Abstract
Objective
The aim of this study was to identify a transcriptomic signature that could be used to classify subjects with autism spectrum disorder (ASD) compared to controls on the basis of blood gene expression profiles. The gene expression profiles could ultimately be used as diagnostic biomarkers for ASD.
Methods
We used the published microarray data (GSE26415) from the Gene Expression Omnibus database, which included 21 young adults with ASD and 21 age- and sex-matched unaffected controls. Nineteen differentially expressed probes were identified from a training dataset (n=26, 13 ASD cases and 13 controls) using the limma package in R language (adjusted p value <0.05) and were further analyzed in a test dataset (n=16, 8 ASD cases and 8 controls) using machine learning algorithms.
Results
Hierarchical cluster analysis showed that subjects with ASD were relatively well-discriminated from controls. Based on the support vector machine and K-nearest neighbors analysis, validation of 19-DE probes with a test dataset resulted in an overall class prediction accuracy of 93.8% as well as a sensitivity and specificity of 100% and 87.5%, respectively.
Conclusion
The results of our exploratory study suggest that the gene expression profiles identified from the peripheral blood samples of young adults with ASD can be used to identify a biological signature for ASD. Further study using a larger cohort and more homogeneous datasets is required to improve the diagnostic accuracy.
Keywords: Autism spectrum disorder, Blood, Microarray analysis, Transcriptome, Machine learning, Decision support techniques
INTRODUCTION
Autism spectrum disorders (ASDs) are devastating neurodevelopmental disorders characterized by deficits in social communication and interaction across multiple contexts as well as restricted, repetitive patterns of interests and behavior. The Centers for Disease Control recently presented that the prevalence of ASD has risen to approximately 1 in 68, and most children are not diagnosed with ASD until after 4 years of age in the United States.1) Because early intensive behavioral and developmental interventions for toddlers and children with autism could improve outcomes,2) there is a scientific need for reliable diagnostic ASD biomarkers that are expressed early in life. Such markers could have a significant impact on diagnosis and treatment.
Although the complex etiologies of ASD are poorly understood, the high heritability of ASD is supported by high concordance rates (from 36% to 95%) in monozygotic twins and higher recurrence risks of 11% and 19% with single-sibling involvement.3–5) Rapid advances in clinical genetic testing technology have increased the diagnostic yield from about 10% a few years ago to about 30%.6) However, because many of these genetic variants show incomplete penetrance and variable phenotypic expression,7) the use of gene expression signature bio-markers may be informative and provide the best model for identifying ASD cases.
In particular, four studies have investigated blood-derived gene expression signatures to differentiate between ASD individuals (toddlers and children) and unaffected controls.8–11) These studies focused on individuals with a mean age of 2.2 to 9.6 years who were at risk for ASD, and there were relatively high predictive accuracies (between 68% and 91%). To date, no study has demonstrated diagnostic prediction using blood-derived gene expression signatures in adult subjects with ASD. Accordingly, whether the gene expression profiles of adult individuals offer information about the ASD risk remains a critical question.
The aim of this study is to apply a transcriptomic approach to identify a gene expression signature with promising performance in the diagnostic prediction of young adults with ASD. Here, we used a published ASD microarray dataset to test the hypothesis. These methods provide researchers with the opportunity to test hypotheses without performing time-consuming, labor-intensive bench work.
METHODS
Acquisition of the Microarray Data
A publicly available microarray dataset (GSE26415) was downloaded from the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) database,12) which was deposited by Kuwano et al.13) The original data included 21 samples of peripheral leukocytes obtained from young adults with ASD and age- and sex-matched controls as well as from healthy women with children with ASD and matched controls. The pre-existing clinical diagnoses of ASD were made by experienced child psychiatrist and developmental pediatrician according to the Diagnostic and Statistical Manual of Mental Disorders, 4th edition, text revision (DSM-IV-TR). In order to corroborate the ASD diagnosis, the Japanese version of the Autism Spectrum Quotient was completed.13) The platform information was GPL6480 (Agilent-014850 Whole Human Genome Microarray 4x44K G4112F). In this study, we utilized 42 microarrays from subjects with ASD (n=21) and their matched controls (n=21) for further analyses. The demographic and clinical characteristics of the study subjects are summarized in Table 1.
