To silence transposition of the DNA transposon Tam3, the BED-zinc finger domain of Tam3 transposase in Antirrhinum retains function at the plasma membrane.
Abstract
Transposable elements (TEs) are considered to be parasites of host genomes because they act as powerful mutagens. If not kept in check, they can cause gene disruption, genome rearrangement, and genomic takeover. Hence, activities of TEs are under the rigid control of hosts. To date, all identified TE regulations have been epigenetic dependent, with the exception of the DNA transposon Tam3. Blocking nuclear translocation of Tam3 transposase (TPase) is consistent with the suppression of Tam3 in Antirrhinum majus. In this article, we discovered that epigenetic-independent regulation of Tam3 is mediated by the BED-zinc finger (Znf-BED) domain of Tam3 TPase. The host targets the N terminus of the Znf-BED domain, which contains two highly conserved aromatic amino acids, to detain Tam3 TPase at the plasma membrane and to silence Tam3. Zinc finger proteins perform broader functions in transcriptional regulation through their DNA binding ability. Our data revealed that the posttranslational epigenetic-independent silencing against TEs was a result of the protein binding ability of the Znf-BED domain.
Transposable elements (TEs), also known as mobile elements, can move and insert into new positions within a genome (Erwin et al., 2014). Plant genomes contain a number of active TEs reported as entities that perturb genome integrity. These have the potential for gene disruption, chromosome breakage, illegitimate recombination, and genome rearrangement (Slotkin and Martienssen, 2007). To maintain the stability and integrity of plant genomes, TE activities are under rigorous control of the hosts.
The mechanisms of silencing TEs in plants include processes such as DNA methylation (Pikaard et al., 2008; Matzke and Mosher, 2014), histone modification (Saze et al., 2012), mRNA degradation (Zhang et al., 2007), and translation inhibition (Iwakawa and Tomari, 2013; Li et al., 2013). These epigenetic regulations range from complete transcriptional silencing by DNA methylation to the elimination of transcripts and translation inhibition by RNA-induced silencing complexes.
Several active transposons have been identified in Antirrhinum, of which Tam3 is a DNA transposon belonging to the hAT (hobo, Ac, Tam3) superfamily (Calvi et al., 1991). hAT family members are widely distributed in multicellular organisms, including plants, animals, and fungi (Rubin et al., 2001). Unlike most other transposons, Tam3 exhibits the unusual feature of activation at low growth temperatures (around 15°C) and inhibition at high temperatures (above 25°C; Harrison and Fincham, 1964; Carpenter et al., 1987). Tam3 has been associated with several loci responsible for anthocyanin pigmentation in Antirrhinum, whose alleles might have caused the flower petal variegations described by Darwin and de Vries (Galun, 2003; Schwarz-Sommer et al., 2003; Hudson et al., 2008). However, the underlying mechanism controlling Tam3 activity is not fully understood. In Antirrhinum, Tam3 transposase (TPase) can be transcribed into mRNA and then translated into protein, which demonstrate identical expression patterns at both low and high temperatures (Uchiyama et al., 2008; Fujino et al., 2011). Thus, transcriptional gene silencing and posttranscriptional gene silencing do not seem to be involved in the suppression of Tam3. Epigenetic regulation can be transmitted to daughter cells through mitotic cell division (Probst et al., 2009). However, when Antirrhinum plants initially grown under high temperatures are transferred to low temperatures, newly formed flowers show variegations in the petals owing to the transposition of Tam3. These indicate that epigenetic control is not the cause of Tam3 inactivation.
The life cycle of a DNA transposon consists of both nuclear and cytoplasmic stages. A DNA transposon in the genome is transcribed into mRNA, which is then exported to the cytoplasm and translated into TPase protein. The TPase is imported back into the nucleus to bind the DNA target site and excise the DNA transposon, which is then ligated into a new target site. In Antirrhinum, temperature controls the sublocalization of Tam3 TPase, which is severely restricted to the plasma membrane (PM) under high temperatures, along with silencing of Tam3 (Fujino et al., 2011). This suggests that mediation of TPase localization is a possible way to limit DNA transposon activity.
In this study, we reveal that epigenetic-independent regulation of Tam3 is mediated by a BED-zinc finger (Znf-BED) domain located in the N-terminal region of Tam3 TPase. The Znf-BED domain is prevalent in TPases of hAT superfamily transposons, including hobo in Drosophila, Ac in maize (Zea mays), and Tam3. The Znf-BED domain is a known DNA binding amino acid motif (Aravind, 2000); here it strongly orientates to localize Tam3 TPase in the PM and was hence designated a PM localization signal. The Znf-BED domain can be divided into two signature parts: an N-terminal containing two conserved aromatic amino acids regions and a C-terminal CCH[H/C] patch. The use of point mutation experiments showed that the N terminus of the Znf-BED domain is the direct binding site targeted by the host to control the distribution of Tam3 TPase in Antirrhinum cells and that this regulation is zinc finger structure dependent. Our data provide an insight into the posttranslational regulation of TEs by Znf-BED domain and also reveal the protein binding property of a Znf-BED domain involved in the detainment of Tam3 TPase at the PM.
RESULTS
Trapping Nuclear Import of Tam3 TPase for Tam3 Inactivation
Antirrhinum line HAM22 carries palidarecurrens::Tam3 (palrec), which contains a 3.6 kb Tam3 insertion in the promoter of the Pallida locus encoding dihydroflavonol-4-reductase that is required for anthocyanin synthesis (Uchiyama et al., 2009; Fig. 1A). The insertion of Tam3 suppressed expression of palrec, resulting in an ivory petal color (Almeida et al., 1989; Fig. 1A). This state was maintained unless the transposition of Tam3 occurred at low temperatures of around 15°C, after which variegation with red spots appeared in the petal (Supplemental Fig. S1A). The Tam3 TPase protein was restricted to the PM at high temperatures around 25°C (Fig. 1B), while a portion of the Tam3 TPase was able to enter the nucleus at permissive temperatures of around 15°C (Supplemental Fig. S1B; Fujino et al., 2011). Only nuclear extracts from plants grown at 15°C are capable of binding the Tam3 sequence, with nuclear extracts from plants grown at 25°C unable to do so (Hashida et al., 2006). Because initiation of the transposition reaction is dependent on the nuclear transport of TPase (Heinlein et al., 1994; Ono et al., 2002), inhibiting the nuclear import of Tam3 TPase is the likely cause of Tam3 stabilization at high temperatures. We determined the localization of Tam3 TPase in onion epidermis cells and tobacco BY2 cells; in these two plants, most Tam3 TPase localized to both the PM and the nucleus and was not influenced by temperature changes (Supplemental Fig. S2).
