Binding of the pentatricopeptide repeat protein EMB2654 to one-half of the plastid rps12 intron is essential for trans-splicing, production of plastid ribosomes, and embryogenesis in Arabidopsis.
Abstract
We report the partial complementation and subsequent comparative molecular analysis of two nonviable mutants impaired in chloroplast translation, one (emb2394) lacking the RPL6 protein, and the other (emb2654) carrying a mutation in a gene encoding a P-class pentatricopeptide repeat protein. We show that EMB2654 is required for the trans-splicing of the plastid rps12 transcript and that therefore the emb2654 mutant lacks Rps12 protein and fails to assemble the small subunit of the plastid ribosome, explaining the loss of plastid translation and consequent embryo-lethal phenotype. Predictions of the EMB2654 binding site match a small RNA “footprint” located on the 5′ half of the trans-spliced intron that is almost absent in the partially complemented mutant. EMB2654 binds sequence specifically to this target sequence in vitro. Altered patterns in nuclease-protected small RNA fragments in emb2654 show that EMB2654 binding must be an early step in, or prior to, the formation of a large protein-RNA complex covering the free ends of the two rps12 intron halves.
Plastids are essential plant organelles that are best known for providing carbon skeletons and energy to the cell via photosynthesis. However, they also play a crucial role in plant development as early as embryogenesis, well before the establishment of the photosynthetic apparatus (Bryant et al., 2011). Plastids derive from cyanobacteria that established an endosymbiotic relationship with eukaryotic cells (Timmis et al., 2004). Although they have lost numerous genes over the last billion years, the plastid genomes of most vascular plants have retained ∼120 genes (Wicke et al., 2011). The majority of the protein-coding genes encode primary components of the photosynthetic apparatus, including major subunits of photosystems I and II, cytochrome b6f, the NDH complex, and ATP synthase. Most of the remaining genes encode components of the transcription and translation machineries, as well as a few key biogenesis genes such as accD, clpP1, matK, ycf1, and ycf2. Proteomics studies have revealed 2 to 3 thousand proteins in the plastid compartment, most of them products of nuclear genes (Friso et al., 2004; Ferro et al., 2010). Assembly of proteins produced by both plastid and nuclear genes are necessary for the biogenesis and function of the chloroplast apparatus, requiring some degree of coordination between the protein synthesis machineries in the cytosol and plastids (Tiller and Bock, 2014).
Transcription in land plant plastids involves two different types of RNA polymerase, nuclear-encoded phage-type RNA polymerases and a plastid-encoded (cyanobacterial-type) RNA polymerase (PEP; Hajdukiewicz et al., 1997; Liere and Börner, 2007). Plastid gene expression also requires a battery of processing factors that perform the extensive posttranscriptional maturation steps of the polycistronic primary transcripts (intron splicing, processing, trimming, and protection of 3′ and 5′ ends, RNA editing) prior to translation (Stern et al., 2010; Barkan, 2011). Pentatricopeptide repeat (PPR) proteins are organellar RNA-binding proteins implicated in these processes that form a large family of ∼450 members in angiosperms (reviewed by Barkan and Small, 2014). They comprise tandem repeats of 35-amino-acid motifs that bind RNA in a highly specific manner, targeting only a single or a limited number of transcripts (Barkan and Small, 2014). These repeats vary in length and amino acid composition, and this variation has been used to define two categories of PPR proteins: P-class PPR proteins are primarily composed of canonical 35-amino-acid motifs, while the PLS-class proteins comprise triplets of motifs of different length and sequence and additional C-terminal domains (Lurin et al., 2004; Cheng et al., 2016). The PLS-class PPR proteins are mostly involved in RNA editing, whereas P-class proteins play important roles in transcript stabilization and intron splicing. Other nucleus-encoded RNA binding proteins such as chloroplast RNA splicing and ribosome maturation proteins (Barkan et al., 2007), plant organellar RNA recognition proteins (Kroeger et al., 2009), and mitochondrial transcription termination factors (Babiychuk et al., 2011) are also implicated in plastid intron splicing, which additionally involves intron maturases (Zoschke et al., 2010). Intron splicing is required to remove the ∼20 introns that were acquired by plastid genes early during the evolution of land plants (Turmel et al., 2006). In Arabidopsis (Arabidopsis thaliana) plastids, six tRNAs and 11 protein-coding genes have introns, but one of them, rps12 (encoding a ribosomal protein), is fragmented, requiring intron trans-splicing to join the disparate parts (Barkan, 2004).
For translation of the polypeptides they encode, plastids use their own protein synthesis machinery (ribosomal 23S, 16S, 5S, and 4.5S RNAs, 37 tRNAs, and 59 ribosomal proteins). Although rRNAs and ribosomal proteins are generally conserved between plastids and bacteria, five plant-specific ribosomal proteins have been described (Yamaguchi and Subramanian, 2000; Yamaguchi et al., 2000; Tiller and Bock, 2014). Plastid translation is essential for cell viability in tobacco (Nicotiana tabacum; Drescher et al., 2000; Kuroda and Maliga, 2003; Kode et al., 2005; Rogalski et al., 2006) and generally also in Arabidopsis (Parker et al., 2014). Interestingly, this is not the case in grasses (Stern et al., 2004), probably because the chloroplast genome in grasses lacks three genes known to be essential in dicots (Konishi et al., 1996; Stern et al., 2004). As a result, many more plastid translation mutants are known from grasses (particularly maize) than from dicots. These plastid translation mutants are recognizable by their characteristic ivory color and the lack of accumulation of plastid rRNAs (Barkan, 1993; Hübschmann et al., 1996; Schmitz-Linneweber et al., 2006; Beick et al., 2008; Williams-Carrier et al., 2008).
In Arabidopsis, nuclear mutants impacting plastid translation were long overlooked because of the severe, frequently lethal phenotype. Several studies focusing on the identification of nuclear genes essential for embryo development (Meinke et al., 1994; Bryant et al., 2011; Candela et al., 2011; Romani et al., 2012; Savage et al., 2013) have noted the high frequency of such genes that encode products presumably involved in plastid translation. It is becoming clear that the essential function provided by plastid translation during embryogenesis is the synthesis of the product of the accD gene, encoding the catalytic subunit of the plastid acetyl-CoA carboxylase (required for malonyl-CoA production for fatty acid biosynthesis). In grasses, a nuclear encoded but plastid-targeted acetyl-CoA carboxylase fully compensates for the lack of the chloroplast accD gene (Konishi et al., 1996). In Brassicas (and some Arabidopsis accessions), the nuclear ACC2 gene can partially complement the loss of accD translation (Babiychuk et al., 2011; Bryant et al., 2011), but this is generally insufficient for survival through embryogenesis in the Arabidopsis genotypes most commonly used in research (Parker et al., 2014).
Mutations that lead to embryo lethality have been named emb (Meinke and Sussex, 1979) and are obviously rather difficult to study. In order to exploit the underused resource constituted by collections of emb mutants, various strategies have been proposed to partially complement the mutation through embryogenesis before allowing the lethal phenotype to develop at the seedling stage where it can be studied (Despres et al., 2001; Babiychuk et al., 2011). In this article, we use this approach to study two nonviable mutants impaired in chloroplast translation, one lacking the RPL6 protein (emb2394) and the other carrying a mutation in a gene encoding a P-class PPR protein (emb2654). We show that EMB2654 is required for the trans-splicing of the plastid rps12 transcript and identify its likely binding site on one of the intron halves. This work offers clues as to how the two intron halves associate and the mechanisms of trans-splicing.
