Abstract
Under stress conditions, such as nutrient starvation, deacylated tRNAs bound within the ribosomal A-site are recognized by the stringent factor RelA, which converts ATP and GTP/GDP to (p)ppGpp. The signaling molecules (p)ppGpp globally rewire the cellular transcriptional program and general metabolism, leading to stress adaptation. Despite the additional importance of the stringent response for regulation of bacterial virulence, antibiotic resistance and persistence, structural insight into how the ribosome and deacylated-tRNA stimulate RelA-mediated (p)ppGpp has been lacking. Here, we present a cryo-EM structure of RelA in complex with the Escherichia coli 70S ribosome with an average resolution of 3.7 Å and local resolution of 4 to >10 Å for RelA. The structure reveals that RelA adopts a unique ‘open’ conformation, where the C-terminal domain (CTD) is intertwined around an A/T-like tRNA within the intersubunit cavity of the ribosome and the N-terminal domain (NTD) extends into the solvent. We propose that the open conformation of RelA on the ribosome relieves the autoinhibitory effect of the CTD on the NTD, thus leading to stimulation of (p)ppGpp synthesis by RelA.
INTRODUCTION
The stringent response (SR) is a central bacterial adaptation mechanism. In response to various environmental stimuli, the RelA/SpoT Homologue (RSH) proteins modulate the intracellular concentration of the alarmone nucleotides guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp), commonly referred to as (p)ppGpp (1–3). Production of (p)ppGpp mediates global rewiring of the cellular transcriptional program and general metabolism, leading to stress adaptation. The SR has been shown to play an important role in regulation of bacterial virulence (4), survival during host invasion (5), as well as antibiotic resistance (6) and persistence (7). Together with the absence of a cytoplasmic (p)ppGpp-mediated SR system in eukaryotes, this makes the RSH enzymes involved in (p)ppGpp metabolism promising new targets for drug development (3,8,9).
Historically, investigations of the SR were focused on the γ-proteobacterium Escherichia coli. In E. coli, the SR is orchestrated by two multi-domain long form RSH enzymes: RelA (10) and SpoT (11). The activity of the two proteins is regulated by different sets of stress signals. SpoT has a strong (p)ppGpp hydrolytic activity and a weak (p)ppGpp synthesis activity (12–14). By contrast, RelA has no hydrolytic activity, but possesses a strong ribosome-dependent (p)ppGpp synthetic activity that is activated by amino acid starvation via sensing of deacylated tRNA in the ribosomal A-site (15–17). It has also been shown that there is another activator of RelA, namely its product ppGpp (18). There are conflicting models for the mechanism of action of RelA. Biochemical studies suggested that (p)ppGpp synthesis causes RelA to ‘hop’ between ribosomes to sample the translational status of the cell (16). Subsequent live cell single molecule tracking experiments suggested that while activation of RelA by starved ribosomes induces RelA dissociation, RelA can perform multiple rounds of catalysis off the ribosome (19). This is hard to reconcile with recent live cell single molecule tracking experiments suggesting that under starvation conditions RelA remains bound to the ribosome and synthesis multiple rounds of (p)ppGpp synthesis (20).
Sequence analysis (2) and biochemical studies (21–23) suggest that RelA has a two domain architecture: An N-terminal domain (NTD) bearing hydrolase (HD) and synthetase (SYNTH) subdomains, the structure of which is available from the Gram-positive bacterium Streptococcus dysgalactiae subsp. equisimilis Rel protein (24). To date, there is no atomic structure of the RelA C-terminal domain (CTD), however, sequence homology suggests the presence of a TGS (Thr-RS, GTPase and SpoT) motif followed by a helical linker region connected to the C-terminal conserved cysteine (CC) and Aspartokinase, Chorismate mutase and TyrA (ACT) subdomains (2). Although the precise function of these subdomains is unclear, the CTD is critical for ribosome binding (25) as well as autoinhibition of the synthetase activity of RelA in the absence of the ribosome (21–23,26). Structural insights into the RSH interaction with the ribosome come from a cryo-electron microscopy (EM) structure of E. coli RelA bound to the Thermus thermophilus 70S ribosome programmed with tRNAfMet in the P-site and tRNAPhe in the A-site (25). The structure reveals that on the ribosome RelA interacts with a distorted A/T-tRNA (25), similar but distinct from the A/T-tRNA observed on the ribosome in the presence of EF-Tu (27–29). The limited resolution (∼11 Å) and conformation flexibility of the bound RelA, however, precluded assignment of the NTD and CTD (25).
Here, we present a cryo-EM structure of E. coli RelA in complex with a translating ribosome stalled with a deacylated-tRNA in the A-site, with an average resolution of 3.7 Å and local resolution of 4 to >10 Å for RelA. The structure reveals that the HD and SYNTH domains within the NTD of RelA are highly flexible and protrude into the solvent where no contact with the ribosome is apparent. In contrast, the CTD of RelA adopts an open conformation on the ribosome that stabilizes an A/T-like conformation of the deacylated tRNA in the A-site, which we term the A/R-tRNA state. The TGS domain of RelA interacts with the CCA-end, suggesting its involvement in discriminating deacylated- from aminoacylated-tRNAs. A long helical linker region extends from the TGS domain, wraps around the A/R-tRNA and positions the CC and ACT domains deeper within the intersubunit cavity, where they interact with H38 of the 23S rRNA. Overall, the structure enables a model to be proposed for how the open conformation of the CTD of RelA on the ribosome leads to stimulation of the (p)ppGpp activity of the NTD.
