Abstract
We have constructed a genome-saturating mutant library of the human gastric pathogen Helicobacter pylori. Microarray tracking of transposon mutants (MATT) allowed us to map the position of 5,363 transposon mutants in our library. While we generally found insertions well distributed throughout the genome, 344 genes had no detectable transposon insertions, and this list is predicted to be highly enriched for essential genes. Comparison to the essential gene set of other bacteria revealed a surprisingly limited overlap with all organisms tested (11%), while 55% were essential in some organisms but not others. We independently verified the essentiality of several gene products, including an HtrA family serine protease, a hypothetical protein with putative phospholipase D activity, and a riboflavin specific deaminase. A limited screen for motility mutants allowed us to estimate that 4.5% of the genome is dedicated to this virulence-associated phenotype.
Helicobacter pylori infects the gastric mucosa of 50% or more of the world population. Infection rates correlate with socioeconomic status and hygiene levels; thus, developing countries and developing populations within developed countries bear the highest burden of infection, as high as 90%. The consequence of this infection ranges from undetected gastritis to ulcer disease (duodenal and gastric) and gastric cancer (gastric adenocarcinoma and mucosal-associated lymphoid tissue lymphoma). The biological mechanisms leading to this diverse disease spectrum remain unknown, but bacterial, host, and environmental components are postulated to be contributing factors. Infection is generally acquired in childhood via person-to-person transmission and persists for life without specific antimicrobial intervention (15).
H. pylori was among the first microorganisms for which a full genome sequence was available (47) and was the first organism for which a second isolate was sequenced (3). This information was used along with comparative genomics to predict a small group of essential genes (73 out of 1,590) in the hopes of identifying novel targets for drug design (11); 33 out of 42 appeared essential by the inability to obtain gene disruption mutants. Another study sought to identify nonessential genes of H. pylori by determining whether insertion mutants of genomic DNA carried on a plasmid in Escherichia coli could be transduced into the H. pylori genome. A study of 204 such clones found that only 43 could be recovered in H. pylori, suggesting that 79% of genes could be essential (28). Both studies relied on a negative result from a transformation experiment to determine essentiality without controlling for differences in transformation efficiencies due to genetic locus or differing amounts of flanking homology. This technical issue prevents extrapolation of these results to the whole genome.
A number of bacterial products have been implicated in disease or establishment of infection, but a comprehensive list of H. pylori virulence determinants does not exist (22, 34). The study of many bacterial pathogens has been facilitated by genetic screens to identify genes involved in virulence-associated phenotypes, such as the ability to adhere to or invade host cells or persist in animal models of infection (18). Unfortunately, H. pylori does not support the replication of common plasmid or transposon vectors, making such studies difficult. Shuttle mutagenesis of H. pylori genomic libraries in E. coli generated libraries of 135 and 912 mutant clones (25, 30, 37, 38), a plasmid integration strategy yielded 1,251 H. pylori mutant clones (8, 9), and, most recently, a library of 639 mutants was made by random insertional mutagenesis of genomic DNA by ligation of an antibiotic resistance cassette (16a). These libraries were screened for clones with defects in adherence, urease activity, motility, competence, growth at low pH, and colonization of a gerbil model of infection. In many of these screens, clones with identical insertion sites were isolated, indicating relatively low library complexity. Thus, only a fraction of the genome has been screened for each of these phenotypes. While H. pylori has a relatively small genome, 1.7 Mb and some 1,500 genes, saturating genetic screens require the generation of larger, nonredundant, stable mutant libraries.
Here we describe the construction of a large library of transposon mutants throughout the H. pylori genome. A whole-genome microarray-based method for mapping the location of transposon insertions has allowed us to demonstrate full genome coverage of our transposon library and to generate a list of open reading frames enriched for essential genes. Independent confirmation of a subset of these genes highlights the emerging principle that the essential gene complement differs among bacterial species. Defining the full complement of essential and nonessential genes of H. pylori provides unique opportunities for understanding the function of genes with no known homology and, genes required for infection and for identifying novel antimicrobial targets.
MATERIALS AND METHODS
Bacterial culture and manipulations.
H. pylori strain G27, which contains the cag pathogenicity island, was used for these studies (14). H. pylori was grown on solid media on horse blood agar (HB) plates, containing 4% Columbia agar base (Oxoid), 5% defibrinated horse blood (HemoStat Labs), 0.2% β-cyclodextrin (Sigma), 10 μg of vancomycin (Sigma) per ml, 5 μg of cefsulodin (Sigma) per ml, 2.5 U of polymyxin B (Sigma) per ml, 5 μg of trimethoprim (Sigma) per ml, and 8 μg of amphotericin B (Sigma) per ml, under microaerobic conditions at 37°C. A microaerobic atmosphere was generated either by using a CampyGen sachet (Oxoid) in a gas pack jar or by incubating the culture in an incubator equilibrated with 10% CO2 and 90% air. For liquid culture, H. pylori was grown in Brucella broth (Difco) containing 10% fetal bovine serum (BB10; Gibco/BRL) with shaking in a microaerobic atmosphere. For antibiotic resistance marker selection, bacterial media were additionally supplemented with 25 μg of chloramphenicol (Cm) per ml or 36 μg of metronidazole (Mtz) per ml. E. coli growth and manipulations were performed as specified by standard laboratory protocols (4).
Construction of H. pylori transposon library.
We modified a commercially available Tn7-based in vitro mutagenesis system (GPS-M; New England Biolabs [NEB]) so it could be used in H. pylori. The pGPS-3 plasmid from NEB was modified by removing the Tn5 neomycin phosphotransferase gene, which is not effective in H. pylori, by BamHI digestion and replacing it with the Campylobacter coli chloramphenicol acetyltransferase gene (Cat), which was PCR amplified from pUOA20 (49) with BamHI linkers by using primers CatBam1 and CatBam2 (Table 1), generating pGPS-cat. The pGPS-cat vector and purified transposase enzyme from NEB were used according to the manufacturer's instructions to mutagenize H. pylori chromosomal DNA, which was purified by CsCl gradient centrifugation (5). This mutagenized DNA was transformed into H. pylori by natural transformation (48), and Cm-resistant clones were selected on solid media. Approximately 10,000 single colonies were directly harvested from the plates and were stored at −80°C to generate the H. pylori GPS mutant library.
TABLE 1.
