The tomato genome encodes 40 ubiquitin-conjugating enzymes (E2), among which members of group III are required for the suppression of host immunity by the AvrPtoB effector and for plant pattern-triggered immunity.
Abstract
Of the three classes of enzymes involved in ubiquitination, ubiquitin-conjugating enzymes (E2) have been often incorrectly considered to play merely an auxiliary role in the process, and few E2 enzymes have been investigated in plants. To reveal the role of E2 in plant innate immunity, we identified and cloned 40 tomato genes encoding ubiquitin E2 proteins. Thioester assays indicated that the majority of the genes encode enzymatically active E2. Phylogenetic analysis classified the 40 tomato E2 enzymes into 13 groups, of which members of group III were found to interact and act specifically with AvrPtoB, a Pseudomonas syringae pv tomato effector that uses its ubiquitin ligase (E3) activity to suppress host immunity. Knocking down the expression of group III E2 genes in Nicotiana benthamiana diminished the AvrPtoB-promoted degradation of the Fen kinase and the AvrPtoB suppression of host immunity-associated programmed cell death. Importantly, silencing group III E2 genes also resulted in reduced pattern-triggered immunity (PTI). By contrast, programmed cell death induced by several effector-triggered immunity elicitors was not affected on group III-silenced plants. Functional characterization suggested redundancy among group III members for their role in the suppression of plant immunity by AvrPtoB and in PTI and identified UBIQUITIN-CONJUGATING11 (UBC11), UBC28, UBC29, UBC39, and UBC40 as playing a more significant role in PTI than other group III members. Our work builds a foundation for the further characterization of E2s in plant immunity and reveals that AvrPtoB has evolved a strategy for suppressing host immunity that is difficult for the plant to thwart.
Ubiquitination as a major posttranslational modification of proteins in eukaryotes has emerged in recent years as an important regulatory mechanism underlying plant innate immunity (Zeng et al., 2006; Fu et al., 2012; Marino et al., 2012; Li et al., 2014). The ubiquitination process involves a consecutive, three-step enzymatic cascade that is catalyzed by three different classes of enzymes: ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin ligase (E3) enzymes. The first step of the process activates ubiquitin, a highly conserved 76-amino acid protein, in an ATP-dependent manner by attaching ubiquitin to an E1 enzyme. The activated ubiquitin is then transferred from the E1 to the Cys residue at the active site of an E2 conjugating enzyme. An E3 ligase then recruits the substrate protein to the E2 to transfer the ubiquitin molecule from E2 to the substrate. Through the action of E1, E2, and E3, ubiquitin is covalently attached usually to the Lys residue of a substrate through an isopeptide bond (Hershko and Ciechanover, 1998). Of the three enzymes involved in ubiquitination, E3 ubiquitin ligases have been the focus of many studies due to their key role in determining substrate specificity for the ubiquitination process. Dozens of E3 enzymes have been implicated so far in either pattern-triggered immunity (PTI) or effector-triggered immunity (ETI; Cheng and Li, 2012; Marino et al., 2012; Li et al., 2014). By contrast, E2 ubiquitin-conjugating enzymes were often mistakenly considered to play an auxiliary role in the ubiquitination process, and few E2 enzymes have been investigated in plants. In fact, E2 enzymes have been found to govern the processivity and topology of polyubiquitin chain formation and, thus, to determine the fate of the modified proteins. In humans and animals, the E2 enzymes have been shown to play a crucial role in regulating both innate and adaptive immunity (Ye and Rape, 2009; Jiang and Chen, 2011).
Similar to humans and animals, plants have evolved a sophisticated innate immune system. It is now known that the plant innate immune system comprises two interlinked layers of defense responses to protect them from attempted pathogen infection. The first layer is termed PTI, which is initiated by the recognition of microbe-associated molecular patterns (MAMPs)/pathogen-associated molecular patterns via cell surface-localized pattern recognition receptors (PRRs; Jones and Dangl, 2006; Macho and Zipfel, 2014). MAMPs are molecules typically associated with a whole class of microbes but are absent in host plants. Several MAMPs and their cognate plant PRR have been identified (Felix et al., 1999; Kunze et al., 2004; Kaku et al., 2006; Kawaharada et al., 2015), among which immunity induced by the recognition of flagellin and the 22-amino acid immunogenic fragment of flagellin, flg22, of bacterial pathogens by the plant PRR FLAGELLIN SENSING2 (FLS2) has been studied extensively. The activation of PTI leads to host responses including production of reactive oxygen species (ROS), activation of mitogen-activated protein kinases, modulation of defense-related gene expression, and deposition of callose at the cell wall (Boller and Felix, 2009; Nguyen et al., 2010). PTI is sufficient to ward off the infection of plants by most microbes; defective plant PRRs or PTI signaling components often result in increased susceptibility of plants to both adapted and nonadapted pathogens (Macho and Zipfel, 2014). To promote colonization, plant pathogens have evolved strategies to evade or suppress PTI by deploying various effectors into the host cell (Zhang et al., 2007; Martin, 2012). However, in a further evolutionary step, some plants have developed intracellular receptors that usually are nucleotide-binding Leu-rich repeat-containing resistance proteins to recognize the presence of specific effectors. Such recognition results in the activation of the second layer of plant defense, termed ETI, which often is accompanied by a rapid programmed cell death (PCD) called the hypersensitive response (HR) at the site of pathogen infection.
Pseudomonas syringae pv tomato (Pst) causes bacterial speck disease on tomato (Solanum lycopersicum) plants that lack genetic resistance. In addition to tomato, Pst also infects the model plant Arabidopsis (Arabidopsis thaliana) and Nicotiania benthamiana, a solanaceous species related to tomato (Katagiri et al., 2002; Wei et al., 2007). Of the various MAMPs associated with Pst, flagellin plays a major role in causing immunity-associated transcriptional changes in tomato (Rosli et al., 2013). To suppress host immune responses, Pst uses a type III secretion system to deliver during the infection process around 30 sequence-diverse effectors into the host cell (Chang et al., 2005; Lindeberg et al., 2006). Many of the effectors have been found to suppress PTI (Li et al., 2005; He et al., 2006; Zhang et al., 2007; Guo et al., 2009). Two of them, AvrPto and AvrPtoB, target FLS2 and the common PTI signaling partner, BRI1-ASSOCIATED RECEPTOR KINASE1 (BAK1), to impede PTI mediated by multiple PRRs (Shan et al., 2008; Martin, 2012). AvrPtoB is a modular protein of which the N-terminal region is recognized by the tomato intracellular Ser/Thr protein kinase Pto that, in conjunction with the nucleotide-binding Leu-rich repeat-containing protein Prf, confers resistance to Pst to tomato (Salmeron et al., 1996; Kim et al., 2002; Abramovitch et al., 2003; Pedley and Martin, 2003). The Pto gene belongs to a clustered small gene family where another member encodes the Fen kinase (Martin et al., 1994; Jia et al., 1997; Pedley and Martin, 2003). Fen does not detect AvrPtoB but recognizes truncated forms or natural variants of AvrPtoB in which the C-terminal domain is absent or ineffective (Abramovitch et al., 2003; Rosebrock et al., 2007). Recognition of AvrPtoB variants by Fen induces a Prf-dependent HR that eventually arrests the growth of pathogens at the site of infection (Abramovitch et al., 2003; Rosebrock et al., 2007). Notably, the C-terminal region of AvrPtoB encodes a structural mimic of a eukaryotic RING/U box domain that has E3 ubiquitin ligase activity (Abramovitch et al., 2006; Janjusevic et al., 2006). AvrPtoB interferes with Fen-mediated ETI by using its C-terminal E3 activity to target Fen for ubiquitination, which leads to degradation of the kinase by the 26S proteasome (Rosebrock et al., 2007). By an unknown mechanism, AvrPtoB also uses its E3 activity to inhibit immunity-associated PCD triggered by various elicitors, including AvrPto/Pto and Avr9/Cf9 (Abramovitch et al., 2003). Additionally, AvrPtoB has been shown to use its E3 activity to ubiquitinate the plant PRRs FLS2 and CERK1 for interference with plant PTI (Göhre et al., 2008; Gimenez-Ibanez et al., 2009). These findings, however, are contrary to the reports that a truncated AvrPtoB lacking the C-terminal E3 activity domain, AvrPtoB1-387, interferes effectively with FLS2- and CERK1-mediated PTI (Xiao et al., 2007; Shan et al., 2008; Zeng et al., 2012).
The ubiquitination of plant immunity-related components by the E3 activity of AvrPtoB is expected to require the cooperation of E2 enzyme(s). There are two possible sources of these cognate E2 enzyme(s). First, it is possible that one or more effectors translocated into the host cell by Pst might serve as the cognate E2. However, of the approximately 30 effectors delivered into the host cell by the type III secretion system, none has been reported to date to function as a ubiquitin E2 enzyme. Additionally and importantly, AvrPtoB was found to be capable of suppressing PCD elicited by HopPsyA on tobacco (Nicotiana tabacum) and Arabidopsis plants when the nonpathogen Pseudomonas fluorescens carrying the cosmid pHIR11, which expresses and secretes effectors AvrPtoB and HopPsyA only, was used for inoculation (Jamir et al., 2004). Furthermore, AvrPtoB uses its E3 activity to suppress immunity-associated PCD and target Fen for degradation when it is transiently coexpressed with PCD elicitor or Fen in N. benthamiana (Abramovitch et al., 2003; Ntoukakis et al., 2009). These observations imply that effectors of Pst do not act as the cognate E2 for AvrPtoB in suppressing host immunity. Instead, they support the notion that the AvrPtoB E3 ligase exploits host E2 enzymes. However, the plant E2 enzyme(s) that work with AvrPtoB in suppressing host immunity have not been identified to date.
A given genome usually encodes dozens of ubiquitin E2 enzymes (Vierstra, 2003; Kraft et al., 2005). The study of this class of ubiquitination-related enzyme in plants, particularly their role in plant innate immunity, is very limited. To address this knowledge gap, we have taken advantage of the recent availability of a high-quality tomato genome sequence and a draft genome sequence of N. benthamiana to identify at genome scale the tomato and N. benthamiana genes that encode ubiquitin E2 enzymes. We have also taken advantage of the fact that AvrPtoB uses plant E2s to subvert host immunity and used the tools for studying gene function at large scale, such as virus-induced gene silencing (VIGS; Chakravarthy et al., 2010; Bombarely et al., 2012; Tomato Genome Consortium, 2012), to pinpoint a subset of tomato E2 genes that is required for plant PTI.
RESULTS
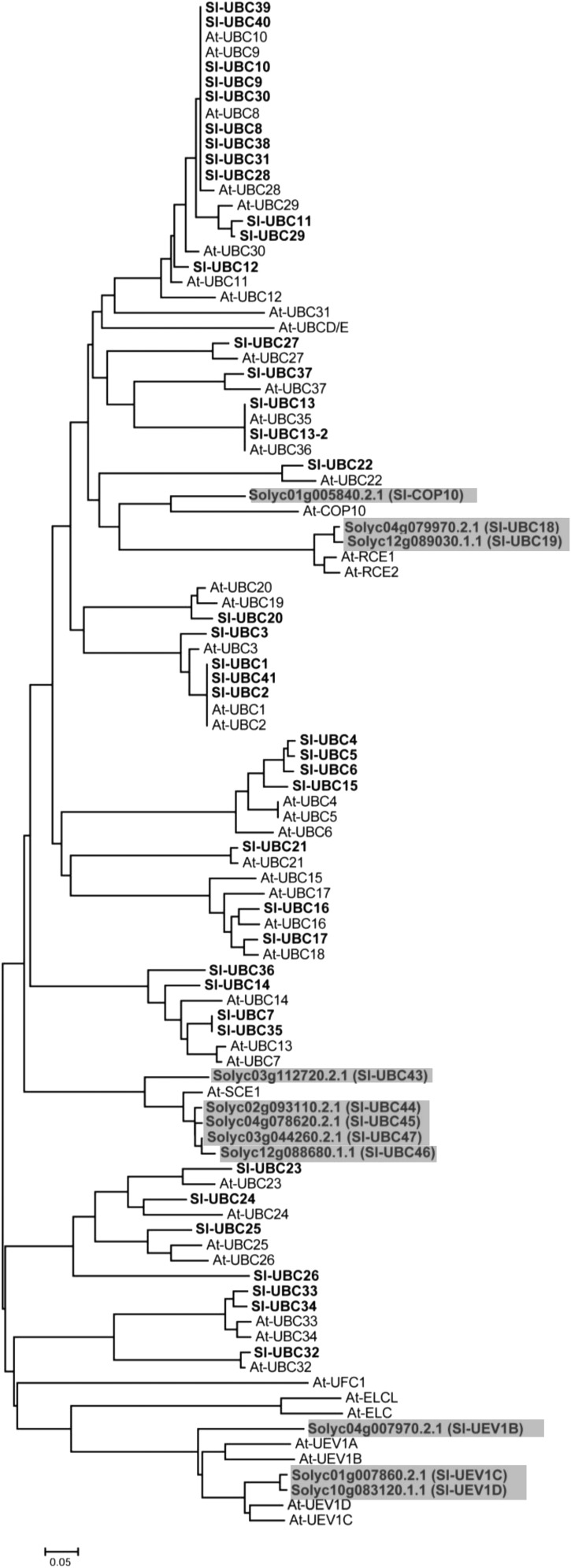
The Tomato Genome Is Predicted to Encode 40 Ubiquitin E2 Proteins That Are Classified into 13 Groups
To study the role of ubiquitin-conjugating enzymes (E2) in plant immunity, we attempted to identify tomato genes encoding ubiquitin E2 enzymes using an array of whole-genome analysis algorithms similar to those described previously (Zeng et al., 2008). The E2 enzymes for ubiquitin and ubiquitin-like modifiers, such as small ubiquitin-like modifier (SUMO) and related to ubiquitin (RUB), possess a 140- to 200-amino acid sequence- and structure-conserved catalytic core called the ubiquitin-conjugating (UBC) domain in which a Cys residue is present at the active site (Vierstra, 2012). Thus, we first screened the tomato genome for genes encoding UBC domain-containing proteins, which led to the identification of 51 tomato genes (Fig. 1). Phylogenetic analysis of tomato and Arabidopsis UBC domain-containing proteins as well as BLAST search of the NCBI database indicated that, among the 51 tomato genes, 40 encode ubiquitin E2 proteins, four encode ubiquitin-conjugating enzyme variant (UEV) proteins (SlUEV1B, SlUEV1C, SlUEV1D, and SlCOP10), two encode putative rubylation E2 enzymes (SlUBC18 and SlUBC19), and five encode putative SUMO E2 enzymes (SlUBC43–SlUBC47; Fig. 1; Table I; Supplemental Table S1; Kraft et al., 2005; Camacho et al., 2009). To be consistent with the nomenclature of E2 enzymes in other plant species, we largely developed names for the tomato UBC domain-containing proteins based on their association with the corresponding Arabidopsis homolog in the phylogenetic tree (Fig. 1; Kraft et al., 2005; Zhao et al., 2013). However, we named the tomato counterpart of Arabidopsis UBC35 and UBC36 as SlUBC13 and SlUBC13-2, respectively, to emphasize that they are UBC13-type ubiquitin E2 enzymes that catalyze exclusively Lys-63-linked ubiquitination (Mural et al., 2013). The phylogenetic analysis also indicated that most of the tomato UBC domain-containing proteins have a corresponding homolog in Arabidopsis. We did not identify the tomato counterpart for Arabidopsis ELC, ELCL, UFC1, UBCD/E, and the E2 variant UEV1A (Fig. 1; Kraft et al., 2005; Wen et al., 2008). To determine the relationship of the tomato UBC domain-containing proteins with those of vertebrates, we also generated the phylogenies of tomato and human UBC domain-containing proteins (van Wijk and Timmers, 2010). Except for tomato UBC20, UBC22, UBC27, UBC32, and COP10, most tomato UBC domain-containing proteins shared highest similarity with another tomato E2 protein in the tree (Supplemental Fig. S1).
