Abstract
Autophagy involves the lysosomal degradation of cytoplasmic contents for regeneration of anabolic substrates during nutritional or inflammatory stress. Its initiation occurs rapidly after inactivation of the protein kinase mammalian target of rapamycin (mTOR) (or mechanistic target of rapamycin), leading to dephosphorylation of Unc-51-like kinase 1 (ULK1) and autophagosome formation. Recent studies indicate that mTOR can, in parallel, regulate the activity of stress transcription factors, including signal transducer and activator of transcription-1 (STAT1). The current study addresses the role of STAT1 as a transcriptional suppressor of autophagy genes and autophagic activity. We show that STAT1-deficient human fibrosarcoma cells exhibited enhanced autophagic flux as well as its induction by pharmacological inhibition of mTOR. Consistent with enhanced autophagy initiation, ULK1 mRNA and protein levels were increased in STAT1-deficient cells. By chromatin immunoprecipitation, STAT1 bound a putative regulatory sequence in the ULK1 5′-flanking region, the mutation of which increased ULK1 promoter activity, and rendered it unresponsive to mTOR inhibition. Consistent with an anti-apoptotic effect of autophagy, rapamycin-induced apoptosis and cytotoxicity were blocked in STAT1-deficient cells but restored in cells simultaneously exposed to the autophagy inhibitor ammonium chloride. In vivo, skeletal muscle ULK1 mRNA and protein levels as well as autophagic flux were significantly enhanced in STAT1-deficient mice. These results demonstrate a novel mechanism by which STAT1 negatively regulates ULK1 expression and autophagy.
Keywords: apoptosis, autophagy, mammalian target of rapamycin (mTOR), proteolysis, signal transducers and activators of transcription 1 (STAT1)
Introduction
Autophagy is a highly conserved eukaryotic stress and survival response that degrades cytoplasmic contents for the recycling of biosynthetic substrates and energy production (for review, see Ref. 1). In macroautophagy, the initial step involves formation of a double membrane vesicle (autophagosome) that can incorporate long-lived cytosolic proteins and organelles (e.g. mitochondria, peroxisomes). Upon fusion with the lysosome, autophagosomal contents are degraded, products are release into the cytosol, and lysosomes are regenerated (2). The initiation of autophagy requires a protein complex that consists of Unc-51-like kinase-1/2 (ULK1/2),4 Fak family kinase-interacting protein of 200 kDa (FIP200, RB1CC1), and autophagy-related 13 and 101 (ATG13, ATG101) (3, 4). Nucleation of autophagosomal membranes requires a second complex that includes Beclin-1 and phosphatidyl-3-kinase class 3 (PI3KCIII, Vps34) (1, 5). During elongation and maturation the protein microtubule-associated protein light chain 3B (LC3B) is lipidated and incorporated into the autophagosomal membrane. Whereas the detection of autophagosomes by electron or fluorescence microscopy are useful markers, the accumulation of lipidated LC3B (LC3BII) in the presence of lysosomal inhibitors (e.g. bafilomycin A, NH4Cl) and the degradation of long-lived proteins are direct measures of autophagic activity or “flux” (6, 7).
Initiation and termination of the autophagic cycle are tightly controlled by mammalian target of rapamycin (mTOR; also known as mechanistic target of rapamycin, MTOR), a ubiquitous protein kinase that promotes anabolic processes (e.g. protein synthesis, mitochondrial biogenesis) and inhibits catabolic processes (e.g. autophagy, lysosomal biogenesis) (8). mTOR nucleates two large protein complexes (9). mTOR complex 1 (mTORC1) stimulates protein synthesis and inhibits autophagy under anabolic conditions, whereas mTOR complex 2 (mTORC2) promotes cytokinesis and cell survival. Under catabolic conditions that inhibit mTORC1 activity (e.g. amino acid, glucose, or mitogen withdrawal) or after pharmacological blockade (e.g. rapamycin, Torin-1), ULK1 is rapidly dephosphorylated permitting the initiation of autophagy (3, 10). Reactivation of mTORC1 terminates autophagy and permits the regeneration of lysosomes (11).
Although numerous studies have addressed the post-translational modifications and protein-protein interactions that regulate autophagosome formation, emerging studies have begun to characterize the transcriptional regulation of genes encoding autophagy proteins (12). Among the transcription factors that were shown to bind autophagy gene promoters, forkhead class O transcription factor 3a (FOXO3a) increased autophagy and atrophy in skeletal muscle (13). The transcription factors SREBP-2, E2F1, and ATF-4 bound and enhanced transcription of the LC3B gene (14–16). Under conditions of amino acid restriction, inactivation of mTOR permitted the nuclear transport of transcription factor EB (TFEB) and the induction of genes involved in autophagosomal maturation and lysosomal function (17). Less is known regarding the regulation of genes encoding proteins in the autophagy initiation complex or mechanisms by which the autophagy transcriptional program is attenuated.
We identified a physical interaction between mTORC1 and signal transducer and activator of transcription-1 (STAT1), a transcription factor that activates a subset of genes involved in programmed cell death (apoptosis) (18–20). Because recent studies indicate an anti-apoptotic role for autophagy, we hypothesized that STAT1 might function as an endogenous and stress-inducible repressor of autophagy gene transcription and autophagy per se. Here, we use in vitro and in vivo models to show that genomic loss of STAT1 increases autophagic flux, ULK1 mRNA levels, and ULK1 gene promoter activity. Furthermore, although apoptosis induced by the mTORC1 inhibitor rapamycin is attenuated in STAT1-deficient cells, it can be restored by inhibiting autophagic activity. RNAi depletion of ULK1 reduced autophagic flux observed in STAT1-deficient cells. Finally, in an in vivo model of autophagy induction during systemic inflammation, ULK1 expression and autophagy are increased in the diaphragms of STAT1-deficient mice. The results indicate an essential role for STAT1 in dampening the autophagic response in vitro and in vivo.
Results
STAT1-deficient Cells Exhibit Increased Autophagic Flux
We used three different models of STAT1 genomic loss to demonstrate a requirement for STAT1 in the suppression of autophagic flux and ULK1 expression. STAT1-deficient U3A cells were generated by random DNA mutagenesis in human fibrosarcoma (2fTGH) cells and subsequent clonal selection for absent responses to interferon (21, 22). For models of STAT1 genomic loss with isogenic controls, we used STAT1 knock-out (−/−) or wild-type (+/+) mice or mouse embryonic fibroblasts (MEFs) (23).
Autophagic flux in STAT1-deficient cells was assessed by evaluating lysosomal proteolysis of long-lived proteins (long-lived protein degradation (LLPD)). LLPD in STAT1-deficient U3A cells was greater than that in control 2fTGH cells (Fig. 1A, column 5 versus column 1). To demonstrate that changes in autophagic flux in U3A cells are due to loss of STAT1 and not due to epigenetic effects of genomic STAT1 mutations, we assessed U3A cells stably reconstituted with recombinant STAT1α (U3A-R cells). LLPD in U3A-R cells was similar to that observed in wild-type 2fTGH cells. To study the induction of autophagy, the mTORC1 inhibitor rapamycin was used. Rapamycin-induced LLPD was also enhanced in U3A cells, suggesting that loss of STAT1 might sensitize cells to the autophagic protein degradation observed upon inactivation of mTORC1 (column 6 versus column 5). There was no independent effect of rapamycin on STAT1 protein levels. STAT1 levels in U3A-R cells were similar to those in 2fTGH cells, and STAT1 was not detected in U3A cells (Fig. 1A, inset). To derive the component of LLPD due to lysosomal degradation, cells were incubated with the lysosomal acidification inhibitor NH4Cl. In each case (Fig. 1A, column 4 versus 2, 8 versus 6, 12 versus 10), NH4Cl abolished rapamycin-induced LLPD, indicating that the effect of rapamycin was likely on the autophagy-lysosomal pathway per se and not on other proteolytic systems. The NH4Cl-sensitive rate of proteolysis was calculated by subtracting LLPD in the presence of NH4Cl from that in its absence and was significantly greater in STAT1-deficient U3A cells exposed to rapamycin (Fig. 1B, column 4 versus 3). The effect of rapamycin on NH4Cl-sensitive LLPD (autophagy) was summarized by subtracting NH4Cl-sensitive LLPD in cells exposed to rapamycin to that in cells exposed to vehicle (Fig. 1C, column 2 versus 1). These results indicate that the loss of STAT1 enhances basal and rapamycin-induced autophagy.
