Abstract
Chlamydia trachomatis is a strict human pathogen producing infections that cause medically important chronic inflammatory diseases, such as blinding trachoma and tubal factor infertility. Isolates exist as serotypes that fall into distinct biologic and pathological groups corresponding to differences in infection tissue tropism and invasion properties. Paradoxically, genome sequencing of several diverse strains has revealed a remarkable level of genomic synteny, suggesting that minor genetic differences determine the pathogen host- and tissue-specific infection characteristics. To better understand the genetic basis of chlamydial pathobiologic diversity, we performed comparative DNA-DNA microarray genomic hybridizations with all 15 C. trachomatis serovariants. We found there are few major genetic differences among the 15 serovars. An exception was the cytotoxin locus located in the plasticity zone, a region that exhibited significant polymorphisms among serovars. We therefore sequenced this region from all 15 serovars. The cytotoxin gene was interrupted by extensive mutations and deletions among the different serovars; however, three basic open reading frame motifs were discovered that correlated with noninvasive oculotropic, urogenitotropic, and invasive serovars. Of interest, only noninvasive genitotropic serovars possessed an intact N-terminal portion of the putative toxin gene. This region contains the UDP-glucose binding domain and the glycosyltransferase domain required for enzymatic activity of the clostridial toxin homologs, suggesting a role in urogenital infection or pathogenesis.
Chlamydia trachomatis is an obligate intracellular prokaryote characterized by a complex intracellular development cycle. Isolates can be differentiated into biovars or serovars based on their in vitro infection properties and type of disease (26) or on antigenic variation of the major outer membrane protein (OmpA), respectively (36). Isolates are serologically classified into 15 OmpA serovariants. Serovars A to C are the etiologic agents of trachoma (15, 26); serovars D to K and L1 to L3 are sexually transmitted pathogens causing cervicitis and urethritis or lymphogranuloma venereum (LGV), respectively. Serovars A to K produce infections restricted to the mucosae, whereas the LGV serovars infect monocytes and disseminate to local draining lymph nodes (26). The serovars associated with mucosal infections produce a wide spectrum of infection outcomes that range from asymptomatic to chronic inflammatory disease (26). Severe inflammatory disease can result in trachoma, the leading cause of blindness, or in pelvic inflammatory disease, a leading cause of tubal factor infertility.
The genomes of several chlamydial strains have been sequenced. Comparative genomics has shown that the small genomes (1.04 to 1.3 Mb) share a striking degree of synteny in that gene order and content are highly conserved (23, 24, 31). One exception is a polymorphic region of the genome termed the plasticity zone (PZ). Read et al. (23) initially hypothesized, based on the significant amount of variation in the PZ among strains, that genes residing in this locus might be chlamydial virulence factors. This was demonstrated by extensive comparative and functional genomic analyses on the C. trachomatis trpBA operon that resides in the PZ (9, 13). The trpBA operon is unique to human C. trachomatis isolates, an observation that intuitively suggested that it might be important in host-pathogen interactions. In support of this hypothesis, C. trachomatis genital and ocular isolates can be unambiguously distinguished from one another by specific mutations in their trpBA operon; specifically, genital isolates have been shown to have an intact operon and encode a functional tryptophan synthase, whereas ocular strains exhibit mutations in the trpA or trpB genes that result in a nonfunctional synthase (12). The ability to synthesize a functional synthase was a potential virulence factor for genital, but not ocular, strains (9). For example, the synthesis of a functional synthase was clearly associated with the ability of genital strains to be rescued by exogenous indole from the inhibitory affects of gamma interferon, which depletes intracellular tryptophan pools through the induction of the tryptophan-catabolizing enzyme indoleamine 2,3-dioxygenase. The ability to synthesize tryptophan from indole could be a major pathogenic mechanism that allows genital strains to avoid host cellular immune responses, particularly in environments where mixed microbial infections predominate and indole producers are found.
Infections of the genital tract and eye with the Chlamydia muridarum and Chlamydia caviae strains closely mimic acute oculogenital C. trachomatis infections of humans (22). Of interest, within the PZ of C. muridarum are three open reading frames (ORFs; TC0437 to -0439) encoding proteins with predicted masses of approximately 360 kDa (24). These proteins have significant homology to the large clostridial toxins (LCTs) and Escherichia coli products referred to as the bacterial adhesin Efa1 (20) and the LifA cytotoxin termed lymphostatin (19). C. trachomatis serovar D has a toxin-like locus that appears to be an ancestral version of a single complete gene, in that the region has been interrupted by frameshift and deletion mutations (31). The C. caviae genome encodes a putative 360-kDa cytotoxin homolog (24). C. muridarum and C. trachomatis serovar D elementary bodies (EBs), but not LGV (L2) EB, are cytotoxic for cultured epithelial cells (2). Moreover, the cytotoxic phenotype of EBs correlated with expression of intact toxin CT438 (C. muridarum) or partial (CT166 of C. trachomatis serovar D) cytotoxin genes. Serovar D ORF CT166 encodes a 70-kDa polypeptide with homology to the enzymatically active site of the LCTs (2). The nontoxic, non- epitheliotropic LGV strain lacked the CT166 ORF and possessed only a small portion of the 3′ end of the cytotoxin gene. Although incomplete, the association of cytotoxicity with intact or partial toxin genes is intriguing, because it could implicate the toxin in the pathogenesis of chlamydial mucosal infections. For this reason, we have extended the genomic analysis of the PZ for all C. trachomatis serovars.
MATERIALS AND METHODS
Chlamydiae.