Figure 1.
Phylogenetic analysis of tomato E2s. The phylogenetic tree shows the UBC domain-containing proteins from tomato and Arabidopsis. The SUMO and RUB E2s as well as UBC-like UEV proteins from tomato and Arabidopsis are included for comparison, with members from tomato being highlighted in gray. The unrooted phylogenetic tree was generated by the neighbor-joining method using the MEGA6 program with 1,000 bootstrap trials (Saitou and Nei, 1987; Tamura et al., 2013). The phylogenetic tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
Table I. Summary of tomato UBC domain-containing proteins, their nomenclature, classification, activity as ubiquitin E2 enzymes, and specificity toward immunity-associated E3 ubiquitin ligases in an in vitro ubiquitination assay.
nd, This protein was not tested for the assay.
| Chromosome Locus | Gene Name | Group No. | Protein in E. coli | E2 Activity | E2/E3 Specificity for AvrPtoB |
|---|---|---|---|---|---|
| Solyc06g072570.2.1 | UBC27 | I | Yes | Yes | No |
| Solyc06g070980.2.1 | UBC1 | II | Yes | Yes | No |
| Solyc03g113100.2.1 | UBC2 | II | Yes | Yes | No |
| Solyc02g067420.2.1 | UBC3 | II | Yes | Yes | No |
| Solyc02g087750.2.1 | UBC41 | II | Yes | Yes | No |
| Solyc12g056100.1.1 | UBC8 | III | Yes | Yes | Yes |
| Solyc08g008220.2.1 | UBC9 | III | Yes | Yes | Yes |
| Solyc05g050230.2.1 | UBC10 | III | Yes | Yes | Yes |
| Solyc03g033410.2.1 | UBC11 | III | Yes | Yes | Yes |
| Solyc07g066080.2.1 | UBC12 | III | Yes | Yes | Yes |
| Solyc10g011740.2.1 | UBC28 | III | Yes | Yes | Yes |
| Solyc02g083570.2.1 | UBC29 | III | Yes | Yes | Yes |
| Solyc03g007470.2.1 | UBC30 | III | Yes | Yes | Yes |
| Solyc01g095490.2.1 | UBC31 | III | Yes | Yes | Yes |
| Solyc08g081950.2.1 | UBC38 | III | Yes | Yes | Yes |
| Solyc06g082600.2.1 | UBC39 | III | Yes | Yes | Yes |
| Solyc06g007510.2.1 | UBC40 | III | Yes | Yes | Yes |
| Solyc12g099310.1.1 | UBC32 | IV | Yes | Yes | No |
| Solyc03g123660.2.1 | UBC33 | IV | Yes | Yes | No |
| Solyc06g063100.2.1 | UBC34 | IV | Yes | Yes | No |
| Solyc05g054550.2.1 | UBC7 | V | Yes | Yes | No |
| Solyc04g011430.2.1 | UBC14 | V | Yes | Yes | No |
| Solyc05g054540.2.1 | UBC35 | V | Yes | Yes | No |
| Solyc09g009720.1.1 | UBC36 | V | Yes | Yes | No |
| Solyc10g012240.2.1 | UBC4 | VI | Yes | Yes | No |
| Solyc01g094810.2.1 | UBC5 | VI | Yes | Yes | No |
| Solyc08g081270.2.1 | UBC6 | VI | Yes | Yes | No |
| Solyc11g071870.1.1 | UBC15 | VI | Yes | Yes | No |
| Solyc11g071260.1.1 | UBC21a | VII | Yes | nd | nd |
| Solyc11g065190.1.1 | UBC20 | VIII | Yes | Yes | No |
| Solyc07g062570.2.1 | UBC13 | IX | Yes | Yes | No |
| Solyc10g007260.2.1 | UBC13-2 | IX | Yes | Yes | No |
| Solyc04g080810.2.1 | UBC16 | X | Yes | Yes | No |
| Solyc02g084760.2.1 | UBC17 | X | Yes | Yes | No |
| Solyc01g111680.2.1 | UBC23a | XI | Yes | nd | nd |
| Solyc02g078210.2.1 | UBC24a | XI | Yes | nd | nd |
| Solyc10g007000.2.1 | UBC25a | XI | Yes | nd | nd |
| Solyc01g079290.1.1 | UBC26a | XI | Yes | nd | nd |
| Solyc10g081160.1.1 | UBC22 | XII | Yes | Yes | No |
| Solyc07g024070.1.1 | UBC37 | XIII | Yes | No | nd |
The recombinant protein of the E2 expressed in E. coli was insoluble.
Sequence analysis indicated that, similar to the domain organization of human E2s, the UBC domain is the only known protein domain detected in all 40 putative tomato E2 proteins except for UBC27 (Supplemental Fig. S2; van Wijk and Timmers, 2010). Tomato UBC27 (SlUBC27) is most homologous to human E2 HIP2 (UBE2K; Supplemental Fig. S1) and Arabidopsis UBC27 (Fig. 1). In addition to the UBC domain, SlUBC27 also contains at its C terminus a ubiquitin-associated (UBA) domain that has been found to mediate protein-protein interactions through binding to ubiquitin molecules (Dikic et al., 2009). Besides the UBC domain, some of the tomato ubiquitin E2 proteins also have N- or C-terminal extensions of amino acids or both (Supplemental Fig. S2). Tomato UBC23, UBC24, UBC25, and UBC26 have long extensions at both N and C termini. By contrast, UBC32, UBC33, UBC34, and UBC37 have a long C-terminal extension only. Further phylogenetic analysis of the 40 putative tomato ubiquitin E2 proteins classified them into 13 groups, among which group III contains 12 highly homologous members and is the largest group (Supplemental Fig. S3). In the Arabidopsis genome, 37 ubiquitin E2 proteins have been identified, and they were classified into 14 and 12 groups, respectively, by different teams (Kraft et al., 2005; Zhao et al., 2013). We numbered the groups of tomato ubiquitin E2s according to the classification of Arabidopsis counterparts by Zhao et al. (2013), with the inclusion of tomato UBC37 as group XIII.
Most Tomato Ubiquitin E2 Proteins Are Enzymatically Active
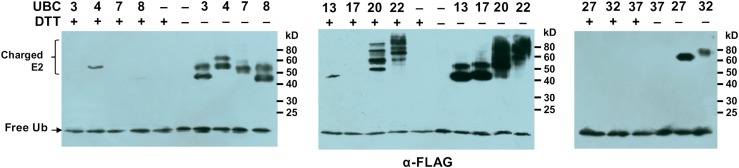
To find out whether the putative tomato E2 genes encode active ubiquitin E2 enzymes, a thioester assay was employed to test their ubiquitin-conjugating activity (Kraft et al., 2005). We observed adequate recombinant protein expression in Escherichia coli for all 40 genes and were able to purify adequate recombinant proteins for 35 tomato E2 genes (Table I; Supplemental Fig. S4). We could not purify sufficient soluble recombinant protein for UBC21, UBC23, UBC24, UBC25, and UBC26 due to their insolubility after expression in E. coli. We also cloned the tomato gene encoding the ubiquitin-activating enzyme (E1), UBA1, and purified the recombinant protein of UBA1 for the thioester assay. In this assay, the E1 enzyme activates the free ubiquitin molecule to form an E1-ubiquitin intermediate. In the presence of an active E2 enzyme, the activated ubiquitin molecule is transferred to the Cys residue at the active site of the E2 to form an E2-ubiquitin adduct through a thioester bond that is sensitive to the thiol-reducing agent dithiothreitol (DTT; Kraft et al., 2005). To examine the enzymatic activity for the 35 putative tomato E2 proteins, we first randomly chose one member from each of the 11 groups to which the 35 E2 proteins belong for the thioester assay (Fig. 2). Except for UBC37, tomato UBC3, UBC4, UBC7, UBC8, UBC13, UBC17, UBC20, UBC22, UBC27, and UBC32 formed adducts with ubiquitin that were lost or significantly reduced in the presence of DTT, indicating that a thioester linkage was formed between ubiquitin and E2 (Fig. 2). These results indicated that these proteins are active ubiquitin E2 enzymes. The thioester assay was then also performed for the other 24 tomato E2 proteins, and all of them were found to possess ubiquitin-conjugating activity (Supplemental Fig. S5A). Additionally, when the Cys residue at the active site of the E2 proteins was mutated to Gly, the E2 proteins lost ubiquitin-conjugating activity (Supplemental Fig. S5B). Therefore, 34 out of the 35 tomato ubiquitin E2 proteins being tested were found to be ubiquitin-conjugating enzymes (E2; Table I).
Figure 2.
Examination of the ubiquitin-conjugating activity of tomato E2s by thioester formation assay. Anti-FLAG western blotting was performed following the thioester assay of different tomato ubiquitin E2s. The reactions of the assay were terminated by adding SDS sample loading buffer in the presence of 100 mm DTT (DTT+) or 4 m urea (DTT−). The formation of DTT-sensitive ubiquitin adducts by tomato E2s is shown as charged E2. The numbers on the right denote the molecular masses of marker proteins in kD. The experiment was repeated two times with similar results.
The Tomato Group III E2 Enzymes Work with AvrPtoB in Catalyzing Ubiquitination
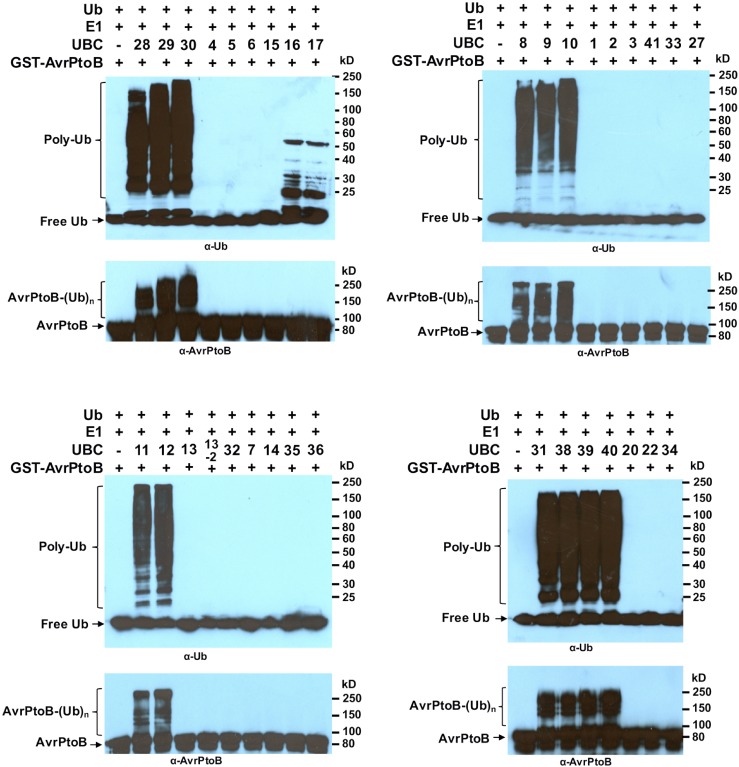
As the first step to pinpoint E2s that are involved in plant immunity, we attempted to identify E2(s) that work with AvrPtoB. Mutations in the RING- and U box-type E3 ligases that affect their in vitro activity also affect in vivo activity (Fang et al., 2000; Xie et al., 2002; Xu et al., 2002; Starita et al., 2013), which suggests that requirements for in vitro and in vivo activity appear to be identical and that the in vitro ubiquitination assay with RING- and U box-type E3 ligases in the absence of a physiological substrate likely truly reflect their in vivo activity (Kraft et al., 2005). Therefore, it is plausible to assume the plant ubiquitin E2 enzyme(s) that work with AvrPtoB in suppressing host immunity also would show specificity toward AvrPtoB in the in vitro ubiquitination reaction. Accordingly, E2-E3 specificity between tomato E2 enzymes and AvrPtoB was examined with an in vitro ubiquitination assay to define the ubiquitin E2 protein that cooperates with AvrPtoB. As shown in Figure 3, in the presence of tomato E1, ubiquitin, AvrPtoB, and the ubiquitination assay buffer, reactions that contained a member of group III E2 enzymes (UBC8–UBC12, UBC28–UBC31, and UBC38–UBC40) produced strong polyubiquitin signal that appeared as a high-Mr smear. Immunoblot analysis using anti-AvrPtoB antibody confirmed the autoubiquitination of AvrPtoB in reactions where a member of group III E2 was present. By contrast, no high-Mr signal was detected in reactions that contained members of other E2s when anti-AvrPtoB antibody was used in the immunoblot. Similarly, except for UBC16 and UBC17, neither of the reactions in which a non-group III E2 was present showed high-Mr signal when anti-ubiquitin antibody was used for immunoblotting.