FIGURE 1.
STAT1 is an endogenous inhibitor of autophagic flux in intact cells. Control human fibrosarcoma (2fTGH) cells, STAT1-deficient human fibrosarcoma (U3A) cells, or STAT1-deficient human fibrosarcoma cells reconstituted with STAT1 (U3A-R) were incubated with vehicle (Ctrl), rapamycin (Rap; 200 ng/ml, 218 nm), 10 mm NH4Cl, or both for 6 h after labeling with [3H]tyrosine as indicated under “Experimental Procedures.” Shown in A are the means of percent increase in free [3H]tyrosine per h (i.e. LLPD) ± S.E. from three independent experiments. The inset is a representative Western blot (composite images from the same blot) for STAT1 or β-ACTIN in 2fTGH, U3A, or U3A-R cells incubated with vehicle or rapamycin for 6 h. Shown in B. are the means of NH4Cl-sensitive LLPD (i.e. amount of LLPD blocked by NH4Cl) as calculated from the same experiments. Shown in C are the means of rapamycin-induced NH4Cl-sensitive LLPD from the same experiments. Column numbers are indicated and referenced in the “Results” section to describe comparisons made. D, 2fTGH or U3A cells were incubated without or with rapamycin for 6 h. Bafilomycin A1 (250 nm) was added 2 h before cell lysis and Western blots for LC3B, phospho-S6 S235/236 (p-S6) and β-TUBULIN. Shown the means of LC3B-II levels normalized to vehicle controls = 1 ± S.E. (E) or the means of bafilomycin-induced-fold increase in LC3B-II levels ± S.E. (ΔLC3B-II) (F), each obtained from six individual experiments. *, p < 0.05 rapamycin, NH4Cl, or bafilomycin versus vehicle control; **, p < 0.05 NH4Cl and rapamycin versus rapamycin alone; †, p < 0.05 U3A versus 2fTGH by Student's t test. G, 2fTGH or U3A cells were incubated without or with 10 nm Torin-1 for 6 h. Bafilomycin A1 (250 nm) was added 2 h before cell lysis and Western blots for LC3B, phospho-S6 Ser-235/236 (p-S6), phospho-p70 S6 kinase Thr-389 (p-S6K), phospho-Akt Ser-473 (p-Akt), STAT1, and β-ACTIN. Shown in H are the means of bafilomycin-induced increase in LC3B-II levels ± S.E. (ΔLC3B-II), each obtained from four individual experiments. *, p < 0.05 Torin-1 versus vehicle control, and U3A versus 2fTGH; †, p < 0.05 U3A versus 2fTGH treated with Torin-1.
Autophagic flux was also assessed by lipidation of LC3B in the absence or presence of the lysosomal inhibitor bafilomycin A1. Bafilomycin increased lipidated LC3B (LC3B-II) levels under all conditions, indicating ongoing autophagic flux (Fig. 1D). In the presence of bafilomycin, mean LC3B-II levels were higher in U3A cells than those in 2fTGH cells (Fig. 1E). The mean bafilomycin-induced increase in LC3B-II levels (ΔLC3B-II, flux) was higher in STAT1-deficient U3A cells than that observed in 2fTGH cells (Fig. 1E); this was true for vehicle- or rapamycin-treated cells. The mean flux, as measured by the mean increase in LC3B-II due to bafilomycin, was also higher in U3A cells than in 2fTGH cells; similarly, rapamycin-induced flux was higher in U3A cells than in 2fTGH cells (Fig. 1F).
We used an additional kinase inhibitor, Torin-1, to confirm a role for STAT1 in the regulation of autophagic activity by mTORC1. Incubation of 2fTGH cells with 10 nm Torin-1 led to a significant increase in autophagic flux (Fig. 1G, lanes 3 and 4 versus 1 and 2; Fig. 1H, column 2 versus 1). Basal and Torin1-induced autophagic flux were increased in U3A cells (Fig. 1G, lanes 5–8, Fig. 1H columns 3 versus 1 and 4 versus 2). Moreover, the augmenting effects of STAT1 deficiency were reversed in STAT1-reconstituted cells (Fig. 1G, lanes 9–12; Fig. 1H, columns 5 and 6). Consistent with a low dose of Torin-1, the phosphorylation of the mTORC1 target p70 S6K, but not the mTORC2 target Akt, was blocked (Fig. 1G). There were no changes observed in total p70 S6 kinase, S6, or AKT levels across conditions (data not shown). STAT1 protein was absent in U3A cells. The magnitude of Torin1-induced autophagy was similar to that caused by Earle's balanced salt solution (EBSS; supplemental Fig. 1A). Taken together, these results demonstrate that loss of STAT1 enhances LC3B lipidation and autophagic flux and that the effect can be reversed by re-expression of STAT1.
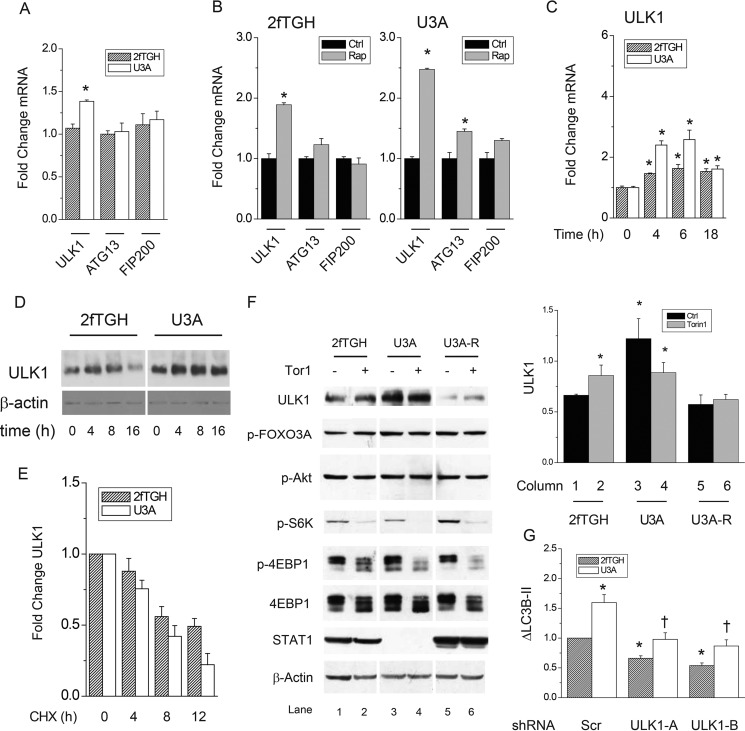
STAT1 Regulates ULK1 mRNA and Protein Levels
We next reasoned that STAT1 might suppress genes that encode proteins in the autophagy initiation complex (i.e. ULK1, ATG13, FIP-200). By quantitative PCR, basal ULK1 mRNA levels were significantly higher in STAT1-deficient U3A when compared with control 2fTGH cells (Fig. 2A). The absence of STAT1 did not alter ATG13 or FIP200 mRNA levels. Loss of STAT1 also enhanced the induction of ULK1 by rapamycin (Fig. 2B). In contrast to ULK1, there was no effect of STAT1 deficiency on FIP200 mRNA levels (Fig. 2B); ATG13 mRNA levels were slightly increased by rapamycin in U3A cells. Unlike genes encoding components of the initiation complex, the expression of mRNAs for Beclin-1 or ATG12 was not altered by STAT1 deficiency or rapamycin (data not shown). There was a significant increase in ULK1 mRNA at 4 and 6 h observed in 2fTGH cells after exposure to rapamycin, and this was enhanced in STAT1-deficient U3A cells (Fig. 2C). Like mRNA levels, baseline and rapamycin-induced ULK1 protein levels were greater in STAT1-deficient U3A (Fig. 2D) cells. The rise in ULK1 levels after incubation with rapamycin for 18 h was sustained in U3A but not 2fTGH cells (Fig. 2D). The degradation of ULK1 in U3A cells incubated with the protein synthesis inhibitor cycloheximide (t½ 8.1 ± 0.8 h) was slightly higher than that observed in 2fTGH cells (t½ 10.8 ± 1.5 h, Fig. 2E). As was the case for autophagic flux (Fig. 1, G and H), incubation with 10 nm Torin1 also increased ULK1 protein levels in 2fTGH cells (Fig. 2F, lane 2 versus 1). Phosphorylation of the mTORC2 target Akt or its substrate FOXO3A, a known inducer of autophagy gene transcription, was unaffected in control or STAT1-deficient cells (Fig. 2F). Consistent with inhibition of mTORC1 by Torin-1, phosphorylation of S6K and 4E-BP1 was reduced. ULK1 protein levels were increased in STAT1-deficient U3A cells, and this was reversed by expression of STAT1 (U3A-R). RNAi depletion of ULK1 reduced autophagic flux in 2fTGH (40% decrease) and U3A (60% decrease) cells (Fig. 2G; supplemental Fig. 1B). Therefore, loss of STAT1 leads to increased ULK1 mRNA and protein expression as well as their induction by mTORC1 inhibitors. AKT and FOXO3A likely do not play a significant role in the changes in autophagic flux observed in STAT1-deficient cells, whereas ULK1 is required.
