The maize C2-Idf mutant has reduced levels of soluble flavonoids, reduced incorporation of tricin into the lignin polymer, and higher total lignin that is enriched in β-β and β-5 interunit linkages.
Abstract
Lignin is a phenolic heteropolymer that is deposited in secondary-thickened cell walls, where it provides mechanical strength. A recent structural characterization of cell walls from monocot species showed that the flavone tricin is part of the native lignin polymer, where it is hypothesized to initiate lignin chains. In this study, we investigated the consequences of altered tricin levels on lignin structure and cell wall recalcitrance by phenolic profiling, nuclear magnetic resonance, and saccharification assays of the naturally silenced maize (Zea mays) C2-Idf (inhibitor diffuse) mutant, defective in the CHALCONE SYNTHASE Colorless2 (C2) gene. We show that the C2-Idf mutant produces highly reduced levels of apigenin- and tricin-related flavonoids, resulting in a strongly reduced incorporation of tricin into the lignin polymer. Moreover, the lignin was enriched in β-β and β-5 units, lending support to the contention that tricin acts to initiate lignin chains and that, in the absence of tricin, more monolignol dimerization reactions occur. In addition, the C2-Idf mutation resulted in strikingly higher Klason lignin levels in the leaves. As a consequence, the leaves of C2-Idf mutants had significantly reduced saccharification efficiencies compared with those of control plants. These findings are instructive for lignin engineering strategies to improve biomass processing and biochemical production.
Lignocellulosic biomass is a renewable natural resource for the carbon-neutral production of biofuels and biochemicals (Vanholme et al., 2013a; Wilkerson et al., 2014; Welker et al., 2015) that is readily available from agricultural crop residues, inedible plant tissues, or dedicated biomass crops (Abramson et al., 2010). Plant secondary cell walls, which make up the bulk of lignocellulosic biomass, are composed mainly of cellulose, hemicelluloses, and lignin. The cell wall polysaccharide fraction, which constitutes roughly 75% of the total lignocellulosic mass, can be converted into fermentable sugars (Wilke et al., 1981; Vanholme et al., 2013a; Marriott et al., 2016). However, the compact structure and complex chemical composition of the plant cell wall negatively affect the enzymatic digestibility of the biomass, a feature known as biomass recalcitrance (Zhao et al., 2012). Several factors limit the enzymatic degradation, such as carbohydrate-lignin cross-linking and, especially, the presence of the aromatic polymer lignin (Chen and Dixon, 2007; Grabber et al., 2008; Van Acker et al., 2013; Vermerris and Abril, 2015; Wang et al., 2015a). Lignin negatively affects the conversion of cell wall polysaccharides by immobilizing hydrolytic enzymes and by blocking enzyme access to the polysaccharides (Chundawat et al., 2011; Vanholme et al., 2012). Therefore, decreasing lignin content and changing lignin composition by genetic engineering or breeding are promising strategies to improve biomass-processing properties.
Lignin is a phenolic heteropolymer that is deposited in secondary-thickened cell walls to provide strength and rigidity to specialized cell types (Boerjan et al., 2003; Vanholme et al., 2010a; Cesarino et al., 2012). Lignin is derived from the oxidative radical-radical coupling of three main hydroxycinnamyl alcohol monomers, the monolignols p-coumaryl, coniferyl, and sinapyl alcohol, that differ in their degree of aromatic ring methoxylation and couple (as their radicals) in a primarily end-wise manner with the growing polymer radical. Once incorporated into the polymer, these monolignols produce p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units (Boerjan et al., 2003). In grasses, a large fraction of the coniferyl and sinapyl alcohol monomers are γ-O-acylated with acetate or p-coumarate, resulting in acetylated and p-coumaroylated units after their polymerization into the lignin polymer (Ralph, 2010).
Besides the canonical monolignols and their acylated analogs, some intermediates of the monolignol biosynthetic pathway, such as p-hydroxycinnamaldehydes and p-hydroxycinnamates, also may be incorporated into native lignins (Ralph et al., 1994, 2004; Ralph, 1996; Lu and Ralph, 2002; Morreel et al., 2004). The remarkable ability of lignin to incorporate a variety of monomers highlights the inherent malleability of lignification, confirms the original combinatorial chemical coupling theory of lignification (Ralph et al., 2008a), and suggests that genetic engineering of novel lignins is a promising strategy to tailor plants with improved processing properties (Vanholme et al., 2012; Wilkerson et al., 2014; Mottiar et al., 2016).
Recent structural characterization of cell walls from monocot species using NMR showed that the flavone tricin [5,7-dihydroxy-2-(4-hydroxy-3,5-dimethoxyphenyl)-4H-chromen-4-one] is part of the native lignin polymer in wheat (Triticum aestivum; del Río et al., 2012; Zeng et al., 2013), coconut (Cocos nucifera) coir (Rencoret et al., 2013), bamboo (Phyllostachys pubescens; Wen et al., 2013), maize (Zea mays; Lan et al., 2015), and sugarcane (Saccharum officinarum; del Río et al., 2015). Tricin is the first flavonoid that is recognized to be an authentic monomer involved in lignification (del Río et al., 2012; Lan et al., 2015). Moreover, tricin is already well recognized as a valuable health-promoting compound due to its antioxidant, antiaging, anticancer, and cardioprotective potential (Zhou and Ibrahim, 2010; Li et al., 2016). Recent findings show that tricin incorporates into the lignin polymer of maize plants via 4′-O-β cross-coupling with normal and acylated monolignols, acting as an initiation site for lignin polymerization because it can only start a lignin chain (Lan et al., 2015, 2016a). Moreover, the initiation role of tricin has helped explain a long-standing enigma of how the lignin chain is initiated in monocots. In dicots and gymnosperms, the lignin polymer is initiated by monolignol dimerization, giving rise to β-ether, phenylcoumaran, and resinol dimers via β-O-4, β-5, and β-β coupling, respectively. In monocots, there is an additional dimeric unit, the tetrahydrofuran structure derived from the β-β coupling of sinapyl p-coumarate (Lan et al., 2015, 2016a). Nevertheless, in monocots, lignin only has a relatively low frequency of β-5 and β-β interunit linkages, which could most easily be explained by the alternative chain initiation, involving either tricin (Lan et al., 2015) or (di)ferulates (Ralph et al., 1995; Grabber et al., 2000; Ralph, 2010) or both. However, this causal relationship has not yet been unambiguously proven.
The flux toward flavonoid biosynthesis and, thus, to the tricin monomer is controlled by chalcone synthase (CHS; Fig. 1). This enzyme converts p-coumaroyl-CoA into naringenin chalcone, which is then isomerized by chalcone isomerase (CHI) to the flavanone naringenin, a common precursor for several flavonoid classes (Shih et al., 2008; Dixon and Pasinetti, 2010). The biochemical route from naringenin toward tricin in grasses has only recently been fully resolved (Lam et al., 2014, 2015). In rice (Oryza sativa), naringenin is desaturated by flavone synthase to form apigenin, after which an extra hydroxyl group is added by flavonoid 3′-hydroxylase to produce luteolin. This hydroxyl group is then methylated by flavonoid O-methyltransferase (FOMT) to form chrysoeriol. Another hydroxyl group is added to the same ring by flavonoid 5′-hydroxylase, and the resulting selgin is then again methylated by FOMT to form tricin (Lam et al., 2015). In maize, two genes have been described to encode CHS: Colorless2 (C2) and Whitepollen1 (Whp1; Coe et al., 1981; Franken et al., 1991). The coding regions of C2 and Whp1 share 94% sequence identity, but the genes have only partially overlapping expression patterns and are independently regulated (Coe et al., 1981; Franken et al., 1991). The C2 gene is expressed in many parts of the plant, including the pericarp, the aleurone layer of the endosperm, tassels, and vegetative organs such as ear husks and leaf sheaths, whereas Whp1 is expressed only in pollen and the aleurone layer of the kernel (Coe et al., 1981; Franken et al., 1991). Based on these expression studies, the C2 gene is the best candidate for having a role in flavonoid biosynthesis in vegetative tissues, such as stems and leaves of maize. As C2 directs the flux toward the biosynthesis of all flavonoids, disruption of C2 would cause the depletion of flavonoids, including tricin, in stems and leaves of maize. Plants carrying the colorless dominant inhibitory c2 mutation, called C2-Idf (inhibitor diffuse), have a duplication of the C2 gene that causes gene silencing (Della Vedova et al., 2005). As a consequence, no C2-specific mRNA can be detected in the C2-Idf mutants (Franken et al., 1991), and these plants do not produce anthocyanins (Coe et al., 1988).
Figure 1.
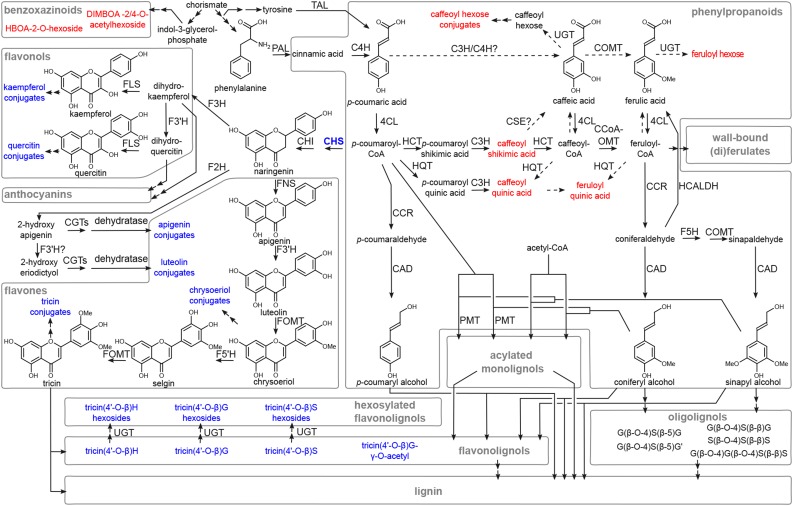
Metabolic map of the phenolic and benzoxazinoid pathways in maize. Solid arrows represent enzymatic conversions for which experimental evidence is available, and dashed arrows represent suggested conversions. Two successive arrows represent two or more metabolic conversions. Metabolic shifts in C2-Idf mutants compared with C2 control plants (Tables II–V) are indicated in color, where red represents a significant increase and blue represents a significant decrease in abundance. Only those metabolic sinks that have been measured in the C2-Idf mutants are shown. Reduced biosynthesis of anthocyanins in C2-Idf mutants has been reported by Coe et al. (1988). The position of the CHS (C2) enzyme is emphasized in a blue boldface font. CAD, Cinnamyl alcohol dehydrogenase; CCoAOMT, caffeoyl-CoA O-methyltransferase; CCR, cinnamoyl-CoA reductase; CGT, C-glycosyltransferase; C3H, p-coumarate 3-hydroxylase; C4H, cinnamate-4-hydroxylase; CHI, chalcone isomerase; 4CL, 4-coumarate:CoA ligase; COMT, caffeic acid O-methyltransferase; CSE, caffeoyl shikimate esterase; F2H, flavanone 2-hydroxylase; F3H, naringenin 3-dioxygenase; F3′H, flavonoid 3′-hydroxylase; F5H, ferulate 5-hydroxylase; F5′H, flavonoid 5′-hydroxylase; FLS, flavonol synthase; FNS, flavone synthase; FOMT, flavonoid O-methyltransferase; HCALDH, hydroxycinnamaldehyde dehydrogenase; HCT, p-hydroxycinnamoyl-CoA:quinate/shikimate p-hydroxycinnamoyltransferase; HQT, hydroxycinnamoyl-CoA:quinate hydroxycinnamoyltransferase; PAL, Phe ammonia lyase; PMT, p-coumaroyl-CoA:monolignol transferase; TAL, Tyr ammonia lyase; UGT, UDP-glucosyltransferase. The relation between TAL and PAL activity has been reported by Rösler et al. (1997) for maize and by Cass et al. (2015) for brachypodium. Enzymes involved in the general phenylpropanoid (PAL, TAL, C4H, 4CL, HCT, C3H, and CCoAOMT) and monolignol-specific (CCR, F5H, COMT, and CAD) pathways are described by Guillaumie et al. (2007). Homologs for the Arabidopsis HCALDH (Nair et al., 2004) are present in maize (Končitíková et al., 2015; Missihoun et al., 2016). PMT has been shown in rice (Withers et al., 2012), brachypodium (Petrik et al., 2014), and maize (Marita et al., 2014). The flavone biosynthesis pathway is shown according to Zhou et al. (2008), Cortés-Cruz et al. (2003), and Lam et al. (2014, 2015). The enzymes involved in the metabolic route via F2H toward apigenin and luteolin conjugates has been described in rice, wheat, and buckwheat (Fagopyrum esculentum; Brazier-Hicks et al., 2009; Du et al., 2010). The biosynthetic routes via HCT and HQT have been described in switchgrass (Escamilla-Treviño et al., 2014) and suggested to be present in maize (Cortés-Cruz et al., 2003). In addition, HCT of sorghum (Sorghum bicolor) has been studied via in vitro activity assays (Walker et al., 2013). The FOMT involved in the conversion of luteolin and selgin into chrysoeriol and tricin, respectively, in maize is described by Zhou et al. (2008) and is identical to the COMT involved in the conversion of 5-hydroxyconiferaldehyde to sinapaldehyde described by Collazo et al. (1992), Vignols et al. (1995), and Piquemal et al. (2002). Question marks indicate biosynthetic steps that are questionable; the reaction from p-coumaric acid toward caffeic acid via a C3H/C4H complex has so far only been shown by in vitro tests using poplar enzymes (Chen et al., 2011). The activity of CSE has been described in switchgrass (Escamilla-Treviño et al., 2014; Ha et al., 2016), but the absence of close homologs with the reported Arabidopsis CSE (Vanholme et al., 2013b; Ha et al., 2016) makes it questionable whether this reaction is present in maize. For nomenclature of the oligolignols, see Supplemental Table S2.
In this study, we examined the consequences of reduced tricin levels on lignin structure and amount, on biomass processing efficiency, and on the carbon flux into the phenylpropanoid pathway using the maize C2-Idf mutant allele (Franken et al., 1991). The C2-Idf allele was in a B; Pl; R-scm2 W22 background (see “Materials and Methods”). The seeds and vegetative tissues of the B; Pl; R-scm2 W22 background (C2 control plants) are purple pigmented, whereas seeds and vegetative tissues of the homozygous C2-Idf plants lack the purple pigment and are yellow and green, respectively. Our findings are instructive for cell wall engineering strategies aimed at improving saccharification and biochemical production.
RESULTS
Phenotypic Characterization of the Maize C2-Idf Mutant
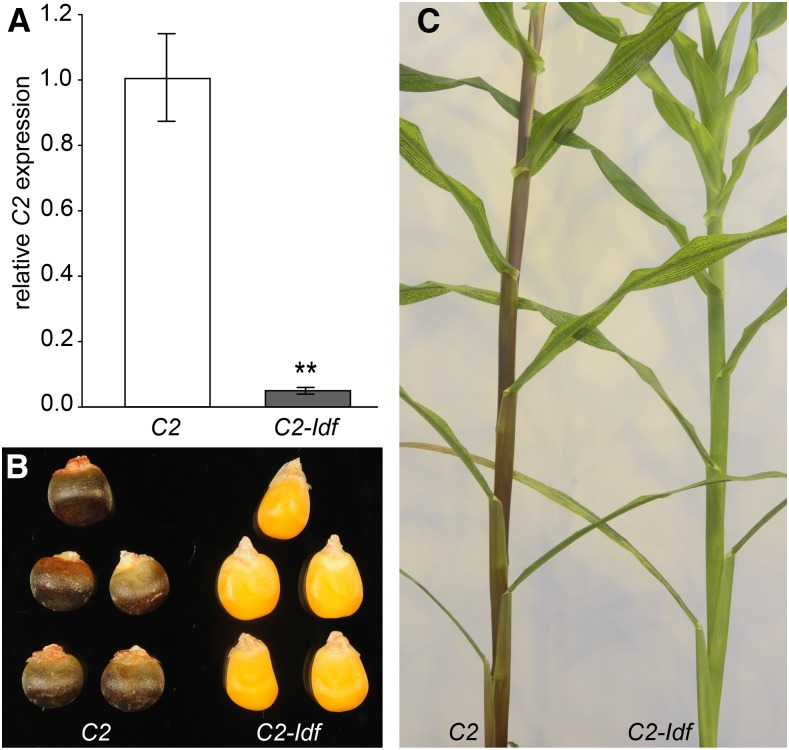
To determine the residual C2 mRNA level in the C2-Idf mutant, a real-time quantitative PCR (RT-qPCR) analysis was conducted. C2 expression was measured in the ninth internode of C2-Idf and control plants at the developmental stage where the plants had 14 unfolded leaves (i.e. growth stage V14). The expression level of C2 in stems of C2-Idf plants was reduced to 5% of the value in control plants (Fig. 2A), confirming the findings of Franken et al. (1991) that C2 expression is highly reduced in the C2-Idf mutants compared with the C2 control plants. Seeds and leaf sheaths of the homozygous C2-Idf plants lacked the purple anthocyanin pigmentation that is typically observed in C2 control plants (Fig. 2, B and C).