Figure 3.
AvrPtoB shows specificity toward group III tomato ubiquitin E2s in in vitro ubiquitination assays. Glutathione S-transferase (GST)-AvrPtoB was tested against purified tomato E2 proteins in in vitro ubiquitination assays to determine the specificity of AvrPtoB toward the E2s. The tomato E2 enzyme used in each reaction is indicated above the lane by its UBC number, and the minus marker (‒) denotes the absence of E2 enzyme in the reaction. AvrPtoB-E2 specificity is confirmed by the presence of a high molecular mass smear of ubiquitinated proteins, detected by the anti-ubiquitin antibody. Polyubiquitinated forms of AvrPtoB were detected by the anti-AvrPtoB antibody. This experiment was repeated two times with similar results. The numbers on the right denote the molecular masses of marker proteins in kD.
In reactions that contain UBC16 or UBC17, the high-Mr signal was present when anti-ubiquitin antibody was used, but the signal was absent when anti-AvrPtoB antibody was used in the immunoblot (Fig. 3). This result suggested that UBC16 and UBC17 might be able to catalyze the formation of ubiquitin chains in the absence of an E3 ligase, which is similar to what was observed for Arabidopsis UBC22 (Kraft et al., 2005). To confirm this, we performed in vitro ubiquitination assays in the presence or absence of AvrPtoB, with UBC28 included as a control (Supplemental Fig. S6A). Unlike UBC28, in which a polyubiquitin signal was detected only in the presence of AvrPtoB (Supplemental Fig. S6A, lanes 1 and 2), the reactions containing UBC16 and UBC17 produced polyubiquitin ladders in the absence of AvrPtoB (lanes 3 and 5). The addition of AvrPtoB to the reaction enhanced the signal but did not alter the pattern of the ladders (lanes 4 and 6). These results indicate that tomato UBC16 and UBC17 are capable of catalyzing polyubiquitination in the absence of an E3 and that AvrPtoB does not have specificity toward them but is able to promote their activity. To confirm this, we performed an additional in vitro ubiquitination assay of UBC16 with or without including AvrPtoB and Fen (as controls) in the reactions. We also included a control reaction in which the same volume (μL) of 40% glycerol as that of AvrPtoB was used (because our stocks of both AvrPtoB and Fen recombinant proteins were stored in 40% glycerol). As indicated in Supplemental Figure S6B, no ubiquitin conjugates were detected in the reaction where UBC16 was absent (lane 3). By contrast, polyubiquitin conjugates were detected in the reaction without an E3 ligase (lane 2). The addition of AvrPtoB or Fen to the reaction increased the strength of polyubiquitin signals but did not alter the pattern of the signal (lanes 1 and 5), suggesting that the enhancement of the activity of UBC16 to catalyze polyubiquitination by AvrPtoB is nonspecific. As expected, the 40% glycerol did not alter UBC16 activity significantly (lane 4).
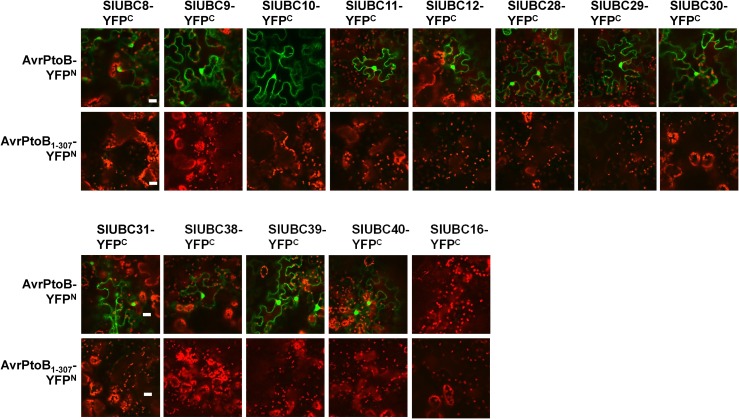
Members of the Group III E2 Enzymes Interact with AvrPtoB in Vivo
The in vitro assays indicated that only group III tomato E2 enzymes act with AvrPtoB to catalyze ubiquitination. To find out whether members of group III E2 enzymes also associate with AvrPtoB in vivo, we employed a bimolecular fluorescence complementation (BiFC) assay to test the interaction of group III E2 with AvrPtoB in N. benthamiana leaves. As shown in Figure 4, full-length AvrPtoB but not the control, the N-terminal AvrPtoB (i.e. the C-terminal E3 domain is removed, AvrPtoB1-307), interacts with all members of group III E2, as green fluorescence was detected when AvrPtoB and a member of group III E2 were coexpressed. By contrast, the control E2 UBC16 failed to interact with AvrPtoB. These data are consistent with the in vitro ubiquitination assays shown in Figure 3 and indicate that the in vivo interaction of AvrPtoB and group III E2s is specific. To further confirm this, we also tested the interaction of AvrPtoB with group III E2s in N. benthamiana protoplasts (Chen et al., 2006). Tomato UBC10 and UBC12 were randomly selected for the assay. As shown in Supplemental Figure S7A, fluorescence was detected in protoplasts where UBC10 or UBC12 was coexpressed with AvrPtoB, indicating that UBC10 and UBC12 interact with AvrPtoB in vivo. No fluorescence was observed in the control experiment where UBC10 or UBC12 was coexpressed with the empty vector (cYFP-EV). Consistently, no fluorescence was observed when UBC16 and AvrPtoB were coexpressed in protoplasts (Supplemental Fig. S7B). However, fluorescence was observed in protoplasts where UBC16 was cotransfected with tomato E1, UBA1, indicating that they interacted as expected (Supplemental Fig. S7B). These results support the finding from the in vitro ubiquitination assay that there is no specificity between UBC16 and UBC17 and AvrPtoB. As mentioned above, we were unable to perform in vitro enzymatic activity assays for tomato UBC21, UBC23, UBC24, UBC25, and UBC26 because we could not purify adequate soluble recombinant proteins. Nevertheless, the BiFC assay can be used to test whether these E2s interact with AvrPtoB in vivo. We thus randomly selected UBC21 and UBC25 to test their interaction with AvrPtoB in the assay. No fluorescence was observed when either UBC21 or UBC25 was cotransfected with AvrPtoB in the protoplasts (Supplemental Fig. S7B). By contrast, fluorescence was detected when UBC21 or UBC25 was coexpressed with UBA1, which suggests that they interact with tomato E1 and, thus, are likely to be true ubiquitin E2 enzymes but without specificity for AvrPtoB. Moreover, the coimmunoprecipitation of UBC29 and UBC30 but not UBC16 (as a control) with AvrPtoB also indicated that AvrPtoB interacts specifically with group III E2 members (Supplemental Fig. S8). Taken together, we conclude that only group III tomato ubiquitin E2 enzymes possess E2-E3 specificity toward AvrPtoB.
Figure 4.
AvrPtoB interacts specifically with tomato group III E2 members in vivo. BiFC was used to detect the interaction of AvrPtoB and group III E2s. Different construct pairs for the BiFC assay were transiently coexpressed in N. benthamiana leaves. Leaves were viewed with the confocal microscope to detect chlorophyll autofluorescence (red color) and green fluorescence (YFP). The control vector expressing N-terminal YFP-fused AvrPtoB1-307 (AvrPtoB1-307-nYFP) and C-terminal YFP-fused SlUBC16 (SlUBC16-cYFP) were used as negative controls for E3 and E2, respectively. The experiment was repeated two times with similar results. Bars = 20 μm.
Group III Ubiquitin E2 Enzymes Are Required for the Suppression of Fen-Mediated PCD by AvrPtoB and AvrPtoB-Promoted Degradation of Fen
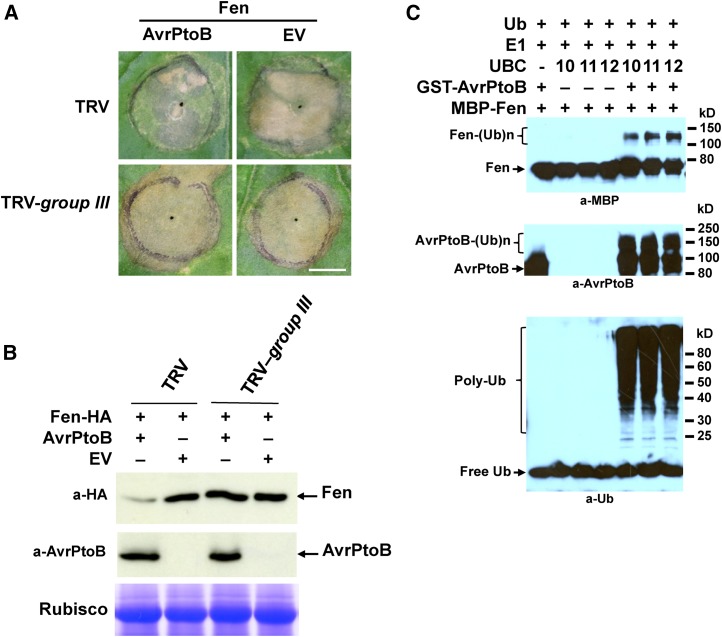
The E2-E3 specificity between the tomato group III E2 enzymes and AvrPtoB suggests that these E2 enzymes are likely required for the E3 activity of AvrPtoB in suppressing host ETI-associated PCD (Abramovitch et al., 2003; Rosebrock et al., 2007). To test this, we used VIGS mediated by the tobacco rattle virus (TRV) vector to knock down the expression of group III E2 genes in N. benthamiana (Caplan and Dinesh-Kumar, 2006; Mural et al., 2013). We used N. benthamiana instead of tomato because it gives higher efficiency and more uniform gene silencing in VIGS and better transient expression of PCD elicitors (Mural et al., 2013). The N. benthamiana genome encodes a corresponding homolog for each of the 40 tomato ubiquitin E2 enzymes (Supplemental Table S1). Additionally, the tomato group III E2 genes and their counterpart from N. benthamiana are highly homologous, sharing approximately 90% to 94% identity in DNA sequence (Supplemental Figs. S9A and S10). The E2 genes also are highly homologous within group III from N. benthamiana, and stretches of DNA sequence longer than 23 nucleotides that are identical among many group III members exist (Supplemental Fig. S9A), which permits the generation of a single TRV vector-based construct that would target all group III E2 genes for silencing (TRV-group III; see “Materials and Methods”). N. benthamiana plants that were infected with TRV-group III displayed greatly reduced expression of group III E2 genes when compared with TRV vector only (TRV)-infected plants, with a relative expression level varying from approximately 60% to 5% depending on the specific gene being analyzed (Supplemental Fig. S9C). By contrast, the expression level of genes encoding nongroup III E2 members UBC16 and UBC17 was not altered in TRV-group III-infected plants, indicating that the silencing of group III E2 genes by TRV-group III is specific (Supplemental Fig. S9C). Compared with TRV-infected plants, N. benthamiana plants infected with TRV-group III displayed slower growth and slightly crinkled leaves, indicating that the group III E2 enzymes also are required for certain aspects of plant development (Supplemental Fig. S9B).
We next tested whether knocking down group III E2 genes affects PCD induced by the overexpression of Fen in N. benthamiana and the degradation of Fen due to being ubiquitinated by AvrPtoB (Rosebrock et al., 2007; Mural et al., 2013). AvrPtoB suppressed PCD induced by the overexpression of Fen on TRV-infected N. benthamiana plants (Fig. 5A). However, knocking down group III E2 genes diminished the suppression of Fen-mediated PCD by AvrPtoB (Fig. 5A). Consistent with the reduced suppression of Fen-mediated PCD by AvrPtoB, no change in the abundance of Fen protein was detected on group III E2 gene-silenced N. benthamiana plants when coexpression of Fen and AvrPtoB was compared with coexpression of Fen and the empty vector (Fig. 5B). By contrast, compared with coexpressing Fen and the empty vector, the abundance of Fen protein was decreased significantly when Fen was coexpressed with AvrPtoB on nonsilenced control plants (Fig. 5B). Consistently, an in vitro ubiquitination assay indicated that members of group III E2 worked with AvrPtoB to ubiquitinate Fen (Fig. 5C). We also coexpressed AvrPtoB and Fen in protoplasts that were prepared from group III E2 gene-silenced and nonsilenced control N. benthamiana plants, respectively. Knocking down group III E2 gene expression in protoplasts led to similar effects on AvrPtoB-promoted degradation of Fen (Supplemental Fig. S11) to what was observed on the leaves of N. benthamiana plants (Fig. 5B). These results indicate that decreased expression of group III E2 genes diminished the suppression of Fen-mediated PCD by AvrPtoB and the AvrPtoB-promoted degradation of Fen. To rule out the possibility that the effect of knocking down group III E2 genes on the suppression of Fen-mediated PCD by AvrPtoB is due to the silencing of group III E2 genes affecting Fen-mediated PCD itself, we tested the induction of PCD by different ETI elicitors as described (Mural et al., 2013) on TRV-infected control plants and TRV-group III-silenced plants. No difference was observed for the occurrence of PCD induced by the elicitors being tested (Supplemental Fig. S12), suggesting that the effect of silencing group III E2 genes on the function of AvrPtoB is specific.