C. trachomatis strains A/HAR-13, B/TW-5/OT, Ba/AP-2, C/TW-3/OT, D/UW-3/Cx, E/Bour, F/IC-Cal-13, G/UW-524/Cx, H/UW-4/Cx, I/UW-4/Ur, J/UW-36/Cx, K/UW-31/Cx, L1/LGV-440, L2/LGV-434, and L3/LGV-404 were grown in HeLa 229 monolayers in T-150 flasks containing high-glucose Dulbecco's modified Eagle's medium (Cellgrow; Mediatech, Inc., Herndon, Va.) supplemented with 10% fetal calf serum containing 1 μg of gentamicin/ml. Infectious EBs were purified by density gradient centrifugation, and the infection-forming units of purified EBs were determined as previously described (8).
Construction of a cDNA microarray for C. trachomatis serovar D.
A spotted microarray was designed based on the genomic sequence of C. trachomatis serovar D (GenBank accession number AE001273) (31). Primer design was performed by Sigma-Genosys (The Woodlands, Tex.) using proprietary software. Primer specifications were included in the design criteria such that the initial 5′ and 3′ primers corresponded to sequences at the beginning and end of ORFs to ensure maximum specificity. Oligonucleotides that failed the amplification or cross-reactivity criteria were redesigned based on the sequence 3 bp closer to the center of the ORF. Primers were synthesized by Sigma-Genosys and shipped in 96-well plates.
PCR amplifications were done in a 96-well format (MPC-3420 skirted 0.2-ml PCR plates; Phenix Research Products, Hayward, Calif.) with 80 ng of target chromosomal DNA in a 100-μl reaction volume. Following PCR, samples were purified by using QIAquick 96 PCR purification kits (QIAGEN, Valencia, Calif.) and run on agarose gels to ensure the amplification was specific for the appropriately sized fragment. Purified PCR products in 96-well plates were transferred to 384-well plates (Uniplate 384, 80 μl; Whatman, Clifton, N.J.), dried in a 42°C oven, and suspended in 6 μl of SSC (3×; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). These source plates were used to array the collection of C. trachomatis ORFs on CMT-GAPS-coated slides (Corning, Corning, N.Y.) by using a VersArray ChipWriter Pro arrayer (Bio-Rad Laboratories, Hercules, Calif.). All ORFs were spotted in triplicate to allow for statistical analyses of fluorescence measurements. All slides were blocked using the succinic anhydride process per the manufacturer's instructions prior to experimental hybridizations.
DNA-DNA microarray analysis of C. trachomatis serovars.
Print runs of the C. trachomatis spotted arrays were tested by direct staining of nucleic acids and DNA-DNA hybridizations with homologous C. trachomatis serovar D DNA. Slides were then dried and examined with a ScanArray 5000 (Perkin-Elmer, Boston, Mass.). C. trachomatis DNAs (30 μg in 1 ml of 10 mM Tris-1 mM EDTA; pH 7.0) were sonicated to produce randomly sheared products of approximately 500 bp. Sheared DNA was labeled with Cy3-dCTP or Cy5-dCTP by random priming of denatured DNA with random hexameric oligonucleotides. Following purification of the labeled DNA, hybridizations were done in Arrayit hybridization cassettes (TeleChem International Inc., Sunnyvale, Calif.) at 42°C for 16 h. Slides were then washed in SSC (2×) at 42°C for 15 min, followed by two washes in SSC (0.1×) for 10 min each. Slides were rapidly dried and scanned, and the subsequent images were analyzed using the QuantArray software package (Perkin-Elmer).
Fluorescence microarray data were compiled and analyzed using signal ratio comparisons and assigning a constant ratio value as a cutoff for the determination of presence or absence. The DNA sequences of the ompA genes are available for all C. trachomatis serovars. Constant ratio value cutoffs were determined to be problematic, as the transition points for the present-absent call were variable between serovars and occasionally between experiments. For this reason the data were reanalyzed using the GACK software algorithms described by Kim et al. (18) that allow for dynamic cutoff determinations. Conservative average cutoff values were used for the trinary analysis. Transition points of log2 ratios of −0.20 and −0.85 were used for the present-slightly divergent cutoff and the slightly divergent-highly divergent cutoff, respectively. The data were then assigned values of −0.5 for divergent genes, 0 for slightly divergent genes, and 0.5 for present genes. We have not used the graded assignment categorization option, as all divergent regions we identified with this technique were subsequently PCR amplified and sequenced. The ompA genes that most diverged from the C. trachomatis serovar D sequence (from serovars A, C, F, G, H, I, J, K, and L3) were categorized as slightly divergent (discussed below).
Sequence analysis of the PZ of C. trachomatis serovars.
Crude genomic DNAs were extracted by mixing 50 μl of EBs (106 to 109 IFU/ml) with 250 μl 0.5 N NaOH at room temperature for 5 min. A 300-μl volume of 1 M Tris-HCl, pH 8.0, was then added to neutralize the mixture. Serial dilutions were used as a template for PCR amplification with Expand high-fidelity polymerase (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. PCR products were generated that spanned the PZ, defined here as the region from CT163 to CT175 in serovar D (31), by using oligonucleotides based on the serovar D genomic sequence. A complete list of oligonucleotides used to generate the PCR products for this study is provided in Table 1. Upon gel purification, the PCR products of several serovars (C, F, G, K, L1, and L3) were cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, Calif.). Isolated clones from each of these serovars were then purified for sequence analysis. For other serovars (Ba, E, H, I, and J), large amounts of PCR-amplified products were purified for sequence analysis with QIAGEN-tip 100 columns (QIAGEN Inc., Valencia, Calif.) according to the manufacturer's specifications. In the cases of serovars H and J, two products approximately 5,500 bp in size were purified for sequencing. (Internal primers JHC450 and JHC451 were designed using preliminary sequence data from amplified clones of H and J.) Purified PCR products and pCR-XL-TOPO clones for each of these serovars were sent to SeqWright (Houston, Tex.) for sequence analyses. PCR-amplified products from serovars A, D, and L2 were also cloned into pCR-XL-TOPO, but their sequence analysis was conducted by our group by using the BigDye terminator cycle sequencing kit and an ABI 377 sequencing apparatus (Applied Biosystems, Foster City, Calif.) per the manufacturer's instructions. DNA sequences were compiled and analyzed using the Sequencher (version 3.1.1; Gene Codes Corp., Ann Arbor, Mich.) and MacVector (version 6.5.3, Oxford Molecular Group, Madison, Wis.) software packages.