FIGURE 2.
STAT1 is an endogenous inhibitor of autophagy genes and ULK1 protein in human fibrosarcoma cells exposed to rapamycin and Torin-1. A, levels of mRNAs of genes encoding autophagy initiation proteins (ULK1, ATG13, FIP200) were measured in control human fibrosarcoma (2fTGH) and STAT1-deficient human fibrosarcoma (U3A) cells by real-time PCR. Shown are the means of -fold change in mRNA levels ± S.E. from three individual experiments with ULK1 mRNA levels in U3A cells = 1. *, p < 0.05 U3A versus 2fTGH by Student's t test. B, 2fTGH or U3A cells were exposed to vehicle (Ctrl) or rapamycin (Rap; 200 ng/ml, 218 nm) for 6 h before measurement of the indicated mRNA levels by real-time PCR. Shown are the means of -fold change in mRNA levels ± S.E. from three individual experiments with ULK1 mRNA levels from vehicle-treated 2fTGH (left panel) or U3A (right panel) cells = 1. Baseline ULK1 mRNA levels were 1.35-fold higher in U3A versus 2fTGH cells. *, p < 0.05 rapamycin versus vehicle control. C, 2fTGH or U3A cells were exposed to rapamycin (Rap; 200 ng/ml, 218 nm) for 0, 4, 6, or 18 h before measurement of ULK1 mRNA levels by real-time PCR. Shown are the means of -fold changes in mRNA levels ± S.E. from three individual experiments with untreated 2fTGH (gray) or untreated U3A (white) cells = 1. *, p < 0.05 rapamycin versus control (0 h). D, 2fTGH or U3A cells were exposed to rapamycin for 0, 4, 8, or 16 h before preparation of whole cell lysates and detection of ULK1 or β-ACTIN protein levels by Western blot analysis. E, 2fTGH or U3A cells were incubated with 50 μm cycloheximide (CHX) for 0, 4, 8, and 12 h before analysis of ULK1 protein levels by Western blots. F, 2fTGH or U3A cells were incubated without or with 10 nm Torin-1 for 6 h before detection of ULK1, phospho-FOXO3A S318/321 (p-FOXO3A), phospho-Akt Ser-473 (p-Akt), phospho-p70 S6 kinase Thr-389 (p-S6K), phospho-4E-BP1 Thr-37/46 (p-4EBP1), total 4E-BP1 (4E-BP1), STAT1, and β-ACTIN by Western blot. Composite images from the same blots are shown. The means of band densities for ULK1 ± S.E. from four different experiments are shown to the right. *, p < 0.05 Torin-1 versus vehicle control, by Student's t test. G, 2fTGH or U3A cells were incubated with negative control shRNA (Scr) or those targeting the ULK1 coding region (ULK1-A) or 3′-UTR (ULK1-B). The means of bafilomycin-induced increase in LC3B-II levels ± S.E. (ΔLC3B-II) were derived from three individual experiments as in Fig. 1E and H. p < 0.05 versus scrambled control in 2fTGH (*) or U3A (†) by Student's t test.
A STAT1-responsive DNA Element Regulates ULK1 Gene Promoter Activity
We next determined whether mutation of a potential STAT1 response element augments activity of an ULK1 promoter fragment cloned into a luciferase reporter construct. We identified a putative DNA-binding site for STAT1 (−124 → −117) and created T→G mutations at the indicated residues (Fig. 3A). Vectors were expressed in STAT1-deficient U3A or control 2fTGH cells. Basal ULK1 promoter activity was significantly increased in U3A cells compared with that in 2fTGH cells (Fig. 3B, columns 5 versus 1). Inhibition of mTORC1 with rapamycin enhanced ULK1 promoter activity in control 2fTGH cells (Fig. 3B, columns 2 versus 1) but less so in U3A cells (Fig. 3B, column 6 versus 5). Mutation of the putative STAT1 binding sequence (ΔSTAT1) significantly increased ULK1 promoter activity in control 2fTGH cells (Fig. 3B, columns 3 versus 1). As expected, mutation of the STAT1 binding sequence had no effect on promoter activity in STAT1-deficient U3A cells (Fig. 3B, columns 7 versus 5). Blockade of mTORC1 with rapamycin did not further increase ULK1 promoter activity when the STAT1 binding sequence was mutated (Fig. 3B, columns 4 versus 3).
FIGURE 3.
STAT1 binds a regulatory DNA sequence in the ULK1 promoter that confers sensitivity to mTOR inhibitors. A, shown is a diagram depicting the 5′-flanking sequence of the ULK1 gene cloned upstream of the firefly luciferase gene (black). The wild-type (WT) and mutated putative STAT1-regulatory sequence (ΔSTAT) is indicated at 126 bp upstream of the transcription start site (+1). RLU, relative luciferase units. B, after transient transfection of the WT or ΔSTAT1 ULK1 promoter constructs, control (2fTGH) or STAT1-deficient (U3A) cells were incubated with vehicle (Ctrl) or rapamycin (Rap; 200 ng/ml, 218 nm) for 6 h before cell lysis and measurement of total protein concentration and luciferase activity. Shown are the means of ULK1 promoter activity normalized to total protein levels ± S.E. from triplicate samples in three individual experiments. Column numbers are indicated and referenced in the “Results” section to describe comparisons made. *, p < 0.05 rapamycin or ΔSTAT1 versus control; †, p < 0.05 U3A versus 2fTGH by Student's t test. C, -fold enrichment of STAT1 at the ULK1 promoter region. Cells were treated with either vehicle, rapamycin (50 nm), or Torin-1 (10 nm) for 2 h. Cells were then assayed for binding of STAT1 to the ULK1 promoter +126 bp upstream of the transcription start site (+1) via chromatin immunoprecipitation (IP). Quantitative PCR was used to detect enrichment of STAT1 at the ULK1 promoter region. *, p < 0.05 IgG versus anti-STAT1.
We next assessed whether STAT1 could bind the putative STAT1 response element (−124 → −117) in the endogenous ULK1 promoter by chromatin immunoprecipitation. In 2fTGH cells, STAT1 binding was enriched by 30% (Fig. 3C). Binding was increased upon incubation of cells with the mTOR inhibitors 10 nm Torin-1 (84% enrichment) or 50 nm rapamycin (62% enrichment) for 2 h (Fig. 3C). There was no binding of STAT1 observed when control primers were used to detect a region of genomic DNA that does not bind transcription factors (data not shown). Thus, a STAT1 response element in the ULK1 promoter binds STAT1 and attenuates the induction of ULK1 promoter activity.
