Abstract
Previous studies demonstrated that Mycoplasma arthritidis strain 158 acquired a high degree of virulence upon lysogenization with bacteriophage MAV1. In the present study, the association between MAV1 and virulence was reexamined by creating new lysogens of 158 and of a relatively avirulent mutant, strain 158-1. In the absence of lysogenization, 158 was more virulent than expected. The virulence of 158 and 158-1 did not increase upon lysogenization. A major antigenic difference between 158 and 158-1 was identified that is unrelated to MAV1 and could account for the difference in virulence.
Mycoplasma arthritidis is a natural pathogen of rats, causing an acute polyarthritis that resolves within 6 to 8 weeks after infection. After recovery, organisms are no longer cultivable from the joints and the animals are protected from future infections (3). Rats and mice can be infected experimentally by injecting live organisms into the caudal vein (15). Arthritic disease is more chronic in mice than in rats, with periods of remission and exacerbation that may persist for the life of the animal (5).
MAV1 is a bacteriophage isolated from M. arthritidis with a 16-kb double-stranded DNA genome (14). It is also thought to be associated with virulence. Previously, the virulence of several strains of M. arthritidis was tested to assess whether the presence of MAV1 DNA correlates with virulence (15). All MAV1 lysogens were highly virulent, and all strains lacking MAV1 DNA were low in virulence. This same study showed that MAV1 lysogenization of M. arthritidis strain 158 generates lysogens (e.g., strain 158L3-1) that are more virulent than the parent. These results were seemingly confirmed by comparing the virulence of strain 158L3-1 to that of what was thought to be strain 158 in a mouse model (11).
A putative virulence factor, vir, was identified in the MAV1 genome sequence. Because vir was predicted to encode a lipoprotein and is one of only two genes transcribed constitutively in the lysogen (the other gene is predicted to encode a phage repressor), vir was proposed to be responsible for the increased arthritogenicity seen in MAV1 lysogens (14). When vir was transformed into nonlysogens of M. arthritidis by using the transposon Tn4001T as a vector, no increase in disease severity occurred (2). Although no change in virulence was noted, all transformants containing vir acquired resistance to infection with MAV1. It appears that the function of Vir is to protect cells from superinfecting phage, possibly by preventing adsorbed phage from injecting their DNA into the host cytoplasm. Because the analysis of the MAV1 genome sequence failed to identify a candidate other than vir as a likely virulence determinant, the issue of whether MAV1 truly is associated with virulence was reexamined in the present study.
Construction of MAV1 lysogens.
Lysogens of the relatively virulent strain M. arthritidis 158 and a low-virulence mutant of 158 (strain 158-1) were constructed for the purpose of assessing the role of MAV1 in pathogenesis. M. arthritidis strain 158 and its subclone 158-1 are described elsewhere (13, 15). Mycoplasmas were propagated in Edgar's agar or Edgar's broth (EB) as described previously (12, 15). Lysogens of 158 and 158-1 were made by stabbing a turbid MAV1 megaplaque (12) onto a lawn of either 158 or 158-1 and placing each agar plug in 1.5 ml of EB. Once the culture reached late-logarithmic phase, the culture was passed through a 0.45-μm Acrodisc syringe filter (Nalgene, Rochester, N.Y.) to obtain single cells, and the eluant was assayed for CFU. A single colony from each plug was picked to ensure that independent lysogens were selected. Megaplaque assays were used to test the resulting lysogens' ability to resist superinfection, indicating lysogenization. Strain 158L5-1 is a lysogen of 158. Strains 158-1L1 and 158-1L2 are lysogens of 158-1. Also used in this study was strain 158L3-1, a previously constructed MAV1 lysogen of strain 158 (15).