RESULTS
Partial Complementation of emb2654 and emb2394
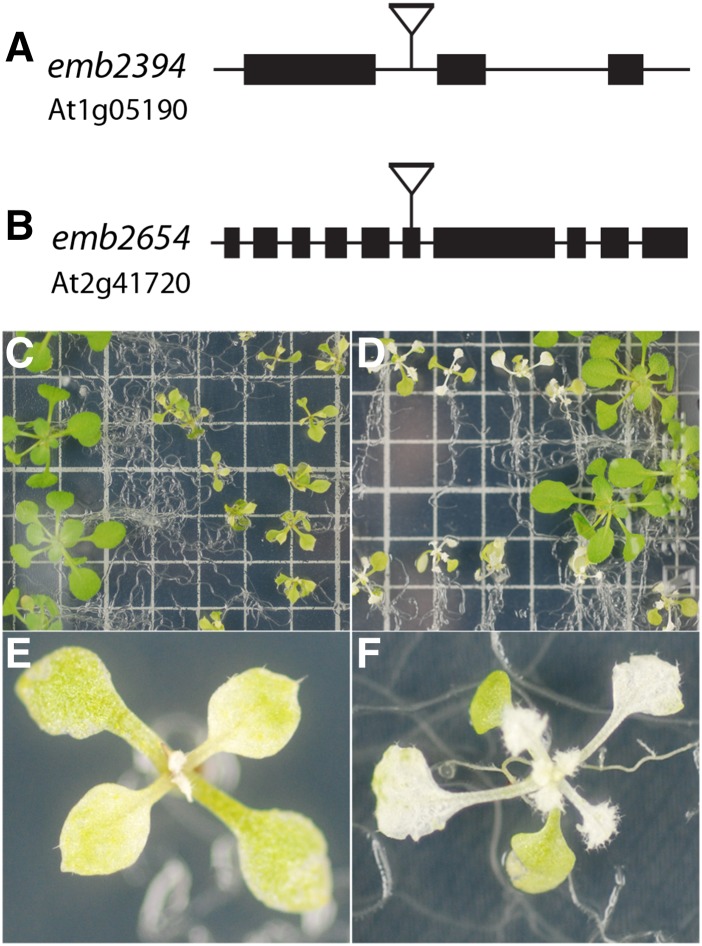
Two lines exhibiting embryo-lethality, emb2394 and emb2654, were chosen from the Seed Genes Project, Essential Genes in Arabidopsis Development (http://www.seedgenes.org). In emb2394, the T-DNA insertion is in the first intron of the RPL6 gene (At1g05190), which encodes the plastid 50S ribosomal protein L6 (Fig. 1). The T-DNA insertion in emb2654 is located in exon 6 of a gene (At2g41720) predicted to encode a P-class PPR protein (Fig. 1). This protein comprises 17 conserved PPR motifs and has been reported as targeted to the chloroplast (Colcombet et al., 2013).
Figure 1.
Gene models and phenotypes of the partially complemented mutants. A and B, The positions of the T-DNA insertions in emb2394 (A) and emb2654 (B) are shown. C to F, The emb2394 (C and E) and emb2654 (D and F) mutants expressing their respective wild-type proteins under control of the seed-specific ABI3 promoter. Large green seedlings in each case are wild-type siblings. Both mutant lines display yellowish green cotyledons and yellow to white leaves with increasingly severe phenotypes until development ceases. Seedlings were grown on half-strength Gamborg B5 medium. Squares on the grid are 1 × 1 cm.
Both lines were partially rescued by complementation of heterozygous lines with a cDNA carrying the wild-type coding sequences under the control of the seed-specific ABSCISIC ACID-INSENSITIVE3 (ABI3) promoter (Despres et al., 2001). Expression driven by the ABI3 promoter allowed development of homozygous mutant embryos as the complementing construct was expressed during embryogenesis. During seedling development, the ABI3 promoter is no longer active, leading to a progressive appearance of phenotypes due to the lack of RPL6 and EMB2654, respectively. The phenotypes observed for the rescued plants are shown in Figure 1. Cotyledons and leaves of the rescued emb2394 line were greenish-yellow, and the leaves serrated (Fig. 1, C and E), a pattern already observed for chloroplast translation mutants (Pinon et al., 2008; Moschopoulos et al., 2012). The rescued emb2654 seedlings had pale green cotyledons and albino leaves (Fig. 1, D and F), suggesting a more severe defect.
EMB2654 Is Necessary for the Accumulation of Plastid-Encoded Proteins
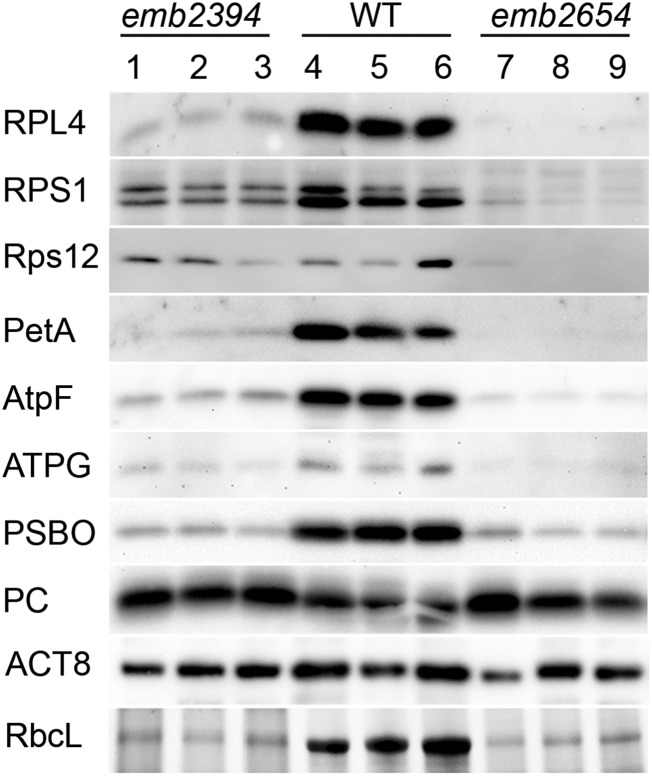
emb2394 is presumably impaired in chloroplast ribosome biogenesis and translation, as the defect is in a gene encoding a known plastid ribosomal protein. The phenotype of the partially rescued emb2654 plants suggested that they may also be deficient in plastid translation, so we performed comparative western blots using antibodies raised against proteins of the large subunit (RPL4) and the small subunit (RPS1, Rps12) of the chloroplast ribosome (Fig. 2). All three proteins are easily detectable in the wild-type samples, but almost undetectable in emb2654. emb2394 shows a much more pronounced deficit in RPL4 than in the small subunit proteins RPS1 and Rps12, consistent with the expected primary defect in large subunit assembly due to the absence of RPL6. We also tested antibodies raised against subunits of the photosynthetic complexes and ATP synthase. The plastid-encoded subunits tested (PetA, AtpF, as well as RbcL, which is shown on an acrylamide gel in the last panel of Fig. 2) were reduced in the two partially complemented mutants, whereas the nuclear-encoded plastocyanin accumulated to normal levels. These results are consistent with both emb2394 and emb2654 being impaired in plastid ribosome biogenesis and plastid translation.
Figure 2.
Western-blot analysis of chloroplast proteins in emb2654 and emb2394. Immunoblots of total proteins from partially complemented seedlings were probed using antibodies raised against various plastid proteins. Several acrylamide gels (three biological replicates for each genotype) were run with identical loadings and blotted onto PVDF membranes. RPL4, RPS1, ATPG (subunit of the ATP synthase), PSBO (subunit of photosystem II), and PC (plastocyanin), are encoded by the nuclear genome. Rps12, PetA (subunit of the cytochrome b6/f), AtpF, and RbcL are encoded by the plastid genome. Due to very different RbcL quantities between wild type (WT) and mutants (RbcL bands are visualized on a stain-free acrylamide gel), the loading was adjusted by comparing nonvarying background bands on gels and ACT8 (actin) was used as a nonplastid control.
