GA induction of genes involved in cell elongation, cell division, and phase transitions requires chromatin-remodeler PICKLE function.
Abstract
PICKLE (PKL) is an ATP-dependent chromodomain-helicase-DNA-binding domain (CHD3) chromatin remodeling enzyme in Arabidopsis (Arabidopsis thaliana). Previous studies showed that PKL promotes embryonic-to-vegetative transition by inhibiting expression of seed-specific genes during seed germination. The pkl mutants display a low penetrance of the “pickle root” phenotype, with a thick and green primary root that retains embryonic characteristics. The penetrance of this pickle root phenotype in pkl is dramatically increased in gibberellin (GA)-deficient conditions. At adult stages, the pkl mutants are semidwarfs with delayed flowering time, which resemble reduced GA-signaling mutants. These findings suggest that PKL may play a positive role in regulating GA signaling. A recent biochemical analysis further showed that PKL and GA signaling repressors DELLAs antagonistically regulate hypocotyl cell elongation genes by direct protein-protein interaction. To elucidate further the role of PKL in GA signaling and plant development, we studied the genetic interaction between PKL and DELLAs using the hextuple mutant containing pkl and della pentuple (dP) mutations. Here, we show that PKL is required for most of GA-promoted developmental processes, including vegetative growth such as hypocotyl, leaf, and inflorescence stem elongation, and phase transitions such as juvenile-to-adult leaf and vegetative-to-reproductive phase. The removal of all DELLA functions (in the dP background) cannot rescue these phenotypes in pkl. RNA-sequencing analysis using the ga1 (a GA-deficient mutant), pkl, and the ga1 pkl double mutant further shows that expression of 80% of GA-responsive genes in seedlings is PKL dependent, including genes that function in cell elongation, cell division, and phase transitions. These results indicate that the CHD3 chromatin remodeler PKL is required for regulating gene expression during most of GA-regulated developmental processes.
Bioactive gibberellins (GAs) play an important role in regulating diverse developmental processes in plants. In Arabidopsis (Arabidopsis thaliana), GAs promote seed germination, vegetative growth, floral induction, and flower and fruit development (Hauvermale et al., 2012). Upon binding to its receptor, the GA-receptor complex activates its signaling pathway by inducing degradation of DELLA proteins, which are conserved nuclear transcription regulators that function as master growth repressors in plants (Ueguchi-Tanaka et al., 2007; Sun, 2011). Recent studies indicate that a major mechanism of DELLA-induced transcription reprogramming is through its direct protein-protein interactions with key transcription factors and chromatin remodeling proteins (Xu et al., 2014; Davière and Achard, 2016). Some of these interactions result in activation of downstream genes, such as type-B ARABIDOPSIS RESPONSE REGULATORS involved in cytokinin signaling (Marín-de la Rosa et al., 2015), ABSCISIC ACID INSENSITIVE3 (ABI3) and ABI5 in ABA signaling (Lim et al., 2013), and Switch/Suc Nonfermenting chromatin-remodeling complexes (Sarnowska et al., 2013). In contrast, other interactions confer antagonistic effects on expression of downstream genes. For example, DELLA inhibits the function of basic helix-loop-helix transcription factors, PHYTOCHROME INTERACTING FACTORs (PIFs), in light signaling (de Lucas et al., 2008; Feng et al., 2008), and BRASSINAZOLE-RESISTANT1 (BZR1), a brassinosteroid signaling activator (Bai et al., 2012b). A recent study also found PKL to be a DELLA-interacting protein (Zhang et al., 2014); PKL is an ATP-dependent CHD3 chromatin remodeling enzyme (Ogas et al., 1999; Ho et al., 2013). The loss-of-function pkl mutant has a low penetrance (∼1%–10%) of the “pickle root” phenotype, i.e. a thick and green primary root that retains embryonic traits (Ogas et al., 1997). Interestingly, under GA-deficient conditions, this pickle root phenotype is dramatically enhanced to over 80% penetrance, suggesting that PKL may mediate GA-induced embryonic-to-vegetative transition (Ogas et al., 1997). In addition, the pkl mutants display semidwarf and delayed flowering phenotypes, which resemble mutants with reduced GA signaling (Ogas et al., 1997; Henderson et al., 2004).
Regulation of chromatin structure plays a pivotal role in changing gene expression during development in eukaryotes (Ho and Crabtree, 2010; Voss and Hager, 2014; Han et al., 2015). In animals, CHD3/CHD4 family proteins reorganize nucleosome structure, which results in either repression or activation of transcription, depending on their interacting proteins. For example, an association of CHD3/CHD4 proteins with the histone deacetylase complex leads to repression of target genes. Like the animal CHD3/4 proteins, PKL also activates and represses transcription of different target genes. However, PKL function appears to correlate with altered levels of the repressive mark H3K27me3 (trimethylation of Lys 27 of histone H3) at the target genes but does not always change histone acetylation (Zhang et al., 2008, 2012; Aichinger et al., 2009, 2011; Xu et al., 2016). It remains unclear whether PKL plays a direct role in generating or removing the H3K27me3 mark. In germinating seeds, PKL represses expression of seed-specific genes to suppress embryonic traits and promote embryonic-to-seedling transition (Ogas et al., 1997, 1999; Dean Rider et al., 2003). PKL also plays a key role in inhibiting auxin response factor ARF7/ARF19-induced lateral root initiation (Fukaki et al., 2006). On the other hand, PKL promotes hypocotyl growth of etiolated seedlings by interacting with transcription factors PIF3 and BZR1, regulators of light and BR signaling, respectively, to activate transcription of target genes (Zhang et al., 2014).
Although the phenotypes of the pkl mutants are similar to GA-response mutants, the specific role of PKL in regulating GA signaling activity remains largely unknown. Previous microarray analysis using RNA isolated from germinating seeds suggested that PKL and GA act in separate pathways, to inhibit expression of seed-specific genes (Zhang et al., 2008). This model was based on the synergistic effects of pkl mutation and uniconazole (a GA biosynthesis inhibitor) on the expression of PKL-responsive genes. Alternatively, it remains possible that there are PKL-dependent and PKL-independent GA-signaling pathways. This is supported by the recent finding that PKL directly binds to DELLA, and chromatin immunoprecipitation-quantitative PCR (qPCR) data suggested an antagonistic effect of DELLA on PKL association with the target genes IAA19 and PRE1 involved in hypocotyl elongation (Zhang et al., 2014). These two hypocotyl elongation genes are direct targets of PIFs and BZR1 (Bai et al., 2012b; Gallego-Bartolomé et al., 2012; Oh et al., 2014), and PKL was shown to activate these genes by binding directly to PIF3 and BZR1, whereas DELLA inhibits PKL interaction with PIF3 and BZR1 (Zhang et al., 2014). These data suggest that PKL and DELLA antagonistically regulate GA-induced hypocotyl elongation.
To elucidate further the role of PKL in GA signaling and plant development, we examined the genetic interaction between PKL and DELLAs by generating and characterizing the hextuple mutant containing pkl and della pentuple (dP) mutations. In this report, we show that PKL plays a key role in most GA-induced and DELLA-repressed developmental processes. Even in the dP background, functional PKL is required for vegetative growth (including hypocotyl, leaf, and inflorescence stem elongation) and phase transitions (juvenile-to-adult leaf and vegetative-to-reproductive phase). Consistent with these results, our RNA-sequencing (RNA-seq) analysis further shows that 80% of GA-responsive genes are PKL dependent, including genes function in cell elongation, cell division, and phase transitions.