MATERIALS AND METHODS
Generation and purification of ErmCL_S10K-SRC
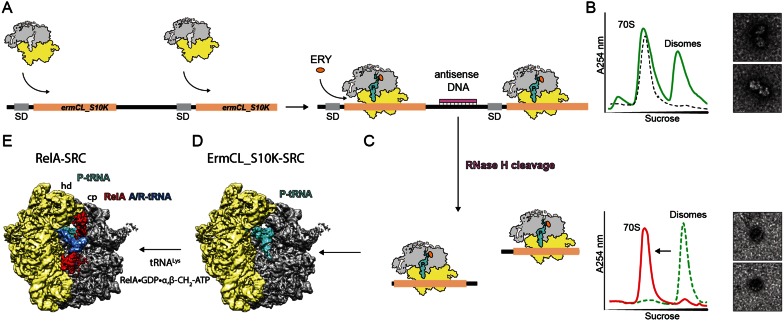
The RelA-stalled ribosomal complex (SRC) was generated based on the disome approach (Figure 1A–D), aspreviously described (30). The 2XermCL construct was modified by mutation of codon 10 AGC (Ser) to AAG (Lys) and synthesized (Eurofins, Martinsried, Germany) such that it contained a T7 promoter followed by a strong ribosome binding site (RBS) spaced by 7 nucleotides (nts) to the ATG start codon of the first ermCL_S10K cistron. A linker of 22 nts separated the stop codon of the first ermCL_S10K cistron and the start codon of the second ermCL_S10K cistron. The linker also comprised the strong RBS 7 nts upstream of the ATG start codon of the second ermCL_S10K cistron, enabling initiation of translation independent from the first ermCL_S10K cistron. With the exception of the S10K mutation, each ermCL_S10K cistron encoded amino acids 1–19 corresponding to the ErmCL leader peptide (Genbank accession number V01278) present on the macrolide resistance plasmid pE194 (31,32). The complete sequence of 2XermCL_S10K construct is: 5′-TAATACGACTCACTATAGGGAGTTTTATAAGGAGGAAAAAATATGGGCATTTTTAGTATTTTTGT AATCAAGACAGTTCATTATCAACCAAACAAAAAATAAAGTTTTATAAGGAGGAAAAAATATGGGCATTTTTAGTATTTTTGTAATCAAGACAGTTCAT TATCAACCAAACAAAAAATAA-3′ (T7 promoter, italics; RBS, bold; ErmCL ORF, underlined with ATC codon in P-site of stalled ribosome shown in bold; Annealing site for complementary DNA oligonucleotide, underlined). In vitro translation of the ermCL_S10K construct was performed using the Rapid Translation System RTS 100 E. coli HY Kit (5PRIME). Translation reactions were analyzed on sucrose density gradients (10%-55% sucrose in buffer A, containing 50 mM HEPES-KOH, pH 7.4, 100 mM KOAc, 25 mM Mg(OAc)2, 6 mM β-mercaptoethanol, 10 μM erythromycin and 1x Complete EDTA-free Protease Inhibitor cocktail (Roche)) by centrifugation at 154 693 ×g (SW-40 Ti, Beckman Coulter) for 3 h at 4°C. For ErmCL_S10K-SRC purification, disome fractions were collected using a Gradient Station (Biocomp) with an Econo UV Monitor (Biorad) and a FC203B Fraction Collector (Gilson). Purified ErmCL_S10K-SRC disomes were concentrated by centrifugation at 88 760 ×g for 4 h at 4°C (TLA120.2 rotor, Beckman Coulter). To obtain monosomes of the ErmCL_S10K-SRC, a short DNA oligonucleotide (5′-ttcctccttataaaact-3′, Metabion) was annealed to the linker between the ermCL_S10K cistrons, generating a DNA–RNA hybrid that could be cleaved by RNase H (NEB) treatment in buffer A at 25°C for 1 h. After cleavage of the disomes, ErmCL_S10K-SRC monosomes were again purified and concentrated by centrifugation at 88 760 × g for 4 h at 4°C (TLA120.2 rotor, Beckman Coulter).
Figure 1.
Generation of a RelA-stalled ribosome complex. (A) The bicistronic 2XermCL_S10K mRNA was translated in the presence of 10 μM erythromycin (ERY) in order to generate (B) ErmCL-S10K_SRC disomes. (C) ErmCL_S10K-SRC disomes were converted to monosomes by antisense DNA-mediated RNase H cleavage, as shown by sucrose density gradient centrifugation and negative stain electron microscopy (EM). (D) A-tRNA deficient ErmCL_S10K-SRCs were used as substrate for (E) RelA binding in the presence of deacylated tRNALys, GDP and α, β-methylene-ATP.
Expression and purification of RelA
RelA (Gene ID: 947244) was cloned into pET46LIC vector with an N-terminal hexahistidine tag (His6) for purification and detection purposes. An enterokinase cleavage site (bold) connects the His6 tag (underlined) to the RelA (MAHHHHHHVDDDDKM). The RelA plasmid was chemically transformed into E. coli BL21 (DE3) competent cells. A volume of 4L LB medium was inoculated at a 1:100 dilution with an overnight culture. RelA expression was induced by addition of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD600 = 0.5 for 1 h at 30°C. Cells were harvested by centrifugation at 5000 × g for 10 min at 4°C (Sorvall, SLC 6000 rotor) and the cell pellet was re-suspended using lysis buffer (300 mM NaCl, 50 mM NaH2PO4, 5 mM imidazole and 1 mM PMSF, pH 8.0). Cells were lysed using the microfluidizer (Microfluidics), followed by centrifugation at 38 724 × g for 30 min at 4°C to remove cellular debris (Sorvall, SS-34 rotor). The cleared lysate was then incubated at 4°C for 20 min with 1.6 ml of Ni-NTA slurry pre-equilibrated with lysis buffer. The lysate–Ni-NTA mixture was centrifuged at 500 ×g for 1 min after which the beads were washed four times with 10 ml of washing buffer (300 mM NaCl, 50 mM NaH2PO4 and 10 mM imidazole, pH8.0). Elution of RelA was carried out using 1 ml of elution buffer (300 mM NaCl, 50 mM NaH2PO4, 250 mM imidazole and 1 mM PMSF, pH 8.0). Eluted RelA was further purified by gel-filtration (Superdex 200 10/300 GL; pre-equilibrated in buffer B (50 mM HEPES, 100 mM KCl, 200 mM NaCl, 10 mM MgCl2, 5 mM β-mercaptoethanol, 2% glycerol and 1 mM PMSF, pH 7.8). Subsequently, purified RelA protein was concentrated via centrifugation through Amicon Ultra-0.5 ml centrifugal filters (30K, Merck Millipore). The activity of the RelA protein was confirmed using the ribosome-dependent ppGpp synthesis assay (Supplementary Figure S1) pasting.
ppGpp synthesis assay
The assays were performed as described in (18) with minor modifications. Ribosomal complexes were formed using heat activated vacant E. coli 70S ribosomes (0.5 μM) programmed with synthetic MF-mRNA 5′-GGCAAGGAGGUAAAAAUGUUCAAA-3′ (Sigma Aldrich) (1 μM), 0.3 mM 3H-GDP, 1 mM ATP, deacylated tRNAfMet and tRNAPhe (Chemical Block Ltd.) (1.5 μM each) (25 mM HEPES-KOH pH 7.5, 15 mM Mg(OAc)2, 95 mM KCl, 5 mM NH4Cl, 0.5 mM CaCl2, 8 mM putrescine and 1 mM spermidine) (33). 3H-ppGpp was separated from 3H-GDP on TLC (PEI cellulose, Macherey-Nagel) in 0.5 M KH2PO4 pH 3.5. UV shadowing of TLC with non-radioactive nucleotides was used to identify the 3H-GDP and 3H-ppGpp spots, the TLC plate was cut and 3H-GDP and 3H-ppGpp fragments of the plate were subjected to scintillation counting individually (Supplementary Figure S1).
