Abstract
Environmental factors, including exogenous exposures and nutritional status, can affect DNA methylation across the epigenome, but effects of exposures on age-dependent epigenetic drift remain unclear. Here, we tested the hypothesis that early-life exposure to bisphenol A (BPA) and/or variable diet results in altered epigenetic drift, as measured longitudinally via target loci methylation in paired mouse tail tissue (3 wks/10 mos old). Methylation was quantified at two repetitive elements (LINE-1, IAP), two imprinted genes (Igf2, H19), and one non-imprinted gene (Esr1) in isogenic mice developmentally exposed to Control, Control+BPA (50 μg/kg diet), Mediterranean, Western, Mediterranean+BPA, or Western+BPA diets. Across age, methylation levels significantly (p<0.050) decreased at LINE-1, IAP, and H19, and increased at Esr1. Igf2 demonstrated Western-specific changes in early-life methylation (p=0.027), and IAP showed marginal negative modification of drift in Western (p=0.058) and Western+BPA (p=0.051). Thus, DNA methylation drifts across age, and developmental nutritional exposures can alter age-related methylation patterns.
Keywords: Epigenetics, DNA Methylation, Drift, Bisphenol A, High Fat Diet, Developmental Origins of Health and Disease
1.1 INTRODUCTION
The epigenome is a dynamic regulatory framework that utilizes epigenetic information to govern the response of cells, tissues, and entire organisms to environmental stressors. Epigenetic control mechanisms operate at several levels, including alterations to DNA itself (e.g. DNA methylation), chromatin remodeling (e.g. histone modifications), and non-coding RNA interactions [1,2]. DNA methylation, which is perhaps the best-studied epigenetic mark, is defined by the addition of a methyl group to the 5′-carbon of cytosine in a cytosine-phospho-guanine (CpG) dinucleotide. Recent evidence indicates that DNA methylation status changes as a function of age in both humans and animal models, and that this change is often gene- or tissue-specific [3–6]. This process of altered DNA methylation across time is termed “epigenetic drift,” and has important implications for gene expression and disease onset throughout the life course [6]. While this process of drift occurs across all individuals, twin studies have shown that genetically identical individuals can have vast divergence in their epigenetic marks as they age [7]. These results suggest that unique environmental exposures throughout life, rather than any inherent genetic predisposition, may lead to a modulation in the rate of age-related drift.
Mounting evidence indicates that exposure to environmental factors during key developmental windows may alter gene regulation and phenotype through changes in epigenetic marks [8]. As such, the epigenome represents a possible mechanism underlying the Developmental Origins of Health and Disease (DOHaD) hypothesis, which states that exposure to nutritional and environmental factors during prenatal and early postnatal periods alters susceptibility to chronic diseases by influencing developmental plasticity [9]. In general, methylation of DNA at specific promoter/enhancer sites is associated with decreased transcription factor binding, as well as decreased transcription [10]. However, gene-specific methyl marks do not accurately predict global methylcytosine levels, which are driven by CpG methylation of non-coding, repetitive DNA elements, including transposons, retrotransposons, and endogenous retroviruses [11,12]. In contrast to the transcriptional effects seen at gene promoters/enhancers, altered methylation of repetitive elements has the potential to affect genetic stability through increased movement of repetitive elements around the genome [13–15]. Based on the differential regulatory effects of site-specific and global methylation levels, it is important to measure both when investigating the biological effects of epigenetic drift. Recent data also indicate that early life exposure to environmental toxicants has the potential to alter age-related global and gene-specific methylation [16,17]. Supporting this idea, we recently demonstrated that developmental lead (Pb) exposure in congenic mice altered DNA methylation levels at imprinted genes, and that exposure was associated with alterations in the rate of epigenetic drift throughout the life-course [18].
Endocrine disrupting chemicals (EDCs) are an important class of environmental factors that have been linked to the developmental origins of adult disease [19]. One such chemical, bisphenol A (BPA), is a commercial monomer that makes up polycarbonate plastic and epoxy resins. BPA is found in a variety of consumer products (e.g. metal can linings, receipt paper, etc.), and has near ubiquitous and continuous human exposure across the world [20]. BPA can directly bind estrogen receptor α, has been shown to activate a variety of growth-related transcription factors, and can also bind effectively to several nuclear receptors involved in cell maturation [21–24]. BPA exposure has also been shown to affect DNA methylation levels across the epigenome [24–27]. In utero doses of BPA in mouse models affect both global and gene promoter-specific methylation, indicating that BPA exposure could alter gene expression during development [24,28,29]. The effects of BPA exposure on epigenetic drift in matched samples have not been previously studied, but based on BPA’s ability to alter the developing epigenome, BPA exposure has the potential to alter drift rates over time.
In addition to chemical exposure, maternal diet can also affect offspring DNA methylation levels [30,31]. The modern “Western High-Fat Diet” (WHFD) is characterized by high saturated and omega-6 polyunsaturated fatty acids, reduced omega-3 fatty acid intake, and increased salt and refined sugar intake [32]. Studies in animal models have shown that alterations in maternal diet, specifically levels of methyl donors, can alter gene-specific and global methylation in offspring, indicating that diet can induce long-lasting, inter-generational changes in methylation [33–35]. Genome-wide studies of the methylome have also noted nutrient-sensitive CpG sites throughout the epigenome, indicating that alterations in diet can affect DNA methylation at specific genomic sites [36]. Based on these results, maternal diet represents an important mediator of the epigenome that has the potential to affect offspring methylation throughout the life course.
To investigate the potential combined effects of diet and BPA exposures on the epigenome, DNA methylation was measured in murine target loci regions -- Long Interspersed Nuclear Element-1 (LINE-1) repeats, Intracisternal A-Particle (IAP) repeats, Insulin-like growth factor 2 (Igf2) differentially methylated region (DMR) 2 [18,37], H19 DMR [18,38], and the promoter region of Estrogen receptor α (Esr1) [39]. These candidate regions fall into three classes – repetitive elements (LINE-1, IAP), imprinted genes (Igf2, H19), and a non-imprinted protein-coding gene (Esr1). Target region classes were chosen based on their potential to reflect global methylation levels, their use as frequent biomarkers in environmental epigenetic studies, and their involvement in growth and metabolism, respectively.
Long interspersed nuclear element-1 (LINE-1) is the most common transposable element in the mouse genome, representing more than 20% of the murine sequence [40]. LINE-1 elements are ancient retrotransposons that replicated in the genome over evolutionary time. Although most LINE-1 elements are no longer active, they have widespread distribution across the genome, making LINE-1 methylation a useful approximation of “global” methylation levels [12]. Intracisternal A-Particle (IAP) retrotransposons are murine, long terminal repeat (LTR)-type genetic elements that also utilize RNA intermediates to retrotranspose around the genome [41]. With the exception of metastable epialleles like the well-studied viable yellow (Avy) IAP element [34], most IAPs are tightly regulated, and have lost their ability to retrotranspose [41]. However, evidence indicates that aging can cause demethylation of IAP promoters, potentially reactivating their retrotransposition competency [41,42]. The IAP assay in this study utilizes a conserved IAP sequence to measure methylation across all IAP retrotransposons present in the murine genome, thereby providing a second, but more genetically “active” approximation of global methylation.
Along with repetitive elements, several imprinted and non-imprinted genes were also investigated. Imprinted genes display parent-of-origin differential methylation and mono-allelic expression [43]. The imprinted genes included in this study, Igf2 and H19, contain differentially methylated regions (DMRs) that exhibit variability in methylation associated with exposure to diet and/or EDCs [37,44–48], making them valuable biomarkers of exposure-induced changes in methylation. In addition to the imprinted genes, we also examined methylation levels at a non-imprinted protein-coding gene – Esr1. Estrogen receptor α is a transcription factor activated by estrogenic ligands, and it mediates estrogen’s involvement in the regulation of growth and development [49]. Evidence indicates that methylation of the Esr1 exon 2 promoter is positively associated with age in unmatched samples of murine small intestine [39]. This fact, combined with Esr1’s biological importance throughout life, makes it an ideal non-imprinted candidate gene for assessing the effects of developmental exposure on epigenetic drift.