TABLE 1.
Primer sequences of oligonucleotides used for cloning, sequence analysis, and serovar classification
| Category of use and primer | Oligonucleotide sequence (5′ → 3′) | Reference |
|---|---|---|
| Cloning and amplification for sequencing | ||
| 00.12 | GCT TTC ATC CAC ATC TTT GAC CG | This study |
| JHC250 | CCC GTT GCA GAG CGG ATG | This study |
| JHC274 | CTC CAT CGC ATC ATA CAG G | This study |
| JHC275 | CCT GTA TGA TGC GAT GGA G | This study |
| JHC450 | CGC GGA TTA GAT CCT TAA GAG TGG | This study |
| JHC451 | GGA GCG CCG CTT CTA CAG AAG GAA AG | This study |
| Serovar classification | ||
| JHC202 | ATG AAA AAA CTC TTG AAA TCG GTA | This study |
| JHC203 | TTA GAA GCG GAA TTG TGC ATT TAC | This study |
| JHC440 | GAG CTG CAA TCT ATG AAG CAA GCT C | This study |
| JHC441 | GCT CCA TAG CAC CTT TAA TAT GCC C | This study |
| JHC442 | TGG TCA GAT CGA CGA GTC CGA GAT C | This study |
| JHC443 | CAT TTC TTT TGA ACT TAC GCA GCA TGA G | This study |
Nucleotide sequence accession number.
The following sequences (in parentheses) have been submitted to GenBank and assigned the following accession numbers: AY647992 (Atox), AY647993 (Batox), AY647994 (Ctox), AY647995 (Dtox), AY647996 (Etox), AY647997 (Ftox), AY647998 (Gtox), AY647999 (Htox), AY648000 (Itox), AY648001 (Jtox), AY648002 (Ktox), AY648003 (L1tox), AY648004 (L2tox), and AY648005 (L3tox) where Atox indicates the cytotoxin gene from serovar A, etc.
RESULTS
Comparative genomic analysis of C. trachomatis by DNA-DNA microarray analysis.
Comparison of the genomes of C. trachomatis isolates (representing each of the 15 human serovars) to the homologous serovar D strain by DNA-DNA microarray is shown in Fig. 1. The microarray results in Fig. 1 are reported in a trinary format as described by Kim et al. (18), i.e., (i) presence of the gene in both test and control strains, (ii) slightly divergent or presence of a related but nonidentical gene in the test strain compared to the control strain, and (iii) highly divergent or absence of a gene in the test strain compared to the control strain. The estimated probability of presence values for the cutoffs were chosen for maximum stringency, as recommended by Kim et al. (18). Overall gene conservation was very high (>99%) compared with similar studies done with Helicobacter pylori (25) and Staphylococcus aureus (14). These results are in agreement with comparisons made using genomic DNA sequences available for C. trachomatis (23, 24, 31), similar microarray analyses of the 14 C. trachomatis serovars using serovar D/UW-3/Cx as the reference strain (5), and with the related human pathogen, Chlamydia pneumoniae (17, 23, 29). The results for the ompA gene comparisons (Fig. 1) between serovars were confirmed by individual hybridizations of amplified ompA genes and comparison to the DNA sequences available in GenBank. These experiments validated the analyses in that serovariants with ompA genes that differed significantly from serovar D gave divergent (trinary) signals in the microarray analysis (Table 2).
FIG. 1.
Comparative genomic hybridization of C. trachomatis reference strains. Chromosomal and plasmid genes (894 and 8 genes, respectively) from C. trachomatis serovar D are shown for the 15 C. trachomatis serovars (A to K and L1 to L3). Each test strain DNA was cohybridized with the control strain DNA (C. trachomatis serovar D), and the fluorescence levels were used in a trinary analysis to determine whether genes were present in the test strain (white), slightly divergent in the test strain (grey), or highly divergent (or absent) in the test strain (black), as defined in Materials and Methods. The expanded section of the figure shows the PZ of all serovars to highlight the region of the genome in which deletions were found. All deletions were found to be centered about the genes encoding the portions of the cytotoxin in serovar D, i.e., CT164 to CT167.
TABLE 2.
Sequence identity of ompA from serovar D/UW-3/Cx in comparison to the other 14 serovarsa
| Serovar | % Homology | Serovar | % Homology |
|---|---|---|---|
| A | 83.97 | H | 84.82 |
| B | 94.32 | I | 84.14 |
| Bab | 94.83 | J | 83.63 |
| C | 84.56 | K | 83.80 |
| E | 93.38 | L1 | 96.18 |
| F | 86.26 | L2 | 92.45 |
| Gb | 82.87 | L3 | 84.56 |
Differences due to insertions in relation to the serovar D ompA sequence were not included in the final nucleotide difference calculations. The sequence for the serovar D ompA was obtained from Stephens et al. (31); sequences for all other ompA genes were obtained from Stothard et al. (33), specifically, accession numbers J03813 (A/Har-13), M17342 (B/TW-5), AF063194 (Ba/Apache-2), M17343 (C/TW3), X52557 (E/Bour 1990), X52080 (F/IC-Cal3), AF063199 (G/UW57), X16007 (H/UW4), AF063200 (I/UW12), AF063202 (J/UW36), AF063204 (K/UW31), M36533 (L1/440), M14738 (L2/434), and X55700 (L3/404).
