Abstract
RitR (formerly RR489) is an orphan two-component signal transduction response regulator in Streptococcus pneumoniae that has been shown to be required for lung pathogenicity. In the present study, by using the rough strain R800, inactivation of the orphan response regulator gene ritR by allele replacement reduced pathogenicity in a cyclophosphamide-treated mouse lung model but not in a thigh model, suggesting a role for RitR in regulation of tissue-specific virulence factors. Analysis of changes in genome-wide transcript mRNA levels associated with the inactivation of ritR compared to wild-type cells was performed by the use of high-density DNA microarrays. Genes with a change in transcript abundance associated with inactivation of ritR included piuB, encoding an Fe permease subunit, and piuA, encoding an Fe carrier-binding protein. In addition, a dpr ortholog, encoding an H2O2 resistance protein that has been shown to reduce synthesis of reactive oxygen intermediates, was activated in the wild-type (ritR+) strain. Microarray experiments suggested that RitR represses Fe uptake in vitro by negatively regulating the Piu hemin-iron transport system. Footprinting experiments confirmed site-specific DNA-binding activity for RitR and identified three binding sites that partly overlap the +1 site for transcription initiation upstream of piuB. Transcripts belonging to other gene categories found to be differentially expressed in our array studies include those associated with (i) H2O2 resistance, (ii) repair of DNA damage, (iii) sugar transport and capsule biosynthesis, and (iv) two-component signal transduction elements. These observations suggest that RitR is an important response regulator whose primary role is to maintain iron homeostasis in S. pneumoniae. The name ritR (repressor of iron transport) for the orphan response regulator gene, rr489, is proposed.
Two-component signal transduction (TCST) systems are used by bacteria to sense their environment. The basic mechanism of TCST involves a membrane-bound sensor histidine kinase which, upon stimulation, autophosphorylates a conserved histidine residue in its intracellular catalytic domain. Transfer of the phosphoryl group to a conserved aspartate in an intracellular response regulator (RR) alters its affinity for DNA, leading to transcriptional activation or repression of specific genes (for reviews, see references 22 and 48). Bacterial pathogens have been shown to express their potential for virulence in response to environmental stimuli, and the growing list of microbial virulence determinants regulated by two-component signal transduction includes biofilm formation and quorum sensing (12), colonization (35), and toxin production (11).
Genome sequencing in Streptococcus pneumoniae revealed 13 TCST pairs along with one orphan response regulator (26, 52, 53). Throup et al. (53) inactivated all 14 response regulators by antibiotic cassette mutagenesis in a type 3 encapsulated strain and demonstrated that 9 of these response regulators were required for murine respiratory tract infection, including the orphan response regulator gene designated rr489 (referred to here as ritR). The ritR locus is not adjacent to a kinase gene, as the other 13 TCST kinase-RR pairs are, and RitR is not known to be phosphorylated by any of the 13 known histidine kinases.
Iron serves as a major factor in microbial virulence both as an essential element and as an element that is able to catalyze the synthesis of reactive oxygen intermediates (ROIs) from H2O2, commonly referred to as the Fenton reaction. The interaction between Fe2+ and H2O2 can be particularly deleterious under iron overload conditions (for reviews, see references 2 and 24). Despite the widespread abundance of iron in nature, the free Fe3+ concentration is estimated to be in the range of 10−18 M at mucosal surfaces (9). Sequestration of iron by host iron-binding proteins, such as hemoglobin, lactoferrin, and transferrin, contributes to its low concentration. In response, bacteria have evolved elaborate mechanisms to capture iron needed for growth while minimizing iron-based toxicity.
The iron-binding transcriptional regulator protein Fur has been shown to regulate iron uptake in both gram-positive and gram-negative bacteria. Fur binds intracellular free Fe2+ directly, increasing its affinity for a conserved DNA motif, the Fur box, a regulatory sequence associated with iron uptake (16). Fur regulation also plays a role in the oxidative stress response (39) and the regulation of other iron-related genes (for reviews, see references 2 and 19). Gram-positive bacteria with a high G+C content, such as corynebacteria and mycobacteria, utilize iron uptake repressors belonging to the DtxR family, which has little or no homology to Fur. In Corynebacterium diphtheriae, DtxR regulates siderophore biosynthesis, iron uptake, and toxin production (51). A potential DtxR family member, SmrB, which resembles TroR (40), a regulator of Mn uptake in Treponema pallidum, has been identified in S. pneumoniae (27); however, a role for this protein in iron homeostasis has not been established.
Three gene clusters, pit, pia, and piu, each specifying a set of four elements of an iron uptake ATP-binding cassette (ABC) transporter, have been implicated in iron uptake by S. pneumoniae (5, 6). In the studies reported here, DNA microarrays were used to learn which genes RitR might regulate. Arrays revealed that RitR was associated with the transcription of genes involved in iron uptake and the oxidative stress response. In DNA footprinting experiments, recombinant glutathione S-transferase (GST)-RitR was shown to bind directly to three sites in the promoter region of the piu operon, thereby providing a direct link between RitR and iron uptake regulation. We also show that ritR is cotranscribed with gnd (encoding 6-phosphogluconate dehydrogenase), suggesting that there is coordinated regulation of iron homeostasis and the pentose phosphate pathway.
MATERIALS AND METHODS
Bacterial strains, transformation, and growth conditions.
Plasmids and bacterial strains used in these studies are listed in Table 1. Oligonucleotide primers and probes are listed in Table 2. S. pneumoniae strains R800 and CP1250 were obtained from D. A. Morrison. Starter cultures were grown overnight in casein hydrolysate broth (38) supplemented with 0.01% glucose (CAT medium). For experimental manipulations, cells were grown in liquid media (brain heart infusion [BHI] or Todd-Hewitt medium supplemented with 0.5% yeast extract) without agitation at 37°C in the presence of 5% CO2. S. pneumoniae cells were plated on Trypticase soy blood agar (TSBA) supplemented with 3% defibrinated sheep blood. Deferrated medium was prepared as described by Brown et al. (6). For Escherichia coli, the conditions used for the growth of cells, transformation, and other experimental manipulations were based on standard techniques (43). E. coli strains DH5αF′ and RR1 were grown in Luria-Bertani medium at 37°C overnight with aeration. The antibiotic concentrations used for plasmid selection in S. pneumoniae were as follows: chloramphenicol, 2 μg/ml; and erythromycin, 1 μg/ml. The antibiotic concentrations used for plasmid selection in E. coli were 34 μg of chloramphenicol per ml and 500 μg of erythromycin per ml. S. pneumoniae strains were transformed as described previously (20).
TABLE 1.
Strains and plasmids used in this study
| Plasmid or strain | Relevant genotype | Derivation, description, and/or phenotype | Reference or source |
|---|---|---|---|
| Plasmids | |||
| pEVP3 | cat; lacZ integrative vector | Cmr LacZ | 38 |
| pRK01 | ermAM tetR tet057 | Weak tet promoter | 47 |
| pRK02 | ermAM tetR tet057-opt | Strong tet promoter | 47 |
| pGEX-2TK | Commercial | Pharmacia | |
| pAU401 | pEVP3::′ritR′ (forward) | From pEVP3 with 5′→3′ ritR targeted displacement and LacZ fusion | This study |
| pAU402 | pEVP3::′ritR′ (reverse) | Like pAU401 but with ritR in the reverse orientation | This study |
| pAU403 | pRK01::′ritR′ (forward) | From pRK01 for low-level tet-inducible ritR expression | This study |
| pAU404 | pRK02::′ritR′ (forward) | From pRK02 for high-level tet-inducible ritR expression | This study |
| pAU410 | pGEX-2TK::′ritR′ | From pGEX-2TK for overexpression of GST-RitR | This study |
| E. coli strains | |||
| BL21DE3 | lacl; phage T7 RNAP | 43 | |
| AU410 | BL21(DE3)(pAU410) | From BL21(DE3) by transformation with pAU410 | This study |
| S. pneumoniae strains | |||
| R800 | S. pneumoniae R6 derivative | 28 | |
| CP1250 | S. pneumoniae Rx derivative | 38 | |
| RU401 | R800 ritR′::(pEVP3)::′ritR 5′→3′ | From R800 by transformation with pEVP3 | This study |
| RU402 | R800 ritR′::(pEVP3)::′ritR 3′→5′ | From R800 by transformation with pEVP3 | This study |
| RU403 | R800 tetRIO57::(pRK01)::′ritR 5′→3′ | From R800 by transformation with pRK01 | This study |
| RU404 | R800 tetRIO57-opt::(pRK02)::′ritR 5′→3′ | From R800 by transformation with pRK02 | This study |
| CPU401 | CP1250 ritR′::(pEVP3)::′ritR 5′→3′ | From CP1250 by transformation with pAU401 | This study |
| CPU402 | CP1250 ritR′::(pEVP3)::′ritR 3′→5′ | From CP1250 by transformation with pAU402 | This study |
TABLE 2.
Oligonucleotide primers and probes used in this study
| Oligonucleotide | Use | Gene target | Sequence |
|---|---|---|---|
| 1F | Cloning with pEVP3/pRK01-02 | ritR-1 | 5′ GATCCGGGAAACGGATTTTATTACTTGAGAAAG 3′ |
| 1R | Cloning with pEVP3/pRK01-02 | ritR-1 | 5′ GATCCGGAAGATCGCCGAAATACGC 3′ |
| 2F | Cloning with pEVP3/pRK01-02 | ritR-2 | 5′ CGGGAAACGGATTTTATTACTTGAGAAAG 3′ |
| 2R | Cloning with pEVP3/pRK01-02 | ritR-2 | 5′ CGGAAGATCGCCGAAATACGC 3′ |
| 3F | Cloning with pGEX2TK | ritR-3 | 5′ GATCCATGGGGAAACGGATTTTATTACTTGAG 3′ |
| 3R | Cloning with pGEX2TK | ritR-3 | 5′ AATTCTATTCTTGCATGGTATATCCAACACC 3′ |
| 4F | Cloning with pGEX2TK | ritR-4 | 5′ CATGGGGAAACGGATTTTATTACTTGAG 3′ |
| 4R | Cloning with pGEX2TK | ritR-4 | 5′ CTATTCTTGCATGGTATATCCAACACC 3′ |
| 5F | Northern blot probe | ritR | 5′ CTTGAGAAAGAACGAAATCTAGCTC 3′ |
| 5R | Northern blot probe | ritR | 3′ CATCTATCCTAAGATTGCGGTAGGTC 5′ |
| 6F | Northern blot probe | gnd | 5′ CAAGCTGGACCTGGTACAGATGC 3′ |
| 6R | Northern blot probe | gnd | 5′ CTACGATTGGTCCATCTTGGCCTTC 3′ |
| 7F | Northern blot probe | cbpF | 5′ CGGTTGGCAATACTTAGAAATTCCTGG 3′ |
| 7R | Northern blot probe | cbpF | 5′ CGTACATATTTCCAACCTGTTGCCATAG 3′ |
| 8F | Northern blot probe | piuB | 5′ GCAGACCTGGCTCCTCTTTCAAG 3′ |
| 8R | Northern blot probe | piuB | 5′ CCAGATTGAAACGATAGGCGATAACTTC 3′ |
| 9F | Northern blot probe | piuA | 5′ GGATACTATTCGCGCTTTAGGATTTG 3′ |
| 9R | Northern blot probe | piuA | 5′ CTTCTTGGATGCTCTTGTCTAGCTTGG 3′ |
| 10F | Northern blot probe | dpr | 5′ GTATGTAGCTCACGTTGCTTTGCACC 3′ |
| 10R | Northern blot probe | dpr | 5′ CGCCTACAAAGATACCGTTTGTCAC 3′ |
| 11F | Northern blot probe | vncS | 5′ CGAACAGGTTTATTTGCAAAG 3′ |
| 11R | Northern blot probe | vncS | 5′ TGCTCCGCTTCCTTTTG 3′ |
| 12F | Northern blot probe | 16S rRNA | 5′ CAAACAGTGACCATCGCTAG 3′ |
| 12R | Northern blot probe | 16S rRNA | 5′ GGAATATCAACATTGAGAACCAC 3′ |
| 13F | Gel shift or footprint analysis | PpiuB (promoter 1) | 5′ GAATAGGATAACAAAGGAGAAAGTCTTG 3′ |
| 13R | Gel shift or footprint analysis | PpiuB (promoter 1) | 5′ GTCCCAATGCTAATAGAGAGAAAGACT 3′ |
Construction of plasmids and mutant strains.