Figure 1.
Phenotype of temporarily inactive Tam3 A. majus line HAM22. A, Structure of the pallidarecurrens::Tam3 allele (palrec) from HAM22 and its flower phenotype at 25°C. Tam3 was embedded in the promoter of palrec, resulting in the silencing of pal. The flower showed an ivory petal color owing to abortion of anthocyanin synthesis. B, Subcellular localization of Tam3 TPase at 25°C in the protoplast of HAM22.
The Znf-BED Domain Is Responsible for Membrane Localization of Tam3 TPase
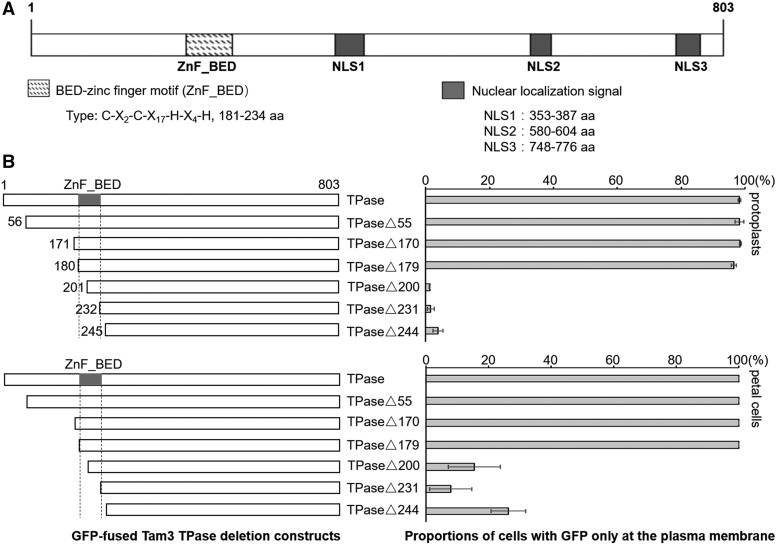
Nuclear localization signals (NLSs) direct the nuclear transport of nuclear proteins following translation in the cytoplasm (Fujino et al., 2011). Tam3 TPase contains at least three experimentally confirmed NLSs (Fujino et al., 2011; Fig. 2A). The nuclear transport of Tam3 TPase was found to be strictly controlled under a high temperature (Figs. 1B and 2B), indicating that other domain(s) in Tam3 TPase function to suppress the nuclear transport of Tam3 TPase. Fujino et al. (2011) previously identified an N-terminal region, located within the 55 to 231 amino acid sequence of Tam3 TPase, which contains a possible nuclear inhibitory domain for arresting the nuclear translocation of itself.
Figure 2.
The Znf-BED motif of Tam3 TPase functions to direct Tam3 TPase to the PM. A, Protein domains of Tam3 TPase. The TPase contains a conserved Znf-BED motif and three experimentally confirmed NLSs. B, Identification of the potential functional domain of Tam3 TPase, which is responsible for directingTam3 TPase to the PM. Left, schematic of the fusion proteins used. Full-length or truncated TPase was fused to the N-terminal region of GFP. Each of these was transformed into protoplasts (top) and petal cells (bottom) of HAM22 to analyze subcellular localization. Numbers in the constructs represent the amino acid position in the Tam3 TPase sequence. Right, two histograms showing the proportions of cells with GFP signal in the PM compared with the total number of cells with green fluorescence (Supplemental Table S1). Data represent means ± sd.
We performed further transient assays to identify the potential functional domain responsible for inhibiting the nuclear import of Tam3 TPase. We constructed a series of GFP-fused Tam3 TPase deletion constructs, including one intact and six truncated sequences (Fig. 2B). These plasmids were transformed into the protoplasts and petal cells of Antirrhinum line HAM22 grown at 25°C. The GFP signal from the series of truncated constructs TPase, TPase∆55, TPase∆170, and TPase∆179 only appeared on the PM (Fig. 2B; Supplemental Table S1). However, constructs with a shorter TPase than TPase∆200 produced green fluorescence in the nucleus of most cells (Fig. 2B; Supplemental Table S1). Interestingly, the region between TPase∆179 and TPase∆244 corresponded to an integral Znf-BED motif (Hashida et al., 2006). These results suggest that the Znf-BED motif served as a functional domain to confine the nuclear import of Tam3 TPase.
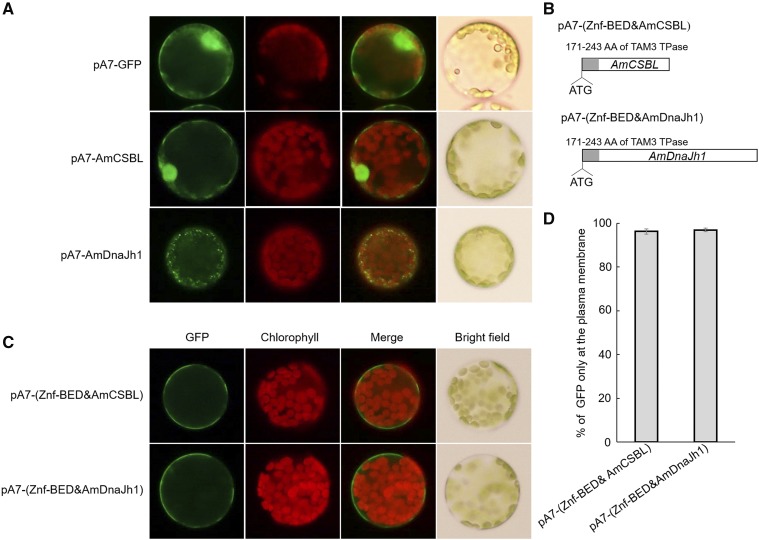
To establish whether the Znf-BED motif could function to confine the nuclear import, we fused the Znf-BED motif into the 5′ terminal of two Antirrhinum majus genes: calcineurin subunit B-like (AmCSBL; KX016023) and dnaJ homolog 1 (AmDnaJh1; KX016024); the proteins encoded by these genes are originally located in the nucleus and mitochondria, respectively (Fig. 3, A and B). When fused with the Znf-BED domain, these two fusion proteins were restricted to the PM (Fig. 3, C and D; Supplemental Table S2), indicating that the Znf-BED motif acted as a strong PM localization signal in Antirrhinum. We initially called this functional segment the nuclear localization inhibitory domain, but hereafter we refer to it as the PM localization signal.
