig. 1). Originally considered separate genes, KIR2DL2 and KIR2DL3 are now known to segregate as alleles of the same gene, termed KIR2DL2/3. Initial studies indicated a simple bipartite system in which KIR2DL2/3 recognizes HLA‐C allotypes with asparagine at residue 80 (the HLA‐C1 motif), and KIR2DL1 recognizes HLA‐C allotypes with lysine at residue 80 (the HLA‐C2 motif). Dimorphism at residue 44 of the KIR molecule causes these specificity differences, where KIR2DL2/3 has lysine and KIR2DL1 has methionine.36, 100, 104, 105 Crystal structures showed that K44 of KIR2DL2 forms a hydrogen bond with the N80 of HLA‐C1.99 By contrast, M44 of KIR2DL1 has no direct contact with HLA‐C but forms part of a charge pocket that accommodates the K80 of HLA‐C2.70 Other residues are involved in the interaction however, and their polymorphism means that there is a range of binding characteristics determined by KIR allotype. The interactions of KIR2DL are further diversified by polymorphism within the subsets of C1‐bearing and C2‐bearing HLA allotypes.34, 103 The basis for these hierarchies may occur due to polymorphism at sites other than position 80, or the distinct repertoires of peptide presented by the different HLA‐C allotypes.106 An additional feature of HLA‐C polymorphism is the differential cell surface expression exhibited by individual allotypes,91 although how this variation impacts NK cell reactivity is yet to be fully defined.
KIR2DL2/3 allotypes form a continuum of binding strength and specificity that includes several inactivated or weakened variants.34, 40, 55, 107, 108, 109 Studies using soluble KIR showed that allotype KIR2DL3*001 is C1‐specific and binds relatively weakly, whereas KIR2DL2*001 binds more strongly and has cross‐reactivity with C2.103 In addition to those directly at the binding site, the substitutions that cause these functional differences may affect the angle or orientation of the binding domain,55, 103 or the efficiency of receptor clustering at the cell surface.99, 108 Allelic polymorphism also influences the functional properties of KIR2DL1, which has evolved to be a highly specific receptor for HLA‐C2.110 Yawata et al. first reported that NK cells educated via KIR2DL1*004 had lower interferon‐γ production than those educated via KIR2DL1*003.111 This occurs because KIR2DL1*004 has weak affinity for HLA‐C2 targets34 as well as reduced capacity for intracellular signal generation.112 A single dimorphism in the transmembrane region determines the signalling capacity; allotypes with R245 transduce a functional inhibitory signal and those with C245 have reduced inhibitory signalling.112 Super‐resolution microscopy has revealed that the nanoscale organization of KIR at cell surfaces depends on the transmembrane sequence, which in turn affects downstream signalling.113 Further modifying the functional range of KIR2DL1 are polymorphisms in the extracellular and cytoplasmic domains that regulate avidity and specificity, as well as the level of cell‐surface expression (Fig. 1).34, 40
Like the inhibitory KIR2D, the interactions between inhibitory KIR3D and their cognate ligands are diversified by their considerable polymorphism. KIR3DL1 recognizes HLA‐A and HLA‐B alleles that encode the Bw4 motif, a region that spans residues 77–83 on the α‐1 helix of the molecule (Fig. 1).35, 114, 115 Recognition of Bw4 by KIR3DL1 is sensitive to polymorphism both within and outside the Bw4 motif, as well as to the sequence of the bound peptide.42, 63, 116, 117, 118 Early work showed that HLA‐Bw4 allotypes with I80 formed more potent ligands for KIR3DL1 than those with T80,119 a functional difference reinforced by associations with disease outcome.27, 58, 120, 121, 122 However, recent high‐resolution studies have identified several I80 Bw4 allotypes that are poorly recognized by KIR3DL1, providing weaker KIR3DL1 ligands than selected T80 Bw4 allotypes.39, 41, 123 Compounding the difficulty of understanding the interactions between KIRD3DL1 and Bw4 is the extensive functional polymorphism of KIR3DL1, which changes its cell‐surface expression25, 38 and capacity to recognize the Bw4 epitope.41, 65, 123
Activating KIR ligand interactions
Although disease associations27, 76, 124, 125 and sequence homology67 with their inhibitory counterparts suggest that activating KIR recognize HLA class I, their cognate ligands have been harder to identify (Fig. 1). As the body of research dedicated to activating KIR has grown, it has become clear that three conserved features of their biology are likely to have hampered the process of ligand discovery; (1) their low affinity for HLA class I,11, 40, 105, 126, 127 (2) narrow specificity11, 126, 127, 128 and (3) high peptide selectivity.10, 11, 64, 127 These features are epitomized by KIR2DS2*001, which binds weakly to C1‐bearing HLA‐C*16:01 and HLA‐C*03:02126, 127 and recognizes HLA‐A*11:01 in a peptide‐dependent manner.10 Another possibility is that activating KIR recognize virus‐induced ligands129, 130 or altered self‐HLA class I molecules caused by viral infection. Recent work has identified open conformers (not bound to β 2‐microglobulin or peptide) of non‐classical HLA class I molecule, HLA‐F, as ligands of KIR3DS1.88, 89 Because HLA‐F can be expressed on activated or HIV‐1‐infected lymphocytes, KIR3DS1‐HLA‐F interaction could be important in the control of the T‐cell response131 and/or HIV‐1 infection.
Activating KIR are less polymorphic than inhibitory KIR (Fig. 4).132 However, the exploration of functional allotypic variation in activating KIR is in its infancy, and several studies point to its potential importance. Examples include the observation that KIR2DS1 allotypes recognize C2‐bearing HLA‐C with a range of avidities40 and that KIR3DS1*014, but not KIR3DS1*013, recognizes the HLA‐Bw4 epitope.65 Further, epidemiological studies suggest that specific variants of KIR2DS5, for which a ligand remains elusive, protect against the development of reproductive disorders.76, 133
Reporter systems could be useful tools to screen for activating KIR ligands and peptide influences of inhibitory KIR.129 The reporter cells are constructed to express the extracellular domains of activating KIR fused to the human CD3ζ cytoplasmic domain. Signalling through these hybrid receptors results in the expression of green fluorescent protein, which can be detected by flow cytometry. This method proved helpful to show that HLA‐F open conformers are ligands for KIR3DS1.88
KIR peptide‐dependence
The emerging field of peptidomics has relevance to the KIR field because binding of KIR to their respective HLA class I ligand is peptide‐dependent.134 There are different mechanisms by which viral infection can rapidly and radically affect the HLA class I peptide repertoire. Some viruses have evolved to evade NK cell immunity through the selection of mutations in MHC‐presented peptides that enhance binding to inhibitory NK cell receptors including the C‐type lectin‐like CD94:NKG2A heterodimer receptor and KIR2DL3.135, 136 Conversely, virus‐induced changes in peptide repertoire may promote beneficial action through KIR by disrupting HLA class I recognition by inhibitory KIR, releasing constraint on NK cells to mount a positive clearance response to infected cells.135 Structural analysis of peptide interaction by NK receptors and better understanding of the mechanisms by which viruses evade the NK cell response could assist the development of novel targeted interventions to exploit the antiviral activities of NK cells.
KIR gene expression
NK and T‐cell KIR repertoire
With the exception of KIR2DL4 and KIR3DL3,137, 138 KIR gene expression is clonally distributed and only a fraction of NK and T cells express a given KIR. KIR repertoire formation is complex. At least six factors are recognized to influence KIR repertoire; (i) transcriptional regulation, (ii) KIR gene content, (iii) allelic variation, (iv) cellular differentiation, (v) self‐HLA class I ligands and (vi) infection (Fig. 3).
KIR repertoire formation is governed by bi‐directional promoter activity and epigenetic silencing. High CpG methylation of the KIR proximal promoter was reported in NK cells and CD8+ T cells lacking expression of the corresponding KIR molecule, and vice versa.139, 140, 141 KIR expression requires the intermediate promoter Pro1;142 however, the strength of KIR proximal promoter antisense activity is probably responsible for clonal KIR distribution in NK and T cells.7, 142, 143, 144 The relative affinity of binding sites for transcription factors involved in sense versus antisense promoter activity determines the probability of generating the sense transcript required for gene activation.142, 143 A recent mouse study suggests that activating receptor‐mediated signalling might regulate this process during NK cell development.145 Polymorphisms in the KIR promotors also impact KIR expression.142, 146, 147
Beside epigenetic and transcriptional regulation, KIR repertoire formation is also dependent on KIR gene content and allelic variations. Each KIR gene is usually present in between zero and three copies in a given individual. As well as when the gene is absent, KIR expression is also abolished in individuals that are homozygous for a null allele, or who only carry the null allele of the gene – e.g. KIR3DL1*004 null allele is present at a mean frequency of 24·2% (SD 10·3) in worldwide populations.56 The frequency of cells positive for a given KIR is tightly linked to KIR gene copy number; a donor with two copies of KIR3DL1 will have a greater frequency of KIR3DL1+ NK cells than donors with only one copy,37, 38, 142 suggesting that each KIR gene copy is regulated independently. Additionally, NK cell KIR expression is related to cellular differentiation. KIR expression is weak or absent in immature NKG2A+ CD56bright NK cells and increases gradually with maturation, reaching its maximum level in NKG2A− CD56dim NK cells.148, 149, 150 Similar observation was made for T cells, in which KIR expression is virtually absent in CD4 and CD8 naive T cells and reaches its maximal level in differentiated effector memory T cells.2, 5, 151, 152 Accordingly, KIR expression is low in less mature cord blood NK and T cells compared with healthy adult control cells.153, 154
Unlike T‐ and B‐cell repertoire formation, there is no evidence for negative selection or deletion of NK cells expressing a combination of receptors that could be harmful or useless.108, 155 Instead, in a process termed education, only NK cells expressing self‐specific HLA class I inhibitory receptors (NKG2A or KIR) become fully functionally competent. The degree to which NK cell education shapes the KIR repertoire has been a matter of debate. Using a mathematical and phenotypic approach, Andersson et al. demonstrated that an adult's NK cell KIR repertoire formation was largely stochastic, in line with probabilistic expression of KIR under bidirectional proximal promoter activity and no selection.156 However, other studies showed a slight but significant bias of the global KIR repertoire in adults toward the expression of KIRs able to recognize self‐HLA class I.111, 157 Paradoxically, the same authors demonstrated that KIR repertoire in newborns was not biased toward self‐HLA class I recognition.158 This observation suggested that KIR repertoire acquisition was indeed stochastic but the slight bias observed in adults might be driven by infections encountered later in life.