RESULTS
PKL Is Required for Most GA-Promoted Processes, Even in the Absence of DELLA Functions
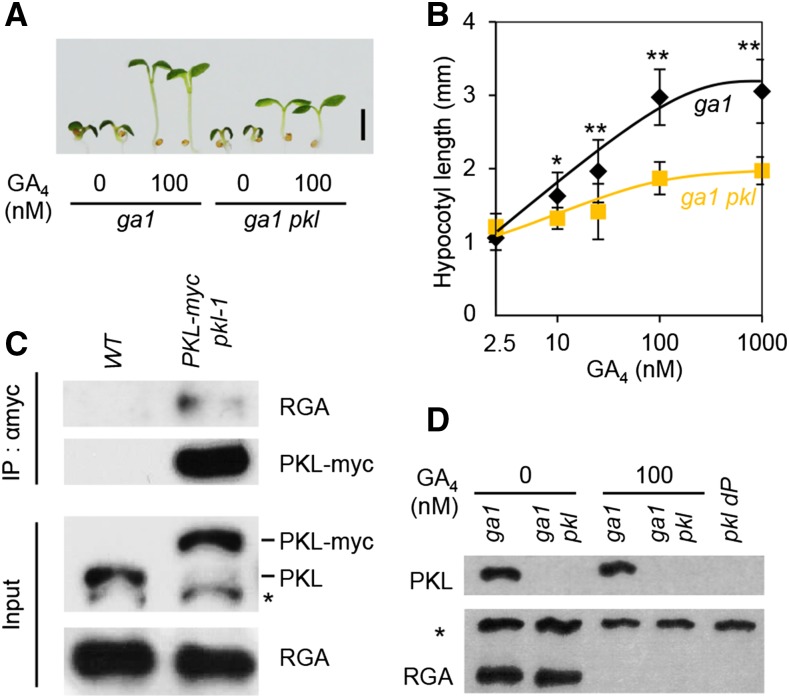
The pkl mutant (SAIL_73_H08; also named epp1-2; Jing et al., 2013) displayed reduced GA responses in hypocotyl elongation (Fig. 1, A and B) and was more sensitive to the effect of GA-biosynthesis inhibitor paclobutrazol (PAC) in preventing seed germination (Supplemental Fig. S1). These results are consistent with previous findings (Henderson et al., 2004) and suggest that PKL plays a positive role in GA signaling. Because PKL and DELLA proteins directly interact in vivo (Fig. 1C; Zhang et al., 2014), we tested whether PKL regulates RGA (an AtDELLA) protein accumulation. However, the endogenous RGA protein levels are similar in ga1 and ga1 pkl (Fig. 1D), indicating that RGA stability is not affected by PKL.
Figure 1.
pkl shows a reduced response to GA. A and B, ga1-13 pkl is less responsive to GA than ga1-13. Plants were grown in the presence of different concentrations of GA4 for 7 d under continuous light. A, Scale bar, 2 mm. B, Average hypocotyl lengths are shown as means ± se (n = 10). *P < 0.05; **P < 0.01 (Student’s t test). C, PKL-myc and RGA interact in vivo. Immunoprecipitation was performed using anti-myc antibody and protein extracts of wild-type (WT) and PPKL:PKL-myc pkl1-1 seedlings that were grown in the presence of PAC. Coimmunoprecipitated RGA was detected using anti-RGA antibody. *Nonspecific band. D, Immunoblot shows comparable accumulation of RGA proteins in ga1 pkl and ga1 seedlings grown as in A.
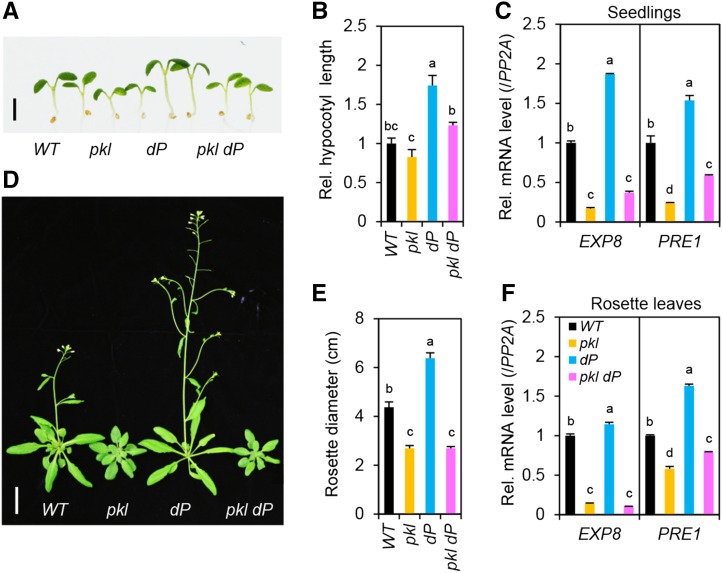
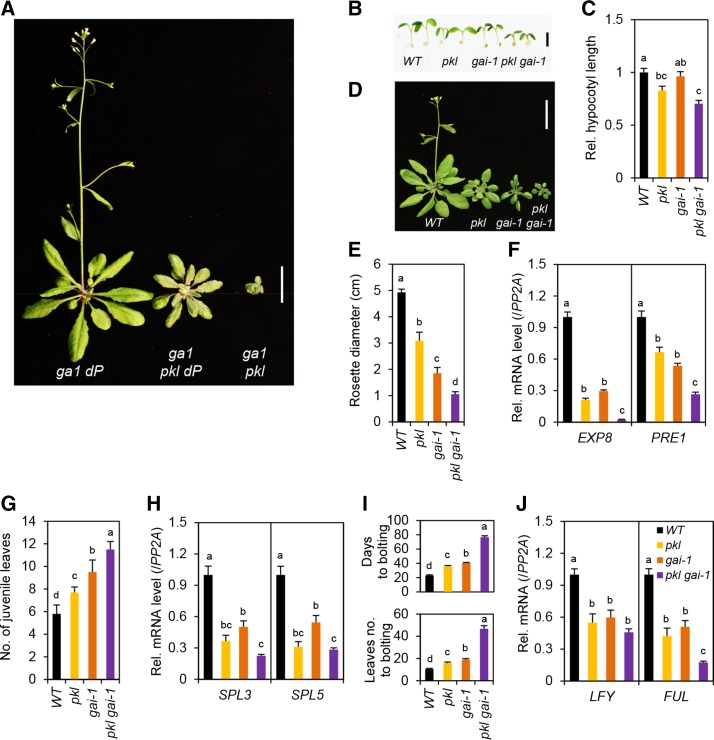
To investigate further the role of PKL in promoting GA signaling, we generated the hextuple mutant pkl dellaP (dP) to examine the genetic interaction between PKL and DELLAs. Previous studies show that DELLAs regulate all GA-mediated developmental processes (Davière and Achard, 2013), and dP display constitutive GA responses, including seed germination, vegetative growth, and floral induction (Park et al., 2013). We found that, in comparison to dP, the hextuple mutant pkl dP displayed reduced vegetative growth, including hypocotyl elongation (Fig. 2, A and B), rosette leaf, and stem growth (Fig. 2, D and E). We also analyzed the effect of pkl and dP on the expression of two DELLA target genes, EXP8 and PRE1, which are DELLA repressed (Bai et al., 2012b; Park et al., 2013). PRE1 is also a direct target of PKL, and its expression is induced by PKL (Zhang et al., 2014). Figure 2, C and F, shows that EXP8 and PRE1 mRNA levels in young seedlings and rosette leaves were reduced by pkl and increased by dP. Consistent with the plant phenotype, transcript levels of these genes in pkl dP were similar to that in pkl (for EXP8) or were intermediate between those in pkl and dP (for PRE1).
Figure 2.
PKL is required for stem and leaf growth in the dellaP mutant (dP). A and B, Hypocotyl elongation of wild type (WT), pkl, dP, and pkl dP. Scale bar = 2 mm in A. B, Hypocotyls of seedlings grown under continuous white light for 7 d were measured (n = 10). C, DELLA-repressed and GA-induced genes (EXP8 and PRE1) in 7-d-old seedlings are repressed in pkl and pkl dP. Relative transcript levels, normalized using PP2A (a constantly expressed gene; Czechowski et al., 2005), were determined by RT-qPCR analysis (three biological replicates). D, Adult plant phenotypes of WT, pkl, dP, and pkl dP. Plants were grown for 34 d under long-day condition. Scale bar = 2 cm. E, Average rosette diameter of 25-d-old LD-grown plants (n = 7). F, Transcript levels of DELLA-repressed genes in rosette leaves of 25-d-old LD-grown plants were quantified by qRT-PCR. Plants were sampled at ZT0 (three biological replicates). In B, C, E, and F, data are means ± se. Different letters above the bars indicate significant differences, P < 0.01.