Figure 2.
Expression levels of C2 (CHS) and visible C2-Idf mutant phenotypes. A, Expression levels of C2 (CHS) in C2 control and C2-Idf mutant internodes were determined by RT-qPCR and are shown relative to the highest C2 expression value within the replicates. Error bars represent se of three biological replicates. **, P < 0.01. B, Photograph of representative C2 control and C2-Idf mutant seeds. C, Photograph of representative plants at the V14 stage, showing the vegetative tissues.
Next, we investigated the effect of CHS down-regulation on plant growth and development. Total plant height, with or without tassel, did not show any significant difference between C2 control and C2-Idf plants (Table I). Also, the length of the ear leaf was not altered significantly. However, the total dry weight of fully senesced C2-Idf plants was increased significantly (+18%). The increase could be attributed to the increase in dry weight of the leaves (+27%), as the dry weight of stems was not altered significantly (Table I). To validate the growth phenotype observed in C2-Idf maize plants, we grew another set of plants. In agreement with the previous results, the dry weight of fully senesced C2-Idf plants was increased significantly compared with the dry weight of C2 control plants, but the difference was larger (+32%; Supplemental Table S1). However, in this second growth experiment, the increase in dry weight was attributable to a significant increase in dry weight of both stems (+34%) and leaves (+25%; Supplemental Table S1).
Table I. Growth parameters of C2 control and C2-Idf mutant maize plants.
Plant and ear height, leaf length, and dry biomass measurements were performed on senesced greenhouse-grown plants. Leaf 7, The seventh leaf that appeared after germination; ear height, the height of the implantation of the ear on the stem; n.s., not significantly different. C2 and C2-Idf values are means ± se (n = 7). Underlined values are significantly different compared with the control: *, 0.05 ≥ P > 0.01; and ***, P < 0.001.
| Growth Parameter | C2 | C2-Idf | Percentage Difference | Student’s t Test |
|---|---|---|---|---|
| Plant height with tassel (cm) | 268.6 ± 16.6 | 259.9 ± 10.3 | −3.2 | n.s. |
| Plant height without tassel (cm) | 225.7 ± 12.0 | 217.1 ± 10.6 | −3.8 | n.s. |
| Ear height (cm) | 123.4 ± 6.45 | 121.1 ± 6.7 | −1.9 | n.s. |
| Leaf 7 length (cm) | 91.3 ± 4.9 | 89.4 ± 2.1 | −2.1 | n.s. |
| Leaf dry weight (g) | 34.9 ± 4.5 | 44.2 ± 6.2 | 26.6 | *** |
| Stem dry weight (g) | 56.5 ± 10.45 | 64.3 ± 9.2 | 13.8 | n.s. |
| Total (leaves + stems) dry weight (g) | 91.4 ± 14.3 | 107.6 ± 150.1 | 17.7 | * |
CHS-Deficient Plants Have an Altered Phenolic Metabolism
Phenolic profiling of mutants in the lignin biosynthetic pathway has provided extensive insight into which branches of the phenylpropanoid biosynthesis the carbon flux is redirected (Vanholme et al., 2012). To investigate whether flavonoid levels were reduced in the C2-Idf mutant, and to reveal which pathways absorb the carbon flux, we performed ultra-high-performance liquid chromatography-mass spectrometry (UHPLC-MS)-based untargeted metabolic profiling (Vanholme et al., 2013b; Sundin et al., 2014) of the ninth internode and the ninth leaf from C2 control and C2-Idf plants. A total of 8,832 peaks could be detected, aligned, and integrated. Only those peaks that were present in all replicates (n = 7) of at least one of the four sample types (two genotypes and two tissues) and that were above a 500-count cutoff, regarded as the noise level, were selected. This selection retained 2,005 and 4,073 peaks in internode and leaf samples, respectively, for further analysis. To pinpoint the metabolites for which the abundance was most affected due to the mutation, a selection with more stringent filters was applied based on significance level (Student’s t test P < 0.001), peak intensity (at least 5,000 counts in either genotype), and fold change between C2-Idf and C2 control samples (at least a 10-fold difference in abundance).
In internodes, the 55 peaks with a significantly lower intensity in C2-Idf plants compared with the C2 control could be assigned to 49 compounds (because of in-source fragmentation and adduct formation, a single compound can give rise to multiple mass-to-charge ratio [m/z] peaks), of which 34 could be structurally characterized (Table II). The 28 peaks with a higher intensity could be assigned to 15 compounds, of which nine compounds could be structurally characterized (Table III). The 34 identified compounds that had reduced abundance in C2-Idf internodes contained apigenin, tricin, or kaempferol as a substructure and could be classified into four metabolic classes: flavones, flavonols, hexosylated flavonolignols, and flavonolignols (Table II). Here, we define flavonolignols as 4′-O-β-linked coupling products of a flavone and one or more phenylpropanoids. This term is in analogy with the existing term flavonolignan that is used for the low-molecular-mass compounds composed of flavonoid and lignan moieties (Begum et al., 2010) and the term oligolignol that is used for oligomers of canonical monolignols (Morreel et al., 2004, 2010a, 2010b). The fact that some flavonolignols were found without further decoration with sugars and their lack of optical activity suggest that these might be considered as lignin oligomers (Lan et al., 2015, 2016a). Alternatively, they were in the process of being further decorated to become part of the hexosylated flavonolignol pool. The identified flavones and flavonols that were reduced in abundance in C2-Idf internodes were decorated with glycerol, pentose, deoxypentose, hexose, hexuronic acid, quinic acid, phenylpropanoids, benzenoids, or a combination of these. Seven of the nine identified compounds that accumulated in C2-Idf plants were phenylpropanoids: four had a caffeate moiety and three had a ferulate moiety (Table III). In addition, two benzoxazinoids had an increased abundance in C2-Idf internodes: 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one-2-O-hexoside and 2,4-dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one-2/4-O-acetylhexoside (Table III).
Table II. List of compounds that are decreased in abundance in C2-Idf internodes.
For each compound, its unique number (No.), mass-to-charge ratio (m/z), retention time (RT), peak area (as mean ± sd), and ratio of the peak area in C2 and C2-Idf are given. inf, Infinite. Tandem mass spectrometry (MS/MS)-based structural characterization is given in Supplemental File S1.
| No. |
m/z |
RT (min) |
Name |
Peak Area, C2 |
Peak Area, C2-Idf |
Ratio, C2/C2-Idf |
|---|---|---|---|---|---|---|
| Flavones | ||||||
| 1 | 533.1310 | 8.60 | Apigenin-6C,8C-dipentoside | 68,464 ± 24,794 | 2,551 ± 1,641 | 27 |
| 2 | 547.1454 | 10.00 | Apigenin-6C-deoxyhexoside-8C-pentoside 1 | 11,323 ± 3,720 | 11 ± 23 | 987 |
| 3 | 547.1457 | 10.63 | Apigenin-6C-deoxyhexoside-8C-pentoside 2 | 116,748 ± 37,294 | 1,482 ± 730 | 79 |
| 4 | 561.1615 | 11.99 | Apigenin-6C,8C-dideoxyhexoside 1 | 6,708 ± 1,755 | 43 ± 41 | 156 |
| 5 | 561.1601 | 12.69 | Apigenin-6C,8C-dideoxyhexoside 2 | 34,529 ± 8,066 | 100 ± 97 | 346 |
| 6 | 563.1405 | 7.08 | Apigenin-6C-hexoside-8C-pentoside | 270,510 ± 91,998 | 2,207 ± 1,168 | 123 |
| 7 | 577.1553 | 8.36 | Apigenin-6C-hexoside-8C-deoxyhexoside 1 | 17,831 ± 5,471 | 17 ± 29 | 1,046 |
| 8 | 577.1557 | 8.88 | Apigenin-6C-hexoside-8C-deoxyhexoside 2 | 75,223 ± 23,574 | 162 ± 167 | 464 |
| 9 | 577.1556 | 9.04 | Apigenin-6C-hexoside-8C-deoxyhexoside 3 | 62,832 ± 19,239 | 103 ± 155 | 613 |
| 10 | 593.1493 | 5.95 | Apigenin-6C,8C-dihexoside | 236,212 ± 79,208 | 1,328 ± 498 | 178 |
| 11 | 491.1213 | 9.94 | Tricin-O-hexoside 1 | 125,244 ± 47,891 | 425 ± 380 | 295 |
| 12 | 491.1208 | 11.40 | Tricin-O-hexoside 2 | 45,980 ± 14,565 | 5 ± 10 | 9,082 |
| 13 | 403.1057 | 15.49 | Tricin + glycerol | 12,669 ± 4,586 | 0 ± 0 | inf |
| 14 | 637.1389 | 10.87 | Tricin + hexuronic acid + pentose | 132,255 ± 47,827 | 1,577 ± 1,258 | 84 |
| 15 | 787.1686 | 12.34 | Tricin + hexuronic acid + pentose + vanillic acid | 54,489 ± 16,951 | 868 ± 475 | 63 |
| 16 | 817.1781 | 11.70 | Tricin + hexuronic acid + pentose + syringic acid | 8,932 ± 2,848 | 29 ± 31 | 310 |
| 17 | 813.1832 | 13.41 | Tricin + hexuronic acid + pentose + ferulic acid | 10,840 ± 2,690 | 54 ± 55 | 199 |
| 18 | 843.1947 | 12.78 | Tricin + hexuronic acid + pentose + sinapic acid | 17,337 ± 4,384 | 405 ± 234 | 43 |
| 19 | 1,009.2540 | 13.66 | Tricin + hexuronic acid + pentose + G(8-O-4)ferulic acid 1 | 18,550 ± 4,836 | 71 ± 83 | 262 |
| 20 | 1,009.2537 | 13.87 | Tricin + hexuronic acid + pentose + G(8-O-4)ferulic acid 2 | 22,561 ± 6,697 | 107 ± 152 | 212 |
| 21 | 1,039.2637 | 13.71 | Tricin + hexuronic acid + pentose + S(8-O-4)ferulic acid | 7,129 ± 1,928 | 2 ± 5 | 4,106 |
| 22 | 801.1826 | 13.91 | Tricin + hexuronic acid + pentose + 182 D | 12,375 ± 5,013 | 7 ± 13 | 1,881 |
| 23 | 827.1988 | 15.24 | Tricin + hexuronic acid + pentose + 208 D | 26,744 ± 5,908 | 376 ± 237 | 71 |
| 24 | 887.2195 | 11.60 | Tricin + hexuronic acid + pentose + 250 D | 21,962 ± 5,950 | 135 ± 50 | 163 |
| 25 | 667.1479 | 9.59 | Tricin + hexuronic acid + hexose 1 | 14,943 ± 4,672 | 55 ± 83 | 272 |
| Flavonols | ||||||
| 26 | 447.0959 | 10.14 | Kaempferol-O-hexoside | 28,991 ± 16,515 | 5 ± 8 | 6,295 |
| 27 | 593.1499 | 8.85 | Kaempferol-O-hexoside + deoxyhexose | 7,781 ± 2,787 | 0 ± 0 | inf |
| Hexosylated flavonolignols | ||||||
| 28 | 687.1894 | 12.17 | Tricin(4′-O-β)G-5/7-O-hexoside 1 | 37,436 ± 13,500 | 51 ± 52 | 731 |
| 29 | 687.1891 | 12.86 | Tricin(4′-O-β)G-5/7-O-hexoside 2 | 27,043 ± 12,014 | 0 ± 0 | inf |
| 30 | 687.1900 | 13.59 | Tricin(4′-O-β)G-5/7-O-hexoside 3 | 8,291 ± 4,223 | 94 ± 57 | 88 |
| 31 | 687.1895 | 14.18 | Tricin(4′-O-β)G-γ/α/4-O-hexoside 1 | 26,948 ± 11,401 | 3 ± 8 | 8,896 |
| 32 | 717.2028 | 11.13 | Tricin(4′-O-β)S-5/7-O-hexoside | 12,748 ± 4,345 | 740 ± 431 | 17 |
| Flavonolignols | ||||||
| 33 | 721.2093 | 16.18 | Tricin(4′-O-β)G(4-O-β)G 1 | 9,700 ± 2,703 | 0 ± 0 | inf |
| 34 | 721.2097 | 16.88 | Tricin(4′-O-β)G(4-O-β)G 2 | 7,333 ± 4.487 | 0 ± 0 | inf |
Table III. List of compounds that are increased in abundance in C2-Idf internodes.
For each compound, its unique number (No.), m/z, retention time (RT), peak area (as mean ± sd), and ratio of the peak area in C2-Idf and C2 are given. *, Compound detected as an in-source fragment. MS/MS-based structural characterization is given in Supplemental File S1.
| No. |
m/z |
RT (min) |
Name |
Peak Area, C2 |
Peak Area, C2-Idf |
Ratio, C2-Idf/C2 |
|---|---|---|---|---|---|---|
| 35 | 353.0908 | 2.84 | 3-O-Caffeoyl quinic acid | 62 ± 104 | 28,607 ± 22,141 | 461 |
| 36 | 353.0808 | 4.07 | 5-O-Caffeoyl quinic acid | 4,775 ± 1,626 | 72,877 ± 48,103 | 15 |
| 37 | 335.0800 | 6.00 | Caffeoyl shikimic acid 1 | 4,478 ± 2,717 | 112,871 ± 86,101 | 25 |
| 38 | 335.0792 | 7.50 | Caffeoyl shikimic acid 2 | 170 ± 319 | 5,571± 6,418 | 33 |
| 39 | 355.1037 | 5.24 | Feruloyl hexose | 1,133 ± 247 | 41,868 ± 16,125 | 37 |
| 40 | 193.0503 | 4.58 | 3-O-Feruloyl quinic acid* | 3,854 ± 824 | 43,075 ± 15,987 | 11 |
| 41 | 367.1060 | 6.59 | 4-O-Feruloyl quinic acid | 1,871 ± 745 | 52,468 ± 25,642 | 28 |
| 42 | 326.0910 | 4.41 | 2-Hydroxy-2H-1,4-benzoxazin-3(4H)-one-2-O-hexoside | 5 ± 14 | 8,934 ± 3,412 | 1,740 |
| 43 | 414.1062 | 7.23 | 2,4-Dihydroxy-7-methoxy-2H-1,4-benzoxazin-3(4H)-one-2/4-O-acetylhexoside | 24 ± 30 | 42,132 ± 1,4371 | 1,746 |
The 79 peaks with a lower intensity in leaves of C2-Idf plants compared with the C2 control could be assigned to 66 compounds, of which 39 could be structurally characterized (Table IV). The 36 peaks with a higher intensity in leaves could be attributed to 34 compounds, of which nine could be (partially) structurally characterized (Table V). The same classes of compounds that had a reduced abundance in the internode of C2-Idf plants also were severely reduced in abundance in their leaves (Table IV). In addition to apigenin- and tricin-containing flavones, three luteolin- and one chrysoeriol-containing flavones also were present in C2 control leaves, but their abundances dropped below 10% in leaves of C2-Idf plants. In the metabolic classes of flavonolignols and hexosylated flavonolignols, coupling products with H units were present, in addition to coupling products with G and S units (Table IV). The nine (partially) identified compounds that accumulated in leaves of C2-Idf plants all contained a caffeate moiety (Table V). The collective metabolic shifts in C2-Idf leaves and stems are visualized in Figure 1.