The presence of MAV1 DNA in the genomes of all lysogens was verified by PCR amplification of the MAV1 vir gene, which encodes a superinfection resistance determinant (2), and the imm gene, which encodes a putative phage repressor. The nucleotide sequences of the primers used in this study are provided in Table 1. Genomic DNA was isolated with the Easy-DNA kit (Invitrogen, Carlsbad, Calif.). Phage genes vir and imm were amplified with primers o.165 and o.166 and primers o.167 and o.168, respectively. PCR amplification of vir and imm from lysogens 158L3-1, 158L5-1, 158-1L1, and 158-1L2 yielded products of 1,320 and 935 bp, respectively. No MAV1 sequences were amplified from nonlysogenic parent strains 158 and 158-1. As a control to demonstrate that the DNA template from the nonlysogens was adequate for PCR amplification, a 390-bp portion of the arcA promoter region (6) was amplified with the arcA forward and arcA reverse primers. The PCR data were confirmed by digesting genomic DNA from each strain with Sau3AI. The 16-kb MAV1 genome is devoid of Sau3AI sites. The largest Sau3AI fragment in nonlysogens 158 and 158-1 was 7.5 kb. As expected, DNAs from 158L3-1, 158L5-1, 158-1L1, and 158-1L2 all contained a 16-kb Sau3AI fragment that was absent in 158 and 158-1.
TABLE 1.
Oligonucleotide sequences used for PCR and DNA sequencing
| Primer | Sequence (5′-3′)a | Nucleotide position(s) of annealing site(s) in 158L3-1 genomeb |
|---|---|---|
| o.165 | GCTAGGATCCGTAATGAGGAATTGGTTGC | 719187-719205, + strand |
| o.166 | GTTGGATCCTGCGAAATCTTTTCAAGG | 720471-720488, − strand |
| o.167 | AGAGGATCCTTTGATGTGAGTCAATACGC | 718824-718843, + strand |
| o.168 | TATGGATCCATCGGGTTTAGCGTAAATGC | 719720-719739, − strand |
| arcA forward | GCATGCTAGCTTCCTTTCATAAGTTATGATGTC | 807687-807709, − strand |
| arcA reverse | GCAGATCTAAAATTTCTTCAAATTCC | 807338-807355, + strand |
| MAV1 sequencing | CAGAGGAAGATGCTAAAAGGTATAG | 722162-722186, + strand |
| left-end MAV1 | TAAGCAGTAGCGTAACTCTTAC | 30249-38270, − strand; 706718-706739, − strand |
| right-end MAV1 | CAGAGGAAGATGCTAAAAGGTATAG | 722162-722186, + strand |
Restriction sites (underlined) are incorporated in the 5′-end region of some primers and would not anneal to the template DNA. These sites were introduced to facilitate cloning of PCR products for experiments that are not relevant to this study.
The nucleotide positions correspond to the completed M. arthritidis genome sequence as determined by collaboration with The Institute for Genomic Research (Rockville, Md.) (B. A. Methe and K. Dybvig, unpublished data).
Parent strains 158 and 158-1 contain sequences homologous to MAV1 DNA.
Analysis of the 158L3-1 genome sequence revealed a stretch of DNA (nucleotide positions 38110 to 38346) homologous to the first 245 bp of the left end of the MAV1 genome. Other than this region and a complete copy of MAV1 DNA integrated at nucleotide position 706579, no MAV1 DNA sequences are present in this lysogen. Compared to the MAV1 genome, the phage remnant at positions 38110 to 38346 had seven randomly distributed base pair substitutions, a 1-bp deletion, a 1-bp insertion, and an 8-bp insertion. This fragment of DNA does not contain any annotated genes. The first MAV1 open reading frame is repB, encoding a putative DNA helicase, which starts at nucleotide position 350 from the left end of the MAV1 genome.
To determine whether nonlysogens 158 and 158-1 also contain a fragment of MAV1 DNA, genomic DNA was subjected to direct sequencing primed with the left-end MAV1 primer. Analysis of the sequences of both strains showed that each possesses the phage remnant in the same genomic position as in 158L3-1. Direct sequencing reactions primed with the right-end MAV1 primer failed to generate a sequence for 158 and 158-1, showing that these strains lack the right terminus of the phage genome. Also, as described above, 158 and 158-1 lack the MAV1 vir and imm genes.