The present study examines longitudinal changes in absolute mean DNA methylation from paired early- (day 21) and late-life (10 months) mouse tail tissue. Matched tail tissue was used due to availability at both weaning and sacrifice, and to eliminate inter-individual confounding. This study investigates whether developmental exposure to BPA and/or altered diet affects the rate of epigenetic drift at these genetic loci. We found clear, gene-specific changes in absolute mean DNA methylation across time in all measured loci. Western diet exposure had a significant modifying effect on the rate of drift at the non-imprinted Esr1 locus. Similarly, exposure to both the Western and Western + BPA diets had a marginally significant modifying effect on age-related methylation at IAP repeats. Effects of BPA exposure in diet did not have a significant effect on the rate of epigenetic drift at LINE-1, IAP, H19, or Esr1, but did have a marginally significant effect on age-related methylation at Igf2. This study demonstrates measurable, gene-specific epigenetic drift, as well as diet-dependent alterations in the rate of drift at a class of repetitive elements and a non-imprinted locus related to murine growth and development.
1.2 RESULTS
1.2.1 Litter parameters
Developmental BPA and/or diet exposure did not significantly alter litter size, sex ratio, or a/a to Avy/a genotypic ratio (n=277). Percent survival was significantly lower in the Control+BPA exposed offspring (survival = 73%) compared to Control (survival = 91%, p=0.006) and Mediterranean+BPA (survival = 85%, p=0.007) exposure groups, but was not significantly different in other comparisons. A subset of a/a non-agouti wild type mouse pups (n=133) was selected for inclusion in longitudinal follow-up up to 10 months of age, which incorporated collection of matched tail tip samples (Table 1).
Table 1. Litter Parameters.
A subset of n=133 mouse pups included in longitudinal follow-up. All pups in the longitudinal subset were maintained until sacrifice at 10 months of age.
| Developmental Exposure Group | N (litter) | Female | Male | Pups (#) |
|---|---|---|---|---|
| Control | 14 | 10 | 12 | 22 |
| Control+BPA | 25 | 9 | 10 | 19 |
| Med HFD | 20 | 11 | 12 | 23 |
| Western HFD | 21 | 11 | 11 | 22 |
| Med+BPA | 15 | 12 | 12 | 24 |
| Western +BPA | 23 | 12 | 11 | 23 |
| Total | 118 | 65 | 68 | 133 |
1.2.2 Exposure and Diet Dependent Changes in PND21 Weanling Mice
For all exposure groups, no significant changes in cross-sectional DNA methylation were found in the LINE-1, IAP, and H19 loci at either time point. However, several significant differences in PND21 cross-sectional methylation were identified at the Esr1 and Igf2 loci (Figure 1). The Esr1 locus demonstrated significant alterations to PND21 methylation when comparing Control, Mediterranean, and Western diets (ANOVA, p = 0.002). Specifically, mice exposed to Western diet showed significantly decreased methylation compared to Control (Tukey’s test, p= 0.027) and Mediterranean (Tukey’s test, p = 0.002), and the Western exposure group demonstrated a significant decrease in methylation compared to the Western + BPA group (Student’s t-test, p=0.005). The Igf2 locus demonstrated significant alterations to PND21 methylation when comparing BPA, Mediterranean + BPA, and Western + BPA diets (ANOVA, p = 0.020). At PND21, Igf2 methylation in the BPA group was significantly lower than methylation in the Western + BPA group (Tukey’s test, p=0.027); a similar, marginally significant decrease in methylation was seen when comparing Mediterranean + BPA and Western + BPA (Tukey’s test, p=0.073).
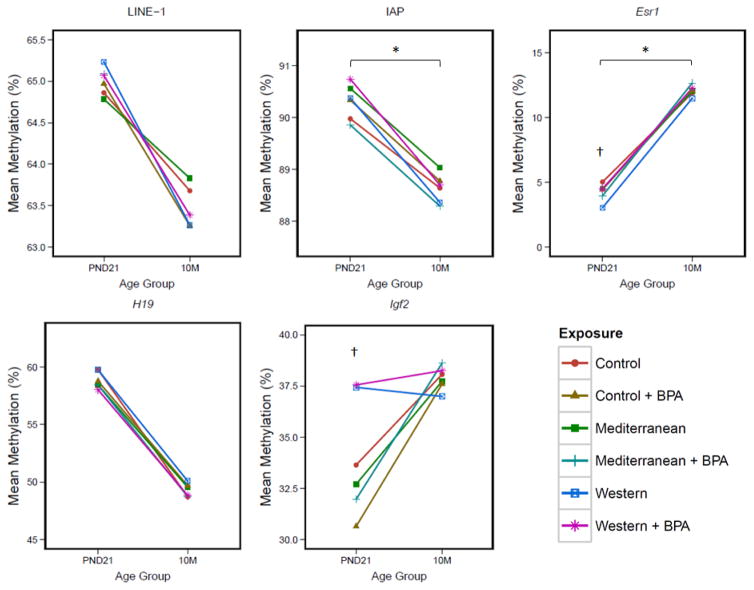
Figure 1. Methylation Drift by Exposure Group.
Visualization of epigenetic drift over time at 5 genetic loci. LINE-1, H19, and IAP demonstrated a negative association between age and % methylation, while Esr1 and Igf2 demonstrated a positive association between age and % methylation. * = age:exposure interaction term p-value <0.10 for at least one exposure group in linear mixed model. † = ANOVA/t-test p-value <0.05 for cross-sectional comparison by exposure group.
1.2.3 DNA Methylation Drifts Over Time
DNA methylation at the LINE-1 and IAP repetitive elements, as well as the Igf2, H19, and Esr1 genes, was quantified from paired PND21 and 10 month tail samples (Table 2). When adjusting for exposure group, sex, and the interaction between age and exposure/sex, LINE-1, IAP, and the H19 locus demonstrated significant decreases in methylation over time (−0.94%, p=0.035; −1.32%, p=2.2E-06; −10.44%, p=6.22E-08; respectively). In contrast, the Igf2 and Esr1 genes demonstrated increased methylation over time; however, only the increase in the Esr1 gene was statistically significant (7.60% increase, p=1.44E-12) (Table 2).
Table 2. Absolute Mean Methylation by Exposure Group.
Linear mixed effect models were used to compare absolute methylation levels over time by exposure group. Age, exposure group, sex, age:exposure, and age:sex were included as terms in all models. Linear mixed models for each gene also included a paired factor to account for matched, within-individual data, as well as a random factor to account for within-litter effects. Separate models were run for each gene; beta coefficients and associated p-values for age predictor from each model are reported.
| Absolute Change in Methylation Levels by Age | |||||
|---|---|---|---|---|---|
| Gene | Paired Tail Samples (N) | PND21 % Methylation (SD) | 10 Month % Methylation (SD) | Adjusted Methylation by Age - Beta coefficient† | p-value |
| LINE-1 | 260 | 65.05 (1.35) | 63.50 (1.22) | −0.943 | 0.035 |
| IAP | 222 | 90.31 (1.37) | 88.62 (1.38) | −1.321 | 2.20E-06 |
| Igf2 | 249 | 34.39 (8.69) | 37.87 (2.87) | 3.275 | 0.112 |
| H19 | 257 | 58.87 (7.06) | 49.31 (3.01) | −10.436 | 6.22E-08 |
| Esr1 | 260 | 4.26 (1.86) | 12.09 (3.00) | 7.604 | 1.44E-12 |
BOLD = p<0.05
Beta coefficient for Age predictor in Linear Mixed Model.