Table 1.
Demographic and clinical characteristics of study subjects
| Characteristic | ASD (n=21) | Control (n=21) |
|---|---|---|
| Demographic | ||
| Gender, male/female | 17/4 | 17/4 |
| Age (yr) | 26.7 (5.5) | 27.0 (5.5) |
| Clinical | ||
| AQ | 30.2 (5.1) | NA |
| WAIS | ||
| VIQ | 96.2 (19.9) | NA |
| PIQ | 87.5 (21.6) | NA |
| FIQ | 91.9 (21.6) | NA |
Values are presented as number only or mean (standard deviation).
ASD, autism spectrum disorder; AQ, autism spectrum quotient; WAIS, Wechsler Adult Intelligence Scale; IQ, intelligence quotients; VIQ, verbal IQ; PIQ, performance IQ; FIQ, full IQ; NA, not applied.
Data Preprocessing and Selection of Differentially Expressed Genes
The raw data in .CEL format were primarily processed using R language (http://www.r-project.org/)14) “limma” package.15) The datasets were imported in R using the “read.maimages” function; the “normexp” function was used for background correction. The adjusted data were transformed with a logarithm for normalization using the quantile method (Supplementary Fig. 1). Filtering was further achieved by building a criterion in which the 95th percentile of the negative probe on each array was set as a standard point of brightness. The control and low expression probes were filtered out when the probes at one-third of the total arrays were 10% less bright than the standard point. The “avereps” function was used to average the replicate spots on each array. Differentially expressed (DE) probes were identified using the moderated t-test from the limma package. p values were adjusted for multiple testing with the Bonferroni correction, and probes were called significant when the adjusted p value was <0.05.
Development of a Prediction Model Using a Machine Learning Algorithm
We applied machine learning to develop a prediction model that used DE probes extracted from the training set, differentiating between individuals with ASD and controls in the test set. Our strategy included two main types of machine learning, unsupervised and supervised learning.
For unsupervised learning, we adopted hierarchical cluster analysis using complete linkage and the Euclidean distance. Cluster analysis and visualization were performed using the “heatmap.2” function in the “gplots” package16) in R.
For supervised learning, we used three different machine learning algorithms, such as the support vector machine (SVM),17) K-nearest neighbors (KNN)18) and linear discriminant analysis (LDA).19) We performed prediction analysis in the subsequent sequential steps. Using the “set.seed” function in R, we randomly divided our data (n=42) into a training dataset (13 ASD and 13 control subjects) and test dataset (8 ASD and 8 control subjects). Each algorithm was trained on the training dataset of 26 randomly selected samples, which were labeled with DE probes. Eight ASD and 8 control subjects in the test data-set were validated. All supervised machine learning analyses were performed using the “MLinterfaces” packages20) in R language.
Supplementary Figure 2 briefly describes the study design. The protocol of this study was reviewed and approved by the institutional review board of Hanyang University Hospital (HYUH IRB-2015-05-008).
RESULTS
Altered Gene Expression Profiling between the ASD and Control Groups
In comparing microarray data for the subjects with ASD (n=13) with those of unaffected controls (n=13) in the training dataset, a total of 19 DE probes were identified (adjusted p value <0.05), including 6 up-regulated probes and 13 down-regulated probes (Supplementary Fig. 3). Among the 19 probes, 15 were annotated as gene symbols using the Bioconductor “hgug4112a.db” package.21) Ten of these genes (or loci) had previously reported associations with ASD (Table 2).
Table 2.