Primers
| Name | Sequence | Length (bp) |
|---|---|---|
| N | ACTTTATTGTCATAGTTTAGATCTATTTTG | 30 |
| N2 | TCAGTTTAAGACTTTATTGTC | 21 |
| S | ATAATCCTTAAAAACTCCATTTCCACCCCT | 30 |
| S2 | CAGTTCCCAACTATTTTGTCC | 21 |
| CEKG2A | GGCCACGCGTCGACTAGTACNNNNNNNNNNAGAG | 34 |
| CEKG2B | GGCCACGCGTCGACTAGTACNNNNNNNNNNACGCC | 35 |
| CEKG2C | GGCCACGCGTCGACTAGTACNNNNNNNNNNGATAT | 35 |
| CEKG4 | GGCCACGCGTCGACTAGTAC | 20 |
| fic1 | ATGCATTTAGACAGGCAGAGT | 21 |
| fic2 | ACTGCGTGATGCCTTTAATGA | 21 |
| oorD1 | ATGGCTAAAATGAGCGCTCCA | 21 |
| oorD2 | TCTCCCTTCTAAAATAGTCTC | 21 |
| porA1 | ATGGCAAAAAGTATTGAATTG | 21 |
| porA2 | AAAAAAGCTCATTTTAGGGCC | 21 |
| 0190-1 | GCTCAGACTTTTTAGTGGGTCGTTTTTG | 28 |
| 0190-2N | ACGCGTCGACTTAAAGCTCTCTTTCAGGAAG | 31 |
| 0190-3N | ATCCACTTTTCAATCTATATCTTTTACCCCACGATTGGCCGC | 42 |
| 0190-4N | CCCAGTTTGTCGCACTGATAACCTTACCAATTCCCTTTCATC | 42 |
| 0226-1 | GCTCTAGAATGGAAGAATCAACAGCGTTT | 29 |
| 0226-2 | ACGCGTCGACTTACCCCATAATGAGCTTGTG | 31 |
| 0226-3 | ATCCACTTTTCAATCTATATCGGTGTAGCACACCACCACCAC | 42 |
| 0226-4 | CCCAGTTTGTCGCACTGATAAGCTCGCCGGTTTTGTTACCGG | 42 |
| 0707-1 | GCTCTAGAGGGGATTGTTTGCAAGAAATA | 29 |
| 0707-2 | ACGCGTCGACCTCATATAACTTACTCATGGC | 31 |
| 0707-3 | ATCCACTTTTCAATCTATATCTTGAGCGTTCAATTCGCTTTC | 42 |
| 0707-4 | CCCAGTTTGTCGCACTGATAAACGCTAAGGATTTGAGCGAGT | 42 |
| 1019-1 | GCTCTAGATTTAAGGAAGTAACCATGATG | 29 |
| 1019-2 | ACGCGTCGACCCACCCCTATCATTTCACCAA | 31 |
| 1019-3 | ATCCACTTTTCAATCTATATCGAATCATGCCACCCAAATCCC | 42 |
| 1019-4 | CCCAGTTTGTCGCACTGATAAAAACCCTAACAAAAAAGAAAC | 42 |
| 1505-1 | GCTCTAGACATGAGACTTTATGAGAGTTT | 29 |
| 1505-2 | ACGCGTCGACAATCTTAAGAGTTTTCTATCC | 31 |
| 1505-3 | ATCCACTTTTCAATCTATATCTTGAACAAATTAAAACGCCCC | 42 |
| 1505-4 | CCCAGTTTGTCGCACTGATAAACCATAAGAACGGACAACCCC | 42 |
| C1 | GATATAGATTGAAAAGTGGAT | 21 |
| C2 | TTATCAGTGCGACAAACTGGG | 21 |
| Rdx1 | CTATCCGGAGTCTTATAA | 18 |
| Rdx2 | TGAATCTCACGCCAAGCA | 18 |
| CAT5Bam | GAGGGTTTCGGGATCCGATAGATTTATGATATAGTGG | 37 |
| CAT3Bam | ATTTGGGCGGGGATCCCGCACTACTCTCGACAGAGAG | 37 |
Screen for motility mutants and detection of flagella.
The H. pylori GPS mutant library was plated from frozen stock to obtain single colonies. Individual clones and the wild-type parent strain were inoculated into plates containing 28 g of Brucella broth/liter, 5% fetal bovine serum, and 0.4% agar. After 4 days, clones that failed to form a 1-cm-diameter halo were picked and retested.
Microarray tracking of transposon site insertion mutants (MATT).
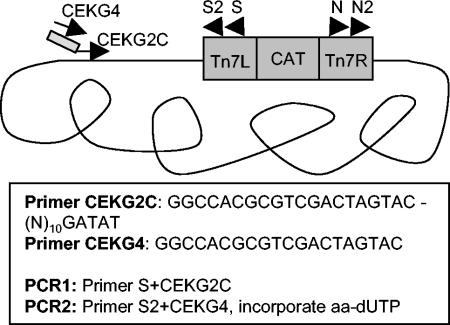
Semirandom PCR was used to amplify DNA-flanking transposon insertions based on a previously established method (12, 33) and is outlined in Fig. 1. Amino-allyl dUTP was incorporated in the second PCR to allow coupling with fluorescent dye for microarray hybridization. Specifically, 100 ng of genomic DNA from a single mutant clone or a pool of mutant clones was PCR amplified in a 20-μl reaction mixture containing 2 μM transposon-specific primer (N or S), 2 μM anchored random primer (CEKG2C or CEKG2A), 0.2 mM deoxynucleoside triphosphates, and 1 U of Taq polymerase with the following cycling conditions: 1 cycle at 94°C for 2 min; 6 cycles at 94°C for 30 s, 42°C for 30 s (−1°C each cycle), and 72°C for 3 min; and 25 cycles at 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min. The reaction products were diluted fivefold, and 1 μl was used as template for a subsequent labeling reaction using a nested transposon specific primer (N2 or S2) and 0.4 mM primer CEKG4; 0.12 mM dATP, dCTP, and dGTP; 0.048 mM dTTP; 0.072 mM amino-allyl-dUTP; and 1 U of Taq polymerase in a 100-μl reaction volume with the following cycling conditions: 1 cycle at 94°C for 30 s and then 30 cycles at 94°C for 30 s, 56°C for 30 s, and 72°C for 2 min. The resulting material was coupled to monofunctional Cy3 or Cy5 dye and hybridized to the H. pylori microarray as described previously (27, 41). Microarrays were analyzed with an Axon scanner with GENEPIX 3.0 software (Axon Instruments, Redwood City, Calif.), and data were further processed with GENEPIX 3.0 software. Microarrays were normalized by the mean of the channel intensity distributions by using the Stanford Microarray Database (SMD) (44). The mean background subtracted signal for channel was collected with SMD. For subsequent analysis, the brightest 1,000 spots from each array were considered as measured by the sum of the mean background-subtracted signal in each channel.
FIG. 1.
MATT method for labeling DNA adjacent to transposon insertions. The first PCR is run with genomic DNA template, primer S reading out from the left arm of the transposon and primer CEKG2C, which contains an anchored degenerate sequence. The second PCR uses the product of the first as template with nested transposon primer S2 and primer CEKG4, complementary to the conserved portion of primer CEKG2C. Amino-allyl dUTP is incorporated in the second PCR to allow conjugation of the resulting product to Cy3 or Cy5 fluorescent dyes for hybridization to the microarray.