The finding of sequences homologous to MAV1 DNA at nucleotide positions 38110 to 38346 in the M. arthritidis genome of the strains under study is of interest. The mam gene is the first gene downstream of these sequences beginning at nucleotide position 38520. MAM is a soluble T-cell mitogen and suspected of being a major M. arthritidis virulence factor, triggering autoimmune disease (3, 4). Many bacterial superantigens are phage encoded (8), and the proximity of mam to the phage remnant at positions 38110 to 38346 suggests that the MAM superantigen may also be of phage origin, from a phage closely related to MAV1.
Identification of MAV1 DNA integration sites in M. arthritidis lysogens.
MAV1 DNA is integrated into host DNA at any of numerous sites consisting of the 7-bp sequence TATTTTT (12), which appears 1,708 times in the genome of 158L3-1. Lysogenization often disrupts a gene's coding region, generating a mutation that may affect the virulence of the mycoplasma. Therefore, the nucleotide position of the bacteriophage in the mycoplasmal chromosome was determined for each lysogen chosen for virulence studies. Mapping of the MAV1 DNA integration site was accomplished by direct genome sequencing primed with the right-end MAV1 primer, which anneals 32 nucleotides from the end of the MAV1 genome (GenBank accession number AF074945). The reaction components and cycling conditions used are described elsewhere (7). Analysis of the resulting sequence identified the junction between the phage and the mycoplasmal chromosome. Genome sequence data from M. arthritidis strain 158L3-1 were obtained from The Institute for Genomic Research through the website at http://www.tigr.org. A comparison of the junction sequence to the genome sequence identified the nucleotide position of the phage DNA in the mycoplasmal genome. The nucleotide position of the integrated MAV1 DNA in each lysogen is provided in Table 2.
TABLE 2.
Library of MAV1 lysogens
| Lysogen | Parent strain | Genomic nucleotide positiona | Disrupted gene product, if any |
|---|---|---|---|
| 158L3-1 | 158 | 706579 | Intergenic |
| 158L4-1 | 158 | 38110 | Intergenic |
| 158L5-1 | 158 | 38110 | Intergenic |
| 158-1L1 | 158-1 | 38110 | Intergenic |
| 158-1L2 | 158-1 | 473548 | Hypothetical protein |
| 158-1L3 | 158-1 | 38110 | Intergenic |
| 158-1L4 | 158-1 | 38110 | Intergenic |
The nucleotide positions correspond to the completed M. arthritidis genome sequence as determined by collaboration with The Institute for Genomic Research (Rockville, Md.) (Methe and Dybvig, unpublished).
Several independent lysogens had MAV1 DNA inserted into nucleotide position 38110, the location of the MAV1 phage remnant. Although lysogens were obtained in which MAV1 DNA was inserted into a traditional TATTTTT insertion site, such lysogens were obtained at a lower frequency than those in which integration occurred at position 38110. The integration sites of three lysogens of strain 158 constructed by Voelker et al. (15) had been previously studied (12). In contrast to the present study, none of the three lysogens had MAV1 DNA integrated at the site of the phage remnant. A comparison of the integration sites in these three lysogens to the complete genome sequence of 158L3-1 reveals that MAV1 DNA had been inserted at nucleotide positions 706571, 697161, and 700131 in lysogens 158L3-1, 158L1-3, and 158L3-2, respectively.
Proposed model of MAV1 DNA integration at nucleotide position 38110.
Perhaps MAV1 DNA was usually integrated at the site of the phage remnant in the present study but not in previous studies because of the method used to select the lysogens. In the present study, lysogens were obtained by filter cloning CFU obtained by stabbing phage-containing plaques. During productive phage infection on agar, there may be substantial quantities of phage DNA in the infected cell and ample opportunity for one of these copies to be integrated into the host chromosome by homologous recombination at the site of the phage remnant. Homologous recombination may occur more readily than site-specific DNA integration because the MAV1 integrase Int may not be produced during productive infection. Once MAV1 DNA has been integrated by homologous recombination with the phage remnant, the infection may be aborted to establish lysogeny as phage gene expression is shut down by the Imm repressor. In contrast to the present study, Voelker et al. obtained lysogens through broth infection, not by stabbing plaques (15). MAV1 infection in broth is usually nonproductive, yielding lysogens with little if any progeny phage. The lack of productive infection may provide little opportunity for homologous recombination between MAV1 DNA and the mycoplasma chromosome because phage DNA may be quickly integrated via the activity of the MAV1 integrase to generate lysogens.