Nineteen probes significantly dysregulated* in the ASD training sample compared with the unaffected control’s training sample
| Probe ID | Gene symbol | Gene name | Location | logFC | p value | Adjusted p value† | Evidence for association with ASD‡ | |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Type | Number of reports | |||||||
| A_32_P9963 | HSF2 | Heat shock transcription factor 2 | 6q22.31 | −0.5288 | 3.99E-08 | 0.0007 | Deletion-duplication of 6q22.31 | 20 |
| A_24_P391104 | RFX1 | Regulatory factor X, 1 (influences HLA class II expression) | 19p13.1 | 0.5379 | 1.16E-07 | 0.0021 | Deletion of 19p13.13-p13.11 | 2 |
| A_23_P214037 | NPM1 | Nucleophosmin (nucleolar phosphoproein B23, numatrin) | 5q35.1 | −0.5139 | 4.34E-07 | 0.0079 | ||
| A_32_P184330 | <NA> | 0.5664 | 4.39E-07 | 0.0080 | ||||
| A_24_P832113 | NPM1 | Nucleophosmin (nucleolar phosphoproein B23, numatrin) | 5q35.1 | −0.4808 | 4.79E-07 | 0.0088 | ||
| A_23_P119683 | MIER2 | Mesoderm induction early response 1, family member 2 | 19p13.3 | 0.3391 | 5.41E-07 | 0.0099 | ||
| A_23_P162807 | MRPS31 | Mitochondrial ribosomal protein S31 | 13q14.11 | −0.4422 | 1.13E-06 | 0.0206 | Deletion-duplication of 13q14.11 | 15 |
| A_23_P88439 | TC2N | Tandem C2 domains, nuclear | 14q32.12 | −0.4056 | 1.25E-06 | 0.0228 | Deletion of 14q32.11-q32.13 | 1 |
| A_32_P188674 | NPM1 | Nucleophosmin (nucleolar phosphoproein B23, numatrin) | 5q35.1 | −0.4742 | 1.30E-06 | 0.0237 | ||
| A_23_P399501 | PKM | Pyruvate kinase, muscle | 15q22 | 0.4023 | 1.32E-06 | 0.0242 | ||
| A_32_P46765 | C12orf29 | Chromosome 12 open reading frame 29 | 12q21.32 | −0.4320 | 1.38E-06 | 0.0252 | Deletion of 12q21.31-q21.33 | 1 |
| A_24_P927883 | JADE2 | Jade family PHD finger 2 | 5q31.1 | −0.3695 | 1.52E-06 | 0.0278 | ||
| A_23_P131676 | ACKR3 | Atypical chemokine receptor 3 | 2q37.3 | −0.4413 | 1.66E-06 | 0.0303 | Deletion of 2q37.1-q37.3 | 3 |
| A_23_P84154 | ARHGAP15 | Rho GTPase activating protein 15 | 2q22.2-q22.3 | −0.3958 | 1.69E-06 | 0.0309 | Rare single gene variant | 6 |
| A_23_P250462 | ATP6AP1 | ATPase, H+ transporting, lysosomal accessory protein 1 | Xq28 | 0.3399 | 2.25E-06 | 0.0412 | Deletion-duplication of Xq27.1-q28 | 1 |
| A_23_P322593 | TAPT1-AS1 | TAPT1 antisense RNA 1 | 4p15.32 | −0.3650 | 2.28E-06 | 0.0417 | Deletion of 4p16.3-p15.32 | 2 |
| A_32_P173058 | TMEM41B | Transmembrane protein 41B | 11p15.4 | −0.3388 | 2.40E-06 | 0.0438 | Deletion-duplication of 11p15.4 | 28 |
| A_23_P117424 | DCAF11 | DDB1 and CUL4 associated factor 11 | 14q11.2 | 0.3174 | 2.54E-06 | 0.0464 | Duplication of 14q11.2-q21.1 | 1 |
| A_32_P90685 | <NA> | −0.4853 | 2.63E-06 | 0.0481 | ||||
ASD, autism spectrum disorder; ID, intellectual disability; NA, not applied; logFC, log2 of fold change.
Adjusted p values <0.05;
Bonferroni correction.
The Simons Foundation Autism Research Initiative (SFARI) Gene 2.0 database (available at http://gene.sfari.org).