Figure 3.
The Znf-BED motif in Tam3 TPase is a strong PM localization signal. A, Subcellular localization of AmCSBL and AmDnaJh1; pA7-GFP was used as a control vector. B, Constructs with Znf-BED motif inserted into the N-terminals of AmCSBL and AmDnaJh1. C, Subcellular localization of Znf-BED motif fused to AmCSBL and AmDnaJh1. D, The proportions of cells with a GFP signal focused on the PM compared with the total number of cells with green fluorescence (Supplemental Table S2). Data represent means ± sd.
The Znf-BED Domain Is Prevalent in the Transposases of hAT DNA Transposons
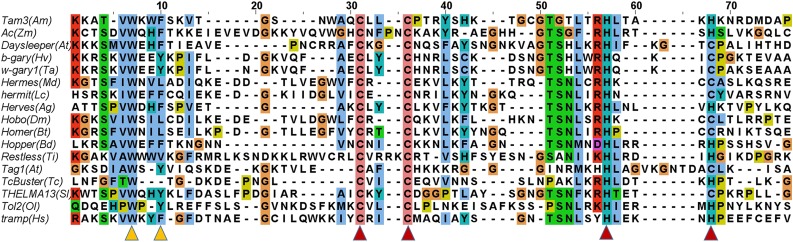
Zinc finger proteins were first discovered in 1985 and have since been recognized as one of the most common regulatory factors in plants, animals, and fungi (Miller et al., 1985). They vary largely in both structure and function. One subclass of the zinc fingers is the Znf-BED, named after the two Drosophila proteins in which it is found: BEAF and DREF (Aravind, 2000). The Znf-BED is a protein domain of about 50 to 60 amino acid residues in length, with two highly conserved regions: aromatic amino acids (Trp and Phe) at the N terminus and a shared pattern of cysteines and histidines predicted to form a zinc finger, characterized by the signature Cx2CxnHx3-5[H/C] (where xn is a variable spacer; Aravind, 2000; Saghizadeh et al., 2009). The Znf-BED is found in one or more copies in cellular regulatory factors and TPases (Aravind, 2000). Here, we found the Znf-BED to be widespread in the hAT superfamily TPase of different species (Fig. 4). These alignments of the Znf-BED domains featured aromatic amino acids at the N terminus as well as the Cys and His signature, the CCH[H/C] motif (Fig. 4). Little is known about the N-terminal motif in Tam3 TPase, while the CCH[H/C] motif is predicted to form a zinc finger. In the N-terminal region, the secondary structure predicted using the PHD program indicated that the T184V185W186K187W188F189 amino acid residues formed a β-strand (Aravind, 2000). Because it has a well-known DNA binding ability, the BED-zinc domain may have a conserved function in regulating the transposition of hAT superfamily transposons.
Figure 4.
Multiple alignment of the Znf-BED domain in transposases of the hAT superfamily. Residues are colored according to the background coloring program of ClustalX Multiple Sequence Alignment. Yellow triangles represent the two conserved aromatic positions at the N-terminal region; red triangles indicate conserved cysteines and histidines predicted to form a zinc finger. Each protein is labeled using its name, followed by species abbreviation in parentheses. Abbreviations: Am, Antirrhinum majus; Zm, Zea mays; At, Arabidopsis thaliana; Hv, Hordeum vulgare; Ta, Triticum aestivum; Md, Musca domestica; Lc Lucilia cuprina; Ag, Anopheles gambiae; Dm, Drosophila melanogaster; Bt, Bactrocera tryoni; Bd, Bactrocera dorsalis; Ti, Tolypocladium inflatum; Tc, Tribolium castaneum; Sl, Silene latifolia; Ol, Oryzias latipes; Hs, Homo sapiens.
The N-Terminal Region Containing the Two Highly Conserved Aromatic Amino Acid Regions of the Znf-BED Domain Is Directly Targeted by the Host for Membrane-Associated Localization of Tam3 TPase
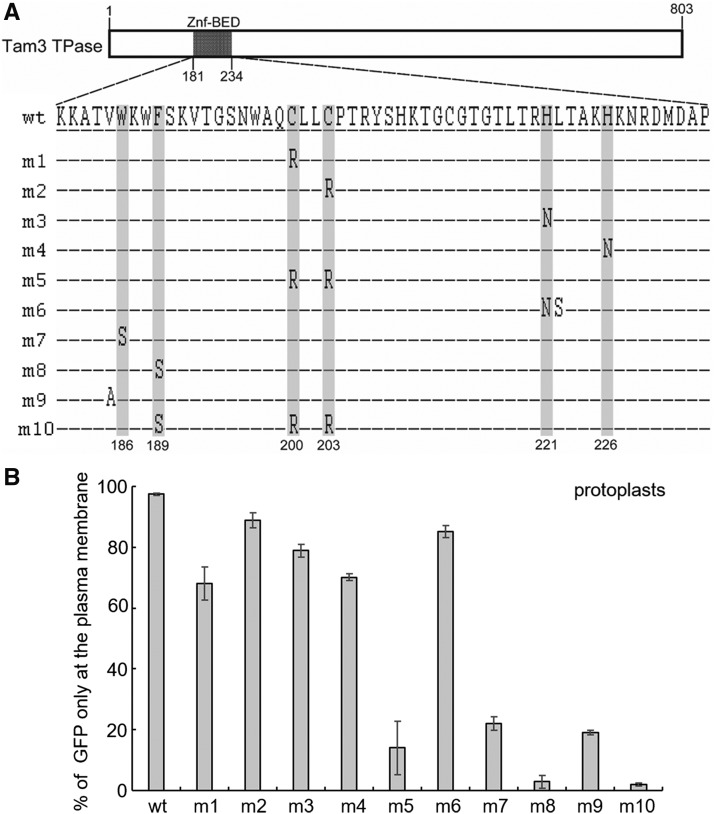
To test the functions of these two motifs, point mutations were introduced into the conserved positions (Fig. 5A). First, C200, C203, H221, and H226 were individually mutated to create m1, m2, m3, and m4, respectively (Fig. 5A). These mutations introduced changes to the subcellular localization of Tam3 TPase (Fig. 5B; Supplemental Table S3). A side-by-side mutation at H221 and L222 contained in m6 also showed a similar result to m3 (Fig. 5, A and B; Supplemental Table S3), suggesting that these amino acids are not the direct action site of the host factor(s). It is feasible that the presence of H209 or C213 compensates for any defect caused by these mutations to the zinc finger structure. However, in m5, where cysteines C200 and C203 were simultaneously changed to R (Fig. 5A), irreparable damage to the zinc finger structure occurred, with most Antirrhinum cells displaying GFP in their nuclei (Fig. 5B; Supplemental Table S3). This indicates that maintenance of the zinc finger structure is indispensable for controlling the PM localization of Tam3 TPase. In N terminus of the Znf-BED domain, m7 and m8 carrying mutations at the two conserved aromatic amino acids, W186 or F189, dramatically reduced the number of the cells with GFP at the PM (Fig. 5, A and B; Supplemental Table S3). In addition to these two positions, m9 revealed that V185 was also functionally necessary for the PM localization of Tam3 TPase (Fig. 5, A and B; Supplemental Table S3). These data suggested that the beta strand was important for the protein binding ability of the Znf-BED domain and was directly targeted by the host to detain Tam3 TPase in the PM. Hence, the m10 construct, which contained mutations in both the beta strand and the CCH[H/C] motif, expressed GFP in the nuclei of most cells, but not in the PMs (Fig. 5, A and B; Supplemental Table S3). These results indicated that protein modifications such as phosphorylation within the Znf-BED domain were not responsible for altering the localization of Tam3 TPase, because not all mutated amino acids were associated with phosphorylation.
