Abstract
The mitotic spindle faithfully separates the genetic material, and has been reverently observed for well over a century. Across eukaryotes, while the mechanisms for moving chromosomes seem quite conserved, mechanisms for assembling the spindle often seem distinct. Two major pathways for spindle assembly are known, one based on centrosomes and the other based on chromatin, and these pathways are usually considered to be fundamentally different. We review observations of spindle assembly in animals, fungi, and plants, and argue that microtubule assembly at a particular location, centrosomes, or chromatin, reflects contingent, cell-type specific factors, rather than reflecting a fundamental distinction in the process of spindle building. We hypothesize that the essential process for spindle assembly is the motor-driven organization of microtubules that accumulate in the form of dense bundles at or near the chromosomes.
Keywords: Centrosomes, Mitosis, Plant spindle, Vertebrate spindle, Yeast spindle
Introduction
To segregate duplicated chromosomes during mitosis, cells assemble a microtubule-based structure called the mitotic spindle. Mitotic spindles of diverse cells share several fundamental features. First, all spindles are bipolar, although the structure present at the poles varies considerably among organisms. Second, all spindles generate forces to segregate the duplicated genetic material. Here, again, the details differ, but the end result is the same: sister chromatids are moved far enough apart for functional separation. Interactions between microtubules and chromosomes, which are required for chromosome motion, typically (but not always) occur at a specific site on each chromosome, the kinetochore. Despite great variation in the number of microtubules involved in the attachment, new results suggest that a common “attachment module” is all but universally present (Johnston et al. 2010). Also evidently conserved ubiquitously, the spindle assembles de novo as the cell enters mitosis. Here, we focus on spindle formation, and compare this process across three kingdoms. Although at first sight, the processes in each taxon appear to be distinct, we argue that spindle assembly is governed by conserved principles.
Centrosome-based spindle assembly: search and capture
In most animal cells, centrosomes play a dominant role in microtubule nucleation and organization (Fig. 1). The centrosome, comprising a pair of centrioles and associated pericentriolar material, duplicates prior to entry into mitosis. At the entry into mitosis, the microtubule nucleating capacity of each of the duplicated centrosomes increases, resulting in an extensive radial array of microtubules extending from each centrosome (Piehl et al. 2004). Centrosomal microtubules are highly dynamic, alternating stochastically between periods of growth and rapid shortening, a behavior called dynamic instability (Mitchison and Kirschner 1984). Because centrosomes are typically located adjacent to the nucleus, the microtubules they nucleate are ideally positioned to interact with kinetochores on duplicated chromosomes following nuclear envelope breakdown.
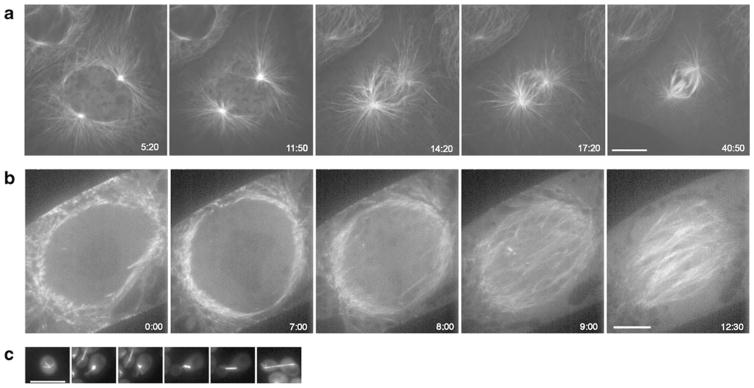
Fig. 1.
Spindle assembly in animal, plant, and yeast cells. a Mammalian epithelial cells (LLC-Pk1) expressing GFP tubulin; note the extensive astral array. b Tobacco cells (BY-2) expressing GFP tubulin. Cells in a and b were imaged using spinning disc confocal microscopy. c Budding yeast cells expressing mCherry-tubulin were imaged using conventional wide-field fluorescence microscopy; note nuclear and astral microtubules are separated by the nuclear envelope. Bars=10 μm
Based on these features of centrosomal microtubules, Kirschner and Mitchison (1986) proposed that spindle formation results from the capture of dynamic astral microtubules by each kinetochore, a model aptly called “Search and Capture” (Kirschner and Mitchison 1986). Microtubules that interact with kinetochores were proposed to become stabilized relative to free, unattached microtubules; over time, capture of additional microtubules would result in the formation of a bundle of stabilized microtubules, the kinetochore fiber. The dynamic instability behavior of microtubules extending from each centrosome and the back-to-back geometry of kinetochores on sister chromatids would favor the biorientation of chromosomes and their congression to the metaphase plate, midway between the centrosomes. According to the original search and capture model, all spindle microtubules were derived from centrosomes.