Unsupervised Machine Learning
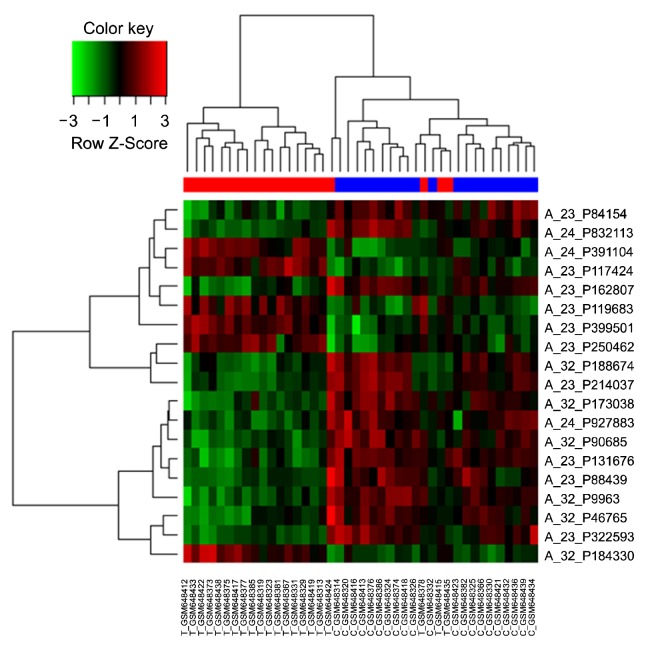
Using the 19-probe expression signature, a hierarchical cluster analysis of all samples (n=42) showed that ASDs were relatively well discriminated from controls (with the sorting of three ASD cases into the control group), suggesting that these probes could be helpful for differentiating between ASDs and controls. Detailed results from the hierarchical cluster analysis are presented in Figure 1.
Fig. 1.
Heat-map overview of the two-way hierarchical clustering analysis of 19 differentially-expressed probes. Each row represents the relative levels of expression for a single probe. The red or green color indicates relatively high or low expression, respectively. In the sample clustering dendrogram, red indicates autism spectrum disorder samples while blue indicates control samples.
Supervised Machine Learning
For the supervised machine learning algorithms, we simply built a classifier using the 19-probe expression signature and assessed its predictive performance. With this 19-probe prediction model, the test dataset was used to validate the prediction of ASD. This validation test revealed that our prediction model successfully distinguished between the individuals with ASD and controls. Both SVM and KNN analysis accurately identified 8 individuals with ASD and 8 controls with the exception of classifying one control as ASD, resulting in a predictive accuracy of 93.8% (sensitivity of 100% and specificity of 87.5%). However, in the LDA analysis, the diagnostic prediction of ASD vs. control samples was 68.8% accurate (Table 3).
Table 3.
Prediction performances of the 19-probe set on the test (validation) set, according to machine learning algorithms
| Accuracy (%) | Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | |
|---|---|---|---|---|---|
| SVM | 93.8 | 100.0 | 87.5 | 88.9 | 100.0 |
| KNN | 93.8 | 100.0 | 87.5 | 88.9 | 100.0 |
| LDA | 68.8 | 62.5 | 75.0 | 71.4 | 66.7 |
SVM, support vector machine; KNN, K-nearest neighbor; LDA, linear discriminant analysis.
DISCUSSION
Our analyses were designed to validate a potential biological signature using peripheral blood microarray data obtained from young Asian adults with ASD in combination with machine learning algorithms. In this exploratory study using previously published microarray data,13) we identified a blood-based gene expression signature that reliably identified young adults with ASD.
These results are consistent with the findings of four previous studies that reported on gene expression signatures with high diagnostic accuracy for toddlers and children with ASD.8–11) The results of this and four previous studies suggest that gene expression profiles from the peripheral blood samples contain a biological signature that could be used to predict the ASD risk in both children and young adults. According to several studies of healthy adults, the expression of most genes within individuals remains temporally stable, and only 1% to 2% of genes display significant changes over time periods of at least one month.22,23) In addition, previous studies observed that the cognitive, behavioral, and emotional symptoms of individuals with ASD generally persist over time.24,25) Therefore, the gene expression patterns underlying these long-standing phenotypes may be constant over time in the transition children to young adults.