Table IV. List of compounds that are decreased in abundance in C2-Idf leaves.
For each compound, its unique number (No.), m/z, retention time (RT), peak area (as mean ± sd), and ratio of the peak area in C2 and C2-Idf are given. inf, Infinite. MS/MS-based structural characterization is given in Supplemental File S1.
| No. |
m/z |
RT (min) |
Name |
Peak Area, C2 |
Peak Area, C2-Idf |
Ratio, C2/C2-Idf |
|---|---|---|---|---|---|---|
| Flavones | ||||||
| 2 | 547.1454 | 10.00 | Apigenin-6C-deoxyhexoside-8C-pentoside | 1,770 ± 3,555 | 1,054 ± 268 | 16 |
| 3 | 547.1457 | 10.63 | Apigenin-6C-deoxyhexose-8C-pentoside | 43,830 ± 10,877 | 2,118 ± 452 | 21 |
| 5 | 561.1601 | 12.69 | Apigenin-6C,8C-dideoxyhexoside 2 | 8,953 ± 2,355 | 538 ± 309 | 17 |
| 6 | 563.1405 | 7.08 | Apigenin-6C-hexoside-8C-pentoside | 289,600 ± 64,693 | 5,024 ± 1,453 | 58 |
| 7 | 577.1553 | 8.36 | Apigenin-6C-hexoside-8C-deoxyhexoside 1 | 31,880 ± 6,590 | 293 ± 133 | 109 |
| 8 | 577.1557 | 8.88 | Apigenin-6C-hexoside-8C-deoxyhexoside 2 | 45,078 ± 9,786 | 587 ± 238 | 77 |
| 9 | 577.1556 | 9.04 | Apigenin-6C-hexoside-8C-deoxyhexoside 3 | 39,304 ± 8,518 | 217 ± 56 | 181 |
| 10 | 593.1493 | 5.95 | Apigenin-6C,8C-dihexoside | 194,461 ± 42,585 | 4,538 ± 1,082 | 43 |
| 44 | 593.1502 | 6.77 | Apigenin-C-hexoside + hexose | 7,521 ± 1,510 | 31 ± 27 | 244 |
| 45 | 447.0955 | 7.02 | Luteolin-C-hexoside | 30,524 ± 9,242 | 1,589 ± 182 | 19 |
| 46 | 579.1343 | 6.15 | Luteolin-6C-hexoside-8C-pentoside | 13,356 ± 4,131 | 2 ± 4 | 8,203 |
| 47 | 593.1507 | 7.13 | Luteolin-C-hexoside + deoxyhexose | 14,903 ± 2,980 | 182 ± 92 | 82 |
| 48 | 651.1179 | 8.50 | Chrysoeriol + hexuronic acid + hexuronic acid | 13,558 ± 2,317 | 1,100 ± 529 | 12 |
| 11 | 491.1213 | 9.94 | Tricin-O-hexoside 1 | 16,812 ± 4,302 | 823 ± 131 | 20 |
| 12 | 491.1208 | 11.40 | Tricin-O-hexoside 2 | 17,680 ± 4,715 | 495 ± 153 | 36 |
| 13 | 403.1057 | 15.49 | Tricin + glycerol | 8,511 ± 2,242 | 5 ± 14 | 1,621 |
| 25 | 667.1479 | 9.59 | Tricin + hexuronic acid + hexose 1 | 11,573 ± 3,063 | 951 ± 423 | 12 |
| 49 | 667.1498 | 9.36 | Tricin + hexuronic acid + hexose 2 | 33,817 ± 6,894 | 2,334 ± 661 | 14 |
| 14 | 637.1389 | 10.87 | Tricin + hexuronic acid + pentose | 20,469 ± 5,449 | 1,185 ± 496 | 17 |
| 17 | 813.1832 | 13.41 | Tricin + hexuronic acid + pentose + ferulic acid | 15,008 ± 3,821 | 1,450 ± 537 | 10 |
| Flavonols | ||||||
| 27 | 593.1499 | 8.85 | Kaempferol-O-hexoside + deoxyhexose | 136,928 ± 31,412 | 199 ± 41 | 688 |
| 49 | 609.1452 | 7.74 | Quercitin-O-hexoside + deoxyhexoside | 20,081 ± 3,690 | 525 ± 196 | 38 |
| Hexosylated flavonolignols | ||||||
| 50 | 657.1798 | 14.74 | Tricin(4′-O-β)H-γ/α/4-Ο-hexoside | 19,127 ± 4,975 | 47 ± 28 | 408 |
| 28 | 687.1894 | 12.17 | Tricin(4′-O-β)G-5/7-O-hexoside 1 | 19,765 ± 5,627 | 0 ± 0 | inf |
| 29 | 687.1891 | 12.86 | Tricin(4′-O-β)G-5/7-O-hexoside 2 | 18,526 ± 5,906 | 3 ± 8 | 5,993 |
| 30 | 687.1900 | 13.59 | Tricin(4′-O-β)G-5/7-O-hexoside 3 | 17,549 ± 5,108 | 4 ± 12 | 3,965 |
| 31 | 687.1895 | 14.18 | Tricin(4′-O-β)G-γ/α/4-O-hexoside 1 | 68,867 ± 17,163 | 11 ± 15 | 6,275 |
| 51 | 687.1901 | 14.80 | Tricin(4′-O-β)G-γ/α/4-O-hexoside 2 | 46,488 ± 10,726 | 58 ± 18 | 806 |
| Flavonolignols | ||||||
| 52 | 495.1309 | 17.65 | Tricin(4′-O-β)H 1 | 11,193 ± 2,943 | 4 ± 4 | 2,653 |
| 53 | 495.1310 | 18.36 | Tricin(4′-O-β)H 2 | 12,388 ± 3,166 | 3 ± 4 | 4,177 |
| 54 | 525.1408 | 18.01 | Tricin(4′-O-β)G 1 | 93,393 ± 24,017 | 17 ± 17 | 5,614 |
| 55 | 525.1411 | 18.77 | Tricin(4′-O-β)G 2 | 75,740 ± 17,902 | 2 ± 5 | 42,988 |
| 56 | 539.1562 | 21.74 | Tricin(4′-O-β)G-α-O-methyl | 6,914 ± 3,117 | 10 ± 27 | 675 |
| 57 | 567.1497 | 22.13 | Tricin(4′-O-β)G-γ-O-acetyl | 9,341 ± 4,185 | 2 ± 6 | 4,472 |
| 33 | 721.2093 | 16.18 | Tricin(4′-O-β)G(4-O-β)G 1 | 14,826 ± 5,233 | 10 ± 12 | 1,558 |
| 34 | 721.2097 | 16.88 | Tricin(4′-O-β)G(4-O-β)G 2 | 20,322 ± 6,742 | 5 ± 14 | 3,873 |
| 58 | 721.2097 | 16.43 | Tricin(4′-O-β)G(4-O-β)G 3 | 9,059 ± 3,083 | 0 ± 0 | inf |
| 59 | 721.2096 | 17.07 | Tricin(4′-O-β)G(4-O-β)G 4 | 10,052 ± 3,231 | 5 ± 8 | 2,131 |
| 60 | 721.2096 | 17.34 | Tricin(4′-O-β)G(4-O-β)G 5 | 22,808 ± 7,483 | 1 ± 3 | 20,696 |
Table V. List of compounds that are increased in abundance in C2-Idf leaves.
For each compound, its unique number (No.), m/z, retention time (RT), peak area (as mean ± sd), and ratio of the peak area in C2-Idf and C2 are given. *, Compound detected as a trimer. MS/MS-based structural characterization is given in Supplemental File S1.
| No. |
m/z |
RT (min) |
Name |
Peak Area, C2 |
Peak Area, C2-Idf |
Ratio, C2-Idf/C2 |
|---|---|---|---|---|---|---|
| 35 | 1,061.2660 | 2.84 | 3-O-Caffeoyl quinic acid* | 104 ± 256 | 5,436 ± 2,952 | 52 |
| 61 | 429.1067 | 4.84 | Caffeoyl hexose glyceric acid | 2,387 ± 480 | 53,779 ± 17,531 | 23 |
| 62 | 445.1372 | 3.97 | Caffeoyl hexose + 104 D | 1,658 ± 496 | 24,959 ± 4,946 | 15 |
| 63 | 457.1377 | 7.52 | Caffeoyl hexose + 116 D 1 | 956 ± 408 | 20,515 ± 5,574 | 21 |
| 64 | 457.1376 | 8.66 | Caffeoyl hexose + 116 D 2 | 2,168 ± 604 | 21,900 ± 5,091 | 10 |
| 65 | 459.1520 | 4.83 | Caffeoyl hexose + 118 D | 1,514 ± 623 | 33,623 ±4,508 | 22 |
| 66 | 475.1467 | 3.72 | Caffeoyl hexose + 134 D | 3,756 ± 1,396 | 72,128 ± 14,663 | 19 |
| 67 | 485.1679 | 11.05 | Caffeoyl hexose + 144 D | 3,247 ± 1,219 | 46,892 ± 6,757 | 14 |
| 68 | 537.1647 | 7.01 | Caffeoyl hexose + 196 D | 2,036 ± 701 | 35,294 ± 6,598 | 17 |
To investigate whether the decrease in tricin levels would lead to alternative initiation of the lignin polymer by homodimerization of canonical monolignols, a targeted analysis was carried out for oligolignols coupled β-O-4, β-5, or β-β in the phenolic profile of internodes of C2-Idf plants. A total of five trilignols and one tetralignol could be identified based on their retention times, m/z, and MS/MS spectra (Supplemental Table S2). However, no significant differences were observed between their abundances in C2 and C2-Idf samples for both leaves and internodes.
C2-Idf Plants Are Affected in Lignin Content and Composition
Next, we investigated the impact of CHS down-regulation, and thus the lack of tricin, on the content and composition of cell wall components. The cell wall residue (CWR), cellulose and lignin contents, and lignin composition were determined in senesced stems and leaves of greenhouse-grown C2-Idf mutant and C2 control plants. No differences were found in the CWR per unit of dry weight in leaves and stems (Table VI). Measurement of the crystalline cellulose content, expressed as a percentage of the CWR, did not show a significant difference between C2-Idf mutant and C2 control plants in stems, whereas in leaves the crystalline cellulose content was reduced significantly (−9%; Table VI).
Table VI. Cell wall composition of senesced greenhouse-grown C2-Idf mutant and C2 control stem and leaf biomass.
Stem and leaf values are given as means ± sd. Lignin monomer composition was determined by thioacidolysis using gas chromatography-mass spectrometry quantitation (n = 7 biological replicates). The accurate tricin level was determined by thioacidolysis using LC-MS quantitation (n = 6 biological/2 technical replicates). Acid-insoluble (Klason), soluble, and total lignin were determined by the Klason method (n = 7 biological/2 technical replicates). Acid-insoluble and total lignin were corrected for the ash content. Ash content was determined on n = 4 biological replicates for C2 control leaf and C2-Idf leaf and stem and n = 3 for C2 control stem. FA, Ferulic acid; FA (EtSH), FA EtSH addition product. Boldface and underlined values indicate significantly increased and decreased values, respectively, compared with those of the control plants. n.s., Not significant; *, 0.05 ≥ P > 0.01; **, 0.01 ≥ P > 0.001; and ***, P ≤ 0.001.
| Cell Wall Parameter | C2 Stem | C2-Idf Stem | Percentage Difference | Student’s t Test | C2 Leaf | C2-Idf Leaf | Percentage Difference | Student’s t Test |
|---|---|---|---|---|---|---|---|---|
| CWR/dry weight (weight %) | 60.72 ± 6.26 | 55.76 ± 4.69 | −8.2 | n.s. | 65.38 ± 2.79 | 64.08 ± 2.5 | −2.0 | n.s. |
| Cellulose/CWR (weight %) | 33.39 ± 2.13 | 33.66 ± 2.04 | −0.8 | n.s. | 22.62 ± 1.89 | 20.65 ± 0.99 | −8.7 | * |
| Ash/CWR (‰) | 0.28 ± 0.25 | 0.88 ± 0.24 | 210 | * | 8.77 ± 0.42 | 7.40 ± 0.78 | −16 | * |
| Acid-insoluble lignin/CWR (%) | 18.99 ± 0.71 | 18.73 ± 0.82 | −1.4 | n.s. | 13.75 ± 1.15 | 18.47 ± 0.74 | 34 | *** |
| Acid-soluble lignin/CWR (%) | 3.31 ± 0.23 | 3.28 ± 0.15 | −0.9 | n.s. | 4.70 ± 1.48 | 5.05 ± 0.34 | 7.4 | n.s. |
| Total lignin/CWR (%) | 22.30 ± 0.82 | 22.01 ± 0.53 | −1.3 | n.s. | 18.45 ± 0.93 | 23.5 ± 0.76 | 27 | *** |
| H/(H + G + S) (mol %) | 1.17 ± 0.12 | 0.89 ± 0.13 | −24 | *** | 2.89 ± 0.24 | 2.66 ± 0.85 | −8.0 | n.s. |
| G/(H + G + S) (mol %) | 23.24 ± 2.63 | 25.63 ± 0.93 | −10 | n.s. | 50.21 ± 1.41 | 49.61 ± 1.33 | −1.2 | n.s. |
| S/(H + G + S) (mol %) | 75.57 ± 2.62 | 73.47 ± 1.02 | −2.8 | n.s. | 46.89 ± 1.58 | 47.72 ± 1.49 | 1.8 | n.s. |
| H/total lignin (μmol mg−1) | 0.025 ± 0.007 | 0.022 ± 0.003 | −12 | * | 0.024 ± 0.004 | 0.030 ± 0.012 | 25 | n.s. |
| G/total lignin (μmol mg−1) | 0.50 ± 0.16 | 0.64 ± 0.05 | 28 | n.s. | 0.41 ± 0.06 | 0.55 ± 0.08 | 34 | ** |
| S/total lignin (μmol mg−1) | 1.63 ± 0.44 | 1.84 ± 0.12 | 13 | n.s. | 0.38 ± 0.06 | 0.53 ± 0.09 | 39 | ** |
| (H + G + S)/total lignin (μmol mg−1) | 2.16 ± 0.59 | 2.54 ± 0.17 | 18 | n.s. | 0.81 ± 0.12 | 1.11 ± 0.17 | 37 | ** |
| S/G | 3.30 ± 0.54 | 2.87 ± 0.14 | −13 | n.s. | 0.94 ± 0.04 | 0.96 ± 0.05 | 2.0 | n.s. |
| FA/total lignin (µmol mg−1) | 0.010 ± 0.001 | 0.011 ± 0.001 | 10 | n.s. | 0.008 ± 0.002 | 0.008 ± 0.001 | 4.8 | n.s. |
| FA (EtSH)/total lignin (µmol mg−1) | 0.014 ± 0.004 | 0.017 ± 0.004 | 21 | n.s. | 0.012 ± 0.002 | 0.011 ± 0.002 | −8.3 | n.s. |
| Tricin/(H + G + S) (mol%) | 1.05 ± 0.29 | 0 ± 0 | −100 | *** | 6.78 ± 1.42 | 0.14 ± 0.04 | −98 | *** |
| Tricin/total lignin (mg g−1) | 7.44 ± 0.85 | 0 ± 0 | −100 | *** | 18.26 ± 2.66 | 0.51 ± 0.13 | −98 | *** |
The ash content was significantly higher in C2-Idf mutant stems (+213%) compared with that of the C2 control (Table VI). In contrast, the ash content was significantly lower in C2-Idf mutant leaves (−16%; Table VI). However, in both stems and leaves, the ash content remained relatively low (i.e. less than 0.1‰ and less than 1‰ of the CWR, respectively). The lignin amount in C2-Idf and C2 control plants was determined using the Klason method. The acid-soluble lignin of stems and leaves was not affected in the C2-Idf mutant, and also the acid-insoluble (Klason) and total lignin of stems did not differ significantly (Table VI). However, the acid-insoluble lignin and total lignin content of leaves of C2-Idf plants were significantly higher by +34% and +27% compared with the values in the C2 control, respectively (Table VI).