MAV1 is not associated with virulence.
Previous studies showed that lysogenization of strain 158 by MAV1 resulted in a profound increase in virulence (11, 15). To further examine whether MAV1 is associated with virulence, multiple lysogens of 158 and 158-1 were compared to the parental nonlysogens for the ability to cause arthritic disease in rats. Only lysogens with MAV1 DNA integrated at sites thought to have little or no impact on virulence, usually intergenic, were chosen for study. Male Lewis rats (Charles River Laboratories Inc., Wilmington, Mass.) weighing an average of 199 g were divided into groups of eight for infection experiments and a control group of five for mock infection. Each rat was injected intravenously in the caudal vein with 200 μl of EB containing 109 CFU of the appropriate mycoplasmal strain or with 200 μl of EB (control animals).
The method used to determine the numerical arthritis scores was similar to those described previously (1, 15). Peripheral joints were measured with a caliper and assigned a score between 0 (no swelling) and 5 (>40% and >70% increase in diameter of the ankle and wrist joints, respectively). Interphalangeal joints and tail vertebrae were assigned a score between 0 and 1.5 on the basis of a visual assessment of swelling. The total arthritis score for each rat was determined, and the average numeric arthritis score per rat was calculated for each group. To assess mobility, animals were assigned the following scores: 0 if they walked normally, 1 if they walked awkwardly, 2 if they refused to walk on one limb, 3 if they failed to walk and crawled, and 4 if they were unable to crawl (1). Mobility, numerical arthritis scores, and changes in weight were analyzed with SigmaStat version 2.03 (SPSS Inc.). Nonparametric data (mobility and numerical arthritis) were ranked and analyzed by Kruskal-Wallis one-way analysis of variance on ranks. All pairwise comparisons were performed with Dunn's method. Parametric data (changes in weight) were analyzed by two-way analysis of variance with the strain of mycoplasma and time as the variables. All pairwise comparisons were performed by the Tukey test. Differences were considered significant at P < 0.05.
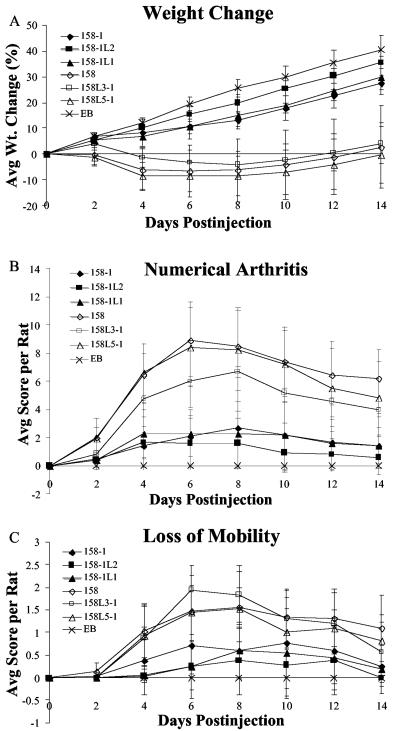
Throughout the 2-week experiment, animals infected with lysogens did not exhibit greater weight loss than rats infected with the respective nonlysogen strains (Fig. 1A). According to statistical analysis, one lysogen, 158-1L2, caused less weight loss than did 158-1. Unlike the other lysogens, which have MAV1 DNA integrated at intergenic sites, a gene encoding a hypothetical protein is disrupted in 158-1L2. If this hypothetical protein is a virulence factor, it would explain why rats infected with this strain lost less weight than those infected with 158-1. However, this attenuation was not observed when loss of mobility and degree of arthritogenicity were compared, demonstrating that the absence of this protein did not affect these hallmarks of arthritic disease. Statistically, groups infected with lysogens did not exhibit an increase in the degree of arthritogenicity (Fig. 1B) or a greater loss of mobility (Fig. 1C) than their parents. Unexpectedly, all strains of 158 parentage were more virulent than all strains derived from 158-1 in all three tests, regardless of lysogenization.