Observations of mitotic cells confirmed that a search and capture mechanism functions in living cells. For example, by imaging spindle formation using differential interference contrast microscopy of highly flattened, newt lung epithelial cells containing unattached chromosomes, Hayden and coworkers (1990) and Rieder and Alexander (1990) showed that individual centrosomal microtubules interact with the kinetochore, resulting in movement of the chromosome toward the pole from which the centrosomal microtubule arose. In addition, kinetochore fiber microtubules were shown to display greater stability than nonkinetochore spindle microtubules, consistent with the search, and capture model (Rieder 1991). The stochastic nature of the search and capture process is also consistent with the observed variability in the duration of mitosis.
Despite the elegance of search and capture, modeling studies demonstrated that dynamic centrosomal microtubules alone are not sufficient to achieve spindle formation within the time frame observed in live cells (Wollman et al. 2005). Using reasonable estimates of the number of centrosomal microtubules and measurements of microtubule dynamic instability parameters for mammalian mitotic cells (Rusan et al. 2001), the modeling has shown that search and capture would require many hours to capture all of the kinetochores, a time frame far greater than the typical duration of spindle formation. This result indicated that, even in centrosome-containing cells, additional mechanisms contribute to spindle assembly.
Chromatin-mediated spindle formation
An even more serious limitation of the search and capture model is the fact that many cells lack centrosomes, yet assemble mitotic spindles; clearly, these cells must use an alternative mechanism for spindle formation. A major breakthrough in our understanding of acentrosomal spindle formation was the discovery that cell-free extracts prepared from xenopus oocytes support spindle formation in vitro. Using this extract system, Heald and coworkers (1996) showed that in extracts lacking centrosomes and kinetochores, microtubules formed around DNA-coated beads and were polarity-sorted into a bipolar spindle. Based on these results, the authors proposed an alternative route for spindle formation based on acentrosomal microtubule formation around chromatin; we will refer to the pathway active in frog egg extracts as “chromatin-mediated” spindle assembly.
Subsequent experiments using the extract system showed that the small GTPase, Ran, which regulates nuclear transport by binding to import receptors (i.e., importin β), is a key regulator of the chromatin-dependent pathway (Carazo-Salas et al. 1999; Kalab et al. 1999; Ohba et al. 1999; Wilde and Zheng 1999; Zhang et al. 1999). By manipulating the level of active Ran in Xenopus extracts, several groups showed that active, GTP-bound Ran was both necessary and sufficient to drive microtubule formation. Because the sole exchange factor for Ran, RCC1, is bound to chromatin, these observations suggest that Ran acts near chromatin to liberate cargo bound to importin β. Farther away from chromosomes, Ran GAP, which is cytoplasmic, would convert Ran to the GDP-bound, inactive form (Kalab and Heald 2008). Consistent with this hypothesis, several microtubule-associated proteins, including TPX2 and NuMA, were subsequently identified that are inactive in promoting microtubule assembly when bound to importin β, and are activated in the presence of GTP-bound Ran. Moreover, imaging cells expressing fluorescent bio-sensors for active Ran, showed that it was present in a gradient around chromosomes in Xenopus extracts and live mammalian cells in mitosis (Kalab et al. 2002, 2006). These results support a model in which Ran regulates spindle microtubule formation by localized activation of spindle assembly factors and molecular motor proteins (Kalab and Heald 2008).
Chromosome-mediated microtubule formation in centrosome containing cells
The identification of a chromatin-mediated spindle assembly pathway, and the established limitations of search and capture, prompted an examination of the possibility that such a pathway exists and functions in centrosome-containing cells. A complication arises because, in vivo, chromatin occurs in the form of chromosomes, which also contain kinetochores; in experiments designed to find the sites of microtubule formation during initial spindle assembly, it can be difficult to tell whether microtubules arise at the kinetochore or elsewhere on the chromosome (i.e., from chromatin). Because either kinetochores or chromatin could contribute, when discussing cells we will therefore refer to the “chromosome-mediated” pathway.
In support of a chromosome-mediated pathway existing in somatic cells, spindle assembly occurs in Drosophila cells containing mutations that disrupt centrosome function and the formation of mitotic asters (Bonaccorsi et al. 1998; Megraw et al. 2001). Direct evidence for the redundancy of centrosomes in cells that normally contain them was obtained in experiments where centrosomes were removed, either by microsurgery or by laser ablation, and functional bipolar spindles formed (Hinchcliffe et al. 2001; Khodjakov et al. 2000). Interestingly, the major defect observed in these cells was a decrease in the fidelity of cytokinesis, likely a result of the severe reduction in astral microtubules that is observed following elimination of centrosomes.