For each transposon pool, amplification from the left side of the transposon using primers S and S2 was labeled with Cy3 (green [G]) and amplification from the right side of the transposon using primers N and N2 was labeled with Cy5 (red [R]). To identify chromosomal loci containing transposon insertions in each pool, we analyzed the data as follows to assign each gene a probability of containing an insertion (P[insertion]). We first averaged the logarithm base 2 of the red/green (log2R/G) signal for duplicate spots for each gene. These values were arranged in chromosomal order. Genes containing a transposon insertion would generate signal in both channels, producing a yellow spot defined as −1 < log2R/G < 1 and having a P[insertion] value of 1. To account for cases where the transposon was at the very end of a gene, we identified adjacent gene spots containing strong signal in opposite channels defined by the following formulae: genei log2R/G > 1 and genei + 1 log2R/G < −1 or genei log2R/G < −1 and genei + 1 log2R/G > 1, where i equals gene order number. In such cases each gene was given a P[insertion] value of 0.5. To calculate the number of insertions per gene, the number of insertions was summed across all 20 pools and was rounded down to the nearest integer.
Fine mapping of transposon site insertions.
Sequencing of the precise site of transposon insertion was performed by direct sequencing of agarose gel-purified PCR products obtained by one of two methods. The motility mutants were analyzed by inverse PCR of HindIII-digested genomic DNA of each clone as described previously (40). The insertions in the fic, por, and oor genes were identified by using a PCR containing a gene-specific primer (Table 1) and primer N or S.
Construction of strains containing two copies of candidate essential genes.
A second copy of the gene of interest was integrated at the rdxA locus as previously described (45). Briefly, the entire gene plus 100 to 300 bp of upstream and downstream sequence was amplified with PCR primers, to which restriction site sequences were appended (XbaI, upstream primer, and SalI, downstream primer). This PCR product was digested, purified, and ligated into pRdxA digested with XbaI and SalI. The resulting recombinant clone was plasmid purified, and 2 μg of the product was used to transform wild-type bacteria by natural transformation and selection on Mtz-containing HB plates.
Construction of gene knockout cassettes.
Gene disruption cassettes were constructed according to the method of Chalker et al. (11). Briefly, N-terminal and C-terminal fragments of each gene were amplified by using the upstream and downstream primers designed for the integration of a second copy of the gene combined with gene-internal primers such that both fragments were 250 to 400 bp in length (Table 1). The gene-internal primers had sequences complementary to the full length of primers C1 and C2, which are used to amplify the C. coli Cat gene, appended to the 5′ end. PCR products from each of three individual PCRs (N terminus, C terminus, and Cat gene) were gel purified. A final PCR was performed with 100 ng of each of the three PCR products as template, and the upstream and downstream primers were used to generate the knockout cassette. This final PCR product was verified by agarose gel electrophoresis, and 10 μl of product was directly used for natural transformation (48) of either wild-type bacteria or a strain containing a second copy of the gene and transformants selected on Cm-containing HB plates.
RESULTS
Construction of H. pylori transposon library.
In order to mutagenize the H. pylori chromosome, we took advantage of a recently developed in vitro mutagenesis system based on the Tn7 transposon (7). This system is particularly attractive because it utilizes a TnsCA225V mutant enzyme complex. This protein can activate TnsAB in the absence of the target site specificity factor TnsD to promote transposition into double-stranded DNA in vitro with a target site selectivity that is not significantly different from random (7). We modified the commercially available GPS-M mutagenesis system (NEB) so it could be used in H. pylori by replacing the Tn5 neomycin phosphotransferase gene in the mini Tn7 transposon with the C. coli Cat gene to create pGPS-Cat. This new vector allowed in vitro transposon mutagenesis of H. pylori chromosomal DNA as described in Materials and Methods. The mutagenized DNA was transformed into H. pylori by natural transformation, and chloramphenicol-resistant clones were selected, generating a library of approximately 10,000 clones containing transposon insertions. We determined that individual clones contained single random insertions by Southern blotting of 24 clones (data not shown).
Phenotypic characterization of the transposon library.
To assess the genomic coverage and functional utility of our library, we screened 200 clones for motility mutants on soft agar. Nine clones had motility defects (Table 2). We cloned the flanking DNA by inverse PCR (40) to determine the site of insertion. We isolated two distinct clones with insertions upstream of the H. pylori homologue of flaG and one insertion within fliA, both of which are genes involved in flagellar regulation or assembly. Mot11 contained an insertion in the putative sensor kinase protein product of atoS. A null mutation in this gene eliminates expression of the major flagellar proteins FlaA and FlaB (6). We also isolated a clone bearing an insertion in the ureB gene. Mutations in this gene have been previously shown to have altered chemotaxis and motility (36), though recent work demonstrates that ureB does not play a direct role in motility (46). The remaining three genes do not have informative homologies. In our very limited screen we find 4.5% of our insertions affect motility, while 3.0% of the sequenced open reading frames present in strain G27 are predicted to function in motility.
TABLE 2.
Motility mutants
| Mutant identity | TIGR identitya | Descriptionb |
|---|---|---|
| mot2 | HP0326 | neuA/flaG |
| mot3 | HP0326 | neuA/flaG |
| mot4 | HP0072 | ureB |
| mot7 | HP1034 | Putative ATP binding |
| mot10 | HP0244 | atoS |
| mot13 | HP1032 | fliA |
| mot15 | HP1479 | H. pylori specificc |
| mot18 | HP1327 | H. pylori specificc |
| mot20 | HP1023 | H. pylori specificc |
TIGR, The Institute for Genomic Research; 26695 locus annotation.
Gene description based on published annotation (47).
no homology to any genes in the database.
Mapping of transposon library insertion sites.
We devised a method (MATT) using a whole-genome H. pylori microarray to monitor the presence of transposon library clones in a pool. The MATT method depends on amplification of the DNA flanking the transposon insertion site by using a combination of specific and nonspecific primers (12, 33). The specific primers hybridize to sequences within the transposon, and the nonspecific primers contain 10 degenerate nucleotides followed by a unique anchor of 4 or 5 nucleotides. We used two different nonspecific primers with different anchor sequences. Primer CEKG2A ended in AGAG and anneals, on average, every 260 bp in the chromosome, based on the published 26695 genome sequence (47). Primer CEKG2C ends in GATAT and anneals, on average, every 1,100 bp in the chromosome. The amplified DNA was coupled to a fluorescent dye and was hybridized to a whole-genome DNA microarray revealing the genomic position of the transposon insertion of individual clones. The amplification method is outlined in Fig. 1.
To test the reproducibility of this method and to further characterize our transposon library, we used MATT to map the insertion site of 20 pools of 300 transposon insertion clones. We performed separate labeling reactions for each side of the transposon and coupled the reaction primed from the right arm of the transposon to Cy5 dye (red) and the reaction primed from the left arm of the transposon to Cy3 dye (green). The two reactions were then combined and hybridized to the microarray. If the transposon landed within the middle of a gene, it should generate signal in the labeling reactions from both sides of the transposon, resulting in a yellow gene spot. If the transposon landed near the 5′ or 3′ end of a gene or in between two genes, one would expect spots from the two chromosomally adjacent genes to give a strong hybridization signal in opposite channels (adjacent red and green spots). We performed and hybridized separate labeling reactions by using the two nonspecific degenerate primers (CEKG2A and CEKG2C). Some genes were detected with both degenerate primers, while many were detected only with one primer (Table 3). We mapped loci in 20 pools, expecting 300 distinct insertions per pool, and detected an average of 268 (standard deviation, ±42) insertions per pool. To calculate the number of insertions for each gene, we summed the probabilities for insertions for each gene across all the pools. We were able to map 5,363 insertion sites of the expected 6,000 (89%). Each gene had between 0 and 19 transposon hits (supplemental Table S1). The raw hybridization data for each array are available at http://genome-www5.stanford.edu.