Chloroplast Transcript Analysis
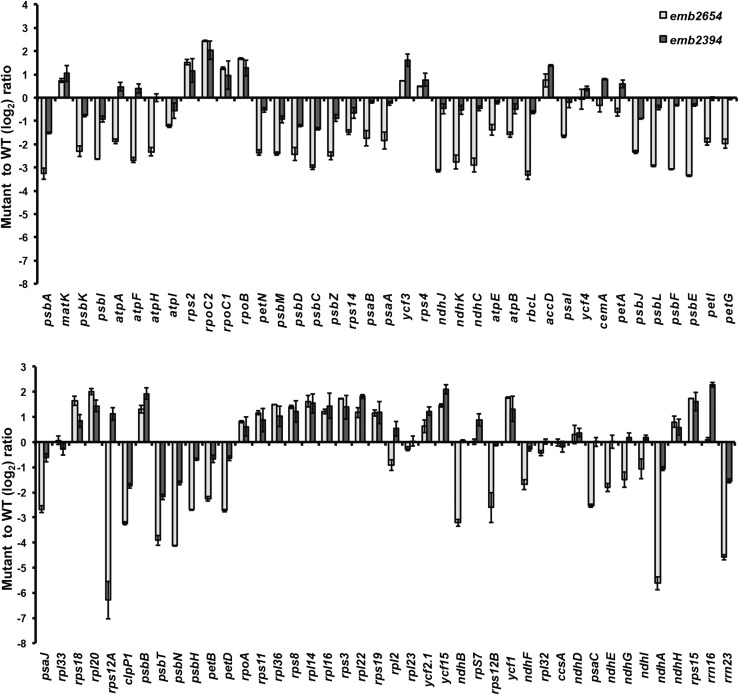
The fact that EMB2654 is a P-class PPR protein presumably involved in RNA processing prompted us to check the accumulation of plastid transcripts in emb2654 using an RT-qPCR screen previously described (Chateigner-Boutin et al., 2008). As a comparison, we also checked plastid transcripts in the rescued emb2394 line. The patterns observed for these mutants (Fig. 3) show similar trends, with an accumulation of the transcripts encoding PEP subunits (rpoA, rpoB, rpoC1, and rpoC2) or ribosomal proteins (rps, rpl transcripts), and a reduction in transcripts from genes relating to the photosynthetic apparatus (rbcL, pet, psa, psb, and ndh transcripts) that are normally transcribed by PEP (Legen et al., 2002). These patterns are reminiscent of mutants deficient in PEP such as clb19 (Chateigner-Boutin et al., 2008), ptac2 and otp70 (Chateigner-Boutin et al., 2011). The differences from wild-type are stronger for emb2654, as would be expected from the stronger visual phenotype of the plants. The most striking departure from the general transcript patterns seen in these mutants is the case of rps12A, which is the most reduced transcript in emb2654 but accumulates to above wild-type levels in emb2394, as is seen for other rps transcripts. This suggested that there may be a specific defect in rps12A expression in emb2654. By rps12A, we mean the gene fragment that includes the first exon of rps12 and the first half of the first intron. This is consistent with the much greater reduction in Rps12 protein in emb2654 than in emb2394 (Fig. 2).
Figure 3.
Plastid transcript levels in emb2654 and emb2394 seedlings. Genome-wide qRT-PCR was performed on chloroplast transcripts from the partially complemented seedlings from both lines (measurements shown here as log2 ratios of gene expression in mutant samples compared to that of phenotypically normal siblings grown in parallel). Both lines display a general accumulation of transcripts related to transcription and translation (transcribed by nuclear-encoded phage-type RNA polymerase), with the noticeable exception of rps12A in emb2654, and a general decrease of the transcripts encoding subunits of the photosynthetic apparatus (transcribed by PEP). The values are means of two biological replicates (bars indicate se).
EMB2654 Is Involved in Trans-Splicing of rps12 Transcripts
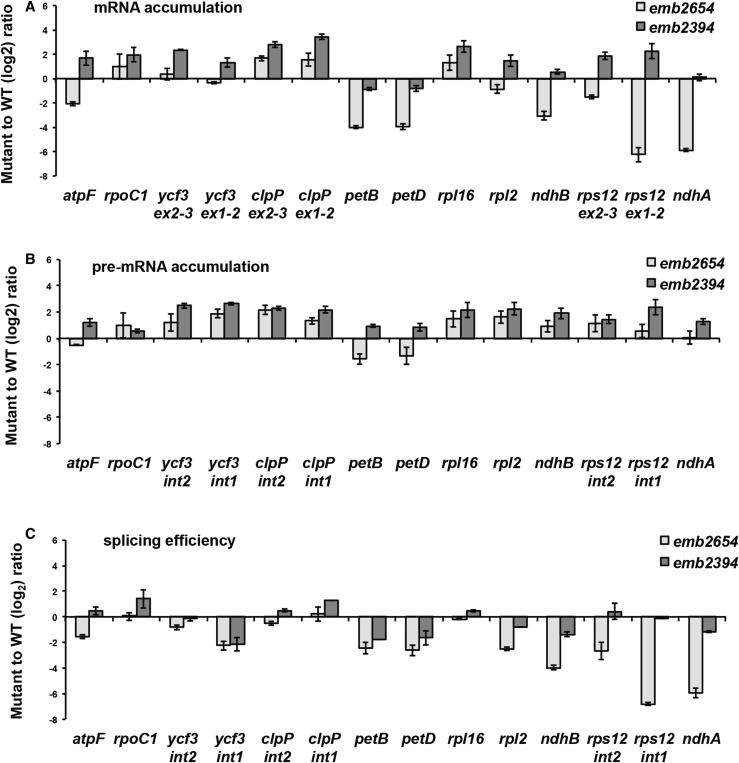
As P-class PPR proteins are often involved in splicing, we searched for splicing defects in emb2654 and in emb2394, used as a control for indirect effects of a loss of plastid translation. This is particularly relevant because the splicing maturase MatK is plastid encoded (Zoschke et al., 2010), and its absence leads to many plastid splicing defects. Defects that are seen in both mutants may be attributed to the lack of chloroplast translation, but any defects seen only in emb2654 could be primary defects. Figure 4 shows the mRNA accumulation in both mutants and that of the corresponding pre-mRNAs. The splicing efficiency (ratio of spliced to unspliced transcripts in the mutant to that of wild type) is also presented. The greatest reduction in splicing efficiency was observed for rps12 intron 1, and this reduction was specific to emb2654. This reduction in splicing efficiency was not due to a problem with accumulation of the pre-mRNA as it accumulated to above wild-type levels. The apparent splicing efficiency of ndhA is also reduced in emb2654 but cannot be attributed to a splicing defect as the pre-mRNA does not accumulate more than in the wild type (Fig. 4). A similar loss of processed ndhA transcripts has been observed in other plastid development mutants, including otp70, clb19 (Chateigner-Boutin et al., 2011), and sot1 (Wu et al., 2016), so it is probably an indirect effect.
Figure 4.
RT-qPCR analysis of intron-containing plastid transcripts in emb2654 and emb2394 seedlings. Transcript levels are compared to phenotypically normal siblings (wild type [WT]) grown in parallel. RT-qPCR was carried out using two sets of primers: One set was designed to specifically amplify spliced RNA (A) and the other to specifically amplify unspliced RNA (B). C, Splicing efficiency as the log2 ratio of spliced to unspliced transcripts in the mutants compared to the wild type. The values are means of three biological replicates for emb2654 and two replicates for emb2394 (bars indicate se).
Prediction of EMB2654 Binding Sites
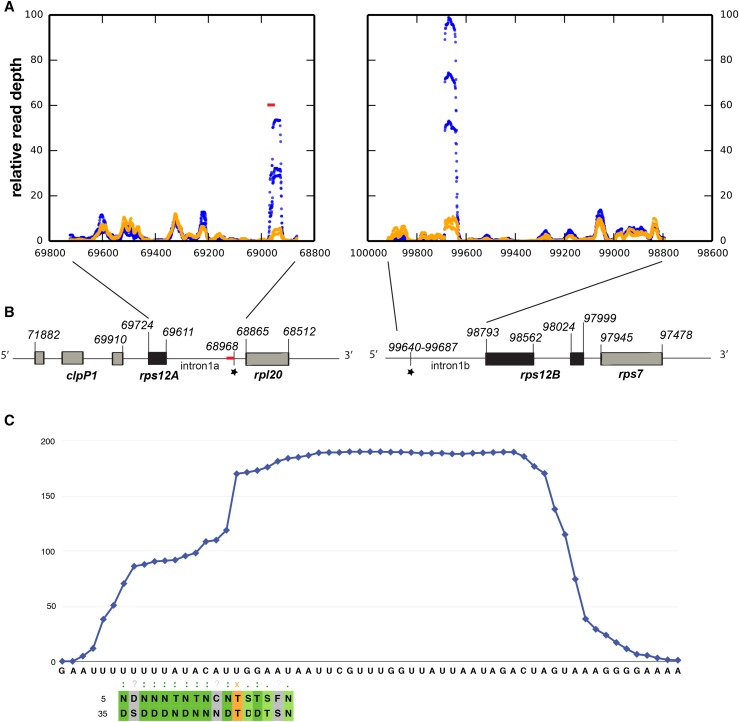
Using the rules proposed to explain RNA sequence recognition by PPR proteins (Barkan et al., 2012), we attempted to predict the sequence recognized by EMB2654. EMB2654 consists of 17 contiguous PPR motifs (Cheng et al., 2016) of which 15 have commonly observed amino acid combinations at the key positions determining base recognition (Barkan et al., 2012; Shen et al., 2016) that allow their base preferences to be predicted. The resulting query sequence (TNTTTATAYNTGRGRNY) was searched for in the Arabidopsis chloroplast genome using the EMBOSS fuzznuc motif recognition software (Rice et al., 2000). Allowing one mismatch, we obtained four hits, two on the direct strand, two on the reverse strand (Supplemental Table S1). The only prediction with no mismatch (123781–123797) was a sequence located in the intergenic sequence between rps15 (123296–123562) and ycf1 (123884–129244) on the reverse strand. These two transcripts are expressed similarly in both mutants and 4-fold higher than in the wild type, suggesting that if they are binding sites for EMB2654, its absence has no detectable effect on their RNA processing. Of the other three matches, the most interesting is located at the 3′ extremity of rps12 intron 1a, close to the rpl20 gene (Fig. 5). Due to the decrease of rps12 splicing efficiency in emb2654, this site was a good candidate for EMB2654 binding.