Although these observations demonstrated that a chromosome-based spindle assembly pathway is present in centrosome-containing cells, it was not known if the elimination of centrosomes, or their inactivation by mutation, activated a “dormant” chromosome-based mechanism, or if the chromosome pathway normally functioned in parallel with centrosome-based spindle formation (O’Connell and Khodjakov 2007; Rieder 2005; Wadsworth and Khodjakov 2004). This question has proved difficult to resolve because centrosomes, when present, typically nucleate hundreds of microtubules, obscuring microtubule formation near chromosomes, if present.
To address this issue, Tulu et al. (2006) treated LLC-Pk1 cells with nocodazole to eliminate microtubules, and then examined microtubule formation following washout. In these cells, centrosomes frequently wander away from the chromosomes, making it feasible to observe microtubule formation at chromosomes and centrosomes separately. Microtubules formed at centrosomes, as expected, but also around chromosomes and kinetochores. Likewise, microtubules form at distal kinetochores of mono-oriented chromosomes on monopolar spindles (induced by monastrol treatment) (Khodjakov et al. 2003) and at kinetochores in flattened Drosophila S2 cells (Maiato et al. 2004). In the latter case, the microtubules that formed at kinetochores subsequently interacted with centrosomal microtubules and were incorporated into the spindle in a dynein-dependent manner (Maiato et al. 2005). Arguably, chromosome-dependent microtubule assembly involves microtubule formation at both chromatin and kinetochores.
Microtubule assembly at kinetochores, which can be conveniently assayed using the nocodazole washout assay (Tulu et al. 2006), requires Ran-regulated microtubule assembly factors, and γ-tubulin (Luders et al. 2006; Torosantucci et al. 2008). In fact, γ-tubulin-containing complexes are recruited to kinetochores by Nup170-160, a subcomplex of the nuclear pore complex (Mishra et al. 2010). Also contributing to microtubule formation at kinetochores is aurora B kinase, a component of the chromosome passenger complex (Maresca et al. 2009). Additional experiments are required to resolve the contributions to chromosome-mediated spindle assembly of microtubule formation at chromatin and kinetochores as well as their regulation.
In mammalian cells, the extent of microtubule formation at centrosomes and chromosomes are apparently interdependent and limited by the availability of tubulin subunits (Tulu et al. 2006). This interdependency was observed in nocodazole washout experiments using cells depleted of TPX2, where centrosomal microtubule formation was enhanced when chromosome-mediated microtubule formation was eliminated. The idea that the tubulin pool limits microtubule assembly might also explain why microtubule formation at kinetochores is so clearly observed in Drosophila S2 cells, and is not as apparent in mammalian cells: S2 cells have few very centrosomal microtubules, so more tubulin would be available for assembly at kinetochores.
These results have established that a chromosome-based pathway functions, even in the continued presence of active centrosomes, and that kinetochores are preferred sites for microtubule assembly, at least in Drosophila and mammalian cells (O’Connell et al. 2009), and finally that centrosomal microtubules alone cannot establish kinetochore fibers (Yang et al. 2007). Given that in a mature spindle, microtubule plus-ends are located at kinetochores (Euteneuer and McIntosh 1981), wherever microtubules originate, plus-ends must be captured eventually by the kinetochore plate.
When kinetochores stay inside the nucleus—the case of yeast
Do kinetochore-derived microtubules facilitate spindle formation in centrosome containing cells? The answer is a resounding “yes” based on recent studies in budding yeast, which provide evidence that kinetochore-derived micro-tubules are likely to assist centrosome-derived microtubules to efficiently “locate” kinetochores for initial interaction. Unlike mammalian cells, yeast cells do not break down their nuclear envelope during mitosis (the so-called “closed mitosis”). Inside the nucleus, the kinetochores are connected to spindle pole bodies (equivalent to the centrosomes of mammalian cells) by microtubules throughout almost the entire cell cycle (Winey and O’Toole 2001). However, during S phase in yeast (which overlaps with mitosis), the kinetochores are disassembled upon centromere DNA replication, causing centromeres to detach from microtubules and translate away from the spindle poles (Kitamura et al. 2007). The kinetochores are reassembled after 1 to 3 min, and they subsequently, as in mammalian cells, capture the lateral sides of spindle pole microtubules for reattachment (Tanaka et al. 2005).