The GenBank-listed sequences for serovars Ba and G truncate the 25 3′-most ends of these genes. It was assumed that these sequences were identical when compared with the D/UW-3/Cx ompA sequence for the above calculations.
Overall diversity, in terms of gene differences (i.e., gene deletion), was very low, with a maximum of eight deleted genes in serovar B. Several serovars had no detectable differences in terms of gene deletions from serovar D (i.e., serovars E, F, G, H, I, J, and K). Without exception, all deleted genes localized to the PZ and were centered about the genes encoding the ORFs with homology to toxin-like genes. In the case of serovar B, a single contiguous deletion was found that encompassed 10 ORFs in the corresponding region of the serovar D genome (i.e., extending from CT162 to trpA). The microarray comparisons allowed for a genome-wide comparison of genetic composition at low resolution. To precisely define the PZ of C. trachomatis, we amplified and sequenced this region from each of the 15 serovars.
Comparative genomic analysis of the PZ of C. trachomatis.
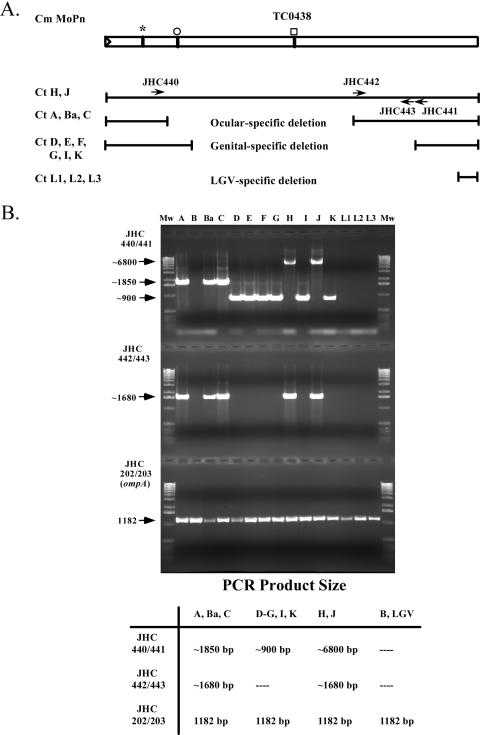
The PZ was defined as the genomic region spanning CT152/ycfV to CT176/dbsB, a 20.3-kbp DNA segment in serovar D (31) initially described by Read et al. (23). Oligonucleotide primers were designed to amplify this region as four overlapping ∼6-kbp fragments (oligonucleotide sequences for these analyses are found in Table 1 and reference 13). The PCR products were generated from each of the serovars (A to L3) and either cloned into pCR-XL-TOPO or analyzed in large-scale PCR amplifications, purified, and sequenced. Amplification patterns for the different serovars exhibited distinct patterns for ocular, genital, and LGV serovars, as shown in Fig. 2. Note that while serovar B was included in this analysis, no products were observed with the cytotoxin-specific primers, as this region of the chromosome is deleted in this serovar. In addition, control PCR amplifications were conducted using ompA-specific primers (primers JHC202 and JHC203). Genital serovars gave one of two amplification patterns (Fig. 2B). While most of the ocular and genital serovars exhibited a large internal deletion of the intact cytotoxin gene (TC0438 of C. muridarum), the genital serovars D, E, F, G, I, and K amplified products were approximately 1 kb smaller than those observed in ocular serovars A, Ba, and C (Fig. 2). Serovars H and J gave amplification patterns that indicated the cytotoxin gene was approximately the size of the intact cytotoxin gene (TC0438) in C. muridarum (Fig. 2A).
FIG. 2.
PCR amplification patterns of the cytotoxin gene in C. trachomatis serovars. Selected oligonucleotides used to amplify the cytotoxin region gave serovar-specific patterns that clustered according to the disease type and biovar. (A) Schematic representation; (B) agarose gel and summary table. An ocular-specific deletion was found for serovars A, Ba, and C by using PCR primer pairs JHC440 and JHC441 (∼1,850 bp) and JHC442 and JHC443 (∼1,680 bp). Two deletion patterns were found for genital serovars. The first group contained serovars D, E, F, G, I, and K and amplified products of ∼900 bp with the primer pair JHC440 and JHC441 and no product with JHC442 and JHC443. The second group consisted of serovars H and J, which produced products of ∼1,680 bp with JHC442 and JHC443 and ∼6,800 bp with JHC440 and JHC441. No products were found for either reaction with the LGV serovars L1, L2, and L3. The symbols above the TC0438 cytotoxin gene from C. muridarum (Cm MoPn) indicate the encoding region for the UDP-glucose binding domain (asterisk), the glycosyltransferase domain (open circle), and the cysteine-protease domain (open square).
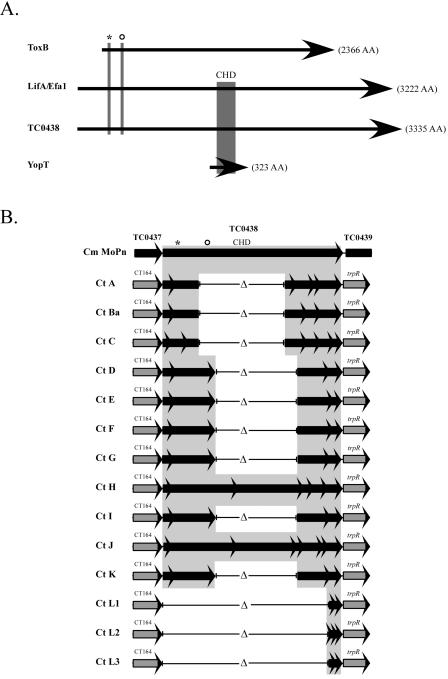
The sequence results for the region CT164 to CT169 (trpR) are shown schematically in Fig. 3B. All serovar sequences aligned with the mouse-adapted C. muridarum species MoPn TC0438 gene, as they shared the strongest degree of identity (at both the nucleotide and predicted protein levels) with this intact putative cytotoxin gene, as opposed to that of the flanking MoPn cytotoxin genes, TC0437 and TC0439. Analysis of the cytotoxin region showed that each serovar contained a unique combination of DNA sequences and ORFs and that none encoded a full-length, in-frame cytotoxin gene. The LGV serovars contained only a small region of homology to the 3′ end of the cytotoxin gene. In contrast to the results for the cytotoxin gene, the sequences for CT164 (5′ flanking gene) and CT169/trpR (3′ flanking gene) were absolutely conserved in all serovars, with the noted exception being serovar B.