Figure 5.
Identification of binding sites for Tam3 TPase-interacting factors for detaining Tam3 TPase at the PM. A, Amino acid sequences of Znf-BED domains in the 10 constructs (m1 to m10) carrying point mutations and wild-type (wt) are depicted. Each construct was fused with the GFP gene at C termini. The gray background represent conserved sites; amino acid positions 186 and 189 for the N-terminal aromatic amino acids; 200, 203, 221, and 226 for the zinc finger. Dashes represent the same amino acid as wild-type. B, Subcellular localization of point mutation constructs. Ten constructs were transformed into protoplasts of Antirrhinum. Proportion of the cells with GFP in the PM to the total number of cells with GFP were counted under a fluorescence microscope (Supplemental Table S3). Data represent means ± sd.
Detainment of Tam3 TPase at the PM through the Protein Binding Ability of Its Znf-BED Domain
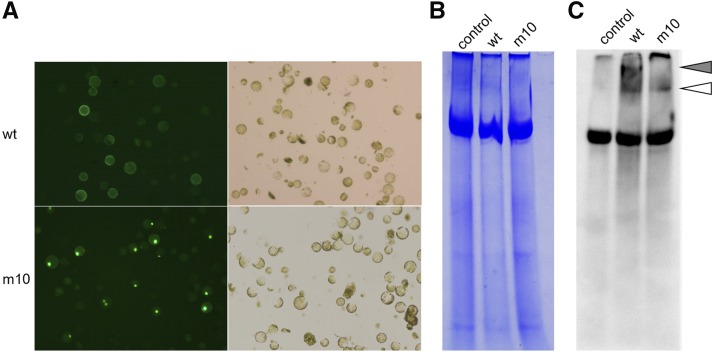
Two possibilities exist for the regulation of Tam3 TPase localization. The first is that membrane protein(s) interact with Tam3 TPase and confine it to the PM. The Znf-BED domain might interact with the NLS and prevent the nuclear localization of Tam3 TPase. However, it seems unlikely that only one Znf-BED domain prevents the function of all three NLSs simultaneously, and Tam3 TPase would be more likely to localize to the cytosol, not the PM. The second possibility involves an alteration of the Tam3 TPase configuration following a temperature shift. However, this would be expected to occur in all plant species, yet apparent preference for PM localization of Tam3 TPase has only been observed in Antirrhinum cells, not in onion or tobacco (Supplemental Fig. S2). To clarify this issue, we conducted native PAGE western-blot analysis to detect the nondenatured protein structure of the two Tam3 TPase-GFP constructs, wild type and m10 (Fig. 5). Wild type and m10 with different Znf-BED domains were transformed into the protoplasts of Antirrhinum (Fig. 6). After incubation for 20 h, we confirmed that the protoplasts with the wild-type construct expressed GFP at the PM, while GFP was expressed in the nuclei of m10 protoplasts (Fig. 6A). The nontransformed protoplast was also prepared as a control (Fig. 6). Active proteins were extracted from these protoplast samples without denaturation and were loaded on a native PAGE gel. Following electrophoresis, western-blot analysis using a monoclonal mouse anti-GFP antibody revealed three specific bands (Fig. 6C). The first band (gray arrowhead) was specific to wild-type samples, the second band (white arrowhead) was shared by wild-type and m10 samples, and the third band was common to all samples (Fig. 6C). The wild-type-specific band reflected the interaction with the host protein(s) through the Znf-BED domain of the Tam3 TPase. The second band indicated the GFP-related protein(s) in both wild type and m10 samples. The common band appeared to be an artifact of GFP-independent binding, because it also appeared in the Coomassie Brilliant Blue (CBB)-stained gel as a major fraction in all samples including nontransformed protoplasts (Fig. 6B).
Figure 6.
Protein binding profile of the Znf-BED domain of Tam3 TPase. A, Mass protoplasts prepared from young Antirrhinum leaves transformed with Tam3 TPase constructs wild-type (wt) and m10 (see Fig. 5). The protoplasts with wild type and m10 showed GFP expression in the PM and nucleus, respectively. Transformation efficiencies for wild-type and m10 constructs were 40 to 50%. Protein preparations were made using these Antirrhinum protoplasts. B, CBB staining of the native PAGE gel for nondenatured proteins extracted from the three protoplast samples, nontransformant protoplasts (control), transiently transformed protoplasts with wild-type construct and m10 construct (m10). Active proteins were extracted using native protein extraction buffer containing 0.20% Triton X-100. C, GFP-Tam3 TPase complex detected by western blotting using an anti-GFP antibody. The duplicate of the gel blot in B was made by loading adjacent lanes with the same samples. Gray arrowhead indicates specific band to wild-type samples, and white arrowhead shows band shared by wild-type and m10 samples.