Although this seems like a cut-and-dried case of search and capture, elegant experiments delaying the kinetochore-reassembly process have demonstrated that microtubules are generated at the kinetochores before they are captured by spindle pole microtubules (Kitamura et al. 2010). These kinetochore-derived microtubules require Stu2 (an orthologue of XMAP215/ch-TOG) for nucleation, and they extend with their plus ends distal to the kinetochores, which is in contrast to the polarity described for Drosophila S2 cells (Maiato et al. 2004). Nascent microtubules extending from the reassembled kinetochores often initiate interaction with the spindle pole microtubules (roughly 20 emanate from each spindle pole; Winey et al. 1995) along their length, either in an antiparallel or parallel manner. This interaction leads to loading of the kinetochores onto the lateral surface of spindle pole microtubules. However, once kinetochores are “loaded” onto spindle microtubules, the kinetochore-derived microtubules rapidly disappear and do not contribute to the metaphase spindle, a result that fits the established understanding of the polarity of microtubules that form the metaphase spindle.
Given that kinetochores associated with microtubules load faster than kinetochores without associated microtubules, it is believed that kinetochore-derived microtubules assist the spindle pole microtubules to efficiently “locate” the kinetochore. In yeast, since a Ran-GTP gradient is not formed during mitosis as its concentration is uniformly high in the nucleus, the kinetochore-based microtubule pathway might represent the dominant mechanism to mediate initial kinetochore capture by spindle microtubules. Taken together, the observations in yeast and other cellular systems show that the chromosome-mediated microtubule pathway is likely to operate in centrosome-containing cells, although the degree to which centrosomal microtubules, kinetochore, and chromatin-based nucleation contribute to kinetochore fiber formation remains unresolved, in particular for mammalian cells.
Peripheral microtubules aid spindle formation
Although not always appreciated, mammalian epithelial cells have an extensive interphase array of microtubules that are not associated with the centrosome. These microtubules also reorganize as cells enter mitosis. By direct observation of early prometaphase cells, Rusan et al. (2001) observed that both individual microtubules and clusters of microtubules are moved inward toward the forming spindle along extending centrosomal microtubules. The inward transport is dynein-dependent. Photoactivation experiments reveal that these microtubules are incorporated into the forming spindle (Tulu et al. 2003). These experiments show that dynamic centrosomal microtubules search for and capture not only kinetochores but also peripheral microtubules, and that peripheral microtubules contribute to spindle formation in centrosome-containing cells.
In addition to the inward motion of peripheral microtubules in early prometaphase cells, small clusters of microtubules are occasionally observed in the peripheral cytoplasm of late prometaphase or metaphase cells (Rusan et al. 2001). The presence of nonspindle microtubules in a mitotic cell presents a potential problem—if chromosomes became associated with these microtubules, then equal chromosome distribution could be compromised. However, almost every time that ectopic microtubules have been detected, they were rapidly moved inward along astral microtubules. The impression from viewing these events is that astral microtubules are continually monitoring the peripheral cytoplasm. If and when other microtubules are encountered, they are rapidly drawn inwards, preventing ectopic chromosome attachment.
A similar clustering and inward motion of noncentrosomal, peripheral microtubules has been observed during meiotic spindle formation in mammalian oocytes (Schuh and Ellenberg 2007). In these large, acentrosomal cells, the interphase microtubules are rapidly reorganized into microtubule organizing centers that continuously interact with each other, then coalesce, resulting in fewer, larger foci that accumulate adjacent to the nuclear envelope. These organizing centers then form a “ball” of microtubules around the chromosomes and eventually sort into a bipolar structure (Schuh and Ellenberg 2008). Together with the observations on somatic mammalian cells, these observations indicate that microtubule motion directed toward the forming spindle is a conserved feature. It may be especially prominent in larger cells that contain extensive interphase microtubule arrays.
Nuclear-envelope mediated spindle formation
Notorious for their lack of centrosomes, mitotic spindles in land plants are frequently lumped together with those of animal oocytes as being “acentrosomal.” Nevertheless, the chromatin-mediated pathway cannot be primary in plants because the spindle assembles during prophase, before nuclear envelope breakdown, thereby precluding chromosome-mediated nucleation (Baskin and Cande 1990). Although plant cells lack defined centrosomes, their nuclear envelope nevertheless nucleates microtubules, and during interphase an array emanates from the nucleus, radiating into the cell (Brown and Lemmon 2007). Similar to centrosomes, nucleation activity of the plant nuclear envelope depends on γ-tubulin complexes, which are scattered over the envelope, rather as if the entire nucleus were a centrosome (Erhardt et al. 2002). In prophase, the orientation of microtubules changes from radial to tangential, and eventually two foci emerge on opposite sides of the intact nucleus (Fig. 1). These foci are the presumptive spindle poles, although after the envelope breaks down, the foci usually fragment into a set of smaller ones that collectively constitute the spindle pole. This suggests that plants use a third mechanism for spindle assembly, namely, a nuclear-envelope mediated pathway.