TABLE 3.
Transposon (Tn) insertions detected by MATT
| Pool identity | No. of Tn insertions detected by MATT fora:
|
|||
|---|---|---|---|---|
| Primer 2A | Primer 2C | Commonb | All hitsc | |
| Pool1 | 211.5 | 202 | 51.5 | 362 |
| Pool2 | 203.5 | 243 | 75.5 | 371 |
| Pool3 | 228 | 104.5 | 52 | 280.5 |
| Pool4 | 237 | 90 | 57 | 270 |
| Pool5 | 195 | 141 | 59.5 | 276.5 |
| Pool6 | 192 | 124.5 | 62 | 254.5 |
| Pool7 | 188 | 117 | 55.5 | 249.5 |
| Pool8 | 181 | 119.5 | 55.5 | 245 |
| Pool9 | 206 | 119.5 | 40 | 285.5 |
| Pool10 | 244.5 | 39.5 | 17 | 267 |
| Pool11 | 180.5 | 172 | 49 | 303.5 |
| Pool12 | 192 | 128.5 | 47.5 | 273 |
| Pool13 | 204 | 122 | 53.5 | 272.5 |
| Pool14 | 195 | 113.5 | 70 | 238.5 |
| Pool15 | 182 | 85 | 33 | 234 |
| Pool16 | 147 | 103 | 46.5 | 203.5 |
| Pool17 | 207.5 | 145.5 | 63 | 290 |
| Pool18 | 168 | 102 | 49.5 | 220.5 |
| Pool19 | 166 | 123 | 50.5 | 238.5 |
| Pool20 | 164.5 | 109.5 | 46.5 | 227.5 |
| Total | 3,893 | 2,504.5 | 1,034.5 | 5,363 |
Gene insertions were detected based on microarray hybridization signal as described in Materials and Methods and were summed for each pool.
Genes with insertions detected in each of the two separate degenerate primer-labeling reactions.
Total insertions detected by the two separate degenerate primer-labeling reactions.
In an attempt to address the sensitivity and specificity of the MATT method for mapping transposon insertions, we performed the analysis described above on a pool of nine clones for which the transposon insertion site had been already mapped by sequencing of the transposon junction. These were Mot2, -4, -7, -10, -13, -15, and -18, an insertion within porB (see below), and an insertion in HP0954. Of 1,660 genes with spots on our microarrays, the genes corresponding to all nine clones gave a positive signal for insertion as well as 29 false positives. This experiment yielded a sensitivity of 100% (9 out of 9) and a specificity of 98% (29 out of 1,651).
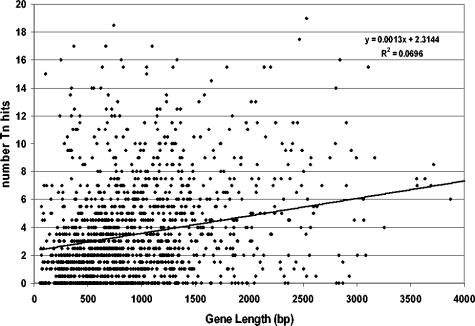
We found transposon insertions distributed throughout the genome. Twenty genes (1.3%) were predicted to have insertions in 15 or more of the 20 pools and may represent hot spots for insertions or primer-specific artifacts as described above. There was a very weak correlation between gene size and the number of transposon insertions detected (Fig. 2). To investigate the role of polarity in the transposon insertions, we compared the frequency of insertions in the first gene of multigene operons to that of single-gene operons. If insertions in the first gene of the operon had polar effects on essential genes downstream, we might expect a higher percentage of genes with zero hits in the first gene of multigene operons. In fact, we observed a higher frequency of genes with zero hits in single-gene operons (73 out of 456; 16%) than in the first genes of multigene operons (63 out of 777; 8%), indicating that polar effects may not play a large role in recovery of insertion mutants with this transposon.
FIG. 2.
Number of transposon (Tn) insertions per gene weakly correlates with gene size in base pairs. A linear regression curve is shown according to y = 0.0013x + 2.3144, where R2 = 0.0696.
Enrichment for essential genes.
We were particularly interested in genes that had no measurable transposon insertions. In principal, this gene class represented genes that cannot be disrupted because they are essential for growth in vitro. Strain G27 contains 1,552 of the 1,660 genes represented on the H. pylori microarray, as assessed by genomic DNA hybridization (41). Of these genes, 344 (23%) had zero transposon hits and, by their annotation, included many that are essential for growth in vitro in other bacteria. We would expect only 89 genes (6%) to have escaped transposon insertion if the distribution of transposon hits followed a Poisson distribution. Thus, we had nearly four times more genes with no transposon insertions than would be expected by chance.
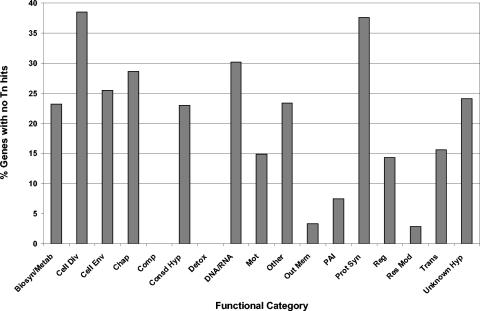
We divided the genes into functional classes and determined the percentage of genes in each class for which we could not observe transposon insertions and, thus, are putative essential genes (Fig. 3). Certain functional classes had a high percentage of essential genes (>25%) and included cell division (38.5%), protein synthesis (37.6%), DNA-RNA synthesis and metabolism (30.2%), chaperones (28.6%), and cell envelope biosynthesis (25.5%), consistent with their involvement in essential processes. Conversely, two categories that contained a large number of genes, outer membrane proteins (60) and restriction modification genes (36), had a very low percentage of essential genes (3.3 and 2.9%, respectively). The H. pylori genome contains a very large family of outer membrane proteins, some of which undergo phase variation in expression (2). While these proteins are thought to be important in the host as adhesins or to counter antigenic stimulation, individually they might not be expected to be required for growth in vitro. Similarly, restriction modification genes may be important for gene regulation and protection of this naturally competent organism from foreign DNA in the host environment where superinfecting strains and organisms would be present, but they would not necessarily be expected to alter fitness during monoculture in vitro. Interestingly, both the conserved hypothetical and unknown (H. pylori-specific) hypothetical classes contained a fairly high percentage of genes with zero insertions (23.0 and 24.0%, respectively), suggesting that there may be a significant number of essential gene products in these two categories.
FIG. 3.