DISCUSSION
Posttranslational Regulation of Tam3 in Antirrhinum
Plants have evolved a set of strategies to control the activity of TEs. Among these, DNA methylation is a stable mark used by hosts to fix transposons at the “off” position. Accumulated evidence previously revealed a negative relationship between DNA methylation and the active state of TEs (Slotkin and Martienssen, 2007). An increased level of DNA methylation in the promoter regions of TEs, such as Ac, Spm, and MuDR, tends to suppress the expression of TPase transcripts, eventually silencing these autonomous elements (Hashida et al., 2005). In plants, the hypermethylation state in these regions can be inherited over several generations (Habu et al., 2001). Although higher temperatures result in an increased level of DNA methylation in both terminal regions of Tam3, DNA methylation is not the cause of Tam3 silencing (Hashida et al., 2003). Moreover, because both transcripts and translational products of Tam3 TPase can be detected in Antirrhinum (Uchiyama et al., 2008; Fujino et al., 2011), mRNA cleavage and translation inhibition do not determine the inactivation of Tam3. Hence, regulation of the Tam3 transposition in Antirrhinum occurs in both epigenetic independent and posttranslational manners.
Mechanisms Involved in Regulating Tam3 Activity Depend on the Znf-BED Domain
Initiation of the transposition of a DNA transposon depends on the nuclear localization of its TPase. The mRNA of Tam3 TPase is translated to protein in the cytoplasm and then translocated into the nucleus where it recognizes and binds specific sites for cut-and-paste processes. In the ovarian somatic cells of Drosophila, Piwi-interacting (pi)RNA-loaded Piwi proteins are selectively transported into the nucleus, where they exert TE silencing (Saito et al., 2009; Ishizu et al., 2011). N-terminally truncated Piwi loaded with piRNAs cannot translocate into the nucleus, and thus cannot silence TEs (Saito et al., 2009; Saito et al., 2010). Although exhibiting the opposite function to TPase, this finding enabled us to speculate that the inhibition of TPase nuclear transport can silence TEs.
In this study, we detected a positive correlation between Tam3 TPase PM localization and Tam3 suppression. When Tam3 was in an inactive state, the localization of Tam3 TPase was confined to the PM (Fig. 1). During cell division, plant cells undergo rupture and restructuring of the nuclear envelope. In the process of nuclear envelope restructuring, it is possible that some cytoplasmic contents, such as soluble proteins and organelle fragments, become incorporated into the nucleus. However, the PM structure is maintained during cell division. Hence, the PM would be suitable for preventing Tam3 TPase from entering the nucleus to strongly control Tam3 activity in Antirrhinum. This is supported by our finding that little variegation caused by Tam3 transposition occurred in the continuous growth of Antirrhinum petals under high temperatures. As long as the Znf-BED domain remained intact, Tam3 TPase was unable to enter the nuclei. However, if the domain was partially or completely destroyed, the majority of Antirrhinum cells expressed GFP in the nucleus (Fig. 2). This suggests that the cis element was indispensable for regulating the membrane-associated localization of Tam3 TPase and that this domain is essential for the epigenetic-independent regulation of Tam3. Mediating the localization of TPase may be a means of arresting TE transposition. Relative to the epigenetic mechanisms of transcriptional gene silencing and posttranscriptional gene silencing, this mechanism is considered an optional function specifically established between the host and TE. It is conceivable that Tam3 TPase plays additional biological roles for the host, given that many TPase-derived genes have important functions in plant development (Bundock and Hooykaas, 2005; Lin et al., 2007; Roccaro et al., 2007). Indeed, the DNA binding ability of Tam3 TPase was indicated by the fact that the Znf-BED domain binds to Tam3 to enable its transposition. Although the main function of TPase is to mediate the transposition of TEs, under certain conditions, such as low temperature for Tam3, they may also function as gene activators or suppressors to enable the host to adapt to stress.
Control of Tam3 Transposition through the Znf-BED Domain Is an Outcome of Coordinated Evolution between the Host and Transposon
The BED-zinc domain is widely present in hAT superfamily transposons of different species (Rubin et al., 2001). The Znf-BED domain is thought to play a conserved role in starting the transposition of TEs belonging to the hAT family (Mack and Crawford, 2001; Hashida et al., 2006; Hickman et al., 2014). To date, no Znf-BED domains in TEs have been shown to interact with host proteins, except for Tam3, which may be involved in bipartite abilities to bind DNA and protein. Tam3 is under strict regulation from the host at the protein level. Unlike in Antirrhinum, we found that the localization of Tam3 TPase in onion epidermis cells and tobacco BY2 cells was not limited to the PM but was also observed in the nucleus with no temperature regulation (Supplemental Fig. S2). This indicates that an Antirrhinum-specific factor(s) or regulatory pathway(s) is involved in the regulation of Tam3. Although hAT superfamily transposons are widely found in multicellular organisms, no reports of horizontal transfer of these elements have been made (Rubin et al., 2001). Therefore, elements of the hAT superfamily are considered to have coevolved vertically with individual host organisms. The distinct behavior of Tam3 in Antirrhinum might be an outcome of coordinate evolution between host and transposon. While the host needs to silence Tam3 to ensure its own safety under normal conditions, Tam3 TPase activity might be beneficial to the host under stress.
Mechanisms Involved in the Detainment of Tam3 TPase at the PM
Zinc finger proteins are one of the most common regulatory factors found in organisms. While the Znf-BED domain is mainly thought to be involved in DNA binding, accumulating evidence suggests that it can also interact with proteins (Diaz-Meco et al., 1996; Becker and Kunze, 1997; Chen et al., 2009). However, the mechanism involved in this protein binding ability is poorly understood. The Znf-BED domain constitutes two parts: an N-terminal region of two highly conserved aromatic amino acids, and a C-terminal CCH[H/C] patch. In TPases of TAG1 and DAYSLEEPER, site-directed mutagenesis of each Cys or His predicted to form a zinc finger led to loss of the DNA binding capacity of the Znf-BED domain (Mack and Crawford, 2001; Bundock and Hooykaas, 2005). Our study also indicated that unrecoverable damage to the CCH[H/C] patch of the Znf-BED domain abolishes the protein binding ability of the Znf-BED domain of Tam3 TPase. Although the zinc finger structure is essential for protein binding activity, our results implied that the N-terminal motif was the direct target site for the host factor(s).