1.2.4 Developmental Exposures Affect Drift Over Time
To examine the potential effects of exposure on methylation, we first examined whether each exposure group had a direct, significant effect on mean methylation compared to Control. LINE-1, IAP, and H19 showed no significant effects of developmental exposures on mean methylation (Table 3). At the Igf2 locus, BPA exposure had a marginally significant negative effect on mean methylation compared to Control (β= −3.81, p=0.066); no other exposure groups had a significant effect on methylation at this gene. In the Esr1 gene, Western diet exposure had a significant negative effect on mean methylation compared to Control (β=1.93, p=0.013); this was the only significant exposure effect in the Esr1 gene (Table 3).
Table 3. Relative Mean Methylation by Exposure Group.
Average methylation by age group was compared across exposure groups using a linear mixed effects model. Age, exposure group, sex, age:exposure, and age:sex were included as terms in all models. Linear mixed models included a paired factor to account for matched, within-individual data, as well as a random factor to account for within-litter effects. Separate models were run for each gene; beta coefficients and associated p-values for the exposure categories from each model are reported. Slope of the age-methylation relationship was tested for heterogeneity across all exposure groups via inclusion of an age:exposure interaction term in the model. Control diet was used as the reference exposure in all comparisons.
| Relative Methylation by Exposure | ||||||||
|---|---|---|---|---|---|---|---|---|
| Methylation by Age | Methylation by Exposure | Slope Heterogeneity | ||||||
| Gene | Developmental Exposure Group | N | PND21 % Methylation (SD) | 10 Month % Methylation (SD) | Methylation by Age Beta Coefficient† | p-value | Age : Exposure Interaction Beta Coefficient† | p-value |
| LINE-1 | Control | 44 | 64.86 (1.14) | 64.00 (1.40) | (Reference) | n/a | (Reference) | n/a |
| Control+BPA | 38 | 64.78 (2.18) | 63.32 (1.32) | −0.079 | 0.855 | −0.596 | 0.316 | |
| Med | 44 | 64.81 (1.53) | 63.83 (1.43) | −0.040 | 0.925 | −0.132 | 0.820 | |
| Western | 43 | 65.18 (1.03) | 63.27 (1.13) | 0.331 | 0.431 | −1.052 | 0.069 | |
| Med+BPA | 47 | 65.08 (1.44) | 63.41 (1.29) | 0.232 | 0.568 | −0.818 | 0.147 | |
| Western+BPA | 46 | 64.97 (1.25) | 63.65 (1.09) | 0.122 | 0.766 | −0.450 | 0.426 | |
| IAP | Control | 38 | 89.98 (1.37) | 88.64 (1.60) | (Reference) | n/a | (Reference) | n/a |
| Control+BPA | 32 | 90.56 (1.02) | 89.03 (1.19) | 0.560 | 0.279 | −0.358 | 0.299 | |
| Med | 38 | 89.86 (1.15) | 88.29 (1.15) | 0.783 | 0.169 | −0.291 | 0.410 | |
| Western | 34 | 90.34 (1.37) | 88.77 (1.37) | 0.484 | 0.356 | −0.645 | 0.058 | |
| Med+BPA | 42 | 90.74 (1.53) | 88.70 (1.53) | 0.206 | 0.689 | −0.325 | 0.335 | |
| Western+BPA | 38 | 90.37 (1.38) | 88.36 (1.38) | 0.788 | 0.135 | −0.649 | 0.051 | |
| Igf2 | Control | 43 | 33.64 (6.23) | 38.07 (2.59) | (Reference) | n/a | (Reference) | n/a |
| Control+BPA | 37 | 30.65 (8.26) | 37.59 (4.58) | −3.811 | 0.066 | 3.355 | 0.221 | |
| Med | 41 | 32.70 (10.69) | 37.83 (2.05) | −0.513 | 0.804 | 0.338 | 0.900 | |
| Western | 41 | 37.43 (11.04) | 36.99 (1.84) | 2.846 | 0.163 | −3.914 | 0.143 | |
| Med+BPA | 44 | 31.97 (8.02) | 38.61 (1.81) | −2.506 | 0.207 | 2.965 | 0.259 | |
| Western+BPA | 45 | 37.55 (8.26) | 38.25 (3.34) | 3.162 | 0.108 | −2.888 | 0.268 | |
| H19 | Control | 38 | 59.77 (7.22) | 48.93 (2.86) | (Reference) | n/a | (Reference) | n/a |
| Control+BPA | 31 | 58.77 (6.43) | 49.61 (2.29) | −0.830 | 0.650 | 1.588 | 0.521 | |
| Med | 42 | 58.43 (7.31) | 49.51 (3.84) | −1.956 | 0.249 | 2.601 | 0.266 | |
| Western | 42 | 59.75 (8.28) | 50.07 (3.23) | 0.426 | 0.806 | 0.814 | 0.730 | |
| Med+BPA | 42 | 58.43 (6.25) | 48.55 (2.67) | −1.047 | 0.532 | 0.688 | 0.766 | |
| Western+BPA | 37 | 58.02 (7.32) | 49.28 (3.03) | −2.334 | 0.173 | 2.776 | 0.236 | |
| Esr1 | Control | 43 | 5.03 (2.25) | 11.90 (1.93) | (Reference) | n/a | (Reference) | n/a |
| Control+BPA | 38 | 4.45 (1.56) | 12.07 (3.00) | −0.587 | 0.458 | 0.758 | 0.468 | |
| Med | 45 | 4.53 (1.84) | 12.07 (2.01) | −0.523 | 0.489 | 0.669 | 0.504 | |
| Western | 42 | 3.08 (1.12) | 11.48 (1.65) | −1.933 | 0.013* | 1.620 | 0.112 | |
| Med+BPA | 46 | 3.96 (1.72) | 12.67 (4.95) | −1.089 | 0.150 | 1.851 | 0.064 | |
| Western+BPA | 46 | 4.51 (1.97) | 12.27 (3.14) | −0.604 | 0.424 | 0.988 | 0.321 | |
Beta coefficient for age predictor in Linear Mixed Model;
BOLD = p<0.10
p<0.05
To further examine the potential effects of exposure on the rate of epigenetic drift, an interaction term between age and categorical exposure was included in the linear mixed model for each gene (Table 3). The Igf2 and H19 genes showed no significant interaction between age and exposure group, indicating that the relationship between age and methylation was not affected by developmental exposures in those genes. On the other hand, at LINE-1 repetitive elements, developmental Western diet exposure had a marginally significant negative effect on the association between age and methylation compared to Control (β= −1.05, p=0.069). Similarly, for IAP, both the Western and Western+BPA diets had marginally significant negative effects on age-related methylation relative to Control (β= −0.645, p=0.058; β= −0.0649, p=0.051). In the Esr1 gene, developmental Mediterranean+BPA diet exposure had a marginally significant positive effect on age-related methylation when compared to Control (β=1.85, p=0.064). Directionality of the interaction between age and exposure was specific to each gene (Table 3, Figure 1).
Given that gene regulation can vary by sex, two additional variables – sex and a sex:age interaction term – were included in the linear mixed model for each gene. Neither sex nor sex:age were significant terms in the mixed models for LINE-1 (p=0.395, p=0.644), IAP (p=0.476, 0.786), Igf2 (p=0.868, p=0.686), and H19 (p=0.294, p=0.639). However, at the Esr1 gene, while the sex categorical variable did not demonstrate a significant effect on methylation, the sex:age interaction term was statistically significant (p=0.003), indicating effect modification of age-related methylation by sex (Table 3).