Generation of the RelA-SRC using the ErmCL_S10K-SRC
The RelA-SRC complex was assembled using a final concentration of 0.125 μM ErmCL_S10K-SRC, 0.625 μM RelA, 0.625 μM tRNALys (Sigma-Aldrich), 500 μM α, β-methylene-ATP (Sigma-Aldrich), 500 μM GDP and 10 μM erythromycin. All components were pre-dissolved in buffer A (as mentioned before but excluding Ery and 1x Complete EDTA-free Protease Inhibitor cocktail). The binding reaction (RelA-SRC sample) was incubated at 37°C for 20 min.
Negative-stain electron microscopy
Ribosomal particles were diluted in buffer A to a final concentration of 0.5 A260/ml. One drop of each sample was deposited on a carbon-coated grid. After 30 s, grids were washed with distilled water and then stained with 2% aqueous uranyl acetate for 15 s. The remaining liquid was removed by touching the grid with filter paper. Micrographs were taken using a Morgagni transmission electron microscope (FEI), 80 kV, wide angle 1K CCD at direct magnifications of 72K.
Cryo-electron microscopy and single particle reconstruction
A total of 4 A260/ml monosomes of the RelA-SRC sample were applied to 2 nm pre-coated Quantifoil R3/3 holey carbon supported grids and vitrified using a Vitrobot Mark IV (FEI Company). Data collection was performed using EM-TOOLS (TVIPS GmbH) on a Titan Krios transmission electron microscope equipped with a Falcon II direct electron detector (FEI Company) at 300 kV at a pixel size of 1.084 Å and a defocus range of 0.7–2.2 μm. Ten frames (dose per frame of 2.5 e−/Å2) were aligned using Motion Correction software (34) and then processed using a frequency-limited refinement protocol that helps prevent overfitting (35), specifically by truncation of high frequencies (in this case, at 8 Å). As reported and expected (35), we find that using this processing regime the 0.143 FSC value provides a good indicator for the true average resolution of the map. Additionally, the local resolution of the map was calculated using ResMap (36). Power-spectra, defocus values, astigmatism and estimation of micrograph resolution were determined using the CTFFIND4 software (37). Micrographs showing Thon rings beyond 3.5 Å resolution were further manually inspected for good areas and power-spectra quality. Data were processed using the SPIDER software package (38), in combination with an automated workflow as described previously (39). After initial, automated particle selection based on the program SIGNATURE (40), initial alignment was performed with 197 090 particles using E. coli 70S ribosome as a reference structure (41). The data set could be sorted into four homogeneous subpopulations using an incremental K-means-like method of unsupervised 3D sorting (42) (Supplementary Figure S2): The ligand-bound subpopulation (24 749 particles; 13%) was defined by the presence of stoichiometric densities for P-tRNA, A/R-tRNA and RelA and could be refined to an average resolution of 3.7 Å (0.143 FSC) and a local resolution extending to 3.5 Å for the core of the 30S and 50S subunit as computed using ResMap (36) (Supplementary Figure S3). The final map of the RelA-SRC was Butterworth-filtered to 4 Å resolution.
Molecular modeling and map-docking procedures
The molecular model of the RelA-SRC was based on the ErmCL-TetM-SRC structure (PDB3J9Y, (43)), which was in turn based on an E. coli ribosome model from (29). The L11 stalk, 30S head domain, H38 and the L1 stalk were fitted as individual rigid bodies into the RelA-SRC map and subsequently manually adjusted and refined in Coot (44). Homology models of the RelA domains TGS (PDB2EKI) and ACT (PDB2KO1) were generated using HHPred (45) and Modeller (46) and could be unambiguously rigid body-fitted into the RelA-SRC cryo-EM density (Supplementary Figure S3). The homology model for the RelA HD-SYNTH domain was based on PDB1VJ7 (24) and fitted into the 12 Å filtered density. In order to yield the best fit, the HD was rotated slightly with respect to the SYNTH. The molecular model for the A/R-tRNA was based on the A/T tRNA of a crystal structure of EF-Tu-bound ribosomes (PDB2WRN, (27)), which was rigid body-fitted into the density. The CCA-end of the A/R-tRNA was tentatively adjusted in Coot (44) to illustrate its rough positioning within the TGS domain of RelA. Alignment of the EF-Tu structure (PDB5AFI) was performed in Chimera on the basis of the 23S rRNA, and has an rmsd of 0.64 with the 23S rRNA of the RelA structure.
Figure preparation
All figures showing electron densities and atomic models were generated using UCSF Chimera (47).
RESULTS
Generation of a RelA-stalled ribosome complex
The RelA-ribosome complex was generated in vitro by addition of purified RelA protein to SRCs. As in our previous studies (30,41,43), the SRCs were obtained using a dicistronic 2Xerm-mRNA encoding two identical Erm-stalling leader peptides (Figure 1A). In vitro translation of the 2Xerm mRNA in the presence of the macrolide antibiotic erythromycin generates disomes, where two ribosomes are stalled on the same mRNA (Figure 1B). The disomes were then separated from non-translating 70S ribosomes using sucrose gradients (Figure 1B). For structural analysis, the disomes were converted back to monosomes by annealing of an antisense DNA oligonucleotide to the linker region between the two erm cistrons (Figure 1C). We first used this disome approach to determine cryo-EM structures of ErmBL-SRCs, revealing that the sample contained a large population of ribosomes bearing A- and P-tRNAs (30) and was thus unsuitable for binding of ligands to the A-site. In contrast, our more recent structure of the ErmCL-SRC contained a single homogenous population of ribosomes with the ErmCL-peptidyl-tRNA (with codon 9 of the mRNA) in the P-site and a free A-site (41) (Figure 1D), which is thus suitable for determination of ErmCL-SRC structures with A-site bound ligands, as exemplified by the ribosome protection protein TetM (43). In the case of the RelA-SRC, we used a variant form of the ErmCL-SRC, termed ErmCL_S10K, where the A-site codon was mutated from AGC (Ser) to AAG (Lys) (Figure 1A). In agreement with previous studies (48,49), mutation of the Ser10 codon, which would be in the A-site of an erythromycin-stalled ErmCL-SRC (41,49), did not noticeably affect the stalling efficiency, and resulted in disome formation in the presence of erythromycin (Figure 1B). The reason for using the ErmCL_S10K construct rather than the wild type ErmCL was simply that, unlike deacylated tRNASer, deacylated tRNALys is commercially available. RelA-SRCs were therefore formed by incubating the ErmCL_S10K-SRC with deacylated tRNALys and purified recombinant E. coli RelA protein (that was shown to be active in ppGpp synthesis, Supplementary Figure S1). Synthesis of ppGpp by RelA has been proposed to lead to dissociation of RelA from the ribosome (16), thus we generated the RelA-SRCs in the presence of GDP and the non-hydrolysable ATP analogue α, β-methylene-ATP (Figure 1E).