In 2004, Guma et al. showed that expression of NKG2C, an activating receptor for HLA‐E, was increased in individuals infected with the human cytomegalovirus (HCMV) and that NKG2C+ cells expressed high level of KIRs.159 It was later demonstrated that these HCMV‐associated NKG2C+ NK cells express self‐specific KIRs and account for the vast majority of adult KIR repertoire deviation toward self HLA class I.160 HCMV‐adapted NK cells lacking NKG2C expression were also reported, most of them expressing activating KIRs.160, 161 Using a large NKG2C‐deficient cohort, it was recently demonstrated that adaptive NK cell responses could occur in the absence of both activating KIRs and NKG2C; however, these adaptive NK cells also largely display repertoire deviation toward self‐specific inhibitory KIR expression.162, 163 Also, self‐specific inhibitory KIR expression is not required for generating adaptive NK cells but is probably necessary for optimal functions. Indeed, HCMV+ TAP‐deficient individuals, with a considerable decrease of HLA class I expression at the cell surface, can develop NKG2C+ adaptive NK cells but these cells remain hypofunctional.164
Studies reporting adaptive NK cell expansion in various viral infections including HIV, hepatitis B virus, hepatitis C virus, chikungunya virus and hantavirus show that adaptive NK cells only occur in HCMV+ individuals, perhaps as the result of opportunistic viral reactivation.165, 166, 167, 168 To date, apart from HCMV, no other viruses have been seen to correlate with the appearance of adaptive NK cells, and associated KIR repertoire deviations, including herpes simplex virus (both HSV‐1 and HSV‐2) and varicella‐zoster virus.160, 169 Instead, it was shown that acute Epstein–Barr virus infection induces NKG2A+ CD57+ expansion lacking KIR expression.170 Altogether, these findings suggest that KIR, and adaptive NK cells, might have mainly evolved to control HCMV. Supporting this hypothesis, HCMV developed several strategies to escape NK cell control.171, 172, 173 Viral proteins induce HLA‐E expression while decreasing classical HLA class I (HLA‐A, B and C) expression at the cell surface of infected cells, allowing the virus to selectively escape NKG2A+ NK cells and interfere with CD8+ T‐cell recognition, respectively.174, 175, 176, 177 NKG2C and inhibitory KIR come into play to counteract this escape strategy of HCMV. NKG2C allows adaptive NK cells to recognize HLA‐E+ HLA class I− infected cells while self‐specific KIR prevent them killing HLA‐E+ HLA class I+ non‐infected cells (Fig. 8). This model provides an explanation for the skewed repertoire in adaptive NK cells seen in HCMV+ individuals towards inhibitory KIR that recognize self HLA class I. Activating KIRs could play a similar role to NKG2C and recognize unknown HCMV‐induced ligands, similarly to Ly49H/m157 in mouse.129, 130
Figure 8.
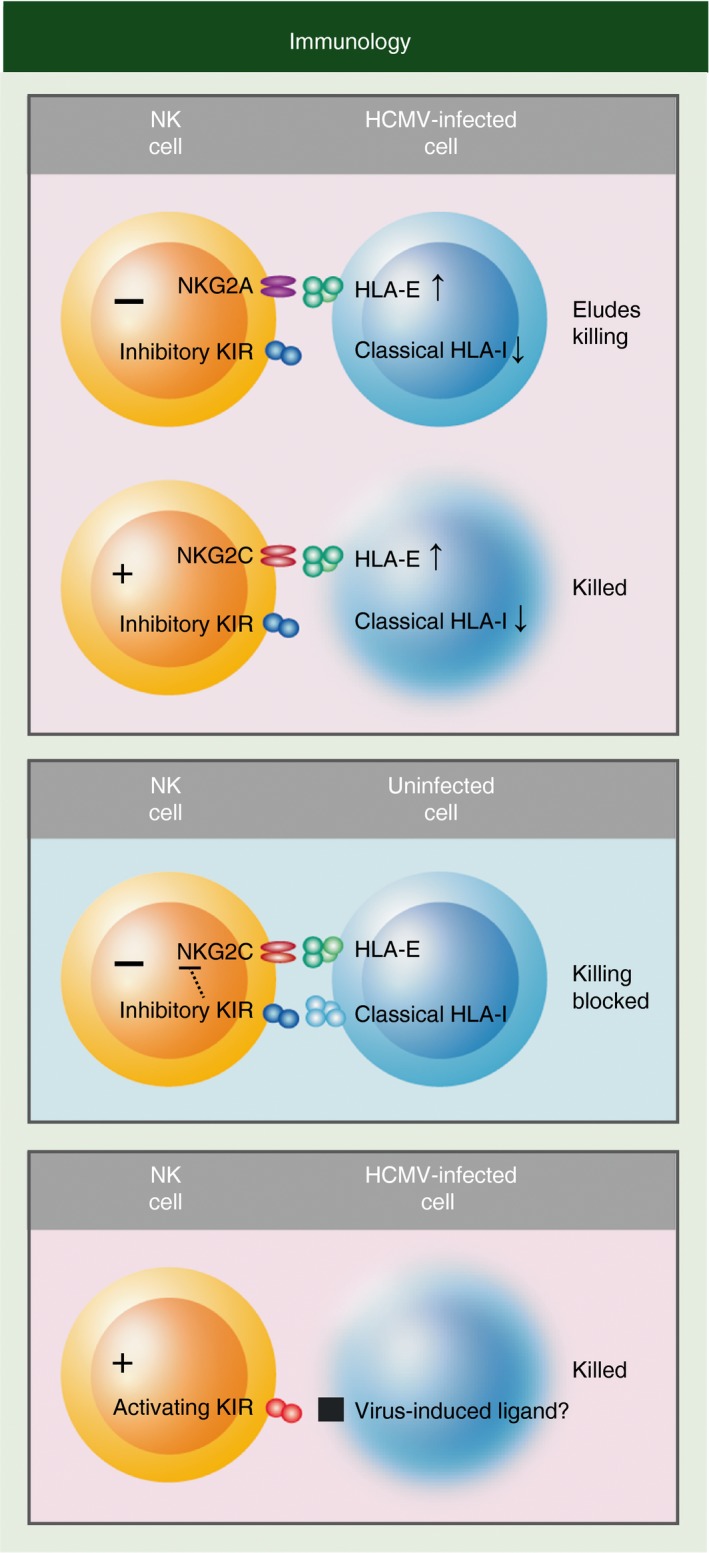
Natural killer (NK) cell strategies in human choriomeningitis virus (HCMV) infection. Viral proteins up‐regulate HLA‐E expression to selectively inhibit NK cells through the NKG2A inhibitory receptor. Concomitantly, the virus down‐regulates classical HLA class I expression to evade CD8+ T cells. NKG2C allows NK cells to detect HLA‐E+ HLA class I‐infected cells. Hence, ‘missing‐self’ (no HLA class I) triggers activation of NK cells already educated/licenced by self‐HLA class I molecules.165, 178 Non‐infected cells are protected from NK cell cytotoxicity by recognition of HLA class I by inhibitory killer‐cell immunoglobulin‐like receptors (KIR), which curbs activation through NKG2C. Activating KIR may directly recognize ‘altered‐self’ ligands that are induced by the virus.15
The role of KIR expression in T cells remains unclear and most of the studies have focused on CD8 T cells, which contain the largest KIR+ subset within the T‐cell compartment. Although NK cell KIR repertoire is considered stable in time, the frequency of KIR+ T cells increases with age, due to accumulation of terminally differentiated T cells.141 Interestingly, HCMV‐specific CD8 T cells almost completely lack KIR expression, and the specificity of KIR‐expressing cells remains largely unknown.2, 5, 7, 151, 179 In contrast, it was shown that KIR+ CD4+ T cells display specificity against HCMV but not Epstein–Barr virus or HSV‐1.4 It is clear that inhibitory and activating KIR can, respectively, dampen and co‐stimulate T‐cell receptor‐mediated activation in CD4 and CD8 T cells.5, 151, 179, 180, 181, 182 Unlike NK cells, self‐specific inhibitory KIR expression does not educate T cells, as they do not display enhanced functional responses upon T‐cell receptor triggering.7 Instead, it was shown that ex vivo KIR+ effector memory CD8+ T cells were hyporesponsive to T‐cell receptor triggering compared with KIR− effector memory CD8+ T cells.7 It was proposed that KIR expression protects from activation‐induced cell death in a ligand‐independent manner, perhaps explaining their accumulation with aging.2, 183
KIR repertoire analysis
KIR repertoire analysis has proved important in the study of NK cell function. Future studies will need to systematically analyse in‐depth KIR repertoire of organ resident NK cells, which are known to display unique KIR repertoires, at least in the uterus and the liver.184, 185, 186 When studying the KIR repertoire all the parameters that are known to influence KIR expression should be considered (Fig. 3), most of all KIR genotypes and cellular differentiation markers. Cytometry panels require many colours to include as many KIR as possible, together with differentiation markers including at least NKG2A, NKG2C and preferably other markers specific to adaptive NK cells (e.g. FCεR1γ, CD57 and NKp30). Strategies for KIR repertoire analysis using high‐dimensional FACS analysis were recently described.187 Several limitations remain in such analysis, such as mis‐binding of antibodies relating to amino acid substitutions in particular KIR allotypes. For example, anti‐KIR2DL3 ECM41 antibody does not recognize KIR2DL3*015 or KIR2DL3*005.187 These allotypes are recognized by antibodies with specificity for receptors encoded by other KIR genes (Table 2). New next‐generation sequencing techniques will allow fast and cost‐effective KIR typing at allelic resolution and will facilitate KIR repertoire interpretation.68 In addition, high‐dimensional mass cytometry (mass cytometry by time‐of‐flight) is a powerful tool for investigating NK cell repertoire diversity through the analysis of many cellular markers simultaneously188 (Fig. 9). The approach is being used to characterize the phenotypes of lymphocytes and their abundance in biopsies of patients, to gain insight into, for example, how receptor genotypes influence disease susceptibility in a cell‐type‐specific manner.188, 189, 190, 191 Once identified, discrete NK cell subpopulations associated with disease could be harnessed for immunotherapeutic strategies or may predict response to treatment. Combined with other single‐cell techniques, e.g. RNA‐Seq, this could be an important step forward to personalized and more cost‐effective treatment.