Figure 5.
RNA-seq analysis of putative “footprints” in the rps12 intron halves. A, RNA-seq was performed on gel-purified 15- to 50-nucleotide RNA fragments from partially complemented emb2654 seedlings (orange) and wild-type siblings (blue). The plots indicate the relative read depth at each nucleotide in the rps12 intron halves. Read depth has been normalized to the average depth across each intron (excluding the region of the putative footprint in each case). Data from three biological replicates are shown. The predicted EMB2654 binding site is shown by a red bar. B, Arrangement of the rps12 genes on Arabidopsis chloroplast DNA. The first exon is in rps12A, which is in the same transcription unit as rpl20 and clpP1. The second and third exons are in rps12B, located approximately 30 kb away and cotranscribed with ndhB and rps7. The position of the predicted EMB2654 binding site is indicated by a red bar, and RNA-seq footprints by black stars. Genome coordinates are indicated. C, Alignment of EMB2654 to its predicted binding site in rps12 intron 1a. The amino acids at the fifth and last positions in each PPR motif are aligned with the RNA sequence. Combinations that correlate with the aligned base (Barkan et al., 2012) are shaded in dark green. Combinations where only the fifth residue correlates with the aligned nucleotide are shaded in light green, combinations of unknown affinity are shaded in gray, and combinations that significantly anticorrelate with the aligned nucleotide are shaded in orange. The blue trace indicates the mean read depth observed in the RNA-seq analysis for wild-type samples in this region, showing the shape and extent of the footprint.
Search for RNA Footprints
It has been shown that many of the chloroplast small RNA fragments revealed by deep sequencing are footprints resulting from the protective action of RNA binding proteins against exonucleases (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012a). To check for the presence of footprints in the rps12 regions in wild type and in partially complemented emb2654 mutants, we performed RNA-seq experiments on gel-purified fractions of small (15–50 nucleotides) RNAs. One footprint whose 5′ extremity is the predicted binding site for EMB2654 was found in rps12A intron 1a (Fig. 5). Another footprint was found on the other half of this intron, rps12 intron 1b. This second footprint was also reported in barley (Zhelyazkova et al., 2012b). The read distribution across the rpl12 intron 1 region in the three biological repeats of emb2654 and wild-type plants shown in Figure 5 revealed a huge decrease for both footprints in the rescued emb2654 samples. A complete loss of the footprint(s) could not be expected, as the mutants are partially complemented. Thus, this result is consistent with EMB2654 binding the extremity of rps12 intron 1a in wild-type plants. We have no evidence that EMB2654 is directly involved in the formation of the footprint on rps12 intron 1b, as there are no predicted binding sites in the vicinity.
Confirmation of Sequence-Specific Binding of EMB2654 to rps12 Intron 1a
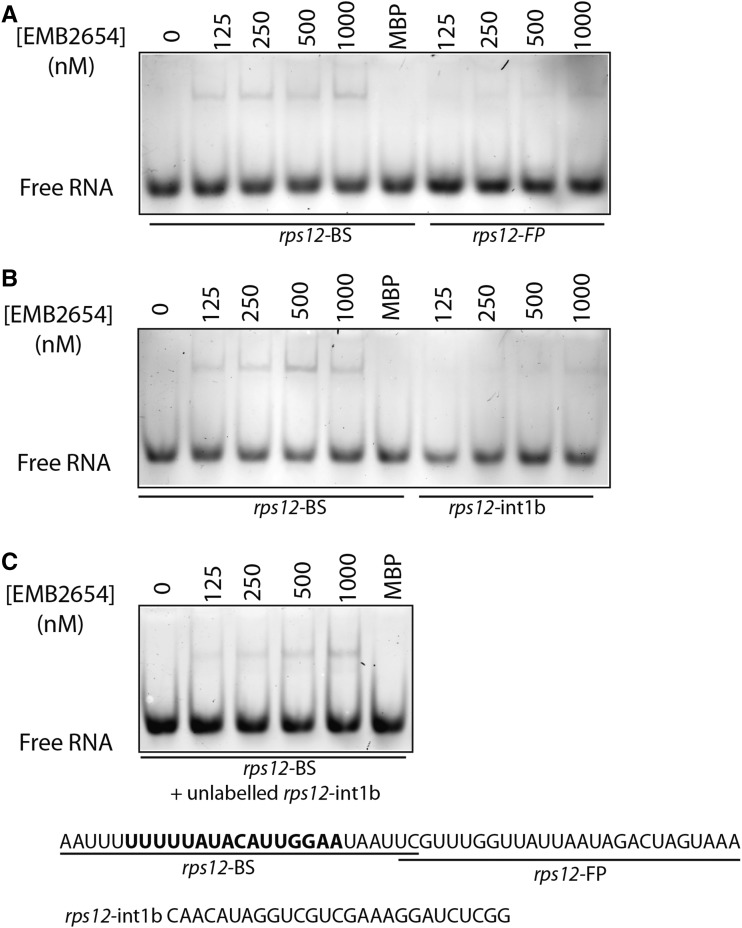
The 44-nucleotide rps12 intron 1a footprint is particularly long compared to other PPR-associated footprints (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012a), and the predicted binding site of EMB2654 is close to the 5′ edge of the footprint and ∼25 nucleotides away from the 3′ edge. This is unusual; if the footprint was formed by blocking 3′-5′ exonuclease activity, as would be expected for a footprint at the 3′ terminus of the RNA, the binding site would typically be within ∼4 nucleotides of the 3′ edge of the footprint (Zhelyazkova et al., 2012a). Hence, to verify where exactly EMB2654 binds in this region, we expressed EMB2654 in Escherichia coli, purified it, and tested its RNA binding ability in vitro, using RNA oligonucleotides matching different regions of the rps12 intron 1a footprint and part of the footprint found on intron 1b (Fig. 6). The results accord with the binding site predictions: EMB2654 binds the 5′ half of the intron 1a footprint (Fig. 6A) but scarcely at all to the 3′ half of the intron 1a footprint or the intron 1b footprint (Fig. 6, A and B). The binding of EMB2654 to the 5′ half of the intron 1a footprint was conserved when in competition with a 10-fold excess of unlabeled intron 1b probe (Fig. 6C).
Figure 6.