Cryo-EM structure of the RelA-SRC
Cryo-EM data were collected on a Titan Krios transmission electron microscope with a Falcon II direct electron detector. From a total of 197 090 ribosomal particles, in silico sorting revealed a large mixture of ribosome populations containing either P-tRNA only (11%), A- and P-tRNAs without an E-tRNA (18%), or fully accommodated A-, P- and E-tRNAs (58%). Despite this heterogeneity, we were able to sort for a small population (24 749 particles, 13%) that contained a P-tRNA, but also the presence of A/R-tRNA and RelA bound within the ribosomal A-site (Supplementary Figure S2). The large heterogeneity of the data set was similar to that observed previously for the complex of E. coli RelA with the T. thermophilus 70S ribosome, where the RelA-containing ribosomes represented 15% of the total population (25). The RelA-containing ribosomes, which we term the RelA-SRC (Figure 1E), could be further refined to final average resolution of 3.7 Å (Fourier shell correlation (FSC) cut-off of 0.143, Supplementary Figure S3A). Although the average and local resolution calculations indicate that the majority of the core of the ribosome is 4.0 Å or better, the resolution of the A/R-tRNA and RelA ranged from 4 Å to >10 Å (Supplementary Figure S3B and C), indicating high flexibility of the A/R-tRNA and RelA within the ribosomal binding site, as observed previously (25).
Novel binding site for RelA on the ribosome
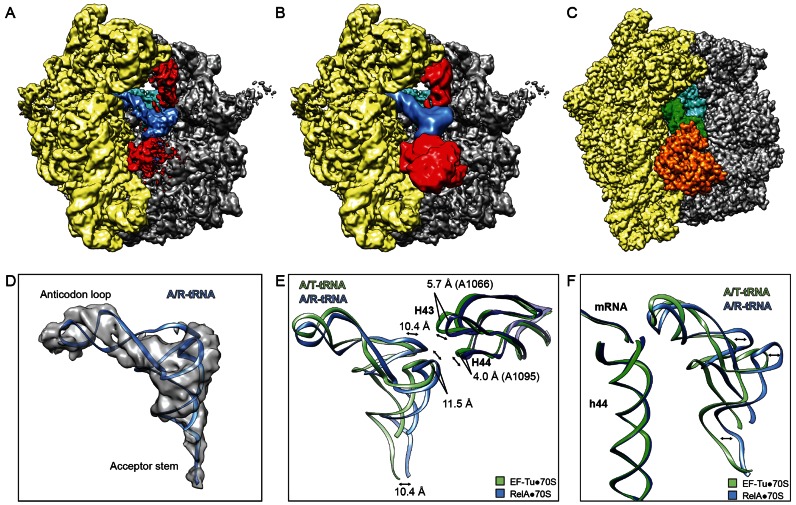
In the RelA-SRC, RelA is bound in the A-site of the ribosome and interacts with a tRNA that has adopted an A/T-tRNA-like conformation (Figure 2A and B), similar but distinct to that observed during decoding when the EF-Tu delivers the aminoacyl-tRNA to the A-site of the ribosome (27–29) (Figure 2C). Since we observed no subpopulations of ribosomes bearing A/T-like tRNA conformations in the absence of RelA but rather only fully accommodated A-tRNAs (Supplementary Figure S2), we reason that this state requires RelA for stabilization and therefore we refer to this binding state of the deacylated tRNA as the A/R-tRNA conformation. Within the limits of the resolution, the anticodon stem loop of the A/R-tRNA in the RelA-SRC appears to be similar to that observed during decoding with EF-Tu (29) (Figure 2D–F). Interaction of the anticodon stem-loop of the A/R-tRNA with the codon of the mRNA is not unexpected in the RelA-SRC, since the stimulation of ribosome-dependent RelA-mediated (p)ppGpp synthesis requires the deacylated tRNA to be cognate to the codon in the A-site (15). In contrast, the most prominent differences between the A/R-tRNA and A/T-tRNA states are in the placement of the elbow and acceptor arm of the tRNA, including the CCA-end, which are shifted by ∼10 Å with respect to one another (Figure 2E and F). Accordingly, the stalk base (H43/H44 of the 23S rRNA) is also shifted by ∼4-6 Å between the RelA and EF-Tu structures (Figure 2E).
Figure 2.
Novel binding site for RelA on the ribosome. (A and B) Overview of the cryo-EM reconstruction of the RelA-SRC filtered to (A) 4 Å and (B) 12 Å resolution showing 30S subunit (yellow), 50S subunit (grey), P-tRNA (cyan), A/R-tRNA (blue) and RelA (red). (C) Overview of EF-Tu-bound E. coli 70S ribosomes with P-tRNA (cyan), A/T-tRNA (green) and EF-Tu (orange) (29). (D) Isolated cryo-electron density with fitted model for the A/R-tRNA (blue). (E and F) Comparison of (E) H43 and H44 of the 23S rRNA and (F) A/R-tRNA of RelA-SRC (blue) and A/T-tRNA of EF-Tu-70S (green) (29).
Stabilization of an A/R-tRNA state by RelA was also observed in the previous cryo-EM reconstruction of the RelA-70S complex (25). In this reconstruction, density assigned to RelA was observed to interact with A/R-tRNA using the external surface of the acceptor arm, whereas no density was reported to extend into the intersubunit region of the ribosome (25). In contrast, at higher resolution additional density is observed in the RelA-SRC for RelA that extends from the CCA-end of the A/R-tRNA into the intersubunit region, namely, passing between the small subunit and acceptor arm of the tRNA toward the large ribosomal subunit and occupying the space between the elbow of the A/R-tRNA and the intersubunit bridge contact between the head (hd) of the small subunit and central protuberance (cp) of the large subunit (Figure 2A). When the cryo-EM map for the RelA-SRC was filtered to 12 Å, additionally density for RelA was also observed extending out of the A-site of the ribosome into the solvent (Figure 2B), indicating the extreme flexibility of this region of RelA. To our knowledge, the extended binding site for RelA observed within the A-site intersubunit crevice of the RelA-SRC is distinct from any other translation factor binding sites so far visualized on the ribosome.