1.3 DISCUSSION
Age-associated changes in level of DNA methylation occurred in all measured genetic loci, with statistically significant changes present at Esr1 and H19 loci and in the LINE-1 and IAP repetitive elements. Consistent with other results in the literature, directionality of drift over time was specific to each gene [6,16]. The non-imprinted gene promoter, Esr1, demonstrated an increase in methylation with age, a result consistent with documented decreases in ERα expression during aging [50]. Meanwhile, repetitive element methylation decreased with age, and the investigated imprinted genes either increased or decreased with age depending upon the locus. These results are also consistent with previous reports [4,6,51], and indicate that epigenetic drift varies in a region-specific manner.
The documented region-specific directionality of drift fits a growing hypothesis in the field – that age-related changes in methylation facilitate development of chronic disease (e.g. cancer) via increased genomic instability and altered regulation of genes related to growth and development [6,52,53]. Decreased methylation of repetitive elements with age has the potential to increase genomic instability through increased transposition of repetitive elements around the genome and dysregulation of expression via cis-chromatin modifying effects [13–15]. Additionally, increased methylation of promoter regions in protein-coding genes is associated with dysregulated transcription [10,54]. Combined, these effects have the potential to produce an epigenetic environment that alters gene expression and may increase the risk of disease states commonly associated with aging.
Given that exposure to exogenous chemicals and altered diet can alter the epigenome [8,24–27,34], we tested the effects of developmental exposure to exogenous chemicals and/or variable diet on the rate of epigenetic drift using an age:exposure interaction term. Age:exposure demonstrated marginal significance at IAP repeats and the Esr1 locus, but not at LINE-1, Igf2, or H19, indicating that exposure is a potential gene-specific effect modifier of epigenetic drift. For IAP repeats, developmental exposure to the Western and Western + BPA diets was associated with a marginally significant decrease in the rate of age-related methylation. At this global locus, the magnitude of this effect did not differ between the Western and Western+BPA diets, suggesting that exposure to Western diet was driving the age:exposure interaction effect. Given that age-related demethylation of IAP promoters has the potential to reactivate retrotransposition competency [41,42], Western HFD, by increasing the rate of age-related methylation loss at IAP elements, may also increase IAP retrotransposition events. This suggests a mechanism by which developmental WHFD influences genomic stability throughout an organism’s life.
Although epigenetic drift at the IAP repetitive element demonstrated effect modification by Western diet exposure, this result was not seen in LINE-1. This suggests that separate classes of repetitive elements exhibit distinct epigenetic responses to environmental factors. Therefore, when studying the effects of the environment on global DNA methylation – both in cross-section and across the life course – multiple classes of repetitive elements should be included in analysis.
At the Esr1 locus, Med+BPA exposure was associated with a marginally significant increase in the rate of age-related methylation. The Med+BPA exposure group had Esr1 methylation levels below Control at PND21, and higher than Control at 10M. This marginal increase in the slope of epigenetic drift at the Esr1 promoter may reflect tighter control of Estrogen Receptor α expression during aging. However, given the lack of significance in the age:exposure interaction terms for the BPA and Mediterranean groups at this gene, it is difficult to determine whether the Med+BPA exposure effect seen here is truly an effect of exposure. Additionally, a protective effect on PND21 survival was observed for the Med+BPA exposure group. Previous studies have observed that nutritional supplementation counteracts negative epigenetic effects on the epigenome [34,55]. Furthermore, in multiple longitudinal human birth cohort studies, maternal adherence to a Mediterranean diet was associated with reduced risk of intrauterine growth restriction, low birth weight and low placental weight [56,57]. Mothers consuming a Mediterranean diet also had higher circulating folate and vitamin B12 concentrations [57]. Folate and vitamin B12 are critical nutrients in the regeneration of S-adenosyl methionine, a major participant in DNA methylation maintenance; this suggests adherence to the Mediterranean diet may impact fetal epigenetic reprogramming. Therefore, it is possible that the developmental Mediterranean diet is providing a protective effect on survival by offsetting BPA-induced changes to epigenetic marks and gene regulation. Future studies should investigate this toxicant-diet interaction more fully.
Sex did not significantly modify drift direction at LINE-1, IAP, Igf2, or H19. However, sex was a significant effect modifier of the relationship between age and methylation at the Esr1 locus. Specifically, as age increased from PND21 to 10 months, there was a significant increase in the effect of sex on methylation at the Esr1 gene. Across all exposures except Med+BPA, male mice demonstrated lower average Esr1 methylation at PND21, but higher average Esr1 methylation at 10 months. This trend, combined with the significant age:sex interaction term, suggests a sex-specific change in regulation of the Esr1 gene during aging. This corroborates the fact that the sexes utilize estrogen for very different processes during reproduction and growth, with females of reproductive age demonstrating higher average serum estrogen than males [49]. Therefore, as the animals reach reproductive age, sex-specific effect modification of age-related Esr1 methylation may occur as a regulatory response to sexually dimorphic estrogen activity.
Given the effect modification of epigenetic drift by exposure group, we also tested the effects of developmental exposure on cross-sectional methylation at PND21 and 10 months. PND21 DNA methylation showed significant changes by exposure group at two candidate regions – Igf2 and Esr1. Igf2 encodes the Insulin-like growth factor 2 protein, an important regulator of cellular glucose transport during development [58,59]. The cross-sectional WHFD-mediated increase in early-life Igf2 methylation may be a biological response to an increased simple sugar load, indicating that developmental exposure to WHFD can affect early-life establishment of epigenetic drift at a gene related to metabolism and growth. Similarly, at the Esr1 gene promoter, WHFD had a significant effect on mean methylation, with a decrease in methylation compared to Control at PND21. Given the directionality of this exposure effect, developmental WHFD exposure may increase transcription of the Estrogen Receptor α (ERα) early in life. Previous studies have shown that ERα is involved in control of lipid metabolism [60], with ERα knockout mice demonstrating increased adipose tissue deposition with aging [61,62]. Developmental exposure to WHFD, which is high in fat and has been linked to obesity, was significantly associated with a decrease in the rate of age-related Esr1 methylation. This suggests that developmental exposure to Western diet alters the epigenetic profile of the Esr1 locus during development, predisposing animals to increased Esr1 transcription in anticipation of the Western diet’s altered nutritional profile. When a mismatch occurs between the developmental and postnatal environment, there is potential for improper regulation of epigenetic marks and disease development [8,9]. The significant effects of exposure on PND21 DNA methylation at the Igf2 and Esr1 loci support this idea, indicating that developmental exposure to environmental factors may not only alter the rate of drift, but also the cross-sectional establishment of DNA methylation at specific genetic loci during development.
At the Igf2 locus, BPA exposure had a marginally significant effect on mean methylation, with a decrease in methylation compared to Control at both PND21 and 10 months. This result, which is not present in the Med+BPA or Western+BPA diets, indicates that BPA exposure alone may alter transcription of the Igf2 gene compared to the Control diet group. A previous study demonstrated decreased Igf2 methylation and increased Igf2 expression in developing embryos as a result of early-life 10 mg/kg/day BPA exposure [46]. Given these past results, the marginally significant effects of 50 μg BPA/kg diet exposure on Igf2 methylation presented in this report may reflect BPA exposure-mediated alterations in Igf2 expression, but further investigation is required. The LINE-1, IAP, and H19 loci did not demonstrate significant cross-sectional effects by exposure, suggesting resistance to exposure-mediated reprogramming effects at these three genetic regions.