Table 2.
Specificities and reported cross‐reaction of killer‐cell immunoglobulin‐like receptors (KIR) antibodies
| Clone | Specificity | Reported cross‐reaction and (comments) | Reference |
|---|---|---|---|
| 143211 | KIR2DL1 | KIR2DS5 | 192 |
| HP‐3E4 | KIR2DL1 | KIR2DS1 and KIR2DS4 | 193 |
| EB6 | KIR2DL1, KIR2DS1 | KIR2DL3*005 | 187, 194, 195 |
| 11PB6 | KIR2DL1, KIR2DS1 | KIR2DL3*005 | 194 |
| HP‐MA4 | KIR2DL1, KIR2DS1 | KIR2DS3 and KIR2DS5 | 192, 193 |
| GL183 | KIR2DL2, KIR2DL3, KIR2DS2 | 196, 197, 198 | |
| DX27 | KIR2DL2, KIR2DL3, KIR2DS2 | 52 | |
| CH‐L | KIR2DL2, KIR2DL3, KIR2DS2 | 199 | |
| 180701 | KIR2DL3 | (Does not recognize KIR2DL3*005 and *015) | 187 |
| ECM41 | KIR2DL3 | (Does not recognize KIR2DL3*005 and *015) | 194 |
| UP‐R1 | KIR2DL5 | 200 | |
| 1F12 | KIR2DS2, KIR2DL3 | 201 | |
| 179315 | KIR2DS4 | 140 | |
| JJC11·6 | KIR2DS4 | KIR2DS3 | 192 |
| FES172 | KIR2DS4 | 202 | |
| DX9 | KIR3DL1 | 203 | |
| 5·133 | KIR3DL1, KIR3DL2 | All KIRs except KIR2DS1 and KIR2DS3 | 192 |
| Z27 | KIR3DL1, KIR3DS1 | 204, 205, 206 | |
| DX30 | KIR3DL1/KIR3DL2 | 207 | |
| DX31 | KIR3DL2 | 207 | |
| NKVFS1 | PanKIR2D | 208 |
Sequence homogeneity between KIR genes at the genomic level translates to high similarity between encoded protein products. As such, analysis of KIR expression is not trivial in primary cells because of the cross‐reaction of available antibodies, in particular between activating and inhibitory isoforms. This can lead to misinterpretation of cell‐staining results. These cross‐reactions must be taken into account when analysing KIR expression. Staining experiments can be supported with control stains, references, representative stains on relevant donors and precise descriptions of strategies for differential staining using sets of antibodies.
Figure 9.
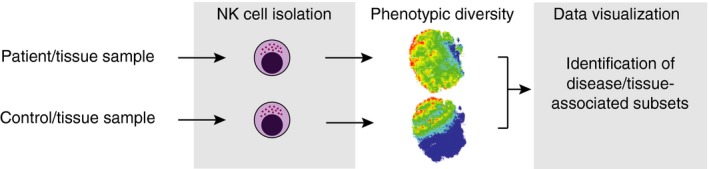
Natural killer (NK) cell analysis by mass cytometry by time‐of‐flight. By interrogation of multiple markers simultaneously, cell phenotypes can be compared between, for example, patients versus controls or between different tissue sites. The approach allows alterations in lymphocyte composition or abundance to be detected that are linked to killer‐cell immunoglobulin‐like receptors (KIR) genotypes and disease.
Although the new methods are increasing scale and resolution of all aspects of KIR study, one obstacle that future repertoire studies will need to overcome is the current lack of antibodies that specifically recognize KIR2DS2, KIR3DS1, KIR2DS3 and KIR2DS5 (Table 2). Although antibody combinations can help to decipher these activating KIRs,187, 192, 201 they do not allow analysis at single KIR level because of antibody cross‐reactivity. Aptamers, short single‐stranded nucleic acid oligomers that bind to a specific target molecule, with their unique features of high binding affinity and specificity, could offer a useful alternative to antibodies in discriminating subtly different forms of KIR.209 Improving KIR repertoire analysis will contribute to our understanding of clinical situations where KIR have a proven or suspected role such as antiviral immune response, transplantation, autoimmunity and reproduction.
KIR model systems
Mouse NK cells do not express KIR but instead Ly49 receptors (C‐type lectin‐like type II transmembrane disulphide‐bonded homodimers), which perform analogous functions.210 For example, like KIR genes, Ly49 genes encode both activating and inhibitory NK receptors that regulate NK cell biology through binding MHC class I molecules. Despite differences between mice and humans with regard to the immune and reproductive systems, the mouse has been used to study how imbalance of NK cell inhibition or activation potential influences pregnancy.211 Humanized mice of homogeneous genetic background might in the future enable study of individual KIR and HLA class I variants in isolation.212 Using a mouse transgenic for an HLA‐B allele that encodes the Bw4 epitope, a recent study investigated the roles of cell‐intrinsic and cell‐extrinsic HLA class I molecules for educating human NK cells.213
Because mice and humans are so divergent, primates are used as in vivo models to study NK cell biology. Some non‐human simian species show a comparable level of diversity and complexity in KIR haplotypes to humans.214 However, there are extensive differences in size and organization between the KIR loci of higher primate species.215 It is evident in primates that different lineages of KIR genes have been expanded concomitantly with species‐specific evolution of MHC class I genes.216 In rhesus macaque, an important animal model of human diseases such as AIDS, binding of certain KIR is influenced by the same MHC epitopes (Bw4 and Bw6) that are important determinants of human KIR interactions.217 In the last decade, progress has been made in characterizing KIR genes in primates and developing specific genotyping assays.218, 219 Specific interactions between KIRs of primates and MHC class I ligands are being identified and monoclonal antibodies against primate KIR proteins are being generated.220, 221 These advances are enabling phenotypic characterization of KIR expression on NK cells and T‐cell subsets in primates and investigation of KIR‐MHC biology in primate models of infectious disease.222, 223, 224
Therapeutic intervention
As KIRs are expressed on effector cells such as adaptive NK cells and effector memory T cells, which have undergone clonal expansion, it is not surprising that KIRs are often found expressed on malignant cells. For instance, in NK and T‐cell large granular lymphocytosis lymphocytes often express activating or inhibitory KIRs in a clonal manner.225, 226, 227, 228 Patients with Sezary syndrome typically display CD4+ T cells expressing KIR3DL2, which might contribute to disease onset.228, 229 For this reason, KIR are considered as a potential therapeutic target because they are only expressed on a small subset of normal lymphocytes, the deletion of which is unlikely to be harmful.230
Genetic information on KIR and HLA is already being used clinically to choose donors for haematopoietic stem cell transplantation for optimal outcome.231 Indeed, it is estimated that a significant reduction of relapse after transplantation for acute myeloid leukaemia can be achieved by choosing donors based on their KIR and HLA class I genotype.232, 233 This could be extended in the future to help, for example, sperm donors with the lowest risk of adverse pregnancy outcomes in assisted reproduction. In‐depth understanding of the molecular pathways that control NK cells will be critical to the therapeutic manipulation and in adoptive transfer strategies of these powerful lymphocytes,234 such as in cancer immunotherapy.16, 29, 235, 236 Immune‐modulation therapies that alter NK and T‐cell function are already in development. For example Lirilumab, the monoclonal antibody that binds KIR, is currently in a phase II clinical trial for lymphoma.237 By blocking the interaction of inhibitory KIR with their HLA class I ligands this antibody facilitates activation of NK cells by impeding inhibitory signalling, potentially promoting destruction of tumour cells.
Conclusion
Analysis of complex genomics represents a significant new frontier for immunology. Challenges lie in determining allelic copies precisely and developing molecular and computational strategies to analyse them at the scale required to definitively relate them to phenotypes. For the KIR gene cluster this process is underway and yielding valuable insights. KIR, therefore, provide a useful model for developing analysis methods for other families of proteins shaped by multi‐allelic copy number variation, immune‐related or otherwise. An important advance has been the development of targeted techniques and bioinformatics tools to precisely type and analyse HLA and KIR copy number and alleles with great accuracy and in high sample numbers. Understanding how KIR variation influences the initiation and progression of disease will be achieved through the application of these novel methods in multidisciplinary projects involving geneticists, statisticians, structural biologists, immunologists and clinicians. The overarching question is how the signals from lymphocyte receptor interactions determined at the genetic level translate to differing functionality and outcomes in settings of infections, pregnancy, autoimmunity and cancer. Understanding the cellular and molecular mechanisms underlying the KIR‐HLA system will contribute to the development of interventions and therapies to ameliorate adverse consequences when these mechanisms go awry.
Summary
The advent of next‐generation sequencing is set to allow the determination of KIR and HLA sequences at super‐resolution. Combining such studies with high‐resolution functional mapping of these polymorphic genes will provide unprecedented insight, not only into the molecular mechanisms that govern the interactions between receptor and ligand, but also into the pathophysiology of infectious and non‐infectious disorders in which KIR and HLA play critical roles.
Disclosures
The authors declare that there is no conflict of interest regarding the publication of this paper.