RelA adopts an extended conformation on the ribosome
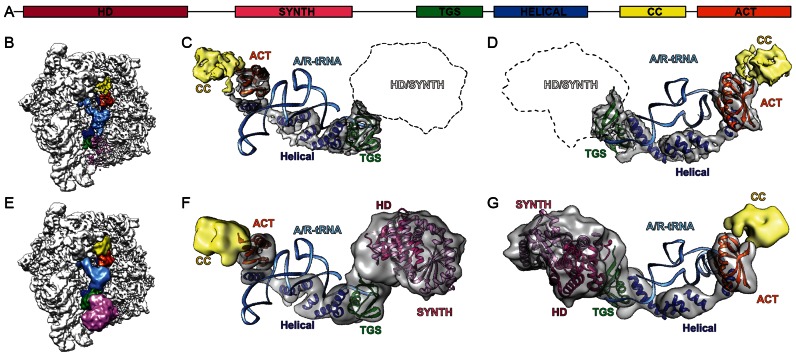
Although de novo model building for RelA was not possible, and despite the absence of a crystal structure for the CTD of RelA, the resolution and density features of the RelA-SRC map enabled homology models for distinct parts of the E. coli RelA to be fitted unambiguously (Figure 3A–D). In particular, homology models were generated and fitted for the TGS and ACT domains located in the CTD of RelA based on nuclear magnetic resonance (NMR) spectroscopy structures of Homo sapiens GTP-binding protein1 TGS domain (PDB2EKI) and of the ACT domain from the Chlorobium tepidum GTP pyrophosphokinase (PDB2KO1), respectively (Figure 3B–D and Supplementary Figure S3D–G). Additionally, we could identify five tubular densities that we assigned to the five predicted α-helices (Supplementary Figure S4) present in the helical linker region that connects the TGS with the CC/ACT subdomains (Figure 3B–D). The poor density for the connections between the helices does not allow us to unambiguously assign the order of the helices, nor the directionality, and therefore polyalanine helices were fitted as placeholders. Moreover, it was not possible to generate a suitable homology model for the CC domain and therefore this domain was left unmodelled (Figure 3B–D). Our model for the CTD of RelA reveals that the TGS domain interacts with the CCA-end of the A/R-tRNA, whereas the helical linker wraps around the acceptor arm, positioning the ACT domain to interact with the elbow region of the A/R-tRNA (Figure 3B–D).
Figure 3.
RelA adopts an extended conformation on the ribosome. (A) Schematic showing the domain organization of E. coli RelA with HD (magenta), SYNTH (pink), TGS (green), HELICAL (blue), CC (yellow) and ACT (orange) subdomains. (B) Overview of RelA-SRC with subdomains colored according to (A) and A/R-tRNA (light blue). (C and D) Complementary views showing isolated electron density of RelA with fitted homology models for TGS (green, PDB2EKI) and ACT (orange, PDB2KO1) subdomains, as well as model poly-Alanine helices fitted to the helical linker region (blue) and density for CC colored in yellow. (E–G) As (B–D) with 12 Å filtered maps and additional fitted homology models for HD (magenta) and SYNTH (pink) based on PDB1VJ7 (24).
Additionally, a homology model could be generated for the HD and SYNTH domains comprising the NTD of RelA based on the X-ray structure of the S. dysgalactiae Rel protein (24). Despite the low resolution of the NTD in the RelA-SRC, the distinct features of the density allowed an good fit (cross-correlation coefficient of 0.82) of the helical bundle of HD domain and the long extended helices within the SYNTH domain (Figure 3E–G). Moreover, only this orientation allows the C-terminal end of the SYNTH domain to be oriented toward the TGS domain of the CTD of RelA. Overall, our model for RelA suggests that the NTD of RelA does not form any stable interactions with the ribosome, with the SYNTH domain extending toward but not contacting the spur (helix 6 of the 16S rRNA) of the small subunit (Figure 3E), whereas in contrast the CTD of RelA snakes through the intersubunit space establishing, in addition to the A/R-tRNA, interactions with both the small and large ribosomal subunits (Figure 3B).
Interactions of the RelA CTD with the ribosome and A/R-tRNA
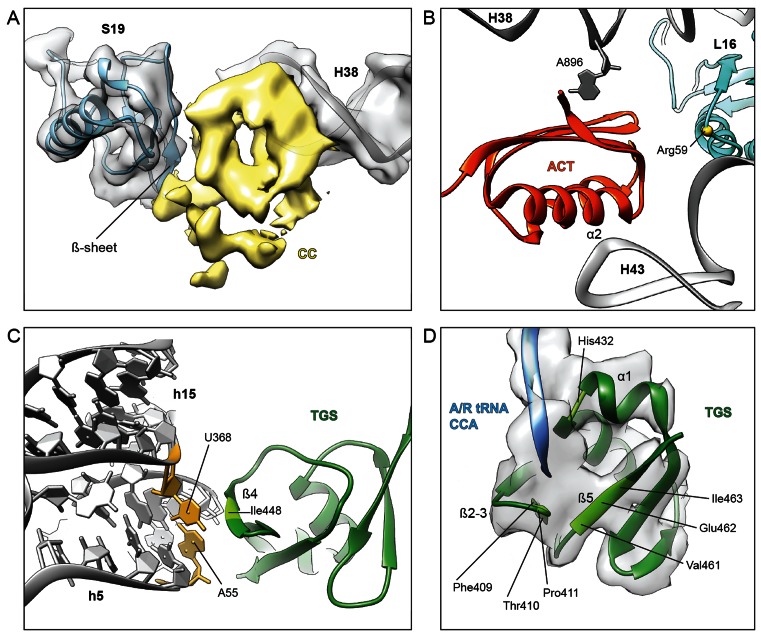
The most extensive interactions between RelA and the ribosome are between the ACT and CC subdomains (Figure 4A and B). The electron density for the CC domain indicates that it contacts components of both the small and large ribosomal subunit, namely, via interaction with the minor groove near the tip of H38 and by contacting the β-sheet of ribosomal protein S19 (Figure 4A). In contrast, the ACT domain contacts only components of the large subunit; specifically, the proximal end of α-helix α2 contacts the tip of H43 of the 23S rRNA, whereas the β-sheet of the ACT domain clearly interacts with A896, which is flipped out of H38 (Figure 4B), as it is in the unbound and vacant 70S structures. Interaction is also observed between the vicinity of Arg59 of ribosomal protein L16 and the terminal end of the β-sheet of the ACT domain (Figure 4B). In addition, α-helix α1 of the ACT domain contacts the elbow region of the A/R-tRNA and α-helix H5 of the helical linker of RelA.
Figure 4.
Interactions of RelA with the ribosome and A/R-tRNA. (A) Cryo-electron density of the CC subdomain of RelA (yellow) suggests interaction with the β-sheet of r-protein S19 (blue) and the minor groove of H38 in the 23S rRNA (grey). (B) Interaction of the C-terminal RelA ACT domain with Arg59 of r-protein L16 (blue) as well as 23S rRNA residues A896 of H38 and the tip of H43. (C) The β4-strand of the TGS domain of RelA (green) approaches the minor groove of h5 of the 16S rRNA, where residues in the vicinity of Ile448 (light green) appear to interact with the base-pair formed by nucleotides A55 and U368 (yellow), which are flipped-out of helices h5 and h15, respectively. (D) Tentative placement of the A/R-tRNA CCA-end (blue) shows vicinity of C74 of the A/R-tRNA with His432 located at the kink in α-helix α1, whereas C75 and A76 enter into a pocket formed by the β1–β2 hairpin and β5-strand of TGS (green).