ritR deletion mutants were constructed by using plasmid pEVP3, provided by D. A. Morrison. For site-specific recombination into ritR, an internal 353-bp DNA fragment from ritR was obtained by PCR by using primer pairs 1F-1R and 2F-2R. The resultant product was cloned by using the melt-anneal method, in which two staggered pairs of the blunt-ended PCR products were melted and annealed to obtain a product with BamHI cohesive ends, as described previously (55). The resultant PCR product was ligated into BamHI-digested pEVP3 plasmid DNA, and constructs containing the insert in the forward orientation (pAU401) and the reverse orientation (pAU402) were obtained. Plasmids pRK01 and pRK02 (47), provided by R. Lutz, were used to place ritR under control of the tet promoter. The same 353-bp DNA fragment described above was ligated into BamHI-cut pRK01 and pRK02 to construct plasmids pAU403 (for low-level tet induction) and pAU404 (for high-level tet induction). pRK01, pRK02, and pEVP3-derived constructs were transformed into E. coli RR1 as an intermediary host prior to transformation into S. pneumoniae R800 (ritR+) or CP1250 (β-Gal−), as needed. Oligonucleotides used as PCR primers or as Northern blot probes are listed in Table 2. To construct a GST-RitR expression strain, AU410 PCR primer pairs 3F-3R and 4F-4R were used to amplify ritR from S. pneumoniae R6 genomic DNA. The resultant products were melted and reannealed to give BamHI and EcoRI overhangs, which were ligated to BamHI/EcoRI-digested pGEX-2TK plasmid DNA (Pharmacia) to obtain pAU410. pAU410 was used to prepare recombinant GST-RitR in amounts needed for DNA footprinting experiments.
Preparation and hybridization of cDNA probes.
RNA was obtained from S. pneumoniae cells grown in 100 ml of BHI liquid medium to an A600 of 0.35 to 0.40, as described by de Sazieu et al. (13). For analysis of transcripts under tet control, 0, 100, and 300 ng of anhydrotetracycline per ml were added at an A600 of approximately 0.2, and cells were grown to an A600 of 0.35 to 0.4. RNA was harvested for cDNA synthesis. cDNA was synthesized by using 5 U of SuperScriptII reverse transcriptase (Invitrogen) in a 100-μl reaction mixture with 100 μg of total RNA as the template. Reaction mixtures were incubated for 20 h at 42°C. To degrade RNA, 14 μl of 1 N NaOH was added, and the reaction mixtures were heated for 15 min at 65°C. The reaction mixture was neutralized with 14 μl of 1 N HCl. The cDNA preparation was purified and concentrated by using a Microcon-30 microfiltration device (Amicon, Bedford, Mass.) and dissolved in 35 μl of water. The cDNA was then partially digested by addition of 12 U of DNase I (Promega), after a pilot incubation to determine the time needed to optimize the yield of 50- to 100-bp fragments for microarray hybridization. The DNase I was denatured at 99°C for 15 min to stop the reactions, and the resultant preparation was washed three times with a Microcon-10 centrifuge column. Digested cDNA was then collected in 30 μl of water and 3′ end labeled with 2 nM biotin-6-ddATP (NEN) and 50 U of deoxynucleotidyl terminal transferase (Promega).
Probes were hybridized to the arrays by using a protocol that specified the use of reagents and laboratory equipment provided by NimbleGen Systems Inc. (Madison, Wis.), as follows. All procedures were performed at 42°C unless otherwise noted. Arrays were loaded onto a hybridization wheel (NimbleGen Systems) in individual sealed chambers and incubated with prehybridization buffer (PHB) (100 μg of salmon sperm DNA per ml, 50 μg of acetylated bovine serum albumin [BSA] per ml, 2× morpholineethanesulfonic acid [MES] buffer) for 15 min and then with hybridization buffer (HB) (PHB plus 3 to 15 μg of biotin-labeled cDNA, 1 nM nonspecific competitor oligonucleotide) for 15 min. Before PHB and HB were loaded into hybridization chambers, the buffers were incubated at 98°C for 5 min, centrifuged at 14,000 rpm for 15 min using an Eppendorf model 5414 microcentrifuge, and kept at 45°C until use. Four hundred microliters of PHB was added to each array in the hybridization wheel and incubated at 42°C for 15 min, and then 400-μl portions of the HB samples were added.
Arrays were hybridized overnight at 42°C on a rotating platform for 16 to 20 h. Arrays were then washed three times with nonstringent wash buffer (NSWB) (6× SSPE, 0.01% Tween 20) (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) and then five times with stringent wash buffer (0.1 M MES buffer [pH 6.5], 26 mM NaCl, 0.01% Tween 20), and this was followed by staining with Cy3-streptavidin. Subsequent washes were performed at the ambient temperature unless otherwise noted. Primary stain buffer (Cy3-streptavidin, 5 mg of acetylated BSA per ml) was then added, and arrays were incubated for 15 min at room temperature. Arrays were then washed three times with 400 μl of NSWB, which was followed by addition of 400 μl of secondary stain buffer (5 μg of biotinylated immunoglobulin G per ml, 100 μg of goat immunoglobulin G per ml) and incubation for 15 min. Following three additional washes with NSWB, slides were briefly washed in 50 ml of NSWB with agitation and then with NimbleGen final wash buffer (proprietary) for 30 s. Arrays were then dried briefly with argon and scanned with an Axon Genepix 4000B scanner at 532 nm and a resolution of 5 μm.
High-density oligonucleotide microarray.
The genome sequence and open reading frame (ORF) predictions for the S. pneumoniae R6 genome were obtained from GenBank accession number NC-003098. The genome sequence file contains 2,038,615 bases and encodes 2,043 predicted open reading frames. A high-density oligonucleotide array for the S. pneumoniae genome was designed for this study containing 40,860 probe pairs, where each pair contains a perfect match (PM) oligonucleotide and a mismatch (MM) oligonucleotide, for a total of 80,142 different 24-nucleotide array feature sequences. The PM sequence is a 24-mer oligonucleotide probe from the S. pneumoniae genome. MM probes were identical except for substitutions at positions 6 and 12 from the 5′ end. To generate MM sequences, bases were systematically substituted by inversion as follows: A → T, C → G, G → C, and T → A. By using the NimbleGen probe assessment and selection software (NimbleGen Systems), potential probe sequences were checked for internal complementarity that would reduce their ability to hybridize optimally. Oligonucleotide features were also checked for sequence redundancy by comparing each probe to the entire S. pneumoniae genome sequence. Twenty distinct high-quality probe pairs were selected to represent each ORF.
Microarray data analysis methods.
Following hybridization, the arrays were scanned, and the median signal intensity for each probe on the array was calculated by using NimbleGen's extraction software. For each probe pair the difference between the PM and MM signal intensities was calculated together with the Tukey biweight mean (21) (biweight constant = 6) from the 20 probe pairs for each ORF. For purposes of normalization of the array intensity data, array-specific scaling factors were calculated by assuming a constant mean signal intensity of 1,000 signal units for each array. The signal for each ORF was obtained by multiplying the raw signal intensity by the array-specific scaling factor. The program Ebarrays (25) was used to identify ORFs with significant changes in expression comparisons of ΔritR versus ritR+ or ΔritR versus tet-induced expression of ritR. The initial parameter settings were as follows: p1 = 0.95, p2 = 0.05, theta1 = 16, theta2 = 1, theta3 = 25. ORFs were selected as significant if the posterior probability associated with differential expression was ≥0.5.
Mouse infection studies.
S. pneumoniae strains R800 (ritR+) and RU402 (ΔritR) were used in thigh and lung infection models as described by Andes and Craig (1). Cyclophosphamide-treated mice were inoculated with 4 × 108 cells intranasally or by injection into the thigh. Organs were harvested at zero time and 48 h and homogenized, and bacterial counts were determined by plating homogenate dilutions on TSBA. The data reported below are the averages of counts obtained from three mice for each time.
ritR-regulated expression of LacZ.
LacZ activity was measured fluorimetrically as described previously (55). S. pneumoniae strain CP401 was grown overnight in CAT medium and diluted 1:5 in 20 ml of fresh BHI medium. One-milliliter samples were collected by centrifugation during growth of the culture, and the resultant cell pellets were washed, resuspended in 100 μl of phosphate-buffered saline containing 1% Triton X-100, and transferred to a 96-well microtiter plate. The test reaction mixture was supplemented with 4 μg of methylumbelliferyl galactoside and incubated for 5 to 10 min at the ambient temperature. Fluorescence was excited at 335 nm, and emitted fluorescence was read at 480 nm with a microtiter plate reader (Titertek Fluoroskan).
Northern blot analysis.
Primers 5F, 5R through 12F, and 12R (Table 2) were used to amplify probes for Northern blot analysis. S. pneumoniae total RNA was prepared as described above and analyzed by using standard methods for electrophoresis, transfer, probing, and detection by autoradiography or by use of a phosphorimager (43).
Iron-related toxicity.
Hydrogen peroxide toxicity was determined as described by Tseng et al. (54), and streptonigrin toxicity was determined as described by Brown et al. (5). Cells were plated on TSBA to measure the surviving cell titer following exposure to the agent.