Acknowledgements
V.B. is supported by the French National Research Agency (ANR) (grant no. NKIR‐ANR‐13‐PDOC‐0025‐01). P.J.N. and H.H. are supported by U.S. National Institutes of Health grant R01 AI17892. J.A.T. is supported by the European Research Council (ERC) and Medical Research Council (MRC).
References
- 1. Battistini L, Borsellino G, Sawicki G, Poccia F, Salvetti M, Ristori G et al Phenotypic and cytokine analysis of human peripheral blood γδ T cells expressing NK cell receptors. J Immunol 1997; 159:3723–30. [PubMed] [Google Scholar]
- 2. Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential expression of leukocyte receptor complex‐encoded Ig‐like receptors correlates with the transition from effector to memory CTL. J Immunol 2001; 166:3933–41. [DOI] [PubMed] [Google Scholar]
- 3. Patterson S, Chaidos A, Neville DC, Poggi A, Butters TD, Roberts IA et al Human invariant NKT cells display alloreactivity instructed by invariant TCR‐CD1d interaction and killer Ig receptors. J Immunol 2008; 181:3268–76. [DOI] [PubMed] [Google Scholar]
- 4. van Bergen J, Kooy‐Winkelaar EM, van Dongen H, van Gaalen FA, Thompson A, Huizinga TW et al Functional killer Ig‐like receptors on human memory CD4+ T cells specific for cytomegalovirus. J Immunol 2009; 182:4175–82. [DOI] [PubMed] [Google Scholar]
- 5. Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D et al Coordinated expression of Ig‐like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol 2004; 173:7223–9. [DOI] [PubMed] [Google Scholar]
- 6. Lanier LL. NK cell recognition. Annu Rev Immunol 2005; 23:225–74. [DOI] [PubMed] [Google Scholar]
- 7. Bjorkstrom NK, Beziat V, Cichocki F, Liu LL, Levine J, Larsson S et al CD8 T cells express randomly selected KIRs with distinct specificities compared with NK cells. Blood 2012; 120:3455–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Remtoula N, Bensussan A, Marie‐Cardine A. Cutting edge: selective expression of inhibitory or activating killer cell Ig‐like receptors in circulating CD4+ T lymphocytes. J Immunol 2008; 180:2767–71. [DOI] [PubMed] [Google Scholar]
- 9. Barclay AN, Hatherley D. The counterbalance theory for evolution and function of paired receptors. Immunity 2008; 29:675–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu J, Xiao Z, Ko HL, Shen M, Ren EC. Activating killer cell immunoglobulin‐like receptor 2DS2 binds to HLA‐A*11. Proc Natl Acad Sci USA 2014; 111:2662–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Graef T, Moesta AK, Norman PJ, Abi‐Rached L, Vago L, Older Aguilar AM et al KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA‐A*11 while diminishing avidity for HLA‐C. J Exp Med 2009; 206:2557–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karre K. Natural killer cell recognition of missing self. Nat Immunol 2008; 9:477–80. [DOI] [PubMed] [Google Scholar]
- 13. Yokoyama WM, Kim S. Licensing of natural killer cells by self‐major histocompatibility complex class I. Immunol Rev 2006; 214:143–54. [DOI] [PubMed] [Google Scholar]
- 14. Orr MT, Lanier LL. Natural killer cell education and tolerance. Cell 2010; 142:847–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trowsdale J, Jones DC, Barrow AD, Traherne JA. Surveillance of cell and tissue perturbation by receptors in the LRC. Immunol Rev 2015; 267:117–36. [DOI] [PubMed] [Google Scholar]
- 16. Benson DM Jr, Caligiuri MA. Killer immunoglobulin‐like receptors and tumor immunity. Cancer Immunol Res 2014; 2:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parham P, Moffett A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat Rev Immunol 2013; 13:133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jiang W, Johnson C, Jayaraman J, Simecek N, Noble J, Moffatt MF et al Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res 2012; 22:1845–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Norman PJ, Cook MA, Carey BS, Carrington CV, Verity DH, Hameed K et al SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics 2004; 56:225–37. [DOI] [PubMed] [Google Scholar]
- 20. Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol 2003; 171:2192–5. [DOI] [PubMed] [Google Scholar]
- 21. Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D et al Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet 2010; 19:737–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Norman PJ, Abi‐Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D et al Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res 2009; 19:757–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert‐Weidenbach K, Corliss B et al Human diversity in killer cell inhibitory receptor genes. Immunity 1997; 7:753–63. [DOI] [PubMed] [Google Scholar]
- 24. Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med 1996; 183:1817–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol 2003; 171:6640–9. [DOI] [PubMed] [Google Scholar]
- 26. Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D et al Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol 2002; 168:2307–15. [DOI] [PubMed] [Google Scholar]
- 27. Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ et al Epistatic interaction between KIR3DS1 and HLA‐B delays the progression to AIDS. Nat Genet 2002; 31:429–34. [DOI] [PubMed] [Google Scholar]
- 28. Martin MP, Nelson G, Lee JH, Pellett F, Gao X, Wade J et al Cutting edge: susceptibility to psoriatic arthritis: influence of activating killer Ig‐like receptor genes in the absence of specific HLA‐C alleles. J Immunol 2002; 169:2818–22. [DOI] [PubMed] [Google Scholar]
- 29. Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A et al Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002; 295:2097–100. [DOI] [PubMed] [Google Scholar]
- 30. Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J et al Combinations of maternal KIR and fetal HLA‐C genes influence the risk of preeclampsia and reproductive success. J Exp Med 2004; 200:957–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig‐like receptor 2DL2/2DL3 D1 domain. J Immunol 2006; 177:5347–57. [DOI] [PubMed] [Google Scholar]
- 32. Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG et al Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol 2007; 37:199–211. [DOI] [PubMed] [Google Scholar]
- 33. Mulrooney TJ, Hou L, Steiner NK, Chen M, Belle I, Ng J et al Promoter variants of KIR2DL5 add to diversity and may impact gene expression. Immunogenetics 2008; 60:287–94. [DOI] [PubMed] [Google Scholar]
- 34. Hilton HG, Norman PJ, Nemat‐Gorgani N, Goyos A, Hollenbach JA, Henn BM et al Loss and gain of natural killer cell receptor function in an African Hunter–Gatherer population. PLoS Genet 2015; 11:e1005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vivian JP, Duncan RC, Berry R, O'Connor GM, Reid HH, Beddoe T et al Killer cell immunoglobulin‐like receptor 3DL1‐mediated recognition of human leukocyte antigen B. Nature 2011; 479:401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Winter CC, Long EO. A single amino acid in the p58 killer cell inhibitory receptor controls the ability of natural killer cells to discriminate between the two groups of HLA‐C allotypes. J Immunol 1997; 158:4026–8. [PubMed] [Google Scholar]
- 37. Beziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S et al Influence of KIR gene copy number on natural killer cell education. Blood 2013; 121:4703–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 2006; 203:633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boudreau JE, Mulrooney TJ, Le Luduec JB, Barker E, Hsu KC. KIR3DL1 and HLA‐B density and binding calibrate NK education and response to HIV. J Immunol 2016; 196:3398–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hilton HG, Guethlein LA, Goyos A, Nemat‐Gorgani N, Bushnell DA, Norman PJ et al Polymorphic HLA‐C receptors balance the functional characteristics of KIR haplotypes. J Immunol 2015; 195:3160–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O'Connor GM et al Killer cell immunoglobulin‐like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med 2016; 213:791–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M. Cutting edge: allele‐specific and peptide‐dependent interactions between KIR3DL1 and HLA‐A and HLA‐B. J Immunol 2007; 178:33–7. [DOI] [PubMed] [Google Scholar]
- 43. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2‐like receptor with increased binding to its HLA‐C ligand. J Immunol 2013; 190:6198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin‐like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet 2006; 7:277–300. [DOI] [PubMed] [Google Scholar]
- 45. Norman PJ, Hollenbach JA, Nemat‐Gorgani N, Guethlein LA, Hilton HG, Pando MJ et al Co‐evolution of human leukocyte antigen (HLA) class I ligands with killer‐cell immunoglobulin‐like receptors (KIR) in a genetically diverse population of sub‐Saharan Africans. PLoS Genet 2013; 9:e1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nemat‐Gorgani N, Edinur HA, Hollenbach JA, Traherne JA, Dunn PP, Chambers GK et al KIR diversity in Māori and Polynesians: populations in which HLA‐B is not a significant KIR ligand. Immunogenetics 2014; 66:597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Guethlein LA, Older Aguilar AM, Abi‐Rached L, Parham P. Evolution of killer cell Ig‐like receptor (KIR) genes: definition of an orang‐utan KIR haplotype reveals expansion of lineage III KIR associated with the emergence of MHC‐C. J Immunol 2007; 179:491–504. [DOI] [PubMed] [Google Scholar]
- 48. Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG et al Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity 2000; 12:687–98. [DOI] [PubMed] [Google Scholar]
- 49. Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin‐like receptor sequences in three ethnic groups. Immunogenetics 2001; 52:195–205. [DOI] [PubMed] [Google Scholar]
- 50. Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S et al Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens 2001; 57:358–62. [DOI] [PubMed] [Google Scholar]
- 51. Hollenbach JA, Nocedal I, Ladner MB, Single RM, Trachtenberg EA. Killer cell immunoglobulin‐like receptor (KIR) gene content variation in the HGDP‐CEPH populations Immunogenetics 2012; 64:719–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valiante NM, Uhrberg M, Shilling HG, Lienert‐Weidenbach K, Arnett KL, D'Andrea A et al Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 1997; 7:739–51. [DOI] [PubMed] [Google Scholar]