The TGS domain of RelA contains one α-helix α1 and four β-strands β1–β4 (Supplementary Figure S4). The β4-strand of the TGS domain of RelA approaches the minor groove of helix 5 (h5 of the 16S rRNA) of the small subunit, where residues in the vicinity of Ile448 appear to interact with the base-pair formed by nucleotides A55 and U368, which are flipped-out of helices h5 and h15, respectively (Figure 4C), as they are in the unbound and vacant 70S structures. The other contacts of the TGS domain are with the CCA-end of the A/R-tRNA (Figure 4D), which adopts a distinct conformation from the A/T-tRNA (Figure 2E and F). Unfortunately, the density does not allow the nucleotides to be unambiguously modelled and therefore only a general description of the interactions can be provided: C74 of the A/R-tRNA is in the vicinity of His432 located at the kink in α-helix α1, whereas C75 and A76 enter into a pocket formed by the β1–β2 hairpin and β5-strand (Figure 4D). This interaction area of the CCA-end encompasses one of the most highly conserved regions of RelA, including His432 in α-helix α1, Phe409-Pro411 in the β1–β2 hairpin and Val461-Ile463 in the β5-strand (Supplementary Figure S5). Although higher resolution will be required to ascertain exactly how these highly conserved amino acids discriminate between deacylated and aminoacylated tRNAs, our structure provides evidence indicating that the TGS domain of RelA is directly involved in this monitoring activity.
DISCUSSION
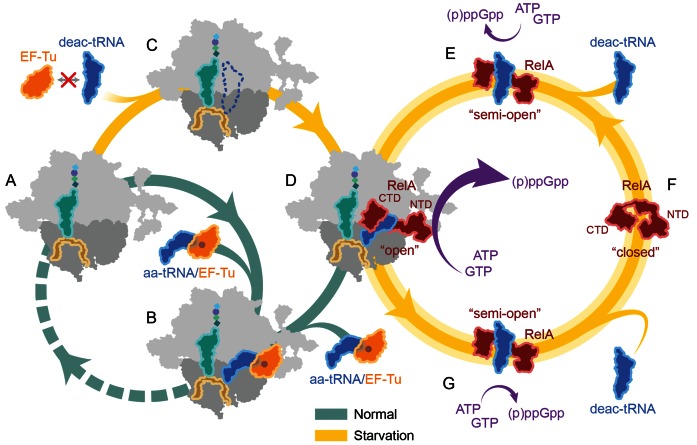
Based on our structural results, together with the available biochemical results from the literature, we present a model for the ribosome-dependent stimulation of RelA-mediated (p)ppGpp synthesis (Figure 5A–G): Under optimal growth conditions, aminoacylated tRNAs are delivered to the ribosome in a ternary complex with EF-Tu and GTP (Figure 5A and B). In contrast, under environmental conditions that evoke the stringent response, such as amino acid starvation, the levels of aminoacylated-tRNAs decrease leading to a concomitant accumulation of deacylated or uncharged tRNAs (50). The absence of aminoacylated tRNAs for the codon displayed in the A-site leads to ribosome stalling during translation elongation (Figure 5C). Although the deacylated tRNAs are not bound by EF-Tu or delivered by EF-Tu to the ribosome, it is possible for deacylated tRNAs to bind non-enzymatically to the A-site of the ribosome (Figure 5C). RelA has been proposed to recognize the deacylated tRNA bound in the A-site and catalyze (p)ppGpp synthesis (15–17). However, given the complex intertwined nature of the interaction between RelA and A/R-tRNA observed in our RelA-SRC structure, as well as the apparent lack of the A/R-tRNA conformation of the deacylated tRNA in the absence of RelA, we favor an alternative hypothesis whereby RelA and deacylated tRNA bind to the ribosome as a pre-formed complex (Figure 5D and E). Indeed, RelA has been shown to interact with deacylated tRNA (but not aminoacylated tRNA) in the absence of the ribosome (26,51,52) (Figure 5E).
Figure 5.
Model for RelA action during the stringent response. (A and B) Under optimal conditions, aminoacyl-tRNAs (aa-tRNAs) are delivered by EF-Tu (orange) to the A-site of the ribosome (green pathway). (C–G) Under starvation conditions (yellow pathway), the interaction of RelA (red) with deacylated A/R-tRNA at the A-site of the ribosome leads to the conversion of RelA from a ‘closed’ to an ‘open’ conformation and thereby stimulating high levels of (p)ppGpp synthesis. For more details, please refer to the text.
Biochemical and genetic evidence suggests that in the absence of the ribosome and deacylated tRNA, RelA exists in an autoinhibited state that produces only low levels of (p)ppGpp (1,3). Specifically, it has been shown that the CTD of RelA is responsible for the observed autoinhibition (21–23,26), leading to the hypothesis that the autoinhibition results from direct interaction between the CTD and NTD of RelA (23), which we term here the ‘closed’ conformation (Figure 5F). While interaction of deacylated tRNA with the CTD of RelA in the absence of the ribosome promotes a slight increase in (p)ppGpp synthesis activity (26,51,52) (Figure 5E), full activity requires the additional presence of the ribosome (15–17,53,54) (Figure 5D).
The structure of RelA-SRC provides a rationale for this increased activity, namely, that the ribosome and A/R-tRNA stabilize an ‘open’ conformation of RelA, which relieves the inhibitory interaction that the CTD imparts on the NTD, and thereby allows the uninhibited NTD to efficiently catalyze the synthesis of (p)ppGpp from GTP/GDP and ATP (Figure 5D). This model is consistent with the observation that deletion of the CTD of RelA prevents interaction of RelA with the ribosome (25) and allows the NTD to synthesize (p)ppGpp in a ribosome-independent manner (21,26,55). Moreover, the RelA-SRC structure indicates that the TGS domain of RelA directly contacts the 3′-end of the deacylated tRNA (Figure 4D), explaining how RelA can monitor and distinguish the aminoacylation state of the A-tRNA (56). It should be noted that ppGpp itself dramatically stimulates the ribosome-dependent RelA-mediated synthesis of itself (18), presumably by acting allosterically through a second, as yet undetermined, binding site on the factor, similar to the allosteric regulation of pppGpp observed recently for the small alarmone synthetase 1 (SAS1) RelQ from Bacillus subtilis (57) and Enterococcus faecalis (58).