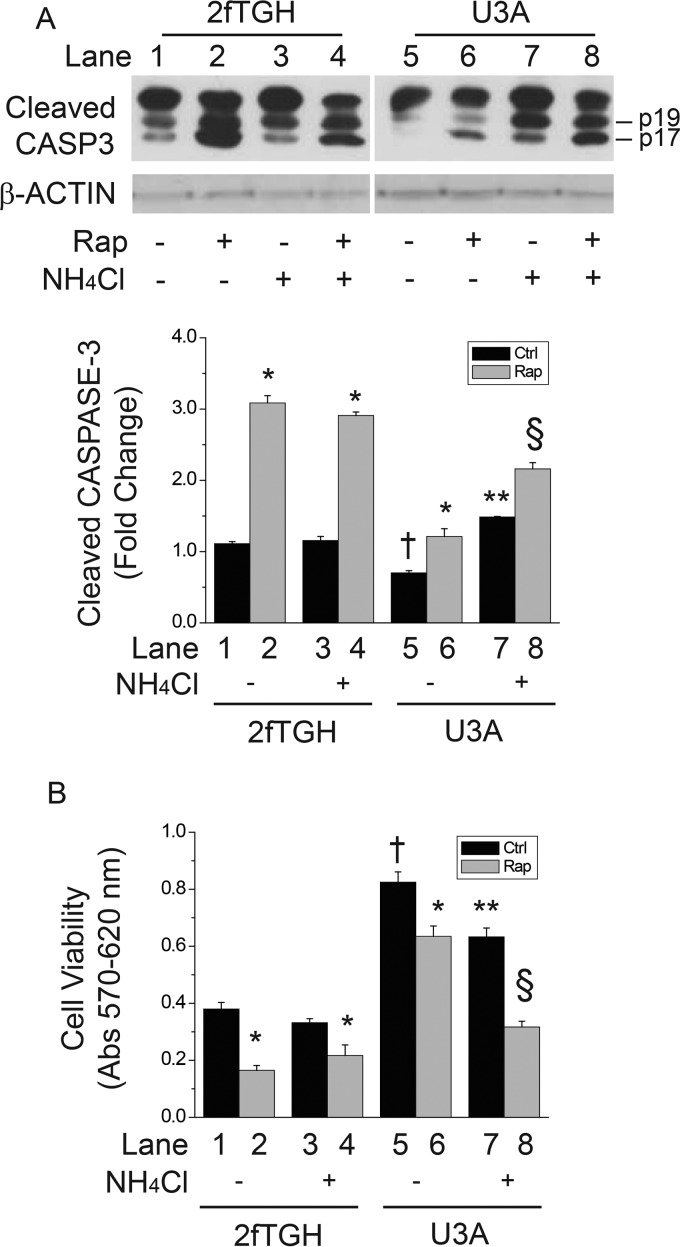
Enhanced Autophagy in STAT1-deficient Cells Attenuates Apoptosis and Promotes Cell Survival
Because STAT1 is an inhibitor of autophagy and ULK1 gene expression, we reasoned that the suppression of apoptosis in STAT1-deficient cells might require autophagic activity. Caspase-3 (CASP3) cleavage products were measured as a marker of apoptosis. As previously shown (20), cleaved CASP3 levels were reduced in STAT1-deficient U3A cells compared with those in 2fTGH controls (Fig. 4A, columns 5 versus 1); rapamycin significantly increased cleaved CASP3 levels in 2fTGH but not in U3A cells (Fig. 4A, columns 1 and 2 versus columns 5 and 6). In U3A cells, the autophagy inhibitor NH4Cl restored basal or rapamycin-induced apoptosis (Fig. 4A, column 7 versus 5 or column 8 versus 6). Administration of NH4Cl in 2fTGH cells (i.e. elevated STAT1) levels had no effect on cleaved CASP3 levels (Fig. 4A, columns 3 and 4 versus columns 2 and 1). To determine whether restoration of apoptosis in U3A cells correlates with reduced viability, we assessed crystal violet staining in cells exposed to rapamycin, NH4Cl, or both for 24 h. Consistent with reduced apoptosis and increased autophagy, cell viability was significantly greater in U3A cells lacking STAT1 when compared with control 2fTGH cells (Fig. 4B, columns 5 versus 1). Rapamycin led to a significant decrease in cell viability in 2fTGH cells (columns 2 versus 1 or 3 versus 4); the magnitude of this reduction was attenuated in STAT1-deficient U3A cells (Fig. 4B, columns 6 versus 5). Consistent with restoration of apoptosis (Fig. 4A), blockade of autophagy with NH4Cl reduced cell viability in STAT1-deficient U3A cells but not in control 2fTGH cells (Fig. 4B, columns 5 and 7 versus columns 1 and 3). As was the case for apoptosis (Fig. 4A), the restoration of cytotoxicity was more prominent in cells incubated with rapamycin (Fig. 4B, columns 6 and 8 versus columns 2 and 4). These data indicate that the reduced apoptosis and cell death observed in STAT1-deficient cells correlates with increased levels of ULK1 and autophagic activity. Furthermore, apoptosis and cell death can be restored in STAT1-deficient cells by blocking autophagy.
FIGURE 4.
Inhibition of autophagy restores apoptosis and cytotoxicity in STAT1-deficient U3A cells. 2fTGH or U3A cells were incubated with vehicle, rapamycin (Rap; 200 ng/ml, 218 nm), 10 mm NH4Cl, or both for 24 h, before detection of cleaved caspase-3 or β-ACTIN levels by Western blot analysis (A) or detection of cells viability by Crystal violet staining (B). Shown in A are composite images from the same gels for cleaved caspase-3 (Cleaved CASP3) or β-ACTIN as well as the means of integrated cleaved CASP3 band density values ± S.E. from three individual experiments. Lane numbers are indicated and referenced in the “Results” section to describe comparisons made. Data in B are the means of sample absorbance at 570 nm minus those at 620 nm ± S.E. from 3 individual experiments. *, p < 0.05 rapamycin versus vehicle control; †, U3A vehicle control versus 2fTGH vehicle control; **, p < 0.05 NH4Cl versus U3A vehicle control; §, p < 0.05 U3A cells exposed to NH4Cl and rapamycin versus those exposed to rapamycin alone.
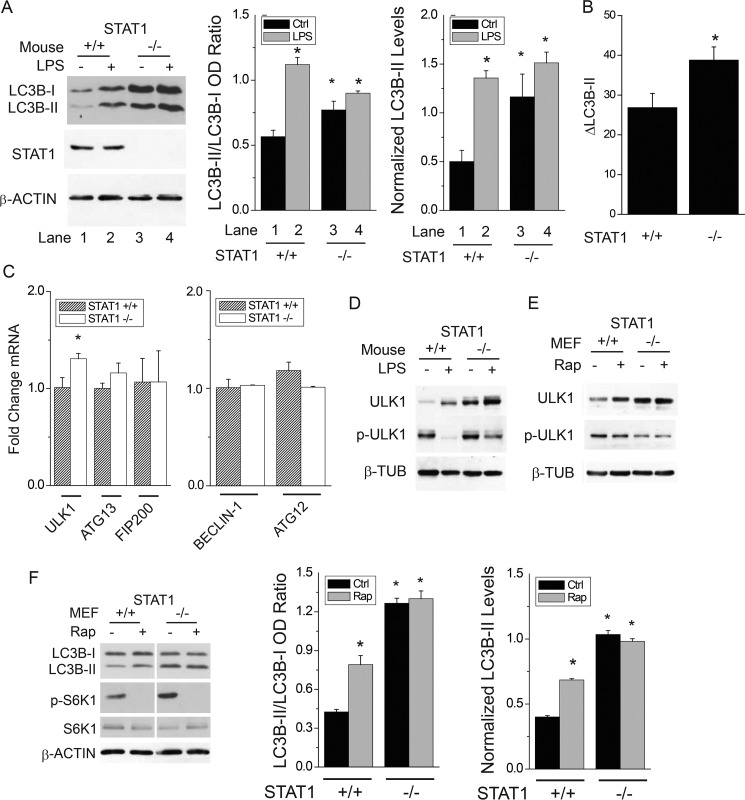
STAT1 Deficiency Increases Autophagy in Skeletal Muscle of Mice
To demonstrate a role for STAT1 in vivo, we used a model of septic inflammation that elicits autophagy and organ dysfunction in mice (24, 25). STAT1−/− mice were exposed to vehicle or bacterial lipopolysaccharide (LPS) for 24 h. As previously shown (25), LPS administration in mice triggered a significant increase in diaphragm autophagy (i.e. increased ratio of LC3B-II to LC3B-I) (Fig. 5A, lanes 2 versus 1 and right panels) of Stat1+/+ mice. Like in cultured cells, loss of Stat1 was associated with increased basal autophagy as indicated by the rise in LC3B protein lipidation (Fig. 5A, lanes 3 versus 1). Injection of LPS did not further increase LC3B protein lipidation in STAT1−/− mice, perhaps due to already elevated basal autophagy in the muscles of these mice. Autophagic flux was increased in the diaphragms of STAT1-deficient mice (Fig. 5B). As was the case for STAT1-deficient cell lines (Fig. 2), Ulk1, but not Beclin-1 or Atg12 (data not shown) mRNA levels, were higher in the diaphragms of STAT1−/− mice than in those from Stat1+/+ mice (Fig. 5C). Similarly, ULK1 protein levels were increased in the diaphragms of STAT1−/− mice (Fig. 5D). Consistent with induction of autophagy, LPS reduced the phosphorylation of ULK1 at Ser-757 in wild-type and knock-out mice (Fig. 5D). As was the case for U3A cells, basal and rapamycin-induced ULK1 protein levels and autophagy were also increased in STAT1−/− MEFs (Fig. 5F); Stat1 mRNA and STAT1 protein were absent in STAT1−/− MEFs (supplemental Fig. 1, C and D). Changes in autophagy attributed to loss of Stat1 or incubation with rapamycin were reduced in MEFs, perhaps due to high levels of autophagy at baseline; moreover, others have demonstrated weak induction of autophagy by rapamycin in MEFs (26). These results indicate that STAT1 is an endogenous suppressor of Ulk1 mRNA and protein expression as well as autophagy in vivo.