Figure 5.
Silencing of the group III E2 genes diminishes the suppression of Fen-mediated PCD by AvrPtoB and AvrPtoB-promoted degradation of Fen in N. benthamiana. A, Knocking down the expression of group III E2 genes diminished the suppression of Fen-mediated PCD by AvrPtoB in N. benthamiana. Transient coexpression of AvrPtoB and Fen-HA in fully expanded N. benthamiana leaves was performed by syringe infiltrating Agrobacterium tumefaciens carrying T-DNA with the AvrPtoB and Fen genes. Transient coexpression of the empty pBTEX vector (EV) and Fen was used as a control. The infiltrated area of each leaf is outlined by the black circle. TRV-group III was used to knock down the expression group III E2 genes. TRV vector only was used as a control. Photographs were taken on day 4 after infiltration. The experiment was repeated two times with similar results. Bar = 1 cm. B, The group III E2s were required for AvrPtoB-promoted degradation of Fen. Transient coexpression of AvrPtoB and Fen-HA in N. benthamiana leaves was performed as described in A. The leaf samples were collected at 36 h after infiltration. Western blotting was performed using anti-HA and anti-AvrPtoB antibody, respectively, to detect Fen-HA and AvrPtoB. Staining of Rubisco subunits by Coomassie Blue demonstrated equal loading. The experiment was repeated two times with similar results. C, Fen was ubiquitinated in the presence of AvrPtoB and a member of group III E2s. The in vitro ubiquitination assays were performed with recombinant E1, E2, AvrPtoB, ubiquitin (Ub), and Fen. Reactions without E2 or AvrPtoB (as denoted with ‒) were used as controls. Polyubiquitinated forms of AvrPtoB [AvrPtoB-(Ub)n] confirmed the presence of E3 ligase activity.
Knocking Down the Group III Ubiquitin E2 Genes Decreases the Induction of PTI-Activated Reporter Genes by flg22
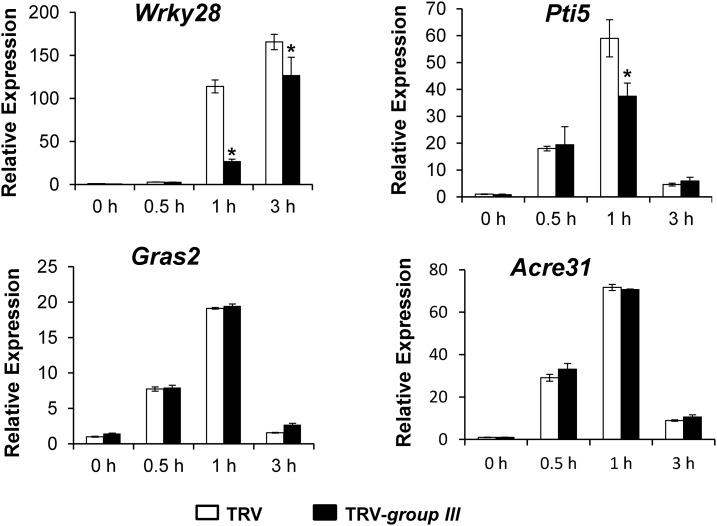
The requirement of group III ubiquitin E2 enzymes for AvrPtoB interference in Fen-mediated PCD prompted us to test whether this group of E2s also is required for plant immunity. We first tested whether the group III E2 enzymes are involved in PTI by examining the induction of PTI-activated reporter genes upon treatment of TRV-group III- and TRV-infected N. benthamiana plants with flg22. We tested the induction of a set of four genes, Wrky28, Pti5, Gras2, and Acre31, that have been developed to assay the activation of PTI in N. benthamiana (Nguyen et al., 2010). Each of the four reporter genes was induced significantly on TRV-infected N. benthamiana plants at 0.5 and 1 h after flg22 treatment (Fig. 6), which indicates the successful activation of PTI by flg22. However, compared with TRV-infected N. benthamiana plants, the induction of Wrky28 and Pti5 was decreased significantly on TRV-group III-infected plants at 1 h after flg22 treatment. By contrast, the induction of Gras2 and Acre31 remained unchanged on TRV-group III-infected plants (Fig. 6).
Figure 6.
Knocking down the group III ubiquitin E2 genes down-regulates the induction of PTI-activated reporter genes by flg22. The expression of the N. benthamiana PTI reporter genes Wrky28, Pti5, Gras2, and Acre31 in 2 μm flg22-treated leaves of group III E2 gene-silenced and nonsilenced TRV control N. benthamiana plants was analyzed by real-time PCR at the indicated time points after flg22 treatment. The x axis of each plot marks different time points (hours) after flg22 treatment. The experiment was performed with three technical repeats in each of the three biological replicates. Error bars indicate sd. Asterisks mark significant reductions of the expression of PTI reporter genes in group III E2 gene-silenced plants compared with nonsilenced TRV control plants (P < 0.05).
The Group III Ubiquitin E2 Enzymes Are Essential for PTI
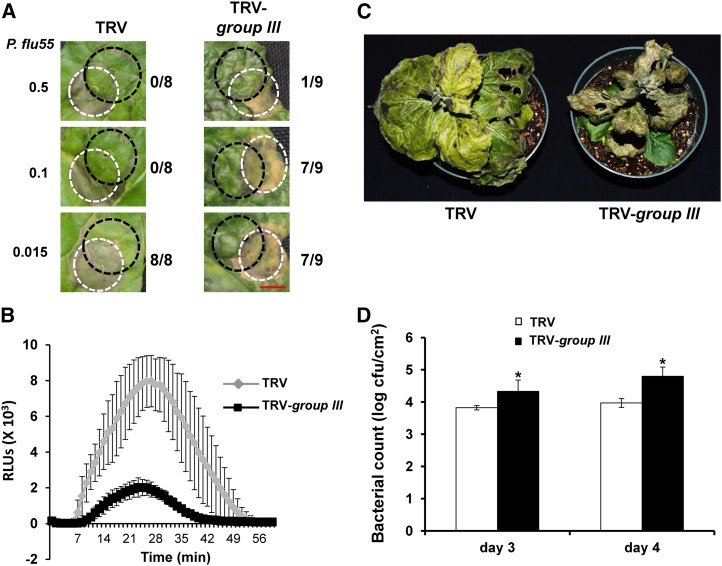
The effect of knocking down group III E2 genes on the induction of PTI-activated reporter genes suggested that these E2 genes are involved in PTI. To verify this, we employed three assays to evaluate the alteration of PTI on TRV-group III-infected N. benthamiana plants. We first performed the cell death suppression assay on TRV-group III- and TRV-infected N. benthamiana plants (Chakravarthy et al., 2010). In the cell death suppression assay, preinduction of PTI by the nonpathogen P. fluorescens 55 (OD600= 0.1 or 0.5; Fig. 7A, black dashed circles) prior to inoculation of the N. benthamiana plants with Pst strain DC3000 (Fig. 7A, white dashed circles) inhibits the HR cell death induced by Pst DC3000 in the overlapping area on TRV control N. benthamiana plant leaves (Fig. 7A; Wei et al., 2007; Chakravarthy et al., 2010). However, our results showed that cell death was observed in the overlapping area on group III E2 gene-silenced plants but not on TRV control N. benthamiana plants (P. fluorescens 55 OD600 = 0.1; Fig. 7A), which indicated a breakdown of PTI induction on group III E2 gene-silenced plants.
Figure 7.
The group III ubiquitin E2s are required for PTI. A, VIGS of group III E2 genes in N. benthamiana compromised PTI-mediated cell death suppression. The group III ubiquitin E2 gene-silenced (TRV-group III) and nonsilenced TRV control (TRV) N. benthamiana plants were first infiltrated with P. fluorescens 55 (P. flu55; marked as black dashed circles) to induce PTI, which was followed by infiltration of the HR-causing strain of Pst, DC3000 (marked as white dashed circles). Numbers at the left side denote the corresponding concentration of P. flu55 (OD600 value) used to activate PTI. The numbers at the right side represent the number of overlapped infiltration areas that displayed cell death and the total number of infiltrated overlapping areas. Photographs were taken on day 4 after infiltration of Pst DC3000. Bar = 1 cm. B, Silencing the group III E2 genes resulted in reduced ROS production induced by flg22 in a chemiluminescence assay. Leaf discs of the group III ubiquitin E2 gene-silenced (TRV-group III) and nonsilenced TRV control (TRV) N. benthamiana plants were incubated with 1 μm flg22 to induce ROS production. RLU, Relative light units. C, Disease symptoms of the group III E2 gene-silenced (TRV-group III) and nonsilenced TRV control (TRV) N. benthamiana plants. Plants were vacuum infiltrated with P. flu55 to induce PTI and then inoculated with Pst strain DC3000ΔhopQ1-1 6 h later. The photograph was taken on day 6 after Pst inoculation. D, Bacterial populations of leaves from plants shown in C. Each treatment represents the mean of four plants, and bars show sd. Experiments were repeated three times with similar results. Asterisks indicate significantly elevated bacterial growth compared with the TRV empty vector (TRV) control plants based on one-way ANOVA (P < 0.01). cfu, Colony-forming units. The experiments in Figure 7 were repeated three times with similar results.
We next measured the ROS production upon flg22 treatment of leaves of group III E2 gene-silenced (TRV-group III-infected) and nonsilenced control (TRV-infected) N. benthamiana plants. Compared with the control plants, ROS production triggered by flg22 treatment was reduced significantly on group III E2 gene-silenced N. benthamiana plants (Fig. 7B). Finally, we examined the effect of knocking down group III E2 genes on restricting the growth of Pst strain DC3000ΔhopQ1-1 on N. benthamiana plants by PTI (Wei et al., 2007). It has been found that preinoculation of N. benthamiana leaves with the nonpathogen P. fluorescens 55 would induce PTI and enhance differences in pathogenic bacterial growth between wild-type and PTI-defective plants (Nguyen et al., 2010; Rosli et al., 2013). Accordingly, group III E2 gene-silenced and nonsilenced control N. benthamiana plants were first vacuum infiltrated with P. fluorescens 55 to induce PTI. These plants were then inoculated with Pst DC3000ΔhopQ1-1 7 h later and monitored for disease development. The group III E2 gene-silenced plants displayed more disease symptoms that were manifested as leaf necrosis than the nonsilenced control N. benthamiana plants (Fig. 7C). The growth of Pst DC3000ΔhopQ1-1 on the group III E2 gene-silenced plants was significantly higher on days 3 and 4 after inoculation than that on the nonsilenced control plants (Fig. 7D). Taken together, these observations indicated that group III E2 genes are required for the induction of PTI.
Silencing of E2s UBC10/12/28/31 Did Not Affect PTI and AvrPtoB-Mediated Degradation of Fen
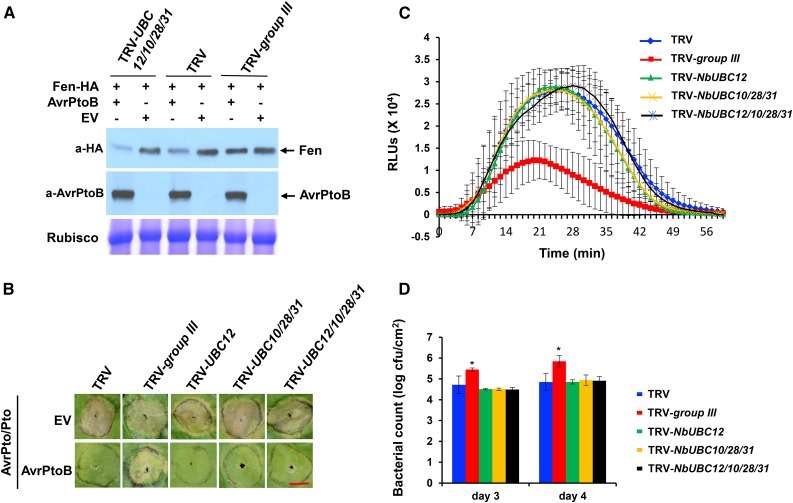
Tomato and N. benthamiana group III ubiquitin E2 contains 12 homologous members (Supplemental Figs. S3 and S9; Supplemental Table S1). The results that silencing of group III E2 genes diminished AvrPtoB-mediated degradation of Fen and that the group III E2 enzymes are involved in PTI prompted us to find out whether certain members of the group possibly contribute more significantly to the observed immunity-related phenotype than other members. As a first step to address this question, we analyzed the expression profile of the group III E2 genes on Rio Grande (RG)-pto11 tomato plants that were inoculated with a Pst strain delivering AvrPtoB. The overall range of changes in the expression of group III E2 genes before and after pathogen treatment was moderate, and except for UBC28 and UBC30, the expression of the group III members was suppressed 6 h post inoculation on plants that were treated with either DC3000ΔavrPtoΔavrPtoB carrying AvrPtoB or mock (Supplemental Fig. S13). Compared with the mock-inoculated plants, however, the expression of UBC8, UBC9, and UBC38 was suppressed, while UBC10, UBC12, UBC28, and UBC31 were induced 6 h post inoculation on plants challenged by DC3000ΔavrPtoΔavrPtoB carrying AvrPtoB at a statistically significant level (P < 0.05). Based on these results, we decided to silence UBC10, UBC12, UBC28, and UBC31 to examine if they play a major role in the AvrPtoB-mediated degradation of Fen and PTI. We specifically silenced UBC12 alone; UBC10, UBC28, and UBC31 together; or UBC10, UBC12, UBC28, and UBC31 together (Supplemental Fig. S14A) in N. benthamiana and examined the AvrPtoB-promoted degradation of Fen and the induction of PTI responses on these plants. Compared with nonsilenced control plants, knocking down the expression of UBC10, UBC12, UBC28, and UBC31 did not result in visible changes in plant growth and development (Supplemental Fig. S14B). Unlike group III-silenced plants, neither knocking down the expression of UBC12 alone nor silencing the three E2 genes UBC10, UBC28, and UBC31 or the genes UBC10, UBC12, UBC28, and UBC31 affected the degradation of Fen caused by AvrPtoB (Fig. 8A), the AvrPtoB-mediated suppression of PCD elicited by AvrPto/Pto (Fig. 8B), and the induction of PTI as indicated by ROS production after flg22 treatment (Fig. 8C) and restriction of the growth of the pathogen (Fig. 8D), which suggests that these four members may not play a major role in PTI and AvrPtoB-mediated suppression of immunity-associated PCD and perhaps the existence of functional redundancy among the members of group III.