- 53. Hedrick PW, Thomson G. Evidence for balancing selection at HLA. Genetics 1983; 104:449–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Greenberg JH, Turner CG, Zegura SL, Campbell L, Fox JA, Laughlin WS et al The settlement of the Americas: a comparison of the linguistic, dental, and genetic evidence [and comments and reply]. Curr Anthropol 1986; 27:477–97. [Google Scholar]
- 55. Gendzekhadze K, Norman PJ, Abi‐Rached L, Graef T, Moesta AK, Layrisse Z et al Co‐evolution of KIR2DL3 with HLA‐C in a human population retaining minimal essential diversity of KIR and HLA class I ligands. Proc Natl Acad Sci USA 2009; 106:18692–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gonzalez‐Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, Silva AL et al Allele frequency net 2015 update: new features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res 2015; 43:D784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J et al HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 2004; 305:872–4. [DOI] [PubMed] [Google Scholar]
- 58. Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F et al Innate partnership of HLA‐B and KIR3DL1 subtypes against HIV‐1. Nat Genet 2007; 39:733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hiby SE, Apps R, Chazara O, Farrell LE, Magnus P, Trogstad L et al Maternal KIR in combination with paternal HLA‐C2 regulate human birth weight. J Immunol 2014; 192:5069–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: allele variant databases. Nucleic Acids Res 2015; 43:D423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Norman PJ, Abi‐Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D et al Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet 2007; 39:1092–9. [DOI] [PubMed] [Google Scholar]
- 62. O'Connor GM, Vivian JP, Widjaja JM, Bridgeman JS, Gostick E, Lafont BA et al Mutational and structural analysis of KIR3DL1 reveals a lineage‐defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol 2014; 192:2875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fadda L, O'Connor GM, Kumar S, Piechocka‐Trocha A, Gardiner CM, Carrington M et al Common HIV‐1 peptide variants mediate differential binding of KIR3DL1 to HLA‐Bw4 molecules. J Virol 2011; 85:5970–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. O'Connor GM, Vivian JP, Gostick E, Pymm P, Lafont BA, Price DA et al Peptide‐dependent recognition of HLA‐B*57:01 by KIR3DS1. J Virol 2015; 89:5213–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O'Connor GM, Yamada E, Rampersaud A, Thomas R, Carrington M, McVicar DW. Analysis of binding of KIR3DS1*014 to HLA suggests distinct evolutionary history of KIR3DS1. J Immunol 2011; 187:2162–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abi‐Rached L, Moesta AK, Rajalingam R, Guethlein LA, Parham P. Human‐specific evolution and adaptation led to major qualitative differences in the variable receptors of human and chimpanzee natural killer cells. PLoS Genet 2010; 6:e1001192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Abi‐Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin‐like receptor and Ly49 from inhibitory homologues. J Exp Med 2005; 201:1319–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Norman PJ, Hollenbach JA, Nemat‐Gorgani N, Marin WM, Norberg SJ, Ashouri E et al Defining KIR and HLA class i genotypes at highest resolution via high‐throughput sequencing. Am J Hum Genet 2016; 99:375–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Single RM, Martin MP, Gao X, Meyer D, Yeager M, Kidd JR et al Global diversity and evidence for coevolution of KIR and HLA. Nat Genet 2007; 39:1114–9. [DOI] [PubMed] [Google Scholar]
- 70. Fan QR, Long EO, Wiley DC. Crystal structure of the human natural killer cell inhibitory receptor KIR2DL1‐HLA‐Cw4 complex. Nat Immunol 2001; 2:452–60. [DOI] [PubMed] [Google Scholar]
- 71. Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ et al Copy number variation of KIR genes influences HIV‐1 control. PLoS Biol 2011; 9:e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. O'Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol 2007; 178:235–41. [DOI] [PubMed] [Google Scholar]
- 73. Song S, Miranda CJ, Braun L, Meyer K, Frakes AE, Ferraiuolo L et al Major histocompatibility complex class I molecules protect motor neurons from astrocyte‐induced toxicity in amyotrophic lateral sclerosis. Nat Med 2016; 22:397–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ahn RS, Moslehi H, Martin MP, Abad‐Santos M, Bowcock AM, Carrington M et al Inhibitory KIR3DL1 alleles are associated with psoriasis. Br J Dermatol 2015; 174:449–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Augusto DG, O'Connor GM, Lobo‐Alves SC, Bass S, Martin MP, Carrington M et al Pemphigus is associated with KIR3DL2 expression levels and provides evidence that KIR3DL2 may bind HLA‐A3 and A11 in vivo . Eur J Immunol 2015; 45:2052–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J et al A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre‐eclampsia. Proc Natl Acad Sci USA 2015; 112:845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Vukcevic D, Traherne JA, Naess S, Ellinghaus E, Kamatani Y, Dilthey A et al Imputation of KIR types from SNP variation data. Am J Hum Genet 2015; 97:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune‐mediated diseases. Nat Rev Genet 2013; 14:661–73. [DOI] [PubMed] [Google Scholar]
- 79. Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol 2008; 20:343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Martin MP, Carrington M. Immunogenetics of HIV disease. Immunol Rev 2013; 254:245–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chen H, Hayashi G, Lai OY, Dilthey A, Kuebler PJ, Wong TV et al Psoriasis patients are enriched for genetic variants that protect against HIV‐1 disease. PLoS Genet 2012; 8:e1002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Garrido‐Rodriguez D, Avila‐Rios S, Garcia‐Morales C, Valenzuela‐Ponce H, Ormsby C, Reyes‐Gopar H et al Killer cell immunoglobulin‐like receptor and human leukocyte antigen gene profiles in a cohort of HIV‐infected Mexican Mestizos. Immunogenetics 2016; 68:703–17. [DOI] [PubMed] [Google Scholar]
- 83. Robinson J, Halliwell JA, Marsh SG. IMGT/HLA and the immuno polymorphism database. Methods Mol Biol 2014; 1184:109–21. [DOI] [PubMed] [Google Scholar]
- 84. Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K et al Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 2012; 40:D13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Horton R, Coggill P, Miretti MM, Sambrook JG, Traherne JA, Ward R et al The LRC haplotype project: a resource for killer immunoglobulin‐like receptor‐linked association studies. Tissue Antigens 2006; 68:450–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Apps R, Del Prete GQ, Chatterjee P, Lara A, Brumme ZL, Brockman MA et al HIV‐1 Vpu Mediates HLA‐C Downregulation. Cell Host Microbe 2016; 19:686–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, Strominger JL et al The selective downregulation of class I major histocompatibility complex proteins by HIV‐1 protects HIV‐infected cells from NK cells. Immunity 1999; 10:661–71. [DOI] [PubMed] [Google Scholar]
- 88. Garcia‐Beltran WF, Holzemer A, Martrus G, Chung AW, Pacheco Y, Simoneau CR et al Open conformers of HLA‐F are high‐affinity ligands of the activating NK‐cell receptor KIR3DS1. Nat Immunol 2016; 17:1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Burian A, Wang KL, Finton KA, Lee N, Ishitani A, Strong RK et al HLA‐F and MHC‐I Open Conformers Bind Natural Killer Cell Ig‐Like Receptor KIR3DS1. PLoS ONE 2016; 11:e0163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A et al Differential natural killer cell‐mediated inhibition of HIV‐1 replication based on distinct KIR/HLA subtypes. J Exp Med 2007; 204:3027–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Apps R, Qi Y, Carlson JM, Chen H, Gao X, Thomas R et al Influence of HLA‐C expression level on HIV control. Science 2013; 340:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Thomas R, Apps R, Qi Y, Gao X, Male V, O'HUigin C et al HLA‐C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA‐C. Nat Genet 2009; 41:1290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Brackenridge S, Evans EJ, Toebes M, Goonetilleke N, Liu MK, di Gleria K et al An early HIV mutation within an HLA‐B*57‐restricted T cell epitope abrogates binding to the killer inhibitory receptor 3DL1. J Virol 2011; 85:5415–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Thananchai H, Makadzange T, Maenaka K, Kuroki K, Peng Y, Conlon C et al Reciprocal recognition of an HLA‐Cw4‐restricted HIV‐1 gp120 epitope by CD8+ T cells and NK cells. AIDS 2009; 23:189–93. [DOI] [PubMed] [Google Scholar]
- 95. Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM et al HIV‐1 adaptation to NK‐cell‐mediated immune pressure. Nature 2011; 476:96–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Jiang W, Johnson C, Simecek N, López‐Álvarez MR, Di D, Trowsdale J. qKAT: a high‐throughput qPCR method for KIR gene copy number and haplotype determination. Genome Med 2016; 8:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Vendelbosch S, de Boer M, Gouw RA, Ho CK, Geissler J, Swelsen WT et al Extensive variation in gene copy number at the killer immunoglobulin‐like receptor locus in humans. PLoS ONE 2013; 8:e67619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Roberts CH, Jiang W, Jayaraman J, Trowsdale J, Holland MJ, Traherne JA. Killer‐cell Immunoglobulin‐like Receptor gene linkage and copy number variation analysis by droplet digital PCR. Genome Med 2014; 6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Boyington JC, Motyka SA, Schuck P, Brooks AG, Sun PD. Crystal structure of an NK cell immunoglobulin‐like receptor in complex with its class I MHC ligand. Nature 2000; 405:537–43. [DOI] [PubMed] [Google Scholar]
- 100. Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA‐C allotype recognition. J Immunol 1998; 161:571–7. [PubMed] [Google Scholar]