Gene nomenclature.
The different gene and locus names associated with the iron transporter subunits mentioned in this paper are cross-referenced in Table 3.
TABLE 3.
Iron uptake systems in S. pneumoniae strains R6 and TIGR4a
| Strain | Cluster no.b | Cluster namec | Gene productd | Locus no.e | Sequence tagf | Enzymatic activityg |
|---|---|---|---|---|---|---|
| R6 | 1 | None | ABC-MSP | SPR0220 | MLVFM | Permease |
| ABC-MSP | SPR0221 | MERKK | Permease | |||
| ABC-NBD | SPR0222 | MSEIE | ATPase | |||
| ABC-SBPx | SPR0223 | MNIEG | SideroBP | |||
| 2 | None | ABC-SBP | SPR0934 | MKNKF | SideroBP | |
| ABC-MSP | SPR0935 | MHAKM | Permease | |||
| ABC-MSP | SPR0936 | MQNLI | Permease | |||
| ABC-NBD | SPR0938 | MKGLW | ATPase | |||
| 3 | Fat | FatD | SPR1684 | MKGRG | Permease | |
| FatC | SPR1685 | MQTKS | Permease | |||
| FecE | SPR1686 | MKLEN | ATPase | |||
| FatB | SPR1687 | MKTSL | SideroBP | |||
| TIGR4 | 1 | Pit | PitA | SP0241 | ? | Permease |
| PitD | SP0241 | ? | Permease | |||
| PitB | SP0242 | MSEIK | ATPase | |||
| PitC | SP0243 | MKKKW | SideroBP | |||
| 2 | Pit2 | PiaA | SP1032 | MKNKF | SideroBP | |
| PiaB | SP1033 | MHAKM | Permease | |||
| PiaC | SP1034 | MQNLI | Permease | |||
| PiaD | SP1035 | MKGLW | ATPase | |||
| 3 | Pit1 | PiuB | SP1869 | MKGRG | Permease | |
| PiuC | SP1870 | MQTKS | Permease | |||
| PiuD | SP1871 | MKLEN | ATPase | |||
| PiuA | SP1872 | MKTSL | SideroBP |
The iron transporter determinants in S. pneumoniae R6 and TIGR4 are organized as three clusters, each consisting of four genes (two heterodimer-forming permease genes, one ABC ATPase gene, and one iron carrier-binding protein gene).
The three iron transporter clusters are designated clusters 1, 2, and 3 in both R6 and TIGR4 in the order of their proximity to the sequencing origin. The three gene clusters preserve synteny between the R6 and TIGR4 genomes that extends to the order of functions of the individual genes within each cluster.
The strongly up-regulated genes in the ritR knockout correspond to R6 cluster 3, where they are annotated as FatD and FatB in GenBank accession no. AY-312585 and as piuB and piuA by Brown et al. (6). Clusters 2 and 3 in the TIGR4 sequence correspond to Pit2 and Pit1, respectively, or to pia and piu, respectively, as described previously (5, 6).
R6 clusters 1 and 2 were designated with ABC prefixes as annotated in GenBank accession no. NC-003028 for TIGR4 and NC-003098 for R6. R6 cluster 3 FatDCB and FecE were named based on their amino acid sequence similarity to V. anguillarum iron transporter elements described by Tettelin et al. (52). TIGR4 clusters 2 and 3 were named PiaABCD and PiuBCDA, respectively (6).
Annotations in GenBank assigned successive numbers to ORF sequences deduced from the DNA sequence data. For R6 the numbers for ORFs range from 1 to 2,046, whereas for TIGR4 the numbers range from 1 to 2,240, and the difference is presumed to be due to pathogenicity islands (6) in the latter organism.
The first five amino acids encoded by each iron transporter ORF was obtained from GenBank and is listed for possible use for verification.
The annotation “SideroBP” indicates siderophore binding protein, as reported in the cited GenBank files. Experimental data reported by Tai et al. (50 ) suggest that the siderophore binding proteins in S. pneumoniae actually bind hemin.
Purification of recombinant GST-RitR.
To construct the GST-RitR expression strain AU410, plasmid pAU410 was introduced by electroporation into E. coli BL21(DE3). As described previously (56), cell extracts from 2 liters of induced AU410 were prepared in 1× phosphate-buffered saline supplemented with 0.1% Triton X-100. The resultant preparation was applied to a 2-ml bed of glutathione (GSH)-conjugated Sepharose 4B resin (Pharmacia) under gravity. GST-RitR was eluted in elution buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 10% glycerol, 10 mM glutathione), and the protein concentration was measured by the Bradford protein assay (Bio-Rad). The final preparation was used at a concentration of 10 mg/ml (189 μM) relative to a BSA standard. The GST-RitR preparation was essentially homogeneous when it was analyzed by polyacrylamide gel electrophoresis (PAGE) (data not shown).
GST-RitR-piuB gel mobility shift.
The piu regulatory region from coordinate −107 to coordinate 110 (217 bp) relative to the transcriptional start site of piuBCDA was amplified by PCR by using primers 13F and 13R. The resultant product was cleaned with a QIAquick PCR purification kit (QIAGEN) and 5′ end labeled in a 25-μl (total volume) mixture with [γ-32P]ATP (specific activity, >6,000 Ci/mmol; 20 μCi) by using 20 U of phage T4 polynucleotide kinase (New England Biolabs). Unincorporated nucleotides were removed by passage through a G50 spin column (Pharmacia). The gel shift reaction mixtures (total volume, 10 μl) contained 1× binding buffer (20 mM HEPES [pH 7.2], 5 mM MgCl2, 1 mM CaCl2, 0.1 mM EDTA, 10% glycerol) supplemented with 10 ng of γ-32P-labeled piuB promoter, 800 ng of poly(dI-dC), and GST-RitR at a series of final concentrations that ranged from 0 to 8 μM. The reaction mixtures were incubated at room temperature for 10 min and loaded onto a 1× TAE (10 mM Tris-acetate [pH 8.0], 1 mM EDTA) nondenaturing 4% polyacrylamide gel, which was run at 16 V/cm (constant voltage) for 1 h. Fractionated RitR-DNA complexes were visualized by autoradiography.
GST-RitR footprint analysis of the piuB promoter region.
PCR primers 13F and 13R were 5′ end labeled in a 25-μl (total volume) mixture with [γ-32P]ATP (specific activity, >6,000 Ci/mmol; 20 μCi) by using 20 U of phage T4 polynucleotide kinase. The labeled primers were cleaned with a QIAquick PCR purification kit (QIAGEN), and unincorporated nucleotides were removed by passage through a G50 spin column (Pharmacia). The piu regulatory region from coordinate −107 to coordinate 110 (217 bp) relative to the transcriptional start site of piuBCDA was amplified by PCR in two reaction mixtures. One mixture contained labeled primer 13F plus unlabeled primer 13R, and the other contained unlabeled primer 13F plus labeled primer 13R. The two PCR products generated were used to produce forward and reverse DNA footprints, respectively. Two picomoles of reverse or forward γ-32P-labeled piu promoter was incubated in 50-μl (total volume) mixtures containing 1× binding buffer supplemented with 1 mM dithiothreitol, 800 ng of dIdC, and a series of GST-RitR concentrations which ranged from 0 to 30 μM. The reaction mixtures were incubated at room temperature for 10 min, after which 0.6 U of DNase RQ1 (Promega), diluted 1:15 from a 1-U/μl stock solution with dilution buffer (Promega) containing 20 mM Tris-HCl (pH 7.0) and 5 mM MgCl2, was added. After 1 min of incubation, the reactions were quenched with stop buffer (200 mM NaCl, 30 mM EDTA, 1% sodium dodecyl sulfate), and the preparation was purified by extraction twice with an equal volume of phenol. The resultant preparation was precipitated with ethanol and resuspended in 4 μl of loading buffer (0.5× Tris-borate-EDTA, 0.05% bromophenol blue, 0.05% xylene cyanol, 8 M urea). Samples were heated at 95°C for 40 s before they were loaded onto a 10% acrylamide denaturing gel in 0.5× Tris-borate-EDTA-urea. A Maxam-Gilbert sequencing ladder was run adjacent to the samples generated from the same PCR fragment that was used in the footprint reactions (46).
Reverse transcriptase mapping of the piuB promoter transcription start site.
The 65-μl primer extension annealing reaction mixture contained 30 μg of RU402 purified total RNA, 1 pmol of primer 13R labeled as described above, and each deoxynucleoside triphosphate at a concentration of 20 mM. The reaction mixture was heated to 95°C for 5 min and cooled to 42°C, after which it was supplemented with 35 μl containing 3 U of SuperScript II reverse transcriptase (Invitrogen), 10 μl of 0.1 M dithiothreitol, and 20 μl of 5× reverse transcriptase buffer (final volume, 100 μl). The resultant reaction mixture was incubated at 42°C for 2 h. A sequencing ladder was synthesized by using a DNA sequencing kit (USB Sequenase, version 2.0) according to the manufacturer's specifications. The 10-μl annealing reaction mixture contained 1 pmol of PCR primer 13R (unlabeled) and 1 pmol of the piuB promoter region. The annealing mixture was brought to 100°C for 5 min and then cooled on ice for 5 min. Sequencing reactions were initiated with addition of 1 μCi of [α-33P]ATP (specific activity, 3,000 Ci/mmol) to each of the four termination reaction mixtures (one containing ddATP, one containing ddGTP, one containing ddCTP, and one containing ddTTP). Electrophoresis and autoradiography were conducted as described above for the footprint analysis. The promoter start site and sequencing ladder reaction mixtures were fractionated in parallel and visualized by autoradiography.
RESULTS
ritR is required for lung infection but not thigh infection.
Throup et al. (53) showed that insertional inactivation (knockout) of ritR led to a loss of the ability of a type 3 encapsulated strain of S. pneumoniae to infect the mouse lung. S. pneumoniae strain R800, a rough avirulent strain (28), was selected for the present studies because both it and its parent strain, R6, have been extensively used in genetic and biochemical investigations. In addition, R800 is easily transformable for recombinant DNA studies (31).
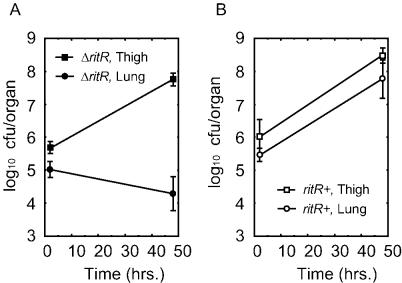
To validate the use of R800 for the present studies, the copy of ritR present in R800 was inactivated by using the insertion-deletion vector pAU402. The resultant strain, RU402, was tested for its ability to infect mouse lungs and thighs as described by Andes and Craig (1). Although R800 is rough and therefore less pathogenic than the type 3 encapsulated strain, its pathogenicity can by enhanced by infecting test mice that have been immunosuppressed by treatment with cyclophosphamide (1). The results (Fig. 1) indicate that inactivation of ritR reduced the ability of S. pneumoniae to proliferate in the mouse lung but did not reduce its ability to infect the thigh. In contrast, R800 (ritR+) cells were able to proliferate in both tissues.