Binding of EMB2654 to sequences within rps12 introns 1a and 1b. REMSA was performed with recombinant EMB2654 protein and RNA oligonucleotides labeled with fluorescein. The rps12-BS probe includes the predicted EMB2654 binding site, while the rps12-int1b probe contains the intron 1b footprint. Numbers above the images refer to the concentration of EMB2654-MBP fusion (nm), except for the lane MBP, in which 1,000 nm of maltose binding protein was used as a control. A and B, No competitor present. C, Competition experiment: EMB2654 binding to rps12-BS in the presence of 10 nm unlabeled rps12-intb probe. The gels were scanned at 488 nm (excitation wavelengths for fluorescein) detected through a 520-nm band-pass filter. EMB2654 shows detectable binding to the rps12-BS probe from 125 nm upwards, but no significant binding to the rps12-FP and rps12-int1b probe.
rps12 Intron 1 Structure
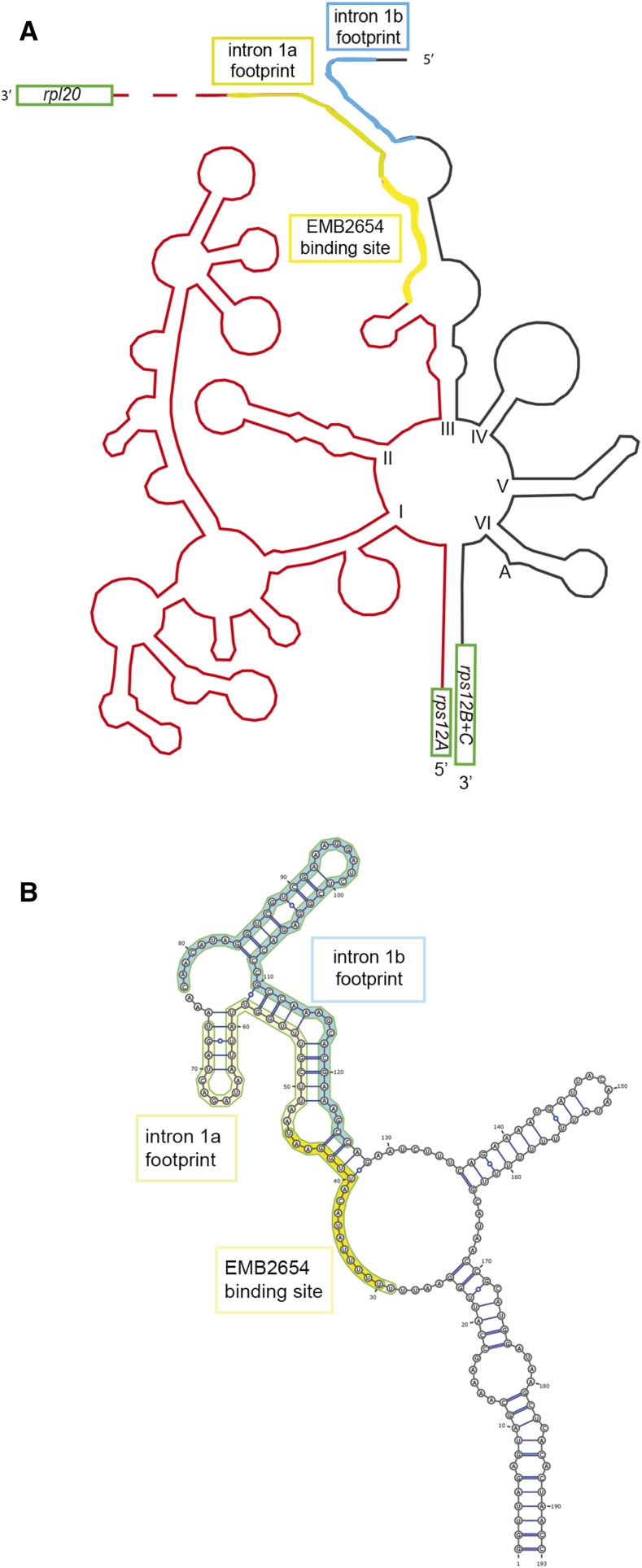
In order to determine the positions of the EMB2654 binding site and the two footprints on the folded intron structure, we used the model proposed for tobacco rps12 intron 1 (Kohchi et al., 1988). For this, the Arabidopsis and tobacco sequences for each half of the intron were respectively aligned, the intron domains identified, and the footprints positioned (Supplemental Fig. S1). The sketch of the folded intron and the positions of the footprints and the EMB2654 binding site are shown in Figure 7. The positions of both footprints are at the same end of the structure, suggesting that they might interact. We used the Vienna Package RNAcofold software (Lorenz et al., 2011) to obtain a likely secondary structure for domain III of the rps12 intron. The predicted structure shows that the two intron termini can indeed form a secondary structure through complementary pairing. The EMB2654 binding site largely overlaps with the longest single-stranded stretch in this predicted structure (Fig. 7).
Figure 7.
Predicted structure of the rps12 intron. A, Sketch of the Arabidopsis rps12 intron, using as a model the structure proposed for the tobacco intron by Kohchi et al. (1988). The Arabidopsis and tobacco sequences are aligned in Supplemental Figure S1. Both the intron 1a footprint (greenish yellow), comprising the EMB2654 binding site (yellow) and intron 1b footprint (blue) are marked. B, Predicted secondary structure of rps12 intron 1 domain III showing the potential interactions between the two intron footprints. This prediction was obtained using RNAcofold (Lorenz et al., 2011) and drawn using VARNA (http://varna.lri.fr/).
DISCUSSION
Genes Essential for Embryogenesis in Arabidopsis
Many embryo-lethal mutations cause an arrest of embryo development at the globular stage and reduced seed pigmentation, characteristics that may be attributed to chloroplast dysfunction. A comprehensive list of 119 nuclear genes encoding chloroplast proteins required for embryo development in Arabidopsis, representing about 30% of known EMB genes, has been compiled (Bryant et al., 2011). The authors defined three main groups of plastid-targeted proteins whose absence is associated with embryo lethality: enzymes involved in the biosynthesis of essential components (amino acids, nucleic acids, fatty acids, and vitamins), proteins involved in translocation and modification of chloroplast proteins, and proteins required for chloroplast gene expression (Bryant et al., 2011). Within the latter class, ribosomal proteins and PPR proteins are well represented. Disruption of a PPR gene should only be embryo lethal if the protein induces essential modifications to transcripts of genes involved in essential functions such as accD or those required for chloroplast translation. The proteins AccD, Ycf1 (thought to be a component of the import apparatus [Kikuchi et al., 2013]), Ycf2, and ClpP (a chloroplast protease and chaperone essential to the biogenesis of thylakoids) are all required for cell viability in tobacco, as shown by directed mutagenesis of the chloroplast genome (Drescher et al., 2000; Shikanai et al., 2001; Kuroda and Maliga, 2003; Kode et al., 2005). Furthermore, these genes have been retained on minimal chloroplast genomes of nonphotosynthetic parasitic plants such as Epifagus virginiana (Wolfe et al., 1992) or the underground orchid Rhizanthella gardnerii (Delannoy et al., 2011), stressing their important role in nonphotosynthetic aspects of plastid biogenesis.
The use of a partial complementation strategy allowed us to characterize two embryo-lethal mutations, emb2394 and emb2654, at a molecular level. Both these mutants are unable to develop beyond the globular stage (87% of embryos blocked at that stage for emb2394 and 99% for emb2654; data from seedgenes.org). The expression of the complementation construct during seed development under the control of the ABI3 promoter (Despres et al., 2001) allowed the embryo to overcome this critical stage. This general approach should hopefully prove useful for the analysis of other lethal mutations in the many other genes required for plastid translation. The line emb2394 carries a putative null mutation in the gene encoding RPL6, a component of the large subunit (50S) of plastid ribosomes. L6 binds directly to 23S rRNA (Davies et al., 1998), is a key player in the later steps of 50S subunit assembly (Shigeno et al., 2016), and is essential in E. coli (Gerdes et al., 2003; Shoji et al., 2011). The well-understood function of RPL6 allowed us to use emb2394 as a control for analysis of a second putative null mutation in the EMB2654 gene encoding a P-class PPR protein.
EMB2654 Is Involved in the Trans-Splicing of rps12 Intron 1
The phenotypes observed for the rescued plants (yellow or albino leaves, stunted growth), as well as the predicted localization of the defective proteins, suggested strong defects in plastid gene expression. The molecular phenotypes (PEP-deficient transcript patterns, reduced levels of plastid-encoded proteins, and ribosomal subunits) are all consistent with defects in the assembly and activity of the plastid translational apparatus. These observations were of course expected for emb2394, lacking RPL6, but the similarities with the molecular phenotypes of emb2654 suggested that it was compromised in a similar way. Some defects were specific to emb2654, notably the extreme reductions in the rps12A transcript (Fig. 3) and the Rps12 protein (Fig. 2). Additional clues were obtained from the analysis of splicing in emb2396 and emb2654. We expected to detect splicing defects in the introns processed by the plastid-encoded maturase MatK. The targets of MatK were determined by RIP-ChIP (Zoschke et al., 2010) and include the group IIA introns of four essential tRNAs (trnV, trnI, trnA, and trnK) and three other introns (atpF, rpl2, and the cis-spliced rps12 intron 2). We did not detect many splicing defects attributable to the lack of MatK in the emb2394 mutant (Fig. 4), but minor defects including atpF, rps12 intron 2, rpl2, and ndhB were observed for emb2654, mostly consistent with a defect in MatK. The primary defect, however, lies in the splicing of rps12 intron 1, which is not a target of MatK. Ribosomal protein S12 is known to be involved in translation initiation in E. coli and is highly conserved between bacteria, mitochondria, and chloroplasts (Toivonen et al., 1999). It is a control element for the translocation of the mRNA:tRNA complex in E. coli, and mutations in S12 have been shown to affect translation accuracy (Cukras et al., 2003). S12 is essential in E. coli (Gerdes et al., 2003; Shoji et al., 2011).