- 101. Winter CC, Long EO. Binding of soluble KIR‐Fc fusion proteins to HLA class I. Methods Mol Biol 2000; 121:239–50. [DOI] [PubMed] [Google Scholar]
- 102. Hilton HG, Moesta AK, Guethlein LA, Blokhuis J, Parham P, Norman PJ. The production of KIR‐Fc fusion proteins and their use in a multiplex HLA class I binding assay. J Immunol Methods 2015; 425:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand‐binding site makes KIR2DL2 a stronger receptor for HLA‐C than KIR2DL3. J Immunol 2008; 180:3969–79. [DOI] [PubMed] [Google Scholar]
- 104. Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA‐C is the inhibitory ligand that determines dominant resistance to lysis by NK1‐ and NK2‐specific natural killer cells. Proc Natl Acad Sci USA 1993; 90:12000–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R et al P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti‐p58 antibodies reconstitute lysis of MHC class I‐protected cells in NK clones displaying different specificities. J Exp Med 1993; 178:597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Fadda L, Borhis G, Ahmed P, Cheent K, Pageon SV, Cazaly A et al Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci USA 2010; 107:10160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2‐like receptor with increased binding to its HLA‐C ligand. J Immunol 2013; 190:6198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Bari R, Thapa R, Bao J, Li Y, Zheng J, Leung W et al KIR2DL2/2DL3‐E(35) alleles are functionally stronger than ‐Q(35) alleles. Sci Rep 2016; 6:23689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Moesta AK, Abi‐Rached L, Norman PJ, Parham P. Chimpanzees use more varied receptors and ligands than humans for inhibitory killer cell Ig‐like receptor recognition of the MHC‐C1 and MHC‐C2 epitopes. J Immunol 2009; 182:3628–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hilton HG, Vago L, Older Aguilar AM, Moesta AK, Graef T, Abi‐Rached L et al Mutation at positively selected positions in the binding site for HLA‐C shows that KIR2DL1 is a more refined but less adaptable NK cell receptor than KIR2DL3. J Immunol 2012; 189:1418–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I‐specific inhibitory receptors and their ligands structure diverse human NK‐cell repertoires toward a balance of missing self‐response. Blood 2008; 112:2369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das Gupta N et al Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood 2009; 114:5182–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Oszmiana A, Williamson DJ, Cordoba SP, Morgan DJ, Kennedy PR, Stacey K et al The size of activating and inhibitory killer Ig‐like receptor nanoclusters is controlled by the transmembrane sequence and affects signaling. Cell Rep 2016; 15:1957–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL et al Conserved and variable residues within the Bw4 motif of HLA‐B make separable contributions to recognition by the NKB1 killer cell‐inhibitory receptor. J Immunol 1997; 158:5237–41. [PubMed] [Google Scholar]
- 115. Gumperz JE, Litwin V, Phillips JH, Lanier LL, Parham P. The Bw4 public epitope of HLA‐B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J Exp Med 1995; 181:1133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA‐B with KIR3DL1. J Immunol 2008; 181:6293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Malnati MS, Peruzzi M, Parker KC, Biddison WE, Ciccone E, Moretta A et al Peptide specificity in the recognition of MHC class I by natural killer cell clones. Science 1995; 267:1016–8. [DOI] [PubMed] [Google Scholar]
- 118. Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA‐B allotypes discriminates among peptides bound to HLA‐B*2705. J Exp Med 1996; 184:1585–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M. NK3‐specific natural killer cells are selectively inhibited by Bw4‐positive HLA alleles with isoleucine 80. J Exp Med 1994; 180:1235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Kuranov AB, Kotter I, Henes JC, Abisheva ST, Steiert I, Riewerts F et al Behçet's disease in HLA‐B*51 negative Germans and Turks shows association with HLA‐Bw4‐80I. Arthritis Res Ther 2014; 16:R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Marra J, Greene J, Hwang J, Du J, Damon L, Martin T et al KIR and HLA genotypes predictive of low‐affinity interactions are associated with lower relapse in autologous hematopoietic cell transplantation for acute myeloid leukemia. J Immunol 2015; 194:4222–30. [DOI] [PubMed] [Google Scholar]
- 122. Foley B, Felices M, Cichocki F, Cooley S, Verneris MR, Miller JS. The biology of NK cells and their receptors affects clinical outcomes after hematopoietic cell transplantation (HCT). Immunol Rev 2014; 258:45–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Mulrooney TJ, Zhang AC, Goldgur Y, Boudreau JE, Hsu KC. KIR3DS1‐specific D0 domain polymorphisms disrupt KIR3DL1 surface expression and HLA binding. J Immunol 2015; 195:1242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hiby SE, Apps R, Sharkey AM, Farrell LE, Gardner L, Mulder A et al Maternal activating KIRs protect against human reproductive failure mediated by fetal HLA‐C2. J Clin Invest 2010; 120:4102–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ivarsson MA, Michaelsson J, Fauriat C. Activating killer cell Ig‐like receptors in health and disease. Front Immunol 2014; 5:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Moesta AK, Graef T, Abi‐Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol 2010; 185:4233–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Stewart CA, Laugier‐Anfossi F, Vely F, Saulquin X, Riedmuller J, Tisserant A et al Recognition of peptide–MHC class I complexes by activating killer immunoglobulin‐like receptors. Proc Natl Acad Sci USA 2005; 102:13224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA‐Cw4 but not HLA‐Cw6 by the NK cell receptor killer cell Ig‐like receptor two‐domain short tail number 4. J Immunol 2001; 166:7260–7. [DOI] [PubMed] [Google Scholar]
- 129. Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science 2002; 296:1323–6. [DOI] [PubMed] [Google Scholar]
- 130. Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV et al Recognition of a virus‐encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci USA 2002; 99:8826–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature 2012; 481:394–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Robinson J, Halliwell JA, McWilliam H, Lopez R, Marsh SG. IPD – the immuno polymorphism database. Nucleic Acids Res 2013; 41:D1234–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Nowak I, Ploski R, Barcz E, Dziunycz P, Kaminski P, Kostrzewa G et al KIR2DS5 in the presence of HLA‐C C2 protects against endometriosis. Immunogenetics 2015; 67:203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Cassidy SA, Cheent KS, Khakoo SI. Effects of peptide on NK cell‐mediated MHC I recognition. Front Immunol 2014; 5:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Das J, Khakoo SI. NK cells: tuned by peptide? Immunol Rev 2015; 267:214–27. [DOI] [PubMed] [Google Scholar]
- 136. Holzemer A, Thobakgale CF, Jimenez Cruz CA, Garcia‐Beltran WF, Carlson JM, van Teijlingen NH et al Selection of an HLA‐C*03:04‐Restricted HIV‐1 p24 gag sequence variant is associated with viral escape from KIR2DL3+ natural killer cells: data from an observational cohort in South Africa. PLoS Med 2015; 12:e1001900; discussion e1001900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rajagopalan S, Long EO. A human histocompatibility leukocyte antigen (HLA)‐G‐specific receptor expressed on all natural killer cells. J Exp Med 1999; 189:1093–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Trompeter HI, Gomez‐Lozano N, Santourlidis S, Eisermann B, Wernet P, Vilches C et al Three structurally and functionally divergent kinds of promoters regulate expression of clonally distributed killer cell Ig‐like receptors (KIR), of KIR2DL4, and of KIR3DL3. J Immunol 2005; 174:4135–43. [DOI] [PubMed] [Google Scholar]
- 139. Santourlidis S, Trompeter HI, Weinhold S, Eisermann B, Meyer KL, Wernet P et al Crucial role of DNA methylation in determination of clonally distributed killer cell Ig‐like receptor expression patterns in NK cells. J Immunol 2002; 169:4253–61. [DOI] [PubMed] [Google Scholar]
- 140. Chan HW, Kurago ZB, Stewart CA, Wilson MJ, Martin MP, Mace BE et al DNA methylation maintains allele‐specific KIR gene expression in human natural killer cells. J Exp Med 2003; 197:245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Li G, Yu M, Weyand CM, Goronzy JJ. Epigenetic regulation of killer immunoglobulin‐like receptor expression in T cells. Blood 2009; 114:3422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi‐directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet 2008; 4:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Davies GE, Locke SM, Wright PW, Li H, Hanson RJ, Miller JS et al Identification of bidirectional promoters in the human KIR genes. Genes Immun 2007; 8:245–53. [DOI] [PubMed] [Google Scholar]
- 144. Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28‐base PIWI‐like RNA in human NK cells. J Immunol 2010; 185:2009–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Freund J, May RM, Yang E, Li H, McCullen M, Zhang B et al Activating receptor signals drive receptor diversity in developing natural killer cells. PLoS Biol 2016; 14:e1002526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wright PW, Li H, Huehn A, O'Connor GM, Cooley S, Miller JS et al Characterization of a weakly expressed KIR2DL1 variant reveals a novel upstream promoter that controls KIR expression. Genes Immun 2014; 15:440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J Immunol 2000; 165:6416–21. [DOI] [PubMed] [Google Scholar]
- 148. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA et al Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK‐cell differentiation uncoupled from NK‐cell education. Blood 2010; 116:3853–64. [DOI] [PubMed] [Google Scholar]