FIG. 1.
Tissue selectivity of S. pneumoniae ΔritR for infectivity. S. pneumoniae RU402 (ΔritR, allele replacement mutant of R800) and R800 (ritR+ R6 derivative) were compared with respect to the ability to infect the mouse lung or thigh. RU402 infected the thigh but not the lung (A), whereas R800 infected both the lung and the thigh (B).
Individual factors that contribute to reduced proliferation of RU402 could include intrinsic effects of reduced ritR expression or host-pathogen interaction factors operative in the mouse lung but not in the thigh. To identify possible intrinsic effects of ritR in S. pneumoniae, comparative transcriptome studies were undertaken, in which RNA from RU402 and R800 were compared by hybridization of the corresponding cDNA to high-density DNA microarrays.
ritR mRNA levels are maximal during exponential growth in vitro.
The timing of ritR expression during the growth cycle of S. pneumoniae indicates when to harvest cells for cDNA microarray probe preparation. Plasmid pEVP3 has been used for insertion-deletion gene expression studies (38). It contains a promoterless E. coli lacZ gene and can be integrated into a selected location by single-crossover site-specific recombination if it is provided with a sequence at the desired integration site. A 353-bp fragment of ritR was ligated into pEVP3 at its BamHI site. Following transformation into E. coli, constructs containing the ritR insert in both the forward and reverse directions were obtained, yielding plasmids pAU401 and pAU402, respectively.
The recipient S. pneumoniae strain, CP1250, is deficient in β-galactosidase activity and can therefore be used in experiments that monitor the activity of E. coli lacZ as a reporter (38). Strain CPU401 was obtained by transformation of the pAU401 construct into CP1250. In addition, the control strain CPU402 was obtained by incorporation of the pAU402 construct into CP1250, integrated in the opposite orientation with respect to pAU401. Integrated constructs were selected on the basis of chloramphenicol resistance, and integration at the expected site in the chromosome was verified by DNA sequencing. Strain CPU401 was grown in BHI liquid medium and sampled, as shown in Fig. 2. LacZ activity in the cell pellets was measured fluorimetrically by using methylumbelliferyl galactoside as the substrate. The results shown in Fig. 2 indicate that LacZ specific activity in the culture increased sharply during early exponential growth until the A600 was 0.3 and remained relatively constant until the onset of the stationary phase at an A600 of about 1.1, whereupon the LacZ specific activity decreased sharply. RNA for microarray studies was therefore harvested in the early exponential phase of growth at an A600 of 0.35 to 0.40, when ritR mRNA and RitR protein would be expected to be maximally abundant.
FIG. 2.
Timing of ritR mRNA synthesis during the growth of S. pneumoniae. A culture of S. pneumoniae CPU401 containing a promoterless E. coli LacZ reporter inserted into the ritR open reading frame was sampled as indicated for determination of turbidity, measured at 600 nm (A600), and for determination of E. coli LacZ activity, measured fluorimetrically at 480 nm (F480) with methylumbelliferyl galactoside as a LacZ substrate. The normalized specific activity of LacZ is plotted as F480/A600. The ritR promoter activity reached the maximal value during exponential growth and decreased as cells entered the stationary phase. To purify RNA for microarray studies, cells were harvested at an A600 of 0.4, as indicated by the arrow, which corresponded to the time at which the ritR mRNA initially reached the maximum level.
Comparative transcriptome analysis of ΔritR and ritR+ strains.
Nothing is known about the regulatory targets of RitR. As an initial approach to this problem, the transcriptional profile of S. pneumoniae RU402 (ΔritR) and that of its parent strain, R800 (ritR+), were compared by using high-density DNA microarrays. Microarrays based on the S. pneumoniae R6 DNA sequence (23) were chemically synthesized as described by Nuwaysir et al. (33). The arrays were probed with cDNA obtained by reverse transcription of total RNA harvested during the early log phase of growth. Array data from three separate experiments were normalized and averaged as described in Materials and Methods.
The statistical program EBarrays, developed by Kendziorski et al. (25), was used to determine if the two gene expression profiles of R800 and RU402 differed from each other in a statistically significant manner. The basis for this determination is the EBarrays-derived value called the probability of differential expression (PDE). In this study a gene was considered differentially expressed if (i) the ΔritR/ritR+ expression profile average log2 ratio was ≥1.0 or ≤−1.0 and (ii) its calculated EBarrys PDE value was ≥0.5. By applying these criteria, 54 genes were found to differ significantly in the ΔritR-ritR+comparison; 17 genes were repressed, and 37 were activated. In this report up-regulated and down-regulated refer to the expression of a given gene in the wild-type R800 strain (ritR+). Results are shown in Table 4. A scatter plot of genes that were differentially expressed in the ΔritR-ritR+ comparison is shown in Fig. 3. Red squares indicate genes which had PDE values of <0.5, and blue squares indicate genes with PDE values of ≥0.5.
TABLE 4.
Comparison of mRNA abundance under wild-type and tet-induced control by ritRa
| Locus | Gene or locus | Function | ΔritR/ritR+
|
ΔritR/<aTc>
|
||
|---|---|---|---|---|---|---|
| log2 | PDE | log2 | PDE | |||
| Activated by ritR+ | ||||||
| spr0336 | RitR | Response regulator | 4.00 | 1.00 | 1.59 | 0.81 |
| spr1620 | ABC-SBP | ABC transporter, substrate-binding protein, sugar transport | 3.31 | 1.00 | 1.04 | 0.28 |
| spr1667 | galT | Galactose-1-phosphate uridylyltransferase | 3.06 | 1.00 | 1.81 | 0.95 |
| spr1097 | nirC | Formate-nitrate transporter | 2.65 | 1.00 | 6.95 | 1.00 |
| spr0686 | rimM | 16S rRNA processing protein RimM | 2.64 | 0.99 | 4.59 | 1.00 |
| spr1430 | dpr | DNA binding, peroxide resistance | 2.51 | 0.98 | 6.21 | 1.00 |
| spr0087 | spr0087 | Hypothetical protein | 2.34 | 0.67 | 1.73 | 0.54 |
| spr0914 | hemH | Ferrochelatase | 2.18 | 0.94 | 4.23 | 1.00 |
| spr1108 | mulY | A/G adenine glycosylase-like | 2.15 | 0.87 | 1.79 | 0.91 |
| spr0750 | ABC-MSP | ABC transporter, ribose/galactose | 2.14 | 0.69 | 3.03 | 1.00 |
| spr0114 | spr0114 | Hypothetical protein | 2.06 | 0.88 | 5.94 | 1.00 |
| spr1201 | spr1201 | Hypothetical protein | 1.91 | 0.52 | 4.62 | 1.00 |
| spr1527 | ABC-SBP | ABC transporter, sugar | 1.89 | 0.74 | 7.87 | 1.00 |
| spr0563 | spr0563 | Hypothetical protein | 1.87 | 0.50 | 6.45 | 1.00 |
| spr0294 | PTS-EII | Phosphotransferase system sugar-specific EII component | 1.84 | 0.67 | 0.41 | 0.06 |
| spr1837 | adhE | Alcohol-acetaldehyde dehydrogenase | 1.80 | 0.64 | 2.56 | 1.00 |
| Repressed by ritR+ | ||||||
| spr1684 | piuB | ABC transporter permease, ferric iron | −8.29 | 1.00 | −4.21 | 1.00 |
| spr1687 | piuA | ABC transporter, substrate-binding ferric iron | −5.88 | 1.00 | −3.55 | 1.00 |
| spr1928 | spr1928 | Hypothetical protein | −4.08 | 1.00 | −3.94 | 1.00 |
| spr1696 | murI | Glutamate racomase | −3.27 | 1.00 | 0.97 | 0.02 |
| spr1814 | RR479 | Response regulator | −3.14 | 1.00 | 3.60 | 1.00 |
| spr0777 | spr0777 | Conserved hypothetical protein | −3.07 | 1.00 | 2.49 | 0.99 |
| spr1198 | Transposase | Degenerate transposase | −2.90 | 1.00 | −0.04 | 0.03 |
| spr0451 | spr0451 | Hypothetical protein | −2.84 | 1.00 | −4.78 | 1.00 |
| spr0502 | spr0502 | Hypothetical protein | −2.81 | 1.00 | 1.21 | 0.25 |
| spr0113 | spr0113 | Hypothetical protein | −2.77 | 1.00 | 2.95 | 1.00 |
| spr0139 | ugd | UDP-glucose dehydrogenase | −2.61 | 1.00 | −2.14 | 1.00 |
| spr0322 | cpsN | dTDP-glucose-4,6-dehydratase | −2.37 | 0.96 | 1.54 | 0.37 |
| spr0893 | spr0893 | Hypothetical protein | −2.35 | 0.93 | 2.27 | 0.96 |
| spr1706 | amiC | ABC transporter, permease, oligopeptide | −2.32 | 0.86 | 1.73 | 0.34 |
| spr0866 | pyrD | Dihydroorotate dehydrogenase | −2.23 | 0.91 | −0.98 | 0.12 |
| spr0597 | rsuA | Ribosomal small-subunit pseudouridine synthase A | −2.23 | 0.96 | 0.32 | 0.05 |
| spr1319 | spr1319 | Hypothetical protein | −2.22 | 0.69 | 0.93 | 0.02 |
| spr1824 | rluD | Ribosomal large-subunit pseudouridine synthase D | −2.20 | 0.94 | −0.50 | 0.05 |
| spr1897 | pstA | ABC transporter, permease, phosphate transport | −2.18 | 0.89 | 0.55 | 0.02 |
| spr0992 | map | Methionine aminopeptidase | −2.15 | 0.81 | 1.74 | 0.34 |
| spr0465 | blpC | Bacteriocin-like peptide, double glycine cleavage type | −2.13 | 0.92 | 1.58 | 0.65 |
| spr1378 | ABC-MSP-tr | ABC transporter, truncation | −2.12 | 0.91 | −2.35 | 1.00 |
| spr0522 | spr0522 | Hypothetical protein | −2.12 | 0.84 | −0.14 | 0.02 |
| spr0698 | spr0698 | Conserved hypothetical protein | −2.12 | 0.86 | 0.10 | 0.02 |
| spr1504 | spr1504 | Hypothetical protein | −2.07 | 0.89 | −2.05 | 1.00 |
| spr1856 | spr1856 | Hypothetical protein | −2.05 | 0.85 | 1.44 | 0.28 |
| spr0042 | IS1167 | Transposase (orf1) | −2.02 | 0.86 | −2.76 | 1.00 |
| spr0363 | recD | Exonuclease V | −2.00 | 0.84 | 0.51 | 0.20 |
| spr1861 | colD | Competence protein | −1.97 | 0.68 | 0.97 | 0.04 |
| spr1816 | spr1816 | Conserved hypothetical protein | −1.96 | 0.58 | 0.87 | 0.02 |
| spr1674 | spr1674 | Conserved hypothetical protein | 1.88 | 0.74 | 1.50 | 0.62 |
| spr0529 | vncS | VncS, histidine kinase | −1.86 | 0.50 | 1.30 | 0.20 |
| spr0870 | dgkA | Diacylglycerol kinase | −1.84 | 0.69 | −1.30 | 0.76 |
| spr1842 | spr1842 | Conserved hypothetical protein | −1.84 | 0.69 | −0.23 | 0.04 |
| spr0757 | parC | Topoisomerase IV subunit A | −1.74 | 0.56 | −1.74 | 0.97 |
| spr1324 | apbE | Thiamine biosynthesis lipoprotein | −1.73 | 0.55 | 5.05 | 1.00 |
| spr1316 | spr1316 | Conserved hypothetical protein | −1.71 | 0.52 | −0.54 | 0.07 |
Array fluorescence signals from three separate ΔritR/ritR+ experiments were averaged and sorted according to their log2 ratios. The probability of differential expression was calculated as described previously (25). Genes that met criteria of a log2 average ratio of ≥1 or ≤−1 and exhibited a PDE of ≥0.5 are shown. Array values based on ritR induction by anhydrotetracycline were calculated by averaging the data from six induced arrays (0, 100, and 300 ng of anhydrotetracycline/ml) for strains RU403 and RU404. The averaged values for the anhydrotetracycline-induced strains RU403 and RU404, <RU403+RU404>, or <aTc> were used to calculate (RU402/<RU403+RU404>) or (ΔritR/<aTc>), i.e., the signal intensity for each gene in the ritR knockout relative to the averaged aTc-induced signal intensity for the same gene. Calculations are shown for comparison with RU402/R800 (ΔritR/ritR+) along with the corresponding PDE values. Other headings are based on the annotation of the R6 sequence (GenBank accession no. NC-003098).