Next, analytical thioacidolysis was used to determine possible shifts in lignin composition of stems and leaves from the C2-Idf mutant. Thioacidolysis followed by gas chromatography-mass spectrometry provides an estimate of the amount of monolignols linked only by β-O-4 bonds. Stem lignin was richer in thioacidolysis-released S units than in G units (S/G = 3.3), whereas in leaves, thioacidolysis-released S and G units were in the same range (S/G = 0.94). In stems, the relative abundances of thioacidolysis-released G and S units did not differ between C2-Idf plants and the C2 controls. In contrast, the relative abundance of thioacidolysis-released H units was 24% lower in C2-Idf stems compared with the C2 controls, although the overall frequency of H units was very low (Table VI). When expressed relative to the Klason lignin content, the amount of thioacidolysis-released H units was lower by 11%. In leaves, no significant differences in the relative abundances of H, G, and S units were observed between C2-Idf and C2 control plants (Table VI). However, the amount of thioacidolysis-released G units, S units, and total H + G + S units per Klason lignin were significantly higher (+34%, +38%, and +36%, respectively) in leaves of C2-Idf plants. No difference in ferulate content was detected (Ralph et al., 2008b; Table VI).
Tricin Is Barely Detectable in C2-Idf Mutants
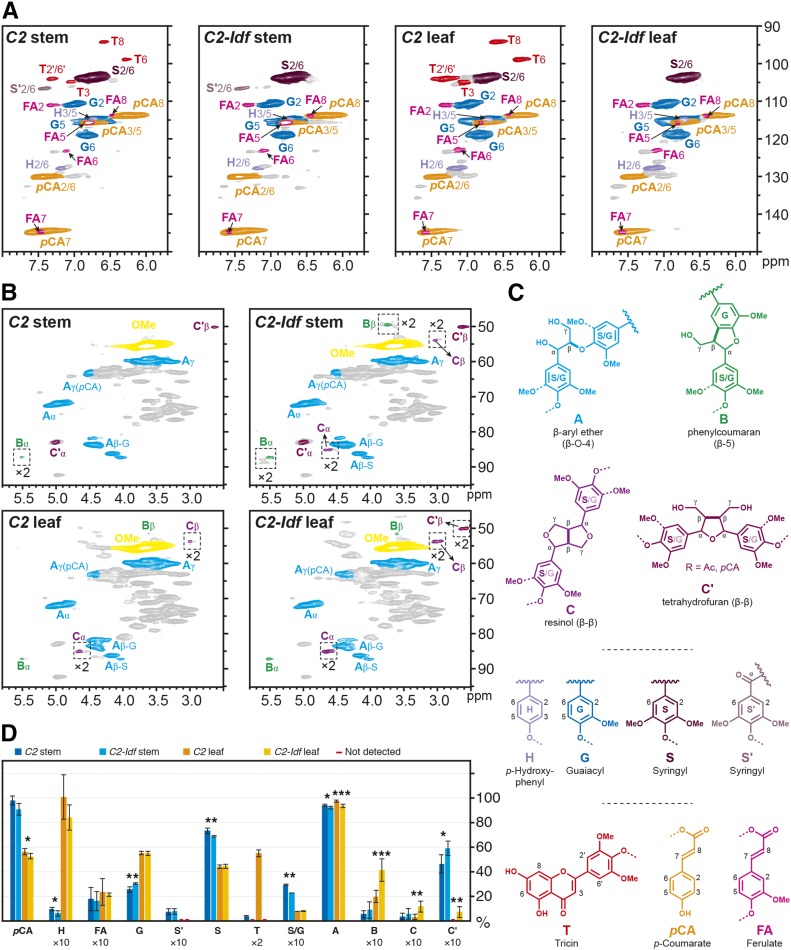
In order to evaluate how the down-regulation of C2 alters lignin structure, 2D-NMR was performed on enzyme lignins from stems and leaves of C2-Idf and C2 control plants. Differences in lignin monomer composition can be visualized from the aromatic regions, whereas the distributions of dimeric units represented by their interunit linkages are deduced from the oxygenated aliphatic regions of the two-dimensional 1H-13C correlation heteronuclear single-quantum coherence (HSQC) spectra (Fig. 3; Supplemental Table S3). The colored contours in Figure 3 reflect the different units in the lignin polymer, with the red contours highlighting the tricin units. The spectra from control samples display the typical signals from the commonly observed H, G, and S units, derived from the polymerization of the canonical monolignols (including their acylated analogs). The signals for H units were observed at low levels compared with those corresponding to G and S units. In addition, four distinctive signals corresponding to tricin appeared in the aromatic region of the HSQC spectra. Interestingly, when the lignin monomeric composition was compared in stems and leaves of the control line, large differences could be observed. Whereas the stem lignin appeared to be rich in S units, with minor amounts of tricin (with an integral of 1.9% based on the total of the S and G components; i.e. S + S′ + G integrals), leaf lignin had a lower S/G ratio and higher levels of tricin (27.6%; Fig. 3; Supplemental Table S3). Note that end groups (like tricin) are significantly and nonlinearly overestimated in such spectra but provide a qualitative view of the altered levels (Mansfield et al., 2012). Leaf lignin also was richer in H units (10.1%) than stem lignin (1%). It should be noted that attributing the entire integrals from contours in the H region has always been in doubt, as other components appear to contaminate this H peak; it is for this reason that we do not sum S + G + H, as the value becomes too unreliable: all comparisons are made based on the total of the S and G components being set at 100%. Most importantly for this study, upon the strong CHS down-regulation found in C2-Idf plants, tricin units were undetectable from both stem and leaf lignins (Fig. 3; Supplemental Table S3) in HSQC spectra. An accurate quantitative analytical method, thioacidolysis of cell wall material followed by liquid chromatography-mass spectrometry (LC-MS) analysis (Lan et al., 2016b), was applied to determine the absolute value of tricin. For the C2 control samples, the content of tricin was 7.44 mg g−1 total lignin for the stem sample and 18.26 mg g−1 for the leaf (Table VI). For the C2-Idf samples, no tricin could be detected in the stem, whereas 0.51 mg g−1 tricin was found in the leaf samples (Table VI). It is also worth pointing out that the percentage of tetrahydrofuran C′ units (β-β interunit linkage) increased from 4.7% to 6% in the stems of C2-Idf maize and, more strikingly, that in leaves, both β-β interunit linkage types C and C′ increased (from 0.3% to 1.2% and from 0% to 0.7%, respectively; Supplemental Table S3).
Figure 3.
HSQC spectra of the enzymatic lignins from stem and leaf of C2-Idf mutant and C2 control plants. A and B, Spectra of the aromatic regions and the side chain regions, respectively. C, The colors of the substructures shown match those of the corresponding signals in the HSQC spectra. The signal intensities in the framed areas indicated by ×2 are at twice the intensity for convenience. D, The bar graph shows the quantification of the lignin units with their characteristic interunit linkages, with some of them, for convenience, being shown as 2- or 10-fold of their original value, as indicated by ×2 and ×10. For statistics, the leaf and stem material of C2-Idf was compared with leaf and stem of the C2 control, respectively. *, 0.05 < P ≤ 0.01; **, 0.01 < P ≤ 0.001; and ***, P < 0.001. For values corresponding to the data points in this graph, see Supplemental Table S3.
Lignin Mr Is Mainly Not Changed in C2-Idf Plants
Gel permeation chromatography (GPC) analysis was performed to determine the Mr of the acetylated lignin isolated from C2-Idf and C2 control plants. The weight-average (Mw) and number-average (Mn) Mr were estimated from the GPC curves (relative values related to polystyrene standards), and the polydispersity index was calculated. The GPC scans of all samples exhibited three peaks, the first of which was out of the range of the polystyrene standard curve and represented an abnormally large Mr for lignin samples. Therefore, we regarded it as an unknown contaminant (Supplemental Fig. S1). The second peak represented the major lignin polymer, and the third originated from lignin oligolignols containing trilignols, tetralignols, and pentalignols with Mr values at around 800 to 1,000. The Mr values of the main peak of the stem lignin samples (second peak) were 8,300 (Mn) and 24,200 (Mw) for the C2-Idf, which was not significantly different from the 8,500 (Mn) and 25,000 (Mw) measured for the C2 control (Supplemental Fig. S1). The lignin polymer from C2-Idf leaves showed 8,400 (Mn), which was not significantly different from the Mr of the lignin from C2 control leaves with 8,000 (Mn), and 22,900 (Mw), which appeared to be moderately (8%) higher than that from the C2 control leaves at 21,100 (Mw; P = 0.03; Supplemental Fig. S1).
C2 Down-Regulation Impacts the Saccharification Efficiency of Leaves But Not Stems
Because tricin units are incorporated into native maize lignin, we further evaluated whether the absence of this monomer, as well as its consequences on lignin structure, affect the saccharification efficiency in stems and leaves of C2-Idf plants. Therefore, senesced stems and leaves of C2-Idf and C2 control plants were saccharified for 48 h, using pretreatments with either acid (1 m HCl, 80°C, 2 h) or alkali (62 mm NaOH, 90°C, 3 h), or without pretreatment, after which enzymatic glucose (Glc) release was measured. Subsequently, the cellulose-to-Glc conversion was calculated based on the measured cellulose content and the obtained saccharification yield.
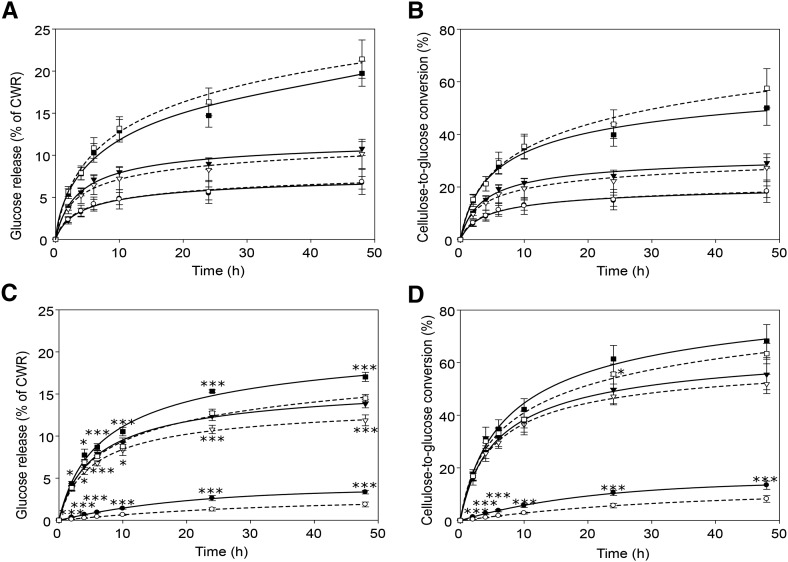
When the stem material was analyzed, no significant differences were found in the saccharification yields expressed on CWR between the C2-Idf plants and the C2 control (Fig. 4, A and B; Supplemental Table S4). Consistent with the fact that the cellulose/CWR did not differ either (Table VI), the cellulose-to-Glc conversion also was found to not differ significantly (Fig. 4B). In contrast, the saccharification yield was reduced significantly in leaves of C2-Idf plants compared with that of the C2 control under all conditions tested (Fig. 4C; Supplemental Table S4). The differences in Glc release were already observed after 2 h of hydrolysis without pretreatment or following acidic pretreatment (Fig. 4C). Following alkaline pretreatment, significant differences were observed from 4 h of hydrolysis onward (Fig. 4C). The cellulose-to-Glc conversion was calculated for all time points in the hydrolysis. At 48 h of saccharification without pretreatment, the cellulose-to-Glc conversion was reduced significantly, by 39%, in leaves of C2-Idf plants compared with the C2 control, whereas no difference was observed with acidic or alkaline pretreatment at this time point (Fig. 4D; Supplemental Table S4).
Figure 4.
Saccharification efficiency of senesced stem (A and B) and leaf (C and D) biomass from C2 control (solid lines) and C2-Idf mutant (dashed lines) plants. The biomass was not pretreated (circles) or was pretreated with either 1 m HCl (triangles) or 62 mm NaOH (squares). The saccharification efficiency is expressed as Glc yield per CWR (A and C) and cellulose-to-Glc conversion (B and D). The numbers corresponding to the data points in these graphs are listed in Supplemental Table S4. Error bars represent sd values of seven biological repeats. *, 0.05 > P ≥ 0.01; and ***, P < 0.001.
DISCUSSION
The flavone tricin was recently reported to be part of the lignin polymer in the cell walls of grasses, and it was suggested to act as an initiation site for lignification (del Río et al., 2012; Lan et al., 2015, 2016a), making it a potential target for bioenergy crop improvement. The biosynthesis of tricin has been described to be accomplished via CHS and CHI activity (Shih et al., 2008; Dixon and Pasinetti, 2010). Based on expression analysis and mutant studies of the two CHS homologs in maize (i.e. C2 and Whp1; Coe et al., 1981; Franken et al., 1991), we deduced that C2 is the most likely gene encoding the flux-determining CHS isozyme in lignifying tissues. To investigate whether altering tricin levels in lignin affects lignin structure, we have studied C2-Idf maize plants that have a natural mutation leading to highly reduced CHS mRNA levels in stems and leaves.
The RT-qPCR-based expression study showed very low residual expression (5%) of the C2 gene in internodes of C2-Idf, corroborating previously reported northern- and western-blot analyses in which no residual C2 expression or C2 protein could be detected in the mutant’s aleurone tissue (Franken et al., 1991). Via phenolic profiling of both internodes and leaves of C2-Idf mutant and C2 control plants, we found that disruption of CHS leads to a severe reduction in the levels of flavones, flavonols, flavonolignols, and hexosylated flavonolignols. The low residual amount of tricin, and flavonoids in general, detected by phenolic profiling suggests that some residual protein is made from the remaining 5% C2 transcript. The majority of the flavones with a reduced abundance in C2-Idf stems and leaves contained an apigenin or tricin substructure. Additionally, we found luteolin and chrysoeriol derivatives as differential flavones. Our data thus support the recent finding that chrysoeriol, instead of tricetin, is the intermediate in tricin biosynthesis in grasses (Lam et al., 2015).
Compounds that were increased significantly in abundance in C2-Idf plants were mainly caffeate and ferulate derived, esterified by quinic acid, shikimic acid, or hexoses. The accumulation of these compounds implicates detoxification routes for phenylpropanoid pathway intermediates that accumulate upon the perturbation of CHS. Remarkably, whereas the identified highly accumulating compounds in C2-Idf leaves were all derived from caffeic acid, mostly hexose conjugates that are further derivatized with small Mr adducts, no caffeoyl hexosides appeared on the list of 15 top-accumulating compounds in internodes. Instead, the most accumulating phenylpropanoids in internodes were mainly quinic and shikimic derivatives of caffeic and ferulic acid. Apparently, the carbon fluxes upon the down-regulation of CHS are different in leaves and internodes.