FIG. 3.
Putative GTPase-inactivating domains present in the cytotoxin gene(s) of C. muridarum (MoPn) and the 15 human C. trachomatis reference serovars. (A) Schematic diagram of the C. muridarum toxin (TC0438) aligned with E. coli LifA/Efa1, C. difficile toxin B, and YopT. The illustration highlights domains that function in the inactivation of GTPases by either covalent or proteolytic modification. Toxin B is 2,366 amino acids (aa) (269.7 kDa), with UDP-glucose binding (UDP-Glc) and glycosyltranseferase (GT) domains located at the protein's N terminus, residues 94 to 137 and 263 to 298, respectively (6, 7). The UDP-Glc domain is shown with an asterisk, and the GT domain is shown with an open circle. YopT is 323 aa (36.3 kDa) and possesses a cysteine protease domain (CPD) within residues 102 to 317. The invariant Cys/His/Asp (CHD, depicted in the diagram by the shaded box) active site is critical for the proteolytic cleavage of Rho GTPases and is contained within this region (27). LifA/Efa1 is 3,222 aa (366 kDa). The protein contains the UDP-Glc (residues 312 to 355) and GT (534 to 569) domains, in addition to the CPD (1445 to 1644). C. muridarum TC0438 encodes a protein of 3,335 aa (376.5 kDa). Similar to LifA/Efa1, TC0438 has both GTPase-inactivating domains, UDP-Glc (residues 314 to 357) and GT (542 to 577), and the CPD (1530 to 1727) (2, 27). (B) Schematic showing the toxin loci for all 15 C. trachomatis reference serovars aligned to the amino acid sequence of the C. muridarium toxin (TC0438). The shaded regions identify ORFs encoding putative proteins that share strong identity with TC0438. The sequences are depicted as partial gene fragments, with arrows representing prematurely truncated ORFs. The noninvasive mucosotropic serovars constitute two genotypes and ocular serovars possess only the UDP-Glu domain, while genital serovars possess both the UDP-Glu domain and the GT domain. All lack the YopT CPD, due either to deletion or to frameshift mutation. Invasive disseminating LGV strains lack sequences corresponding to any of the domains.
These results suggest there is selective pressure for the loss of portions of the cytotoxin gene by these isolates and that the disruption of the gene can be achieved by multiple mechanisms, including deletion and accumulation of nonsense mutations that disrupt the continuity of the intact ORF. The divergent deletion patterns for ocular and genital isolates may be a reflection of the evolutionary relationships between the serovars (i.e., all ocular serovars had a common ancestor with a specific deletion of the cytotoxin gene). Alternately, the resultant smaller ORFs may encode proteins, imparting functional differences between the three pathological groups. Of note in this regard, all genital isolates have a large ORF that could theoretically encode both the UDP-glucose binding and the glycosyltransferase domains (Fig. 3). While the ocular serovars A, Ba, and C encode a predicted protein that contains the UDP-glucose binding domain, they all lack the glycosyltransferase domain. Finally, the LGV serovars have the encoding regions for both domains deleted from their genomes.
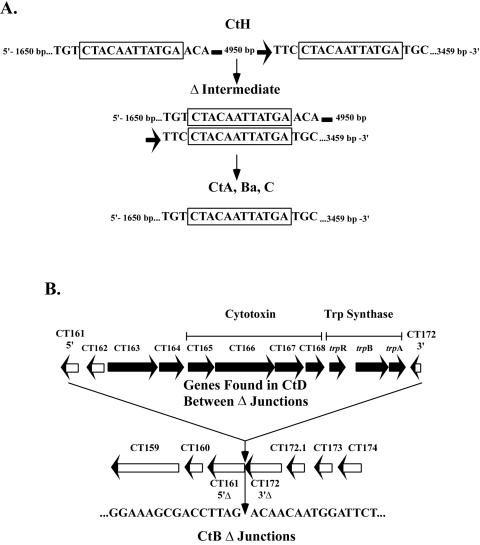
Examination of the cytotoxin sequences suggested some mechanistic explanations for the observed deletions. Sequence analysis of serovars A, Ba, and C identified a common deletion junction. Alignment of the ocular deletion junctions to that of the serovar H cytotoxin region revealed a pair of 12-bp direct repeats. Deletion of the region between these direct repeats (via alignment of the 12-bp microhomology) and recombination (35) would result in the loss of 3,477 bp, including one of the 12-bp repeat elements (Fig. 4A). Genital serovars that contain the large internal deletions do not share similar microhomology end points, as seen with the ocular strains, yet all deletion junctions map to the same location for each of the genital serovars as well. In addition, further analysis does not reveal imperfect microhomologies at the deletion junctions that could account for the deletion event (1). Moreover, the fact that all of the genital serovars exhibited the same deletion end points strongly suggests they derive from the same ancestral deletion event. If the assumption is made that the ancestral full-length gene was common between ocular and genital isolates, it seems likely that similar deletions would be found in genital isolates. One possible explanation is that genital isolates have retained functional portions of the ancestral gene (such as the glycosyltransferase domain) that are required for their tissue-specific pathogenesis yet are found between the direct repeats, whereas ocular isolates not requiring this activity lost these portions. The large deletion found in serovar B encompasses many of the genes found in the PZ, including the entire cytotoxin region as well as the entire tryptophan synthase operon (trpBA), as shown in Fig. 4B. Of interest, this deletion junction is not associated with identifiable direct repeat elements. In total, the deletion includes 10 complete and 2 partial ORFs, a total of 9,662 bp, using the sequenced serovar D genomic region for comparison (Fig. 4B).