FIG. 3.
Scatter plot analysis of the array data. The normalized average fluorescence signal for RU402 on the y axis is plotted versus the normalized average fluorescence signal for R800 on the x axis. Genes that were significantly differentially expressed (PDE, ≥0.5) appear as off-diagonal blue squares, whereas genes that were differentially expressed and had a PDE of <0.5 are represented by red squares. Genes above the diagonal were repressed in the presence of ritR, whereas genes below the diagonal were activated.
mRNA levels repressed by ritR.
There were the following noteworthy changes in mRNA levels that were repressed by ritR. (i) piuB and piuA (average log2 ratios, −8.29 and −5.28, respectively) exhibited the highest degree of differential expression. The genes encoding PiuB and PiuA form part of a four-gene iron transport operon, and these proteins have been identified as a permease and a ferrophore-binding protein, respectively (5, 6). The two internal genes in the piu operon, piuC and piuD, were not differentially expressed. According to Tai et al. (50), PiuA binds iron in the form of hemin. (ii) The average log2 ratios for the histidine kinase gene vncS and the response regulator gene rr479 were −1.86 and −3.14, respectively. Since VncS was the only histidine kinase which met the PDE significance criterion, its link to RitR was investigated further by Northern blot analysis, as described below. (iii) Ugd (UDP glucose dehydrogenase), which catalyzes the conversion of UDP-d-glucose to UDP-d-glucuronic acid and is required for capsular polysaccharide biosynthesis in S. pneumoniae type 3 (3), was also repressed (average log2 ratio, −2.61). These data suggest that RitR represses iron uptake by limiting the availability of at least two of the four components that comprise the piu iron uptake transport system and, in addition, negatively affects transcript levels of proteins belonging to other functional categories.
mRNA levels activated by ritR.
There were the following noteworthy changes in mRNA levels that were activated by ritR. (i) ritR had the highest average log2 ratio, 4.0, indicating that it had indeed been activated in R800 (ritR+) relative to RU402 (ΔritR). These observations confirmed that ritR had been effectively knocked out in RU402. (ii) There were changes in homologs of Dpr, an iron storage-peroxide resistance protein, and in AdhE, an iron-binding alcohol dehydrogenase, both of which are implicated in H2O2 resistance in E. coli (15, 42) (average log2 ratios, 2.51 and 1.8, respectively). Also activated were (iii) HemH, which is responsible for catalyzing the addition of Fe2+ to the protoporphyrin ring, the final step in hemin synthesis (average log2 ratio, 2.18) (for a review, see reference 34); (iv) MutY, an iron-sulfur cluster binding A/G glycosylase used in DNA A/G mismatch repair (17) (average log2 ratio, 2.15); and (v) the loci spr1620 and spr1527, coding for two sugar-binding proteins (average log2 ratios, 3.31 and 1.89, respectively), and the locus spr0750, coding for a membrane-spanning permease component of a putative ribose/galactose ABC sugar transporter (average log2 ratio, 2.14). These data suggest that ritR is linked to the activation of genes required for the oxidative stress response, heme biosynthesis, and sugar uptake.
Comparative transcriptome analysis of gene expression by ritR under tet control.
To measure changes in mRNA levels as a function of ritR expression, ritR was placed under control of an inducible tet promoter. Anhydrotetracycline was used to activate transcription over a range of concentrations that produced ritR transcript levels that varied from undetectable to maximally expressed. The 353-nucleotide fragment of ritR used previously to construct plasmids pAU401 and pAU402 was cloned into the BamHI site of plasmids pRK01 and pRK02. pRK01 contains a weak low-background tetracycline-inducible promoter, P57, whereas pRK02 contains the stronger P57-opt promoter and can be used to induce transcription levels severalfold compared with those obtainable with pRK01 (47). The resultant pRK01 and pRK02 constructs, designated pAU403 and pAU404, respectively, were integrated into the S. pneumoniae R800 chromosome following selection on TSBA containing erythromycin to obtain strains RU403 and RU404. RU403 and RU404 were exposed during the early log phase to 0, 100, and 300 ng of anhydrotetracycline per ml for 1 h before total RNA was harvested. The resultant RNA preparations were reverse transcribed and hybridized to high-density DNA microarrays as described above.
Due to the variability of differentially expressed genes, possibly as a result of the degree of RitR activation at different cellular concentrations, the data for the six microarrays that were probed with cDNA from tet-induced strains RU403 and RU404 were averaged and compared to the mutant RU402 (ΔritR) probed array data. PDE values were assigned to the averaged and sorted log2 ratios for comparison. Of 290 genes with PDE values of ≥0.5 and log2 average ratios of ≥1 or ≤−1, 245 were found to be activated in R800 (ritR+), and 45 were repressed. A partial list corresponding to the 54 entries (of the 290 genes) that were also present in the RU402-versus-R800 comparison is shown in Table 4. The complete list of 290 entries for the tet induction series can be accessed at http://weisblumlab.pharmacology.wisc.edu.
The results from the array hybridizations based on tet-induced ritR expression paralleled those from the ΔritR-probed arrays. Most PDE values and average log2 ratios of activated genes from both the RU402-versus-R800 and tet-induced ritR array experiments correlated; there were two notable exceptions, spr0294, a phosphotransferase system sugar-specific EII component, and spr1620, a sugar-binding protein-ABC transporter component (Table 4). In contrast, genes that were repressed in the RU402-versus-R800 comparison varied considerably with respect to both the PDE and log2 ratio values of the tet-induced ritR array experiments. Notable exceptions were piuB, piuA, ugd, rr479, and the genes encoding several hypothetical proteins. Interestingly, some genes, such as rr479, were activated in the tet-inducible cDNA-probed arrays but were previously repressed in the RU402-versus-R800 comparison. The difference observed might be explained by the absence of regulation by RitR at the nonphysiological levels attainable in RU403 and RU404 cells carrying the tet-inducible ritR construct.
Linkage between ritR and the pentose phosphate pathway.
The immediate upstream and downstream neighbors of ritR in the S. pneumoniae chromosome are gnd (encoding 6-phosphoguconate dehydrogenase) and cpbF (encoding choline-binding protein F), respectively. Northern blot analysis was used to test for transcriptional linkage in the gnd-ritR-cbpF region. 5′-32P-labeled probes specific for ritR, gnd, and cpbF were used to probe Northern blots of RNA harvested from R800 cells. Results shown in Fig. 4 indicate that the gnd and ritR probes hybridized to a 2.2-kb fragment, whereas the cpbF probe hybridized to a shorter 1-kb fragment. Since 2.2 kb correlates with the predicted length of the combined gnd and ritR ORFs, we inferred that ritR is transcriptionally linked to the upstream gene gnd but not to the downstream gene cpbF, which is transcribed independently. Transcriptional activation of the gnd-ritR gene cluster would therefore up-regulate gnd, which encodes a limiting enzyme of the pentose phosphate pathway.
FIG. 4.
Northern blot analysis of ritR and neighboring genes. The RNA fraction from S. pneumoniae R800 (ritR+) contains a 2.2-kb transcript that hybridizes with either gnd or ritR but not with cbpF probes, indicating that gnd and ritR are cotranscribed independent of the 1-kb cbpF transcript located immediately downstream.
Knocking out ritR raises the question as to whether the gnd transcript remains intact. The array hybridization data showed no significant difference between the levels of gnd-specific cDNA in R800 and RU402 (average log2 ratio, −0.6), suggesting that gnd was not differentially transcribed in the mutant RU402 strain. Moreover, strain CP401, with E. coli lacZ inserted into ritR, has a high level of LacZ activity, suggesting that the full-length gnd-lacZ transcript is present following pEVP3 integration and that gnd is also transcribed. These data suggest that the gnd transcript is present in the RU401 and RU402 ritR mutants.
Northern blot analysis of regulation by ritR.