FIGURE 5.
STAT1 is an endogenous inhibitor of autophagy in skeletal muscles from mice exposed to bacterial lipopolysaccharide. A, wild-type (+/+) mice or those homozygous for allelic loss of STAT1 (−/−) were exposed to saline or intraperitoneal E. coli lipopolysaccharide (LPS), 5 μg/kg, and euthanized 24 h later. LC3B in diaphragm homogenates was detected by Western blot analysis and quantified by band densitometry (gel). Shown are the means of LC3B-II:LC3B-I ratio or LC3B-II normalized to β-ACTIN levels (± S.E.) from the diaphragms of four individual mice. B, wild-type (+/+) or STAT1 knock-out (−/−) mice were exposed to vehicle or colchicine (0.4 mg/kg/24 h i.p.) for 48 h before detection of LC3B-II in the diaphragm by Western blot. The mean change in LC3B-II attributed to colchicine (ΔLC3B-II, autophagic flux) ±S.E. is shown (n = 4 per group). C, mRNA levels for autophagy genes were detected from the diaphragms of mice in panel A by real-time PCR. Shown are the means of -fold-change mRNA levels versus wild-type saline-treated controls (± S.E.) for four individual mice. D, ULK1, phospho-ULK1 T757 (p-ULK1; mTORC1 phosphorylation site) or β-TUBULIN from diaphragm protein homogenates were assessed by Western blot. Gels are representative of four individual mice. E and F, STAT1 wild-type (+/+) or knock-out (−/−) MEF cells were exposed to rapamycin for 6 h before preparation of whole cell lysates and detection of ULK1, phospho-ULK1 T757 (pULK1), and β-TUBULIN (E) or LC3B, phospho-p70 S6 kinase Thr-389 (p-S6K), S6K, and β-ACTIN (F) protein levels by Western blot analysis. *, p < 0.05 versus vehicle-treated wild-type.
Discussion
The current study reveals a previously undescribed mechanism by which STAT1 dampens the expression of ULK1 and autophagic flux in cultured cells and in vivo. We used cell and animal models of genomic loss of STAT1 to avoid potential off-target effects of molecular (e.g. RNAi, overexpression systems) or pharmacological inhibitors. STAT1-deficient cells and mice exhibited increased autophagy and ULK1 expression (Figs. 1, 2, and 5). The enhanced autophagy and ULK1 expression observed in STAT1-deficient U3A cells were not due to genetic or epigenetic modifications independent of STAT1, as they were reversed by expression of recombinant STAT1α (Figs. 1 and 2). Similar effects of STAT1 loss on ULK1 and autophagy were also observed in MEFs with targeted genomic knock-out of STAT1. Moreover, enhanced autophagy was required for the inhibition of apoptosis observed in STAT1-deficient cells (Fig. 4), and STAT1 was an endogenous suppressor of Ulk1 expression and skeletal muscle autophagy in mice (Fig. 5). Although the transcriptional control of autophagosome nucleation genes (e.g. BECN1, BNIP3, LC3B) has been evaluated (12, 27–29), less is known regarding the regulation of genes encoding proteins in the initiation complex. We demonstrate that the ULK1 gene 5′-flanking region contains a functional DNA sequence that binds STAT1, and that binding is enhanced by mTORC1 inhibitors (Fig. 3). Thus, the increased autophagy, ULK1 expression, and promoter activity in STAT1-deficient cells reflect absent suppression of ULK1 transcription by STAT1. In contrast, nucleation/elongation genes (e.g. BECN1, ATG12) were not altered in STAT1-deficient cells in vitro or in vivo.
Several studies have investigated the role of transcription factors in autophagy and gene expression. Of the few that demonstrated direct regulation of autophagy gene promoters, transcriptional induction was generally associated with 1.5–3-fold changes in levels of mRNA, protein, and autophagic flux. For instance, like STAT1, the nuclear import of TFEB is enhanced by inactivation of mTORC1; however, whereas STAT1 controls ULK1 expression, TFEB activates genes that encode regulators of autophagosomal maturation (17, 30). Other examples include the forkhead transcription factor FOXO3a, which activates the autophagy transcriptional program by binding the promoters of autophagy nucleation or maturation genes, including Atg12, Lc3b, and Gabarapl1 (7). The NF-κB transcription factor binds to and activates the Becn1 promoter in T-cells (29). Unlike TFEB, FOXO3a, or NF-κB, STAT1 is a novel suppressor of ULK1 transcription. The effect of STAT1 was likely independent of Akt or FOXO3a activity as we observed no differences in their phosphorylation in U3A- or Torin-1-treated cells. A possible mechanism(s) for inhibition of transcription by STAT1 include the following: (i) transcriptional repression (31–35), (ii) interaction with histone acetylases or deacetylases (36–39), or (iii) post translational modification of STAT1 and/or repression of a co-activator (40, 41). Further functional analysis of the ULK1 5′-flanking region as well as the transcriptional complexes and post-translational modifications that regulate ULK1 gene transcription and autophagy, are the subject of ongoing investigations.
Simultaneous control of ULK1 phosphorylation and transcription by mTORC1 and STAT1 is consistent with our previous work. When inactivated, mTORC1 associates with latent (i.e. unphosphorylated) STAT1 in a macromolecular complex (18), enhances its nuclear import (20), and augments the subsequent induction of STAT1-dependent apoptosis genes. Mechanisms of latent STAT1 signaling and its regulation of apoptosis genes are reviewed in Yang and Stark (42). Because autophagy appears to counteract apoptosis, the attenuating effect of latent STAT1 on ULK1 gene transcription observed in the current study is consistent with this functional paradigm and represents a feed forward inhibitory mechanism that limits autophagic activity. Further evidence of an anti-apoptotic role for autophagy arises from the ability of NH4Cl, an autophagy inhibitor, to restore rapamycin-induced apoptosis in STAT1-deficient cells (Fig. 4). In agreement, one study revealed that loss of STAT1 was associated with increased autophagy and reduced myocardial damage in a mouse model of ischemia/reperfusion (43). Also of note, the relative roles of ULK1 and STAT1 might differ with respect to autophagy and cell survival when cells are exposed to interferons. For instance, STAT1 was required for IFN-α-induced BECN1 expression, autophagy, and apoptosis in leukemia cells (44). In a separate study, ULK1 was required for the induction of interferon-sensitive genes by type-I interferons in a mechanism that appeared to involve p38 MAPK and not STAT1 or autophagy (45). Despite the pleiotropic effects of STAT1 and ULK1, our data consistently demonstrate an attenuating effect of STAT1 on ULK1 expression and autophagy in cultured cells and in murine skeletal muscle.
To demonstrate that STAT1 regulates autophagy in vivo, we assessed skeletal muscle autophagy in Stat1-deficient mice exposed to systemic bacterial lipopolysaccharide (25). Transcriptional regulation of autophagy was previously shown to control skeletal muscle atrophy (7). Importantly, atrophy involves both autophagic and proteasomal proteolysis, and mTORC1 was thought to contribute by post-transcriptional mechanisms (46). Our new data now implicate STAT1 and its regulation by mTOR in the transcriptional control of autophagy genes in the mouse diaphragm (Fig. 5). Interestingly, although STAT1 deficiency increased proteasomal and autophagic proteolysis to a similar extent (Fig. 1A), the majority of rapamycin-induced proteolysis in STAT1-deficient cells was due to lysosomal degradation (Fig. 1, B and C). Other conditions in which autophagy influences skeletal muscle protein metabolism include sepsis and prolonged mechanical ventilation (25, 47). Future studies can further characterize the mechanisms by which STAT1 controls skeletal muscle function.