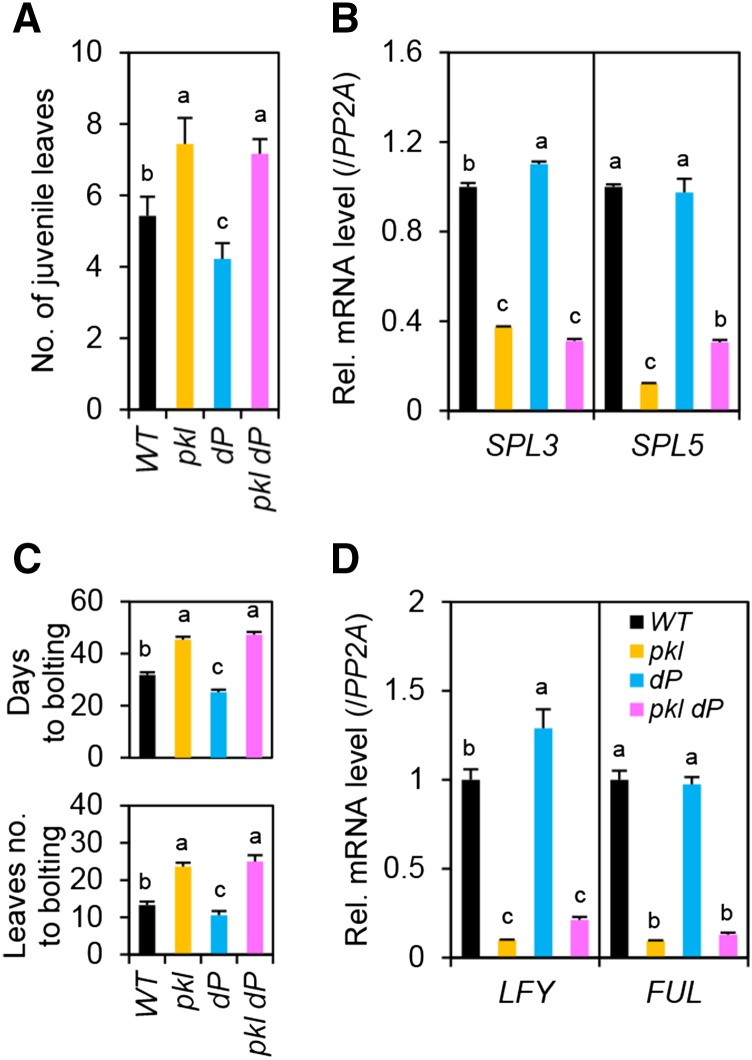
GA induces juvenile-to-adult phase transition as indicated by the appearance of the abaxial trichomes (Chien and Sussex, 1996; Telfer et al., 1997). GA also promotes vegetative-to-inflorescence phase transition (Wilson et al., 1992). Both of these GA-induced phase transitions are repressed by DELLAs (Silverstone et al., 1997; Dill and Sun, 2001; Gan et al., 2007; Park et al., 2013). We found that pkl is epistatic to dP in juvenile-to-adult transition (Fig. 3A) and floral induction (Fig. 3C). These phase transitions are controlled by key transcription factors: SQUAMOSA PROMOTER BINDING PROTEIN-LIKEs (SPLs) for juvenile-to-adult phase transition, and SPLs, FRUITFUL (FUL, also known as AGL8), and LEAFY (LFY) for floral induction (Weigel et al., 1992; Hempel et al., 1997; Huijser and Schmid, 2011). Previous studies showed that GA promotes these phase transitions by inducing transcription of SPLs (Galvão et al., 2012; Porri et al., 2012) and LFY (Blazquez et al., 1998), and DELLAs inhibit their expression (Galvão et al., 2012; Park et al., 2013). A recent study also showed that PKL promotes juvenile-to-adult transition by reducing expression of miR156, which inhibits expression of SPLs (Xu et al., 2016). By RT-qPCR analysis, we showed that SPL3 and LFY mRNA levels were slightly induced by dP, whereas SPL3, SPL5, LFY, and FUL mRNA levels were repressed by pkl (Fig. 3, B and D). In pkl dP, transcript levels of these phase transition genes were either similar to those in pkl (for SPL3, LFY, and FUL) or intermediate between those in pkl and dP (for SPL5). Taken together, these results indicate that even in the absence of any functional DELLAs, PKL is required to promote vegetative growth and phase transitions including juvenile-to-adult transition and flowering.
Figure 3.
PKL is required for developmental transitions in dellaP. A, Delayed juvenile-to-adult phase transition in pkl and pkl dP. Juvenile leaves were counted in LD-grown plants (n = 9). B, Expression of SPL3 and SPL5. Nineteen-day-old LD-grown plants were sampled at ZT12 for qRT-PCR (n = 3 biological replicates). C, Late flowering in pkl and pkl dP. Bolting days and rosette leaves at bolting were quantified in the plants grown under LD condition (SE, n = 9). D, Expression of LFY and FUL. Plants were grown and sampled as in B (three biological replicates). In A to D, data are means ± se. Different letters above the bars indicate significant differences, P < 0.01. WT, Wild type.
dellaP Rescued Seed Germination Defect and Embryonic Root Phenotype of pkl
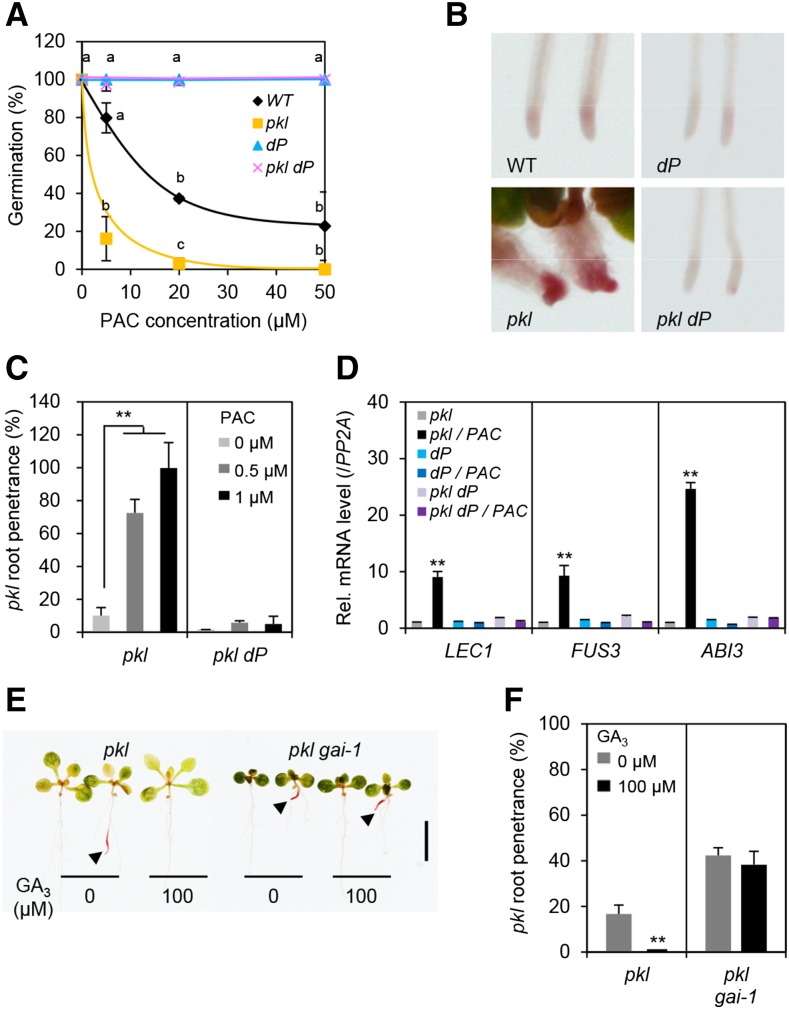
Although many GA-mediated processes depend on PKL even in the dP background, we found that dellaP (dP) rescued seed germination (Fig. 4A), and pickle root defects of pkl (Fig. 4, B and C). Previous studies showed that the PAC-treated pkl mutant seedlings express high levels of seed-specific genes, such as LEC1, FUS3, and ABI3 (Ogas et al., 1999; Aichinger et al., 2009). Consistent with the lack of pickle root phenotype in pkl dP, transcript levels of LEC1, FUS3, and ABI3 were not elevated in this hextuple mutant (Fig. 4D).