Gene expression microarrays primarily measure messenger RNA for thousands of identified genes.26) The microarrays specifically evaluate the sequence of DNA that is transcribed to RNA in the genome at a given time. Prediction models using multivariate gene expression have been widely adopted for screening, diagnosis, and prognosis.27,28) Several previous transcriptome-wide studies of gene expression in ASD subjects have used post mortem brain tissue29–31) or peripheral blood samples.8–11,13) Among them, the gene expression profiles using peripheral blood have shown that RNA expression is disrupted across hundreds of genes in individuals with ASD. Blood-based analyses of gene expression profiles are encouraging because blood samples are easily obtainable from living individuals and are likely to contain ASD-relevant signatures.
Although the connection between blood and brain transcriptomic profiles is not well known, growing evidence suggests that measurements performed in tissues that are not primarily involved in the disease process may uncover disease signatures.10) Sullivan et al.32) have established a shared gene expression profile between whole blood and brain tissues suggesting that the cautious and thoughtful use of peripheral gene expression may be a useful surrogate for gene expression in the brain. Further research will be required to determine whether the dysregulated signatures in peripheral blood are actual indicators of the brain pathophysiology in ASD. Our results could also provide further evidence of the emerging consensus that peripheral blood is a potential source of biological signatures that are responsible for dysregulation of the brain and other unreachable tissues.33)
The gene list in our study partially overlaps with previously reported candidate genes and loci associations for ASD (Table 2). These various transcriptomic changes would be representative of the genomic alteration in the ASD. Blood-derived gene expression studies of subjects with ASD repeatedly demonstrate dysregulation of immune/inflammation genes.34) Regulatory factor X1 (RFX1; transcription factor regulating a wide variety of genes involved in immunity)35) expression was significantly increased in the ASD group in our study. Substantial percentages of patients with ASD show peripheral markers of mitochondrial energy metabolism dysfunction.36) We found the mitochondrial ribosomal protein S31 (MRPS31) expression was significantly reduced in the ASD group. In particular, we identified a probe (A_23_P399501, pyruvate kinase muscle isozyme [PKM]) that has the best ability for detecting whether a sample was collected from a patient with ASD (Supplementary Fig. 4). The PKM expression level was significantly higher in ASD subjects than in controls. Pyruvate kinase is an enzyme involved in glycolysis. Its primary function is to catalyze the transfer of a phosphate group from phosphoenolpyruvate to adenosine diphosphate as the last step of glycolysis, generating one molecule of pyruvate and one molecule of adenosine triphosphate.37) A previous study also demonstrated that the plasma pyruvate levels were higher in children with autism than in controls.38) These results suggested that the PKM expression level in peripheral blood may serve as a biomarker to distinguish ASD from controls.
Our study has several limitations, mostly stemming from small sample size and lack of phenotypic information of the original data. In particular, most of ASD subjects in this study exhibited normal intelligence quotients (IQ; mean full scale IQ, 91.9), this probably does not represent the broader ASD population. Unfortunately, it is not well understood about the connection between the peripheral blood and the brain transcriptomic profiles and the influence of age factor for gene expression in subjects with ASD. The results of our study should be cautiously interpreted. If further analysis is performed on a more homogeneous dataset and validated in an independent, large cohort of cases and controls, the accuracy of the results should be higher. These strategies for class prediction analyses will help identify robust biomarkers for both the diagnosis of ASD and individualized treatment options for patients and their families.39)
In conclusion, this study reveals a blood-based gene expression signature that has promising accuracy in distinguishing between young adults with ASD and age- and sex-matched unaffected controls. The ability of the 19 DE probes to correctly predict ASD samples compares favorably with the results of four previous studies on ASD diagnosis in toddlers and children. This transcriptomics approach may shed light on an important aspect of clinical biomarker discovery, offering high predictive accuracy for detecting ASD.
Supplementary Information
Acknowledgments
We thank Dr. Yuki Kuwano for generously depositing the microarray data series (GSE26415) online.