Figure 8.
Functional redundancy exists among the group III E2 members. A, No effect on the degradation of Fen caused by AvrPtoB was observed on N. benthamiana plants in which the expression of group III E2 genes NbUBC10, NbUBC12, NbUBC28, and NbUBC31 was knocked down. The experiment was performed as shown in Figure 5B and was repeated two times with similar results. B, Knocking down the expression of NbUBC12 alone, NbUBC10, NbUBC28, and NbUBC31, or NbUBC10, NbUBC12, NbUBC28, and NbUBC31 did not diminish the suppression of AvrPto/Pto-elicited PCD by AvrPtoB in N. benthamiana. Transient coexpression of AvrPtoB and AvrPto/Pto in fully expanded N. benthamiana leaves was performed by syringe infiltrating A. tumefaciens carrying T-DNA with the AvrPtoB and AvrPto/Pto genes. Transient coexpression of the empty pBTEX vector (EV) and AvrPto/Pto was used as a control. Photographs were taken on day 4 after infiltration of Pto/AvrPto and AvrPtoB. Bar = 1 cm. C, Silencing NbUBC12 alone, NbUBC10, NbUBC28, and NbUBC31, or NbUBC10, NbUBC12, NbUBC28, and NbUBC31 failed to reduce ROS production induced by flg22 in a chemiluminescence assay. Leaf discs of the VIGS-treated N. benthamiana plants were incubated with 1 μm flg22 to induce ROS production. RLU, Relative light units. D, Bacterial populations of the Pst strain DC3000ΔhopQ1-1 on leaves of various VIGS-treated plants. Plants were vacuum infiltrated with P. fluorescens 55 to induce PTI and then inoculated with the Pst strain DC3000ΔhopQ1-1 6 h later. Each treatment represents the mean of four plants, and bars show sd. Experiments were repeated three times with similar results. Asterisks indicate significantly increased bacterial growth on group III-silenced plants compared with the nonsilenced control plants based on one-way ANOVA (P < 0.01). cfu, Colony-forming units.
The Group III E2s UBC11, UBC28, UBC29, UBC39, and UBC40 Likely Play a More Important Role in PTI
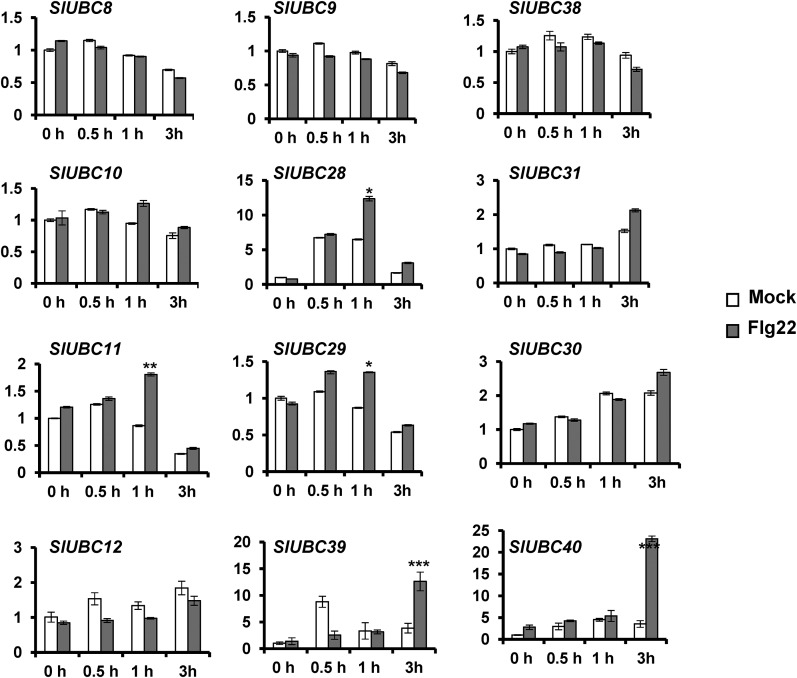
We next examined the expression patterns of the group III E2 genes on tomato plants that were infiltrated with flg22 or water (mock). As shown in Figure 9, the genes UBC8, UBC9, UBC10, UBC12, UBC29, UBC30, UBC31, and UBC38 showed similar expression patterns in mock- and flg22-infiltrated plants at 0 h (before the treatment) and 0.5, 1, and 3 h after infiltration. By contrast, UBC11, UBC28, and UBC29 showed more than 1.5-fold higher induction in flg22-treated plants than in the mock-infiltrated control plants at 1 h after infiltration, and UBC39 and UBC40 showed over 3-fold more induction after flg22 treatment than the mock-treated plants at 3 h after infiltration. These results suggest that UBC11, UBC28, UBC29, UBC39, and UBC40 are involved in, and perhaps play a more significant role in, PTI than other group III members and that functional redundancy exists among these members, such that knocking down UBC28 in UBC10/12/28/31-silenced plants described above did not affect PTI and AvrPtoB-mediated degradation of Fen. Since we were unable to silence UBC28 without off-targeting UBC10 and UBC31 in VIGS due to high sequence identity among them, we then silenced UBC11, UBC29, UBC39, and UBC40 together (Supplemental Fig. S15) and examined ROS production and activation of the PTI reporter gene on these plants after flg22 treatment (Supplemental Fig. S16, A and B). Unlike silencing UBC10/12/28/31, knocking down these E2 genes reduced host PTI, as manifested by diminished ROS production and the induction of PTI-activated reporter gene (Supplemental Fig. S16, A and B). However, the diminishment of ROS production and the activation of PTI reporter gene on the UBC11/29/39/40-silenced plants were to a lesser extent than on the group III-knocked down plants. We next performed a bacterial growth assay to examine the effect of knocking down UBC11/29/39/40 on the growth of Pst strain DC3000ΔhopQ1-1 on N. benthamiana plants. We observed a slight, but not significant, increase in bacterial growth in UBC11/29/39/40-silenced plants (Supplemental Fig. S16C). Additionally, the degradation of Fen by AvrPtoB appears to be unaffected on the UBC11/29/39/40-silenced plants (Supplemental Fig. S16D).
Figure 9.
Expression profile of group III E2 genes in flg22-treated tomato leaves detected by real-time PCR. Leaves of tomato RG-pto11 plants were infiltrated with 2 µm flg22 for PTI activation or with water as mock treatment. The expression of group III E2 genes at 0, 0.5, 1, and 3 h post flg22 infiltration was analyzed by quantitative reverse transcription (qRT)-PCR using the tomato EF1α gene (SlEF1a) as the internal reference. The y axis depicts the relative expression of the gene tested. The experiment was performed with three technical repeats in each of the three biological replicates. Error bars indicate sd. Asterisks mark the fold change of E2 genes induced in flg22-treated plants compared with mock-treated plants: *, greater than 1.5-fold; **, greater than 2-fold; and ***, greater than 3-fold.
DISCUSSION
To reveal the role of ubiquitin-conjugating enzymes (E2) in plant innate immunity, we have identified from the tomato genome 51 genes encoding UBC domain-containing proteins. Among them, 40 were predicted to encode ubiquitin E2 proteins and were classified into 13 groups. While most tomato members share highest similarity with another tomato protein in the phylogeny of tomato and human UBC domain-containing proteins, many tomato UBC domain-containing proteins find their closest homolog in Arabidopsis based on the phylogenetic tree of the UBC domain-containing proteins encoded by these two genomes, suggesting that these proteins are conserved in different plant species. These data, along with the confirmation that 34 out of the 40 genes encode active ubiquitin-conjugating enzymes, validate the algorithms we used for the identification of tomato ubiquitin E2 proteins at the genome scale. To date, genome-wide identification of plant ubiquitin E2 enzymes has been reported only for Arabidopsis and rice (Oryza sativa; Kraft et al., 2005; Bae and Kim, 2014). The tomato genome is one of the smallest diploid genomes in the family Solanaceae, and its genome sequence serves as a reference for other plant species in the family (Rensink et al., 2005; Matsukura et al., 2008). The identification of a complete set of tomato ubiquitin E2 enzymes thus provides the foundation for functional characterization of this class of enzyme in tomato and other crops in the Solanaceae.
We were unable to purify the recombinant proteins for tomato UBC21, UBC23, UBC24, UBC25, and UBC26 from E. coli due to insolubility, which is consistent with a previous finding that the recombinant protein for the Arabidopsis counterpart of tomato UBC21, UBC24, and UBC25 is insoluble in E. coli or in insect cells (Kraft et al., 2005; Zhao et al., 2013). The Arabidopsis UBC26 (AtUBC26) was unable to be induced in E. coli under various conditions tested by one team (Zhao et al., 2013), while another team was able to induce and purify the recombinant protein but found it to be inactive in vitro (Kraft et al., 2005). Although in vitro activity assays were unable to be performed for these five tomato ubiquitin E2s, UBC21 and UBC25 interacted in vivo with the tomato E1 ubiquitin-activating enzyme, UBA1, suggesting that they likely also are true ubiquitin E2 enzymes. Similar to the finding about Arabidopsis UBC37 (AtUBC37; Kraft et al., 2005), we were unable to detect ubiquitin-conjugating activity for tomato UBC37 in the thioester assay. This could be due to several reasons: the UBC37 protein may not fold properly in E. coli; it may require cofactor(s) for its enzymatic activity that are not present in the in vitro thioester assay; or it may conjugate a ubiquitin-like protein but not the ubiquitin. Another possibility is that it needs to interact with another protein for its ubiquitin-conjugating activity, which is especially likely considering that it has a long C-terminal amino acid extension.
Five tomato UBC domain-containing proteins (UBC43–UBC47) were predicted to encode SUMO E2 enzymes, and they grouped with the Arabidopsis SUMO E2 SCE1 and human SUMO E2 UBE2I (UBC9) in the phylogenetic analysis. In humans and animals, UBE2I is the only known SUMO E2 enzyme (Flotho and Melchior, 2013). In Arabidopsis, only the SUMO E2 enzyme SCE1 was identified, but rice, poplar (Populus spp.), tomato, sorghum (Sorghum bicolor), and maize (Zea mays) were found to encode multiple SUMO E2s based on their genome sequence (Novatchkova et al., 2012; Augustine et al., 2016). The biological significance of the multiple SUMO E2s that are predicted in many plants, especially crop genomes, is unknown at present. Determining whether the predicted plant SUMO E2 proteins are bona fide SUMO E2 enzymes will be the first step to address the question.
In this study, we utilized AvrPtoB to facilitate the identification of E2s that are involved in plant immunity. AvrPtoB worked specifically with group III E2s in catalyzing ubiquitination in vitro. Functional characterization indicated that the group III E2s are required for AvrPtoB E3 ligase activity-mediated degradation of Fen and the suppression of immunity-associated PCD by AvrPtoB. Importantly, plant PTI also was significantly affected when the group III E2 genes were knocked down. By contrast, immunity-associated PCD induced by several ETI-related elicitors was not affected on group III-silenced plants. Group III contains 12 highly homologous E2 members; thus, functional redundancy among these members likely exists. Except for UBC28 and UBC30, the expression of group III E2 genes was suppressed 6 h after inoculation on both tomato plants that were inoculated with mock and tomato plants that were inoculated with a Pst strain delivering AvrPtoB. However, the expression of UBC10, UBC12, UBC28, and UBC31 in the Pst-inoculated plants was higher than in the mock-inoculated control plants 6 h after inoculation at a statistically significant level (P < 0.05). Specific knocking down of UBC10, UBC12, UBC28, and UBC31 together, nevertheless, did not impair AvrPtoB-promoted degradation of Fen and PTI. Further expression profiling indicated that UBC11, UBC28, UBC29, UBC39, and UBC40 were significantly induced either 1 or 3 h after treatment when the tomato plants were challenged by flg22, suggesting that they may play a more important role than other group III members in PTI. Indeed, unlike silencing UBC10/12/28/31, knocking down UBC11, UBC29, UBC39, and UBC40 together affected plant PTI, although the effect was to a lesser extent than that of group III-silenced plants. This result, combined with the observation that UBC28 was induced both on plants challenged by a Pst strain delivering AvrPtoB and on plants challenged by flg22 yet knocking down UBC10, UBC12, UBC28, and UBC31 together did not affect plant PTI, suggest that functional redundancy exists among UBC11, UBC28, UBC29, UBC39, and UBC40.