The trapping of protein nuclear translocation can be mediated by the nuclear pore complex or by protein modification (Greber and Gerace, 1992; Zhou et al., 2010). However, retention of Tam3 TPase at the PM seems to be regulated differently. In humans, the endoplasmic reticulum (ER) membrane protein NSIG-1 binds the NH2-terminal membrane domain of the SREBP cleavage-activating protein (SCAP) and facilitates retention of the SCAP/SREBP complex in the ER (Yang et al., 2002). NSIG-1-mediated ER retention of SCAP is sterol dependent. Binding with sterols causes a conformational change of SCAP, which increases the affinity of SCAP for INSIG-1 (Yang et al., 2002). The SREBP/SCAP/INSIG-1 complex becomes trapped in the ER by an as-yet-unknown mechanism (Yang et al., 2002). In the absence of sterols, the SCAP/SREBP complex is free to exit the ER and reach the Golgi complex (DeBose-Boyd et al., 1999; Nohturfft et al., 2000). The mutant SCAP(TM1-6) contains a Y298C substitution and fails to be retained in the ER upon sterol addition (Yang et al., 2002). We have demonstrated that Antirrhinum stores Tam3 TPase in the PM under nonstressed conditions. This study revealed that the Znf-BED domain in Tam3 TPase acts as a PM localization signal to interact with host factor(s). In this process, we assume that the host factor(s) binds mature Tam3 TPase proteins in the Golgi complex, and the protein complex is then transported to the PM via a transport vesicle. It appears that N-terminal aromatic amino acids in the Znf-BED domain have protein binding ability, while the zinc finger structure is necessary for controlling the PM localization of Tam3 TPase.
CONCLUSION
Our data imply that the function of the Znf-BED domain of Tam3 TPase undergoes strong selection from the host. Besides the DNA binding ability, the Znf-BED domain, which also serves as a PM localization signal, is targeted by host factor(s) to detain Tam3 TPase at the PM. This is closely related to Tam3 activity. Further experiments identified amino acids in the Znf-BED domain that are involved in the interaction between the host and Tam3 TPase. The N-terminal region comprises two highly conserved aromatic amino acids that appear to be the direct target site, while the zinc finger conformation seems to be indispensable for TPase detainment. This work provides insights into the mechanism and function of a Znf-BED domain involved in protein binding and TE regulation.
MATERIALS AND METHODS
Plant Materials
The Tam3-active Antirrhinum majus line HAM22, originally from John Innes Centre stock JI: 1 line (pallidarecurres), was used in this study. This line was initially grown for 1.5 months at 25°C in a greenhouse and was subsequently transferred into 15°C or 25°C growth chambers for at least 2 weeks. Onions were gifted from the Sapporo Experimental Station of Sumika Agrotech, Japan.
Plasmid Construction
The 2409-bp Tam3 TPase coding sequence was amplified from HAM22 genomic DNA by PCR using Tks Gflex DNA polymerase (Takara Bio). The amplified segment was induced into a pMD-20T vector using a ligation mix following the manufacturer’s instructions (Takara Bio). Following verification by sequencing, the target segment was inserted into XhoI and SpeI sites of the pA7-GFP vector, at the N terminus of the GFP gene. Truncated Tam3 TPase constructs (TPase∆55, TPase∆170, TPase∆179, TPase∆200, TPase∆231, and TPase∆244) were constructed by PCR and inserted into the same position of the pA7-GFP vector as the construct TPase.
To confirm whether the Znf-BED domain could guide anonymous proteins to the PM, AmCSBL and AmDnaJh1 coding sequences lacking stop codons were amplified using HAM22 cDNA products as template. The fragments were also cloned into XhoI and SpeI sites of the vector pA7-GFP, generating the constructs pA7-AmCSBL and pA7-DnaJh1, respectively. Overlapping PCR was adopted to add the sequence of the BED-zinc domain of Tam3 TPase to the N terminus of these two genes (Ge and Rudolph, 1997). In the first round PCR reaction of pA7-(Znf-AmCSBL), product 1 was amplified by primers T3TPase∆170 (XhoI)-F 5′-CCGCTCGAGCGGATGGCCTCTACATCAAGACC-3′ and Znf_BED&AmCSBL-R 5′-GATGAAGCACTACCCATTGTACCGTCTGGTTGTC-3′ using plasmids of construct TPase∆170 as template; product 2 was amplified by primers Znf_BED&AmCSBL-F 5′-GACAACCAGACGGTACAATGGGTAGTGCTTCATC-3′ and AmCSBL (SpeI)-R 5′-GGACTAGTCCGTCAATGGGGACTTCCATC-3′ using plasmids of construct pA7-AmCSBL as template. In the second round PCR reaction, product 3 was amplified by primers T3TPase∆170 (XhoI)-F and AmCSBL (SpeI)-R using a mixture of amplification product 1 and 2 as template. Product 3 was inserted into XhoI and SpeI sites of the pA7-GFP vector to generate construct pA7-(Znf-AmCSBL). Construct pA7-(Znf-DnaJh1) was constructed in a similar way. The primers used are listed in Supplemental Table S4.
Point Mutation
Point mutation constructs were created using overlapping PCR (Ge and Rudolph, 1997). Primers were designed to include the desired changes. The presence of a desired mutation(s) was confirmed for all constructs by sequencing. Primers used are listed in Supplemental Table S4.
Transient Expression Assay
Transient expression assays of GFP in Antirrhinum petal and onion epidermal cells were performed using the Helium Biolistic gene transformation system, IDERA GIE-III (Tanaka). Tissues were placed on the surface of 0.5× Murashige and Skoog medium. Approximately 2.0 µg plasmid DNA was precipitated onto 1.0 µm gold microcarriers (Bio-Rad). Bombardment delivery into prepared tissue was performed according to the methods described by Uchiyama et al. (2008).
Protoplast Transient Transformation
Protoplasts were prepared from young Antirrhinum leaves and fresh tobacco BY2 cells. PEG-calcium transfections were performed in accordance with Yoo et al. (2007). For each construct, 20 µg of plasmid DNA was used. Following transient transformation, all samples were cultivated at 25°C for 20 h.