This study demonstrates measurable exposure-based modification to the rate of epigenetic drift, but the potential biological effects of this modification remain unclear without concurrent, longitudinal measurements of gene expression. Longitudinal measures of gene expression would provide a validation of DNA methylation results, demonstrating whether age- and exposure-related alterations to the epigenome have measurable physiological effects. As such, future studies investigating the effects of early-life toxicant exposure on epigenetic drift could expand the interpretability of their results by examining the effects of exposure and age on longitudinal gene expression, and/or examining DNA methylation and expression levels in other biological tissues of interest including blood – which may be accessed at multiple time points – and target tissues such as liver and brain.
By using matched tail tissue in this longitudinal study, drift rates reflected defined changes within organisms in the study population rather than changes in time between two separate populations. This matched design, combined with the controlled developmental exposure, isolates the effects of exposure for each organism in the study population, allowing for a direct test of the hypothesis that environmental factors can modify the rates of epigenetic drift. Despite the longitudinal design, an inherent limitation of this study is the inability of bisulfite sequencing to differentiate between 5-methylcyosine (5-mC) and 5-hydroxymethylcytosine (5-hmC). Although hydroxymethylation is not expected to be a major epigenetic mark in tail tissue, recent study showed that aging affects global hydroxymethylation in healthy hepatic tissue, with a general trend towards increasing 5-hmC levels in older mice [63]. This reported increase in global hepatic 5-hmC levels over time is at odds with the previously reported loss of global 5-mC in cancer cells [4,6,51], suggesting that these separate epigenetic marks can change in different ways during aging. As a result, future epigenetic drift studies must better characterize the effects of aging on 5-hmC levels at specific CpG sites in the genome, as well as across tissue types.
1.4 CONCLUSION
We measured longitudinal DNA methylation in tail tissue collected from isogenic mice at PND21 and again at 10 months of age, then quantified the magnitude of epigenetic drift from these samples at five genetic loci – two repetitive elements, two imprinted genes, and one non-imprinted gene. The use of matched tail tissue from an isogenic mouse colony allowed for strict control of genetic, environmental, and dietary measures, as well as removal of potential confounding. This study demonstrates clear, gene-specific directionality of epigenetic drift during aging, supporting the growing hypothesis that epigenetic drift plays an important role in the link between aging and cancer [6,52,53]. In addition, we showed several diet- and sex-dependent alterations to the rate of drift at both imprinted and non-imprinted genes. These alterations indicate that developmental exposure to altered diet or BPA can affect methylation changes during the life course. Diet-dependent changes in DNA methylation were also evident in two investigated loci at PND21, demonstrating the effect of developmental exposure on early-life establishment of epigenetic marks. To improve the generalizability of these results, the dynamics of epigenetic drift at the studied gene regions should be further evaluated in human cohorts.
1.5 MATERIALS AND METHODS
1.5.1 Mouse Colony
Mice included in longitudinal analysis were a/a offspring sourced from a genetically invariant Avy/a mouse colony maintained by sibling mating and forced heterozygosity for more than 220 generations [64]. Within this colony, the Avy allele is passed through the heterozygous male line, which has a genetically constant background 93% identical to C57BL/6J strain [64,65]. Two weeks prior to mate-pairing with Avy/a males, six week old wild type a/a dams were placed on one of six experimental diet groups: (1) Control (modified AIN-93G), (2) Control + 50 μg BPA/kg diet, (3) Mediterranean HFD chow, (4) Mediterranean + 50 μg BPA/kg diet, (5) Western HFD chow, and (6) Western HFD + 50 μg BPA/kg diet (Figure 2). Dietary exposure was continued through pregnancy and lactation, at which point treatment group pups were shifted over to a modified AIN-93G Control diet containing 7% corn oil rather than 7% soybean oil (Harlan Teklad). The 50 ug/kg diet BPA exposure level was chosen based on previous studies, which demonstrated both increased global methylation and sex-specific phenotypic effects at 50 μg/kg BPA [28,66]. BPA (0.01 g) was mixed with sucrose (9.99 g) in glass containers to achieve a 0.1% BPA mixture. To achieve the 50 μg/kg BPA concentration, 0.1% BPA/sucrose mixture was included at 0.05 g/kg diet in custom Control/HFD diets by the manufacturer (Harlan Teklad). Western HFD and Mediterranean HFD mixtures were designed based on the U.S. junk food diet and the human Cretan diet, respectively [67–71]. Protein was kept constant between the three base diets, but vitamin levels, lipid ratios, and carbohydrate types were altered to mimic human consumption (Table 4) [71].
Figure 2. Diagram of Exposure Timing.
F0 dams were assigned to one of six dietary BPA/HFD exposure groups two weeks prior to mating. Exposure continued throughout conception, gestation, and through lactation until weaning at post-natal day 21 (PND21). After weaning, offspring were transferred to an ad libitum Control diet, which continued until sacrifice at 10 months of age. Matched tail tips were collected at both PND21 and 10 months.
Table 4. Comparison of Nutrient Content by Diet.
Three base diets were included as developmental exposures in this study – Control, Mediterranean HFD, and Western HFD. For all three base diets, protein was kept constant, but vitamin levels, lipid ratios, and carbohydrate types were altered to mimic human consumption. BPA was added to each diet to produce three additional developmental dietary exposure groups – Control+BPA, Med+BPA, and Western+BPA. Apart from BPA addition, nutrient content was not altered from the base diet levels in these three groups.
| Nutrient Content in 3 Base Diets | |||
|---|---|---|---|
| Diet Nutrients | Control | Mediterranean | Western |
| Kcal/g | 3.98 | 4.53 | 4.72 |
| %Calories from Fat | 16% | 42% | 40% |
| PUFE : SFA : MUFA | 1 : 0.2 : 0.5 | 1 : 1.3 : 5.6 | 1 : 1.9 : 1.6 |
| Protein (casein) | 20 | 19 | 19 |
| Carb Content (g/100 g chow) | |||
| Cornstarch | 40 | 23 | 14 |
| Sucrose | 10 | 9.2 | 25.5 |
| Cellulose | 5 | 8 | 2 |
| Vitamin A (IU) | 4000 | 8000 | 4000 |
| Vitamin C (mg) | 0 | 500 | 0 |
| Vitamin D (IU) | 1000 | 1000 | 400 |
| Vitamin E (IU) | 75 | 75 | 25 |
| Folic Acid (mg) | 2 | 4 | 1 |
| Sodium (mg) | 1039 | 1039 | 7000 |
| Potassium (mg) | 3600 | 8000 | 3600 |
| Magnesium (mg) | 513 | 850 | 513 |
1.5.2 Exposure and Tissue Collection
At postnatal day 21 (PND21), offspring were tail tipped, and collected tail tissue was frozen at −80°C. For each exposure group, a subset of PND 21 a/a wild-type pups were maintained until 10 months of age – Control: n = 22, Control+BPA: n=19, Mediterranean (Med): n=23, Mediterranean+BPA: n=24, Western: n=22, Western+BPA: n=23 (Table 1). At 10 months of age, remaining mice were sacrificed, and tail tissue was again collected. The offspring with tail tips collected at both PND21 and 10 months of age represent the population used to measure epigenetic drift in this paper. All animals in this study were stored in polycarbonate-free cages with ad libitum access to food and drinking water, and were maintained in accordance with Institute for Laboratory Animal Research (ILAR) guidelines [72]. The study protocol was approved by the University of Michigan Committee on Use and Care of Animals (UCUCA).