In addition to group III members, other tomato E2s also may play an important role in plant immunity. The pathogenicity protein βC1 encoded by the DNA-β of certain monopartite Begomovirus spp. is important for disease symptom development and for the suppression of host gene silencing by the virus (Eini et al., 2009). βC1 was found to interact with the tomato E2 ubiquitin-conjugating enzyme SlUBC3 to down-regulate the host ubiquitin proteasome pathway, which contributes to the induction of DNA-β-specific symptoms in host plants (Eini et al., 2009). It is likely that the interaction of βC1 with SlUBC3 impairs the ubiquitin-conjugating activity of this E2 enzyme and, thus, results in perturbation of the host ubiquitination system, suggesting that SlUBC3 plays a role in host resistance to βC1-related viruses. Additionally, the tomato E2s UBC13 (i.e. Fni3) and UBC13-2 were shown previously to catalyze exclusively the unconventional, Lys-63-linked ubiquitination and to be required for immunity-associated PCD induced by Fen and several other ETI elicitors (Mural et al., 2013). More recently, two gene expression profiling studies of tomato immunity against Pst using high-throughput RNA sequencing identified multiple Flagellin-induced repressed by effectors genes and genes induced specifically during ETI, among which UBC12, UBC13, UBC13-2, UBC25, and UBC28 were found to be induced significantly during ETI, and UBC25 also was induced significantly in PTI (Rosli et al., 2013; Pombo et al., 2014). These findings not only corroborate the discovery that tomato UBC13 (Fni3) and UBC13-2 play an important role in ETI (Mural et al., 2013) but also suggest that tomato E2s other than the group III members, such as UBC25, also may play an important role in plant immunity. Functional characterization of these E2s in tomato immunity would verify this postulation and provide additional insights into the regulation of plant immunity by E2s. On the other hand, nearly a dozen E3 ubiquitin ligases have been implicated in PTI to date (Zeng et al., 2004; Lu et al., 2011; Cheng and Li, 2012; Ishikawa et al., 2014; He et al., 2015). However, it is unclear which ubiquitin E2 enzyme(s) work with these E3 ligases in the regulation of PTI. Therefore, determining the ubiquitin E2 enzymes with which each of these known E3 ligases act in PTI will reveal E2s that are key to plant immunity other than the ones identified in this study.
It has been suggested that AvrPtoB acquired its C-terminal E3 ubiquitin ligase domain to suppress host ETI during the coevolution of Pst and tomato, which represents a typical example of the arms race in host-pathogen interactions (Rosebrock et al., 2007; Munkvold and Martin, 2009). However, the E2 enzymes that act with AvrPtoB E3 activity in suppressing plant immunity have remained unknown ever since the C-terminal domain of AvrPtoB was revealed to be an E3 ubiquitin ligase a decade ago (Janjusevic et al., 2006). We identified here the tomato group III E2 enzymes as the only cognate E2s of AvrPtoB in catalyzing ubiquitination and showed that functional redundancy exists among the group III members. Evolutionarily, exploiting a group of plant E2s with functional redundancy is apparently to the advantage of AvrPtoB, as change or deletion of a single or even multiple members of them will not be able to affect the suppression of host immunity by its E3 activity. Furthermore, the discoveries that group III E2 enzymes also are essential for PTI and affect plant growth and development imply that it would be difficult for the plant to alter or delete the group III E2 enzymes in order to thwart the strategy of exploiting this group of E2s by AvrPtoB. Thus, the selection pressure may have forced plants to evolve the Pto gene encoding a protein that is recalcitrant to the E3 ligase activity of AvrPtoB and capable of recognizing two different domains in the N terminus of AvrPtoB to activate ETI (Martin, 2012; Mathieu et al., 2014). However, a previous study of the avrPtoB gene from the Pst strain T1 (Lin et al., 2006) and a more recent study of Pst race 1 strains collected from the fields of California indicated that Pst may have evolved a mechanism to reduce the accumulation of AvrPtoB to escape host detection by Pto while preserving a degree of their virulence activity (Kunkeaw et al., 2010), which continues the theme of an arms race in the coevolution of Pst and tomato.
It is conceivable that members of the group III E2 enzymes also work with plant E3 ligases, including those that are involved in plant growth and development. In support of this, knocking down group III E2 genes resulted in growth and developmental changes in N. benthamiana plants. The intimate connection between plant growth and defense has been increasingly recognized (Lozano-Durán and Zipfel, 2015). In fact, various components regulating plant immunity also have been found to play important roles in plant growth and development. The Arabidopsis bak1 mutant is semidwarf (Chinchilla et al., 2007), and silencing BAK1 in N. benthamiana results in a dwarf stature and crinkled leaves (Chakravarthy et al., 2010). Additionally, BAK1 is associated with grain filling and leaf development in rice (Khew et al., 2015). The receptor-like kinase ERECTA not only affects the development of aerial organs but also is involved in plant disease resistance (Godiard et al., 2003). The E3 ligase SPL11/PUB13 regulates defense and flowering time in rice and Arabidopsis, respectively (Li et al., 2012; Liu et al., 2012). However, the immunity phenotypes we observed in the group III gene-silenced plants are unlikely to be caused indirectly by growth and developmental changes, as the diminishment of the AvrPtoB-promoted degradation of Fen due to knocking down group III genes was observed in both leaves and protoplasts of N. benthamiana plants.
The identification of UBC11, UBC12, UBC13, UBC13-2, UBC25, UBC28, UBC29, UBC39, and UBC40 in either PTI and/or ETI in this and previous studies warrants further investigation of them in plant immunity (Mural et al., 2013; Rosli et al., 2013; Pombo et al., 2014). Since the connections of these E2s with E3 ligases in plant immunity have not been established, screening for and characterization of E3 ligases that work with these E2s in PTI and/or ETI and subsequently pinpointing and characterizing the substrates they modify should be the next experiments in elucidating the roles of E2s in the plant immune system. Additionally, components other than E3 ligases that interact with the these E2s also might contribute to the regulation of plant immunity. The membrane-anchored ubiquitin fold (MUB) proteins were found to interact specifically with the Arabidopsis counterparts of tomato group III E2 enzymes (Dowil et al., 2011). Considering the critical importance of group III E2s for PTI as shown here and that the PRRs that recognize MAMPs in PTI are localized to the plasma membrane, it would be intriguing to find out whether the MUB proteins cooperate with the group III E2 enzymes, particularly the E2s UBC11, UBC28, UBC29, UBC39, and UBC40, and their cognate E3 ubiquitin ligases to modify certain PRRs and, thus, play a role in regulating PTI.
MATERIALS AND METHODS
Growth of Bacteria and Plant Materials
Agrobacterium tumefaciens strains GV3101 and GV2260 and strains of Pseudomonas syringae pv tomato and Pseudomonas fluorescens 55 were grown at 28°C on Luria-Bertani and King’s B medium, respectively, with appropriate antibiotics. Nicotiana benthamiana and tomato (Solanum lycopersicum) RG-pto11 (pto11/pto11, Prf/Prf) seeds were germinated, and plants were grown on autoclaved soil in a growth chamber with 16 h of light (approximately 300 μmol m−2 s−1 at the leaf surface of the plants), 24°C/23°C day/night temperature, and 50% relative humidity.
DNA Manipulation and Plasmid Construction
All DNA manipulations were performed using standard techniques (Sambrook and Russell, 2001). The open reading frames (ORFs) of the tomato E1 gene SlUBA1 and 24 tomato E2 genes were amplified from tomato cDNA using the Q5 High-Fidelity DNA Polymerase (New England Biolabs). The ORFs of 16 other tomato E2 genes were amplified from the plasmid DNA of tomato cDNA clones. The E2 mutant SlUBC13C89G was as described (Mural et al., 2013), and SlUBC8C85G and SlUBC10C85G E2 mutant ORFs were prepared by site-directed mutagenesis. The amplified ORFs were cloned into the pGEX-4T-1 vector or into the pENTR/SD/D-TOPO entry vector by Gateway cloning (Life Technologies), followed by construction into the pDEST15 or pDEST17 vector using Gateway LR reaction according to instructions provided by the manufacturer (Life Technologies). While the cloned cDNA sequence matched the genome annotation for most of the tomato ubiquitin E2 genes, errors in the annotation for tomato genes UBC25 and UBC38 were detected when compared with cloned cDNA. Constructs for VIGS were prepared by Gateway cloning using the pENTR/SD/D-TOPO entry vector and the pTRV2 vector (Caplan and Dinesh-Kumar, 2006) according to protocols provided by the manufacturer (Life Technologies). The insert of the TRV-group III construct used for silencing the group III E2 genes in N. benthamiana was cloned by overlapping PCR of six individual fragments from the NbUBC9, NbUBC11, NbUBC12, NbUBC28, NbUBC30, and NbUBC39 genes. Based on the high percentage of identity in their DNA sequence, a DNA fragment from NbUBC9 was designed for silencing the genes NbUBC8, NbUBC9, and NbUBC38; a DNA fragment from NbUBC28 was designed for silencing genes NbUBC10, NbUBC28, and NbUBC31; a fragment from NbUBC11 was designed for silencing NbUBC11 and NbUBC29; and a fragment from NbUBC39 was designed for silencing NbUBC39 and NbUBC40. Examination of sequences was conducted during the selection of DNA fragments for building the TRV-group III construct to avoid off-target silencing of E2 genes from other groups, as verified by the results shown in Supplemental Figure S9C. To test the functional redundancy of group III members, optimal fragments used for building the VIGS constructs to silence NbUBC12 alone, the E2 genes NbUBC10, NbUBC28, and NbUBC31, the four E2 genes NbUBC12, NbUBC10, NbUBC28, and NbUBC31, or the E2 genes NbUBC11, NbUBC29, NbUBC39, and NbUBC40 were first predicted by the VIGS tool (Fernandez-Pozo et al., 2015) and then further manually selected to minimize the identity of DNA sequence to other members of group III (more than four nucleotides are mismatched within any 21-nucleotide DNA stretch when compared with other group III members). The constructs used for the BiFC assay were prepared using the vectors pA7-nYFP, pA7-cYFP, pSPYNE173, and pSPYCE(M) (Waadt et al., 2008; Mural et al., 2013). All genes used for the transfection of protoplasts were cloned into a pTEX 35S cauliflower mosaic virus promoter expression cassette with hemagglutinin (HA) tag at the C terminus (Mural et al., 2013). Primers used for recombinant DNA cloning are listed in Supplemental Table S2.
Identification of Tomato UBC Domain-Containing Proteins
To identify UBC domain-containing proteins from the tomato genome (Tomato Genome Consortium, 2012), the following five-step procedure was used. (1) All relevant domain families consistent with the annotation terms Ubiquitin-Protein Ligase (EC 6.3.2.19), Protein Ubiquitinylation (GO:0016567), and Small Conjugating Protein Ligase Activity (GO:0019787) were identified in the Structural Classification of Proteins hierarchy via the results of Pethica et al. (2012). (2) The International Tomato Annotation Group (ITAG) version 2.3 protein annotations of the tomato genome were processed through the SUPERFAMILY pipeline (Gough et al., 2001; Oates et al., 2015), which provided hidden Markov model assignment to each specific domain family through the use of HMMER3 (Eddy, 2011). (3) For every resulting protein annotation likely containing a UBC domain, each whole sequence was searched against the TAIR10 protein annotations for Arabidopsis (Arabidopsis thaliana) with NCBI BLASTP, which provided the gene name of the most homologous Arabidopsis protein sequence for each candidate in tomato. (4) Coverage of EST and UniGene models for each candidate was assigned from data obtained from the ITAG genome database. (5) Prioritizing results was based on E-value scores for family-level assignment to a domain, whether the protein was found to match a known UBC from plants, and whether there was sufficient evidence from ITAG that the transcript was expressed (Zeng et al., 2008; Tomato Genome Consortium, 2012).
Sequence Alignment and Phylogenetic Analysis
For sequence alignment, sequences of interest in the FASTA format were entered into the ClustalX 2.1 program and aligned using the ClustalX algorithm (Larkin et al., 2007). The phylogenetic analysis was then performed with the MEGA6 program using the aligned sequences (Tamura et al., 2013). To build an unrooted phylogenetic tree using MEGA6, the evolutionary history was inferred using the neighbor-joining method with 1,000 bootstrap trials. The evolutionary distances were computed using the p-distance method, in which the evolutionary distance unit represents the number of amino acid (or nucleotide) substitutions per site (Nei and Kumar, 2000). Branches corresponding to partitions reproduced in less than 50% of bootstrap replicates were collapsed in the tree.
Expression and Purification of Proteins
GST- and 6×His-tagged fusion proteins were expressed in Escherichia coli strain BL21 (DE3) and purified with Glutathione Sepharose 4 Fast Flow beads (GE Healthcare) and Ni-NTA agarose (Qiagen), respectively, by following the protocol provided by the manufacturer. Except for UBC12, UBC4, and UBC36, which were expressed as 6×His-fusion proteins, all 32 other E2s were fused with GST for purification of their recombinant proteins. The purified proteins were further desalted and concentrated in the protein storage buffer (50 mm Tris-HCl, pH 8, 50 mm KCl, 0.1 mm EDTA, 1 mm DTT, and 0.5 mm PMSF) using the Amicon Centrifugal Filter (Millipore). The desalted and concentrated recombinant protein was stored at −80°C in the presence of a final concentration of 40% glycerol until being used. The concentration of purified protein was determined using protein assay agent (Bio-Rad). The quality of purified proteins was analyzed by 10% SDS-PAGE (Supplemental Fig. S4).
Thioester Assay of E2 Ubiquitin-Conjugating Activity
The ubiquitin-conjugating activity assay of tomato E2s was performed as described with modifications (Mural et al., 2013). Briefly, in a 20-µL reaction, 40 ng of tomato E1 GST-Sl-UBA1 was preincubated with 2 μg of FLAG-ubiquitin in the assay buffer (20 mm Tris-HCl, pH 7.5, 10 mm MgCl2, and 1 mm ATP) at 28°C for 10 min. To the reaction, an optimal amount (50–250 ng) of GST- or 6×His-fused E2 protein was added and continued for 15 min. The reaction was then split equally and terminated by the addition of SDS sample loading buffer with either 100 mm DTT or 4 m urea sample buffer. The reactions were immunoblotted with mouse monoclonal anti-FLAG M2-peroxidase-conjugated antibody (Sigma-Aldrich) before being detected using an ECL kit (Pierce, now Thermo Fisher).