Figure 4.
dellaP rescued seed germination defect and pickle root phenotype of pkl. A, dP rescued seed germination of pkl in response to PAC treatment (average of three biological replicates). Different letters above the curves indicate significant differences, P < 0.01. The data for pkl and wild type (WT) shown in this figure are the same as those in Supplemental Figure S1. B and C, Pickle root penetrance is reduced by dP but enhanced by PAC. B, Sudan red staining of wild type, dP, pkl, and pkl dP. Roots of 9-d-old seedlings grown under the continuous white light (Wc) in the presence of 0.5 µm PAC are shown. C, Seedlings were grown on Murashige and Skoog media containing 0, 0.5, or 1 µm PAC under Wc. D, Relative transcript levels of seed-specific genes in pkl, dP, and pkl dP seedlings grown in 0 and 0.5 µm PAC under Wc. three biological replicates). E and F, Pickle root penetrance was reduced by GA, but enhanced by gai-1. Representative Fat-red-stained seedlings of pkl and pkl gai-1 grown in 0 and 100 µm GA3 media under Wc. E, scale bar = 5 mm. F, Quantification of pickle root penetrance in pkl and pkl gai-1 grown as in E (average of three biological replicates, n > 30). In A, C, D, and F, the data are means ± se. In A, different letters above the bars indicate significant differences, P < 0.01. In C, D, and F, **P < 0.01.
pkl and gai-1 Additively Repress Plant Growth and Developmental Phase Transitions
Our analysis of the hextuple mutant pkl dP indicates that most of GA-promoted growth and developmental processes are repressed by DELLAs and activated by PKL. In addition, most of the data in Figures 2 and 3 showed that pkl was epistatic to dP in rosette leaf growth and phase transition, under wild-type GA conditions, although the hypocotyl length of pkl dP was intermediate between those of pkl and dP. However, ga1 pkl was more dwarfed than pkl (Fig. 1A), likely due to elevated accumulation of DELLAs in the GA-deficient background. To examine pkl and dP interaction further, we compared the phenotypes of the heptuple mutant ga1 pkl dP to ga1 pkl and ga1 dP. As shown in Figure 5A, this heptuple mutant displayed an intermediate phenotype when compared to ga1 pkl and ga1 dP, indicating that pkl and dP do not display a simple epistatic relationship. Based on these results and the fact that DELLA and PKL directly interact, we predicted that pkl and gai-1 (a gain-of-function, semidwarf mutant; Koornneef et al., 1985; Peng et al., 1997) should additively inhibit GA responses. Indeed, we found that pkl and gai-1 additively inhibit the growth of hypocotyl and rosette leaf (Fig. 5, B–E), juvenile-to-adult phase transition (Fig. 5G), and floral induction (Fig. 5I). In addition, gai-1 increased the penetrance of the pickle root phenotype in pkl gai-1 even after GA treatment (Fig. 4, E and F). Consistent with the phenotypes of pkl gai-1, transcript levels of EXP8, PRE1, SPLs, and FUL were further reduced in this double mutant in comparison to the pkl and gai-1 single mutants (Fig. 5, F, H, and I).
Figure 5.
PKL and DELLAs antagonistically regulate plant development. A, ga1 pkl dP displayed intermediate phenotypes in comparison to ga1 pkl and ga1 dP. The photo shows representative 32-d-old plants that were grown under LD (bar = 2 cm). B to E, pkl and gai-1 additively repress growth and developmental transitions. B and C, Additive inhibition of hypocotyl elongation in gai-1 pkl. Seedlings were grown under continuous white light for 7 d (n = 10). B, Scale bar = 2 mm. D, Phenotypes of 32-d-old wild type (WT), pkl, gai-1, and pkl gai-1 under LD (bar = 2 cm). E, Additive inhibition of vegetative growth in gai-1 pkl. Twenty-five-day-old LD-grown plants were quantified for rosette diameter (n = 6). F, Expression of EXP8 and PRE1 in 19-d-old LD-grown plants. Plants were sampled at ZT0 (three biological replicates). G, Additive delay of juvenile-to-adult phase transition in gai-1 pkl grown under LD condition (n = 10). H, Expression of SPL3 and SPL5 grown and sampled as in F (three biological replicates). I, Additive delay of flowering in gai-1 pkl. Bolting days and rosette leaves at bolting were quantified in the plants grown under LD (n = 8). J, Expression of LFY and FUL grown and sampled as in F (three biological replicates). In C and E to J, the data are means ± se. Different letters above the bars indicate significant differences; P < 0.01.
Expression of 80% of GA-Responsive Genes Is PICKLE Dependent
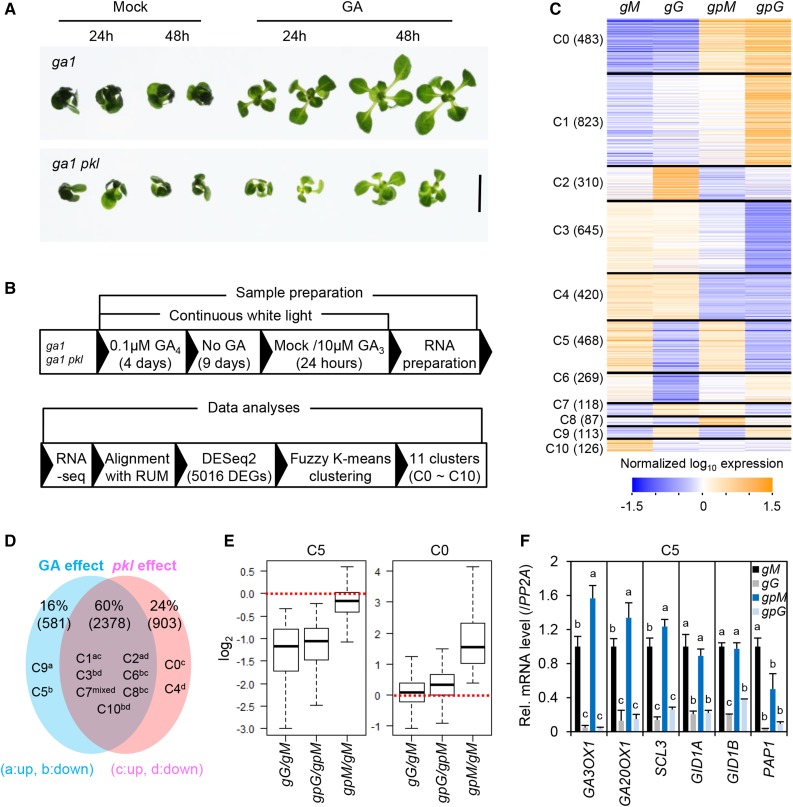
Based on the pkl dP mutant phenotype, we predicted that PKL plays a key role in regulating GA-responsive genes that are important for vegetative growth and phase transitions. To test this hypothesis, global transcriptome analysis was performed by RNA-seq using shoots of 13-d-old ga1-13 and ga1-13 pkl that were mock-treated or 10 µm GA3-treated for 24 h. This GA treatment of ga1 in the presence or absence of the pkl mutation allowed us to identify genes regulated either by GA, PKL, or both. The GA-deficient mutant ga1-13 is a null allele in the Col-0 background (Eriksson et al., 2006) and is a severe dwarf (Fig. 6A). To avoid the indirect effect of the pickle root on gene expression, we eliminated this phenotype by germinating the ga1 and ga1 pkl seeds in the presence of 0.1 µm GA4; this low concentration of GA4 was used during the pretreatment because GA4 can be rapidly deactivated in the plant. After 4 d, these seedlings were then transferred to media without GA for 9 d to deplete the residual GA4 (Fig. 6B). The resulting 13-d-old seedling (ga1 and ga1 pkl) displayed GA-deficient, dark-green dwarf phenotypes, with ga1 pkl showing an even more severe phenotype than ga1 (Fig. 6A). Subsequently, these 13-d-old ga1 and ga1 pkl seedlings were then treated with mock or 10 µm GA3 for 24 h; GA3 was used here because it is more stable in vivo, as it cannot be inactivated efficiently in the plant. The 24 h time point for GA treatment was chosen because at earlier time points (1–3 h), the growth-relevant genes such as EXPs, were only slightly induced by GA (Supplemental Fig. S2). We obtained two biological replicas of the four samples (mock-treated ga1 [gM], GA-treated ga1 [gG], mock-treated ga1 pkl [gpM], and GA-treated ga1 pkl [gpG)]), isolated mRNAs, and analyzed by RNA-seq analysis. RNA-seq data were analyzed using two-factor modeling implemented in the DESeq2 package (Love et al., 2014) to identify genes that are regulated either by GA treatment, pkl mutation, or both GA treatment and pkl mutation. We identified 581 genes that are only responsive to GA, 903 genes that are only affected by pkl, and 2252 genes that are responsive to both GA and pkl (Fig. 6D). Importantly, 80% of GA-responsive genes displayed PKL-dependent expression, suggesting that PKL plays a key role in regulating GA responses.
Figure 6.