1.5.3 DNA isolation
Genomic DNA was isolated from PND21 tail tissue (≤3mm) using a phenol-chloroform-isoamyl alcohol protocol [73]. Genomic DNA was isolated from 10 month tail tissue (3 mm) using the Maxwell Mouse Tail DNA Purification Kit (Promega, Cat. #AS1120). Yield and purity of all DNA was measured using a NanoDrop spectrophotometer, and then genomic DNA was bisulfite converted using the Zymo Research 96-well EZ-methylation kit (Zymo Research, Cat. #D5004). Briefly, bisulfite conversion was accomplished through the addition of sodium bisulfite to 0.5–1 μg of genomic DNA, thereby converting unmethylated cytosines to uracil. During polymerase chain reaction (PCR) amplification, uracils are replaced with thymines, making any remaining cytosines a direct, quantitative measure of methylation [74]. PCR amplification was performed on bisulfite converted DNA using HotStarTaq master mix (Qiagen, Cat. #203443), RNAse-free water, forward primer (9 pmol), and biotinylated reverse primer (9 pmol). Total PCR volume was 30 μL per sample, and gel electrophoresis was used to verify PCR product identity.
1.5.4 DNA Methylation Measurement
Specific PCR amplification for regions of interest (Igf2, H19, Esr1, IAP, and LINE-1) was performed on bisulfite converted DNA with primers designed using the PyroMark Assay Design software 2.0 and mm9 mouse genome. DNA methylation levels were quantified using the PyroMark Q96 MD instrument (Qiagen). Pyrosequencing samples were run in duplicate, and the average of the duplicates provided the final methylation percentages. Sample duplicates with coefficient of variation (%CV) > 10% were discarded and re-run. Pyrosequencing assay information, including primer sequences, chromosomal location, annealing temperature, and sequences to analyze are available in Table 5. In an effort to reduce plate-to-plate batch effects, matched samples were run on the same plate for all PCR amplification and pyrosequencing runs. All pyrosequencing plates included 0% and 100% bisulfite converted methylation controls, as well as a no template control, to ensure proper functioning of the instrument and to provide background standards of methylation for each gene.
Table 5. PCR conditions.
Information for each assay, including genomic location, primer sequences (5′-3′), sequence to analyze, amplicon length, annealing temperature, number of cycles, and number of CpG sites measured.
| Primer/Sequence to Analyze | Igf21 | H191 | LINE-1 | IAP | Esr12 |
|---|---|---|---|---|---|
| Location | chr7:149839707–149839926 strand = reverse |
chr7:149767589–149767843 | Repetitive Element | Repetitive Element | chr10:4712147–4712203 |
| Forward PCR Primer | TTTTTTAATA TGATATTTGG AGATAGTT |
GGGGGGTTAT AAATGTTATT AGGGGGGTAGG |
AAGGGGTTTGTG TTTTAGATTAGG |
GTGTTATTTTTTGA TTGGTTGTAGTTT |
TTTGGAAGTTGT AGTTTTTGGTTA GT |
| Reverse PCR Primer | biotin- CCACATAATTTAAT TCACTAATAATT ACTA |
biotin- AACCCCTAAC CTCATAAAAC CCATAACTAT AAAATCA |
biotin- AACTCCCCACCA AATCCTAAAACC TCTA |
biotin- ACCAAAAATATCT TATAACTACTTATA CT |
biotin- ACAAAACACAA ATAACCCAACTC |
| Sequencing Primer | AATATGATATTT GGCGATAGTT |
GTGTAAAGAT TAGGGTTGT |
AGTTTGTTTTTTT ATGTATTATAGT |
ATTTTTTGATTGGT TGTAGTTTA |
GGAGAGGAGTA TGTAAAG |
| Sequence to Analyze | YGYGGGAYGT TTGYGTAGAG GTTTGTTTGT TTTTTTGYGT GTTYGTYGGG GTYGT |
GYGGTYAGTG AAGTTTYGTA TATYG |
TTTAGGTTTY GYGYGATTGG ATTGGGGTAG AYGTTGTGTT TTATTTATTA GAGGTTT |
TYGGTYGAGT TGAYGTTAYG GGGAAAGTAG AGTATAAGTA GTTA |
TTGGAGAATT YGGGAGYGTT TGGGTGYGTT TTTTGGAGTT GGGTTATTTG TGTTTT |
| Amplicon Length (bp) | 220 | 255 | 132 | 87 | 131 |
| Annealing Temperature (°C) | 56 | 55 | 61 | 56 | 55 |
| Number of Cycles | 50 | 40 | 44 | 45 | 40 |
| Number of CpG Sites | 8 | 4 | 4 | 4 | 3 |
1.5.5 Data Analysis
Matched tail tissue was collected at postnatal day 21 and 10 months of age from a total of 133 a/a offspring. The effect of developmental BPA/HFD exposure on sex ratio and litter survival rate was determined by Fisher’s exact test, with Control as the reference group. Litter number, sex ratio, and litter survival rate were compared between exposure groups using a combined statistical approach involving both 3-way ANOVAs and Independent Student’s T-tests. This same approach was used to compare cross-sectional PND21 or 10 month methylation data by exposure group. Separate 3-way ANOVAs were performed to compare Control vs. Mediterranean vs. Western and Control+BPA vs. Med+BPA vs. Western+BPA exposure groups. Separate Student’s t-tests were performed to individually compare methylation between base diets and their associated BPA exposure diet (e.g. Control vs. Control + BPA). For all ANOVAs, Tukey’s post-hoc test was used to determine the significance of each group-to-group comparison. Mixed effect linear models were used to compare absolute methylation levels over time by exposure group. Age, exposure group, and sex were included as explanatory variables in all models. Linear mixed models for each candidate region also included a paired factor to account for matched, within-individual data, as well as a random factor to account for within-litter effects. Homogeneity of relative age-related methylation was compared by exposure group via inclusion of an age:exposure interaction term in all mixed models. An interaction term between age and sex was also included in an effort to identify and/or control for potential modifying effects of sex on methylation levels.
Mixed models were fit using the following format: Methylation ~ Age + Sex + Exposure + Age:Exposure + Age:Sex + [1|ID] + [1|Litter]. For all models, the methylation outcome variable was defined as mean methylation across all amplicon CpG sites for two passing replicates. The lme4 package within the statistical program R was used for all linear mixed models (R version 3.2.3, http://www.rproject.org). Alpha significance levels were set at p≤0.05 for all statistical comparisons.
Highlights.
Age had a significant effect on DNA methylation at investigated candidate genes.
Epigenetic drift directionality and magnitude was specific to each candidate gene.
Developmental exposure to Western diet modified the rate of epigenetic drift.
Western diet was associated with increased early-life DNA methylation at Igf2.