The rps12 gene is split between two different locations on the chloroplast genome. The first exon, rps12A, lies between clpP1 and rpl20, these three genes forming a cotranscription unit, while exons 2 and 3 (rps12B) are cotranscribed with rps7 and ndhB (Fig. 5). Thus, maturation of the mRNA requires multiple processing steps, including cleavage of the precursors prior to trans-splicing to generate mature rps12 mRNA. It was reported in rice and in barley (Kanno and Hirai, 1993; Hübschmann et al., 1996) that the rps12A precursor is first cleaved between clpP1 and rps12 exon 1 and then between rps12 exon 1 and rpl20. Spliced rps12 remains linked to rps7 as a dicistronic mRNA in liverwort and barley chloroplasts (Kohchi et al., 1988; Hübschmann et al., 1996) showing the absence of a processing site between rps12B and rps7. Cis-splicing of intron 2 is the first step in uniting the three rps12 exons (Hübschmann et al., 1996), a step that requires the plastid-encoded maturase, MatK (Zoschke et al., 2010). Trans-splicing of the rps12A and rps12B transcripts is required to join exon 1 to exons 2 and 3. This step is the most severely affected of all splicing events in emb2654, being reduced about 70-fold (Fig. 4) in the partially complemented mutant material compared to wild type. The reduced efficacy (∼6-fold) of rps12 intron 2 splicing also observed in emb2654 (Fig. 4) does not appear to explain the much greater reduction in the efficacy of splicing of rps12 intron 1. In barley mutants lacking plastid translation (and thus MatK), where intron 2 is not spliced at all, trans-splicing of intron 1 still occurs, albeit at a reduced rate (Hübschmann et al., 1996). It seems probable therefore that the primary defect explaining the embryo-lethal phenotype of emb2654 is the failure to trans-splice intron 1 of rps12, leading to a failure to synthesize Rps12 and to assemble ribosomal 30S subunits and a consequential loss of plastid translation.
EMB2654 joins three other proteins known to be involved in this trans-splicing reaction. The CAF2/CRS2 complex is required for splicing of many plastid group II introns, including rps12 intron 1 (Ostheimer et al., 2003). PPR4, the only PPR protein in plants known to carry both PPR motifs and an RNA recognition motif domain is, like EMB2654, specifically implicated in splicing of rps12 intron 1 (Schmitz-Linneweber et al., 2006). Although these proteins were originally identified in maize, putative homologs of all three proteins exist in Arabidopsis. The Arabidopsis CAF2 and CRS2 homologs have similar roles in plastid intron splicing to their maize counterparts (Asakura and Barkan, 2006; Asakura et al., 2008). The function of the putative Arabidopsis homolog of PPR4 (At5g04810) is unknown, but the seed development phenotypes of plants heterozygous for mutations in this gene are consistent with it being essential for embryogenesis (Savage et al., 2013), so a conserved role in rps12 splicing is possible. There may be many more factors involved in this trans-splicing event; in Chlamydomonas, 14 genetic loci were identified as being required for trans-splicing of psaA transcripts (Goldschmidt-Clermont et al., 1990).
Possible Roles of EMB2654 in Trans-Splicing
Bioinformatic prediction of the target preferences of EMB2654 based on the known correlations between the amino acids at key positions within PPR motifs and aligned RNA nucleotides (Barkan et al., 2012) highlighted a sequence near the end of rps12 intron 1a. Binding to this site was confirmed by the discovery of a small RNA “footprint” overlapping this sequence, indicative of nuclease protection by a bound protein (or proteins; Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012a) and by in vitro electrophoretic mobility shift assays (Fig. 6). Despite the discovery of many RNA binding proteins implicated in plant organellar intron splicing (for review, see de Longevialle et al., 2010), their roles have remained rather unclear, in large part because the sequences they bind to have not been identified. For example, it is not known how PPR4, CAF2, or CRS2 bind to rps12 intron 1 or even which intron half they associate with (Schmitz-Linneweber et al., 2006). Therefore, knowing where EMB2654 binds to the intron sequence is potentially an important breakthrough for understanding how splicing proceeds.
The first intron of rps12 can be folded into six typical group II domains (I–VI), radiating from a central wheel (Michel and Dujon, 1983). A folded structure was proposed for the Marchantia and tobacco rps12 intron 1 (Kohchi et al., 1988) that is reproduced for the Arabidopsis sequence in Figure 7. Domains I, II, and one strand of domain III are downstream of the rps12A exon and the other strand of domain III, as well as domains IV to VI, are upstream of the first exon of rps12B (Fig. 7). We aligned the sequences of both parts of rps12 intron 1 with those of tobacco and could place all the domains on the sequence (see Supplemental Fig. S1). As noted for tobacco and liverwort (Kohchi et al., 1988), the ends of the two intron halves have complementary sequences that can base-pair (Fig. 7). The EMB2654 binding site is close to the 3′ end of intron 1a, overlapping with the longest predicted single-stranded region. What might the role of this binding be in the splicing process? Other PPR proteins binding to the 3′ ends of mRNAs in plant organelles help protect and stabilize the RNA by preventing degradation by 3′-5′ exonuclease activity (Pfalz et al., 2009). We think that this is unlikely in this instance, because the RNA is not destabilized in the absence of EMB2654 (in fact it accumulates to a much higher level) and because the binding site is not right at the extremity of the RNA; there are at least 27 nucleotides 3′ of the last nucleotide predicted to be contacted by EMB2654 in the small RNA footprint that accumulates in wild type. Instead, we think that the changes in the patterns of small RNA footprints reveal that the function of EMB2654 is to promote the formation of a protein/RNA complex that covers the termini of both intron halves, necessitating their interaction. In the absence of EMB2654, not only is protection of the sequence bound by the protein lost, but also protection of the sequence immediately 3′ (within the intron 1a footprint) that is not bound by EMB2654 (as shown in Fig. 6). Furthermore, protection of the strong footprint at the 5′ end of intron 1b is also lost, suggesting that it is dependent upon binding of EMB2654 intron 1a. Hence, binding of EMB2654 to intron 1a must be an early step in the formation of the splicing complex, required for formation of the structures that protect the termini of both intron halves, and that are presumably subsequently involved in splicing. It is unlikely that these structures are only formed of RNA; probably other proteins, such as PPR4 (Schmitz-Linneweber et al., 2006), are involved too. Unfortunately, due to the presence of an additional RNA-binding RNA recognition motif domain and some rather nonstandard PPR motif sequences, we are unable to predict where PPR4 might bind. Subsequently, more general factors such as the CAF2/CRS2 complex are likely to bind (Ostheimer et al., 2003). The general strategy employed here of using partial complementation of emb mutants combined with computational prediction of binding sites and RNA-seq analysis to reveal changes in protein/RNA associations is a powerful and widely usable method to characterize the molecular functions of the numerous RNA-binding proteins with essential roles in plastid biogenesis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) T-DNA insertion lines CS24054 (emb2654) and CS16176 (emb2394) were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). The heterozygous lines were sown on half-strength Gamborg B5 medium supplemented with 0.5% Suc in a 22°C controlled environment room with 16 h photoperiod. Plants were transferred to soil after 2 weeks and genotyped by PCR.
Partial Complementation of emb2654 and emb2394
A 2411-bp fragment of the (ABI3; AT3G24650) promoter was amplified using a HindIII restriction site forward primer 5′-GCATCAAGCTTCAACAAACGACTAGTACTGATATATACATC and a AvrII restriction site reverse primer 5′-GCATCCCTAGGCGTTGAAGTGGAAATGAAACAATAAACTAG, cloned into the pH7WG expression vector and transferred into One Shot ccdB Survival 2 T1R Competent Cells (Invitrogen) according to the manufacturer’s instructions.