Inhibition of autophagy by NH4Cl had no effect on apoptosis or cytotoxicity in cells expressing STAT1 (Fig. 4). This suggests that autophagy may be particularly important for cell survival when STAT1 activity is reduced. In agreement, the expression and activity of endogenous inhibitors of STAT1 such as isoforms of suppressor of cytokine signaling (SOCS) or protein inhibitor of activated STAT (PIAS) can be elevated in human cancers or during systemic inflammation (48–51) and may promote cell survival by enhancing autophagy. Moreover, recent studies indicate an inhibitory role for STAT1 in the transcription of genes involved in oxidative phosphorylation and mitochondrial biogenesis, which are both implicated in skeletal muscle dysfunction and cancer (52, 53). Future studies will evaluate the molecular links between STAT1 signaling, the regulation of autophagy, and metabolism under conditions of metabolic stress or neoplastic transformation.
Experimental Procedures
Cell Culture and Reagents
Human fibrosarcoma 2fTGH cells, their STAT1-deficient counterparts (U3A), and U3A cells stably expressing STAT-1α (STAT1-reconstituted U3A; U3A-R) were provided by Dr. George Stark (Cleveland Clinic). U3A cells were generated by random DNA mutagenesis in 2fTGH cells and subsequent clonal selection for absent responses to interferon (21, 22). We obtained immortalized wild-type control (+/+) and STAT1-knock-out (−/−) MEFs from Dr. A. Koromilas (McGill University) that were generated from mice with targeted disruption of the STAT1 locus (originally donated by Dr. J. Durbin) (23). All cell lines were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Invitrogen). Selection for 2fTGH and U3A cells was maintained with 125 μg/ml hygromycin (BioShop). Incubations with the mTORC1 allosteric inhibitor rapamycin (Biomol; 200 ng/ml, 218 nm), the mTOR kinase active site inhibitor Torin1 (Millipore; 10 nm), the lysosomal and/or endosomal acidification inhibitors bafilomycin A1 (Enzo; 250 nm) and NH4Cl (Sigma; 10 mm), and l-tyrosine-[ring-3,5-3H] ([3H]tyrosine; PerkinElmer Life Sciences) were performed in subconfluent cultures for the indicated times. mTORC1 inhibitors were used to induce autophagy, and lysosomal acidification inhibitors were used to inhibit autophagy. Torin-1 was used at a concentration that preferentially inhibits mTORC1. For ULK1 depletion by RNAi, the lentiviral vector pLKO.1 containing DNA encoding short hairpin RNAs targeting the coding (ULK1-A) or 3′-UTR (ULK1-B) of ULK1 or no known human gene (scrambled negative control) (MISSION shRNA, Sigma) was introduced into HEK293T with lentiviral packaging vectors pRSV-REV, pMDLg/pRRE, and pCMV-VSV-G using calcium chloride. Viruses were collected 72 h later, and target cells were infected with the collected viruses in the presence of Polybrene for 3 days before measuring autophagic flux or ULK1 mRNA levels.
Animal Procedures
All procedures were approved by the Animal Ethics Committees of McGill University or Université de Montréal and were in accordance with the guidelines of the Canadian Council of Animal Care. Adult (8–12 weeks old) wild-type or STAT1−/− BALB/C mice were injected i.p. with a single dose of phosphate-buffered saline (PBS) (control) or Escherichia coli lipopolysaccharide (5 mg/kg serotype 055:B5; Sigma), a known inducer of autophagy in skeletal muscle (25). Animals were euthanized with sodium pentobarbital after 24 h before rapid excision of the diaphragm. Each muscle was weighed, flash-frozen in liquid nitrogen, and stored at −80 °C for later use.
Preparation of Cell Lysates and Detection of Proteins
Endogenous or recombinant proteins in whole cell lysates or tissue homogenates were detected by Western blot analysis. Whole cell lysates from 2fTGH, U3A, or MEFs were generated after washing twice in PBS and adding lysis buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 50 mm NaF, 1 mm EDTA, 1 mm Na3VO4, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mm PMSF, 1% Nonidet P-40, and 10% glycerol). Mouse diaphragm homogenates were prepared in the same buffer. Total proteins, 20–50 μg, were separated by SDS-PAGE and transferred to PVDF membranes (Bio-Rad) before analysis by Western blot with antibodies directed against the following proteins: LC3B (#2775), phospho-ULK1 Ser-757 (#6888), phospho-S6 Ser-235/236 (#2211), phospho-p70S6K1 Thr-389 (#9205), p70S6K1 (#9202), phospho-4E-BP1 Thr-37/46 (#9459), 4E-BP1 (#9644), phospho-FOXO3A Ser-318/321 (#9465), phospho-Akt Ser-473 (#4060), cleaved CASPASE-3 (#9661), β-ACTIN (#3700) (Cell Signaling), ULK1 (#A7481, Sigma), β-TUBULIN (#H7, Developmental Studies Hybridoma Bank), or STAT1 (#SC-345, Santa Cruz). For detection of cleaved CASPASE-3, CHAPS (final concentration 0.3%) was added to the lysis buffer, and 80 μg of protein were used for Western blots. After incubation with antibodies linked to horse-radish peroxidase (The Jackson Laboratory, Bar Harbor, ME) and ECL reagent (Amersham Biosciences), membranes were exposed to X-ray film. Band densitometry was performed after imaging using an AlphaImager device (Alpha Innotech) and analysis by Alpha Ease FC software (version 4.1.0). Band density was obtained using the spot density and auto-background functions. The means of band density values from multiple experiments were compared by Student's t test.
Determination of Lysosomal Proteolysis
The long-lived protein degradation assay was adapted from a previous study (7). 2fTGH, U3A, or U3A-R cells were incubated with [3H]tyrosine, 4 μCi/ml, for 24 h. Cells were then incubated with medium containing 2 mm unlabeled tyrosine for 2 h to prevent reincorporation of [3H]tyrosine and to permit proteolysis of short-lived proteins. Medium containing the autophagy inducer rapamycin or the lysosomal inhibitor ammonium chloride (NH4Cl, 10 mm) was added. At 0, 4, 8, and 16 h after the addition of inhibitors, 200 μl of medium from each sample was collected and stored at 4 °C. At 16 h, cells were dissolved in 0.2 n NaOH and combined with scintillation fluid (Fisher Scintisafe 30%). Proteins in media aliquots were precipitated with 10% TCA. The acid-soluble supernatant containing free [3H]tyrosine was combined with scintillation fluid and counted in a scintillation counter (Beckman). The percentage proteolysis per hour was calculated as the counts per min (cpm) released in the medium divided by the total cpm incorporated (i.e. cells plus released) × 100. The rate of protein degradation was calculated from the slope of the line representing cumulative radioactivity loss over time.
Transfection of Plasmids and Promoter Analysis
For ULK1 reporter analysis, cells were transiently transfected with 10 μl of Lipofectamine LTX (Invitrogen) and 2.0 μg of wild-type ULK1 promoter constructs cloned into luciferase-expressing PGL3 plasmids or those containing a mutation in the STAT1 binding sequence (ΔSTAT1). The plasmid construct with the human ULK1 promoter linked to luciferase cDNA (i.e. pGL3-ULK1) was obtained from Dr. Kenichi Yoshida (54). The structure of the cloned promoter sequence and its STAT1-binding site are shown in Fig. 4A. pGL3-ULK1 containing a mutation in the STAT1-binding sequence (ΔSTAT1) was constructed using the QuikChange site-directed mutagenesis kit (Stratagene) and the following primer sequences: ULK1 ΔSTAT1 forward, 5′-ccgcggctctggtgggtctccgttgg-3′; reverse, 5′-ccaacggagacccaccagagccgcgg-3′. Mutations were verified by automated sequencing. The Renilla PRL-TK plasmid (Promega) was co-transfected with PGL3 plasmids to normalize for transfection efficiency. Firefly and Renilla luciferase activities were detected by luminometry using the Dual Luciferase Assay kit (Promega). Values for firefly luciferase were divided by the corresponding Renilla luciferase values to obtain relative luciferase units.