Genes with no transposon (Tn) insertions are enriched in some functional classes but not in others. The percentage of genes with no transposon insertions in each functional class is indicated. Abbreviations: Biosyn/Metab, biosynthesis and metabolism; Cell Div, cell division; Cell Env, cell envelope; Chap, chaperone; Comp, DNA competence; Detox, detoxification; DNA/RNA, DNA and RNA metabolism; Mot, motility; Out Mem, outer membrane; PAI, pathogenicity island; Prot Syn, protein synthesis; Reg, regulation; Res Mod, restriction modification; Trans, transporters; Unknown Hyp, unknown hypothetical.
Precise localization of insertion site predicted by MATT.
Our analysis of genes that escaped transposon insertion indicates that while there may certainly be some that are not essential for in vitro growth, as many as 75% of the genes with zero insertions should be essential. Comparing our results with those in the published literature, however, we found that we observed insertions in several genes previously predicted to be essential in H. pylori. Of the 34 genes predicted to be essential by Chalker et al. (11), we detected insertions in 15 of the genes (44%). We investigated three loci where we had conflicting data: fic, porGDAB, and oorDABC, all of which were predicted to be essential in the previous study. fic (HP1159) encodes a putative cyclic AMP-induced cell filamentation gene that is not essential in E. coli. We went back to the pools that were predicted to contain insertions in this gene and performed PCR with primers reading in from the 5′ and 3′ ends of the gene combined with primers reading out from either end of the transposon. We sequenced PCR products from three pools predicted to contain insertions in fic to determine the exact site of the transposon insertion. We found insertions at base pairs 83 and 336 of the coding sequence. The third insertion actually mapped to base pair 210 of the upstream open reading frame, consistent with the fact that the MATT PCR product was larger than the predicted size of the fic gene. porGDAB (HP1108 to HP1111) encodes subunits of a predicted pyruvate flavodoxin oxidoreductase. While we detected no insertions in the porG gene, we detected insertions in each of the other subunits. We sequenced an insertion at base pair 332 of the porB coding sequence. oorDABC (HP0588 to HP0591) encodes subunits of a putative 2-oxoglutarate oxidoreductase. We detected no insertions in oorABC but predicted several insertions in oorD. PCR analysis from the pools predicted to contain insertions in oorD with gene-specific primers revealed no PCR products that were the same size or smaller than the oorD coding sequence, indicating that the insertions we predicted to lie in this gene in fact lie in adjacent sequences and supporting the entire oor locus being essential. These results indicate that in a minority of cases we overestimated the existence of insertions within a given open reading frame, because our hybridization signal was generated from an insertion in an adjacent gene. In all cases, however, we were able to detect insertions closely linked to the locus, where we predicted an insertion by MATT. For these three loci, two previously predicted to be essential are not essential in our strain while one is essential, in spite of the fact that we predicted an insertion in one of the four genes in the operon.
Confirmation of predicted essential genes.
As stated above, 344 genes were predicted by MATT to have escaped transposon insertion, and we expect 74% of these genes to be essential. Named (annotated) genes accounted for 187 or 54% of our list of putative essential genes. While many of the genes in this list seemed to obviously encode essential functions, such as the cell division protein FtsZ or subunits of RNA polymerase, there were also apparent false positives, such as six flagellar biosynthesis genes, two pathogenicity island genes, and one urease subunit gene. Therefore, we directly tested the essentiality of four genes which either had no obvious annotation or were annotated to be genes whose essentiality was at best ambiguous compared to experimental data from other bacterial species. Additionally, we tested one gene where we detected a single transposon insertion, while the same gene was expected to be essential in H. pylori (Table 4).
TABLE 4.
Transformations with knockout cassettes
| Name | Description | Chloramphenicol-resistant transformants
|
|
|---|---|---|---|
| 1 Copya | 2 Copyb | ||
| HP0190 | Conserved hypothetical | 0 | + |
| HP0226 | Conserved hypothetical integral membrane protein | + | + |
| HP0707 | mraW, S-adenosyl-methyltransferase | + | + |
| HP1019 | htrA, serine protease | 0 | + |
| HP1505 | ribG, putative riboflavin-specific deaminase | 0 | + |
Transformation into wild-type strain G27 with one copy of gene of interest.
Transformation into strain G27 containing a second copy of the gene at the rdxA locus.
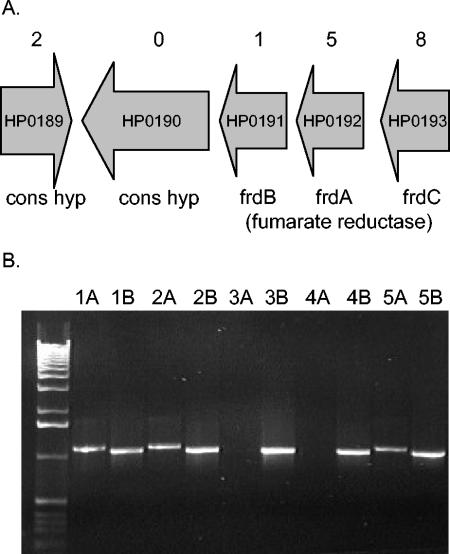
For example, gene HP0190 encodes a conserved hypothetical protein containing homology to phospholipase D enzymes which, by our analysis, might be essential in H. pylori. It is predicted to be the fourth gene in an operon encoding three subunits of fumarate reductase (Fig. 4A). We detected transposon insertions in each of the fumarate reductase subunits, consistent with the experimental observation in E. coli that these genes are not essential. Similarly, two insertions were predicted for HP0189, which is predicted to encode a conserved hypothetical integral membrane protein and is transcribed in the opposite direction of HP0190. To test the essentiality of this gene, we attempted to directly mutate it by transformation with a deletion-insertion knockout cassette containing a chloramphenicol resistance marker. If HP0190 was essential, we would expect no transformation, that is, no chloramphenicol-resistant colonies. To rule out cloning or transformation difficulties, we created a strain with a second copy of HP0190 integrated at the rdxA locus by using pRdxA as described by Smeets et al. (45) and repeated the transformation in the strain with two copies. In this case, one would expect transformants (gene insertion) even if the gene were essential. Transformation of the cassette directed to knock out HP0190 into the wild-type strain yielded no chloramphenicol-resistant colonies, but transformation into the strain with two copies of the gene yielded transformants (Table 4). We performed PCR to demonstrate that the knockout cassette could integrate at either the wild-type or rdxA locus (Fig. 4B). In this experiment, two of five transformants had the knockout cassette integrated in the gene copy at the native locus while three of five showed integration at the rdxA locus. For the clones where the native locus was knocked out, HP0190 could be sufficiently expressed at the rdxA locus to perform its essential function. HP0190 is predicted to be the fourth gene in an operon. While we included 150 bp of sequence upstream of the translation initiation codon in the integration construct, transcription likely is driven by the native promoter of the rdxA locus. We conclude that the lack of transformation in the strain with one copy of the gene was due to the fact that HP0190 is essential and not because of polar effects on another gene(s) or peculiarities of recombination at this locus.
FIG. 4.