FIG. 4.
Characterization of the deletion junctions of the cytotoxin genes of the ocular serovars. (A) The cytotoxin gene of C. trachomatis serovar H (CtH) has two direct 12-bp repeats separated by 4,956 bp. Recombination between these repeats would result in the sequence of the cytotoxin loci found in serovars A (CtA), Ba (CtBa), and C (CtC), with loss of one of the 12-bp repeats. (B) A large deletion in the cytotoxin region of the genome of serovar B results in the loss of 9,662 bp (in comparison to the D/UW-3/Cx genome sequence), including 10 complete ORFs, the 5′ portion of CT161, and the 3′ portion of CT172. This deletion encompasses the entire cytotoxin locus and the tryptophan synthase regulon.
DISCUSSION
Comparative genome hybridization (CGH) is a powerful tool for the comparison of bacterial genomes among related groups of strains or serovars. Analysis of the 15 C. trachomatis serovars demonstrated that these organisms comprise a closely related group of strains, perhaps not surprising considering the high degree of synteny found between C. trachomatis serovar D and the more distantly related species C. muridarum, C. caviae, and C. pneumoniae. We found a maximal difference of 10 deleted genes (0.9%) in the comparison of all serovars to serovar D, an extremely low level of divergence compared with similar studies involving S. aureus (14), H. pylori (25), and Vibrio cholerae (11). It is important to note that the CGH analysis was a one-way comparison, in that we could only detect loss of genes with respect to the homologous serovar D genomic content, and that the test serovars may encode novel genes not found in serovar D. While the resolution of CGH is rather low, variations between the test and target (serovar D) DNAs can be identified when divergence is in the vicinity of 90% identity. Using ompA as a reference gene, serovars sharing 92.45% identity (L2) or greater (Table 2) with the control serovar D sequence are conserved-highly homologous (Fig. 1). Conversely, the remaining serovars exhibiting conserved identity in the range of 82.87% (serovar G) to 86.26% (serovar F) were classified as slightly divergent. It is important to note that changes involving small deletions, insertions, or nonsense mutations are not detectable in hybridization analyses of whole genes and may result in important strain differences. This point is demonstrated by the comparative genomic analyses reported by Caldwell et al. (9) that distinguish ocular serovars from genital serovars of C. trachomatis based on single nucleotide changes and polymorphisms that lead to the inactivation or altered enzymatic activity of tryptophan synthase. While considering the limitations, the overall level of homology is nonetheless striking, particularly because the serovars tested comprise three groups of disease-associated strains: those causing trachoma, those causing sexually transmitted diseases, and those causing LGV. These findings suggest that the tissue tropism or virulence traits involved in pathogenic outcome of disease are controlled by a small number of genetic differences.
Our findings are similar to those recently reported by Brunelle et al. (5). In that study, the authors attempted to identify genes that have a high degree of polymorphism by using a competitive binding array. The authors mixed their test genomic DNAs with that of serovar D genomic DNA, followed by hybridization to a serovar D-based, PCR-amplified microarray slide. The approach was designed to identify genes that were more divergent from the test strain (serovar D) and thus may play a role in differences in tissue tropism and pathogenesis. It is interesting that CT166 was identified in all nongenital serovars, which is obviously in agreement with our results. However, while both studies employed a competitive binding-based assay, our microarray approach used a larger percentage of the entire genome, in that ∼99% of all putative ORF DNA sequences were spotted on the microarray. The D/UW-3 genome consists of 1,050,012 bp with 893 chromosomal ORFs (3) and 8 plasmid ORFs. If one were to assume that 90% of the genome encoded the 901 ORFs, that would account for ∼945,000 bp of the genome. Our microarray analyses attempted to look at these ORFs in their entirety. Conversely, Brunelle et al. (5) amplified products for their hybridizations that consisted of, on average, 60% of each ORF tested in their study. The authors stated that this was done in an effort to construct primer pairs specific for, and amplification of, the largest portion of each ORF analyzed. Of the total 894 chromosomal ORFs identified in D/UW-3 by Stephens et al. (30), 28 were removed from their study due to a high background signal. As such, assuming an average ORF size of 1,057 bp, Brunelle et al. (5) assayed ∼549,242 bp, or ∼52.3% of the entire genome. While our assay may not have been as sensitive as theirs, it encompassed a larger portion of the entire genome. Moreover, in our study ompA was identified as divergent from D/UW-3 in all serovars with the exception of serovars B, Ba, E, L1, and L2. Conversely, Brunelle et al. (5) stated that all serovars diverged from D/UW-3 with the exceptions B, Ba, E, H, I, J, and L1. It is possible that the PCR-amplified product used for hybridization in the latter study contains a smaller, internal portion of the ompA ORF. As such, the hypervariable domain sequences (36) would play a larger roll in the hybridization analyses, resulting in the differing results.