To verify some of the key microarray observations, DNA probes representing piuB, piuA, dpr, ritR, and vncS were PCR amplified, 5′ end labeled with γ-32P, and analyzed by Northern blot hybridization to fractionated total RNA isolated from RU402 and R800 (Fig. 5A). Also included were RNA samples from the tet-induced RU403 and RU404 strains grown in the presence of 0, 100, or 300 ng of anhydrotetracycline per ml (Fig. 5B). Results shown in Fig. 5A indicate that the level of ritR mRNA in RU402 cells was undetectable, whereas the piuB and piuA mRNA levels were detectable only in RU402 cells; the dpr mRNA levels were reciprocally regulated relative to piuB. Figure 5B confirms that the expression of ritR was induced by anhydrotetracycline compared to wild-type levels and that the piuA mRNA levels decreased as the RitR levels increased. Moreover, Fig. 5B indicates that the vncS mRNA levels paralleled those of induced expression of ritR, despite its PDE value, 0.2 (log2 ratio, 1.3), in our tet-induced ritR array data, indicating a possible linkage between the expression of these two genes. This observation is in contrast to the RU402-versus-R800 array data, where vncS exhibited a log2 ratio of −1.86, indicating repression by RitR. This could have been due to differences in the phosphorylation state of RitR in the two sets of arrays, with RitR insufficiently regulated at higher levels in the tetracycline-induced arrays compared with a wild-type level of regulation in the RU402-versus-R800 array experiments. Consistent with the findings of Brown et al. (5), Fig. 5C shows that piuB and piuA Northern probes both hybridized to a 4-kb fragment, which is the predicted size of the piuBCDA polycistonic mRNA, confirming that these genes are indeed cotranscribed as part of a four-gene operon. This observation contrasts with the apparent absence of detectable differential expression of the two internal genes in the piuBCDA operon, piuC and piuD, in the array studies. Site-specific processing or degradation of the piu polycistronic mRNA or a lack of sufficient cDNA for these two genes may account for this observed inconsistency.
FIG. 5.
Northern blot analysis of S. pneumoniae mRNA. (A) mRNA levels for ritR (lanes 1 and 2), piuB (lanes 3 and 4), piuA (lanes 5 and 6), and dpr (lanes 7 and 8). ritR mRNA levels in S. pneumoniae R800 ritR+ (+) and RU402 ΔritR (Δ) were compared. ritR mRNA is absent in ΔritR strain RU402 (lane 2). Repression of puiB and piuA transcription in R800 and the reciprocal relationship between piuA and piuB with respect to dpr are shown. (B) mRNA levels were measured as described above for panel A, except that RNA samples from S. pneumoniae RU403 (lower-activity tet promoter-driven ritR) and RU404 (higher-activity tet promoter-driven ritR), as a function of added anhydrotetracycline, were used. Note the dependence of ritR mRNA abundance and the corresponding repression of piuA on anhydrotetracycline. VncS, whose function is unknown, was also tested and appeared to be strongly dependent on ritR expression compared to the wild-type (wt) control (R800) with no anhydrotetracycline. Lanes 1, 2, and 3 contained RNA extracted from RU403 cells treated with 0, 100, and 300 ng of anhydrotetracycline per ml. Lanes 4, 5, and 6 contained RNA extracted from RU404 cells treated with 0, 100, and 300 ng of anhydrotetracycline per ml. (C) Northern blot analysis of RNA from S. pneumoniae R800 (ritR+) and RU402 (ΔritR) with probes specific for piuB and piuA. Probes for piuB and piuA were obtained by PCR with primer pairs 8F-8R and 9F-9R, respectively (Table 2). Inactivation of ritR led to overexpression of the piuBCDA operon, as shown in lanes 2 and 3. The 4-kb fragment is the fragment expected for the full-length piuBCDA mRNA. Faster-moving fragments are presumed to represent degradation products of the piuBCDA message. Except for a 300-bp fragment (lane 1), no hybridization was detected in the RNA obtained from R800 cells, indicating the extent of repression of piuBCDA mRNA by ritR. The 300-bp RNA may be a regulatory RNA, and its origin is unknown. kD, kilodaltons.
Streptonigrin susceptibility.
The action of streptonigrin requires the presence of iron (58), and susceptibility to inhibition by streptonigrin can be used as an indirect measurement of intracellular iron concentration (4, 6). Tai et al. (50) reported that PiuA binds hemin with high affinity and hemoglobin at a lower affinity. This observation suggests a possible hemophore-mediated iron uptake role for the Piu transporter. Indeed, if RitR represses the piu operon as the data from the array and Northern blot experiments indicate, increased susceptibility to streptonigrin would be expected in the presence of hemin or hemoglobin in RU402 (ΔritR) due to the derepression of the piu operon.
To address this question, RU402 and R800 cells were grown in iron-depleted medium supplemented with Fe2+, Fe3+, hemin, or hemoglobin, and susceptibility to streptonigrin was tested. Results shown in Fig. 6A and B indicate that there was little difference in susceptibility to streptonigrin between RU402 and R800 in deferrated medium. In contrast, supplementing the deferrated medium with exogenous Fe2+ and with hemin reduced the viability of RU402 relative to that of R800 by 5 and 3 orders of magnitude, respectively. In contrast, supplementation of the medium with Fe3+ or hemoglobin, which S. pneumoniae is also able to use as sources of iron (49), resulted in no observable difference in survival. These data imply that ritR represses the acquisition of hemin and free Fe2+ in vitro and not the acquisition of hemoglobin or Fe3+.
FIG. 6.
Susceptibility to streptonigrin and H2O2 treatment: ritR+ versus ΔritR. (A) S. pneumoniae R800 (ritR+) cells were preincubated in the presence or absence of 50 μM iron, followed by addition of 2.5 μg of streptonigrin/ml. The number of viable cells varied little in response to iron in any of the four forms tested (Fe2+, Fe3+, hemin, and hemoglobin). The data indicate the log percentage of surviving cells compared to the no-iron (No addn) control. Hgb, hemoglobin. (B) The conditions used were the same as those described above for panel A, except that S. pneumoniae RU402 (ΔritR) cells were used. Losses of viability were seen for addition of Fe2+ (5 orders of magnitude) and for addition of hemin (3.5 orders of magnitude), whereas Fe3+ or hemoglobin had no detectable effect. (C) Effect of hydrogen peroxide on survival of S. pneumoniae. The number of surviving CFU in the culture was determined for S. pneumoniae R800 (ritR+) or RU402 (ΔritR) challenged with 40 mM H2O2 for various times.
Hydrogen peroxide susceptibility.
Dpr, a conserved iron storage protein which confers H2O2 resistance, and AdhE, an iron-binding alcohol dehydrogenase, have both been implicated in adaptation to H2O2 toxicity (2, 15, 42). Based on these reports, an expected consequence of insertional inactivation of ritR would be elevated susceptibility to H2O2 owing to the decreased levels of dpr and adhE mRNA, as observed in the microarray studies (Table 4 and Fig. 3). In addition, iron overload due to the derepression of the piu operon could also contribute to an increase in H2O2 toxicity by increasing the intracellular free iron concentration and the potential for synthesis of ROIs.
To address this question, S. pneumoniae R800 (ritR+) and RU402 (ΔritR) cells were challenged with 40 mM H2O2 as described by Tseng et al. (54). Each treated culture was sampled after exposure to H2O2, as indicated in Fig. 6C, and surviving cells were counted by plating on TSBA. The resultant killing curves, shown in Fig. 6C, indicate that R800 cells survived 10 min longer than RU402 cells under the conditions that were used. In contrast, use of paraquat at a concentration of 60 mM resulted in no observable differences (data not shown). Consistent with this observation, the microarray data showed that S. pneumoniae Fe and Mn superoxide dismutases were not differentially expressed in either set of array experiments (data not shown), indicating that ritR affects genes that either reduce Fe-catalyzed synthesis of ROIs or remediate their genotoxic effects but do not affect gene regulation or detoxification associated with superoxide.
GST-RitR-piu promoter reverse transcriptase mapping and gel mobility shift.
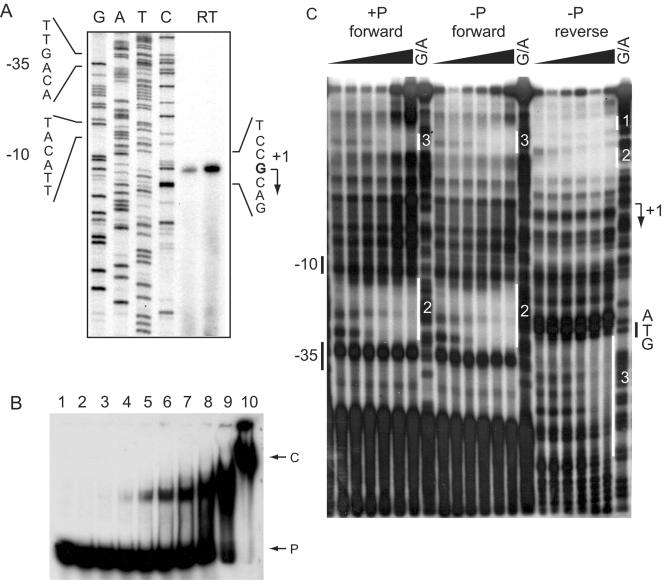
Total RNA from RU402 enriched for piu mRNA (due to derepression of piu) was used to determine the transcription start site for the piu operon by reverse transcriptase mapping. 5′-32P-labeled DNA oligomer 13R, complementary to the noncoding strand within the piuB ORF, served as the primer. The results of the primer extension (Fig. 7) indicate that transcription in the piuBCDA promoter region is initiated at coordinate −42 (G) relative to the ATG of the piuB ORF. The ability of RitR to shift the mobility of the piuB promoter region was tested by using a 5′-32P-labeled 217-bp DNA fragment upstream of piuB between coordinates −107 and 110 relative to the transcriptional start site of piuBCDA. The labeled DNA was incubated with GST-RitR, and the resultant complexes were analyzed by PAGE. A GST-RitR concentration of 60 nM was enough to show evidence of retardation (Fig. 7B). Preincubation of GST-RitR with 50 mM acetyl phosphate or phosphoramidate did not alter the affinity of GST-RitR for the piu promoter (data not shown). These data suggested that GST-RitR bound specifically to the piu promoter.
FIG. 7.
Characterization of the piu promoter transcription start site, gel retardation analysis, and RitR footprint. (A) The transcription start site for the piuBCDA message was determined by reverse transcriptase (RT) mapping by using total RNA extracted from S. pneumoniae RU402 (ΔritR). The results indicate that the piuB message starts with G. (B) Gel retardation analysis of the piuB promoter. A 217-nucleotide DNA sequence containing the piu promoter region in 32P-labeled form was incubated with different concentrations of GST-RitR, and the resultant complexes were fractionated by PAGE. Lanes 1 to 10 contained 0, 0.030, 0.060, 0.125, 0.25, 0.50, 1.0, 2.0, 4.0, and 8.0 μM GST-RitR, respectively. The first appearance of retardation occurred at 0.060 μM GST-RitR (lane 3). P, probe; C, complex. (C) Footprint analysis of RitR bound to the piuB promoter. Two preparations of the piuB promoter region were obtained by PCR, each labeled with 32P at one 5′ end. GST-RitR was bound to each of the two PCR products. Following incubation with DNase I and PAGE fractionation of the resultant fragments, the footprints were visualized by autoradiography. +P, RitR preincubated with 50 mM acetyl phosphate; −P, no acetyl phosphate added; G/A, G/A ladder. Three distinct binding sites for RitR are labeled. The relationship of the RitR DNA-binding regions to the piu promoter DNA sequence is shown in Fig. 8A.