In addition, the phenolic profiling revealed that, in C2 control stems and leaves, tricin was coupled 4′-O-β to p-coumaryl, coniferyl, and sinapyl alcohols in dimeric and trimeric structures. We recently reported all these flavonolignols (which were previously termed tricin oligolignols) to be present in maize internodes (Lan et al., 2016a). Also, when specifically searching for oligolignols without tricin, we found trilignols and tetralignols coupled via all three types of bonds (β-O-4, β-β, and β-5), as expected for combinatorial coupling (Morreel et al., 2010a; Ralph, 2010). This shows that not all monolignols couple in muro to (di)ferulate or tricin nucleation sites but that they also couple mutually to initiate lignin chains. The presence of hexosylated flavonolignols suggests that the aglycone dimers are made in the cytoplasm prior to being glycosylated. Indeed, it was shown in Arabidopsis (Arabidopsis thaliana) leaf protoplasts that monolignols are not only coupled in the cell wall, as commonly accepted, but also inside the cell, where the monolignol radicals couple combinatorially to produce a plethora of dimers and small lignin oligomers that are subsequently glycosylated and stored in the vacuole (Dima et al., 2015). Our data indicate that tricin also couples through radical coupling in the cytoplasm, as glycosylated derivatives were detected for various tricin-monolignol coupling products.
NMR data (Fig. 3; Supplemental Table S3) showed that phenylcoumaran (β-5; B), resinol (β-β; C), and tetrahydrofuran (also β-β, but from acylated monolignols; C′) units were elevated in leaves from the C2-Idf mutant. However, in stems, the increase was only significant for the tetrahydrofuran units C′. The increased occurrence of β-β units is likely the direct consequence of the lack of tricin in the lignin of these mutants. Tricin can only start a lignin chain and can only do so via 4′-O-β coupling with a monolignol or acylated monolignol (Lan et al., 2015, 2016a). In the absence of tricin, the canonical monomers (or their acylated counterparts) will initiate the polymerization more frequently by dimer formation, via β-β, β-5, and β-O-4 coupling, if at least one of the monomers is coniferyl alcohol (or its conjugates), or only via β-β coupling, if the monomer is sinapyl alcohol (or its conjugates). Whereas β-O-4 and β-5 units A and B also are made during lignin chain elongation, the β-β units C and C′ can only be part of a starting structure and, in dicots, overwhelmingly derive from sinapyl alcohol and, in grasses, from either sinapyl alcohol or its p-coumarate conjugate (Lan et al., 2015, 2016a). Therefore, the increased frequencies of β-β units C and C′, along with the reduced signals from β-O-4 units A, are signs of increased monolignol dimerization in the absence of tricin. This finding is in agreement with the fact that the level of β-β units had an opposite trend to the level of tricin units in a series of wheat straw lignin fractions that differed in Mr (Lan et al., 2016b). On the other hand, we cannot exclude the possibility that the shifts in coupling frequencies are a consequence of the increased flux toward monolignols or acylated monolignols in C2-Idf mutants, as determined by the increased Klason lignin content in these plants, rather than the absence of tricin per se.
Based on our NMR data of the C2 control, the lignin from maize leaves contained a higher fraction of tricin units than the lignin of maize stem (with relative integrals of 28% versus 2% indicating a major difference). However, as noted above, end units are nonlinearly overestimated in HSQC spectra, so analytical thioacidolysis was used to provide accurate quantitation of the level of incorporation (Lan et al., 2016b). Also upon accurate quantitation, the tricin content was higher in leaves (about 18 mg g−1) than in stems (about 7 mg g−1). The higher levels of tricin in leaves may be attributable to the biological function of tricin in protecting plants against insect attack (Bing et al., 2007) and protecting DNA against radiation damage (Stapleton and Walbot, 1994; van de Staaij et al., 2002). In line with the higher tricin content in leaves, the consequences of the C2-Idf mutation were more pronounced in leaves than in stems; the relative fraction of condensed β-5, β-β, and β-β′ units was more elevated in C2-Idf leaves.
Total lignin content was 27.5% higher in C2-Idf leaves compared with that in C2 leaves. It is likely that the increased lignin content is simply the result of the redirection of the carbon flux toward monolignols upon the blockage of flavonoid biosynthesis. Such a redirection of the flux would have a particularly strong effect in the C2 background that has a high flux toward flavonoids. Recently, it was reported that stems of C3H1-RNAi (for RNA interference) maize plants (which produce less lignin) have increased levels of tricin, consistent with a redirection of the carbon flux to the synthesis of flavonoids instead of the canonical monolignols (Fornalé et al., 2015). In the leaves of maize C2-Idf mutants, the reverse takes place: a substantial redirection of the carbon flux from the biosynthesis of flavonoids to the biosynthesis of the canonical monolignols. Although this scenario seems obvious, it is not generally observed. For instance, the redirection of the flux was observed in leaves but not in stems, possibly because lignifying cells of leaves contain more flavonoids (as leaves are more exposed to environmental stresses) than lignifying cells in the stem, which are largely protected from environmental stresses by the leaf sheaths. Alternatively, the flux was (partially) redirected to lignin in leaves but to other metabolic sinks in stems (e.g. to caffeoyl and feruloyl shikimates), as revealed via phenolic profiling. Furthermore, the Arabidopsis chs loss-of-function mutant showed no alterations in lignin content and composition (Li et al., 2010; Vanholme et al., 2010b), and CHS suppression in flax (Linum usitatissimum) even led to lignin reduction (Zuk et al., 2016). In addition, although an increase in lignin content was observed in fruits of strawberry (Fragaria × ananassa) silenced for CHS via agroinfiltration (Ring et al., 2013), this increase was later assigned to the agroinfiltration method and not to the silencing of CHS, because injection of Agrobacterium tumefaciens carrying the control vector already increased lignin content (Yeh et al., 2014). Taken together, these combined observations suggest different metabolic plasticity in different plant species and even in different organs upon modified CHS expression.
We also observed a significant decrease in cellulose content as a percentage of CWR in leaves of C2-Idf plants that might logically be a direct consequence of the relative increase in lignin content. A series of studies using chemical inhibitors, mutants, or transgenic plants have suggested that cellulose and noncellulosic components of secondary cell walls might be regulated in a compensatory fashion, but the underlying mechanisms remain to be understood. Transgenic poplars (Populus spp.) deficient in different steps of the lignin biosynthetic pathway have been reported to have less lignin and relatively more cellulose in their cell walls (Jouanin et al., 2000; Li et al., 2003a; Leplé et al., 2007). However, such an inverse relation between lignin and cellulose was not observed in a set of Arabidopsis mutants altered in lignin biosynthesis, in which a reduction in lignin content seemed to be compensated for by a relative increase in hemicellulose content (Vanholme et al., 2012; Van Acker et al., 2013). In grasses, whose cell wall composition and structure differ significantly from those of dicot plants, the same relation (i.e. more hemicellulose in low-lignin plants) has been reported. Stems of low-lignin transgenic switchgrass (Panicum virgatum) down-regulated for COMT had increased amounts of xylan, the predominant hemicellulose component in switchgrass, both when plants were cultivated in the greenhouse (Fu et al., 2011) and in the field (Baxter et al., 2014). In brachypodium (Brachypodium distachyon), the reduced lignin content of a mutant for the lignin-specific laccase BdLAC5 was compensated for by a slight increase in the content of hemicellulose, whereas the amount of cellulose remained unchanged (Wang et al., 2015b). The brown midrib mutants isolated in maize show a characteristic brown coloration of the leaf midrib that is associated with reduced lignin levels and altered lignin composition (Sattler et al., 2010). Compositional analyses of different maize brown midrib mutants showed no differences in cellulose content but increased levels of hemicellulose in both stalk and stover (Lechtenberg et al., 1972; El-Tekriti et al., 1976; Cone and Engels, 1993). Also, the inverse relation between lignin and cellulose amount has been described in grasses. Down-regulation of CCoAOMT in maize led to reduced lignin content and increased cellulose levels, with no significant differences in hemicellulose levels (Li et al., 2013). The brittle culm (bc) mutants in rice show reduced tissue mechanical strength due to significant reductions in cellulose content, which is often accompanied by increased levels of lignin, as in the case of bc1, bc10, and bc12, each of which is affected in a different aspect of cellulose deposition (Li et al., 2003b; Zhou et al., 2009; Zhang et al., 2010). Similarly, the reduced cellulose content in mature stems of the spontaneous maize mutant brittle stalk2 (bk2), disrupted in a gene orthologous to the BC1 gene in rice, goes along with a marked enrichment in lignin deposition, resulting in brittleness of all aerial plant organs (Sindhu et al., 2007). In agreement with its corresponding mutant in rice, bk2 and wild-type plants were indistinguishable in all aspects of plant growth and development (Ching et al., 2006; Sindhu et al., 2007). In brachypodium, the dwarf spaghetti1 (spa1) mutant presents a unique phenotype combining brittleness with increased elasticity of the internodes (Timpano et al., 2015). The cell walls of spa1, whose causative mutation remains elusive, contain less crystalline cellulose and higher levels of lignin and hemicellulose. However, these compensatory effects are not general. For example, the T-DNA insertion mutant flexible culm1 in rice, which is deficient in a CAD, shows reduced secondary cell wall thickness and decreased levels of both lignin and cellulose (Li et al., 2009).
Additionally, our results showed that C2-Idf plants have an increase in vegetative dry weight, which is a desirable trait for lignocellulosic crops (Feltus and Vandenbrink, 2012). The reason for this increase is not clear. In previous work, the dry weight and stem height measurements of an Arabidopsis chs mutant did not differ from those of control plants (Li et al., 2010; Vanholme et al., 2010b). In a different study using the same chs mutant, reduced growth was observed versus control plants when grown under adverse environmental growth conditions, such as high light intensity and nitrogen starvation (Misyura et al., 2013), indicating that the growth phenotype is dependent on the environment. C2-Idf maize plants might allocate more carbon to biomass production by saving on plant defense compounds (among which flavonoids are made via CHS). Alternatively, some of the differentially abundant compounds might have growth-stimulating properties. However, although the donor line with the C2-Idf mutation was backcrossed seven times to the W22 background, we cannot exclude the possibility that residual linkage drag is at the basis of the increased dry weight.
We have shown that stems of C2-Idf and C2 control plants have a similar saccharification efficiency, whereas the leaves of C2-Idf plants show a significant decrease in Glc release with and without pretreatment compared with leaves of C2 control plants. However, the reduction in Glc release in C2-Idf leaves was less apparent when the saccharification efficiency was expressed as cellulose-to-Glc conversion. This is because C2-Idf leaves also had a lower cellulose content per CWR. Therefore, at least part of the lower Glc release is due to the reduced cellulose content in the leaves of C2-Idf plants. It has been established that saccharification efficiency is negatively correlated with lignin amount in both monocot and dicot species (Chen and Dixon, 2007; Sattler et al., 2010; Xu et al., 2011; Bouvier d’Yvoire et al., 2013; Jung et al., 2013; Van Acker et al., 2013; Vanholme et al., 2013a) and that a lower degree of polymerization of lignin eases its extraction from the biomass (Eudes et al., 2012; Vanholme et al., 2012; Shen et al., 2013). The alkaline pretreatment hydrolyzes ester bonds, thereby releasing (di)ferulates that acylate the hemicellulose (and of which a part is also coupled to lignin) and p-coumarates that additionally acylate the lignin backbone (Ralph, 2010). Consequently, the alkaline pretreatment results in partial extraction of the lignin from the biomass. The acid pretreatment used, on the other hand, hydrolyzes hemicellulosic glycosidic bonds (Gómez et al., 2014). As (di)ferulates link hemicelluloses with lignin, this acid pretreatment also will solubilize part of lignin. Thus, both alkaline and acid pretreatments temper the influence of the higher lignin content on the saccharification efficiency. Because the cellulose-to-Glc conversion was not different between C2-Idf and C2 control leaf samples upon acidic or alkaline pretreatment, whereas it was different when no pretreatment was applied, we contend that the 27% increase in total lignin content in leaves of C2-Idf plants is the main cause of the reduced cellulose-to-Glc conversion when no pretreatment was used prior to saccharification (Fig. 4D). The saccharification data on stems did not show any significant difference in Glc release, regardless of the pretreatment, in agreement with the observation that lignin content was increased only in leaves. In addition, these results indicate that the shifts in lignin composition in C2-Idf stems were too subtle to influence the saccharification efficiency.
Recent findings highlight the metabolic malleability of plant lignification, indicating that lignin can be engineered to dramatically reduce its adverse impact for industrial purposes (Vanholme et al., 2012; Mottiar et al., 2016). For example, the incorporation of coniferyl ferulate into poplar and gramineous feedstock shows (slightly) reduced lignification and allows more efficient delignification and enzymatic hydrolysis of cell walls. The cell walls lignified with coniferyl ferulate were more readily hydrolyzed with fibrolytic enzymes both with and without alkaline pretreatment (Grabber et al., 2008; Wilkerson et al., 2014). As a second example, incorporation of acylated monolignols (mainly coniferyl and sinapyl p-coumarate) into the lignin of Arabidopsis resulted in an increased solubilization of these lignins in alkaline medium (Sibout et al., 2016). Our data point to an overall positive relation between the abundance of tricin and saccharification efficiency in leaves. However, this relation is an indirect consequence of the lower lignin amount in C2 relative to C2-Idf. To test whether tricin and the resulting reduced frequency of β-β, β-β′, and β-5 interunit linkages and the increased frequency of β-O-4 interunit linkages have a direct effect on cell wall properties, plants with higher amounts of tricin in their lignin than C2 plants are needed. The incorporation of tricin into dicot lignins that normally do not produce tricin, or its overproduction in grasses through genetic engineering, may be a promising strategy to test the direct influence of tricin levels in the lignin on lignocellulose recalcitrance. The production of tricin in species that normally do not produce this metabolite has already been achieved in the dicot Arabidopsis (Lam et al., 2015), but its incorporation into lignins and the effect on saccharification have not yet been investigated.
Given the growing consensus that lignin valorization is essential for the environmental sustainability and economics of the lignocellulosic biorefinery (Ragauskas et al., 2014; Rinaldi et al., 2016), high-lignin biomass crops may prove particularly useful if significant value can be extracted from the lignin component. Because flavonoids protect plants from biotic and abiotic stresses (Stapleton and Walbot, 1994; Falcone Ferreyra et al., 2012), it is unlikely that high-lignin and low-flavonoid C2-Idf or chs knockout mutants would flourish under field conditions. However, RNA interference-based genetic engineering approaches could be used to mimic C2-Idf or chs knockout in lignifying cells only (Smith et al., 2013). Leaving the carbon flux toward flavonoids in nonlignifying cell types unaltered, such plants should not necessarily be more sensitive to stresses than their wild-type counterparts. CHS is likely the most elegant step to engineer low-flavonoid and high-lignin characteristics, because CHS is the branch point toward flavonoids; the substrate that accumulates upon CHS deprivation (p-coumaroyl-CoA) can readily be used by HCT in the general phenylpropanoid- and monolignol-specific pathways to produce lignin. The strategy to down-regulate CHS only in cells that produce monolignols is challenging, because the exact site of monolignol production is not yet known. Depending on whether lignifying cells produce the majority of the monolignols themselves or whether monolignols are produced by neighboring parenchyma cells and then transported toward the cell wall of lignifying cells (Pesquet et al., 2013; Smith et al., 2013), different cell-specific promoters are needed for the silencing approach. In the first case, secondary cell wall-specific promoters (such as promoters of secondary cell wall cellulose biosynthesis or lignin-specific genes; Petrik et al., 2016) could be used to drive the expression of silencing constructs. The thermal conversion of lignocellulosics to renewable gas and bio-oils has been heavily researched, and the benefits for industrial implementation have become clear (Yaman, 2004; Saidi et al., 2014). Feedstocks with high lignin levels might be a promising source of aromatic/phenolic compounds to produce bio-based products from lignin, as proposed in a lignin-first biorefinery approach (Van den Bosch et al., 2015; Rinaldi et al., 2016) and as demonstrated recently with high-level monomer production via hydrogenolysis (Shuai et al., 2016).