Chromatin Immunoprecipitation
Protein-DNA complexes of 2fTGH cells were cross-linked with 1% formaldehyde (Sigma). After 10 min, unreacted formaldehyde was quenched with 200 mm glycine, and the cells were washed twice in PBS. The cells were then collected in SDS lysis buffer (Millipore) and sonicated for 5 s using a Microson Ultra Cell Disruptor (Misonix). Chromatin immunoprecipitation was performed using 2 μg of either an anti-Stat1α antibody (#SC-345x, Santa Cruz) or purified rabbit IgG (#ab171870, Abcam). The protein-DNA complexes were precipitated using magnetic Protein A beads (Millipore). After removing unbound chromatin, samples were extracted in elution buffer (1% SDS, 0.1 m NaHCO3) and heated overnight at 65 °C to reverse cross-links before DNA purification (Purelink Quick PCR Purification kit, Life Technologies) and real-time PCR using the following primers (Invitrogen): ULK1 promoter forward 5′-tgttcgtcacgcccggctct-3′ and reverse 5′-ggatccgactccgactccga-3′ (190 bp); negative control forward 5′-atggttgccactggggatct-3′ and reverse 5′-tgccaaagcctaggggaaga-3′ (174 bp).
Real-time PCR for Gene Expression Analysis
Total cellular RNA was isolated (RNAspin kit, GE Healthcare), and cDNAs were generated by reverse transcription (Invitrogen Superscript II Reverse Transcriptase kit) using 2 μg of total RNA as substrate. SYBR Green-based real-time PCR was performed using 2 μl of cDNA in 25 μl of universal PCR mix (Invitrogen). The following human gene specific primers (Invitrogen) were synthesized and used in the quantification of transcripts; amplicon lengths are shown in parentheses: ULK1 forward 5′-ggacaccatcaggctcttcc-3′, reverse 5′-gaagccgaagtcagcgatct-3′ (169 bp); ATG13 forward 5′-cccaggacagaaaggacctg-3′; reverse 5′-aaccaatctgaacccgttgg-3′ (133 bp); FIP200 forward 5′- cagtgctgggacggatacaa-3′, reverse 5′- tccaatgcaagctgtgtcct-3′ (186 bp); Beclin-1 forward 5′-aacctcagccgaagactgaa-3, reverse 5′gacgttgagctgagtgtcca-3′ (132 bp); ATG12 forward 5′-ggcagtagagcgaacacgaa-3′, reverse 5′gggaaggagcaaaggactga-3′ (116 bp); STAT1 forward 5′-tccatcctttggtacaacatgc-3′, reverse 5′-cagagagggagcaggtgttt-3′ (311 bp); β-ACTIN forward 5′-agaaaatctggcaccacacc-3′, reverse 5′-ggggtgttgaaggtctcaaa-3′ (142 bp). Murine gene-specific primers include the following: ATG12 forward 5′-ggcctcggaacagttgttta-3′, reverse 5′-cagcaccgaaatgtctctga-3′ (200 bp); Beclin-1 forward 5′-cttggaggaggagaggctga-3′, reverse 5′-tgtggaaggtggcattgaag-3′ (263 bp); ULK1 forward 5′-cactgcgtggctcacctaag, reverse 5′-agccaacagggtcagcaaat-3′ (140 bp); Fip200 forward 5′-accgtgcacctgctattcct-3′, reverse 5′-catcatggacaagcccttca-3′ (183 bp); ATG13 forward 5′-gtgggcaccctcactctttc-3′, reverse 5′-gggatagggacggtcaacaa-3′ (126 bp); β-ACTIN forward 5′-aaccgtgaaaagatgacccag-3′, reverse 5′-cacagcctggatggctacgta-3′ (75 bp). Primers were verified by performing dissociation curves. Real-time PCR reactions were carried out using the ABI 7500 Real Time PCR System. Results are expressed as -fold induction in mRNA levels as calculated by the ΔΔCT method (55).
Detection of Cell Viability in Vitro
Cells were washed twice with PBS before incubation for 15 min with 0.2% crystal violet (Sigma) dissolved in 25% methanol. Excess stain was removed by washing 4 times with PBS, and cells were solubilized in 1% SDS for 15 min. The absorbance of each sample, 100 μl, was determined at 570 and 620 nm using a SpectraMax M2 microplate reader (Molecular Devices).
Statistical Analysis
Student's t test was used to test for statistical significance between groups. p values <0.05 were considered statistically significant.
Author Contributions
A. S. K. and S. N. H. conceived and coordinated the study and wrote the paper. A. A. G., B. N., and A. M. J. S. performed and analyzed the experiments in Figs. 1–5. A. E. K., S. W., Y. B., N. M., and S. T. Q. supported and performed the experiments in Fig. 5. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. J. Durbin for permission to use STAT1 knock-out mice and embryonic fibroblasts, Dr. K. Yoshida for providing the ULK1 promoter construct, and Dr. G. Stark for providing STAT1-deficient (U3A) and control (2fTGH) cells. We gratefully acknowledge the generous support of the J. T. Costello Memorial Research Fund, the Richard and Edith Strauss Canada Foundation, the Lloyd Carr-Harris Foundation, and Fonds de Recherche du Québec-Santé.
This work was supported, in whole or in part, by NCI, National Institutes of Health Grant R01CA125436-01A1 (to A. S. K.). This work was also supported by Canada Research Chairs (950-213853; to A. S. K.), Fonds de Recherche du Québec-Santé (31052; to A. S. K.), Canadian Institutes of Health Research (to S. N. H., S. T. Q., and Y. B.), and a Meakins Christie Post-doctoral Fellowship (to B. N. and A. M. J. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Fig. 1.
- ULK1/2
- Unc-51-like kinase-1/2
- LC3B
- light chain 3B
- mTOR
- mammalian target of rapamycin
- mTORC1
- mTOR complex 1
- FOXO3a
- forkhead class O transcription factor 3a
- TFEB
- transcription factor EB
- MEF
- mouse embryonic fibroblast
- LLPD
- long-lived protein degradation
- U3A-R
- reconstituted U3A.
References
- 1.Mizushima N., and Komatsu M. (2011) Autophagy: renovation of cells and tissues. Cell 147, 728–741 [DOI] [PubMed] [Google Scholar]
- 2.Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., Hailey D. W., Oorschot V., Klumperman J., Baehrecke E. H., and Lenardo M. J. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., and Kim D. H. (2009) ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan E. Y., Longatti A., McKnight N. C., and Tooze S. A. (2009) Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol. Cell. Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell R. C., Tian Y., Yuan H., Park H. W., Chang Y. Y., Kim J., Kim H., Neufeld T. P., Dillin A., and Guan K. L. (2013) ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 15, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klionsky D. J., Abdalla F. C., Abeliovich H., Abraham R. T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J. A., Ahn H. J., Ait-Mohamed O., Ait-Si-Ali S., Akematsu T., Akira S., et al. (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao J., Brault J. J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S. H., and Goldberg A. L. (2007) FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 6, 472–483 [DOI] [PubMed] [Google Scholar]
- 8.Laplante M., and Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zoncu R., Efeyan A., and Sabatini D. M. (2011) mTOR: from growth signal integration to cancer, diabetes and ageing. Nat. Rev. Mol. Cell Biol. 12, 21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim J., Kundu M., Viollet B., and Guan K. L. (2011) AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breitkreutz A., Choi H., Sharom J. R., Boucher L., Neduva V., Larsen B., Lin Z. Y., Breitkreutz B. J., Stark C., Liu G., Ahn J., Dewar-Darch D., Reguly T., Tang X., Almeida R., Qin Z. S., Pawson T., Gingras A. C., Nesvizhskii A. I., and Tyers M. (2010) A global protein kinase and phosphatase interaction network in yeast. Science 328, 1043–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M. C., and Kroemer G. (2013) Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 23, 310–322 [DOI] [PubMed] [Google Scholar]
- 13.Sanchez A. M., Candau R. B., and Bernardi H. (2014) FoxO transcription factors: their roles in the maintenance of skeletal muscle homeostasis. Cell. Mol. Life Sci. 71, 1657–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Polager S., Ofir M., and Ginsberg D. (2008) E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 27, 4860–4864 [DOI] [PubMed] [Google Scholar]
- 15.Seo Y. K., Jeon T. I., Chong H. K., Biesinger J., Xie X., and Osborne T. F. (2011) Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 13, 367–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I., and Harris A. L. (2010) Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29, 4424–4435 [DOI] [PubMed] [Google Scholar]
- 17.Settembre C., and Ballabio A. (2011) TFEB regulates autophagy: an integrated coordination of cellular degradation and recycling processes. Autophagy 7, 1379–1381 [DOI] [PubMed] [Google Scholar]