All of the deletion events detected in the C. trachomatis serovars are found in the PZ and include ORFs related to the chlamydial cytotoxin-like gene. Genomic sequencing of a number of chlamydial species has provided a wealth of information about these cryptic organisms, including the presence of three high-molecular-weight cytotoxin-like genes in the mouse-adapted species C. muridarum (MoPn) (23), a single large cytotoxin in the guinea pig strain C. caviae (GPIC) (24), and the presence of disrupted, partial cytotoxin-like ORFs in serovar D, a human pathogen (31). The chlamydial toxin gene homologs have homology to the LCTs A and B, predominantly in the regions shown to be involved in glycosyltransferase and UDP-glucose binding activities (6). This homology is restricted to the N-terminal third of the proteins. The truncated nature of the ORFs in serovar D suggested that these regions were not expressed and were most likely the remnants of a prior, intact gene. Despite the disruption of the putative ancestral intact cytotoxin gene, the smaller toxin ORF, CT166 of serovar D, is still transcribed and expressed in a developmental cycle-specific manner (2).
The cytotoxin locus from the C. trachomatis reference strains is highly polymorphic in that each of the genes in serovars A to K has a unique locus. Only serovars H and J have retained the central portion of the gene, encoding the region described as a putative cysteine protease domain (27), but this region also contains premature stop mutations. Of interest, we identified two smaller cytotoxin alleles from the serovar H DNA PCR amplifications that had deletions within the central region of the gene (data not shown). This suggests that the serovar H population is nonclonal and that multiple inactivated cytotoxin loci can be found in a single reference strain population. Together, these findings imply that the deletion and nonsense mutations found in the cytotoxin gene have arisen recently and that different mutational events can be found in a single population. These findings contrast the situation in the LGV reference strains in which the extensive deletion of the cytotoxin gene is nearly identical in serovars L1, L2, and L3. As a result, the chlamydial serovars can be classified into their pathological groups based on their toxin genotype. The genitotropic strains (D to K) all encode an intact N-terminal-encoding toxin ORF that includes both the UDP-glucose binding and glycosyltransferase domains. The LGV (L1 to L3) strains lack both of these domains, while the ocular serovars (A, Ba, and C) only encode the UDP-glucose binding domain. The obvious exception to this genotypic classification is the oculotropic serovar B, because this entire region is deleted from the chromosome.
The chlamydial cytotoxin has distinct homology to the E. coli genes encoding Efa1/LifA. Efa1 has been shown to influence attachment of Shiga toxin-producing E. coli (STEC) 0111:H- to Chinese hamster ovary cells (20) and is required for colonization of bovine intestine by STEC serotypes 05 and 0111 (32). LifA, found in enteropathogenic E. coli, inhibits the proliferation of human peripheral blood lymphocytes (19). The efa1 gene is also found in all non-O157 STEC serotypes (19, 20), whereas the O157:H7 strains contain a truncated version of the gene (16, 21). This truncated version of efa1 lacks a large portion of the central region of the gene analogous to the deletions in the cytotoxin gene in chlamydial strains. Transposon mutants of efa1 adhere poorly to cultured Chinese hamster ovary cells and bovine intestinal cells while, notably, miniTn5Km2 insertions immediately upstream of the truncated efa1 gene in an O157:H7 strain also reduce bacterial adherence to Caco-2 cells (34). The functional nature of the truncated version of Efa1 suggests that the analogous truncated chlamydial protein may also have biological activity.
Two primary mechanisms of pathogen-encoded, enzyme-mediated GTPase inactivation have been described: glycosyltransferase (6, 7) and proteolytic cleavage (28). Each can independently result in the disruption of the host cytoskeleton. Multiple bacterial virulence factors have been described that can inactivate as well as activate GTPases (4). It is interesting that Efa1/LifA and the C. muridarum cytotoxins (TC0437 to TC0439) encode both the glycosyltransferase and cysteine protease domains in the same protein (Fig. 3A). The reason for this functional redundancy is not yet understood, especially in the case of C. muridarum, where not one but three large cytotoxin-like genes are present in tandem. Curiously, C. trachomatis has apparently lost the need for either function when the cellular target is ocular or invasive in nature. The implied need for a functional glycosyltranseferase domain in genitotropic serovars also remains unanswered. The fact that the LGV and ocular serovars lack the glycosyltranseferase domain does not suggest they cannot modulate host-encoded GTPase proteins. Carabeo et al. (10) recently reported a requirement for the GTPase Rac protein in cellular invasion and actin recruitment by the LGV serovar L2. Moreover, those authors noted the presence of a type III secretion apparatus that may play a role in this activity, although the presence of possible effector proteins (i.e., SopE- or SptP-like homologs) has yet to be determined.
The biologic role of the chlamydial cytotoxins has received little attention to date. We have focused on the human C. trachomatis serovars, in which the nature of the cytotoxin gene is highly polymorphic. There are, however, chlamydial species that infect animals and encode intact cytotoxins: the C. caviae strain GPIC infects guinea pigs and encodes a single cytotoxin, the C. muridarum strain MoPn infects mice and encodes three cytotoxins, arranged in tandem in the genome. The central C. muridarum cytotoxin (TC0438) is the most closely related to the sequence of the intact human cytotoxin described here. The strict association of complete and partial toxin genes with chlamydial strains that exhibit mucosal and epithelial tropism, regardless of host species, presents a strong indirect argument that the toxin is an important virulence factor for mucosotropic chlamydial pathogens. Understanding the function of the toxin in mediating mucosal infection tropism and pathogenesis is an important future research goal.
Acknowledgments
We are grateful for the technical contributions of Bill Whitmire in the isolation of the EBs used for this study, Lorne Rose for data analysis, Kelly Matteson for secretarial assistance in the preparation of the manuscript, and Brenda Rae Marshall for editorial assistance.