MATERIALS AND METHODS
Plant Material
All analyses were carried out with homozygous C2-Idf mutant and C2 control maize (Zea mays) plants. Seed stocks of these lines were obtained from the James Birchler laboratory (University of Missouri). The C2-Idf allele was in a B; Pl; R-scm2 W22 background, where B stands for the B-I allele conditioning pigmentation (Goff et al., 1990), Pl stands for the Pl-Rhoades (Pl-Rh) allele (the Pl-Rh allele is highly expressed and produces strong pigmentation in most tissues of the maize plant; Hollick et al., 1995), and R-scm2 stands for an R allele that conditions anthocyanin pigmentation in the embryo and endosperm of the kernel (Styles et al., 1973). The C2; B; Pl; R-scm2 W22 (C2 control) was generated by crossing a full-color C2; b; pl; R-scm2 W22 inbred stock with a C2; B; Pl; r-g W22 inbred stock. The two W22 stocks originally trace to materials from the University of Wisconsin. From the cross of these two W22 lines, selection for pigment in the kernel as well as strong pigment in the plant was conducted at each generation of selfing until a true breeding stock with pigment in kernels and plants was selected. This stock carries a normal allele of the C2 gene. C2-Idf; B; Pl ;R-scm2 W22 (C2-Idf) was generated as follows: the C2-Idf allele (Maize Genetics Cooperation Stock Center ear 95-1318-3⊗) was backcrossed to the b; pl; R-scm2 W22 stock for seven generations to introgress it into W22 and then selfed to produce a true breeding line C2-Idf; b; pl; R-scm2 W22. Next, this C2-Idf; b; pl; R-scm2 W22 stock was crossed with the B; Pl; r-g W22 stock mentioned above and then self-pollinated over several generations with selection for a true breeding line that was C2-Idf; B; Pl; R-scm2 W22. The C2-Idf mutant and C2 maize plants were grown in a greenhouse that was kept at 26°C/22°C and 16-h/8-h (day/night) rhythm using a combination of high-pressure sodium vapor lamps (RNP-T/LR/600W/S/230/E40; Radium) and metal halide lamps with quartz burners (HRI-BT/600W/D230/E40; Radium). For expression analysis, six biological replicates of each genotype were grown until the V14 stage. Internode 9 was dissected with removal of the leaf sheath and collected in liquid nitrogen. The samples were then kept at −70°C and subsequently milled with a Grindomix GM200 using precooled stainless-steel jars (Retsch). These samples were pooled two-by-two to obtain three biological replicates. For metabolomics analysis, seven biological replicates of each genotype were grown until the V14 stage, and internode and leaf 9 were harvested and kept at −70°C and subsequently milled as above. Another set of seven biological replicates of each genotype was grown to maturity for growth phenotyping and all cell wall analysis (lignin, cellulose, NMR, GPC, and saccharification). The classification of the growth stages was according to Ritchie et al. (1996), Nafziger (2009), and O’Keeffe (2009). The leaves and internodes were counted from the bottom, as is common for maize. We kept a record of leaf development so as not to miss out on leaves that shriveled up during plant development. No distinction was made between juvenile and adult leaves. Whole leaves were sampled (i.e. including leaf blade, midrib, ligule, and leaf sheath).
Growth Analysis
The final plant height was measured from soil level using a foldable measuring stick. After senescence, stems and leaves from whole plants, excluding the top and bottom 15 cm, were harvested separately for each individual plant and weighed separately. Each sample was milled separately using a cutting mill (Fristch) fitted with a 0.5-mm sieve and used for cell wall characterization and saccharification assays. From the total of seven replicates, six samples were used for NMR analysis (see below).
Gene Expression Analysis
For expression analyses of the CHS (C2) gene, primers at the 3′ end of CHS were designed using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) with standard settings. The primers were 5′-CAGAAGGCGATCAAGGAGTG-3′ and 5′-GGTACATCATGAGGCGGTTC-3′. The RNA was extracted using an RNeasy kit (Qiagen), and DNase treatment was performed using DNA-free (Ambion, Life Technologies). Extracted RNA was quantified using a Nanodrop ND-1000 spectrophotometer (Thermo Scientific), and 400 ng of RNA was used for cDNA synthesis using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). A 10× diluted cDNA sample was used for RT-qPCR using a sensiFAST SYBR No-ROX kit (Bioline) on a LightCycler 480 (Roche). Samples were run in technical triplicates on the LC480 with the following protocol: one activation cycle of 10 min at 95°C; 45 amplification cycles of 10 s at 95°C, 10 s at 60°C, and 10 s at 72°C; and one melting curve cycle measuring from 65°C to 95°C. Fluorescence values were exported from the LightCycler program, whereupon Ct values, normalization factors, and primer efficiencies were calculated according to Ramakers et al. (2003) using ZmEF1a and Zm18S as reference genes. Primers were 5′-AGTCCGTTGAGATGCACCATG-3′ and 5′-CACATACCCACGCTTCAGATCC-3′ for ZmEF1a and 5′-ACCTTACCAGCCCTTGACATATG-3′ and 5′-GACTTGACCAAACATCTCACGAC-3′ for Zm18S, as described by Voorend et al. (2016).
Metabolic Profiling
Ground material (250 mg) was extracted after shaking with 7.5 mL of methanol (HPLC grade) at 70°C for 10 min. After centrifugation, methanol was evaporated and samples were suspended in 300 µL of water:cyclohexane (2:1) for extraction. A 15-µL sample of the aqueous phase was subjected to UHPLC-MS on a Waters Acquity UHPLC system (Waters) connected to a Synapt HDMS Q-TOF mass spectrometer (Micromass), basically as described previously (Vanholme et al., 2013b). In brief, chromatographic separation was performed on a Waters Acquity BEH C18 column (2.1 mm × 150 mm, 1.7 µm) with a gradient elution, with the mobile phase composed of water containing 1% acetonitrile and 0.1% formic acid (A) and acetonitrile containing 1% water and 0.1% formic acid (B; all v/v). During the gradient elution, a flow rate of 350 µL min−1 was applied, with initialization at 0 min, 5% B; 30 min, 50% B; and 33 min, 100% B. Negative mode mass spectrometry setting, chromatogram integration, and alignment via Progenesis QI software (Waters) were as described previously (Vanholme et al., 2013b). The integrated peak area of the m/z values (peaks) was normalized with the function Normalize to External Standard, with dry weight (mg) of the pellet remaining after methanol extraction used as the external standard. To select peaks for which metabolites were significantly increased or decreased in C2-Idf compared with control plants, the following criteria were used: (1) present in all samples; (2) average normalized abundance higher than 5,000 counts; (3) 10-fold increased/decreased peak area in C2-Idf versus control; and (4) Student’s t test P < 0.001. Annotation of compounds matching these criteria was based on accurate m/z (±0.02 D), isotope distribution, and MS/MS similarities. For MS/MS acquisition, a pooled sample of all seven biological replicates per sample type was run on the UHPLC-MS device in both negative and positive ionization mode. The obtained deconvoluted spectra were structurally elucidated based on their similarity with commercially available standards and previously identified metabolites that have already been described in the literature (Supplemental File S1). For targeted analysis of the oligolignols, compounds were identified via m/z (±0.02 D), retention time, and MS/MS similarity.
Statistical Analysis
Statistical analyses consisted of Student’s t tests for comparisons of C2-Idf mutant and C2 control samples for lignin, cellulose, CWR, and saccharification assays. These analyses were carried out in the software package SPSS Statistics 22 (IBM). For metabolic profiling, the normalized (by dry weight) peak area was first transformed (arcsinh) and then subjected to Student’s t test (two-tailed distribution based on F test) performed in Microsoft Excel Professional Plus 2013.
Purified CWR Analysis
Aliquots (10 mg) of milled stem or leaf material were subjected to a sequential extraction to obtain a purified CWR: water (98°C), ethanol (76°C), chloroform (59°C), and acetone (54°C), 30 min each. The remaining CWR was dried under vacuum.
Lignin and Ash Analysis
Klason lignin was determined using a modified Klason lignin analysis method as described previously (Cullis et al., 2004). Briefly, ground samples were solvent extracted (water [3 × 40 mL], 80% ethanol [3 × 40 mL], and acetone [1 × 40 mL]) and freeze dried for 2 d. Once dried, 0.2 g of dried and extractive-free samples was weighed into test tubes and treated with 3 mL of 72% sulfuric acid for 2 h, with stirring every 10 min. The samples were then diluted to 4% sulfuric acid with 112 mL of nanopure water. The solutions were transferred to flasks and autoclaved at 121°C for 60 min. The samples were filtered through medium-coarseness crucibles (Pyrex) and washed with 200 mL of nanopure water. The retentate was dried at 105°C and then weighed to determine acid-insoluble residue content. Some of the filtrate was collected for acid-soluble lignin analysis. Acid-soluble lignin was determined by measuring the absorbance of the filtrate by a UV spectrophotometer at 205 nm and calculated using an extinction coefficient of 110 L g−1 cm−1. Ash content was determined largely as described by the National Renewable Energy Laboratory in its laboratory analytical procedure “Determination of Ash in Biomass” (http://www.nrel.gov/docs/gen/fy08/42622.pdf). In short, acid-insoluble residue was prepared from about 0.3 g of CWR as described above and filtered through a filter paper (Sartorius) using a Buchner filter system (Merck Millipore). Next, the filter paper with the retentate was transferred to a porcelain crucible and placed in a furnace (LT 15/13; Nabertherm) using the following settings: 105°C for 12 min, to 250°C at 10°C min−1, 250°C for 30 min, to 575°C at 20°C min−1, 575°C for 180 min, and finally drop to 105°C. The ash content was determined by weighing the crucible and ash on an analytical balance (XPE105; Mettler-Toledo) and correcting for the empty crucible and the ash content of an empty filter paper. The acid-insoluble lignin (Klason lignin) was then calculating by subtracting the ash content from the acid-insoluble residue. The lignin composition was investigated via thioacidolysis similar to Rolando et al. (1992), but on 10 mg of CWR and a 1-mL reaction mixture (0.025 mL of boron trifluoride etherate + 0.1 mL of ethanethiol + 0.875 mL of dioxane).
Cellulose Analysis
Aliquots (5 mg) of milled stem or leaf material were subjected to a sequential extraction to obtain a purified CWR, as described above. Cellulose was measured on 5 mg of dry stems with sulfuric acid and anthrone according to Foster et al. (2010). The CWR was incubated with 2 m trifluoroacetic acid and 20 μL of inositol (5 mg mL−1) for 2 h at 99°C while shaking (750 rpm). After incubation, the remaining pellet was washed three times with water and twice with acetone and dried under vacuum. H2SO4 (175 µL of 72% [v/v]) was added to the dried pellet. The samples were incubated for 30 min at room temperature, whereupon the samples were vortexed with an additional incubation time of 15 min at room temperature. A total of 825 µL of water was added, and the samples were vortexed again. The samples were centrifuged at 10,000 rpm for 10 min. Ten microliters was diluted with 90 µL of water on a 96-well plate. To each sample, 200 µL of freshly prepared anthrone reagent (2 mg anthrone mL−1 72% H2SO4) was added. After incubation at 80°C for 30 min, the samples were cooled on the bench and the absorption was measured at 625 nm.
Tricin Content Analysis
Tricin was determined by analytical thioacidolysis essentially as published (Lapierre et al., 1986) but using LC-MS detection as described (Lan et al., 2016b).
Preparation of Enzyme Lignin for NMR and GPC
Isolated maize stem and leaf samples (1 g, ground to 60–80 mesh) were successively extracted with water and 80% ethanol (5 × 45 mL for each) under ultrasonication to remove the soluble extractives. The extractive-free materials (approximately 500 mg) were ball milled for 1 h (5-min interval break every 5 min). The ball-milled material was then treated with crude cellulases (Trichoderma viride; Calbiochem) at 35°C (3 × 48 h) to saccharify most of the carbohydrates, producing a lignin-enriched fraction containing essentially all of the original lignin. The enzyme lignins from maize stem and leaf were obtained as brown powders after lyophilization.
GPC Analysis
The obtained enzyme lignin was first acetylated (10 mg) in 0.5 mL of pyridine:acetate anhydride (2:1, v/v; 10 mL) at room temperature for 12 h. Acetylated maize lignin was obtained after coevaporation of the solvent with ethanol under reduced pressure at 45°C until pyridine and acetic anhydride were completely removed. The acetylated product was dissolved in N,N-dimethylformamide (0.1 m lithium bromide) for GPC acquisition. Analytical GPC was performed on a Shimadzu LC20 device with a photodiode array detector (SPD-M20A). Lithium bromide (0.1 m) in N,N-dimethylformamide was used as an eluent at 40°C with a flow rate of 0.3 mL min−1. Polystyrene (low molecular) Standard ReadyCal Set M(p) 250−70,000 (P/N 76552; Fluka) was used to calibrate and calculate the Mr distribution of the lignin samples. Separation was performed using a series of three columns, first a TSKgel guard column with α-stationary phase (6 mm × 4 cm, 13 μm; TOSOH Bioscience), then a TSKgel Alpha-2500 column (7.8 mm × 30 cm, 13 μm; TOSOH Bioscience), followed by a TSKgel Alpha-M column (7.8 mm × 30 cm, 13 μm; TOSOH Bioscience).
NMR Characterization of Lignin Samples
Enzyme lignins (20–30 mg) were dissolved in 1 mL of dimethyl sulfoxide-d6:pyridine-d5 (4:1, v/v) in an NMR tube. NMR spectra were recorded on a Bruker Biospin (Billerica) AVANCE 700-MHz spectrometer fitted with a cryogenically cooled 5-mm 1H-optimized resonance (1H/31P/13C/15N; QCI) gradient probe with inverse geometry (proton coils closest to the sample). Bruker’s Topspin 3.5 (Mac) software was used to process spectra. The central dimethyl sulfoxide-d6 solvent peaks were used as internal references (δC:δH, 39.51:2.49). The HSQC experiment used a Bruker standard pulse sequence, hsqcetgpsisp.2, with the following parameters: acquired from 10 to 0 ppm in F2 (1H) with 200-ms acquisition time, 200 to 0 ppm in F1 (13C) with 8-ms acquisition time of 32 scans; the d24 delay was set to 0.86 ms (1/8J, J = 145 Hz); and the relaxation delay was 1 s. The total acquisition time for a sample was approximately 6 h. Processing used typical matched Gaussian apodization (GB = 0.001, LB = −0.3) in F2 and squared cosine-bell apodization in F1. The quantitation of aromatic and side chain distributions was performed based on the volume integration of contours in HSQC spectra. For aromatics, the percentages of H units, ferulate (FA), p-coumarate (pCA), and tricin (T) were calculated based on the total of G, S, and oxidized syringyl (S′) units (i.e. on the total prototypical G + S aromatic lignin units). For the aliphatic region, the percentages of cinnamaldehyde (X2) and benzaldehyde (X3) end groups were calculated based on the sum of the normal units: β-aryl ether (β-O-4; A), phenylcoumaran (β-5; B), resinol (β-β; C), and tetrahydrofuran (β-β; C′). The correlations from G2, S2/6, S′2/6 (logically halved for S and S′), H2/6 (halved), FA2, pCA2/6 (halved), T6 in the aromatic region, and Aα, Bα, Cβ, C′β, X2γ, and X3α in the aliphatic region were used for integral calculations. Note that these are uncorrected integrals and are only comparatively accurate, not absolute quantifications; end units, in particular (and including tricin), are overquantitated compared with polymer internal units in these spectra (Mansfield et al., 2012).
Saccharification Assay
Aliquots (10 mg) of dry stem or leaf material were used. The biomass was pretreated with 1 mL of either 1 m HCl (80°C, 2 h) or 62 mm NaOH (90°C, 3 h) while shaking (850 rpm). The supernatant was removed, and the pellet containing pretreated material was washed three times with water to obtain a neutral pH. Subsequently, the saccharification assay was performed as described by Van Acker et al. (2013, 2016). The enzyme mix added contained cellulase from Trichoderma reesei ATCC 26921 and cellobiase from Aspergillus niger (Sigma-Aldrich) in a 5:3 ratio. Both enzymes were first desalted over an Econo-Pac 10DG column (Bio-Rad) stacked with Bio-Gel P-6DG gel (Bio-Rad) according to the manufacturer’s guidelines. The desalted cellobiase was 350-fold diluted prior to mixing with desalted cellulase. The enzyme mix was further diluted 10-fold, and the activity of the diluted enzyme mix was measured with a filter paper assay (Xiao et al., 2004). To each sample, the enzyme mix with an activity of 0.014 filter paper units, dissolved in acetic acid buffer (pH 4.8), was added. Aliquots (20 µL) were taken after 0, 2, 6, 10, 24, and 48 h of incubation at 50°C and 10-fold diluted with acetic acid buffer (pH 4.8). The concentration of Glc was measured as described by Van Acker et al. (2013, 2016).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GPC comparison of lignin from C2-Idf mutant and C2 control stem and leaf.