Eighty percent of GA-regulated genes depend on PKL for their response to GA. A, Growth response to GA in ga1 and ga1 pkl. Plants were grown as described in B. Representative plants are shown. Scale bar = 5 mm. B, Summary of plant growth conditions, mutants/treatments for sample preparation, and RNA-seq analysis (two biological replicates for each genotype/treatment). C and D, Heat maps and Venn diagram of coregulated gene clusters identified by fuzzy K-means clustering. Clusters are categorized based on their responses to GA and/or pkl mutation. gG versus gM for GA-responsive genes in PKL background; gpG versus gpM for GA-responsive genes in the pkl mutant background; gpM versus gM for pkl-responsive genes without GA treatment; gpG versus gG for pkl-responsive genes after GA treatment. A, Up-regulated by GA; B, Down-regulated by GA; C, Up-regulated in pkl; D, Down-regulated in pkl. D, Averaged expression pattern of genes in each cluster. Normalized log values of uniquely aligned read counts are shown for all samples, where gM, gG, gpM, and gpG indicate mock-treated ga1, GA-treated ga1, mock-treated ga1 pkl, and GA-treated ga1 pkl, respectively, with two replicates. E, Standard box plots for the C5 cluster (GA-repressed and PKL-independent) and the C0 cluster (GA-independent and PKL-dependent). gG/gM indicates the effect of GA on cluster gene expression in the ga1 background; gpG/gpM indicates the effect of GA on gene expression in ga1 pkl; gpM/gM indicates the effect of pkl on gene expression in the ga1 background. The thick line within each box indicates the median. For the C5 cluster, the means are −1.38 (gG/gM), −1.22 (gpG/gpM), and −0.21 (gpM/gM). For the C0 cluster, the means are 0.10 (gG/gM), 0.32 (gpG/gpM), and 1.90 (gpM/gM). F, RT-qPCR confirms the effect of GA and pkl on the expression of genes in the C5 cluster. Data are means ± se (three biological replicates). Different letters above the bars indicate significant differences; P < 0.01.
We then used fuzzy K-means clustering (Gasch and Eisen, 2002) as described in “Materials and Methods,” to further identify patterns of coregulation among these GA- and/or PKL-responsive genes. We identified 11 clusters (>80 members with membership value > 0.5) showing similar expression patterns over the four samples (Fig. 6, C and D; Supplemental Data S1). Gene Ontology (GO) terms enriched in each cluster are shown in Supplemental Table S1 and Supplemental Data S2. The GA-responsive, PKL-independent genes belong to the C5 and C9 clusters; The C5 genes were repressed by GA, whereas C9 genes were induced by GA (Fig. 6, C and E). The C5 cluster (GA-repressed genes) contains known early DELLA-induced genes (Zentella et al., 2007), including GA biosynthesis genes (GA3ox1 and GA20ox1), GA receptors (GID1A, GID1B), GA signaling component (SCL3), and PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1), also known as MYB75 that activates transcription of genes encoding anthocyanin biosynthesis enzymes (Loreti et al., 2008; Supplemental Data S1). RT-qPCR analysis further confirmed that expression of these genes are down-regulated by GA but are not affected by pkl (Fig. 6F).
The PKL-responsive, GA-independent genes belong to the C0 and C4 clusters (Fig. 6, C and E). The C0 cluster genes are upregulated by the pkl mutation and contain 28 seed-specific genes (Supplemental Table S1), which is consistent with the role of PKL in repressing expression of these genes during postembryonic development. The C4 cluster (downregulated in pkl) is highly enriched in GO terms in plastid localization and photosynthesis, suggesting a role of PKL in regulating chloroplast function. When we compared our RNA-seq data with a previous microarray dataset for PKL-responsive genes using RNA from 14-d-old wild type and pkl seedlings (Zhang et al., 2012), the C0 and C4 genes showed the highest overlap among all gene clusters (Supplemental Fig. S3A).
The GA- and PKL-responsive genes (in C1–C3, C6–C8, and C10 clusters) show different expression combinations (Fig. 6, C and D). The C1 cluster genes are up-regulated by both GA and pkl, the C3 and C10 genes are down-regulated by both GA and pkl. Interestingly, the C3 cluster is enriched for genes for chloroplast function (Supplemental Table S1), similar to the C4 cluster. The C2 genes are up-regulated by GA but down-regulated by pkl. In addition, we found that GA induction of the C2 genes depends on PKL function (Fig. 6C). The C6 and C8 clusters are down-regulated by GA but up-regulated by pkl. However, C6 genes are only dramatically down-regulated by GA in the absence of pkl mutation, whereas C8 genes are only significantly down-regulated by GA in the presence of pkl. The C7 cluster genes showed different GA responses in the presence or absence of the pkl mutation: up-regulated by GA in ga1, up-regulated by pkl but down-regulated by GA in ga1 pkl. Interestingly, 80.5% of C7 genes function in translation, including genes encoding ribosomes or function in ribosome biogenesis (Supplemental Table S1; Supplemental Fig. S4). These expression patterns suggest that GA induces plant growth in part by promoting protein translation and that PKL represses expression of these genes. It is unclear why GA treatment represses their expression in the pkl mutant background.
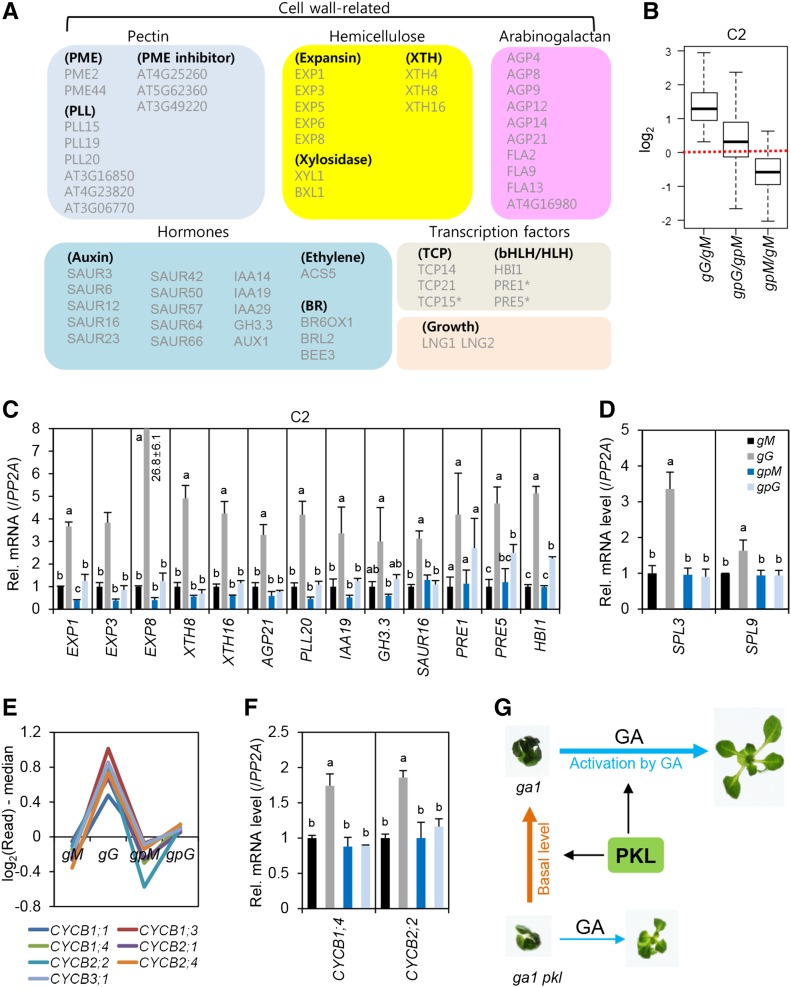
When we compared our RNA-seq data to a previous transcriptome dataset for DELLA-responsive genes (Gallego-Bartolomé et al., 2011), the C2 and C5 genes showed the highest overlap among all gene clusters (Supplemental Fig. S3B). Among the five clusters of GA- and PKL-responsive genes, we found that the C2 cluster is enriched for genes that are involved in GA-mediated vegetative growth (cell expansion, cell division) and phase transitions (Fig. 7). Therefore, our further analysis focused on this gene cluster.
Figure 7.
PKL enhances basal-level expression and GA-responsive induction of C2 cluster genes. A, A summary of growth-related genes that are enriched in the C2 cluster. PME, Pectin methylesterase; PLL, pectin lyase; AGP, arabinogalactan; XTH, xyloglucan endotransglucosylase/hydrolase; AGP, arabinogalactans; FLA, FASCICLIN-LIKE ARABINOGALACTAN. Asterisk indicates that the genes showed C2 cluster-like expression pattern but were excluded from the clustering process due to low expression levels. B, Standard box plot for the GA-induced and PKL-dependent C2 cluster genes. The means are 1.47 (gG/gM), 0.41 (gpG/gpM), and −0.62 (gpM/gM). C and D, Relative transcript levels of 13 selected growth-related C2 genes and two phase-transition genes SPL3 and SPL9 were confirmed by qRT-PCR (n = 3 biological replicates). E and F, GA activation of CYCB expression depends on PKL. E, RNA-seq data showing coregulation of seven CYCB genes. F, RT-qPCR analysis confirmed the effects of GA and pkl on the expression of two CYCB genes. G, A model for the role of PKL in promoting seedling growth. PKL maintains the basal expression and GA induction of C2 genes. C, D, and F, Data are means ± se. Different letters above the bars indicate significant differences, P < 0.01.