FIG. 1.
Development of arthritis in rats injected with M. arthritidis strains 158-1, 158-1L2, 158-1L1, 158, 158L3-1, and 158L5-1 and with EB as a control. Weight change (A), average arthritis score (B), and mobility score (C) per rat over a 2-week period are shown. Each point represents the mean measurement of eight rats (five for EB) with the standard deviation.
The results shown in Fig. 1 agree with prior literature with regard to the finding that 158L3-1 is highly virulent. Our finding that 158 was also highly virulent disagrees with previous reports. Accordingly, new stocks of 158 were obtained from L. R. Washburn. In our experiments, the virulence of these new stocks of 158 was comparable to that of 158L3-1 (data not shown). There is no obvious explanation for why 158 was highly virulent in our experiments but was low in virulence in the studies of Voelker et al. (15). Care was taken to ensure that all lysogens had integrated MAV1 DNA sequences and that all nonlysogens, including 158, lacked MAV1 DNA. We have no evidence that MAV1 is associated with virulence.
Antigenic differences unrelated to MAV1 correlate with virulence.
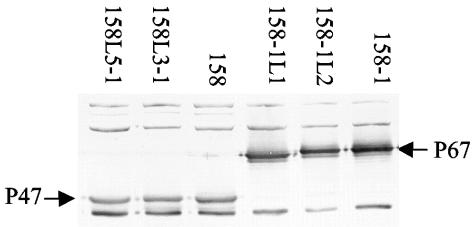
Western analysis of total proteins from 158, 158-1, and their lysogens revealed a major antigenic difference (Fig. 2). Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide (10%) gel electrophoresis, transferred to a nitrocellulose membrane, and reacted with sera from Lewis rats infected with 158L3-1 as described previously (10). Virulent strain 158 and its lysogens all had an immunodominant protein of about 47 kDa (P47). Relatively avirulent strain 158-1 and its lysogens, which are also avirulent, lacked P47 and in its place had a larger major antigen of 67 kDa (P67). Interestingly, the Western assay results were the same regardless of whether the source of serum was animals that had been experimentally infected with 158 or with 158-1 (data not shown). P47 is clearly not MAV1 encoded but may be associated with virulence.
FIG. 2.
Immunoblot assay of M. arthritidis strains used in animal experiments reacted with anti-158 rat polyclonal sera.
The Western assay results indicate that serum from infected animals reacts more strongly with P47 and P67 than with most other M. arthritidis antigens. Because serum from animals infected with 158, lacking P67, reacted just as strongly with P67 as did serum from animals infected with 158-1, strain 158 must produce an antigen with epitopes that cross-react with P67. Similarly, serum from animals infected with 158-1 reacts very strongly with P47. Although any of several antigens may have epitopes in common with P47 and P67, a straightforward interpretation is that P47 and P67 are closely related. Regardless of which of these two proteins is produced during infection of the animal host, the serum would react strongly with either protein on an immunoblot. Because strain 158 produces P47 while lacking P67 and strain 158-1, a subclone of 158, produces P67 but not P47, it is possible that these proteins are phase and/or size variable, as are numerous mycoplasmal surface proteins (9). In support of the possibility of size variation, we noted that P67 was reproducibly smaller, perhaps 66 kDa, in 158-1L1 than in 158-1 and 158-1L2 (Fig. 2). Since it is a major antigen, it is reasonable to speculate that P47 has a role in pathogenesis.
In addition to the study of Voelker et al., we question the results of a study done in our laboratory by Tu et al. in which the virulence of 158 and 158L3-1 was examined in the mouse (11). The stocks used for the virulence experiments by Tu et al. were examined retrospectively for the presence of P47 or P67. The stock that was reported to be strain 158 and to be low in virulence in the mouse was found to produce P67, not P47. Therefore, the previous report in which we concluded that 158L3-1 was more virulent than 158, lending support to the role of MAV1 in virulence, was in error in that the wrong strain was used for comparison to 158L3-1. The nonlysogen used in this previous report was low-virulence strain 158-1, not highly virulent strain 158. Thus, not only are strain 158 and its lysogens more virulent than strain 158-1 in the rat as reported here, but 158-1 is also low in virulence in the mouse.