Supplemental Table S1. Growth parameters of C2-Idf mutant and C2 control plants.
Supplemental Table S2. Targeted search for oligolignols isolated from stem and leaf of C2-Idf and C2 control plants.
Supplemental Table S3. Relative quantification of lignin units and linkages based on volume integrals from the HSQC spectra of stem and leaf of C2-Idf mutant and C2 control plants.
Supplemental Table S4. Saccharification efficiency of senesced stem and leaf biomass from C2-Idf mutant and C2 control plants.
Supplemental File S1. MS/MS spectra of the characterized metabolites.
Supplementary Material
Acknowledgments
We thank Dr. James Birchler (University of Missouri) for the seeds of C2 control and C2-Idf mutant maize plants, Arturo González Quiroga (Laboratory for Chemical Technology, Ghent University) for technical assistance in measuring the ash content, and Annick Bleys for help in preparing the article.
Glossary
- RT-qPCR
real-time quantitative PCR
- UHPLC-MS
ultra-high-performance liquid chromatography-mass spectrometry
- MS/MS
tandem mass spectrometry
- CWR
cell wall residue
- H
p-hydroxyphenyl
- G
guaiacyl
- S
syringyl
- HSQC
heteronuclear single-quantum coherence
- LC-MS
liquid chromatography-mass spectrometry
- GPC
gel permeation chromatography
- Mw
weight-average Mr
- Mn
number-average Mr
Footnotes
This work was supported by Petrobras and the Agency for Innovation by Science and Technology (IWT) through the IWT-SBO project BIOLEUM (grant no. 130039) and the IWT-FISCH-SBO project ARBOREF; by the Department of Energy Great Lakes Bioenergy Research Center (Office of Science grant no. DE-FC02-07ER64494 to W.L., R.A.S., and J.R.); by the China Scholarship Council (Ph.D. scholarship at the University of Wisconsin, Madison, to W.L.); by the Research Foundation Flanders (postdoctoral fellowship to R.V.); and by FAPESP (BIOEN Young Investigator Award grant no. 2015/02527–1to I.C.).
Articles can be viewed without a subscription.
References
- Abramson M, Shoseyov O, Shani Z (2010) Plant cell wall reconstruction toward improved lignocellulosic production and processability. Plant Sci 178: 61–72 [Google Scholar]
- Baxter HL, Mazarei M, Labbe N, Kline LM, Cheng Q, Windham MT, Mann DGJ, Fu C, Ziebell A, Sykes RW, et al. (2014) Two-year field analysis of reduced recalcitrance transgenic switchgrass. Plant Biotechnol J 12: 914–924 [DOI] [PubMed] [Google Scholar]
- Begum SA, Sahai M, Ray AB (2010) Non-conventional lignans: coumarinolignans, flavonolignans, and stilbenolignans. Fortschr Chem Org Naturst 93: 1–70 [DOI] [PubMed] [Google Scholar]
- Bing L, Hongxia D, Maoxin Z, Di X, Jingshu W (2007) Potential resistance of tricin in rice against brown planthopper Nilaparvata lugens (Stål). Acta Ecol Sin 27: 1300–1306 [Google Scholar]
- Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Bouvier d’Yvoire M, Bouchabke-Coussa O, Voorend W, Antelme S, Cézard L, Legée F, Lebris P, Legay S, Whitehead C, McQueen-Mason SJ, et al. (2013) Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium distachyon. Plant J 73: 496–508 [DOI] [PubMed] [Google Scholar]
- Brazier-Hicks M, Evans KM, Gershater MC, Puschmann H, Steel PG, Edwards R (2009) The C-glycosylation of flavonoids in cereals. J Biol Chem 284: 17926–17934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass CL, Peraldi A, Dowd PF, Mottiar Y, Santoro N, Karlen SD, Bukhman YV, Foster CE, Thrower N, Bruno LC, et al. (2015) Effects of PHENYLALANINE AMMONIA LYASE (PAL) knockdown on cell wall composition, biomass digestibility, and biotic and abiotic stress responses in Brachypodium. J Exp Bot 66: 4317–4335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarino I, Araújo P, Júnior A, Mazzafera P (2012) An overview of lignin metabolism and its effect on biomass recalcitrance. Braz J Bot 35: 303–311 [Google Scholar]
- Chen F, Dixon RA (2007) Lignin modification improves fermentable sugar yields for biofuel production. Nat Biotechnol 25: 759–761 [DOI] [PubMed] [Google Scholar]
- Chen HC, Li Q, Shuford CM, Liu J, Muddiman DC, Sederoff RR, Chiang VL (2011) Membrane protein complexes catalyze both 4- and 3-hydroxylation of cinnamic acid derivatives in monolignol biosynthesis. Proc Natl Acad Sci USA 108: 21253–21258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224: 1174–1184 [DOI] [PubMed] [Google Scholar]
- Chundawat SPS, Beckham GT, Himmel ME, Dale BE (2011) Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng 2: 121–145 [DOI] [PubMed] [Google Scholar]
- Coe EH, McCormick SM, Modena SA (1981) White pollen in maize. J Hered 72: 318–320 [Google Scholar]
- Coe EH Jr, Neuffer MG, Hoisington DA (1988) The genetics of corn. In Sprague GF, Dudley JW, eds, Corn and Corn Improvement, Ed 3 American Society of Agronomy, Madison, WI, pp 81–258 [Google Scholar]
- Collazo P, Montoliu L, Puigdomènech P, Rigau J (1992) Structure and expression of the lignin O-methyltransferase gene from Zea mays L. Plant Mol Biol 20: 857–867 [DOI] [PubMed] [Google Scholar]
- Cone JW, Engels FM (1993) The influence of ageing on cell wall composition and degradability of three maize genotypes. Anim Feed Sci Technol 40: 331–342 [Google Scholar]
- Cortés-Cruz M, Snook M, McMullen MD (2003) The genetic basis of C-glycosyl flavone B-ring modification in maize (Zea mays L.) silks. Genome 46: 182–194 [DOI] [PubMed] [Google Scholar]
- Cullis IF, Saddler JN, Mansfield SD (2004) Effect of initial moisture content and chip size on the bioconversion efficiency of softwood lignocellulosics. Biotechnol Bioeng 85: 413–421 [DOI] [PubMed] [Google Scholar]
- Della Vedova CB, Lorbiecke R, Kirsch H, Schulte MB, Scheets K, Borchert LM, Scheffler BE, Wienand U, Cone KC, Birchler JA (2005) The dominant inhibitory chalcone synthase allele C2-Idf (inhibitor diffuse) from Zea mays (L.) acts via an endogenous RNA silencing mechanism. Genetics 170: 1989–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río JC, Lino AG, Colodette JL, Lima CF, Gutiérrez A, Martínez ÁT, Lu F, Ralph J, Rencoret J (2015) Differences in the chemical structure of the lignins from sugarcane bagasse and straw. Biomass Bioenergy 81: 322–338 [Google Scholar]
- del Río JC, Rencoret J, Prinsen P, Martínez ÁT, Ralph J, Gutiérrez A (2012) Structural characterization of wheat straw lignin as revealed by analytical pyrolysis, 2D-NMR, and reductive cleavage methods. J Agric Food Chem 60: 5922–5935 [DOI] [PubMed] [Google Scholar]
- Dima O, Morreel K, Vanholme B, Kim H, Ralph J, Boerjan W (2015) Small glycosylated lignin oligomers are stored in Arabidopsis leaf vacuoles. Plant Cell 27: 695–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Pasinetti GM (2010) Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154: 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Chu H, Chu IK, Lo C (2010) CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol 154: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Tekriti RA, Lechtenberg VL, Bauman LF, Colenbrander VF (1976) Structural composition and in vitro dry matter disappearance of brown midrib corn residue. Crop Sci 16: 387–389 [Google Scholar]
- Escamilla-Treviño LL, Shen H, Hernandez T, Yin Y, Xu Y, Dixon RA (2014) Early lignin pathway enzymes and routes to chlorogenic acid in switchgrass (Panicum virgatum L.). Plant Mol Biol 84: 565–576 [DOI] [PubMed] [Google Scholar]
- Eudes A, George A, Mukerjee P, Kim JS, Pollet B, Benke PI, Yang F, Mitra P, Sun L, Çetinkol ÖP, et al. (2012) Biosynthesis and incorporation of side-chain-truncated lignin monomers to reduce lignin polymerization and enhance saccharification. Plant Biotechnol J 10: 609–620 [DOI] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Rius SP, Casati P (2012) Flavonoids: biosynthesis, biological functions, and biotechnological applications. Front Plant Sci 3: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feltus FA, Vandenbrink JP (2012) Bioenergy grass feedstock: current options and prospects for trait improvement using emerging genetic, genomic, and systems biology toolkits. Biotechnol Biofuels 5: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornalé S, Rencoret J, Garcia-Calvo L, Capellades M, Encina A, Santiago R, Rigau J, Gutiérrez A, Del Río JC, Caparros-Ruiz D (2015) Cell wall modifications triggered by the down-regulation of coumarate 3-hydroxylase-1 in maize. Plant Sci 236: 272–282 [DOI] [PubMed] [Google Scholar]
- Foster CE, Martin TM, Pauly M (2010) Comprehensive compositional analysis of plant cell walls (lignocellulosic biomass) Part II: Carbohydrates. J Vis Exp 37: e1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken P, Niesbach-Klösgen U, Weydemann U, Maréchal-Drouard L, Saedler H, Wienand U (1991) The duplicated chalcone synthase genes C2 and Whp (white pollen) of Zea mays are independently regulated: evidence for translational control of Whp expression by the anthocyanin intensifying gene in. EMBO J 10: 2605–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Mielenz JR, Xiao X, Ge Y, Hamilton CY, Rodriguez M Jr, Chen F, Foston M, Ragauskas A, Bouton J, et al. (2011) Genetic manipulation of lignin reduces recalcitrance and improves ethanol production from switchgrass. Proc Natl Acad Sci USA 108: 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Klein TM, Roth BA, Fromm ME, Cone KC, Radicella JP, Chandler VL (1990) Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J 9: 2517–2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez LD, Vanholme R, Bird S, Goeminne G, Trindade LM, Polikarpov I, Simister R, Morreel K, Boerjan W, McQueen-Mason SJ (2014) Side by side comparison of chemical compounds generated by aqueous pretreatments of maize stover, Miscanthus and sugarcane bagasse. BioEnergy Res 7: 1466–1480 [Google Scholar]
- Grabber JH, Hatfield RD, Lu F, Ralph J (2008) Coniferyl ferulate incorporation into lignin enhances the alkaline delignification and enzymatic degradation of cell walls. Biomacromolecules 9: 2510–2516 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem 48: 6106–6113 [DOI] [PubMed] [Google Scholar]
- Guillaumie S, San-Clemente H, Deswarte C, Martinez Y, Lapierre C, Murigneux A, Barrière Y, Pichon M, Goffner D (2007) MAIZEWALL: database and developmental gene expression profiling of cell wall biosynthesis and assembly in maize. Plant Physiol 143: 339–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CM, Escamilla-Trevino L, Yarce JCS, Kim H, Ralph J, Chen F, Dixon RA (2016) An essential role of caffeoyl shikimate esterase in monolignol biosynthesis in Medicago truncatula. Plant J 86: 363–375 [DOI] [PubMed] [Google Scholar]
- Hollick JB, Patterson GI, Coe EH Jr, Cone KC, Chandler VL (1995) Allelic interactions heritably alter the activity of a metastable maize pl allele. Genetics 141: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanin L, Goujon T, de Nadaï V, Martin MT, Mila I, Vallet C, Pollet B, Yoshinaga A, Chabbert B, Petit-Conil M, et al. (2000) Lignification in transgenic poplars with extremely reduced caffeic acid O-methyltransferase activity. Plant Physiol 123: 1363–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Vermerris W, Gallo M, Fedenko JR, Erickson JE, Altpeter F (2013) RNA interference suppression of lignin biosynthesis increases fermentable sugar yields for biofuel production from field-grown sugarcane. Plant Biotechnol J 11: 709–716 [DOI] [PubMed] [Google Scholar]
- Končitíková R, Vigouroux A, Kopečná M, Andree T, Bartoš J, Šebela M, Moréra S, Kopečný D (2015) Role and structural characterization of plant aldehyde dehydrogenases from family 2 and family 7. Biochem J 468: 109–123 [DOI] [PubMed] [Google Scholar]
- Lam PY, Liu H, Lo C (2015) Completion of tricin biosynthesis pathway in rice: cytochrome P450 75B4 is a unique chrysoeriol 5′-hydroxylase. Plant Physiol 168: 1527–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam PY, Zhu FY, Chan WL, Liu H, Lo C (2014) Cytochrome P450 93G1 is a flavone synthase II that channels flavanones to the biosynthesis of tricin O-linked conjugates in rice. Plant Physiol 165: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J (2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Morreel K, Lu F, Rencoret J, Del Río JC, Voorend W, Vermerris W, Boerjan W, Ralph J (2016a) Maize tricin-oligolignol metabolites and their implications for monocot lignification. Plant Physiol 171: 810–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Rencoret J, Lu F, Karlen SD, Smith BG, Harris PJ, Del Río JC, Ralph J (2016b) Tricin-lignins: occurrence and quantitation of tricin in relation to phylogeny. Plant J http://dx.doi.org/10.1111/tpj.13315 [DOI] [PubMed] [Google Scholar]
- Lapierre C, Monties B, Rolando C (1986) Thioacidolysis of poplar lignins: identification of monomeric syringyl products and characterization of guaiacyl-syringyl lignin fractions. Holzforschung 40: 113–118 [Google Scholar]
- Lechtenberg VL, Muller LD, Bauman LF, Rhykerd CL, Barnes RF (1972) Laboratory and in vitro evaluation of inbred and F2 populations of brown midrib mutants of Zea mays L. Agron J 64: 657–660 [Google Scholar]
- Leplé JC, Dauwe R, Morreel K, Storme V, Lapierre C, Pollet B, Naumann A, Kang KY, Kim H, Ruel K, et al. (2007) Downregulation of cinnamoyl-coenzyme A reductase in poplar: multiple-level phenotyping reveals effects on cell wall polymer metabolism and structure. Plant Cell 19: 3669–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL (2003a) Combinatorial modification of multiple lignin traits in trees through multigene cotransformation. Proc Natl Acad Sci USA 100: 4939–4944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Pu Y, Yoo CG, Ragauskas AJ (2016) The occurrence of tricin and its derivatives in plants. Green Chem 18: 1439–1454 [Google Scholar]
- Li X, Bonawitz ND, Weng JK, Chapple C (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22: 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Chen W, Zhao Y, Xiang Y, Jiang H, Zhu S, Cheng B (2013) Downregulation of caffeoyl-CoA O-methyltransferase (CCoAOMT) by RNA interference leads to reduced lignin production in maize straw. Genet Mol Biol 36: 540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang Y, Yao J, Chen G, Li X, Zhang Q, Wu C (2009) FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Mol Biol 69: 685–697 [DOI] [PubMed] [Google Scholar]
- Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, et al. (2003b) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15: 2020–2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Ralph J (2002) Preliminary evidence for sinapyl acetate as a lignin monomer in kenaf. Chem Commun (Camb) 1: 90–91 [DOI] [PubMed] [Google Scholar]
- Mansfield SD, Kim H, Lu F, Ralph J (2012) Whole plant cell wall characterization using solution-state 2D NMR. Nat Protoc 7: 1579–1589 [DOI] [PubMed] [Google Scholar]
- Marita JM, Hatfield RD, Rancour DM, Frost KE (2014) Identification and suppression of the p-coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea mays L. Plant J 78: 850–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott PE, Gómez LD, McQueen-Mason SJ (2016) Unlocking the potential of lignocellulosic biomass through plant science. New Phytol 209: 1366–1381 [DOI] [PubMed] [Google Scholar]