Microscopic Observation
Fluorescence signals were observed using a UV-fluorescence microscope (Olympus) equipped with a U-MNIBA filter set (470–490 nm excitation filter, 505 nm dichroic mirror, 515–550 nm barrier filter) for GFP, and a U-MWIG filter set (520–550 nm excitation filter, 560 nm dichroic mirror, 580 nm barrier filter) for chloroplast autofluorescence.
Active Protein Extraction and Native PAGE Western Blot
After determining fluorescence, protoplasts were harvested by centrifugation at 100g for 2 min. Active protein was extracted with 50 µL protein extraction buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 0.2% Triton X-100, and protease inhibitor cocktail [Sigma-Aldrich]) for 1-mL protoplasts (approximately 1 × 106 cells). The extracts were gently shaken for 30 min at 0°C and centrifuged at 13,000 rpm for 15 min at 4°C, and the supernatants were collected for native PAGE western-blot analysis. Proteins were prepared in a nonreducing and nondenaturing sample buffer (50 mm Tris-HCl, pH 6.5, 10% glycerol, 12.5 mm EDTA, and 0.02% bromophenol blue), then separated on 6% PAGE gel without sodium dodecyl sulfate on ice. The running buffer was also sodium dodecyl sulfate-free. Gel electrophoresis was performed at 20 mA, then the proteins were transferred to a polyvinylidene fluoride membrane. Independently, the same samples were electrophoresed in adjacent lanes to check the running profile after CBB staining. The membrane was treated for blocking with 5% (w/v) ECL prime blocking agent (GE), and incubated overnight at 4°C with monoclonal mouse anti-GFP antibody diluted 1000-fold (anti-GFP, Monoclonal Antibody [mFX75], Wako Pure Chemical Industry). After washing three times with TBS, 0.1% Tween 20, the membrane was incubated for 2 h with secondary anti-mouse immunoglobulin antibody conjugated to horseradish peroxidase (GE), diluted 3000-fold, and then stained with ECL Western Blotting Detection Reagent (GE). Signals were detected using the chemical luminescent detection system (Ez-Capture MG, ATTO).
Protein Alignments
TPase coding sequences of the hAT family were acquired from GenBank and EMBL databases. Sequence alignments were performed by ClustalX Multiple Sequence Alignment (Thompson et al., 1994, 1997). Jalview software was employed to perform alignment editing for the visualization of final results (Waterhouse et al., 2009).
Accession Numbers
Sequence data from this article can be found in GenBank/EMBL database libraries under accession numbers CAA38906 (Tam3), CAA29005 (Ac), CAB68118 (Daysleeper), AM087608 (b-gary), AM087609 (w-gary1), AAC37217 (Hermes), U22467 (hermit), AAS21248 (Herves), 85002 (Hobo), AAD03082 (Homer), AAL93203 (Hopper), CAA93759 (Restless), AF051562 (Tag1), ABF20545 (TcBuster), AAP59878 (THELMA13), BAA87039 (Tol2), CAA76545 (tramp), KX016023 (AmCSBL), and KX016024 (AmDnaJh1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phenotype of Tam3 temporarily active Antirrhinum line at 15°C.
Supplemental Figure S2. Subcellular localization of Tam3 TPase in tobacco BY2 cells and onion epidermis cells.
Supplemental Table S1. Measurements of the rate of PM-located GFP signal of intact and truncated Tam3 TPase in protoplasts and petal cells of HAM22.
Supplemental Table S2. Measurements of the rate of PM-located GFP signal of pA7-(Znf-AmCSBL) and pA7-(Znf-DnaJh1) in protoplasts of HAM22.
Supplemental Table S3. Measurements of the rate of PM-located GFP signal of intact and mutated Tam3 in protoplasts HAM22.
Supplemental Table S4. Primers list of plasmid constructions.
Supplementary Material
Acknowledgments
We thank Drs. Sunlu Chen and Taichi E. Takasuka for valuable comments on the manuscript.
Glossary
- TE
transposable element
- PM
plasma membrane
- NLS
nuclear localization signal
- CBB
Coomassie Brilliant Blue
- ER
endoplasmic reticulum
References
- Almeida J, Carpenter R, Robbins TP, Martin C, Coen ES (1989) Genetic interactions underlying flower color patterns in Antirrhinum majus. Genes Dev 3: 1758–1767 [DOI] [PubMed] [Google Scholar]
- Aravind L. (2000) The BED finger, a novel DNA-binding domain in chromatin-boundary-element-binding proteins and transposases. Trends Biochem Sci 25: 421–423 [DOI] [PubMed] [Google Scholar]
- Becker H-A, Kunze R (1997) Maize Activator transposase has a bipartite DNA binding domain that recognizes subterminal sequences and the terminal inverted repeats. Mol Gen Genet 254: 219–230 [DOI] [PubMed] [Google Scholar]
- Bundock P, Hooykaas P (2005) An Arabidopsis hAT-like transposase is essential for plant development. Nature 436: 282–284 [DOI] [PubMed] [Google Scholar]
- Calvi BR, Hong TJ, Findley SD, Gelbart WM (1991) Evidence for a common evolutionary origin of inverted repeat transposons in Drosophila and plants: hobo, Activator, and Tam3. Cell 66: 465–471 [DOI] [PubMed] [Google Scholar]
- Carpenter R, Martin C, Coen ES (1987) Comparison of genetic behaviour of the transposable element Tam3 at two unlinked pigment loci in Antirrhinum majus. Mol Gen Genet 207: 82–89 [Google Scholar]
- Chen T, Li M, Ding Y, Zhang LS, Xi Y, Pan WJ, Tao DL, Wang JY, Li L (2009) Identification of zinc-finger BED domain-containing 3 (Zbed3) as a novel Axin-interacting protein that activates Wnt/β-catenin signaling. J Biol Chem 284: 6683–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBose-Boyd RA, Brown MS, Li W-P, Nohturfft A, Goldstein JL, Espenshade PJ (1999) Transport-dependent proteolysis of SREBP: rRlocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99: 703–712 [DOI] [PubMed] [Google Scholar]
- Diaz-Meco MT, Municio MM, Sanchez P, Lozano J, Moscat J (1996) Lambda-interacting protein, a novel protein that specifically interacts with the zinc finger domain of the atypical protein kinase C isotype lambda/iota and stimulates its kinase activity in vitro and in vivo. Mol Cell Biol 16: 105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin JA, Marchetto MC, Gage FH (2014) Mobile DNA elements in the generation of diversity and complexity in the brain. Nat Rev Neurosci 15: 497–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Hashida SN, Ogawa T, Natsume T, Uchiyama T, Mikami T, Kishima Y (2011) Temperature controls nuclear import of Tam3 transposase in Antirrhinum. Plant J 65: 146–155 [DOI] [PubMed] [Google Scholar]