While it is easy in our model to envisage how multiple rounds of (p)ppGpp synthesis by RelA could occur on the ribosome, as reported recently (20), we cannot exclude alternative models where (p)ppGpp synthesis leads to dissociation of RelA from the ribosome (16,19). Moreover, it remains unclear as to the order and timing of release of RelA and deacylated-tRNA from the ribosome following synthesis of (p)ppGpp (59), with one report suggested (p)ppGpp synthesis leads to release of deacylated-tRNA but not RelA, whereas another observed release of RelA, but not deacylated tRNA upon (p)ppGpp synthesis (16). From a structural viewpoint, it is hard to envisage how the intertwined CTD of RelA could dissociate from the ribosome without prior or concomitant dissociation of the deacylated A/R-tRNA. As mentioned, RelA has been shown to interact with deacylated tRNA in the absence of the ribosome and that this interaction occurs with the CTD and promotes a slight increase in (p)ppGpp synthesis activity (26,51,52). Consistently, fluorescence resonance energy transfer experiments indicate that the distance between the NTD and CTD of RelA increases upon binding of deacylated tRNA (60). This suggests to us that following dissociation from the ribosome, but still in the presence of deacylated tRNA, RelA can adopt a ‘semi-open’ conformation that can retain (p)ppGpp synthesis activity for an extended period of time (Figure 5G), consistent with the interpretation of single molecule experiments examining RelA action in living cells (19). Nevertheless, RelA has low affinity for deacylated tRNA in the absence of the ribosome (51), suggesting that either the RelA-tRNA complex becomes stabilized by rebinding to the ribosome and stimulating higher levels of (p)ppGpp synthesis, or the RelA-tRNA complex disassembles, allowing RelA to adopt the less active closed conformation (Figure 5F). It seems probable that the former pathway of rebinding would be favoured under conditions of starvation where a high ratio of deacylated tRNAs over aminoacylated tRNAs exists in the cell. In contrast, the latter pathway of disassembly would be favored as the nutrient deprivation is alleviated and the ratio is reversed, such that the increased intracellular levels of charged aminoacyl-tRNA promote A-site binding and thereby allows translation to resume (Figure 5B).
ACCESSION NUMBERS
The coordinates and cryo-EM map for the RelA-SRC have been deposited in the Protein Data Bank and EM DataBank under accession codes 5L3P and EMD-4001, respectively.
NOTE ADDED IN PROOF
During production of our work, a cryo-EMreconstruction of a E. coli RelA-70S complex was published by Ramakrishnan and coworkers (61). Our findings are in perfect agreement with their results and lead to the same conclusion: The ribosome and A/R-tRNA stabilize an ‘open’ conformation of RelA, which relieves the inhibitory interaction that the CTD imparts on the NTD, and thereby allows the uninhibited NTD to efficiently catalyze the synthesis of (p)ppGpp from GTP/GDP and ATP.
Supplementary Material
Acknowledgments
The authors would like to thank Gemma Atkinson for help with the Weblogo and Heidimarie Sieber and Charlotte Ungewickell for expert technical assistance.
Footnotes
Present address: Agata L. Starosta, Centre for Bacterial Cell Biology, Institute for Cell and Molecular Biosciences, University of Newcastle, Newcastle upon Tyne, UK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [FOR-1805 and GRK1721 to D.N.W. and R.B., SPP-1879 to D.N.W, SFB-646 to R.B.]; Swedish Research council Vetenskapsrådet [2013-4680 to V.H.]. Funding for open access charge: DFG [FOR-1805 and SPP-1879 to D.N.W].
Conflict of interest statement. None declared.
REFERENCES
- 1.Potrykus K., Cashel M. (p)ppGpp: still magical. Annu. Rev. Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson G.C., Tenson T., Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T., Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dalebroux Z.D., Svensson S.L., Gaynor E.C., Swanson M.S. ppGpp conjures bacterial virulence. Microbiol. Mol. Biol. Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geiger T., Francois P., Liebeke M., Fraunholz M., Goerke C., Krismer B., Schrenzel J., Lalk M., Wolz C. The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 2012;8:e1003016. doi: 10.1371/journal.ppat.1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole K. Bacterial stress responses as determinants of antimicrobial resistance. J. Antimicrob. Chemother. 2012;67:2069–2089. doi: 10.1093/jac/dks196. [DOI] [PubMed] [Google Scholar]
- 7.Maisonneuve E., Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050. [DOI] [PubMed] [Google Scholar]
- 8.Wexselblatt E., Kaspy I., Glaser G., Katzhendler J., Yavin E. Design, synthesis and structure-activity relationship of novel Relacin analogs as inhibitors of Rel proteins. Eur. J. Med. Chem. 2013;70:497–504. doi: 10.1016/j.ejmech.2013.10.036. [DOI] [PubMed] [Google Scholar]
- 9.Wexselblatt E., Oppenheimer-Shaanan Y., Kaspy I., London N., Schueler-Furman O., Yavin E., Glaser G., Katzhendler J., Ben-Yehuda S. Relacin, a novel antibacterial agent targeting the Stringent Response. PLoS Pathog. 2012;8:e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cashel M., Gallant J. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature. 1969;221:838–841. doi: 10.1038/221838a0. [DOI] [PubMed] [Google Scholar]
- 11.Laffler T., Gallant J. spo T, a new genetic locus involved in the stringent response in E. coli. Cell. 1974;1:27–30. [Google Scholar]
- 12.Xiao H., Kalman M., Ikehara K., Zemel S., Glaser G., Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- 13.Seyfzadeh M., Keener J., Nomura M. spoT-dependent accumulation of guanosine tetraphosphate in response to fatty acid starvation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1993;90:11004–11008. doi: 10.1073/pnas.90.23.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vinella D., Albrecht C., Cashel M., D'Ari R. Iron limitation induces SpoT-dependent accumulation of ppGpp in Escherichia coli. Mol. Microbiol. 2005;56:958–970. doi: 10.1111/j.1365-2958.2005.04601.x. [DOI] [PubMed] [Google Scholar]
- 15.Haseltine W.A., Block R. Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U.S.A. 1973;70:1564–1568. doi: 10.1073/pnas.70.5.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wendrich T.M., Blaha G., Wilson D.N., Marahiel M.A., Nierhaus K.H. Dissection of the mechanism for the stringent factor RelA. Mol. Cell. 2002;10:779–788. doi: 10.1016/s1097-2765(02)00656-1. [DOI] [PubMed] [Google Scholar]
- 17.Jenvert R.-M., Schiavone L. Characterization of the tRNA and ribosome-dependent pppGpp-synthesis by recombinant stringent factor from Escherichia coli. FEBS J. 2005;272:685–695. doi: 10.1111/j.1742-4658.2004.04502.x. [DOI] [PubMed] [Google Scholar]
- 18.Shyp V., Tankov S., Ermakov A., Kudrin P., English B.P., Ehrenberg M., Tenson T., Elf J., Hauryliuk V. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012;13:835–839. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.English B.P., Hauryliuk V., Sanamrad A., Tankov S., Dekker N.H., Elf J. Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E365–E373. doi: 10.1073/pnas.1102255108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Bouveret E., Zhang Y., Liu K., Wang J.D., Weisshaar J.C. Effects of amino acid starvation on RelA diffusive behavior in live Escherichia coli. Mol. Microbiol. 2016;99:571–585. doi: 10.1111/mmi.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber G., Metzger S., Aizenman E., Roza S., Cashel M., Glaser G. Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 1991;266:3760–3767. [PubMed] [Google Scholar]