REFERENCES
- 1.Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years--autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warren Z, McPheeters ML, Sathe N, Foss-Feig JH, Glasser A, Veenstra-Vanderweele J. A systematic review of early intensive intervention for autism spectrum disorders. Pediatrics. 2011;127:e1303–e1311. doi: 10.1542/peds.2011-0426. [DOI] [PubMed] [Google Scholar]
- 3.Constantino JN, Zhang Y, Frazier T, Abbacchi AM, Law P. Sibling recurrence and the genetic epidemiology of autism. Am J Psychiatry. 2010;167:1349–1356. doi: 10.1176/appi.ajp.2010.09101470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozonoff S, Young GS, Carter A, Messinger D, Yirmiya N, Zwaigenbaum L, et al. Recurrence risk for autism spectrum disorders: a Baby Siblings Research Consortium study. Pediatrics. 2011;128:e488–e495. doi: 10.1542/peds.2010-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg RE, Law JK, Yenokyan G, McGready J, Kaufmann WE, Law PA. Characteristics and concordance of autism spectrum disorders among 277 twin pairs. Arch Pediatr Adolesc Med. 2009;163:907–914. doi: 10.1001/archpediatrics.2009.98. [DOI] [PubMed] [Google Scholar]
- 6.Schaefer GB, Mendelsohn NJ. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med. 2013;15:399–407. doi: 10.1038/gim.2013.32. [DOI] [PubMed] [Google Scholar]
- 7.Codina-Solà M, Rodríguez-Santiago B, Homs A, Santoyo J, Rigau M, Aznar-Laín G, et al. Integrated analysis of whole-exome sequencing and transcriptome profiling in males with autism spectrum disorders. Mol Autism. 2015;6:21. doi: 10.1186/s13229-015-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glatt SJ, Tsuang MT, Winn M, Chandler SD, Collins M, Lopez L, et al. Blood-based gene expression signatures of infants and toddlers with autism. J Am Acad Child Adolesc Psychiatry. 2012;51:934–944.e932. doi: 10.1016/j.jaac.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu VW, Lai Y. Developing a predictive gene classifier for autism spectrum disorders based upon differential gene expression profiles of phenotypic subgroups. N Am J Med Sci (Boston) 2013;6 doi: 10.7156/najms.2013.0603107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong SW, Collins CD, Shimizu-Motohashi Y, Holm IA, Campbell MG, Lee IH, et al. Characteristics and predictive value of blood transcriptome signature in males with autism spectrum disorders. PLoS One. 2012;7:e49475. doi: 10.1371/journal.pone.0049475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pramparo T, Pierce K, Lombardo MV, Carter Barnes C, Marinero S, Ahrens-Barbeau C, et al. Prediction of autism by translation and immune/inflammation coexpressed genes in toddlers from pediatric community practices. JAMA Psychiatry. 2015;72:386–394. doi: 10.1001/jamapsychiatry.2014.3008. [DOI] [PubMed] [Google Scholar]
- 12.Edgar R, Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol. 2006;24:1471–1472. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuwano Y, Kamio Y, Kawai T, Katsuura S, Inada N, Takaki A, et al. Autism-associated gene expression in peripheral leucocytes commonly observed between subjects with autism and healthy women having autistic children. PLoS One. 2011;6:e24723. doi: 10.1371/journal.pone.0024723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warnes GR, Bolker B, Bonebakker L, Gentleman R, Liaw WHA, Lumley T, et al. gplots: various R programming tools for plotting data. R package version. 2012;2(4) [Google Scholar]
- 17.Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20:273–297. doi: 10.1007/BF00994018. [DOI] [Google Scholar]
- 18.Shakhnarovich G, Indyk P, Darrell T. Nearest-neighbor methods in learning and vision: theory and practice. Cambridge, MA: MIT Press; 2006. [Google Scholar]
- 19.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936;7:179–188. doi: 10.1111/j.1469-1809.1936.tb02137.x. [DOI] [Google Scholar]
- 20.Carey V, Gentleman R, Mar J, Vertrees cfJ, Gatto L. R package version 1.52.0. 2016. MLInterfaces: uniform interfaces to R machine learning procedures for data in Bioconductor containers. [Google Scholar]