Little is known about prophase spindle assembly in plants (Ambrose and Cyr 2007; Murata et al. 2005; Bannigan et al. 2008). Interestingly, plants have an unmistakable TPX2 ortholog that has been implicated in spindle assembly (Vos et al. 2008). Unusually, plant TPX2 contains a signal for nuclear export and, in both Arabidopsis thaliana and Nicotiana tobacum, the protein appears to be exported specifically during late prophase. Furthermore, anti-TPX2 IgG, microinjected into Tradescantia virginiana stamen hair cells in prophase, prevents or delays nuclear envelope breakdown. Plant TPX2 has been hypothesized to act at the outside of the nuclear envelope to promote local microtubule assembly, although the disposition of the cognate GEF and GAP remain to be defined. In general, the Ran-mediated pathway for nuclear import and export appears to operate in plant cells (Meier 2007; Meier and Brkljacic 2009), although far fewer of the interactions have been mapped.
After the nuclear envelope breaks down, both search and capture from a de facto spindle pole as well as a chromosome-mediated pathway presumably both contribute to spindle maturation, but little is known about either. Plant kinetochores nucleate microtubules actively when metaphase cells are released from microtubule inhibition (Baskin and Cande 1990). Furthermore, living A. thaliana tissue culture cells (Chan et al. 2005) and tobacco BY-2 cells (Ambrose and Cyr 2008) expressing GFP-tubulin occasionally fail to form a prophase spindle but nevertheless form a functional mitotic spindle in prometaphase. While the latter studies establish that there is no requirement for a bipolar microtubule array to form during prophase, they leave open the existence, let alone the importance, of chromosome-mediated nucleation for spindle assembly in plant cells. This is because in these observations, abundant microtubules already surrounded the nucleus before the envelope broke down and, after that, only sorting out and kinetochore capture might be required to form a bipolar spindle. Furthermore, if plant TPX2 has been co-opted to drive assembly on the nuclear envelope, then this protein might be unavailable for chromosome-mediated assembly.
Evidently, more work is needed to clarify the steps of spindle formation in land plants. Nevertheless, the data available support the view that there is more than one alternative to centrosomes.
Hearing the theme among the variations
In cells that contain them, centrosomes and the microtubules they nucleate are a visually dominant element of spindle formation (Fig. 1). Nevertheless, the centrosomal array is dispensable for spindle formation. When present, centrosomal microtubules will participate in spindle formation, through interactions with kinetochores and with any kinetochore- or chromosome-derived microtubule bundles. Centrosomal microtubules also mediate inward motion of peripheral microtubules and set up a spindle midzone in which antiparallel microtubules from the opposite centrosome interact, an interaction that contributes to the establishment of spindle bipolarity and length. However, even this classic function of the centrosomal array can be replaced by microtubules generated from chromosomes (Ferenz et al. 2009).
Perhaps the essential function of centrosomal microtubules is in spindle positioning and cytokinesis. Land plant cells, which are naturally acentrosomal, have evolved a system completely separate from the mitotic spindle for orienting cell division (Smith 2001). In animal cells from which centrosomes have been removed, spindle position and cytokinesis, in contrast to chromosome segregation, are frequently defective (Hinchcliffe et al. 2001; Khodjakov et al. 2000). Furthermore, in a range of cell types, dynamic astral microtubules mediate asymmetric spindle positioning through interactions with the cell cortex (Siller and Doe 2009). Both central spindle and astral microtubules generate signals for the assembly of the contractile ring (Bringmann and Hyman 2005). Astral microtubules might be especially important to confine the equatorial signal in large cells (von Dassow 2009). These observations are consistent with the idea that centrosomal microtubules function as a transport system that integrates spindle components into a unified cellular structure (Wadsworth and Khodjakov 2004). While microtubules generated around chromosomes participate in chromosome movement and can fully replace the function of centrosomal microtubules if the need arises, the microtubules assembled around chromosomes seldom, if ever, reproduce astral functions.
Dynamic centrosomal microtubules extend individually into the cytoplasm and the most important functions of these microtubules may relate to this organization. In contrast, microtubules that assemble near chromosomes or nuclear envelope form dense arrays and bundles. When in close proximity, microtubules can be cross-linked by spindle assembly factors and microtubule-based motor proteins that contain two microtubule-binding sites, and then dynamically cross-linked microtubules can be polarity sorted and focused into poles. In diverse cells, microtubule density is regulated by γ-tubulin-dependent microtubule formation along preexisting microtubules (Janson et al. 2005; Murata et al. 2005), a process that requires the augmin complex (Goshima et al. 2008). Without the ability to form dense bundles, which happens when mammalian cells lose the function of the augmin complex, or the microtubule severing protein, katanin, they either fail to generate bipolar spindles (Srayko et al. 2006), or the bipolar spindles that do form are nonfunctional (Uehara et al. 2009).