- 22.Gropp M., Strausz Y., Gross M., Glaser G. Regulation of Escherichia coli RelA requires oligomerization of the C- terminal domain. J. Bacteriol. 2001;183:570–579. doi: 10.1128/JB.183.2.570-579.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mechold U., Murphy H., Brown L., Cashel M. Intramolecular regulation of the opposing (p)ppGpp catalytic activities of Rel(Seq), the Rel/Spo enzyme from Streptococcus equisimilis. J. Bacteriol. 2002;184:2878–2888. doi: 10.1128/JB.184.11.2878-2888.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 25.Agirrezabala X., Fernandez I.S., Kelley A.C., Carton D.G., Ramakrishnan V., Valle M. The ribosome triggers the stringent response by RelA via a highly distorted tRNA. EMBO Rep. 2013;14:811–816. doi: 10.1038/embor.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jain V., Saleem-Batcha R., China A., Chatterji D. Molecular dissection of the mycobacterial stringent response protein Rel. Protein Sci. 2006;15:1449–1464. doi: 10.1110/ps.062117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmeing T.M., Voorhees R.M., Kelley A.C., Gao Y.G., Murphy F.V.t., Weir J.R., Ramakrishnan V. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuette J.C., Murphy F.V. IV, Kelley A.C., Weir J.R., Giesebrecht J., Connell S.R., Loerke J., Mielke T., Zhang W., Penczek P.A., et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28:755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fischer N., Neumann P., Konevega A.L., Bock L.V., Ficner R., Rodnina M.V., Stark H. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by C-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 30.Arenz S., Ramu H., Gupta P., Berninghausen O., Beckmann R., Vazquez-Laslop N., Mankin A.S., Wilson D.N. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat. Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iordanescu S. Three distinct plasmids originating in the same Staphylococcus aureus strain. Arch. Roum. Pathol. Exp. Microbiol. 1976;35:111–118. [PubMed] [Google Scholar]
- 32.Narayanan C.S., Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985;13:7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antoun A., Pavlov M.Y., Tenson T., Ehrenberg M.M. Ribosome formation from subunits studied by stopped-flow and Rayleigh light scattering. Biol. Proced. Online. 2004;6:35–54. doi: 10.1251/bpo71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., Cheng Y. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheres S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kucukelbir A., Sigworth F.J., Tagare H.D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohou A., Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 39.Becker T., Franckenberg S., Wickles S., Shoemaker C.J., Anger A.M., Armache J.P., Sieber H., Ungewickell C., Berninghausen O., Daberkow I., et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J.Z., Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 41.Arenz S., Meydan S., Starosta A.L., Berninghausen O., Beckmann R., Vazquez-Laslop N., Wilson D.N. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol. Cell. 2014;56:446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loerke J., Giesebrecht J., Spahn C.M. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- 43.Arenz S., Nguyen F., Beckmann R., Wilson D.N. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-A resolution. Proc. Natl. Acad. Sci. U.S.A. 2015;112:5401–5406. doi: 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Soding J., Biegert A., Lupas A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eswar N., Eramian D., Webb B., Shen M.Y., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 47.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 48.Mayford M., Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J. Mol. Biol. 1989;206:69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez-Laslop N., Thum C., Mankin A.S. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 50.Yegian C.D., Stent G.S., Martin E.M. Intracellular condition of Escherichia coli transfer RNA. Proc. Natl. Acad. Sci. U.S.A. 1966;55:839–846. doi: 10.1073/pnas.55.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avarbock D., Avarbock A., Rubin H. Differential regulation of opposing RelMtb activities by the aminoacylation state of a tRNA.ribosome.mRNA.RelMtb complex. Biochemistry. 2000;39:11640–11648. doi: 10.1021/bi001256k. [DOI] [PubMed] [Google Scholar]
- 52.Avarbock A., Avarbock D., Teh J.S., Buckstein M., Wang Z.M., Rubin H. Functional regulation of the opposing (p)ppGpp synthetase/hydrolase activities of RelMtb from Mycobacterium tuberculosis. Biochemistry. 2005;44:9913–9923. doi: 10.1021/bi0505316. [DOI] [PubMed] [Google Scholar]
- 53.Rojiani M.V., Jakubowski H., Goldman E. Effect of variation of charged and uncharged tRNA(Trp) levels on ppGpp synthesis in Escherichia coli. J. Bacteriol. 1989;171:6493–6502. doi: 10.1128/jb.171.12.6493-6502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Payoe R., Fahlman R.P. Dependence of RelA-mediated (p)ppGpp formation on tRNA identity. Biochemistry. 2011;50:3075–3083. doi: 10.1021/bi1015309. [DOI] [PubMed] [Google Scholar]
- 55.Svitil A.L., Cashel M., Zyskind J.W. Guanosine tetraphosphate inhibits protein synthesis in vivo - a possible protective mechanism for starvation stress in Escherichia-coli. J. Biol. Chem. 1993;268:2307–2311. [PubMed] [Google Scholar]
- 56.Sprinzl M., Richter D. Free 3′-OH group of the terminal adenosine of the tRNA molecule is essential for the synthesis in vitro of guanosine tetraphosphate and pentaphosphate in a ribosomal system from Escherichia coli. Eur. J. Biochem. 1976;71:171–176. doi: 10.1111/j.1432-1033.1976.tb11103.x. [DOI] [PubMed] [Google Scholar]
- 57.Steinchen W., Schuhmacher J.S., Altegoer F., Fage C.D., Srinivasan V., Linne U., Marahiel M.A., Bange G. Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl. Acad. Sci. U.S.A. 2015;112:13348–13353. doi: 10.1073/pnas.1505271112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gaca A.O., Kudrin P., Colomer-Winter C., Beljantseva J., Liu K., Anderson B., Wang J.D., Rejman D., Potrykus K., Cashel M., et al. From (p)ppGpp to (pp)pGpp: Characterization of Regulatory Effects of pGpp Synthesized by the Small Alarmone Synthetase of Enterococcus faecalis. J. Bacteriol. 2015;197:2908–2919. doi: 10.1128/JB.00324-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richter D. Stringent factor from Escherichia coli directs ribosomal binding and release of uncharged tRNA. Proc. Natl. Acad. Sci. U.S.A. 1976;73:707–711. doi: 10.1073/pnas.73.3.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain V., Saleem-Batcha R., Chatterji D. Synthesis and hydrolysis of pppGpp in mycobacteria: a ligand mediated conformational switch in Rel. Biophys. Chem. 2007;127:41–50. doi: 10.1016/j.bpc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Brown A., Fernández I.S., Gordiyenko Y., Ramakrishnan V. Ribosome-dependent activation of stringent control. Nature. 2016 doi: 10.1038/nature17675. doi:10.1038/nature17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.