HP0190 is an essential gene. (A) Genomic region surrounding HP0190. Gene annotation is according to Tomb et al. (47). Numbers above each open reading frame indicate the number of transposon insertions detected by MATT. (B) A knockout cassette can integrate at either locus containing HP0190 in a strain containing a second copy at the rdxA locus. The number above each lane indicates independent chloramphenicol-resistant clones. The letters represent the primer pair used for amplification. A lanes (e.g. 1A, 2A, etc.) depict PCR results with primers rdx1 and cat2, which will only give a product if integration occurred at the rdxA locus. B lanes indicate PCR with primers HP0190-1 and cat2, which give a product for integration at the native locus and the rdxA locus. Reactions were fractionated on a 0.8% agarose gel, and DNA was visualized by ethidium bromide staining. The far left lane contains a 1-kb ladder DNA standard (Gibco-BRL). Clones 1, 2, and 5 have integrated the knockout cassette into the gene copy at the rdxA locus. Clones 3 and 4 have integrated the knockout cassette into the gene copy at the native HP0190 locus.
We analyzed the remaining four predicted open reading frames by using the same strategy. Two were confirmed to be essential. HP1019 is predicted to encode an htrA-type serine protease and had no observable transposon insertions by MATT. While in E. coli htrA (degP) is only required for growth at high temperature, we could only obtain HP1019 knockout transformants from the strain containing two copies of the gene. Interestingly, in this case the knockout cassette could only integrate into the gene copy at the rdxA locus, indicating that for this gene, insufficient gene expression could be driven from the rdxA locus to complement the essential function of the gene. We also confirmed that HP1505 is an essential gene. HP1505 encodes a riboflavin deaminase/reductase (ribD) that has been characterized by cross-complementation of E. coli riboflavin biosynthesis mutants (17). While this gene is not required for growth in E. coli when riboflavin is present, it was reported that riboflavin-deficient mutants of H. pylori could not be constructed even when additional riboflavin was used to supplement the medium (17). We detected a single transposon insertion in this gene by MATT, but when we attempted to directly knock out this gene we were unable to recover transformants unless a second copy of the gene was present. In this case we detected integration of the knockout cassette at both the wild-type and rdxA locus. Looking back at the data from pool 4, the single pool for which we detected an insertion in gene HP1505, we also detected an insertion in the adjacent gene HP1506. Insertions in HP1506 were detected in several pools; thus, transposon insertion in this gene was the likely source of the false-positive signal we detected for gene HP1505.
Two of the five genes we tested were not essential, as evidenced by the fact that we could recover transformants of the gene knockout cassette in the wild-type strain (containing a single copy of the gene). These genes were HP0226, which encodes a putative conserved hypothetical integral membrane protein, and HP0707, which shows a high degree of similarity to a conserved family of S-adenosyl-methyltransferases (mraW). Interestingly, mraW homologues are found in both gram-positive and gram-negative bacteria (but not in Archaea), often in a cluster of genes involved in cell division, such as ftsZ (10). While mraW is essential in E. coli (10, 20) and Mycobacterium tuberculosis (43), it is not essential in Bacillus subtilis, as measured by targeted disruption (16), or in Haemophilus influenzae, as measured by genomic footprinting (1).
Of the five genes tested, three (60%) only gave transformants in a strain containing two copies of the gene, indicating they are essential, while two gave transformants, indicating they are not essential genes. We biased our experiment towards investigating genes with the highest likelihood of being false positives in our screen for essential genes. In spite of this, our results support our hypothesis that genes with no insertions are highly enriched for essential genes.
DISCUSSION
Phylogenetically, H. pylori groups with the δ/ɛ-proteobacteria, which is the oldest subdivision of the proteobacteria (23). The plasmid and transposon vectors useful in the α-, β-, and γ-protoebacteria have not been effective in this organism, hampering its genetic analysis. Previous attempts at transposon mutagenesis employed shuttle mutagenesis strategies in E. coli (9, 30, 39). Perhaps due to restriction barriers or instability of H. pylori DNA clones in E. coli, these libraries have tended to be of low complexity, with less than one genome's equivalent of mutant clones. We exploited H. pylori's natural competence for DNA uptake and its propensity for homologous recombination by double crossovers in combination with a high-efficiency, random in vitro mini Tn7-based transposon system (7) to generate a library of 10,000 insertional mutants. Characterization of this transposon library revealed that the transposon insertions are well distributed throughout the chromosome. As the H. pylori genome contains roughly 1,500 genes, this represents the first comprehensive mutant library described for this organism, with approximately sixfold genome coverage.
We developed a method to map the genomic locus of transposition insertion for pools of clones. This represents a variation on a similar method developed by Sassetti et al. (TraSH) to identify conditionally essential genes in mycobacteria (42). The major difference in the two methods lies in the amplification of the transposon-flanking sequences for hybridization to microarrays. Our method uses semirandom PCR directly on genomic DNA from pools of clones, while the other method uses transcription of size-selected restriction-digested genomic DNA to which a T7 promoter has been ligated.
We tested the fidelity of our method in pilot experiments containing clones with previously sequenced insertion boundaries and by retrieving clones with predicted insertions from several pools of random clones. Our pilot experiment with a very small pool of nine clones gave a very high sensitivity (100%) with relatively high specificity (98%). This level of sensitivity was not matched in the overall screen, where only 89% of the expected insertions were mapped. This may result from regions of the genome with a dearth of binding sites for the two anchors used in our amplification strategy. While the sensitivity could be improved, possibly with the use of additional nonspecific primers, for this analysis we simply analyzed additional pools of mutants. Our experiments mapping predicted transposon clone insertion sites revealed that we consistently identified the correct locus. This empirically verified the utility of this method and suggested that the specificity of the overall screen was not very different from that of the pilot pool.
Using our mapping method, we were able to localize the site of transposon insertions for 5,363 random clones, over three genome equivalents. We found insertions distributed throughout the chromosome consistent with the lack of sequence specificity documented for the TnsABCA255V enzyme. Still, we did detect a small number of genes that appear to have a high frequency of insertions. Further detailed analysis of the transposon junction of these insertions will be required to determine if these represent hot spots for transposition or an artifact of our detection method. We observed little bias against small genes or indication of polar effects. The lack of polar effects was confirmed globally by failing to find an increased frequency in genes with zero transposon insertions of the first gene of predicted operons. Furthermore, two of the confirmed essential genes lay downstream of nonessential genes in which we detected transposon insertions.