Based on these observations, we speculate that the sine qua non for spindle formation is the assembly of a dense array of short microtubules centered around chromosomes (Fig. 2). As a corollary, the source of these microtubules—nuclear envelope, acentrosomal organizing centers, chromatin, or kinetochores—is of secondary importance. In most cases, formation of these arrays requires Ran, and the exceptions—spindle assembly during meiosis I of vertebrate oocytes and spindle assembly within the confines of small yeast nuclei—likely involve other nuclear factors and microtubule nucleators or stabilizers (Dumont et al. 2007). Consistent with our proposal, during meiosis in Caenorhabditis elegans, lateral microtubule bundles that do not make direct contact with kinetochores, mediate spindle formation and chromosome congression (Wignall and Villeneuve 2009). What appears to be essential is that given a collection of dense microtubule bundles, a bipolar structure invariably emerges. Perhaps dense bundles provide a suitable substrate for the push and pull of the sorting engines? If so, spindle assembly can then be separated into two major processes: microtubule coalescence and sorting. Each process is driven by more than one mechanism to ensure the faithful delivery of genetic inheritance from mother to daughter cells.
Fig. 2.
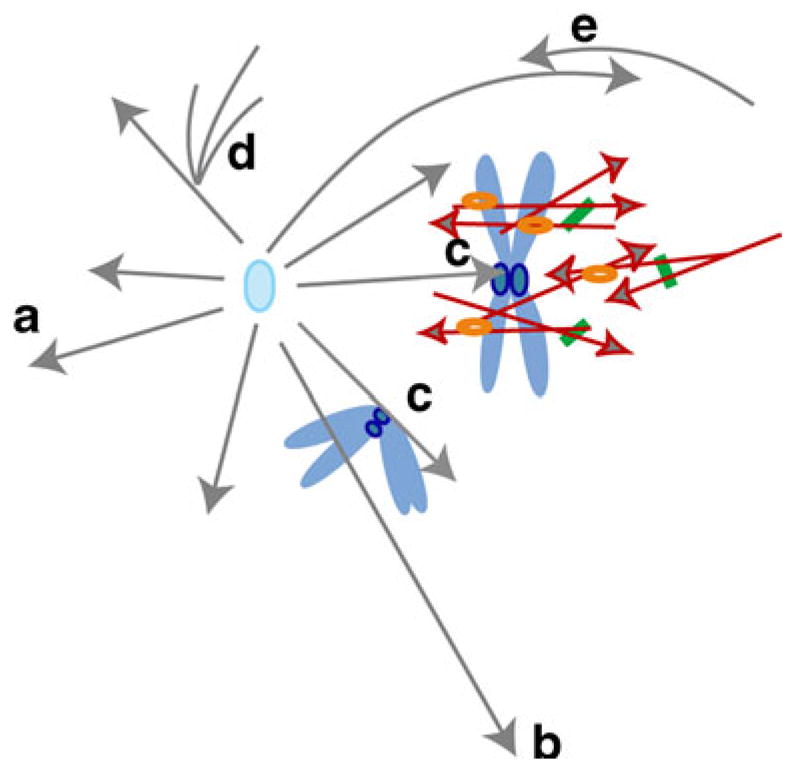
Schematic depiction of the contribution of astral and chromosome-associated microtubules to spindle formation. Only one centrosome of a spindle is shown for clarity. Astral microtubules (gray) mediate interactions with the cell cortex for spindle positioning (a) and cytokinesis (b); interact with kinetochores (c); with peripheral microtubules (d) and with microtubules from the opposite centrosome (e). Chromosome-associated microtubules (red) form dense arrays; molecular motors (orange) and microtubule-associated proteins (green) mediate the motion and bundling of these microtubules. Arrowheads depict microtubule plus-ends
Acknowledgments
The authors thank Dr. Laura Francis for her comments on the manuscript, and all the members of our laboratories for discussions. We gratefully acknowledge research support from NIH (W.-L. L., GM076094; P.W., GM 059057).
Footnotes
Conflict of Interest The authors declare that they have no conflict of interest.
Contributor Information
Patricia Wadsworth, Department of Biology, Morrill Science Center, University of Massachusetts, Amherst, MA 01003, USA.
Wei-Lih Lee, Department of Biology, Morrill Science Center, University of Massachusetts, Amherst, MA 01003, USA.
Takashi Murata, National Institute for Basic Biology, Myodaiji-cho, Okazaki, Aichi, Japan.