In Vitro Ubiquitination Assay
The in vitro ubiquitination assay was performed similar to that described previously (Mural et al., 2013). Briefly, 3 µg of ubiquitin, 40 ng of E1 (GST-SlUBA1), an optimal amount (50–250 ng) of GST- or 6×His-fused E2, and 2 µg of GST-AvrPtoB E3 ligase were added to a 30-μL reaction in the presence of ubiquitination assay buffer (50 mm Tris-HCl, pH 7.5, 5 mm ATP, 5 mm MgCl2, 2 mm DTT, 3 mm creatine phosphate, and 5 µg mL−1 creatine phosphokinase). For the ubiquitination of MBP-Fen, 200 ng of MBP-Fen was added to the reaction of in vitro ubiquitination assay. The reactions were incubated at 30°C for 1.5 h and then terminated by the addition of SDS sample loading buffer with 100 mm DTT. The reaction products were resolved by 10% SDS-PAGE and analyzed by immunoblotting using mouse monoclonal anti-ubiquitin M2-peroxidase-conjugated (horseradish peroxidase) antibody (Sigma-Aldrich) to identify the polyubiquitin signal. Polyubiquitinated forms of AvrPtoB and MBP-Fen were detected using rabbit anti-AvrPtoB antibody and anti-MBP antibody, respectively, as described (Rosebrock et al., 2007).
BiFC Assay
The BiFC assay that is based on split yellow fluorescent protein (YFP) was used to test the interaction of various E2-E3 pairs in N. benthamiana leaves and protoplasts (Chen et al., 2006; Waadt et al., 2008). For the assay using N. benthamiana leaves, N-terminal YFP-fused AvrPtoB (AvrPtoB-nYFP) and C-terminal YFP-fused E2 (E2-cYFP) proteins were transiently coexpressed in the leaves of N. benthamiana plants. N-terminal YFP-fused AvrPtoB1-307 (AvrPtoB1-307-nYFP) and C-terminal YFP-fused SlUBC16 (SlUBC16-cYFP) proteins were used as negative controls of E3 and E2, respectively. The leaves were imaged at 48 h after infection. For the assay using protoplasts, the empty vectors expressing the N terminus and C terminus of YFP (nYFP-EV and cYFP-EV) were used as negative controls. Protoplasts were prepared from leaves of wild-type N. benthamiana plants as described (Rosebrock et al., 2007). Approximately 1 × 104 protoplasts that were suspended in a volume of 200 μL were then cotransfected with 10 µg of plasmid DNA of each individual of the construct pair to be tested. The cotransfected protoplast was imaged 21 h after transfection using an Olympus FV500 Inverted (Olympus IX-81) confocal microscope with the following excitation and emission wavelengths: YFP, 514.5 nm (excitation) and 525 to 555 nm (emission); chlorophyll autofluorescence, 640.5 nm (excitation) and 663 to 738 nm (emission).
Coimmunoprecipitation
The coimmunoprecipitation assay of HA-tagged AvrPtoB and 10Myc-tagged E2 was performed as described previously with some modifications (Moffett et al., 2002). Protein extracts from agroinfiltrated N. benthamiana leaves were prepared by grinding 0.8 g of leaf tissue in 1.5 mL of extraction buffer (25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 10% glycerol, 1 mm DTT, and 0.15% Nonidet P-40) in the presence of plant protease inhibitor cocktail (Sigma-Aldrich). Extracts were spun for 5 min at 12,000 rpm two times, and the supernatant was added to 25-µL anti-HA (3F10) agarose beads (Roche). Extracts were incubated with shaking at 4°C for 3 h followed by washing three times with the washing buffer (25 mm Tris-HCl, pH 7.5, 1 mm EDTA, 150 mm NaCl, 10% glycerol, 0.15% Nonidet P-40, and 1× protease inhibitor cocktail), and the pellet was resuspended in 100 μL of 1× SDS-PAGE loading buffer. Immunoprecipitated samples were separated by 10% SDS-PAGE and analyzed by immunoblot using anti-HA and anti-MYC antibodies (Santa Cruz).
VIGS
Gene silencing was induced using the TRV vectors (Caplan and Dinesh-Kumar, 2006) as described (Mural et al., 2013).
Effectiveness of VIGS of Group III E2 Genes
Determination of the effectiveness of knocking down group III E2 genes, NbUBC12 alone, the three E2 genes NbUBC10, NbUBC28, and NbUBC31, the four E2 genes NbUBC12, NbUBC10, NbUBC28, and NbUBC31, or the E2 genes NbUBC11, NbUBC29, NbUBC39, and NbUBC40 by VIGS was performed as described (Mural et al., 2013).
Transient Expression of Recombinant Proteins in N. benthamiana Plants and Protoplasts
To study the suppression of PCD by AvrPtoB on the group III E2 gene-silenced and nonsilenced control N. benthamiana plants, AvrPtoB was transiently coexpressed with the PCD elicitors Fen and AvrPto/Pto, respectively, on plants approximately 4 weeks after VIGS infection as described (Abramovitch et al., 2003). The final OD600 values of A. tumefaciens strain GV2260 harboring AvrPtoB and the PCD elicitor used for transient coexpression are as follows: AvrPtoB (OD600 = 0.01) and Fen (OD600 = 0.6); AvrPtoB (OD600 = 0.4) and AvrPto/Pto (OD600 = 0.4 for each). To analyze the degradation of Fen in the presence of AvrPtoB, A. tumefaciens strain GV2260 harboring the genes Fen-HA (OD600 = 0.1) and AvrPtoB (OD600 = 0.2) was transiently coexpressed in the leaves of N. benthamiana plants. To monitor the degradation of Fen in the presence of AvrPtoB in protoplasts, protoplasts were prepared from the leaves of N. benthamiana plants as described (Rosebrock et al., 2007). An amount of 6 × 104 protoplasts in a volume of 200 μL was then cotransfected with 10 μg of each plasmid DNA of pTEX-AvrPtoB-HA and pTEX-Fen-HA. Protein was extracted at 21 h post transfection by adding 80 µL of lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 5 mm EDTA, pH 8, 1% Triton X-100, 10% glycerol, and 0.2% protease inhibitor) to the protoplast pellets, followed by adding 80 µL of 2× sample loading buffer with 2% 2-mercaptoethanol included. The total proteins were analyzed by immunoblot using anti-HA antibody.
Real-Time PCR
Four-week-old tomato RG-pto11 plants (pto11/pto11, Prf/Prf) were vacuum infiltrated with a suspension of Pst strain DC3000ΔavrPtoΔavrPtoB expressing AvrPtoB under the control of a Pst hrp-inducible promoter (Lin et al., 2006) as described (Anderson et al., 2006). Leaf samples were collected at 0 and 6 h post inoculation. Total RNA was isolated using the RNeasy Plant Mini Kit with DNase treatment (Qiagen) following the protocol provided by the manufacturer. The first-strand cDNA was synthesized using SuperScript III reverse transcriptase and oligo(dT) primer (Life Technologies) according to the instructions from the manufacturer. qRT-PCR was performed using gene-specific primers and SYBR Green (Life Technologies) on the LightCycler 480 Instrument II (Roche). To determine whether the group III E2 genes are induced during PTI, leaves of 4-week-old RG-pto11 tomato plants were infiltrated with 2 µm flg22 or water only (mock) using a 1-mL needleless syringe (BD). To analyze the expression of the PTI reporter genes Wrky28, Pti5, Gras2, and Acre31 (Nguyen et al., 2010), leaves of N. benthamiana plants were infiltrated with 2 µm flg22 using a 1-mL needleless syringe. All primers used in qRT-PCR are shown in Supplemental Table S2, and either SlEF1a or NbEF1α was used as an internal reference accordingly.
Bacterial Population Assay
The bacterial population assay was conducted as described (Nguyen et al., 2010) with minor modifications. Briefly, N. benthamiana plants about 4 weeks after VIGS infection were first vacuum infiltrated with P. fluorescens 55 by submersion of the aerial parts of the plant in a suspension of P. fluorescens 55 (5 × 107 colony-forming units mL−1) containing 0.002% Silwet L-77 and 10 mm MgCl2. Seven hour later, the plants were inoculated with Pst DC3000ΔhopQ1-1 (2 × 105 colony-forming units mL−1) in the presence of 0.002% Silwet L-77 and 10 mm MgCl2 by vacuum infiltration. Inoculated plants were maintained in a growth chamber and monitored daily for symptom development. The assessment of bacterial populations on days 3 and 4 after inoculation of Pst DC3000ΔhopQ1-1 was conducted as described (Anderson et al., 2006).
Cell Death Suppression Assay
The cell death suppression assay was performed as described (Nguyen et al., 2010).
ETI Elicitor-Triggered Cell Death Assay
The cell death assay was performed as described previously (Mural et al., 2013).
ROS Assay
A luminol-horseradish peroxidase assay was used to quantify ROS induction as described previously (Cai et al., 2011) with small modifications. In brief, leaf discs of 4 mm in diameter were punched out with a cork borer from N. benthamiana plants and floated adaxial side up in 200 µL of distilled, deionized water at room temperature overnight in wells of a 96-well Nunc white plate (Thermo Scientific). On the second day, the distilled, deionized water was replaced with 100 µL of ROS testing buffer containing 1 µm flg22 peptide, 34 mg mL−1 luminol (Sigma-Aldrich), and 20 μg mL−1 horseradish peroxidase (Sigma-Aldrich). Luminescence was measured using a Bioteck Synergy II plate reader. Twelve leaf discs from three group III ubiquitin E2 gene-silenced and three nonsilenced control N. benthamiana plants were tested in parallel.
Accession Numbers
Sequence data of the Arabidopsis, N. benthamiana, and human E2 genes that were used in this article can be found in the GenBank data library with the accession numbers provided in Supplemental Table S1. Sequence data of the tomato E2 genes from this article can be found in GenBank under the following accession numbers: SlUBC1 (KY246895), SlUBC2 (KY246896), SlUBC3 (KY246897), SlUBC4 (KY246898), SlUBC5 (KY246899), SlUBC6 (KY246900), SlUBC7 (KY246901), SlUBC8 (KY246902), SlUBC9 (KY246903), SlUBC10 (KY246904), SlUBC11 (KY246905), SlUBC12 (KY246906), SlUBC13 (KY246927), SlUBC13-2 (KY246928), SlUBC14 (KY246908), SlUBC15 (KY246909), SlUBC16 (KY246910), SlUBC17 (KY246911), SlUBC20 (KY246912), SlUBC21 (KY246913), SlUBC22 (KY246914), SlUBC23 (KY246915), SlUBC24 (KY246916), SlUBC25 (KY246917), SlUBC26 (KY246918), SlUBC27 (KY246919), SlUBC28 (KY246920), SlUBC29 (KY246921), SlUBC30 (KY246922), SlUBC31 (KY246923), SlUBC32 (KY246924), SlUBC33 (KY246925), SlUBC34 (KY246926), SlUBC35 (KY246907), SlUBC36 (KY246934), SlUBC37 (KY246929), SlUBC38 (KY246930), SlUBC39 (KY246931), SlUBC40 (KY246932), and SlUBC41 (KY246933).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny of the human and tomato UBC domain-containing proteins.
Supplemental Figure S2. Schematic representation of domain organization in the tomato UBC domain-containing proteins.
Supplemental Figure S3. Tomato ubiquitin E2 enzymes are classified into 13 subgroups.
Supplemental Figure S4. Purified tomato E2 proteins as shown by SDS-PAGE.
Supplemental Figure S5. Examination of the ubiquitin-conjugating activity of tomato E2 proteins by thioester formation assay.
Supplemental Figure S6. AvrPtoB shows no specificity toward the tomato ubiquitin E2 enzymes SlUBC16 and SlUBC17.
Supplemental Figure S7. AvrPtoB interacts with tomato group III E2 members but not with E2s from other groups in BiFC assays.
Supplemental Figure S8. Members of group III E2 interacted with AvrPtoB in coimmunoprecipitation assay.
Supplemental Figure S9. Knocking down group III E2 genes in N. benthamiana by VIGS.
Supplemental Figure S10. Homologs of the group III E2 genes from N. benthamiana share high nucleotide sequence identity with their counterpart of tomato.
Supplemental Figure S11. Silencing group III E2 genes diminished AvrPtoB-promoted degradation of Fen in N. benthamiana protoplasts.
Supplemental Figure S12. Silencing of group III E2 genes does not influence multiple ETI elicitor-triggered PCD.
Supplemental Figure S13. Effect of AvrPtoB on the expression of tomato group III E2 genes.
Supplemental Figure S14. Specific silencing of E2 genes NbUBC12 alone, NbUBC10, NbUBC28, and NbUBC31, and NbUBC10, NbUBC12, NbUBC28, and NbUBC31 in N. benthamiana by VIGS.
Supplemental Figure S15. Specific silencing of E2 genes NbUBC11, NbUBC29, NbUBC39, and NbUBC40 together in N. benthamiana by VIGS.
Supplemental Figure S16. UBC11, UBC28, UBC29, UBC39, and UBC40 of group III play a more important role in PTI.
Supplemental Table S1. List of UBC domain-containing proteins from tomato, Arabidopsis, N. benthamiana, and human.
Supplemental Table S2. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank James Giovannoni for sharing the tomato cDNA clones containing the ORFs of 16 tomato ubiquitin E2 genes as well as Christian Elowsky and You Zhou (Biotechnology Center, University of Nebraska-Lincoln) for helping with the confocal microscope.