- 149. Yu J, Mao HC, Wei M, Hughes T, Zhang J, Park IK et al CD94 surface density identifies a functional intermediary between the CD56bright and CD56dim human NK‐cell subsets. Blood 2010; 115:274–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Beziat V, Descours B, Parizot C, Debre P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE 2010; 5:e11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. van der Veken LT, Diez Campelo M, van der Hoorn MA, Hagedoorn RS, van Egmond HM, van Bergen J et al Functional analysis of killer Ig‐like receptor‐expressing cytomegalovirus‐specific CD8+ T cells. J Immunol 2009; 182:92–101. [DOI] [PubMed] [Google Scholar]
- 152. van Bergen J, Thompson A, van der Slik A, Ottenhoff TH, Gussekloo J, Koning F. Phenotypic and functional characterization of CD4 T cells expressing killer Ig‐like receptors. J Immunol 2004; 173:6719–26. [DOI] [PubMed] [Google Scholar]
- 153. Warren HS, Rana PM, Rieger DT, Hewitt KA, Dahlstrom JE, Kent AL. CD8 T cells expressing killer Ig‐like receptors and NKG2A are present in cord blood and express a more naive phenotype than their counterparts in adult blood. J Leukoc Biol 2006; 79:1252–9. [DOI] [PubMed] [Google Scholar]
- 154. Le Garff‐Tavernier M, Beziat V, Decocq J, Siguret V, Gandjbakhch F, Pautas E et al Human NK cells display major phenotypic and functional changes over the life span. Aging Cell 2010; 9:527–35. [DOI] [PubMed] [Google Scholar]
- 155. Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA et al Human NK cell education by inhibitory receptors for MHC class I. Immunity 2006; 25:331–42. [DOI] [PubMed] [Google Scholar]
- 156. Andersson S, Fauriat C, Malmberg JA, Ljunggren HG, Malmberg KJ. KIR acquisition probabilities are independent of self‐HLA class I ligands and increase with cellular KIR expression. Blood 2009; 114:95–104. [DOI] [PubMed] [Google Scholar]
- 157. Schonberg K, Sribar M, Enczmann J, Fischer JC, Uhrberg M. Analyses of HLA‐C‐specific KIR repertoires in donors with group A and B haplotypes suggest a ligand‐instructed model of NK cell receptor acquisition. Blood 2011; 117:98–107. [DOI] [PubMed] [Google Scholar]
- 158. Schonberg K, Fischer JC, Kogler G, Uhrberg M. Neonatal NK‐cell repertoires are functionally, but not structurally, biased toward recognition of self HLA class I. Blood 2011; 117:5152–6. [DOI] [PubMed] [Google Scholar]
- 159. Guma M, Angulo A, Vilches C, Gomez‐Lozano N, Malats N, Lopez‐Botet M. Imprint of human cytomegalovirus infection on the NK cell receptor repertoire. Blood 2004; 104:3664–71. [DOI] [PubMed] [Google Scholar]
- 160. Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT et al NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 2013; 121:2678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F et al Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig‐like receptor in patients transplanted with NKG2C–/– umbilical cord blood. J Immunol 2014; 192:1471–9. [DOI] [PubMed] [Google Scholar]
- 162. Liu LL, Landskron J, Ask EH, Enqvist M, Sohlberg E, Traherne JA et al Critical role of CD2 co‐stimulation in adaptive natural killer cell responses revealed in NKG2C‐deficient humans. Cell Rep 2016; 15:1088–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Muntasell A, Pupuleku A, Cisneros E, Vera A, Moraru M, Vilches C et al Relationship of NKG2C copy number with the distribution of distinct cytomegalovirus‐induced adaptive NK cell subsets. J Immunol 2016; 196:3818–27. [DOI] [PubMed] [Google Scholar]
- 164. Beziat V, Sleiman M, Goodridge JP, Kaarbo M, Liu LL, Rollag H et al Polyclonal expansion of NKG2C+ NK cells in TAP‐deficient patients. Front Immunol 2015; 6:507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Beziat V, Dalgard O, Asselah T, Halfon P, Bedossa P, Boudifa A et al CMV drives clonal expansion of NKG2C+ NK cells expressing self‐specific KIRs in chronic hepatitis patients. Eur J Immunol 2012; 42:447–57. [DOI] [PubMed] [Google Scholar]
- 166. Petitdemange C, Becquart P, Wauquier N, Beziat V, Debre P, Leroy EM et al Unconventional repertoire profile is imprinted during acute chikungunya infection for natural killer cells polarization toward cytotoxicity. PLoS Pathog 2011; 7:e1002268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lindgren T, Ahlm C, Mohamed N, Evander M, Ljunggren HG, Bjorkstrom NK. Longitudinal analysis of the human T cell response during acute hantavirus infection. J Virol 2011; 85:10252–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Brunetta E, Fogli M, Varchetta S, Bozzo L, Hudspeth KL, Marcenaro E et al Chronic HIV‐1 viremia reverses NKG2A/NKG2C ratio on natural killer cells in patients with human cytomegalovirus co‐infection. AIDS 2010; 24:27–34. [DOI] [PubMed] [Google Scholar]
- 169. Bjorkstrom NK, Svensson A, Malmberg KJ, Eriksson K, Ljunggren HG. Characterization of natural killer cell phenotype and function during recurrent human HSV‐2 infection. PLoS ONE 2011; 6:e27664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Azzi T, Lunemann A, Murer A, Ueda S, Beziat V, Malmberg KJ et al Role for early‐differentiated natural killer cells in infectious mononucleosis. Blood 2014; 124:2533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R et al Modulation of natural killer cells by human cytomegalovirus. J Clin Virol 2008; 41:206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Fielding CA, Aicheler R, Stanton RJ, Wang EC, Han S, Seirafian S et al Two novel human cytomegalovirus NK cell evasion functions target MICA for lysosomal degradation. PLoS Pathog 2014; 10:e1004058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Lodoen MB, Lanier LL. Viral modulation of NK cell immunity. Nat Rev Microbiol 2005; 3:59–69. [DOI] [PubMed] [Google Scholar]
- 174. Ulbrecht M, Martinozzi S, Grzeschik M, Hengel H, Ellwart JW, Pla M et al Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA‐E and prevents NK cell‐mediated lysis. J Immunol 2000; 164:5019–22. [DOI] [PubMed] [Google Scholar]
- 175. Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S et al Surface expression of HLA‐E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287:1031. [DOI] [PubMed] [Google Scholar]
- 176. Khan N, Bruton R, Taylor GS, Cobbold M, Jones TR, Rickinson AB et al Identification of cytomegalovirus‐specific cytotoxic T lymphocytes in vitro is greatly enhanced by the use of recombinant virus lacking the US2 to US11 region or modified vaccinia virus Ankara expressing individual viral genes. J Virol 2005; 79:2869–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Miller‐Kittrell M, Sparer TE. Feeling manipulated: cytomegalovirus immune manipulation. Virol J 2009; 6:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Hoglund P, Glas R, Ohlen C, Ljunggren HG, Karre K. Alteration of the natural killer repertoire in H‐2 transgenic mice: specificity of rapid lymphoma cell clearance determined by the H‐2 phenotype of the target. J Exp Med 1991; 174:327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Bonorino P, Leroy V, Dufeu‐Duchesne T, Tongiani‐Dashan S, Sturm N, Pernollet M et al Features and distribution of CD8 T cells with human leukocyte antigen class I‐specific receptor expression in chronic hepatitis C. Hepatology 2007; 46:1375–86. [DOI] [PubMed] [Google Scholar]
- 180. Snyder MR, Nakajima T, Leibson PJ, Weyand CM, Goronzy JJ.. Stimulatory killer Ig‐like receptors modulate T cell activation through DAP12‐dependent and DAP12‐independent mechanisms. J Immunol 2004; 173:3725–31. [DOI] [PubMed] [Google Scholar]
- 181. Guerra N, Michel F, Gati A, Gaudin C, Mishal Z, Escudier B et al Engagement of the inhibitory receptor CD158a interrupts TCR signaling, preventing dynamic membrane reorganization in CTL/tumor cell interaction. Blood 2002; 100:2874–81. [DOI] [PubMed] [Google Scholar]
- 182. Henel G, Singh K, Cui D, Pryshchep S, Lee WW, Weyand CM et al Uncoupling of T‐cell effector functions by inhibitory killer immunoglobulin‐like receptors. Blood 2006; 107:4449–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Ugolini S, Arpin C, Anfossi N, Walzer T, Cambiaggi A, Forster R et al Involvement of inhibitory NKRs in the survival of a subset of memory‐phenotype CD8+ T cells. Nat Immunol 2001; 2:430–5. [DOI] [PubMed] [Google Scholar]
- 184. Marquardt N, Beziat V, Nystrom S, Hengst J, Ivarsson MA, Kekalainen E et al Cutting edge: identification and characterization of human intrahepatic CD49a+ NK cells. J Immunol 2015; 194:2467–71. [DOI] [PubMed] [Google Scholar]
- 185. Sharkey AM, Gardner L, Hiby S, Farrell L, Apps R, Masters L et al Killer Ig‐like receptor expression in uterine NK cells is biased toward recognition of HLA‐C and alters with gestational age. J Immunol 2008; 181:39–46. [DOI] [PubMed] [Google Scholar]
- 186. Ivarsson MA, Stiglund N, Marquardt N, Westgren M, Gidlof S, Bjorkstrom NK. Composition and dynamics of the uterine NK cell KIR repertoire in menstrual blood. Mucosal Immunol, 2016; doi: 10.1038/mi.2016.50. [DOI] [PubMed] [Google Scholar]
- 187. Beziat V, Traherne J, Malmberg JA, Ivarsson MA, Bjorkstrom NK, Retiere C et al Tracing dynamic expansion of human NK‐cell subsets by high‐resolution analysis of KIR repertoires and cellular differentiation. Eur J Immunol 2014; 44:2192–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC et al Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Bjorkstrom NK, Ljunggren HG, Michaelsson J. Emerging insights into natural killer cells in human peripheral tissues. Nat Rev Immunol 2016; 16:310–20. [DOI] [PubMed] [Google Scholar]
- 190. van Unen V, Li N, Molendijk I, Temurhan M, Hollt T, van der Meulen‐de Jong AE et al Mass cytometry of the human mucosal immune system identifies tissue‐ and disease‐associated immune subsets. Immunity 2016; 44:1227–39. [DOI] [PubMed] [Google Scholar]
- 191. Strauss‐Albee DM, Horowitz A, Parham P, Blish CA. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol 2014; 193:4871–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Czaja K, Borer AS, Schmied L, Terszowski G, Stern M, Gonzalez A. A comprehensive analysis of the binding of anti‐KIR antibodies to activating KIRs. Genes Immun 2014; 15:33–7. [DOI] [PubMed] [Google Scholar]
- 193. De Miguel M, López‐Botet M. Characterization of monoclonal antibodies specific for receptors of the KIR family. Inmunología 2002; 21:187–93. [Google Scholar]
