Abstract
Expression of the bla1 and bla2 genes in an archetypal Bacillus anthracis strain is insufficient for penicillin resistance. In a penicillin-resistant clinical isolate, both genes are highly transcribed, but bla1 is the major contributor to high-level resistance to ampicillin. Differential expression of the bla genes is dependent upon strain background.
Ciprofloxacin, doxycycline, and penicillin G are currently recommended by the Centers for Disease Control and Prevention and the Food and Drug Administration as therapeutics for inhalation and cutaneous anthrax (16). Prototypical Bacillus anthracis strains are susceptible to all three of these antibiotics. There have been no reports of naturally occurring ciprofloxacin- or doxycycline-resistant B. anthracis strains, but naturally occurring penicillin-resistant B. anthracis isolates have been reported (5, 19, 20, 32), and surveys of clinical and soil-derived strains have revealed penicillin G resistance in up to 16% of isolates tested (3, 6, 8, 10, 21, 25, 26).
Bacterial resistance to β-lactam antibiotics is most commonly attributed to the synthesis of β-lactamases, which are enzymes that hydrolyze amides, amidines, and other carbon-nitrogen bonds in cyclic amides (29). The presence of two β-lactamase genes, bla1 and bla2, in the penicillin-susceptible Sterne strain of B. anthracis was reported previously (7). Sterne is a well-studied attenuated strain that serves as a live vaccine for animals (33). Sterne is toxigenic due to the presence of pXO1, which carries the toxin genes, but is capsule deficient because it lacks pXO2, which harbors the capsule biosynthesis genes (Table 1). With the exception of being pXO2 negative, Sterne is considered to be a prototypical B. anthracis strain (34). The bla genes of Sterne, located approximately 900 kb apart on the chromosome, encode functional β-lactamases. Escherichia coli clones containing either gene exhibit penicillinase activity. Bla1 appears to be a group 2a penicillinase. Bla2 is similar to a group 3 Bacillus cereus metalloenzyme (23). Nevertheless, the low level of bla gene expression in Sterne is insufficient for resistance to ampicillin (7, 23), and induction of β-lactamase activity has not been observed following growth in sublytic levels of penicillin G or cefoxitin (Y. Chen and T. M. Koehler, unpublished data).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Plasmids | ||
| E. coli | ||
| pBC | Cmr | Stratagene |
| pUC18::Ωkm-2 | pUC18 carrying an Ω element with kanamycin resistance marker aph3 (Ω-km-2); Kmr | 28 |
| pJRS322 | pUC18 carrying an Ω element with spectinomycin resistance marker aad9 (Ω-sp); Spr | 31 |
| pUTE618 | Ω-km-2 from pUC18::Ωkm-2 cloned into the EcoRI site of pBC, source of Ω-km-2 for gene replacement, Cmr Kmr | This work |
| pUTE619 | Ω-sp from pJRS322 cloned into the EcoRI site of pBC, source of Ω-sp for gene replacement, Cmr Spr | This work |
| Bifunctional | ||
| pHT304-18Z | Contains promoterless lacZ; Apr in E. coli, Emr in B. anthracis | 1 |
| pUTE29 | Source of replication origin of pBC16; Apr in E. coli | 9 |
| pUTE546 | Ω-sp from pJRS322 cloned into the BamHI site of pHT304-18Z; Apr in E. coli, Spr in B. anthracis | This work |
| pUTE547 | Pbla1-lacZ; amplicon generated using primers Pbla1-1 and Pbla1-2 (Table 2) cloned into the PstI/BamHI sites of pUTE546; Apr in E. coli, Spr in B. anthracis | This work |
| pUTE548 | Pbla2-lacZ; amplicon generated using primers Pbla2-1 and Pbla2-2 (Table 2) cloned into the PstI/BamHI sites of pUTE546; Apr in E. coli, Spr in B. anthracis | This work |
| pUTE583 | Replication origin from pUTE29 and erm from pHT304-18Z cloned into pBC; gene replacement vector harbors a SacI site for insertion of an Ω element flanked by DNA sequences directly upstream and downstream of the target gene coding region; Cmr in E. coli, Emr in B. anthracis | This work |
| Strains | ||
| B. anthracis | ||
| UM44b | Sterne strain derivative; Pens | 35 |
| 32 | pXO1, pXO2; Penr | 21 |
| UT223 | 32 cured of pXO1; Penr | This work |
| UT247 | UT223, Δbla1 Kmr | This work |
| UT248 | UT223, Δbla2 Spr | This work |
| UT249 | UT223, Δbla1 Δbla2 Kmr Spr | This work |
| E. coli | ||
| TG1 | Cloning host, Aps | 2 |
| GM2163 | Host for isolation of unmethylated DNA for electroporation into B. anthracis, dam dcm mutant | 27 |
Apr, ampicillin resistant; Aps, ampicillin susceptible; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant; Pens, penicillin susceptible; Penr, penicillin resistant; Spr, spectinomycin resistant.
Weybridge (Sterne) strain derivative.