Acknowledgments
This work was supported by Public Health Service grant AR44252 from the National Institutes of Health. Preliminary sequence data were obtained from The Institute for Genomic Research through the website at http://www.tigr.org.
We thank Portia Caldwell for technical assistance and Trenton R. Schoeb for providing advice on animal care and behavior.
Editor: J. T. Barbieri
REFERENCES
- 1.Butler, S. H., J. Weil-Fugazza, F. Godefroy, and J. M. Besson. 1985. Reduction of arthritis and pain behaviour following chronic administration of amitriptyline or imipramine in rats with adjuvant-induced arthritis. Pain 23:159-175. [DOI] [PubMed] [Google Scholar]
- 2.Clapper, B., A.-H. T. Tu, A. Elgavish, and K. Dybvig. 2004. The vir gene of bacteriophage MAV1 confers resistance to phage infection on Mycoplasma arthritidis. J. Bacteriol. 186:5715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, B. C. 1991. The immunobiology of Mycoplasma arthritidis and its superantigen. Curr. Top. Microbiol. Immunol. 174:107-119. [DOI] [PubMed] [Google Scholar]
- 4.Cole, B. C., and C. L. Atkin. 1991. The Mycoplasma arthritidis T-cell mitogen, MAM: a model superantigen. Immunol. Today 12:271-276. [DOI] [PubMed] [Google Scholar]
- 5.Cole, B. C., J. R. Ward, K. S. Jones, and J. F. Cahill. 1971. Chronic proliferative arthritis of mice induced by Mycoplasma arthritidis. I. Induction of disease and histopathological characteristics. Infect. Immun. 4:344-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dybvig, K., C. T. French, and L. L. Voelker. 2000. Construction and use of derivatives of transposon Tn4001 that function in Mycoplasma pulmonis and Mycoplasma arthritidis. J. Bacteriol. 182:4343-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heiner, C. R., K. L. Hunkapiller, S.-M. Chen, J. I. Glass, and E. Y. Chen. 1998. Sequencing multimegabase-template DNA with BigDye terminator chemistry. Genome Res. 8:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49:93-105. [DOI] [PubMed] [Google Scholar]
- 9.Razin, S., D. Yogev, and Y. Naot. 1998. Molecular biology and pathogenicity of mycoplasmas. Microbiol. Mol. Biol. Rev. 62:1094-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simmons, W. L., and K. Dybvig. 2003. The Vsa proteins modulate susceptibility of Mycoplasma pulmonis to complement killing, hemadsorption, and adherence to polystyrene. Infect. Immun. 71:5733-5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu, A.-H., J. R. Lindsey, T. R. Schoeb, A. Elgavish, H. Yu, and K. Dybvig. 2002. Role of bacteriophage MAV1 is a mycoplasmal virulence factor for the development of arthritis in mice and rats. J. Infect. Dis. 186:432-435. [DOI] [PubMed] [Google Scholar]
- 12.Voelker, L. L., and K. Dybvig. 1998. Characterization of the lysogenic bacteriophage MAV1 from Mycoplasma arthritidis. J. Bacteriol. 180:5928-5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voelker, L. L., and K. Dybvig. 1996. Gene transfer in Mycoplasma arthritidis: transformation, conjugal transfer of Tn916, and evidence for a restriction system recognizing AGCT. J. Bacteriol. 178:6078-6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voelker, L. L., and K. Dybvig. 1999. Sequence analysis of the Mycoplasma arthritidis bacteriophage MAV1 genome identifies the putative virulence factor. Gene 233:101-107. [DOI] [PubMed] [Google Scholar]
- 15.Voelker, L. L., K. E. Weaver, L. J. Ehle, and L. R. Washburn. 1995. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect. Immun. 63:4016-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]