PKL Is Required for GA Induction of C2 Cluster Genes for Cell Expansion, Cell Division, and Phase Transitions
The C2 cluster genes are up-regulated by GA but are unresponsive or much less responsive to GA in the pkl background (Figs. 6C and 7B), indicating that PKL plays a key role in GA-induced expression of these genes. The C2 cluster includes a large number of genes that function in cell expansion. These include genes encoding cell wall modification enzymes (e.g. EXPs, AGPs [arabinogalactans], XTHs [xyloglucan endotransglucosylases/hydrolases], AGPs [arabinogalactans], and PLLs [pectin lyases]; Cosgrove, 2005), genes for hormone metabolism, transport, and signaling (e.g. GH3.3, AUX1, IAAs, smf SAURs; Ljung, 2013; Ren and Gray, 2015), growth-related transcription factors PREs, HBI1, and TCPs (S. Lee et al., 2006; Kieffer et al., 2011; Bai et al., 2012a), and novel cell-growth regulators LONGIFOLIA1 (LNG1) and LNG2 (Y.K. Lee et al., 2006; Fig. 7A). The C2 cluster genes also include genes that function in cell division (CYCBs for B-type cyclins; Fig. 6E), and phase transitions (SPLs). RT-qPCR analysis further confirmed that transcript levels of 17 C2 cluster genes were up-regulated by GA, whereas the pkl mutation abolished or greatly reduced the inductive effect of GA (Fig. 7, C, D, and F).
In the severe GA-deficient ga1 background, PKL appears to play a role in maintaining the basal expression of the C2 cluster gene because pkl mutation further reduced transcript levels of C2 genes (Figs. 6C and 7B). This is consistent with the more severely dwarfed phenotype of ga1 pkl in comparison to that of ga1 (Fig. 6A).
Two of the C2 cluster genes, IAA19 and PRE1, were recently shown to be induced by PKL via its direct interaction with PIF3 and BZR1, which are two key transcription factors in light and BR signaling pathways, respectively (Zhang et al., 2014). We compared our C2 cluster gene list with previously published gene list for direct targets of PIF4 and BZR1 (Oh et al., 2012) and found that 48% of the C2 genes are BZR1 direct targets, 36% of C2 genes are PIF4 direct targets, and 22% of C2 genes are common targets of BZR1 and PIF4 (Supplemental Fig. S5). These results indicate that PKL plays a key role in GA, BR, and light-regulated seedling growth.
DISCUSSION
Our mutant studies and RNA-seq analysis indicate that the chromatin remodeler PKL plays an important role in most of GA-regulated processes, including vegetative growth and developmental phase transitions (Fig. 7G). In the GA-deficient ga1 background, PKL also plays a role in maintaining a basal level of expression of the C2 genes. Although a previous study showed that DELLAs interfere with PKL function by direct protein-protein interaction (Zhang et al., 2014), we show that removing all DELLA functions (with dP mutations) is insufficient to rescue the semidwarf and late-flowering phenotypes of pkl. Our RNA-seq results are consistent with the pkl dP mutant phenotype. The GA- and PKL-induced C2 cluster genes include many genes involved in cell elongation, cell division, and phase transitions, and expression of these genes can only be induced by GA when PKL is functional. The C2 cluster is also enriched for direct target genes of PIF4 and BZR1; these transcription factors have been shown previously to bind PKL and DELLAs (de Lucas et al., 2008; Feng et al., 2008; Bai et al., 2012b; Zhang et al., 2014). Taken together, these results suggest that PKL may activate the growth-related C2 genes by direct interaction with PIFs and BZR1 and that DELLAs inhibit expression of C2 genes by antagonistic interactions with PKL, PIFs, and BZR1. Among the C2 genes, PRE1 and IAA19 were shown to be direct targets of PKL by chromatin immunoprecipitation-qPCR, and elevated DELLA levels led to reduced PKL binding to the promoters of PRE1 and IAA19 (Zhang et al., 2014). Similar analysis will need to be performed to determine whether other C2 genes are direct PKL and DELLA targets.
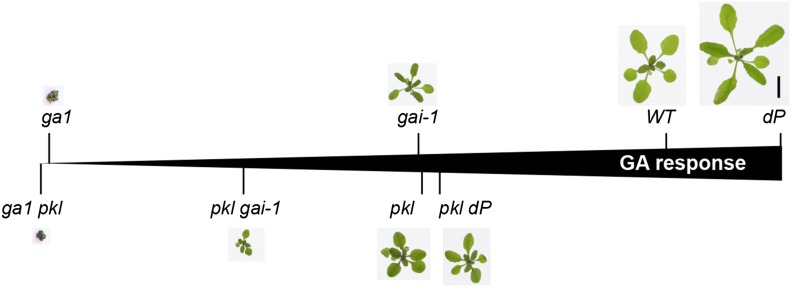
Our genetic analysis further showed that pkl and dP do not always display a simple epistatic relationship (Fig. 8). In the wild-type GA background, pkl appeared to be epistatic to dP in stem elongation and phase transitions. However, in the ga1 background, dP partially rescued these pkl defects. The effect of dP in the wild-type GA background is less dramatic than that in the ga1 background, likely because DELLAs accumulate to low levels under wild-type GA conditions. Considering that the pkl allele (epp1-2) used in our genetic analysis is a null allele, our data suggest that DELLAs repress vegetative growth and phase transitions by antagonizing PKL as well as other unknown factors that are independent of PKL. Both PKL-dependent and PKL-independent pathways are required for full activation of GA-induced vegetative growth and phase transitions. This idea was further supported by the additive effects of gai-1 (a gain-of-function DELLA mutation) and pkl in repressing these developmental processes. In contrast, in germinating seeds, transcriptional regulation including chromatin remodeling may be unique in comparison to later developmental stages (vegetative growth and phase transition). We found that dP completely rescued germination and pickle root phenotypes of pkl. These results suggest that DELLAs inhibit germination and enhance embryonic cell fate by inhibiting both PKL and other factors that are PKL independent. However, unlike vegetative growth and phase transitions, de-repression of these PKL-independent factors by removing DELLAs (in the dP background) is sufficient to compensate for the pkl defect during seed germination. Taken together, DELLAs repress all developmental stages examined by inhibiting both PKL-dependent and PKL-independent pathways. However, in the dP background, PKL-dependent pathway is only essential for GA-induced vegetative growth and phase transition but is dispensable for GA-induced seed germination. Identification of PKL-independent DELLA-interacting factors will help to elucidate the mechanisms involved.
Figure 8.
The effect of pkl on GA responses during vegetative growth. Photos show representative 19-d-old plants that were grown under LD conditions. Scale bar (1 cm) is shown in the dP mutant image. WT, Wild type.
PKL has been shown previously to promote embryonic-to-vegetative transition by suppressing the expression of seed-specific genes (Dean Rider et al., 2003). This is consistent with our RNA-seq data showing that the PKL-repressed genes in the C0 cluster are enriched for seed-specific genes. We also found that the PKL-induced genes in the C3 and C4 clusters are enriched for genes involved in photosynthesis and chloroplast development, suggesting that PKL plays a role in regulating chloroplast function.
Our data and previous studies indicate that the CHD3 chromatin remodeler PKL plays a key role in regulating gene expression in response to multiple signals throughout plant development. However, the molecular mechanism of PKL function is less clear. Recent studies suggest that PKL represses gene expression by increasing the repressive H3K27me3 mark, and in one case PKL also reduces the active H3K27ac2 mark (Zhang et al., 2008, 2012, 2014; Aichinger et al., 2009; Xu et al., 2016). However, PKL has also been linked to activation of gene expression through reduction of H3K27me3 at the target gene promoters. Furthermore, a CHD3 protein, CHR729 in rice, has been shown to bind both the repressive H3K27me3 mark and the active H3K4me2 mark via two distinct domains, i.e. chromodomain and plant homeodomain, respectively (Hu et al., 2012). It will be important to examine the functions of plant homeodomain and chromodomain of PKL in planta. Elucidation of whether PKL plays a direct role in recruiting or removing repressive and/or active histone marks on target gene chromatin will help illuminate how PKL modulates gene expression during development.