DNA footprint analysis of the piuBCDA promoter by GST-RitR.
The site(s) for the binding of RitR upstream of piuB was characterized by a DNA footprint analysis in which the forward and reverse 217-bp DNA fragments, corresponding to coordinates −107 and 110 relative to the transcriptional start site of piuBCDA, were incubated with a series of GST-RitR concentrations. The resultant complexes were treated with DNase I and analyzed by PAGE and autoradiography. Results shown in Fig. 7C and summarized in Fig. 8A indicate that GST-RitR binds selectively to three sites in the piu promoter regulatory region. One binding site is located upstream of the −35 box, the second binding site is located between the −35 and −10 regulatory elements, and the third binding site is located just within the piuB ORF. The three binding sites can be aligned, as shown in Fig. 8B, to give the AT-rich consensus sequence (A/T)NATTAN(A/T)(A/T)(A/T)R(A/T)YRR, centered on an invariant ATTA core motif. The pattern of GST-RitR binding together with data from the DNA microarray and the Northern blot analysis suggests that RitR acts by binding to the promoter region of piuB to repress transcription of the piu operon. No discernible differences were noted if GST-RitR was preincubated with acetyl phosphate.
FIG. 8.
Annotated DNA sequence of the piuB promoter region. (A) DNA sequence of the piuB promoter region with the transcriptional regulatory elements annotated as follows: −35, −10, +1 (start of transcription), RBS (ribosome-binding site for translation initiation), and the first 14 amino acids of PiuB. Three binding sites for RitR and their relationship to the piu promoter are underlined and labeled #1, #2, and #3. (B) Alignment of the three binding site sequences, showing the resultant consensus that was used as a search key to locate additional similar sequences in the S. pneumoniae genome.
DISCUSSION
Iron uptake regulation in S. pneumoniae by ritR.
Iron uptake and its regulation are critical factors in bacterial pathogenesis (2, 7, 19). A notable exception is the case of Borrelia burgdorferi, which may not require iron (41). The major mechanism of iron uptake regulation in prokaryotes is based on transcriptional control mediated by Fur or DtxR family iron-binding transcriptional regulators (19). In the present studies, by using a combination of genetic and biochemical techniques, we described a unique form of iron transporter regulation occurring in S. pneumoniae, in which an orphan OmpR family response regulator, RitR, acts as a repressor of mRNA synthesis specific for iron transporter components. The studies reported here were based on an examination of the piu gene cluster, one of three gene clusters in the S. pneumoniae genome identified by Brown et al. (6), each of which specifies the four components of an S. pneumoniae iron transporter. piuB and piuA, part of the piuBCDA four-gene cluster, encode a permease subunit and hemin periplasmic binding protein, respectively. One of the most striking observations in this report came from our DNA microarray studies in which elevated piuB and piuA mRNA levels in RU402 (ΔritR) indicated that one role of RitR is to repress transcription of these genes. The finding that piuC and piuD were not differentially expressed could reflect selective degradation of the piuBCDA message at internal sites in the transcript. Interestingly, a similar effect was attributed to differential instability of the four-gene transcript in the Vibrio anguillarum fat Fe transporter operon, which the piu transporter closely resembles (10, 57). In addition, further experiments showed that RitR binds physically at three sites upstream of piuB to a specific AT-rich sequence. Collectively, our data suggest a role in iron regulation for RitR in S. pneumoniae.
Uptake of iron in the form of hemin.
Tai et al. (50) reported that PiuB binds hemin with a high affinity in vitro, suggesting that this is the form in which PiuBCDA acquires and possibly transports iron. Indeed, compared with R800, RU402 (ΔritR) exhibited 5 orders of magnitude difference for Fe2+ and 3 orders of magnitude difference for hemin (but not hemoglobin or Fe3+) with respect to streptonigrin susceptibility. Interestingly, a heme oxygenase determinant does not seem to be present in the pneumococcal genome (52). Given this observation, we can only assume that iron somehow enters the cell in the free ionic form until sufficient data become available to pinpoint the precise mechanism(s) of iron transport. In addition, we cannot rule out the possibility that the repression of iron storage protein genes (e.g., dpr [log2 ratios of 2.5 and 6.2 in the R800-versus-RU402 and tetracycline-induced arrays, respectively] [Table 4]) could affect intracellular iron concentrations and thus streptonigrin susceptibility. Isotopic Fe2+ uptake studies are needed to resolve these issues.
The existence of two additional iron transporter gene clusters, namely, pit and pia, may provide other options for iron acquisition and for interpreting the streptonigrin effect shown in Fig. 6A and B (5, 6). Interestingly, a RitR binding site has also been identified upstream of the pia gene cluster between its predicted −35 and −10 promoter elements, suggesting an additional iron-regulatory role for RitR (data not shown). The additive effect of piaA and piuA in pathogenesis is consistent with the observations of Brown et al. (8), who noted that mice immunized with recombinant PiuA and/or PiaA were protected against a systemic challenge.
Regulation of RitR.
The present studies suggest that RitR represses the piu iron transporter, but the precise mechanism by which RitR itself is regulated is unknown. Four possible mechanisms of regulation are (i) control of the activation of RitR by phosphorylation, (ii) control of the amount of RitR that is synthesized, (iii) interaction of RitR with another protein (e.g., phosphatase), and (iv) modulation of RitR activity by noncovalent ligand-protein interaction. Neither a cognate kinase for RitR nor a mechanism by which S. pneumoniae senses iron has been found. Additionally, at this time it is not clear whether a cognate kinase even exists or if phosphorylation plays any role in the regulatory activity of RitR. Indeed, we were unable to alter the affinity of RitR for the piu promoter when we attempted to phosphorylate it by addition of small phosphate donors (Fig. 7). Even though RitR appears to contain a conserved aspartate residue, these data bring into question whether this response regulator has evolved another mechanism of regulation (e.g., growth rate-dependent regulation); however, a role for phosphorylation of RitR cannot be ruled out at this time. Interestingly, C. diphtheriae has a TCST system, ChrAS, that has been reported to sense heme through the histidine kinase ChrS and to initiate transcription of hmnO (encoding heme oxygenase) following activation of the response regulator ChrA (45). It is tempting to speculate that the orphan RitR works in a fashion similar to ChrA through 1 of the 13 histidine kinases encoded in the S. pneumoniae genome, sensing iron-hemin in the host and signaling for derepression of the piu transporter.
Coregulation of gnd-ritR and H2O2 toxicity.
gnd and ritR are cotranscribed as a single 2-kb mRNA (Fig. 4). Given this observation, elevated expression of gnd-ritR would be expected to repress iron uptake while stimulating expression of the pentose phosphate pathway, which is responsible for (i) generating NADPH, which provides reducing power for detoxification of reactive oxygen intermediates, and (ii) synthesizing nucleotides needed for repair of Fe-catalyzed damage of DNA (46, 59).
S. pneumoniae is able to produce high levels of H2O2, which can reach millimolar concentrations. This effect has been shown to aid the pneumococcus in colonization of the nasopharynx by inhibiting the growth of competitors, such as Haemophilus influenzae (36). A more direct use of H2O2 as a weapon by S. pneumoniae is suggested by its cytotoxic effect on epithelial cells (14). The observations reported in our studies are relevant to the findings reported by Pericone et al. (37), who suggested that catalase-deficient S. pneumoniae is able to survive toxic concentrations of H2O2 in the presence of intracellular iron concentrations exceeding the concentration in E. coli. Possible contributing factors that were suggested included (i) an enhanced ability to repair DNA damaged by ROIs and (ii) the presence of an H2O2 resistance protein, such as Dpr, that sequesters iron. Both of these mechanisms are consistent with our experimental findings.
The enhanced ability to repair DNA damaged by ROIs could be mediated by gnd-ritR activation. The pentose phosphate could provide the cell with NADPH for detoxification of Fe-catalyzed synthesis of ROIs and synthesis of nucleotides for excision repair, while RitR could contribute to a reduction in Fe-catalyzed ROI synthesis by reducing iron uptake during exponential growth under high-iron conditions. In support of this interpretation, the resistance of Deinococcus radiodurans to UV light and H2O2 was shown by Zhang et al. to depend on an intact pentose phosphate pathway (59). In a comparison of a zwf+ (Zwischenferment, glucose-6-phosphate dehydrogenase) strain and a Δzwf strain of D. radiodurans, Zhang et al. noted 1-order-of-magnitude reduced survival of the Δzwf mutant in 0.03% (10 mM) H2O2. In contrast, only a minimal difference was seen in a comparison between an fda+ (aldolase) strain and a Δfda strain in which the Embden-Meyerhoff pathway was interrupted. Moreover, our array data indicate that mutY RNA levels are activated in the presence of RitR. The S. pneumoniae mutY gene encodes an A/G glycosylase responsible for removal of 7,8-dihydro-8-oxyoguanine (44), a major product of oxidative damage to DNA (18, 29). Collectively, these data support our conjecture that S. pneumoniae could possess a greater ability to repair DNA than E. coli possesses that enables it to survive in the presence of high concentrations of H2O2. Experiments to test whether the pentose phosphate pathway plays a role in DNA repair in S. pneumoniae similar to the role that it plays in D. radiodurans are currently in progress.
The second H2O2 resistance factor suggested by Pericone et al. (37) is the protection of DNA by Dpr, a protein that sequesters intracellular Fe2+ and prevents H2O2 toxicity. Indeed, our array data show that there is a marked increase during exponential growth in the mRNA levels of the putative H2O2 resistance orthologs adhE, encoding an iron-binding alcohol dehydrogenase, and dpr. This observation indicates a possible role for these genes in S. pneumoniae so that it can avoid DNA damage caused by interactions with Fe-catalyzed ROIs. Interestingly, neither Pericone et al. (37) nor workers in our lab (data not shown) could obtain a dpr deletion mutant, suggesting that the Dpr protein is required for viability of S. pneumoniae. Concomitant with repression of iron transporter synthesis by RitR, elevated dpr expression would be consistent with a model in which Dpr sequesters iron that enters the cell despite repression of iron transporter synthesis. However, it is not clear at this time whether RitR directly regulates dpr or adhE or whether their activation is an indirect effect of excess iron entering the cell in RU402 through another signaling system.
Sugar transport-ugd regulation by ritR.