Tobias I. Baskin, Department of Biology, Morrill Science Center, University of Massachusetts, Amherst, MA 01003, USA
References
- Ambrose JC, Cyr R. Mitotic spindle assembly and function. In: Verma DPS, Hong Z, editors. Cell division control in plants. Springer; Berlin: 2007. pp. 141–167. [Google Scholar]
- Ambrose JC, Cyr R. Mitotic spindle organization by the preprophase band. Mol Plant. 2008;1:950–960. doi: 10.1093/mp/ssn054. [DOI] [PubMed] [Google Scholar]
- Bannigan A, Lizotte-Waniewski M, Riley M, Baskin TI. Emerging molecular mechanisms that power and regulate the anastral mitotic spindle of flowering plants. Cell Motil Cytoskel. 2008;65:1–11. doi: 10.1002/cm.20247. [DOI] [PubMed] [Google Scholar]
- Baskin TI, Cande WZ. The structure and function of the mitotic spindle in flowering plants. Annu Rev Plant Physiol Plant Mol Biol. 1990;41:277–315. [Google Scholar]
- Bonaccorsi S, Giansanti MG, Gatti M. Spindle self-organization and cytokinesis during male meiosis in asterless mutants of Drosophila melanogaster. J Cell Biol. 1998;142:751–761. doi: 10.1083/jcb.142.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Hyman A. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. The pleiomorphic plant MTOC: an evolutionary perspective. J Integr Plant Biol. 2007;49:1142–1153. [Google Scholar]
- Carazo-Salas RE, Guarguaglini G, Gruss OJ, Segref A, Karsenti E, Mattaj IW. Generation of GTP-bound ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature. 1999;400:178–181. doi: 10.1038/22133. [DOI] [PubMed] [Google Scholar]
- Chan J, Calder G, Fox S, Lloyd C. Localization of the microtubule end binding protein EB1 reveals alternative pathways of spindle development in arabidopsis suspension cells. Plant Cell. 2005;17:1737–1748. doi: 10.1105/tpc.105.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret M-E, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac M-H. A centriole and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol. 2007;176:295–305. doi: 10.1083/jcb.200605199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt M, Stoppin-Mellet V, Campagne S, Canaday J, Mutterer J, Fabian T, Sauter M, Muller T, Peter C, Lambert A-M, Schmit AC. The plant Spc98p homologue colocalizes with γ-tubulin at microtubule nucleation sites and is required for microtubule nucleation. J Cell Sci. 2002;115:2423–2431. doi: 10.1242/jcs.115.11.2423. [DOI] [PubMed] [Google Scholar]
- Euteneuer U, McIntosh JR. Structural polarity of kinetochore microtubules in PtK1 cells. J Cell Biol. 1981;89:338–345. doi: 10.1083/jcb.89.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenz N, Paul R, Fagerstrom C, Mogilner A, Wadsworth P. Dynein antagonizes Eg5 by crosslinking and sliding antiparallel microtubules. Curr Biol. 2009;19:1833–1838. doi: 10.1016/j.cub.2009.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G, Mayer M, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden JH, Bowser SS, Rieder CL. Kinetochores capture astral microtubules during chromosome attachment to the mitotic spindle: direct visualization in live newt lung cells. J Cell Biol. 1990;111:1039–1045. doi: 10.1083/jcb.111.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Janson ME, Setty TG, Paoletti A, Tran PT. Efficient formation of bipolar microtubule bundles requires microtubule-bound gamma-tubulin complexes. J Cell Biol. 2005;169:297–308. doi: 10.1083/jcb.200410119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K, Joglekar A, Hori A, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Pu RT, Dasso M. The Ran GTPase regulates mitotic spindle assembly. Curr Biol. 1999;9:481–484. doi: 10.1016/s0960-9822(99)80213-9. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Copenagle L, Gordon MB, Compton DA, Kapoor T. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J Cell Biol. 2003;160:671–683. doi: 10.1083/jcb.200208143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW, Mitchison T. Beyond self-assembly: from microtubules to morphogenesis. Cell. 1986;45:329–342. doi: 10.1016/0092-8674(86)90318-1. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Komoto S, Kitamura Y, Antony C, Tanaka TU. Kinetochores generate microtubules with distal plus-ends: their roles and limited lifetime in mitosis. Dev Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Patel UK, Stearns T. GCP-WD is a γ-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat Cell Biol. 2006;8:137–147. doi: 10.1038/ncb1349. [DOI] [PubMed] [Google Scholar]
- Maiato H, Rieder CL, Khodjakov A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol. 2004;167:831–840. doi: 10.1083/jcb.200407090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED. Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol. 2009;19:1210–1215. doi: 10.1016/j.cub.2009.05.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw TL, Kao L-R, Kaufman T. Zygotic development without functional centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/s0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- Meier I. Composition of the plant nuclear envelope: theme and variations. J Exp Bot. 2007;58:27–34. doi: 10.1093/jxb/erl009. [DOI] [PubMed] [Google Scholar]