Editor: D. L. Burns
REFERENCES
- 1.Albertini, A. M., M. Hofer, M. P. Calos, and J. H. Miller. 1982. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell 29:319-328. [DOI] [PubMed] [Google Scholar]
- 2.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belland, R. J., G. Zhong, D. D. Crane, D. Hogan, D. Sturdevant, J. Sharma, W. L. Beatty, and H. D. Caldwell. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 100:8478-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boquet, P., and E. Lemichez. 2003. Bacterial virulence factors targeting Rho GTPases: parasitism or symbiosis? Trends Cell Biol. 13:238-246. [DOI] [PubMed] [Google Scholar]
- 5.Brunelle, B. W., T. L. Nicholson, and R. S. Stephens. 2004. Microarray-based genomic surveying of gene polymorphisms in Chlamydia trachomatis. Genome Biol. 5:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busch, C., F. Hofmann, R. Gerhard, and K. Aktories. 2000. Involvement of a conserved tryptophan residue in the UDP-glucose binding of large clostridial cytotoxin glycosyltransferases. J. Biol. Chem. 275:13228-13234. [DOI] [PubMed] [Google Scholar]
- 7.Busch, C., F. Hofmann, J. Selzer, S. Munro, D. Jeckel, and K. Aktories. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 273:19566-19572. [DOI] [PubMed] [Google Scholar]
- 8.Caldwell, H. D., J. Kromhout, and J. Schachter. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect. Immun. 31:1161-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caldwell, H. D., H. Wood, D. Crane, R. Bailey, R. B. Jones, D. Mabey, I. Maclean, Z. Mohammed, R. Peeling, C. Roshick, J. Schachter, A. W. Solomon, W. E. Stamm, R. J. Suchland, L. Taylor, S. K. West, T. C. Quinn, R. J. Belland, and G. McClarty. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J. Clin. Investig. 111:1757-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carabeo, R. A., S. Grieshaber, A. Hasenkrug, C. A. Dooley, and T. Hackstadt. 2004. Requirement for the Rac GTPase in Chlamydia trachomatis invasion of non-phagocytic cells. Traffic 5:418-425. [DOI] [PubMed] [Google Scholar]
- 11.Dziejman, M., E. Balon, D. Boyd, C. M. Fraser, J. F. Heidelberg, and J. J. Mekalanos. 2002. Comparative genomic analysis of Vibrio cholerae: genes that correlate with cholera endemic and pandemic disease. Proc. Natl. Acad. Sci. USA 99:1556-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feener, E. P., W. C. Shen, and H. J. Ryser. 1990. Cleavage of disulfide bonds in endocytosed macromolecules. A processing not associated with lysosomes or endosomes. J. Biol. Chem. 265:18780-18785. [PubMed] [Google Scholar]
- 13.Fehlner-Gardiner, C., C. Roshick, J. H. Carlson, S. Hughes, R. J. Belland, H. D. Caldwell, and G. McClarty. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J. Biol. Chem. 277:26893-26903. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald, J. R., D. Sturdevant, S. M. Mackie, S. R. Gill, and J. A. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8621-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grayston, J. T., and S. Wang. 1975. New knowledge of chlamydiae and the diseases they cause. J. Infect. Dis. 132:87-105. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, T., K. Makino, M. Ohnishi, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayam, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 17.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 18.Kim, C. C., E. A. Joyce, K. Chan, and S. Falkow. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:0065.1-0065.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68:2148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 21.Perna, N. T., G. R. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. S. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 22.Rank, R. G. 1999. Models of immunity, p. 239-295. In R. S. Stephens (ed.), Chlamydia: intracellular biology, pathogenesis, and immunity. ASM Press, Washington, D.C.
- 23.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Read, T. D., G. S. Muers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. F. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, Peterson, J., M. J. Beanan, O. White, S. L. Salzberg, R. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydiophilia caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salama, N., K. Guillemin, T. K. McDaniel, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schachter, J. 1978. Chlamydial infections. N. Engl. J. Med. 298:428-434. [DOI] [PubMed] [Google Scholar]
- 27.Shao, F., P. M. Merrit, Z. Bao, R. W. Innes, and J. E. Dixon. 2002. A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109:575-588. [DOI] [PubMed] [Google Scholar]
- 28.Shao, F., V. O. Panayiotis, B. Zhaoquin, K. E. Bowers, C. A. Fierke, and J. E. Dixon. 2003. Biochemical characterization of the Yersinia YopT protease: cleavage site and recognition elements in Rho GTPases. Proc. Natl. Acad. Sci. USA 100:904-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shirai, M., H. Hirakawa, M. Kimoto, M. Tabuchi, F. Kishi, K. Ouchi, T. Shiba, K. Ishii, M. Hattori, S. Kuhara, and T. Nakazawa. 2000. Comparison of whole genome sequences of Chlamydia pneumoniae J138 from Japan and CWL029 from USA. Nucleic Acids Res. 28:2311-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stephens, R., S. Kalman, C. Fenner, and R. Davis. 1998. Chlamydia genome project. [Online.] http://chlamydia-www.berkeley.edu:4231/.
- 31.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 32.Stevens, M. P., P. M. van Diemen, G. Frankel, A. D. Phillips, and T. S. Wallis. 2002. Efa1 influences colonization of the bovine intestine by Shiga toxin-producing Escherichia coli serotypes O5 and O111. Infect. Immun. 70:5158-5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stothard, D. R., G. A. Toth, and B. E. Batteiger. 2003. Polymorphic membrane protein H has evolved in parallel with the three disease-causing groups of Chlamydia trachomatis. Infect. Immun. 71:1200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Saskawa. 2000. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 10:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Welcker, A. J., J. de Montigny, S. Potier, and J. Souciet. 2000. Involvement of very short DNA tandem repeats and the influence of the RAD52 gene on the occurence of deletions in Saccharomyces cerevisiae. Genetics 156:549-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan, Y., Y.-X. Zhang, N. G. Watkins, and H. D. Caldwell. 1989. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect. Immun. 57:1040-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]