We found an almost fourfold higher frequency of genes with no detectable insertions than we would have expected by chance. We hypothesized that the major reason we failed to detect insertions in a given gene is that it encodes a protein required for viability, i.e., an essential gene. This same logic has been employed to query a number of bacterial genomes for essential and nonessential genes, using a variety of approaches to map the site of transposon insertion. These include direct sequencing of the transposon junction for Mycoplasma genitalium (26) and Salmonella enterica serovar Typhimurium (31), genomic footprinting of H. influenzae (1) and E. coli (20), and microarray hybridization in Mycobacterium tuberculosis (43). These studies have estimated the number of essential genes to range from 265 to 620, representing 16 to 51% of the predicted open reading frames. In addition to transposon mutagenesis analysis, the essential gene set has been approached by other methods, including bioinformatic predictions based on pathway analysis (35), antisense RNA for Staphylococcus aureus (19, 29), and targeted gene deletion for Saccharomyces cerevisiae, B. subtilis, and E. coli (21, 32, 51). While targeted gene deletion is the most rigorous test, complete genome analysis by this method requires considerable resources and has been achieved to completion only for the model eukaryote S. cerevisiae (21) and for a subset of the B. subtilis genome (32). The only organism for which both transposon mapping and target gene disruption data have been compiled for a large fraction of genes is E. coli. In this case, 27% of genes gave conflicting results for essentiality by the two methods (20). One reason for this discrepancy is that transposon-based methods involve growing a pool of mutants in competition with each other. Thus, mutations that confer a growth disadvantage as well as those that are absolutely essential will both be underrepresented. Regardless, transposon mapping methods clearly identify the majority of truly essential genes as well as identify genes that confer some growth disadvantage.
When we attempted to compare data for the genes that we predicted to be essential to similar data from other microbes, the analysis was complicated by different nomenclature used for the same genes in different organisms. Of the 187 named genes in our list, 102 are essential in E. coli, H. influenzae, or M. tuberculosis. Of these, only 20 genes are essential in all four organisms, while 46 genes are essential in three species and 36 genes are essential in two species. It seems noteworthy that pairwise comparison of the H. pylori essential gene set with other microorganisms shows the most overlap with the gram-positive M. tuberculosis (66 genes). Perhaps this is not surprising, considering that H. pylori resides in the δ/ɛ branch of the proteobacteria while E. coli and H. influenzae reside in the γ branch and are thought to be the more divergent from the gram-positive bacteria, which include mycobacteria (23).
Of the genes we tested directly, HP0190 encodes a conserved hypothetical protein with putative phospholipase D activity. While this activity has been measured in a variety of bacteria (13), mutants of these genes in Pseudomonas aeruginosa (50) and Yersinia pestis (24) are required for full virulence in some hosts but not for survival in vitro. Thus, we were quite surprised to find it essential for H. pylori growth in rich media. HP1019 encodes a putative serine protease of the HtrA subfamily. These highly conserved enzymes, found in both prokaryotes and eukaryotes, have chaperone activity in addition to protease activity, and some homologues have lost protease activity and retain only chaperone properties. E. coli contains three homologues, none of which are required for viability, though degP is required for growth at high temperature. The single H. pylori homologue, HP1019, contains the protease catalytic triad and is most similar to degP. In H. pylori this gene appears to be essential. In H. influenzae there are two family members; the one with highest similarity to HP1019 is not essential as measured by genomic footprinting, while the second member could not be assessed by this method (1). In contrast, M. tuberculosis contains three family members, but the one most similar to HP1019 was also found to be essential by mapping of transposon mutants (43). Whether the protease or chaperone activity or both are essential in H. pylori remains an open question. HP1019 may be essential in H. pylori due to lack of redundancy in the genome or may suggest that, even under optimal growth temperature, it requires HP1019 function for the assembly of some essential factor which, in other organisms, is only needed at higher temperatures. HP1505 encodes a riboflavin biosynthesis protein that we had not expected to be essential for growth in rich media but, consistent with other attempts to disrupt this pathway in H. pylori (17), no mutants could be recovered unless a second copy of the gene was present. Finally, we investigated an S-adenosyl-methyltransferase encoded by HP0707 that is essential in both M. tuberculosis and E. coli (10, 20, 43) but not in B. subtilis (16), and it is predicted to be nonessential in H. influenzae based on genetic footprinting (1). We could directly show this gene is not essential in H. pylori in spite of detecting no transposon insertions in this gene. These results show that outside of perhaps a few highly conserved processes, such as protein synthesis, the essential gene set varies from organism to organism, even for nutrient-replete growth in vitro. Thus, bioinformatics approaches must be coupled with experimental methods such as transposon mutagenesis to identify those truly essential genes in the most rigorous manner.
We verified both the essentiality and nonessentiality of several genes. These analyses revealed that only a minority of genes that we predicted to be essential do not prove to be essential when tested by an independent measure. When picking genes to test, we chose genes where we had no information based on homology or for which the gene was not essential in at least some organisms where homologues had been studied and, thus, biased our sample towards those most likely to be false positives. Still, two out of four genes were found to be essential, indicating that the list of genes with zero transposon hits is very highly enriched for truly essential genes. Conversely, in a minority of cases we predict the presence of insertions in essential genes, because the actual transposon insertion lies in an adjacent gene. Thus, while the genes with zero hits include a certain number of false positives, they underestimate the essential gene set in H. pylori.
Our data represent the first comprehensive analysis of the full set of essential genes in H. pylori and provide a number of potential targets for new antimicrobial design. In addition, the methods for saturation mutagenesis of the chromosome and for monitoring the behavior of pools of transposon mutants opens the door for genetic analysis of virulence-associated phenotypes in this important human pathogen. Indeed, the general method can be productively employed for the detection of essential genes in any selective environment, including that of infected animals.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from The Pew Charitable Trusts (N.R.S.), a New Development fund from the Fred Hutchinson CR (N.R.S.)., and National Institutes of Health grants AI38459 and CA92229 (S.F.).
We thank members of the Falkow lab and the Salama lab for helpful discussions. We thank David Baldwin, Chris Cosma, and Delia Pinto-Santini for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm, R. A., J. Bina, B. M. Andrews, P. Doig, R. E. Hancock, and T. J. Trust. 2000. Comparative genomics of Helicobacter pylori: analysis of the outer membrane protein families. Infect. Immun. 68:4155-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 397:176-180. [DOI] [PubMed] [Google Scholar]
- 4.Ausebel, F., R. Brent, R. Kingston, D. Moore, J. Seidman, J. Smith, and K. Stuhl (ed.). 1997. Short protocols in molecular biology, 3rd ed. John Wiley & Sons, New York, N.Y.