- 194. Falco M, Romeo E, Marcenaro S, Martini S, Vitale M, Bottino C et al Combined genotypic and phenotypic killer cell Ig‐like receptor analyses reveal KIR2DL3 alleles displaying unexpected monoclonal antibody reactivity: identification of the amino acid residues critical for staining. J Immunol 2010; 185:433–41. [DOI] [PubMed] [Google Scholar]
- 195. Moretta A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R et al Existence of both inhibitory (p58) and activatory (p50) receptors for HLA‐C molecules in human natural killer cells. J Exp Med 1995; 182:875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Moretta A, Ciccone E, Tambussi G, Bottino C, Viale O, Pende D et al Surface molecules involved in CD3‐negative NK cell function. A novel molecule which regulates the activation of a subset of human NK cells. Int J Cancer Suppl 1989; 4:48–52. [DOI] [PubMed] [Google Scholar]
- 197. Moretta A, Tambussi G, Bottino C, Tripodi G, Merli A, Ciccone E et al A novel surface antigen expressed by a subset of human CD3– CD16+ natural killer cells. Role in cell activation and regulation of cytolytic function. J Exp Med 1990; 171:695–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Biassoni R, Cantoni C, Falco M, Verdiani S, Bottino C, Vitale M et al The human leukocyte antigen (HLA)‐C‐specific “activatory” or “inhibitory” natural killer cell receptors display highly homologous extracellular domains but differ in their transmembrane and intracytoplasmic portions. J Exp Med 1996; 183:645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Ferrini S, Cambiaggi A, Meazza R, Sforzini S, Marciano S, Mingari MC et al T cell clones expressing the natural killer cell‐related p58 receptor molecule display heterogeneity in phenotypic properties and p58 function. Eur J Immunol 1994; 24:2294–8. [DOI] [PubMed] [Google Scholar]
- 200. Estefania E, Flores R, Gomez‐Lozano N, Aguilar H, Lopez‐Botet M, Vilches C. Human KIR2DL5 is an inhibitory receptor expressed on the surface of NK and T lymphocyte subsets. J Immunol 2007; 178:4402–10. [DOI] [PubMed] [Google Scholar]
- 201. David G, Morvan M, Gagne K, Kerdudou N, Willem C, Devys A et al Discrimination between the main activating and inhibitory killer cell immunoglobulin‐like receptor positive natural killer cell subsets using newly characterized monoclonal antibodies. Immunology 2009; 128:172–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C et al p46, a novel natural killer cell‐specific surface molecule that mediates cell activation. J Exp Med 1997; 186:1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA‐B molecules. J Exp Med 1994; 180:537–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R et al Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol 2001; 166:2992–3001. [DOI] [PubMed] [Google Scholar]
- 205. Carr WH, Rosen DB, Arase H, Nixon DF, Michaelsson J, Lanier LL. Cutting edge: KIR3DS1, a gene implicated in resistance to progression to AIDS, encodes a DAP12‐associated receptor expressed on NK cells that triggers NK cell activation. J Immunol 2007; 178:647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Trundley A, Frebel H, Jones D, Chang C, Trowsdale J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur J Immunol 2007; 37:780–7. [DOI] [PubMed] [Google Scholar]
- 207. Leong CC, Chapman TL, Bjorkman PJ, Formankova D, Mocarski ES, Phillips JH et al Modulation of natural killer cell cytotoxicity in human cytomegalovirus infection: the role of endogenous class I major histocompatibility complex and a viral class I homolog. J Exp Med 1998; 187:1681–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208. Spaggiari GM, Contini P, Carosio R, Arvigo M, Ghio M, Oddone D et al Soluble HLA class I molecules induce natural killer cell apoptosis through the engagement of CD8: evidence for a negative regulation exerted by members of the inhibitory receptor superfamily. Blood 2002; 99:1706–14. [DOI] [PubMed] [Google Scholar]
- 209. Zhang H, Zhou L, Zhu Z, Yang C. Recent progress in aptamer‐based functional probes for bioanalysis and biomedicine. Chemistry 2016; 22:9886–900. [DOI] [PubMed] [Google Scholar]
- 210. Rahim MM, Makrigiannis AP. Ly49 receptors: evolution, genetic diversity, and impact on immunity. Immunol Rev 2015; 267:137–47. [DOI] [PubMed] [Google Scholar]
- 211. Moffett A, Colucci F. Co‐evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol Rev 2015; 267:283–97. [DOI] [PubMed] [Google Scholar]
- 212. Belkin D, Torkar M, Chang C, Barten R, Tolaini M, Haude A et al Killer cell Ig‐like receptor and leukocyte Ig‐like receptor transgenic mice exhibit tissue‐ and cell‐specific transgene expression. J Immunol 2003; 171:3056–63. [DOI] [PubMed] [Google Scholar]
- 213. Boudreau JE, Liu XR, Zhao Z, Zhang A, Shultz LD, Greiner DL et al Cell‐extrinsic MHC Class I molecule engagement augments human NK cell education programmed by cell‐intrinsic MHC Class I. Immunity 2016; 45:280–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Kruse PH, Rosner C, Walter L. Characterization of rhesus macaque KIR genotypes and haplotypes. Immunogenetics 2010; 62:281–93. [DOI] [PubMed] [Google Scholar]
- 215. Averdam A, Petersen B, Rosner C, Neff J, Roos C, Eberle M et al A novel system of polymorphic and diverse NK cell receptors in primates. PLoS Genet 2009; 5:e1000688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Guethlein LA, Norman PJ, Hilton HH, Parham P. Co‐evolution of MHC class I and variable NK cell receptors in placental mammals. Immunol Rev 2015; 267:259–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Rosner C, Kruse PH, Hermes M, Otto N, Walter L. Rhesus macaque inhibitory and activating KIR3D interact with Mamu‐A‐encoded ligands. J Immunol 2011; 186:2156–63. [DOI] [PubMed] [Google Scholar]
- 218. Moreland AJ, Guethlein LA, Reeves RK, Broman KW, Johnson RP, Parham P et al Characterization of killer immunoglobulin‐like receptor genetics and comprehensive genotyping by pyrosequencing in rhesus macaques. BMC Genom 2011; 12:295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Bimber BN, Evans DT. The killer‐cell immunoglobulin‐like receptors of macaques. Immunol Rev 2015; 267:246–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Older Aguilar AM, Guethlein LA, Hermes M, Walter L, Parham P. Rhesus macaque KIR bind human MHC class I with broad specificity and recognize HLA‐C more effectively than HLA‐A and HLA‐B. Immunogenetics 2011; 63:577–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Hermes M, Weil S, Groth A, Dressel R, Koch J, Walter L. Characterisation of mouse monoclonal antibodies against rhesus macaque killer immunoglobulin‐like receptors KIR3D. Immunogenetics 2012; 64:845–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Hermes M, Albrecht C, Schrod A, Brameier M, Walter L. Expression patterns of killer cell immunoglobulin‐like receptors (KIR) of NK‐cell and T‐cell subsets in Old World monkeys. PLoS ONE 2013; 8:e64936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Albrecht C, Malzahn D, Brameier M, Hermes M, Ansari AA, Walter L. Progression to AIDS in SIV‐infected rhesus macaques is associated with distinct KIR and MHC class I polymorphisms and NK Cell dysfunction. Front Immunol 2014; 5:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Walter L, Ansari AA. MHC and KIR polymorphisms in Rhesus Macaque SIV infection. Front Immunol 2015; 6:540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Zambello R, Teramo A, Barila G, Gattazzo C, Semenzato G. Activating KIRs in chronic lymphoproliferative disorder of NK cells: protection from viruses and disease induction? Front Immunol 2014; 5:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Zambello R, Trentin L, Ciccone E, Bulian P, Agostini C, Moretta A et al Phenotypic diversity of natural killer (NK) populations in patients with NK‐type lymphoproliferative disease of granular lymphocytes. Blood 1993; 81:2381–5. [PubMed] [Google Scholar]
- 227. Saez‐Borderias A, Romo N, Ruiz‐Cabello F, Canton J, Tielemans D, Langerak AW et al Natural killer cell receptor expression reflects the role of human cytomegalovirus in the pathogenesis of a subset of CD4+ T‐cell large granular lymphocytosis. Hum Immunol 2011; 72:226–8. [DOI] [PubMed] [Google Scholar]
- 228. Lundell R, Hartung L, Hill S, Perkins SL, Bahler DW. T‐cell large granular lymphocyte leukemias have multiple phenotypic abnormalities involving pan‐T‐cell antigens and receptors for MHC molecules. Am J Clin Pathol 2005; 124:937–46. [PubMed] [Google Scholar]
- 229. Thonnart N, Caudron A, Legaz I, Bagot M, Bensussan A, Marie‐Cardine A. KIR3DL2 is a coinhibitory receptor on Sezary syndrome malignant T cells that promotes resistance to activation‐induced cell death. Blood 2014; 124:3330–2. [DOI] [PubMed] [Google Scholar]
- 230. Battistella M, Janin A, Jean‐Louis F, Collomb C, Leboeuf C, Sicard H et al KIR3DL2 (CD158k) is a potential therapeutic target in primary cutaneous anaplastic large‐cell lymphoma. Br J Dermatol 2016; 175:325–33. [DOI] [PubMed] [Google Scholar]
- 231. Ruggeri L, Mancusi A, Urbani E, Velardi A. Identifying NK alloreactive donors for haploidentical hematopoietic stem cell transplantation. Methods Mol Biol 2016; 1393:141–5. [DOI] [PubMed] [Google Scholar]
- 232. Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M et al HLA‐C‐dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med 2012; 367:805–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SG et al Donor killer cell Ig‐like receptor B haplotypes, recipient HLA‐C1, and HLA‐C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 2014; 192:4592–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Guillerey C, Smyth MJ. NK cells and cancer immunoediting. Curr Top Microbiol Immunol 2016; 395:115–45. [DOI] [PubMed] [Google Scholar]
- 235. Liu LL, Pfefferle A, Yi Sheng VO, Bjorklund AT, Beziat V, Goodridge JP et al Harnessing adaptive natural killer cells in cancer immunotherapy. Mol Oncol 2015; 9:1904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer 2016; 16:7–19. [DOI] [PubMed] [Google Scholar]
- 237. Kohrt HE, Thielens A, Marabelle A, Sagiv‐Barfi I, Sola C, Chanuc F et al Anti‐KIR antibody enhancement of anti‐lymphoma activity of natural killer cells as monotherapy and in combination with anti‐CD20 antibodies. Blood 2014; 123:678–86. [DOI] [PMC free article] [PubMed] [Google Scholar]