- 18.Fielhaber J. A., Han Y. S., Tan J., Xing S., Biggs C. M., Joung K. B., and Kristof A. S. (2009) Inactivation of mammalian target of rapamycin increases stat1 nuclear content and transcriptional activity in α4- and protein phosphatase 2A-dependent fashion. J. Biol. Chem. 284, 24341–24353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fielhaber J. A., Dydensborg A. B., Triantafillopoulos A., Bouchard M., Qureshi S. T., and Kristof A. S. (2010) Rapamycin enhances lipopolysaccharide-induced apoptosis and lung injury. Am. J. Respir. Crit. Care Med. 181, A3542 [Google Scholar]
- 20.Fielhaber J. A., Tan J., Joung K. B., Attias O., Huegel S., Bader M., Roux P. P., and Kristof A. S. (2012) Regulation of Karyopherin-α1 and nuclear import by mTOR. J. Biol. Chem. 287, 14325–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellegrini S., John J., Shearer M., Kerr I. M., and Stark G. R. (1989) Use of a selectable marker regulated by α interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9, 4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller M., Laxton C., Briscoe J., Schindler C., Improta T., Darnell J. E. Jr., Stark G. R., and Kerr I. M. (1993) Complementation of a mutant cell line: central role of the 91-kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J. 12, 4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durbin J. E., Hackenmiller R., Simon M. C., and Levy D. E. (1996) Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–450 [DOI] [PubMed] [Google Scholar]
- 24.Mofarrahi M., Guo Y., Haspel J. A., Choi A. M., Davis E. C., Gouspillou G., Hepple R. T., Godin R., Burelle Y., and Hussain S. N. (2013) Autophagic flux and oxidative capacity of skeletal muscles during acute starvation. Autophagy 9, 1604–1620 [DOI] [PubMed] [Google Scholar]
- 25.Mofarrahi M., Sigala I., Guo Y., Godin R., Davis E. C., Petrof B., Sandri M., Burelle Y., and Hussain S. N. (2012) Autophagy and skeletal muscles in sepsis. PLoS ONE 7, e47265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., and Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., Sugimoto T., Haneda M., Kashiwagi A., and Koya D. (2010) Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Invest. 120, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., Burden S. J., Di Lisi R., Sandri C., Zhao J., Goldberg A. L., Schiaffino S., and Sandri M. (2007) FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471 [DOI] [PubMed] [Google Scholar]
- 29.Copetti T., Bertoli C., Dalla E., Demarchi F., and Schneider C. (2009) p65/RelA modulates BECN1 transcription and autophagy. Mol. Cell. Biol. 29, 2594–2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J. C., Wolff N. C., Tran T. A., Zou L., Xie X. J., Corey D. R., and Brugarolas J. (2011) Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramana C. V., Chatterjee-Kishore M., Nguyen H., and Stark G. R. (2000) Complex roles of Stat1 in regulating gene expression. Oncogene 19, 2619–2627 [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan P., and Maric M. (2011) Signal transducer and activator of transcription 1 negatively regulates constitutive γ interferon-inducible lysosomal thiol reductase expression. Immunology 132, 209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu B., Gross M., ten Hoeve J., and Shuai K. (2001) A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. U.S.A. 98, 3203–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramana C. V., Grammatikakis N., Chernov M., Nguyen H., Goh K. C., Williams B. R., and Stark G. R. (2000) Regulation of c-myc expression by IFN-γ through Stat1-dependent and -independent pathways. EMBO J. 19, 263–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sibinga N. E., Wang H., Perrella M. A., Endege W. O., Patterson C., Yoshizumi M., Haber E., and Lee M. E. (1999) Interferon-γ-mediated inhibition of cyclin A gene transcription is independent of individual cis-acting elements in the cyclin A promoter. J. Biol. Chem. 274, 12139–12146 [DOI] [PubMed] [Google Scholar]
- 36.Korzus E., Torchia J., Rose D. W., Xu L., Kurokawa R., McInerney E. M., Mullen T. M., Glass C. K., and Rosenfeld M. G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279, 703–707 [DOI] [PubMed] [Google Scholar]
- 37.Klampfer L., Huang J., Swaby L. A., and Augenlicht L. (2004) Requirement of histone deacetylase activity for signaling by STAT1. J. Biol. Chem. 279, 30358–30368 [DOI] [PubMed] [Google Scholar]
- 38.Nusinzon I., and Horvath C. M. (2003) Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. U.S.A. 100, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang X., Gao J. S., Guan Y. J., McLane K. E., Yuan Z. L., Ramratnam B., and Chin Y. E. (2007) Acetylation-dependent signal transduction for type i interferon receptor. Cell 131, 93–105 [DOI] [PubMed] [Google Scholar]
- 40.Krämer O. H., Baus D., Knauer S. K., Stein S., Jäger E., Stauber R. H., Grez M., Pfitzner E., and Heinzel T. (2006) Acetylation of Stat1 modulates NF-κB activity. Genes Dev. 20, 473–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krämer O. H., Knauer S. K., Greiner G., Jandt E., Reichardt S., Gührs K. H., Stauber R. H., Böhmer F. D., and Heinzel T. (2009) A phosphorylation-acetylation switch regulates STAT1 signaling. Genes Dev. 23, 223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J., and Stark G. R. (2008) Roles of unphosphorylated STATs in signaling. Cell Res. 18, 443–451 [DOI] [PubMed] [Google Scholar]
- 43.McCormick J., Suleman N., Scarabelli T. M., Knight R. A., Latchman D. S., and Stephanou A. (2012) STAT1 deficiency in the heart protects against myocardial infarction by enhancing autophagy. J. Cell. Mol. Med. 16, 386–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu S., Cao L., Yu Y., Yang L., Yang M., Liu K., Huang J., Kang R., Livesey K. M., and Tang D. (2013) Inhibiting autophagy potentiates the anticancer activity of IFN1@/IFNα in chronic myeloid leukemia cells. Autophagy 9, 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saleiro D., Mehrotra S., Kroczynska B., Beauchamp E. M., Lisowski P., Majchrzak-Kita B., Bhagat T. D., Stein B. L., McMahon B., Altman J. K., Kosciuczuk E. M., Baker D. P., Jie C., Jafari N., Thompson C. B., et al. (2015) Central role of ULK1 in type I interferon signaling. Cell Rep. 11, 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sandri M. (2010) Autophagy in skeletal muscle. FEBS Lett. 584, 1411–1416 [DOI] [PubMed] [Google Scholar]
- 47.Hussain S. N., Mofarrahi M., Sigala I., Kim H. C., Vassilakopoulos T., Maltais F., Bellenis I., Chaturvedi R., Gottfried S. B., Metrakos P., Danialou G., Matecki S., Jaber S., Petrof B. J., and Goldberg P. (2010) Mechanical ventilation-induced diaphragm disuse in humans triggers autophagy. Am. J. Respir. Crit. Care Med. 182, 1377–1386 [DOI] [PubMed] [Google Scholar]
- 48.Culig Z. (2013) Suppressors of cytokine signalling-3 and -1 in human carcinogenesis. Front. Biosci. 5, 277–283 [DOI] [PubMed] [Google Scholar]
- 49.Hoefer J., Schäfer G., Klocker H., Erb H. H., Mills I. G., Hengst L., Puhr M., and Culig Z. (2012) PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am. J. Pathol. 180, 2097–2107 [DOI] [PubMed] [Google Scholar]
- 50.Liu B., and Shuai K. (2008) Targeting the PIAS1 SUMO ligase pathway to control inflammation. Trends Pharmacol. Sci. 29, 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ilangumaran S., Ramanathan S., and Rottapel R. (2004) Regulation of the immune system by SOCS family adaptor proteins. Semin. Immunol. 16, 351–365 [DOI] [PubMed] [Google Scholar]
- 52.Meier J. A., and Larner A. C. (2014) Toward a new STATe: the role of STATs in mitochondrial function. Semin. Immunol. 26, 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pitroda S. P., Wakim B. T., Sood R. F., Beveridge M. G., Beckett M. A., MacDermed D. M., Weichselbaum R. R., and Khodarev N. N. (2009) STAT1-dependent expression of energy metabolic pathways links tumour growth and radioresistance to the Warburg effect. BMC Med. 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kusama Y., Sato K., Kimura N., Mitamura J., Ohdaira H., and Yoshida K. (2009) Comprehensive analysis of expression pattern and promoter regulation of human autophagy-related genes. Apoptosis 14, 1165–1175 [DOI] [PubMed] [Google Scholar]
- 55.Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.