The emb2654 cDNA was amplified with attB recombination sites (forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTACCGTTACCAATTTCAAG and reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACAACAATGTGCCTTCGTG). Likewise, the emb2394 cDNA was amplified with attB recombination sites (forward primer 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCTTCCTCACTCGTCTCATCT and reverse primer 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCATTTCTTCTTTCCAGCTTTTCC). PCR was performed with Phusion high-fidelity DNA polymerase (BioLabs) according to manufacturer’s instructions. The PCR products were purified using QIAquick gel extraction kit (Qiagen), cloned to pDONR207 (Invitrogen) using Gateway BP Clonase II enzyme mix (Invitrogen), and transformed into Escherichia coli competent cells (DH5α). The plasmids with correct sequences were cloned into the Gateway-compatible expression vector pH7WG containing the ABI3 promoter and T35S terminator using Gateway LR Clonase II enzyme mix (Invitrogen) according to manufacturer’s instructions and transformed into DH5α competent cells. Positive clones were confirmed by PCR, and emb2654 and emb2394 mutants were transformed with the corresponding construct using Agrobacterium tumefaciens C58C1(pCH32) (Tai et al., 1999) by floral dip (Clough and Bent, 1998). Transformed seedlings were double-selected on Gamborg B5 media containing 0.5% Suc, Basta (to select for the original mutation-causing T-DNA insertion), and hygromycin B (to select for the complementing ABI3-driven construct). Selected seedlings were subsequently genotyped in order to confirm that plants homozygous for the emb mutations could be easily recognized by the visual phenotype (Fig. 1). For the experiments described here, T3 seeds collected from EMB/emb T2 plants were used, with the homozygous mutant plants selected visually after germination on agar plates and transferred to liquid medium in a hydroponics system.
Protein Extraction and Western-Blot Analysis
T3 generations of rescued mutants were sown on Gamborg B5 medium as previously described. Leaves of 3-week-old partially complemented ABI3pro:EMB2654 and ABI3pro:EMB2394 seedlings were snap frozen in liquid nitrogen and ground. The total protein was acetone precipitated, resuspended and separated by SDS-PAGE, transferred to PVDF membrane (Bio-Rad) and incubated with specific antibodies raised against the plastid proteins ATPG (ATP synthase subunit; McCormac and Barkan, 1999), RPL4, RPS1, Rps12, PSBO (photosystem II subunit), PC (plastocyanin), PetA (cytochrome b6/f subunit), AtpF (Agrisera), and ACT8 (actin; Sigma-Aldrich). Signals were detected using a BM chemiluminescence Western Blotting kit (Roche), and visualized using a GE Healthcare ImageQuant RT ECL analyzer.
qPCR Analysis of Plastid Transcripts
Leaf material of 3-week-old T3 seedlings were snap frozen in liquid nitrogen, and total RNA was extracted using the RNeasy plant mini kit (Qiagen) and further treated with RNase-free DNase (Qiagen) according to the manufacturer’s instructions. DNA-free RNA (1.5 μg) was used for first-strand cDNA synthesis using the SuperScript III first-strand synthesis system for RT-PCR (Invitrogen) according to the manufacturer’s instructions. The qPCR assay was performed in a LightCycler480 instrument (Roche Diagnostics) using LightCycler 480 SYBR Green I Master Mix (Roche Diagnostics) in a 5-µL reaction volume with the following thermal cycling program: 95°C for 10 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s. The primers used for the splicing test and the full transcriptome analysis were previously described (Chateigner-Boutin et al., 2008). Two biological repeats were analyzed and each sample run in triplicate. Data were analyzed using the LightCycler 480 software release 1.5.0 (Roche Diagnostics).
Sequencing
Leaf material of 3-week-old seedlings of partially complemented emb2654 plants containing ABI3pro:EMB2654 and wild-type grown on Gamborg B5 medium were snap-frozen in liquid nitrogen and their total RNA extracted using miRNeasy Mini Kit (Qiagen). Gel purification, sequencing of small RNAs, and sequence analysis was done as described in Wu et al. (2016).
Protein Expression in E. coli and RNA Electromobility Shift Assay (REMSA)
The EMB2654 cDNA lacking the region encoding the N-terminal plastid targeting sequence was amplified using the primers EMB2654-attB-F (5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGATTCGGG-TTGAGAACGACCGG) and At2G41720-attB-R (5′-GGGGACCAC-TTTGTACAAGAAAGCTGGGTCTTACAACAATGTGCCTCGTG) and cloned into the expression vector pETG-41K (EMBL). The protein containing an N-terminal 6× His tag was expressed in the E. coli C41 (BL21) strain and purified using Bio-Rad Nuvia resin. REMSA was performed as described previously (Schallenberg-Rüdinger et al., 2013; Kindgren et al., 2015) with a few modifications. Briefly, 10 μL binding buffer consisting of 1× THE (34 mm Tris, 66 mm HEPES, and 0.1 mm EDTA, pH 8.3) with 200 mm NaCl, 5 mm DTT, 5 mg/mL heparin, and 0.1 mg/mL BSA were mixed with 5 μL protein dilution and incubated at room temperature for 10 min. 5′-Fluorescein-labeled or unlabeled probes (Sigma-Aldrich) were heated at 94°C for 2 min and cooled on ice for 5 min. Denatured probes (final concentration 1 nm each) were then added to the binding reaction for a total reaction volume of 25 μL. The reactions were incubated at 25°C for 15 min, loaded onto a 5% native acrylamide, and run at 4°C. The gels were imaged with a Typhoon Trio imager (GE Healthcare). Fluorescein-labeled oligonucleotides were excited at 488 nm and detected through a 520-nm band-pass filter.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of rps12 intron 1a and 1b from N. tabacum and Arabidopsis.
Supplemental Table S1. Potential binding sites for EMB2654.
Supplemental Table S2. Primers used in this work.
Acknowledgments
We thank Prof. Alice Barkan (University of Oregon) for the gift of the anti-ATPG antibody.
Glossary
- PEP
plastid-encoded (cyanobacterial-type) RNA polymerase
Footnotes
This work was supported by grants from the Australian Research Council to I.S. (CE140100008 and FL140100179) and K.A.H. (DE120101117). H.R. was supported by a PhD stipend from the Deutscher Akademischer Austauschdienst.
Articles can be viewed without a subscription.