Acknowledgments
Funding: This work was supported by the University of Michigan (UM) NIEHS/EPA Children’s Environmental Health and Disease Prevention Center P20 ES018171/RD834800 and P01 ES022844/RD83543601, the Michigan Lifestage Environmental Exposures and Disease (M-LEEaD) NIEHS Core Center (P30 ES017885), as well as the UM NIEHS Institutional Training Grant T32 ES007062 (JJK, EHM, LM, CF), NIH Grant K99/R00 ES022221 (CF), and F31 ES025101 (EHM). The authors have no conflicts of interest and declare no competing financial interests.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–63. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 2.Bernal AJ, Jirtle RL. Epigenomic disruption: the effects of early developmental exposures. Birth Defects Res A Clin Mol Teratol. 2010;88:938–44. doi: 10.1002/bdra.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madrigano J, Baccarelli A, Mittleman MA, Sparrow D, Vokonas PS, Tarantini L, Schwartz J. Aging and epigenetics: longitudinal changes in gene-specific DNA methylation. Epigenetics. 2012;7:63–70. doi: 10.4161/epi.7.1.18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teschendorff AE, West J, Beck S. Age-associated epigenetic drift: implications, and a case of epigenetic thrift? Hum Mol Genet. 2013;22:R7–R15. doi: 10.1093/hmg/ddt375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Florath I, Butterbach K, Müller H, Bewerunge-Hudler M, Brenner H. Cross-sectional and longitudinal changes in DNA methylation with age: an epigenome-wide analysis revealing over 60 novel age-associated CpG sites. Hum Mol Genet. 2014;23:1186–201. doi: 10.1093/hmg/ddt531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Issa JP. Aging and epigenetic drift: a vicious cycle. J Clin Invest. 2014;124:24–9. doi: 10.1172/JCI69735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–9. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Waterland RA, Jirtle RL. Early nutrition, epigenetic changes at transposons and imprinted genes, and enhanced susceptibility to adult chronic diseases. Nutrition. 2004;20:63–8. doi: 10.1016/j.nut.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Bateson P, Barker D, Clutton-Brock T, Deb D, D’Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, et al. Developmental plasticity and human health. Nature. 2004;430:419–21. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 10.Medvedeva YA, Khamis AM, Kulakovskiy IV, Ba-Alawi W, Bhuyan MS, Kawaji H, Lassmann T, Harbers M, Forrest AR, Bajic VB, et al. Effects of cytosine methylation on transcription factor binding sites. BMC Genomics. 2014;15:119. doi: 10.1186/1471-2164-15-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang AS, Estécio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson HH, Marsit CJ, Kelsey KT. Global Methylation in Exposure Biology and Translational Medical Science. Environmental Health Perspectives. 2011;119:1528–1533. doi: 10.1289/ehp.1103423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez J, Frigola J, Vendrell E, Risques RA, Fraga MF, Morales C, Moreno V, Esteller M, Capella G, Ribas M, Peinado MA. Chromosomal instability correlates with genome-wide DNA demethylation in human primary colorectal cancers. Cancer Res. 2006;66:8462–9468. doi: 10.1158/0008-5472.CAN-06-0293. [DOI] [PubMed] [Google Scholar]
- 15.Ross JP, Rand KN, Molloy PL. Hypomethylation of repeated DNA sequences in cancer. Epigenomics. 2010;2:245–69. doi: 10.2217/epi.10.2. [DOI] [PubMed] [Google Scholar]
- 16.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huen K, Yousefi P, Bradman A, Yan L, Harley KG, Kogut K, Eskenazi B, Holland N. Effects of age, sex, and persistent organic pollutants on DNA methylation in children. Environ Mol Mutagen. 2014;55:209–22. doi: 10.1002/em.21845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faulk C, Liu K, Barks A, Goodrich JM, Dolinoy DC. Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics. 2014;9:934–41. doi: 10.4161/epi.29024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newbold RR, Padilla-Banks E, Jefferson WN. Environmental estrogens and obesity. Mol Cell Endocrinol. 2009;304:84–9. doi: 10.1016/j.mce.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calafat AM, Ye X, Wong LY, Reidy JA, Needham LL. Exposure of the U.S. population to bisphenol A and 4-tertiary-octylphenol: 2003–2004. Environ Health Perspect. 2008;116:39–44. doi: 10.1289/ehp.10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson CS, Bulayeva NN, Wozniak AL, Alyea RA. Xenoestrogens are potent activators of nongenomic estrogenic responses. Steroids. 2007;72:124–34. doi: 10.1016/j.steroids.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sui Y, Ai N, Park SH, Rios-Pilier J, Perkins JT, Welsh WJ, Zhou C. Bisphenol A and its analogues activate human pregnane X receptor. Environ Health Perspect. 2012;120:399–405. doi: 10.1289/ehp.1104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krüger T, Long M, Bonefeld-Jørgensen EC. Plastic components affect the activation of the aryl hydrocarbon and the androgen receptor. Toxicology. 2008;246:112–23. doi: 10.1016/j.tox.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 24.Singh S, Li SS. Epigenetic effects of environmental chemicals bisphenol a and phthalates. Int J Mol Sci. 2012;13:10143–53. doi: 10.3390/ijms130810143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolstenholme JT, Rissman EF, Connelly JJ. The role of Bisphenol A in shaping the brain, epigenome and behavior. Horm Behav. 2011;59:296–305. doi: 10.1016/j.yhbeh.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XF, Zhang LJ, Feng YN, Chen B, Feng YM, Liang GJ, Li L, Shen W. Bisphenol A exposure modifies DNA methylation of imprint genes in mouse fetal germ cells. Mol Biol Rep. 2012;39:8621–8. doi: 10.1007/s11033-012-1716-7. [DOI] [PubMed] [Google Scholar]
- 27.Manikkam M, Tracey R, Guerrero-Bosagna C, Skinner MK. Plastics derived endocrine disruptors (BPA, DEHP and DBP) induce epigenetic transgenerational inheritance of obesity, reproductive disease and sperm epimutations. PLoS One. 2013;8:e55387. doi: 10.1371/journal.pone.0055387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anderson OS, Nahar MS, Faulk C, Jones TR, Liao C, Kannan K, Weinhouse C, Rozek LS, Dolinoy DC. Epigenetic responses following maternal dietary exposure to physiologically relevant levels of bisphenol A. Environ Mol Mutagen. 2012;53:334–42. doi: 10.1002/em.21692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JH, Sartor MA, Rozek LS, Faulk C, Anderson OS, Jones TR, Nahar MS, Dolinoy DC. Perinatal bisphenol A exposure promotes dose-dependent alterations of the mouse methylome. BMC Genomics. 2014;15:30. doi: 10.1186/1471-2164-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–9. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myles IA. Fast food fever: reviewing the impacts of the Western diet on immunity. Nutr J. 2014;13:61. doi: 10.1186/1475-2891-13-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Niculescu MD, Craciunescu CN, Zeisel SH. Dietary choline deficiency alters global and gene-specific DNA methylation in the developing hippocampus of mouse fetal brains. FASEB J. 2006;20:43–9. doi: 10.1096/fj.05-4707com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollingsworth JW, Maruoka S, Boon K, Garantziotis S, Li Z, Tomfohr J, Bailey N, Potts EN, Whitehead G, Brass DM, Schwartz DA. In utero supplementation with methyl donors enhances allergic airway disease in mice. J Clin Invest. 2008;118:3462–9. doi: 10.1172/JCI34378. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Parle-McDermott A, Ozaki M. The impact of nutrition on differential methylated regions of the genome. Adv Nutr. 2011;2:463–71. doi: 10.3945/an.111.001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (Igf2) locus. Hum Mol Genet. 2006;15:705–16. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 38.Stouder C, Deutsch S, Paoloni-Giacobino A. Superovulation in mice alters the methylation pattern of imprinted genes in the sperm of the offspring. Reprod Toxicol. 2009;28:536–41. doi: 10.1016/j.reprotox.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Maegawa S, Hinkal G, Kim HS, Shen L, Zhang L, Zhang J, Zhang N, Liang S, Donehower LA, Issa JP. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 2010;20:332–40. doi: 10.1101/gr.096826.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sookdeo A, Hepp CM, McClure MA, Boissinot S. Revisiting the evolution of mouse LINE-1 in the genomic era. Mob DNA. 2013;4:3. doi: 10.1186/1759-8753-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horie K, Saito ES, Keng VW, Ikeda R, Ishihara H, Takeda J. Retrotransposons influence the mouse transcriptome: implication for the divergence of genetic traits. Genetics. 2007;176:815–27. doi: 10.1534/genetics.107.071647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–73. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sasaki H, Ishihara K, Kato R. Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation. J Biochem. 2000;127:711–5. doi: 10.1093/oxfordjournals.jbchem.a022661. [DOI] [PubMed] [Google Scholar]