MATERIALS AND METHODS
Plants Materials, Growth Conditions, and Statistical Analysis
Plants were grown in the growth room at 22°C under long-day (LD; 16-h-light/8-h-dark cycle) condition. All the plants used in this study are in Columbia (Col-0) background, and notably gai-1 and gai-t6 are introgressed lines obtained by backcrossing with Col-0 six times (Oh et al., 2007). pkl (SAIL_73_H08; epp1-2) and ga1-13 (SALK_109115) were obtained from the Arabidopsis Biological Resource Center. The dellaP mutant (rga-29: SALK_089146; gai-t6, rgl1: SALK_136162; rgl2: SALK_027654; rgl3-3: CS16355) and pPKL-PKL-myc pkl-1 transgenic line were described previously (Zhang et al., 2012; Park et al., 2013). Most of the experiments in this study used pkl (SAIL_73_H08; epp1-2). The only exception is the coimmunoprecipitation experiment, in which pkl1-1 allele was used. To generate pkl dellaP and ga1-13 dellaP, pkl and ga1-13 were first crossed with dellaP and then each of F1 plants was backcrossed with dellaP. Resulting F1 plants were genotyped to obtain the plants that were heterozygous for ga1-13 and pkl and either heterozygous or homozygous for each of della mutants, and hextuple mutants were obtained in the F2 generation. The ga1-13 pkl dP heptuple mutant was generated by crossing pkl dP with ga1 dP. All the primers used to test T-DNA insertions are listed in Supplemental Table S2. All statistical analyses were performed with the statistical package JMP Pro 10.0.2 (SAS Institute) using Student’s t tests.
Characterization of Plant Growth and Developmental Transitions
Seeds were sterilized by incubating with 0.5% hypochlorite and 0.1% Tween 20 for 10 min and rinsing with water four times. Sterilized seeds were stratified at 4°C for 4 d. Seeds in ga1-13 background were stratified in the presence of 50 µm GA4 and washed six times before planting. After stratification, seeds were planted on growth medium (GM) (0.5× Murashige and Skoog medium, 1% Suc, 0.05% MES, and 0.7% plant agar). For germination test, seeds (60 per assay) were planted on GM with different concentration of PAC and grown under continuous white light for 6 d. Protrusion of radicle was scored as germination. For hypocotyl elongation test, plants were grown on GM horizontally either in the presence or absence of different concentration of GA4 for 7 d under continuous white light and hypocotyl length of seedlings was measured using ImageJ software (https://imagej.nih.gov/ij/). For rosette diameter measurement, pictures of the plants grown under LD condition for 25 d were taken and analyzed using ImageJ software.
For the analysis of juvenile-to-adult transition during the vegetative stage, leaves lacking the trichomes on the abaxial side of 1-month-old LD-grown plants were counted under the dissecting microscope (Park et al., 2013). Flowering time was determined by scoring the number of rosette leaves and the required number of days until the plants had visible floral buds.
Lipid Staining for Pickle Root Phenotype Analysis
Accumulation of triacylglycerol in seedlings was visualized by staining using Sudan red. Briefly, staining solution was freshly prepared and filtered through two layers of filter paper (0.5% Sudan red and 60% isopropanol; Aichinger et al., 2009). Seedlings were dipped in the staining solution for 30 min, washed with water extensively, and visualized.
Coimmunoprecipitation and Immunoblot Analysis
To test PKL-RGA interaction in vivo, we used seedlings of pPKL-PKL-myc pkl-1 and Col-0 grown on GM with 0.5 µm PAC under the continuous white light condition for 9 d. Samples were ground in liquid nitrogen, and total proteins were extracted using nondenaturing extraction buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm β-mercaptoethanol, 1% Triton X-100, 50 µm MG-132, and 1× Protease inhibitor cocktail [Roche]). The lysate was cleared by passing through four layers of Miracloth and centrifugation at 4°C and 15,000 rpm for 10 min. Protein complex immunoprecipitated with anti-myc antibody, and protein A-agarose beads were analyzed by SDS-PAGE, and RGA was detected by immunoblotting using a rabbit anti-RGA serum (DU176; Silverstone et al., 2001). Endogenous PKL was detected using a rabbit anti-PKL serum (Li et al., 2005).
RNA Extraction, Reverse Transcription, and Quantitative PCR
Total RNA was isolated using the Quick-RNA MiniPrep kit (Zymo Research). In brief, 100 mg of fresh Arabidopsis (Arabidopsis thaliana) tissues were ground in extraction buffer and processed following the manufacturer’s protocol. Reverse transcription was performed using M-MLV RTase (Promega) using anchored oligo dT. For qPCR, the FastStart Essential DNA Green Master mix was used on a LightCycler 96 Instrument (Roche Applied Science). Relative transcript levels were normalized using PP2A (a constantly expressed gene; Czechowski et al., 2005).
RNA-Seq Analysis
One microgram of total RNA was processed for stranded mRNA sequencing, using the Illumina HiSeq2000 platform (Illumina) at the Duke Center for Genomic and Computational Biology. Minimum 22 million high-quality RNA-seq reads (50-bp single end) per sample were aligned to Arabidopsis genome and gene model (TAIR10) sequences, with RNA-seq Unified Mapper (Grant et al., 2011). On average, 92.28% (±0.81%) of reads were unambiguously mapped, while 3.53% (±0.81%) were aligned multiple times. We counted reads mapped to each gene model. After filtering out gene models with low expression (i.e. total unambiguously mapped read counts across all samples < 2), as well as a small number of gene models with more than 20% of their read counts derived from ambiguous mapping, 23,338 gene models were subjected to downstream analyses. Differently expressed gene models (DEGs) were identified using DESeq2 (Love et al., 2014), based on a stringent cutoff (adjusted P value < 0.001) recommended for a small-sample RNA-seq experiment (Soneson and Delorenzi, 2013). Total 5016 DEGs showed significantly different expression due to either the effect of GA, pkl mutation, or their interactions. Among them, coregulated DEG clusters were identified using fuzzy K-means clustering (Gasch and Eisen, 2002), based on regulated log values for read counts normalized across all samples per each DEG. We used PlantGSEA (Yi et al., 2013) and BinGO (Maere et al., 2005) to identify and visualize networks of GO terms enriched in each coregulated DEG cluster. The RNA-seq data have been deposited to the NCBI Sequence Read Archive database (BioProject ID: PRJNA359514).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: PKL (At2g25170), RGA (At2g01570), GAI (At1g14920), RGL1 (At1g66350), RGL2 (At3g03450), RGL3 (At5g17490), PP2A (At1g13320), LEC1 (At1g21970), FUS3 (At3g26790), ABI3 (At3g24650), EXP8 (At2g40610), PRE1 (At5g39860), SPL3 (At2g33810), SPL5 (At3g15270), LFY (At5g61850), FUL (At5g60910), GA3OX1 (At1g15550), GA20OX1 (At4g25420), SCL3 (At1g50420), GID1A (At3g05120), GID1B (At3g63010), PAP1 (At1g56650), EXP1 (At1g69530), EXP3 (At2g37640), XTH8 (At1g11545), XTH16 (At3g23730), AGP21 (At1g55330), PLL20 (At3g07010), IAA19 (At3g15540), GH3.3 (At2g23170), SAUR16 (At4g38860), PRE5 (At3g28857), HBI1 (At2g18300), SPL9 (At2g42200), CYCB1;4 (At2g26760), CYCB2;2 (At4g35620), and PP2A (At1g13320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. pkl is more sensitive to PAC than the wild type in seed germination.
Supplemental Figure S2. GA induction time course of EXPs.
Supplemental Figure S3. Comparison of RNA-seq data with previous microarray datasets.
Supplemental Figure S4. Ribosome-related C7 cluster genes were up-regulated by GA in ga1 but down-regulated by GA in ga1 pkl.
Supplemental Figure S5. The C2 cluster genes are highly enriched in PIF4- and BZR1-target genes.
Supplemental Table S1. Summary of GO terms enriched among genes in each cluster.
Supplemental Table S2. List of primers and their uses.
Supplemental Data 1. RNA-seq analyses on the effect of the pkl mutation, GA treatment, and their interactions (see “Materials and Methods” and “Results” sections in the main text for detail).
Supplemental Data S2. GO terms in biological process, molecular function, and cellular component categories that are overrepresented, with false discovery rate (see “Materials and Methods” in the main text) smaller than 0.05, in each cluster.
Supplementary Material
Glossary
- RNA-seq
RNA-sequencing
- PAC
paclobutrazol
- GO
Gene Ontology
Footnotes
This work was supported by the U.S. Department of Agriculture (2014-67013-21548 to T.-p.S.), the National Institutes of Health (R01 GM100051 to T.-p.S.), the National Science Foundation (MCB-0918954 to J.O. and MCB 1616827 to D.-H.O. and M.D.), the National Research Foundation of Korea (2012R1A2A1A01003133 to G.C.), and the Next Generation BioGreen21 Program (PJ011379) of the Rural Development Administration, Republic of Korea (M.D.).
Articles can be viewed without a subscription.