References
- Asakura Y, Barkan A (2006) Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol 142: 1656–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakura Y, Bayraktar OA, Barkan A (2008) Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA 14: 2319–2332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiychuk E, Vandepoele K, Wissing J, Garcia-Diaz M, De Rycke R, Akbari H, Joubès J, Beeckman T, Jänsch L, Frentzen M, et al. (2011) Plastid gene expression and plant development require a plastidic protein of the mitochondrial transcription termination factor family. Proc Natl Acad Sci USA 108: 6674–6679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (1993) Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2004). Intron splicing in plant organelles. In Daniell H., Chase C.D., eds., Molecular Biology and Biotechnology of Plant Organelles. Springer, Dordrecht, The Netherlands, pp 295–322. [Google Scholar]
- Barkan A. (2011) Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins KP (2007) The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA 13: 55–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet 8: e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Small I. (2014). Pentatricopeptide repeat proteins in plants. Ann Rev Plant Biol 65: 415–442. [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol Cell Biol 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant N, Lloyd J, Sweeney C, Myouga F, Meinke D (2011) Identification of nuclear genes encoding chloroplast-localized proteins required for embryo development in Arabidopsis. Plant Physiol 155: 1678–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H, Pérez-Pérez JM, Micol JL (2011) Uncovering the post-embryonic functions of gametophytic- and embryonic-lethal genes. Trends Plant Sci 16: 336–345 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, de Longevialle AF, Fujii S, Small I (2011) OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript rpoC1. Plant J 65: 532–542 [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, de la Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, et al. (2008) CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. Plant J 56: 590–602 [DOI] [PubMed] [Google Scholar]
- Cheng S, Gutmann B, Zhong X, Ye Y, Fisher MF, Bai F, Castleden I, Song Y, Song B, Huang J, et al. (2016) Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J 85: 532–547 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Lopez-Obando M, Heurtevin L, Bernard C, Martin K, Berthomé R, Lurin C (2013) Systematic study of subcellular localization of Arabidopsis PPR proteins confirms a massive targeting to organelles. RNA Biol 10: 1557–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukras AR, Southworth DR, Brunelle JL, Culver GM, Green R (2003) Ribosomal proteins S12 and S13 function as control elements for translocation of the mRNA:tRNA complex. Mol Cell 12: 321–328 [DOI] [PubMed] [Google Scholar]
- Davies C, Bussiere DE, Golden BL, Porter SJ, Ramakrishnan V, White SW (1998) Ribosomal proteins S5 and L6: High-resolution crystal structures and roles in protein synthesis and antibiotic resistance. J Mol Biol 279: 873–888 [DOI] [PubMed] [Google Scholar]
- Delannoy E, Fujii S, Colas des Francs-Small C, Brundrett M, Small I (2011) Rampant gene loss in the underground orchid Rhizanthella gardneri highlights evolutionary constraints on plastid genomes. Mol Biol Evol 28: 2077–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despres B, Delseny M, Devic M (2001) Partial complementation of embryo defective mutations: A general strategy to elucidate gene function. Plant J 27: 149–159 [DOI] [PubMed] [Google Scholar]
- Drescher A, Ruf S, Calsa T Jr., Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104 [DOI] [PubMed] [Google Scholar]
- de Longevialle AF, Small ID, Lurin C (2010) Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol Plant 3: 691–705 [DOI] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Giacomelli L, Ytterberg AJ, Peltier J-B, Rudella A, Sun Q, Wijk KJV (2004) In-depth analysis of the thylakoid membrane proteome of Arabidopsis thaliana chloroplasts: New proteins, new functions, and a plastid proteome database. Plant Cell 16: 478–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. (2003) Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185: 5673–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt-Clermont M, Girard-Bascou J, Choquet Y, Rochaix JD (1990) Trans-splicing mutants of Chlamydomonas reinhardtii. Mol Gen Genet 223: 417–425 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T, Hess WR, Börner T (1996) Impaired splicing of the rps12 transcript in ribosome-deficient plastids. Plant Mol Biol 30: 109–123 [DOI] [PubMed] [Google Scholar]
- Kanno A, Hirai A (1993) A transcription map of the chloroplast genome from rice (Oryza sativa). Curr Genet 23: 166–174 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kindgren P, Yap A, Bond CS, Small I (2015) Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. Plant Cell 27: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kode V, Mudd EA, Iamtham S, Day A (2005) The tobacco plastid accD gene is essential and is required for leaf development. Plant J 44: 237–244 [DOI] [PubMed] [Google Scholar]
- Kohchi T, Umesono K, Ogura Y, Komine Y, Nakahigashi K, Komano T, Yamada Y, Ozeki H, Ohyama K (1988) A nicked group II intron and trans-splicing in liverwort, Marchantia polymorpha, chloroplasts. Nucleic Acids Res 16: 10025–10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T, Shinohara K, Yamada K, Sasaki Y (1996) Acetyl-CoA carboxylase in higher plants: Most plants other than gramineae have both the prokaryotic and the eukaryotic forms of this enzyme. Plant Cell Physiol 37: 117–122 [DOI] [PubMed] [Google Scholar]
- Kroeger TS, Watkins KP, Friso G, van Wijk KJ, Barkan A (2009) A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc Natl Acad Sci USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Maliga P (2003) The plastid clpP1 protease gene is essential for plant development. Nature 425: 86–89 [DOI] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Liere K, Börner T (2007). Transcription and transcriptional regulation in chloroplasts. In Bock R., ed, Cell and Molecular Biology of Plastids, Springer-Verlag, Heidelberg, Germany, pp 121–174 [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL (2011) ViennaRNA package 2.0. Algorithms Mol Biol 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac DJ, Barkan A (1999) A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11: 1709–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC (1994) Leafy cotyledon mutants of Arabidopsis. Plant Cell 6: 1049–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Sussex IM (1979) Embryo-lethal mutants of Arabidopsis thaliana. A model system for genetic analysis of plant embryo development. Dev Biol 72: 50–61 [DOI] [PubMed] [Google Scholar]
- Michel F, Dujon B (1983) Conservation of RNA secondary structures in two intron families including mitochondrial-, chloroplast- and nuclear-encoded members. EMBO J 2: 33–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moschopoulos A, Derbyshire P, Byrne ME (2012) The Arabidopsis organelle-localized glycyl-tRNA synthetase encoded by EMBRYO DEFECTIVE DEVELOPMENT1 is required for organ patterning. J Exp Bot 63: 5233–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostheimer GJ, Williams-Carrier R, Belcher S, Osborne E, Gierke J, Barkan A (2003) Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J 22: 3919–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N, Wang Y, Meinke D (2014) Natural variation in sensitivity to a loss of chloroplast translation in Arabidopsis. Plant Physiol 166: 2013–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Bayraktar OA, Prikryl J, Barkan A (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME (2008) Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development 135: 1315–1324 [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A (2000) EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Rogalski M, Ruf S, Bock R (2006) Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P (2012) Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J 72: 922–934 [DOI] [PubMed] [Google Scholar]
- Ruwe H, Schmitz-Linneweber C (2012) Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res 40: 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage LJ, Imre KM, Hall DA, Last RL (2013) Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS One 8: e73291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg-Rüdinger M, Kindgren P, Zehrmann A, Small I, Knoop V (2013) A DYW-protein knockout in Physcomitrella affects two closely spaced mitochondrial editing sites and causes a severe developmental phenotype. Plant J 76: 420–432 [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Zhang D, Guan Z, Liu Y, Yang Z, Yang Y, Wang X, Wang Q, Zhang Q, Fan S, et al. (2016) Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat Commun 7: 11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeno Y, Uchiumi T, Nomura T (2016) Involvement of ribosomal protein L6 in assembly of functional 50S ribosomal subunit in Escherichia coli cells. Biochem Biophys Res Commun 473: 237–242 [DOI] [PubMed] [Google Scholar]
- Shikanai T, Shimizu K, Ueda K, Nishimura Y, Kuroiwa T, Hashimoto T (2001) The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol 42: 264–273 [DOI] [PubMed] [Google Scholar]
- Shoji S, Dambacher CM, Shajani Z, Williamson JR, Schultz PG (2011) Systematic chromosomal deletion of bacterial ribosomal protein genes. J Mol Biol 413: 751–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR (2010) Chloroplast RNA metabolism. Annu Rev Plant Biol 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A (2004) Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Tai TH, Dahlbeck D, Clark ET, Gajiwala P, Pasion R, Whalen MC, Stall RE, Staskawicz BJ (1999) Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc Natl Acad Sci USA 96: 14153–14158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller N, Bock R (2014) The translational apparatus of plastids and its role in plant development. Mol Plant 7: 1105–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis JN, Ayliffe MA, Huang CY, Martin W (2004) Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat Rev Genet 5: 123–135 [DOI] [PubMed] [Google Scholar]
- Toivonen JM, Boocock MR, Jacobs HT (1999) Modelling in Escherichia coli of mutations in mitoribosomal protein S12: Novel mutant phenotypes of rpsL. Mol Microbiol 31: 1735–1746 [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C (2006) The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol 23: 1324–1338 [DOI] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D (2011) The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol 76: 273–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams-Carrier R, Kroeger T, Barkan A (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14: 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD (1992) Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA 89: 10648–10652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Liu S, Ruwe H, Zhang D, Melonek J, Zhu Y, Hu X, Gusewski S, Yin P, Small ID, et al. (2016) SOT1, a pentatricopeptide repeat protein with a small MutS-related domain, is required for correct processing of plastid 23S-4.5S rRNA precursors in Arabidopsis thaliana. Plant J 85: 607–621 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 50 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28466–28482 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, von Knoblauch K, Subramanian AR (2000) The plastid ribosomal proteins. Identification of all the proteins in the 30 S subunit of an organelle ribosome (chloroplast). J Biol Chem 275: 28455–28465 [DOI] [PubMed] [Google Scholar]
- Zhelyazkova P, Hammani K, Rojas M, Voelker R, Vargas-Suárez M, Börner T, Barkan A (2012a) Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res 40: 3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T (2012b) The primary transcriptome of barley chloroplasts: Numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24: 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R, Nakamura M, Liere K, Sugiura M, Börner T, Schmitz-Linneweber C (2010) An organellar maturase associates with multiple group II introns. Proc Natl Acad Sci USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]