- 21.Carlson M. R package version 3.2.2. 2016. hgug4112a.db: Agilent “Human Genome, Whole” annotation data (chip hgug4112a) [Google Scholar]
- 22.Karlovich C, Duchateau-Nguyen G, Johnson A, McLoughlin P, Navarro M, Fleurbaey C, et al. A longitudinal study of gene expression in healthy individuals. BMC Med Genomics. 2009;2:33. doi: 10.1186/1755-8794-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell C, Vernon SD, Karem KL, Nisenbaum R, Unger ER. Assessment of normal variability in peripheral blood gene expression. Dis Markers. 2002;18:201–206. doi: 10.1155/2002/462465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Freeman BJ, Rahbar B, Ritvo ER, Bice TL, Yokota A, Ritvo R. The stability of cognitive and behavioral parameters in autism: a twelve-year prospective study. J Am Acad Child Adolesc Psychiatry. 1991;30:479–482. doi: 10.1097/00004583-199105000-00020. [DOI] [PubMed] [Google Scholar]
- 25.Soke GN, Philofsky A, Diguiseppi C, Lezotte D, Rogers S, Hepburn S. Longitudinal changes in Scores on the Autism Diagnostic Interview--Revised (ADI-R) in pre-school children with autism: Implications for diagnostic classification and symptom stability. Autism. 2011;15:545–562. doi: 10.1177/1362361309358332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B, Xu F, Baba Y. An evaluation of linear RNA amplification in cDNA microarray gene expression analysis. Mol Genet Metab. 2006;87:71–79. doi: 10.1016/j.ymgme.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Goh LK, Chen G, Verma S, Tan CH, Lee TS. Analysis of blood-based gene expression signature in first-episode psychosis. Psychiatry Res. 2012;200:52–54. doi: 10.1016/j.psychres.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 28.van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 29.Purcell AE, Jeon OH, Zimmerman AW, Blue ME, Pevsner J. Postmortem brain abnormalities of the glutamate neurotransmitter system in autism. Neurology. 2001;57:1618–1628. doi: 10.1212/WNL.57.9.1618. [DOI] [PubMed] [Google Scholar]
- 30.Nagarajan RP, Hogart AR, Gwye Y, Martin MR, LaSalle JM. Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics. 2006;1:e1–e11. doi: 10.4161/epi.1.4.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Campbell DB, D’Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan PF, Fan C, Perou CM. Evaluating the comparability of gene expression in blood and brain. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:261–268. doi: 10.1002/ajmg.b.30272. [DOI] [PubMed] [Google Scholar]
- 33.Baron CA, Liu SY, Hicks C, Gregg JP. Utilization of lymphoblastoid cell lines as a system for the molecular modeling of autism. J Autism Dev Disord. 2006;36:973–982. doi: 10.1007/s10803-006-0134-x. [DOI] [PubMed] [Google Scholar]
- 34.Rossignol DA, Frye RE. A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry. 2012;17:389–401. doi: 10.1038/mp.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herrero Sanchez C, Reith W, Silacci P, Mach B. The DNA-binding defect observed in major histocompatibility complex class II regulatory mutants concerns only one member of a family of complexes binding to the X boxes of class II promoters. Mol Cell Biol. 1992;12:4076–4083. doi: 10.1128/MCB.12.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palmieri L, Persico AM. Mitochondrial dysfunction in autism spectrum disorders: cause or effect? Biochim Biophys Acta. 2010;1797:1130–1137. doi: 10.1016/j.bbabio.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Gupta V, Bamezai RN. Human pyruvate kinase M2: a multifunctional protein. Protein Sci. 2010;19:2031–2044. doi: 10.1002/pro.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giulivi C, Zhang YF, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304:2389–2396. doi: 10.1001/jama.2010.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YJ, Oh SH, Park C, Hong M, Lee AR, Yoo HJ, et al. Advanced pharmacotherapy evidenced by pathogenesis of autism spectrum disorder. Clin Psychopharmacol Neurosci. 2014;12:19–30. doi: 10.9758/cpn.2014.12.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.