- Galun E. (2003) Transposable Elements: A Guide to the Perplexed and the Novice with Appendices on RNAi, Chromatin Remodeling and Gene Tagging. Springer Science & Business Media, New York [Google Scholar]
- Ge L, Rudolph P (1997) Simultaneous introduction of multiple mutations using overlap extension PCR. Biotechniques 22: 28–30 [DOI] [PubMed] [Google Scholar]
- Greber UF, Gerace L (1992) Nuclear protein import is inhibited by an antibody to a lumenal epitope of a nuclear pore complex glycoprotein. J Cell Biol 116: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habu Y, Kakutani T, Paszkowski J (2001) Epigenetic developmental mechanisms in plants: molecules and targets of plant epigenetic regulation. Curr Opin Genet Dev 11: 215–220 [DOI] [PubMed] [Google Scholar]
- Harrison BJ, Fincham JRS (1964) Instability at the Pal locus in Antirrhinum majus. 1. Effects of environment on frequencies of somatic and germinal mutation. Heredity 19: 237–258 [Google Scholar]
- Hashida SN, Kishima Y, Mikami T (2005) DNA methylation is not necessary for the inactivation of the Tam3 transposon at non-permissive temperature in Antirrhinum. J Plant Physiol 162: 1292–1296 [DOI] [PubMed] [Google Scholar]
- Hashida SN, Kitamura K, Mikami T, Kishima Y (2003) Temperature shift coordinately changes the activity and the methylation state of transposon Tam3 in Antirrhinum majus. Plant Physiol 132: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashida SN, Uchiyama T, Martin C, Kishima Y, Sano Y, Mikami T (2006) The temperature-dependent change in methylation of the Antirrhinum transposon Tam3 is controlled by the activity of its transposase. Plant Cell 18: 104–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M, Brattig T, Kunze R (1994) In vivo aggregation of maize Activator (Ac) transposase in nuclei of maize endosperm and petunia protoplasts. Plant J 5: 705–714 [DOI] [PubMed] [Google Scholar]
- Hickman AB, Ewis HE, Li X, Knapp JA, Laver T, Doss AL, Tolun G, Steven AC, Grishaev A, Bax A, et al. (2014) Structural basis of hAT transposon end recognition by Hermes, an octameric DNA transposase from Musca domestica. Cell 158: 353–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A, Critchley J, Erasmus Y (2008) The genus Antirrhinum (snapdragon): A flowering plant model for evolution and development. Cold Spring Harb Protoc 2008: pdb.emo100 [DOI] [PubMed] [Google Scholar]
- Ishizu H, Nagao A, Siomi H (2011) Gatekeepers for Piwi-piRNA complexes to enter the nucleus. Curr Opin Genet Dev 21: 484–490 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y (2013) Molecular insights into microRNA-mediated translational repression in plants. Mol Cell 52: 591–601 [DOI] [PubMed] [Google Scholar]
- Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. (2013) MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153: 562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318: 1302–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack AM, Crawford NM (2001) The Arabidopsis TAG1 transposase has an N-terminal zinc finger DNA binding domain that recognizes distinct subterminal motifs. Plant Cell 13: 2319–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15: 394–408 [DOI] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A (1985) Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J 4: 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ (2000) Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes. Cell 102: 315–323 [DOI] [PubMed] [Google Scholar]
- Ono A, Kim S-H, Walbot V (2002) Subcellular localization of MURA and MURB proteins encoded by the maize MuDR transposon. Plant Mol Biol 50: 599–611 [DOI] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Ream T, Wierzbicki AT (2008) Roles of RNA polymerase IV in gene silencing. Trends Plant Sci 13: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Dunleavy E, Almouzni G (2009) Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol 10: 192–206 [DOI] [PubMed] [Google Scholar]
- Roccaro M, Li Y, Sommer H, Saedler H (2007) ROSINA (RSI) is part of a CACTA transposable element, TamRSI, and links flower development to transposon activity. Mol Genet Genomics 278: 243–254 [DOI] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA (2001) Structure and evolution of the hAT transposon superfamily. Genetics 158: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghizadeh M, Akhmedov NB, Yamashita CK, Gribanova Y, Theendakara V, Mendoza E, Nelson SF, Ljubimov AV, Farber DB (2009) ZBED4, a BED-type zinc-finger protein in the cones of the human retina. Invest Ophthalmol Vis Sci 50: 3580–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC (2009) A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizu H, Komai M, Kotani H, Kawamura Y, Nishida KM, Siomi H, Siomi MC (2010) Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev 24: 2493–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Tsugane K, Kanno T, Nishimura T (2012) DNA methylation in plants: relationship to small RNAs and histone modifications, and functions in transposon inactivation. Plant Cell Physiol 53: 766–784 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Davies B, Hudson A (2003) An everlasting pioneer: The story of Antirrhinum research. Nat Rev Genet 4: 657–666 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Fujino K, Ogawa T, Wakatsuki A, Kishima Y, Mikami T, Sano Y (2009) Stable transcription activities dependent on an orientation of Tam3 transposon insertions into Antirrhinum and yeast promoters occur only within chromatin. Plant Physiol 151: 1557–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama T, Saito Y, Kuwabara H, Fujino K, Kishima Y, Martin C, Sano Y (2008) Multiple regulatory mechanisms influence the activity of the transposon, Tam3, of Antirrhinum. New Phytol 179: 343–355 [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Espenshade PJ, Wright ME, Yabe D, Gong Y, Aebersold R, Goldstein JL, Brown MS (2002) Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell 110: 489–500 [DOI] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Zhang B, Wang Q, Pan X (2007) MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol 210: 279–289 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Mao H, Li S, Cao S, Li Z, Zhuang S, Fan J, Dong X, Borkan SC, Wang Y, et al. (2010) HSP72 inhibits Smad3 activation and nuclear translocation in renal epithelial-to-mesenchymal transition. J Am Soc Nephrol 21: 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.