References
- Aichinger E, Villar CB, Di Mambro R, Sabatini S, Köhler C (2011) The CHD3 chromatin remodeler PICKLE and polycomb group proteins antagonistically regulate meristem activity in the Arabidopsis root. Plant Cell 23: 1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichinger E, Villar CB, Farrona S, Reyes JC, Hennig L, Köhler C (2009) CHD3 proteins and polycomb group proteins antagonistically determine cell identity in Arabidopsis. PLoS Genet 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Fan M, Oh E, Wang ZY (2012a) A triple helix-loop-helix/basic helix-loop-helix cascade controls cell elongation downstream of multiple hormonal and environmental signaling pathways in Arabidopsis. Plant Cell 24: 4917–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai MY, Shang JX, Oh E, Fan M, Bai Y, Zentella R, Sun T, Wang Z-Y (2012b) Brassinosteroid, gibberellin, and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14: 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazquez MA, Green R, Nilsson O, Sussman MR, Weigel D (1998) Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien JC, Sussex IM (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière JM, Achard P (2013) Gibberellin signaling in plants. Development 140: 1147–1151 [DOI] [PubMed] [Google Scholar]
- Davière JM, Achard P (2016) A pivotal role of DELLAs in regulating multiple hormone signals. Mol Plant 9: 10–20 [DOI] [PubMed] [Google Scholar]
- Dean Rider S Jr., Henderson JT, Jerome RE, Edenberg HJ, Romero-Severson J, Ogas J (2003) Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J 35: 33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O (2006) GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Taniguchi N, Tasaka M (2006) PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF7 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 48: 380–389 [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Alabadí D, Blázquez MA (2011) DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana. PLoS One 6: e23918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Grau-Enguix F, Abbas M, Locascio A, Thomas SG, Alabadí D, Blázquez MA (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109: 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvão VC, Horrer D, Küttner F, Schmid M (2012) Spatial control of flowering by DELLA proteins in Arabidopsis thaliana. Development 139: 4072–4082 [DOI] [PubMed] [Google Scholar]
- Gan Y, Yu H, Peng J, Broun P (2007) Genetic and molecular regulation by DELLA proteins of trichome development in Arabidopsis. Plant Physiol 145: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Eisen MB (2002) Exploring the conditional coregulation of yeast gene expression through fuzzy k-means clustering. Genome Biol 3: research0059.1–research0059.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant GR, Farkas MH, Pizarro AD, Lahens NF, Schug J, Brunk BP, Stoeckert CJ, Hogenesch JB, Pierce EA (2011) Comparative analysis of RNA-Seq alignment algorithms and the RNA-Seq unified mapper (RUM). Bioinformatics 27: 2518–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Wu MF, Cui S, Wagner D (2015) Roles and activities of chromatin remodeling ATPases in plants. Plant J 83: 62–77 [DOI] [PubMed] [Google Scholar]
- Hauvermale AL, Ariizumi T, Steber CM (2012) Gibberellin signaling: A theme and variations on DELLA repression. Plant Physiol 160: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel FD, Weigel D, Mandel MA, Ditta G, Zambryski PC, Feldman LJ, Yanofsky MF (1997) Floral determination and expression of floral regulatory genes in Arabidopsis. Development 124: 3845–3853 [DOI] [PubMed] [Google Scholar]
- Henderson JT, Li HC, Rider SD, Mordhorst AP, Romero-Severson J, Cheng JC, Robey J, Sung ZR, de Vries SC, Ogas J (2004) PICKLE acts throughout the plant to repress expression of embryonic traits and may play a role in gibberellin-dependent responses. Plant Physiol 134: 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Crabtree GR (2010) Chromatin remodelling during development. Nature 463: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho KK, Zhang H, Golden BL, Ogas J (2013) PICKLE is a CHD subfamily II ATP-dependent chromatin remodeling factor. Biochim Biophys Acta 1829: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Liu D, Zhong X, Zhang C, Zhang Q, Zhou DX (2012) CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc Natl Acad Sci USA 109: 5773–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Jing Y, Zhang D, Wang X, Tang W, Wang W, Huai J, Xu G, Chen D, Li Y, Lin R (2013) Arabidopsis chromatin remodeling factor PICKLE interacts with transcription factor HY5 to regulate hypocotyl cell elongation. Plant Cell 25: 242–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer M, Master V, Waites R, Davies B (2011) TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J 68: 147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Loenen MEP, van Rijn L, Zeevaart JAD (1985) A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant 65: 33–39 [Google Scholar]
- Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, Chung WI (2006) LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development 133: 4305–4314 [DOI] [PubMed] [Google Scholar]
- Lee S, Lee S, Yang KY, Kim YM, Park SY, Kim SY, Soh MS (2006) Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol 47: 591–600 [DOI] [PubMed] [Google Scholar]
- Li HC, Chuang K, Henderson JT, Rider SD Jr., Bai Y, Zhang H, Fountain M, Gerber J, Ogas J (2005) PICKLE acts during germination to repress expression of embryonic traits. Plant J 44: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Park J, Lee N, Jeong J, Toh S, Watanabe A, Kim J, Kang H, Kim DH, Kawakami N, et al. (2013) ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25: 4863–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K. (2013) Auxin metabolism and homeostasis during plant development. Development 140: 943–950 [DOI] [PubMed] [Google Scholar]
- Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P (2008) Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol 179: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M (2005) BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21: 3448–3449 [DOI] [PubMed] [Google Scholar]
- Marín-de la Rosa N, Pfeiffer A, Hill K, Locascio A, Bhalerao RP, Miskolczi P, Grønlund AL, Wanchoo-Kohli A, Thomas SG, Bennett MJ, et al. (2015) Genome wide binding site analysis reveals transcriptional coactivation of cytokinin-responsive genes by DELLA proteins. PLoS Genet 11: e1005337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogas J, Cheng J-C, Sung ZR, Somerville C (1997) Cellular differentiation regulated by gibberellin in the Arabidopsis thaliana pickle mutant. Science 277: 91–94 [DOI] [PubMed] [Google Scholar]
- Ogas J, Kaufmann S, Henderson J, Somerville C (1999) PICKLE is a CHD3 chromatin-remodeling factor that regulates the transition from embryonic to vegetative development in Arabidopsis. Proc Natl Acad Sci USA 96: 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Hu J, Yusuke J, Jung B, Paik I, Lee H-S, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19: 1192–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. eLife 3: e03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Nguyen KT, Park E, Jeon JS, Choi G (2013) DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis. Plant Cell 25: 927–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Ren H, Gray WM (2015) SAUR proteins as effectors of hormonal and environmental signals in plant growth. Mol Plant 8: 1153–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska EA, Rolicka AT, Bucior E, Cwiek P, Tohge T, Fernie AR, Jikumaru Y, Kamiya Y, Franzen R, Schmelzer E, et al. (2013) DELLA-interacting SWI3C core subunit of switch/sucrose nonfermenting chromatin remodeling complex modulates gibberellin responses and hormonal cross talk in Arabidopsis. Plant Physiol 163: 305–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Jung H-S, Dill A, Kawaide H, Kamiya Y, Sun TP (2001) Repressing a repressor: Gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martínez EC, Sun TP (1997) The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics 146: 1087–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneson C, Delorenzi M (2013) A comparison of methods for differential expression analysis of RNA-seq data. BMC Bioinformatics 14: 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. (2011) The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol 21: R338–R345 [DOI] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Motoyuki A, Matsuoka M (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58: 183–198 [DOI] [PubMed] [Google Scholar]
- Voss TC, Hager GL (2014) Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 15: 69–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville CR (1992) Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol 100: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hu T, Smith MR, Poethig RS (2016) Epigenetic regulation of vegetative phase change in Arabidopsis. Plant Cell 28: 28–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Liu Q, Yao T, Fu X (2014) Shedding light on integrative GA signaling. Curr Opin Plant Biol 21: 89–95 [DOI] [PubMed] [Google Scholar]
- Yi X, Du Z, Su Z (2013) PlantGSEA: A gene set enrichment analysis toolkit for plant community. Nucleic Acids Res 41: W98–W103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Bishop B, Ringenberg W, Muir WM, Ogas J (2012) The CHD3 remodeler PICKLE associates with genes enriched for trimethylation of histone H3 lysine 27. Plant Physiol 159: 418–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Jing Y, Jiang Z, Lin R (2014) The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26: 2472–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Rider SD Jr., Henderson JT, Fountain M, Chuang K, Kandachar V, Simons A, Edenberg HJ, Romero-Severson J, Muir WM, et al. (2008) The CHD3 remodeler PICKLE promotes trimethylation of histone H3 lysine 27. J Biol Chem 283: 22637–22648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.