- 44.Hoyo C, Murtha AP, Schildkraut JM, Jirtle RL, Demark-Wahnefried W, Forman MR, Iversen ES, Kurtzberg J, Overcash F, Huang Z, Murphy SK. Methylation variation at IGF2 differentially methylated regions and maternal folic acid use before and during pregnancy. Epigenetics. 2011;6:928–36. doi: 10.4161/epi.6.7.16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKay JA, Xie L, Harris S, Wong YK, Ford D, Mathers JC. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Mol Nutr Food Res. 2011;55:1026–35. doi: 10.1002/mnfr.201100008. [DOI] [PubMed] [Google Scholar]
- 46.Susiarjo M, Sasson I, Mesaros C, Bartolomei MS. Bisphenol a exposure disrupts genomic imprinting in the mouse. PLoS Genet. 2013;9:e1003401. doi: 10.1371/journal.pgen.1003401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee HS, Barraza-Villarreal A, Biessy C, Duarte-Salles T, Sly PD, Ramakrishnan U, River J, Herceg Z, Romieu I. Dietary supplementation with polyunsaturated fatty acid during pregnancy modulates DNA methylation at IGF2/H19 imprinted genes and growth of infants. Physiol Genomics. 2014;46:851–7. doi: 10.1152/physiolgenomics.00061.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaRocca J, Binder AM, McElrath TF, Michels KB. The impact of first trimester phthalate and phenol exposure on IGF2/H19 genomic imprinting and birth outcomes. Environ Res. 2014;133:396–406. doi: 10.1016/j.envres.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bondesson M, Hao R, Lin CY, Williams C, Gustafsson J. Estrogen receptor signaling during vertebrate development. Biochim Biophys Acta. 2015;1849:142–51. doi: 10.1016/j.bbagrm.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson ME, Westberry JM. Regulation of oestrogen receptor gene expression: new insights and novel mechanisms. J Neuroendocrinol. 2009;21:238–42. doi: 10.1111/j.1365-2826.2009.01830.x. [DOI] [PubMed] [Google Scholar]
- 51.Maegawa S, Gough SM, Watanabe-Okochi N, Lu Y, Zhang N, Castoro RJ, Estecio MR, Jelinek J, Liang S, Kitamura T, et al. Age-related epigenetic drift in the pathogenesis of MDS and AML. Genome Res. 2014;24:580–91. doi: 10.1101/gr.157529.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Nousemehr H, Bell GG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–6. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Decottignies A, d’Adda di Fagagna F. Epigenetic alterations associated with cellular senescence: a barrier against tumorigenesis or a red carpet for cancer? Semin Cancer Biol. 2011;21:360–6. doi: 10.1016/j.semcancer.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 54.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–67. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernal AJ, Dolinoy DC, Huang D, Skaar DA, Weinhouse C, Jirtle RL. Adaptive radiation-induced epigenetic alterations mitigated by antioxidants. The FASEB Journal. 2013;27:665–71. doi: 10.1096/fj.12-220350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chatzi L, Mendez M, Garcia R, Roumeliotaki T, Ibarluzea J, Tardon A, et al. Mediterranean diet adherence during pregnancy and fetal growth: INMA (Spain) and RHEA (Greece) mother-child cohort studies. British Journal of Nutrition. 2012;107:135–145. doi: 10.1017/S0007114511002625. [DOI] [PubMed] [Google Scholar]
- 57.Timmermans S, Steegers-Theunissen RP, Vujkovic M, den Breeijen H, Russcher H, Lindemans J, et al. The Mediterranean diet and fetal size parameters: the Generation R Study. British Journal of Nutrition. 2012;108:1399–1409. doi: 10.1017/S000711451100691X. [DOI] [PubMed] [Google Scholar]
- 58.O’Dell S, Day I. Molecules in focus Insulin-Like growth factor II (IGF-II) The International Journal of Biochemistry & Cell Biology. 1998;30:767–71. doi: 10.1016/s1357-2725(98)00048-x. [DOI] [PubMed] [Google Scholar]
- 59.Fowden AL, Sibley C, Reik W, Constancia M. Imprinted genes, placental development and fetal growth. Horm Res. 2006;65(Suppl 3):50–8. doi: 10.1159/000091506. [DOI] [PubMed] [Google Scholar]
- 60.Foryst-Ludwig A, Kintscher U. Metabolic impact of estrogen signalling through ERalpha and ERbeta. J Steroid Biochem Mol Biol. 2010;122:74–81. doi: 10.1016/j.jsbmb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 61.Ohlsson C, Hellberg N, Parini P, Vidal O, Bohlooly-Y M, Rudling M, Lindberg MK, Warner M, Angelin B, Gustafsson JA. Obesity and disturbed lipoprotein profile in estrogen receptor-alpha-deficient male mice. Biochem Biophys Res Commun. 2000;278:640–5. doi: 10.1006/bbrc.2000.3827. [DOI] [PubMed] [Google Scholar]
- 62.Cooke PS, Heine PA, Taylor JA, Lubahn DB. The role of estrogen and estrogen receptor-alpha in male adipose tissue. Mol Cell Endocrinol. 2001;178:147–54. doi: 10.1016/s0303-7207(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 63.Tammen SA, Dolnikowski GG, Ausman LM, Liu Z, Kim KC, Friso S, Choi SW. Aging alters hepatic DNA hydroxymethylation, as measured by liquid chromatography/mass spectrometry. J Cancer Prev. 2014;19(4):301–8. doi: 10.15430/JCP.2014.19.4.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterland RA. Do maternal methyl supplements in mice affect DNA methylation of offspring? J Nutr. 2003;133:238. doi: 10.1093/jn/133.1.238. [DOI] [PubMed] [Google Scholar]
- 65.Weinhouse C, Anderson OS, Bergin IL, Vandenbergh DJ, Gyekis JP, Dingman MA, Yang J, Dolinoy DC. Dose-dependent incidence of hepatic tumors in adult mice following perinatal exposure to bisphenol A. Environ Health Perspect. 2014;122:485–91. doi: 10.1289/ehp.1307449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson OS, Peterson KE, Sanchez BN, Zhang Z, Mancuso P, Dolinoy DC. Perinatal bisphenol A exposure promotes hyperactivity, lean body composition, and hormonal responses across the murine life course. FASEB J. 2013;27:1784–92. doi: 10.1096/fj.12-223545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kafatos A, Verhagen H, Moschandreas J, Apostolaki I, Van Westerop JJ. Mediterranean diet of Crete: foods and nutrient content. J Am Diet Assoc. 2000;100:1487–93. doi: 10.1016/s0002-8223(00)00416-8. [DOI] [PubMed] [Google Scholar]
- 68.Trichopoulou A, Katsouyanni K, Gnardellis C. The traditional Greek diet. Eur J Clin Nutr. 1993;47(Suppl 1):S76–81. [PubMed] [Google Scholar]
- 69.Block G, Dresser CM, Hartman AM, Carroll MD. Nutrient sources in the American diet: quantitative data from the NHANES II survey. I. Vitamins and minerals. Am J Epidemiol. 1985;122:13–26. doi: 10.1093/oxfordjournals.aje.a114072. [DOI] [PubMed] [Google Scholar]
- 70.Block G, Dresser CM, Hartman AM, Carroll MD. Nutrient sources in the American diet: quantitative data from the NHANES II survey. II. Macronutrients and fats. Am J Epidemiol. 1985;122:27–40. doi: 10.1093/oxfordjournals.aje.a114084. [DOI] [PubMed] [Google Scholar]
- 71.Marchlewicz EH, Djuric Z, Barks J, Tang L, Song P, Peterson KE, Dolinoy DC. Maternal diet during pregnancy and post-pregnancy weight predict offspring liver triglyceride levels at postnatal day 10. J Nutr. In submission. [Google Scholar]
- 72.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. 8. Washington (DC): National Academies Press (US); 2011. [PubMed] [Google Scholar]
- 73.Sambrook J, Russell DW. Purification of nucleic acids by extraction with phenol:chloroform. CSH Protoc 2006. 2006 doi: 10.1101/pdb.prot4455. [DOI] [PubMed] [Google Scholar]
- 74.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic Acids Res. 2001;29:E65–5. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]