Glossary
- PTI
pattern-triggered immunity
- ETI
effector-triggered immunity
- MAMP
microbe-associated molecular pattern
- PRR
pattern recognition receptor
- ROS
reactive oxygen species
- PCD
programmed cell death
- HR
hypersensitive response
- VIGS
virus-induced gene silencing
- DTT
dithiothreitol
- BiFC
bimolecular fluorescence complementation
- ORF
open reading frame
- ITAG
International Tomato Annotation Group
- qRT
quantitative reverse transcription
Footnotes
This work was supported by start-up funds from the University of Nebraska-Lincoln, the U.S. Department of Agriculture National Institute of Food and Agriculture (grant no. 2012-67014-19449 to L.Z.), and the National Science Foundation (grant no. IOS-1460221to L.Z.).
Articles can be viewed without a subscription.
References
- Abramovitch RB, Janjusevic R, Stebbins CE, Martin GB (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci USA 103: 2851–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramovitch RB, Kim YJ, Chen S, Dickman MB, Martin GB (2003) Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JC, Pascuzzi PE, Xiao F, Sessa G, Martin GB (2006) Host-mediated phosphorylation of type III effector AvrPto promotes Pseudomonas virulence and avirulence in tomato. Plant Cell 18: 502–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine RC, York SL, Rytz TC, Vierstra RD (2016) Defining the SUMO system in maize: SUMOylation is up-regulated during endosperm development and rapidly induced by stress. Plant Physiol 171: 2191–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H, Kim WT (2014) Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem Biophys Res Commun 444: 575–580 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bombarely A, Rosli HG, Vrebalov J, Moffett P, Mueller LA, Martin GB (2012) A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol Plant Microbe Interact 25: 1523–1530 [DOI] [PubMed] [Google Scholar]
- Cai R, Lewis J, Yan S, Liu H, Clarke CR, Campanile F, Almeida NF, Studholme DJ, Lindeberg M, Schneider D, et al. (2011) The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathog 7: e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2009) BLAST+: architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J, Dinesh-Kumar SP (2006) Using viral vectors to silence endogenous genes. Curr Protoc Microbiol Chapter 16: Unit 16I.6. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Velásquez AC, Ekengren SK, Collmer A, Martin GB (2010) Identification of Nicotiana benthamiana genes involved in pathogen-associated molecular pattern-triggered immunity. Mol Plant Microbe Interact 23: 715–726 [DOI] [PubMed] [Google Scholar]
- Chang JH, Urbach JM, Law TF, Arnold LW, Hu A, Gombar S, Grant SR, Ausubel FM, Dangl JL (2005) A high-throughput, near-saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA 102: 2549–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li X (2012) Ubiquitination in NB-LRR-mediated immunity. Curr Opin Plant Biol 15: 392–399 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JDG, Felix G, Boller T (2007) A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 448: 497–500 [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains: from structures to functions. Nat Rev Mol Cell Biol 10: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowil RT, Lu X, Saracco SA, Vierstra RD, Downes BP (2011) Arabidopsis membrane-anchored ubiquitin-fold (MUB) proteins localize a specific subset of ubiquitin-conjugating (E2) enzymes to the plasma membrane. J Biol Chem 286: 14913–14921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. (2011) Accelerated profile HMM searches. PLOS Comput Biol 7: e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eini O, Dogra S, Selth LA, Dry IB, Randles JW, Rezaian MA (2009) Interaction with a host ubiquitin-conjugating enzyme is required for the pathogenicity of a geminiviral DNA beta satellite. Mol Plant Microbe Interact 22: 737–746 [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Fernandez-Pozo N, Rosli HG, Martin GB, Mueller LA (2015) The SGN VIGS tool: user-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol Plant 8: 486–488 [DOI] [PubMed] [Google Scholar]
- Flotho A, Melchior F (2013) Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82: 357–385 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Yan S, Saleh A, Wang W, Ruble J, Oka N, Mohan R, Spoel SH, Tada Y, Zheng N, et al. (2012) NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486: 228–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP (2009) AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol 19: 423–429 [DOI] [PubMed] [Google Scholar]
- Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, Grimsley NH, Marco Y (2003) ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J 36: 353–365 [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, de Torres M, Mansfield JW, Robatzek S (2008) Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol 18: 1824–1832 [DOI] [PubMed] [Google Scholar]
- Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313: 903–919 [DOI] [PubMed] [Google Scholar]
- Guo M, Tian F, Wamboldt Y, Alfano JR (2009) The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact 22: 1069–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Shan L, Lin NC, Martin GB, Kemmerling B, Nürnberger T, Sheen J (2006) Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 125: 563–575 [DOI] [PubMed] [Google Scholar]
- He Q, McLellan H, Boevink PC, Sadanandom A, Xie C, Birch PRJ, Tian Z (2015) U-box E3 ubiquitin ligase PUB17 acts in the nucleus to promote specific immune pathways triggered by Phytophthora infestans. J Exp Bot 66: 3189–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Yamaguchi K, Sakamoto K, Yoshimura S, Inoue K, Tsuge S, Kojima C, Kawasaki T (2014) Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat Commun 5: 5430. [DOI] [PubMed] [Google Scholar]
- Jamir Y, Guo M, Oh HS, Petnicki-Ocwieja T, Chen S, Tang X, Dickman MB, Collmer A, Alfano JR (2004) Identification of Pseudomonas syringae type III effectors that can suppress programmed cell death in plants and yeast. Plant J 37: 554–565 [DOI] [PubMed] [Google Scholar]
- Janjusevic R, Abramovitch RB, Martin GB, Stebbins CE (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311: 222–226 [DOI] [PubMed] [Google Scholar]
- Jia Y, Loh YT, Zhou J, Martin GB (1997) Alleles of Pto and Fen occur in bacterial speck-susceptible and fenthion-insensitive tomato cultivars and encode active protein kinases. Plant Cell 9: 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Chen ZJ (2011) The role of ubiquitylation in immune defence and pathogen evasion. Nat Rev Immunol 12: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc Natl Acad Sci USA 103: 11086–11091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F, Thilmony R, He SY (2002) The Arabidopsis thaliana-Pseudomonas syringae interaction. The Arabidopsis Book 1: e0039, doi/10.1199/tab.0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaharada Y, Kelly S, Nielsen MW, Hjuler CT, Gysel K, Muszyński A, Carlson RW, Thygesen MB, Sandal N, Asmussen MH, et al. (2015) Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature 523: 308–312 [DOI] [PubMed] [Google Scholar]
- Khew CY, Teo CJ, Chan WS, Wong HL, Namasivayam P, Ho CL (2015) Brassinosteroid insensitive 1-associated kinase 1 (OsI-BAK1) is associated with grain filling and leaf development in rice. J Plant Physiol 182: 23–32 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Lin NC, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109: 589–598 [DOI] [PubMed] [Google Scholar]
- Kraft E, Stone SL, Ma L, Su N, Gao Y, Lau OS, Deng XW, Callis J (2005) Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol 139: 1597–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkeaw S, Tan S, Coaker G (2010) Molecular and evolutionary analyses of Pseudomonas syringae pv. tomato race 1. Mol Plant Microbe Interact 23: 415–424 [DOI] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Li B, Lu D, Shan L (2014) Ubiquitination of pattern recognition receptors in plant innate immunity. Mol Plant Pathol 15: 737–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Ahn IP, Ning Y, Park CH, Zeng L, Whitehill JG, Lu H, Zhao Q, Ding B, Xie Q, et al. (2012) The U-box/ARM E3 ligase PUB13 regulates cell death, defense, and flowering time in Arabidopsis. Plant Physiol 159: 239–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, Zhou JM (2005) Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci USA 102: 12990–12995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin NC, Abramovitch RB, Kim YJ, Martin GB (2006) Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl Environ Microbiol 72: 702–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeberg M, Cartinhour S, Myers CR, Schechter LM, Schneider DJ, Collmer A (2006) Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact 19: 1151–1158 [DOI] [PubMed] [Google Scholar]
- Liu J, Li W, Ning Y, Shirsekar G, Cai Y, Wang X, Dai L, Wang Z, Liu W, Wang GL (2012) The U-box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol 160: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C (2015) Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci 20: 12–19 [DOI] [PubMed] [Google Scholar]
- Lu D, Lin W, Gao X, Wu S, Cheng C, Avila J, Heese A, Devarenne TP, He P, Shan L (2011) Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 332: 1439–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C (2014) Plant PRRs and the activation of innate immune signaling. Mol Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Marino D, Peeters N, Rivas S (2012) Ubiquitination during plant immune signaling. Plant Physiol 160: 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. (2012) Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB. In Martin F, Kamoun S, eds, Effectors in Plant-Microbe Interactions. Wiley-Blackwell, Oxford, pp 121–154 [Google Scholar]
- Martin GB, Frary A, Wu T, Brommonschenkel S, Chunwongse J, Earle ED, Tanksley SD (1994) A member of the tomato Pto gene family confers sensitivity to fenthion resulting in rapid cell death. Plant Cell 6: 1543–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu J, Schwizer S, Martin GB (2014) Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS Pathog 10: e1004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsukura C, Aoki K, Fukuda N, Mizoguchi T, Asamizu E, Saito T, Shibata D, Ezura H (2008) Comprehensive resources for tomato functional genomics based on the miniature model tomato Micro-Tom. Curr Genomics 9: 436–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett P, Farnham G, Peart J, Baulcombe DC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. EMBO J 21: 4511–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkvold KR, Martin GB (2009) Advances in experimental methods for the elucidation of Pseudomonas syringae effector function with a focus on AvrPtoB. Mol Plant Pathol 10: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mural RV, Liu Y, Rosebrock TR, Brady JJ, Hamera S, Connor RA, Martin GB, Zeng L (2013) The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and Suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25: 3615–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S (2000) Molecular Evolution and Phylogenetics. Oxford University Press, Oxford [Google Scholar]
- Nguyen HP, Chakravarthy S, Velásquez AC, McLane HL, Zeng L, Nakayashiki H, Park DH, Collmer A, Martin GB (2010) Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol Plant Microbe Interact 23: 991–999 [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Tomanov K, Hofmann K, Stuible HP, Bachmair A (2012) Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytol 195: 23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntoukakis V, Mucyn TS, Gimenez-Ibanez S, Chapman HC, Gutierrez JR, Balmuth AL, Jones AME, Rathjen JP (2009) Host inhibition of a bacterial virulence effector triggers immunity to infection. Science 324: 784–787 [DOI] [PubMed] [Google Scholar]
- Oates ME, Stahlhacke J, Vavoulis DV, Smithers B, Rackham OJ, Sardar AJ, Zaucha J, Thurlby N, Fang H, Gough J (2015) The SUPERFAMILY 1.75 database in 2014: a doubling of data. Nucleic Acids Res 43: D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41: 215–243 [DOI] [PubMed] [Google Scholar]
- Pethica RB, Levitt M, Gough J (2012) Evolutionarily consistent families in SCOP: sequence, structure and function. BMC Struct Biol 12: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo MA, Zheng Y, Fernandez-Pozo N, Dunham DM, Fei Z, Martin GB (2014) Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biol 15: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, Lee Y, Liu J, Iobst S, Ouyang S, Buell CR (2005) Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics 6: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebrock TR, Zeng L, Brady JJ, Abramovitch RB, Xiao F, Martin GB (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448: 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin GB (2013) Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol 14: R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Salmeron JM, Oldroyd GE, Rommens CM, Scofield SR, Kim HS, Lavelle DT, Dahlbeck D, Staskawicz BJ (1996) Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86: 123–133 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Shan L, He P, Li J, Heese A, Peck SC, Nürnberger T, Martin GB, Sheen J (2008) Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4: 17–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Pruneda JN, Lo RS, Fowler DM, Kim HJ, Hiatt JB, Shendure J, Brzovic PS, Fields S, Klevit RE (2013) Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci USA 110: E1263–E1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijk SJ, Timmers HT (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J 24: 981–993 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2003) The ubiquitin/26S proteasome pathway, the complex last chapter in the life of many plant proteins. Trends Plant Sci 8: 135–142 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2012) The expanding universe of ubiquitin and ubiquitin-like modifiers. Plant Physiol 160: 2–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J (2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Wei CF, Kvitko BH, Shimizu R, Crabill E, Alfano JR, Lin NC, Martin GB, Huang HC, Collmer A (2007) A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J 51: 32–46 [DOI] [PubMed] [Google Scholar]
- Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W (2008) Arabidopsis UEV1D promotes lysine-63-linked polyubiquitination and is involved in DNA damage response. Plant Cell 20: 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, He P, Abramovitch RB, Dawson JE, Nicholson LK, Sheen J, Martin GB (2007) The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J 52: 595–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Q, Guo HS, Dallman G, Fang S, Weissman AM, Chua NH (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419: 167–170 [DOI] [PubMed] [Google Scholar]
- Xu W, Marcu M, Yuan X, Mimnaugh E, Patterson C, Neckers L (2002) Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc Natl Acad Sci USA 99: 12847–12852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M (2009) Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol 10: 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L, Velásquez AC, Munkvold KR, Zhang J, Martin GB (2012) A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J 69: 92–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Park CH, Venu RC, Gough J, Wang GL (2008) Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol Plant 1: 800–815 [DOI] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16: 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Vega-Sánchez ME, Zhu T, Wang GL (2006) Ubiquitination-mediated protein degradation and modification: an emerging theme in plant-microbe interactions. Cell Res 16: 413–426 [DOI] [PubMed] [Google Scholar]
- Zhang J, Shao F, Li Y, Cui H, Chen L, Li H, Zou Y, Long C, Lan L, Chai J, et al. (2007) A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1: 175–185 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Tian M, Li Q, Cui F, Liu L, Yin B, Xie Q (2013) A plant-specific in vitro ubiquitination analysis system. Plant J 74: 524–533 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.