- Meier I, Brkljacic J. Adding pieces to the puzzling plant nuclear envelope. Curr Opin Plant Biol. 2009;12:752–759. doi: 10.1016/j.pbi.2009.09.016. [DOI] [PubMed] [Google Scholar]
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BMA, Dasso M. The Nup107-160 complex and g-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Murata T, Sonobe S, Baskin TI, Hyodo S, Hasezawa S, Nagata T, Horio T, Hasebe M. Microtubule-dependent microtubule nucleation based on recruitment of gamma-tubulin in higher plants. Nat Cell Biol. 2005;7:961–968. doi: 10.1038/ncb1306. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Khodjakov AL. Cooperative mechanisms of mitotic spindle formation. J Cell Sci. 2007;120:1717–1722. doi: 10.1242/jcs.03442. [DOI] [PubMed] [Google Scholar]
- O’Connell CB, Loncarek J, Kalab P, Khodjakov A. Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J Cell Biol. 2009;187:43–51. doi: 10.1083/jcb.200903076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba T, Nakamura M, Nishitani H, Nishimoto T. Self-organization of microtubule asters induced in Xenopus egg extracts by GTP-bound Ran. Science. 1999;284:1356–1362. doi: 10.1126/science.284.5418.1356. [DOI] [PubMed] [Google Scholar]
- Piehl M, Tulu US, Wadsworth P, Cassimeris L. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle using GFP tagged EB1. Proc Natl Acad Sci USA. 2004;101:1584–1588. doi: 10.1073/pnas.0308205100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL. The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma. 1991;84:145–158. doi: 10.1007/BF00293368. [DOI] [PubMed] [Google Scholar]
- Rieder CL. Kinetochore fiber formation in animal somatic cells: dueling mechanisms come to draw. Chromosoma. 2005;114:310–318. doi: 10.1007/s00412-005-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Fagerstrom C, Yvon AC, Wadsworth P. Cell cycle dependent changes in microtubule dynamics in living cells expressing GFP-alpha tubulin. Mol Biol Cell. 2001;12:971–980. doi: 10.1091/mbc.12.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- Smith LG. Plant cell division: building walls in the right places. Nat Rev Mol Cell Biol. 2001;2:33–39. doi: 10.1038/35048050. [DOI] [PubMed] [Google Scholar]
- Srayko M, O’Toole ET, Hyman AA, Mueller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16:1944–1949. doi: 10.1016/j.cub.2006.08.029. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Torosantucci L, De Luca M, Guarguaglini G, Lavia P, Degrassi F. Localized RanGTP accumulation promotes microtubule nucleation at kinetochores in somatic mammalian cells. Mol Biol Cell. 2008;19:1873–1882. doi: 10.1091/mbc.E07-10-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulu US, Rusan N, Wadsworth P. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome containing cells. Curr Biol. 2003;13:1894–1899. doi: 10.1016/j.cub.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P. Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol. 2006;16:536–541. doi: 10.1016/j.cub.2006.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara R, Nozawa R, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. PNAS. 2009;106:6998–7003. doi: 10.1073/pnas.0901587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dassow G. Concurrent cues for cytokinetic furrow induction in animal cells. Trends Cell Biol. 2009;19:165–173. doi: 10.1016/j.tcb.2009.01.008. [DOI] [PubMed] [Google Scholar]
- Vos JW, Pieuchot L, Evrard J-L, Janski N, Bergdoll M, de Ronde D, Perrez LH, Sardon T, Vernos I, Schmit A-C. The plant TPX2 protein regulates prospindle assembly before nuclear enveolpe breakdown. THe Plant Cell. 2008;20:2783–2797. doi: 10.1105/tpc.107.056796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadsworth P, Khodjakov A. E pluribus unum: toward a universal mechanism for spindle assembly. Trends Cell Biol. 2004;14:413–419. doi: 10.1016/j.tcb.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A, Zheng Y. Stimulation of microtubule aster formation and spindle assembly by the small GTPase Ran. Science. 1999;284:1359–1358. doi: 10.1126/science.284.5418.1359. [DOI] [PubMed] [Google Scholar]
- Winey M, O’Toole ET. The spindle cycle in budding yeast. Nat Cell Biol. 2001;3:E23–E27. doi: 10.1038/35050663. [DOI] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, Mastronarde DN, Giddings TH, McDonald KL, McIntosh JR. Three dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R, Cytrynbaum EN, Jones JT, Meyer T, Scholey JM, Mogilner A. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr Biol. 2005;15:828–832. doi: 10.1016/j.cub.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for normal chromosome motion and congression independent of the spindle assembly checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Hughes M, Clarke PR. Ran-GTP stabilises microtubule asters and inhibits nuclear assembly in Xenopus egg extracts. J Cell Sci. 1999;112:2453–2461. doi: 10.1242/jcs.112.14.2453. [DOI] [PubMed] [Google Scholar]