Three components belonging to three separate sugar transport systems were observed to be strongly up-regulated in the presence of RitR during log-phase growth. These components included two sugar-binding proteins and a permease belonging to a putative ribose/galactose sugar transporter (Table 4). Interestingly, d-ribose can be metabolized only via the pentose phosphate pathway (46), which is directly associated with ritR because this gene is cotranscribed with gnd. Additionally, ugd was strongly down-regulated in the presence of ritR. In Salmonella enterica serovar Typhimurium, Ugd is regulated directly by three different TCST systems: PmrA/B, which senses Fe3+; PhoP/Q, which senses low concentrations of Mg2+; and RcsC/B/YojN, which senses a variety of signals, including osmolarity and temperature (32). Ugd catalyzes the conversion of UDP-d-glucose to UDP-d-glucuronic acid, a necessary step in the synthesis of capsular polysaccharides in S. pneumoniae type 3 (3), and this process has been shown to be negatively regulated by tyrosine kinase (CpsD)-mediated phosphorylation (30). A possible interpretation of these observed regulatory changes is that while up-regulation of RitR can potentially activate the pentose phosphate pathway through gnd, it can also regulate sugar uptake and provide the pentose phosphate pathway with substrates while repressing alternative pathways for sugar utilization (e.g., capsule biosynthesis through ugd). Interestingly, a RitR consensus binding sequence is present in the promoter region of ugd located 151 bp from upstream of its start codon, suggesting a direct role for RitR regulation of this gene.
Compared to other organisms, little is known about iron homeostasis and oxidative stress regulation in S. pneumoniae. In this report we show that RitR represses the piu iron uptake operon and is linked to the expression of genes involved in the oxidative stress response. In addition, we show that ritR is cotranscribed with gnd, which encodes a critical enzyme of the pentose phosphate pathway that produces nucleotides and reducing power for the cell. In the absence of an identified cognate kinase, it will be of interest to learn what factors regulate RitR. At this time we can only point to the linkage of ritR to gnd.
Acknowledgments
This research was supported by NIH research grants K08 404/06 and AI01767-01A1 to David R. Andes. DNA microarrays were constructed by NimbleGen Systems Inc., Madison, Wis., with support from SBIR grant 5R44HG002193-03 from the NIH, which also supported Jeremy D. Glasner.
Tyson Park provided expert technical assistance. For numerous helpful discussions we thank the staff of the University of Wisconsin Gene Expression Center, including Sandra Splinter Bon-Durant, John Luecke, and Nelson Wayne Davis. We also thank Donald Morrison for helpful discussions and for plasmid pEVP3 and M. Stieger for plasmids pRK01 and pRK02. We thank Christina Kendziorski for the use of her program EBarrays to identify differentially expressed genes.
REFERENCES
- 1.Andes, D., and W. A. Craig. 2002. Pharmacodynamics of the new fluoroquinolone gatifloxacin in murine thigh and lung infection models. Antimicrob. Agents Chemother. 46:1665-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 3.Arrecubieta, C., R. Lopez, and E. Garcia. 1994. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J. Bacteriol. 176:6375-6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolzan, A. D., and M. S. Bianchi. 2001. Genotoxicity of streptonigrin: a review. Mutat. Res. 488:25-37. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. S., S. M. Gilliland, J. Ruiz-Albert, and D. W. Holden. 2002. Characterization of pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect. Immun. 70:4389-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, J. S., and D. W. Holden. 2002. Iron acquisition by Gram-positive bacterial pathogens. Microbes Infect. 4:1149-1156. [DOI] [PubMed] [Google Scholar]
- 8.Brown J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullen, J. J. 1981. The significance of iron in infection. Rev. Infect. Dis. 3:1127-1138. [DOI] [PubMed] [Google Scholar]
- 10.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885-18891. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, J. K., and J. I. Rood. 2000. Glutamate residues in the putative transmembrane region are required for the function of the VirS sensor histidine kinase from Clostridium perfringens. Microbiology 146:517-525. [DOI] [PubMed] [Google Scholar]
- 12.Cvitkovitch, D. G., Y. H. Li, and R. P. Ellen. 2003. Quorum sensing and biofilm formation in streptococcal infections. J. Clin. Investig. 12:1626-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Saizieu, A., U. Certa, J. Warrington, C. Gray, W. Keck, and J. Mous. 1998. Bacterial transcript imaging by hybridization of total RNA to oligonucleotide arrays. Nat. Biotechnol. 16:45-48. [DOI] [PubMed] [Google Scholar]
- 14.Duane, P. G., J. B. Rubins, H. R. Weisel, and E. N. Janoff. 1993. Identification of hydrogen peroxide as a Streptococcus pneumoniae toxin for rat alveolar epithelial cells. Infect. Immun. 61:4392-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echave, P., J. Tamarit, E. Cabiscol, and J. Ros. 2003. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278:30193-30198. [DOI] [PubMed] [Google Scholar]
- 16.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fromme, J. C., A. Banerjee, S. J. Huang, and G. L. Verdine. 2004. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature 427:652-656. [DOI] [PubMed] [Google Scholar]
- 18.Grollman, A. P., and M. Moriya. 1993. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 9:246-249. [DOI] [PubMed] [Google Scholar]
- 19.Hantke, K. 2002. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 20.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoaglin, D. C., R. Mosteller, and J. W. Tukey. 2000. Understanding robust and exploratory data analysis. John Wiley and Sons, New York, N.Y.
- 22.Hoch, J. A., and T. J. Silhavy (ed.) 1995. Two-component signal transduction. American Society for Microbiology, Washington, D.C.
- 23.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D.-J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P.-M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imlay, J. A. 2003. Pathways of oxidative damage. Annu. Rev. Microbiol. 57:395-418. [DOI] [PubMed] [Google Scholar]
- 25.Kendziorski, C. M., M. A. Newton, H. Lan, and M. N. Gould. 2003. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 22:3899-3914. [DOI] [PubMed] [Google Scholar]
- 26.Lange R, C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, K. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 27.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 28.Lefevre, J. C., J. P. Claverys, and A. M. Sicard. 1979. Donor deoxyribonucleic acid length and marker effect in pneumococcal transformation. J. Bacteriol. 138:80-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michaels, M. L., J. Tchou, A. P. Grollman, and J. H. Miller. 1992. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry 31:10964-10968. [DOI] [PubMed] [Google Scholar]
- 30.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 35:1431-1442. [DOI] [PubMed] [Google Scholar]
- 31.Morrison, D. A. 1997. Streptococcal competence for genetic transformation: regulation by peptide pheromones. Microb. Drug Resist. 3:27-37. [DOI] [PubMed] [Google Scholar]
- 32.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 33.Nuwaysir, E. F., W. Huang, T. J. Albert, J. Singh, K. Nuwaysir, A. Pitas, T. Richmond, T. Gorski, J. P. Berg, J. Ballin, M. McCormick, J. Norton, T. Pollock, T. Sumwalt, L. Butcher, D. Porter, M. Molla, C. Hall, F. Blattner, M. R. Sussman, R. L. Wallace, F. Cerrina, and R. D. Green. 2002. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 12:1749-1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panek, H., and M. R. O'Brian. 2002. A whole genome view of prokaryotic haem biosynthesis. Microbiology 148:2273-2282. [DOI] [PubMed] [Google Scholar]
- 35.Panthel, K., P. Dietz, R. Haas, and D. Beier. 2003. Two-component systems of Helicobacter pylori contribute to virulence in a mouse infection model. Infect. Immun. 71:5381-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pericone, C. D., K. Overweg, P. W. Hermans, and J. N. Weiser. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 68:3990-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pericone, C. D., S. Park, J. A. Imlay, and J. N. Weiser. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the Fenton reaction. J. Bacteriol. 185:6815-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pestova, E. V., and D. A. Morrison. 1998. Isolation and characterization of three Streptococcus pneumoniae transformation-specific loci by use of a lacZ reporter insertion vector. J. Bacteriol. 180:2701-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 40.Posey, J. E., J. M. Hardham, S. J. Norris, and F. C. Gherardini. 1999. Characterization of a manganese dependent regulatory protein, TroR, from Treponema pallidum. Proc. Natl. Acad. Sci. USA 96:10887-10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posey, J. E., and F. C. Gherardini. 2000. Lack of a role for iron in the Lyme disease pathogen. Science 288:1651-1653. [DOI] [PubMed] [Google Scholar]
- 42.Pulliainen, A. T., S. Haataja, S. Kahkonen, and J. Finne. 2003. Molecular basis of H2O2 resistance mediated by streptococcal Dpr. Demonstration of the functional involvement of the putative ferroxidase center by site-directed mutagenesis in Streptococcus suis. J. Biol. Chem. 278:7996-8005. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Samrakandi, M. M., and F. Pasta. 2000. Hyperrecombination in Streptococcus pneumoniae depends on an atypical mutY homologue. J. Bacteriol. 182:3353-3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitt, M. P. 1999. Identification of a two-component signal transduction system from Corynebacterium diphtheriae that activates gene expression in response to the presence of heme and hemoglobin. J. Bacteriol. 181:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sprenger, G. A. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324-330. [DOI] [PubMed] [Google Scholar]
- 47.Stieger, M., B. Wohlgensinger, M. Kamber, R. Lutz, and K. Keck. 1999. Integrational plasmids for the tetracycline-regulated expression of genes in Streptococcus pneumoniae. Gene 226:243-251. [DOI] [PubMed] [Google Scholar]
- 48.Stock, A. M., V. L., Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 49.Tai, S. S., C. J. Lee, and R. E. Winter. 1993. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun. 61:5401-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tai, S. S., C. Yu, and J. K. Lee. 2003. A solute binding protein of Streptococcus pneumoniae iron transport. FEMS Microbiol. Lett. 220:303-308. [DOI] [PubMed] [Google Scholar]
- 51.Tao, X., N. Schiering, H. Y. Zeng, D. Ringe, and J. R. Murphy. 1994. Iron, DtxR, and the regulation of diphtheria toxin expression. Mol. Microbiol. 14:191-197. [DOI] [PubMed] [Google Scholar]
- 52.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2000. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 53.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 54.Tseng, H. J., A. G. McEwan, J. C. Paton, and M. P. Jennings. 2002. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect. Immun. 70:1635-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulijasz, A. T., A. Grenader, and B. Weisblum. 1996. A vancomycin-inducible lacZ reporter system in Bacillus subtilis: induction by antibiotics that inhibit cell wall synthesis and by lysozyme. J. Bacteriol. 178:6305-6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ulijasz, A. T., B. K. Kay, and B. Weisblum. 2000. Peptide analogues of the VanS catalytic center inhibit VanR binding to its cognate promoter. Biochemistry 39:11417-11424. [DOI] [PubMed] [Google Scholar]
- 57.Wertheimer, A. M., W. Verweij, Q. Chen, L. M. Crosa, M. Nagasawa, M. E. Tolmasky, L. A. Actis, and J. H. Crosa. 1999. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect. Immun. 67:6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeowell, H. N., and J. R. White. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob. Agents Chemother. 22:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y. M., J. K. Liu, and T. Y. Wong. 2003. The DNA excision repair system of the highly radio-resistant bacterium Deinococcus radiodurans is facilitated by the pentose phosphate pathway. Mol. Microbiol. 48:1317-1323. [DOI] [PubMed] [Google Scholar]