- Missihoun TD, Kotchoni SO, Bartels D (2016) Active sites of Reduced Epidermal Fluorescence1 (REF1) isoforms contain amino acid substitutions that are different between monocots and dicots. PLoS ONE 11: e0165867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misyura M, Colasanti J, Rothstein SJ (2013) Physiological and genetic analysis of Arabidopsis thaliana anthocyanin biosynthesis mutants under chronic adverse environmental conditions. J Exp Bot 64: 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Dima O, Kim H, Lu F, Niculaes C, Vanholme R, Dauwe R, Goeminne G, Inzé D, Messens E, et al. (2010a) Mass spectrometry-based sequencing of lignin oligomers. Plant Physiol 153: 1464–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morreel K, Kim H, Lu F, Dima O, Akiyama T, Vanholme R, Niculaes C, Goeminne G, Inzé D, Messens E, et al. (2010b) Mass spectrometry-based fragmentation as an identification tool in lignomics. Anal Chem 82: 8095–8105 [DOI] [PubMed] [Google Scholar]
- Morreel K, Ralph J, Kim H, Lu F, Goeminne G, Ralph S, Messens E, Boerjan W (2004) Profiling of oligolignols reveals monolignol coupling conditions in lignifying poplar xylem. Plant Physiol 136: 3537–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD (2016) Designer lignins: harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Nafziger E. (2009) Corn. In Illinois Agronomy Handbook. Crop Science Extension & Outreach, Urbana, IL, pp 2.1–2.26 [Google Scholar]
- Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. Plant Cell 16: 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keeffe K. (2009) Maize Growth & Development. NSW Department of Primary Industries, New South Wales, Australia [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik DL, Cass CL, Padmakshan D, Foster CE, Vogel JP, Karlen SD, Ralph J, Sedbrook JC (2016) BdCESA7, BdCESA8, and BdPMT utility promoter constructs for targeted expression to secondary cell-wall-forming cells of grasses. Front Plant Sci 7: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG, et al. (2014) p-Coumaroyl-CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon. Plant J 77: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal J, Chamayou S, Nadaud I, Beckert M, Barrière Y, Mila I, Lapierre C, Rigau J, Puigdomenech P, Jauneau A, et al. (2002) Down-regulation of caffeic acid o-methyltransferase in maize revisited using a transgenic approach. Plant Physiol 130: 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragauskas AJ, Beckham GT, Biddy MJ, Chandra R, Chen F, Davis MF, Davison BH, Dixon RA, Gilna P, Keller M, et al. (2014) Lignin valorization: improving lignin processing in the biorefinery. Science 344: 1246843. [DOI] [PubMed] [Google Scholar]
- Ralph J. (1996) An unusual lignin from kenaf. J Nat Prod 59: 341–342 [Google Scholar]
- Ralph J. (2010) Hydroxycinnamates in lignification. Phytochem Rev 9: 65–83 [Google Scholar]
- Ralph J, Brunow G, Harris PJ, Dixon RA, Schatz PF, Boerjan W (2008a) Lignification: are lignins biosynthesized via simple combinatorial chemistry or via proteinaceous control and template replication? In Daayf F, Lattanzio V, eds, Recent Advances in Polyphenol Research, Vol 1. Wiley-Blackwell, Oxford, pp 36–66 [Google Scholar]
- Ralph J, Grabber JH, Hatfield RD (1995) Lignin-ferulate crosslinks in grasses: active incorporation of ferulate polysaccharide esters into rye-grass lignins. Carbohydr Res 275: 167–178 [Google Scholar]
- Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung HJG (1994) Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc 116: 9448–9456 [Google Scholar]
- Ralph J, Kim H, Lu F, Grabber JH, Leplé JC, Berrio-Sierra J, Derikvand MM, Jouanin L, Boerjan W, Lapierre C (2008b) Identification of the structure and origin of a thioacidolysis marker compound for ferulic acid incorporation into angiosperm lignins (and an indicator for cinnamoyl CoA reductase deficiency). Plant J 53: 368–379 [DOI] [PubMed] [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, et al. (2004) Lignins: natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem Rev 3: 29–60 [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RHL, Moorman AFM (2003) Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66 [DOI] [PubMed] [Google Scholar]
- Rencoret J, Ralph J, Marques G, Gutiérrez A, Martínez Á, del Río JC (2013) Structural characterization of lignin isolated from coconut (Cocos nucifera) coir fibers. J Agric Food Chem 61: 2434–2445 [DOI] [PubMed] [Google Scholar]
- Rinaldi R, Jastrzebski R, Clough MT, Ralph J, Kennema M, Bruijnincx PCA, Weckhuysen BM (2016) Paving the way for lignin valorisation: recent advances in bioengineering, biorefining and catalysis. Angew Chem Int Ed Engl 55: 8164–8215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring L, Yeh SY, Hücherig S, Hoffmann T, Blanco-Portales R, Fouche M, Villatoro C, Denoyes B, Monfort A, Caballero JL, et al. (2013) Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol 163: 43–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie SW, Hanway JJ, Thompson HE (1996) How a Corn Plant Develops. Cooperative Extension Service, Iowa State University of Science and Technology, Ames, IA [Google Scholar]
- Rolando C, Monties B, Lapierre C (1992) Thioacidolysis. In Lin SY, Dence CW, eds, Methods in Lignin Chemistry. Springer-Verlag, Berlin, pp 334–349 [Google Scholar]
- Rösler J, Krekel F, Amrhein N, Schmid J (1997) Maize phenylalanine ammonia-lyase has tyrosine ammonia-lyase activity. Plant Physiol 113: 175–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saidi M, Samimi F, Karimipourfard D, Nimmanwudipong T, Gates BC, Rahimpour MR (2014) Upgrading of lignin-derived bio-oils by catalytic hydrodeoxygenation. Energy Environ Sci 7: 103–129 [Google Scholar]
- Sattler SE, Funnell-Harris DL, Pedersen JF (2010) Brown midrib mutations and their importance to the utilization of maize, sorghum, and pearl millet lignocellulosic tissues. Plant Sci 178: 229–238 [Google Scholar]
- Shen H, Poovaiah CR, Ziebell A, Tschaplinski TJ, Pattathil S, Gjersing E, Engle NL, Katahira R, Pu Y, Sykes R, et al. (2013) Enhanced characteristics of genetically modified switchgrass (Panicum virgatum L.) for high biofuel production. Biotechnol Biofuels 6: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih CH, Chu H, Tang LK, Sakamoto W, Maekawa M, Chu IK, Wang M, Lo C (2008) Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta 228: 1043–1054 [DOI] [PubMed] [Google Scholar]
- Shuai L, Amiri MT, Questell-Santiago YM, Héroguel F, Li Y, Kim H, Meilan R, Chapple C, Ralph J, Luterbacher JS (2016) Formaldehyde stabilization facilitates lignin monomer production during biomass depolymerization. Science 354: 329–333 [DOI] [PubMed] [Google Scholar]
- Sibout R, Le Bris P, Legée F, Cézard L, Renault H, Lapierre C (2016) Structural redesigning Arabidopsis lignins into alkali-soluble lignins through the expression of p-coumaroyl-CoA:monolignol transferase PMT. Plant Physiol 170: 1358–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G (2007) Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol 145: 1444–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L (2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton AE, Walbot V (1994) Flavonoids can protect maize DNA from the induction of ultraviolet radiation damage. Plant Physiol 105: 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles ED, Ceska O, Seah KT (1973) Developmental differences in action of R and B alleles in maize. Can J Genet Cytol 15: 59–72 [Google Scholar]
- Sundin L, Vanholme R, Geerinck J, Goeminne G, Höfer R, Kim H, Ralph J, Boerjan W (2014) Mutation of the inducible ARABIDOPSIS THALIANA CYTOCHROME P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol 166: 1956–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timpano H, Sibout R, Devaux MF, Alvarado C, Looten R, Falourd X, Pontoire B, Martin M, Legée F, Cezard L, et al. (2015) Brachypodium cell wall mutant with enhanced saccharification potential despite increased lignin content. BioEnergy Res 8: 53–67 [Google Scholar]
- Van Acker R, Vanholme R, Piens K, Boerjan W (2016) Saccharification protocol for small-scale lignocellulosic biomass samples to test processing of cellulose into glucose. Bio Protoc 6: e1701 [Google Scholar]
- Van Acker R, Vanholme R, Storme V, Mortimer JC, Dupree P, Boerjan W (2013) Lignin biosynthesis perturbations affect secondary cell wall composition and saccharification yield in Arabidopsis thaliana. Biotechnol Biofuels 6: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Bosch S, Schutyser W, Vanholme R, Driessen T, Koelewijn SF, Renders T, De Meester B, Huijgen WJJ, Dehaen W, Courtin C, et al. (2015) Reductive lignocellulose fractionation into soluble lignin-derived phenolic monomers and dimers and processable carbohydrate pulps. Energy Environ Sci 8: 1748–1763 [Google Scholar]
- van de Staaij J, de Bakker NVJ, Oosthoek A, Broekman R, van Beem A, Stroetenga M, Aerts R, Rozema J (2002) Flavonoid concentrations in three grass species and a sedge grown in the field and under controlled environment conditions in response to enhanced UV-B radiation. J Photochem Photobiol B 66: 21–29 [DOI] [PubMed] [Google Scholar]
- Vanholme B, Desmet T, Ronsse F, Rabaey K, Van Breusegem F, De Mey M, Soetaert W, Boerjan W (2013a) Towards a carbon-negative sustainable bio-based economy. Front Plant Sci 4: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Cesarino I, Rataj K, Xiao Y, Sundin L, Goeminne G, Kim H, Cross J, Morreel K, Araujo P, et al. (2013b) Caffeoyl shikimate esterase (CSE) is an enzyme in the lignin biosynthetic pathway in Arabidopsis. Science 341: 1103–1106 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Demedts B, Morreel K, Ralph J, Boerjan W (2010a) Lignin biosynthesis and structure. Plant Physiol 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme R, Morreel K, Darrah C, Oyarce P, Grabber JH, Ralph J, Boerjan W (2012) Metabolic engineering of novel lignin in biomass crops. New Phytol 196: 978–1000 [DOI] [PubMed] [Google Scholar]
- Vanholme R, Ralph J, Akiyama T, Lu F, Pazo JR, Kim H, Christensen JH, Van Reusel B, Storme V, De Rycke R, et al. (2010b) Engineering traditional monolignols out of lignin by concomitant up-regulation of F5H1 and down-regulation of COMT in Arabidopsis. Plant J 64: 885–897 [DOI] [PubMed] [Google Scholar]
- Vermerris W, Abril A (2015) Enhancing cellulose utilization for fuels and chemicals by genetic modification of plant cell wall architecture. Curr Opin Biotechnol 32: 104–112 [DOI] [PubMed] [Google Scholar]
- Vignols F, Rigau J, Torres MA, Capellades M, Puigdomènech P (1995) The brown midrib3 (bm3) mutation in maize occurs in the gene encoding caffeic acid O-methyltransferase. Plant Cell 7: 407–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorend W, Nelissen H, Vanholme R, De Vliegher A, Van Breusegem F, Boerjan W, Roldán-Ruiz I, Muylle H, Inzé D (2016) Overexpression of GA20-OXIDASE1 impacts plant height, biomass allocation and saccharification efficiency in maize. Plant Biotechnol J 14: 997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C (2013) Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A-dependent transferases and synthases. Plant Physiol 162: 640–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Dudareva N, Morgan JA, Chapple C (2015a) Genetic manipulation of lignocellulosic biomass for bioenergy. Curr Opin Chem Biol 29: 32–39 [DOI] [PubMed] [Google Scholar]
- Wang Y, Bouchabke-Coussa O, Lebris P, Antelme S, Soulhat C, Gineau E, Dalmais M, Bendahmane A, Morin H, Mouille G, et al. (2015b) LACCASE5 is required for lignification of the Brachypodium distachyon Culm. Plant Physiol 168: 192–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker CM, Balasubramanian VK, Petti C, Rai KM, DeBolt S, Mendu V (2015) Engineering plant biomass lignin content and composition for biofuels and bioproducts. Energies 8: 7654–7676 [Google Scholar]
- Wen JL, Sun SL, Xue BL, Sun RC (2013) Recent advances in characterization of lignin polymer by solution-state nuclear magnetic resonance (NMR) methodology. Materials (Basel) 6: 359–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CR, Yang RD, Sciamanna AF, Freitas RP (1981) Raw materials evaluation and process development studies for conversion of biomass to sugars and ethanol. Biotechnol Bioeng 23: 163–183 [Google Scholar]
- Wilkerson CG, Mansfield SD, Lu F, Withers S, Park JY, Karlen SD, Gonzales-Vigil E, Padmakshan D, Unda F, Rencoret J, et al. (2014) Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344: 90–93 [DOI] [PubMed] [Google Scholar]
- Withers S, Lu F, Kim H, Zhu Y, Ralph J, Wilkerson CG (2012) Identification of grass-specific enzyme that acylates monolignols with p-coumarate. J Biol Chem 287: 8347–8355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Storms R, Tsang A (2004) Microplate-based filter paper assay to measure total cellulase activity. Biotechnol Bioeng 88: 832–837 [DOI] [PubMed] [Google Scholar]
- Xu B, Escamilla-Treviño LL, Sathitsuksanoh N, Shen Z, Shen H, Zhang YH, Dixon RA, Zhao B (2011) Silencing of 4-coumarate:coenzyme A ligase in switchgrass leads to reduced lignin content and improved fermentable sugar yields for biofuel production. New Phytol 192: 611–625 [DOI] [PubMed] [Google Scholar]
- Yaman S. (2004) Pyrolysis of biomass to produce fuels and chemical feedstocks. Energy Convers Manage 45: 651–671 [Google Scholar]
- Yeh SY, Huang FC, Hoffmann T, Mayershofer M, Schwab W (2014) FaPOD27 functions in the metabolism of polyphenols in strawberry fruit (Fragaria sp.). Front Plant Sci 5: 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Helms GL, Gao X, Chen S (2013) Quantification of wheat straw lignin structure by comprehensive NMR analysis. J Agric Food Chem 61: 10848–10857 [DOI] [PubMed] [Google Scholar]
- Zhang M, Zhang B, Qian Q, Yu Y, Li R, Zhang J, Liu X, Zeng D, Li J, Zhou Y (2010) Brittle Culm 12, a dual-targeting kinesin-4 protein, controls cell-cycle progression and wall properties in rice. Plant J 63: 312–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Zhang L, Liu D (2012) Biomass recalcitrance. Part I. The chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioprod Biorefin 6: 465–482 [Google Scholar]
- Zhou JM, Fukushi Y, Wollenweber E, Ibrahim RK (2008) Characterization of two O-methyltransferase-like genes in barley and maize. Pharm Biol 46: 26–34 [Google Scholar]
- Zhou JM, Ibrahim RK (2010) Tricin: a potential multifunctional nutraceutical. Phytochem Rev 9: 413–424 [Google Scholar]
- Zhou Y, Li S, Qian Q, Zeng D, Zhang M, Guo L, Liu X, Zhang B, Deng L, Liu X, et al. (2009) BC10, a DUF266-containing and Golgi-located type II membrane protein, is required for cell-wall biosynthesis in rice (Oryza sativa L.). Plant J 57: 446–462 [DOI] [PubMed] [Google Scholar]
- Zuk M, Działo M, Richter D, Dymińska L, Matuła J, Kotecki A, Hanuza J, Szopa J (2016) Chalcone synthase (CHS) gene suppression in flax leads to changes in wall synthesis and sensing genes, cell wall chemistry and stem morphology parameters. Front Plant Sci 7: 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.