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 6.Beier, D., and R. Frank. 2000. Molecular characterization of two-component systems of Helicobacter pylori. J. Bacteriol. 182:2068-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biery, M. C., F. J. Stewart, A. E. Stellwagen, E. A. Raleigh, and N. L. Craig. 2000. A simple in vitro Tn7-based transposition system with low target site selectivity for genome and gene analysis. Nucleic Acids Res. 28:1067-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bijlsma, J. J., A. L. M. Lie, I. C. Nootenboom, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. Identification of loci essential for the growth of Helicobacter pylori under acidic conditions. J. Infect. Dis. 182:1566-1569. [DOI] [PubMed] [Google Scholar]
- 9.Bijlsma, J. J., C. M. Vandenbroucke-Grauls, S. H. Phadnis, and J. G. Kusters. 1999. Identification of virulence genes of Helicobacter pylori by random insertion mutagenesis. Infect. Immun. 67:2433-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrion, M., M. J. Gomez, R. Merchante-Schubert, S. Dongarra, and J. A. Ayala. 1999. mraW, an essential gene at the dcw cluster of Escherichia coli codes for a cytoplasmic protein with methyltransferase activity. Biochimie 81:879-888. [DOI] [PubMed] [Google Scholar]
- 11.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chun, K. T., H. J. Edenberg, M. R. Kelley, and M. G. Goebl. 1997. Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13:233-240. [DOI] [PubMed] [Google Scholar]
- 13.Cole, R., and P. Proulx. 1975. Phospholipase D activity of gram-negative bacteria. J. Bacteriol. 124:1148-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, and R. Rappuoli. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. USA 90:5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Covacci, A., J. L. Telford, G. Del Giudice, J. Parsonnet, and R. Rappuoli. 1999. Helicobacter pylori virulence and genetic geography. Science 284:1328-1333. [DOI] [PubMed] [Google Scholar]
- 16.Daniel, R. A., A. M. Williams, and J. Errington. 1996. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J. Bacteriol. 178:2343-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.deJong, R., D. Bakker, A. H. M. vanVliet, E. J. Kuipers, C. M. J. E. Vandenbroucke-Grauls, and J. G. Kusters. 2003. Direct random insertion mutagenesis of Helicobacter pylori. J. Microbiol. Methods 52:93-100. [DOI] [PubMed] [Google Scholar]
- 17.Fassbinder, F., M. Kist, and S. Bereswill. 2000. Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol. Lett. 191:191-197. [DOI] [PubMed] [Google Scholar]
- 18.Finlay, B. B., and S. Falkow. 1997. Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61:136-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, K. G. C. P. King, M. McCarthy, C. Malone, B. Misiner, D. Robbins, Z. Tan, Z. Y. Zhu Zy, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 22.Guillemin, K. J., and N. R. Salama. 2002. Helicobacter pylori functional genomics, p. 291-320. In B. Wren and N. Dorrell (ed.), Functional microbial genomics, vol. 33. Academic Press, London, United Kingdom. [Google Scholar]
- 23.Gupta, R. S., and E. Griffiths. 2002. Critical issues in bacterial phylogeny. Theor. Popul. Biol. 61:423-434. [DOI] [PubMed] [Google Scholar]
- 24.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and A. Forsberg. 2002. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 25.Hofreuter, D., S. Odenbreit, G. Henke, and R. Haas. 1998. Natural competence for DNA transformation in Helicobacter pylori: identification and genetic characterization of the comB locus. Mol. Microbiol. 28:1027-1038. [DOI] [PubMed] [Google Scholar]
- 26.Hutchison, C. A., S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 27.Israel, D. A., N. Salama, U. Krishna, U. M. Rieger, J. C. Atherton, S. Falkow, and R. M. Peek, Jr. 2001. Helicobacter pylori genetic diversity within the gastric niche of a single human host. Proc. Natl. Acad. Sci. USA 98:14625-14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenks, P. J., L. C. Chevalier, C. Ecobichon, and A. Labigne. 2001. Identification of nonessential Helicobacter pylori genes using random mutagenesis and loop amplification. Res. Microbiol. 152:725-734. [DOI] [PubMed] [Google Scholar]
- 29.Ji, Y., G. Woodnutt, M. Rosenberg, and M. K. Burnham. 2002. Identification of essential genes in Staphylococcus aureus using inducible antisense RNA. Methods Enzymol. 358:123-128. [DOI] [PubMed] [Google Scholar]
- 30.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197:813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knuth, K., H. Niesalla, C. J. Hueck, and T. M. Fuchs. 2004. Large-scale identification of essential Salmonella genes by trapping lethal insertions. Mol. Microbiol. 51:1729-1744. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Debarbouille, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauel, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. Seegers, J. Sekiguchi, A. Sekowska, S. J. Seror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manoil, C. 2000. Tagging exported proteins using Escherichia coli alkaline phosphatase gene fusions. Methods Enzymol. 326:35-47. [DOI] [PubMed] [Google Scholar]
- 34.Montecucco, C., and R. Rappuoli. 2001. Living dangerously: how Helicobacter pylori survives in the human stomach. Nat. Rev. Mol. Cell Biol. 2:457-466. [DOI] [PubMed] [Google Scholar]
- 35.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakamura, H., H. Yoshiyama, H. Takeuchi, T. Mizote, K. Okita, and T. Nakazawa. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect. Immun. 66:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odenbreit, S., M. Till, and R. Haas. 1996. Optimized BlaM-transposon shuttle mutagenesis of Helicobacter pylori allows the identification of novel genetic loci involved in bacterial virulence. Mol. Microbiol. 20:361-373. [DOI] [PubMed] [Google Scholar]
- 38.Odenbreit, S., M. Till, D. Hofreuter, G. Faller, and R. Haas. 1999. Genetic and functional characterization of the alpAB gene locus essential for the adhesion of Helicobacter pylori to human gastric tissue. Mol. Microbiol. 31:1537-1548. [DOI] [PubMed] [Google Scholar]
- 39.Odenbreit, S., B. Wieland, and R. Haas. 1996. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J. Bacteriol. 178:6960-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pang, K. M., and D. A. Knecht. 1997. Partial inverse PCR: a technique for cloning flanking sequences. BioTechniques 22:1046-1048. [DOI] [PubMed] [Google Scholar]
- 41.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. USA 98:12712-12717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 44.Sherlock, G., T. Hernandez-Boussard, A. Kasarskis, G. Binkley, J. C. Matese, S. S. Dwight, M. Kaloper, S. Weng, H. Jin, C. A. Ball, M. B. Eisen, P. T. Spellman, P. O. Brown, D. Botstein, and J. M. Cherry. 2001. The Stanford Microarray Database. Nucleic Acids Res. 29:152-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smeets, L. C., J. J. Bijlsma, S. Y. Boomkens, C. M. Vandenbroucke-Grauls, and J. G. Kusters. 2000. comH, a novel gene essential for natural transformation of Helicobacter pylori. J. Bacteriol. 182:3948-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, S., and D. E. Berg. 2004. Motility of urease-deficient derivatives of Helicobacter pylori. J. Bacteriol. 186:885-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. (Erratum, 389:412.) [DOI] [PubMed] [Google Scholar]
- 48.Wang, Y., K. P. Roos, and D. E. Taylor. 1993. Transformation of Helicobacter pylori by chromosomal metronidazole resistance and by a plasmid with a selectable chloramphenicol resistance marker. J. Gen. Microbiol. 139:2485-2493. [DOI] [PubMed] [Google Scholar]
- 49.Wang, Y., and D. E. Taylor. 1990. Chloramphenicol resistance in Campylobacter coli: nucleotide sequence, expression, and cloning vector construction. Gene 94:23-28. [DOI] [PubMed] [Google Scholar]
- 50.Wilderman, P. J., A. I. Vasil, Z. Johnson, and M. L. Vasil. 2001. Genetic and biochemical analyses of a eukaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol. 39:291-303. [DOI] [PubMed] [Google Scholar]
- 51.Yu, B. J., B. H. Sung, M. D. Koob, C. H. Lee, J. H. Lee, W. S. Lee, M. S. Kim, and S. C. Kim. 2002. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 20:1018-1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.