B. anthracis 32 is a highly β-lactamase-positive clinical isolate for which the penicillin MIC is between 64 and 128 μg/ml (21, 32). PCRs employing strain 32 DNA and oligonucleotide primers corresponding to bla1 and bla2 sequences from the penicillin-susceptible UM44 (Sterne) strain (Tables 1 and 2) resulted in amplicons of the expected sizes for the bla1 and bla2 genes (data not shown), indicating that strain 32 harbors both bla genes. Although all experiments described here were performed in facilities that met Centers for Disease Control and Prevention guidelines, in the interest of maximum safety, we created a toxin-deficient derivative of strain 32 by curing it of pXO1 by culturing the strain in a medium containing a sublethal concentration of novobiocin (14). Results of PCR experiments testing for the presence of the toxin genes (17) and immunoblots testing for the presence of the anthrax toxin protein-protective antigen and lethal factor in culture supernates (15) revealed a cured strain, UT223, that was pXO1 negative and pXO2 positive (Table 1). UT223, like its parent, produced high-level β-lactamase activity and was able to grow in media containing ampicillin (100 μg/ml). Nucleotide sequences of amplified DNAs indicated that the coding sequence of bla1 from the β-lactamase-positive strain UT223 is identical to that of bla1 from UM44. The coding sequences of the bla2 genes from UT223 and UM44 differ by one nucleotide, which is predicted to result in a single-amino-acid substitution (Y62C). Both bla2 genes conferred ampicillin resistance to E. coli, indicating that the sequence difference does not dramatically affect protein function.
TABLE 2.
Primers used in this study
| Primer paira | Sequence (5′-3′) | PCR amplification product (bp) |
|---|---|---|
| BLA1-1 | GGTACCAAATGGCTCATTGG | bla1 upstream region (1,024) |
| BLA1-2 | CCCGGATCCTAATATACCAACACACATC | |
| BLA1-3 | CCCGGATCCATTGCAGAGGCGGCTGAAG | bla1 downstream region (923) |
| BLA1-4 | GTCGACAGTCGCTATTGATACACC | |
| BLA1-5 | CATTGCAAGTTGAAGCGAAA | bla1 internal region (680) |
| BLA1-6 | TGTCCCGTAACTTCCAGCTC | |
| Pbla1-1 | GGTACCTCGAGTCGGATTGATATGAAGACA | bla1 promoter region (645) |
| Pbla1-2 | GGATCCCATTCCTTTCAAATACTTAATACC | |
| BLA2-1 | CTGCAGGGTATATTCATTCCGG | bla2 upstream region (806) |
| BLA2-2 | CCCGGATCCACCCTTTCTTTCAAACG | |
| BLA2-3 | CCCGGATCCTGGTCATGGAGAAGTAGG | bla2 downstream region (1,015) |
| BLA2-4 | GAGCTCATGGTTCCTCTCCCGTC | |
| BLA2-5 | TTGTCGATTCTTCTTGGGATG | bla2 internal region (483) |
| BLA2-6 | CCCCTACTTCTCCATGACCA | |
| Pbla2-1 | GGTACCTCGAGAAAGCAAAATATGAAGTTTGCG | bla2 promoter region (711) |
| Pbla2-2 | GGATCCCACCCTTTCTTTCAAACGAA | |
| 16S-1 | CACACTGGGACTGAGACACG | 16S rRNA gene internal region (902) |
| 16S-2 | AAGGGGCATGATGATTTGAC |
DNA oligonucleotides were purchased from Integrated DNA, Technologies (Coralville, Iowa). PCRs were performed by using a PE-Applied Biosystems (Norwalk, Conn.) PCR system 9700.
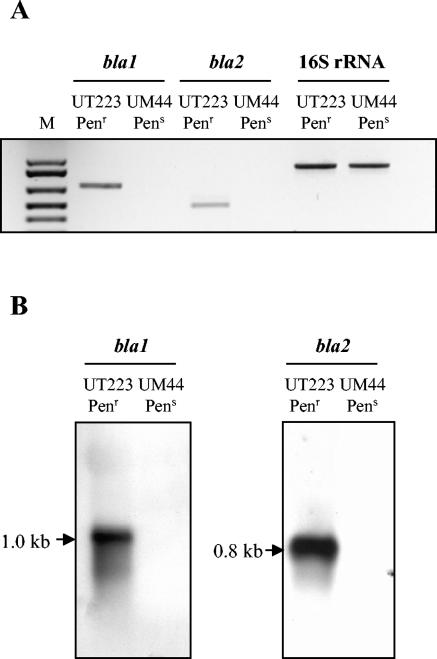
To compare levels of bla gene expression in the penicillin-resistant and -susceptible strains, RNA was isolated from UT223 and UM44 cultures grown to exponential phase and subjected to reverse transcription-PCR and Northern hybridization (7). Both bla genes were transcribed in UT223 but not in UM44 (Fig. 1A). Transcript sizes corresponded to gene lengths: 1.0 kb for bla1 and 0.8 kb for bla2 (Fig. 1B). Thus, the bla genes of UT223 are expressed as monocistronic transcripts.
FIG. 1.
bla gene transcription in penicillin-resistant (Penr) and penicillin-susceptible (Pens) B. anthracis strains. Total RNA was extracted from B. anthracis strains during exponential growth in Luria-Bertani medium. (A) Reverse transcription-PCR analysis of bla transcripts. Lane M, molecular size markers. (B) Northern blot analysis of bla transcripts. Internal DNA probes for bla1 and bla2 were generated by using primers listed in Table 2. Sizes of transcripts are indicated.
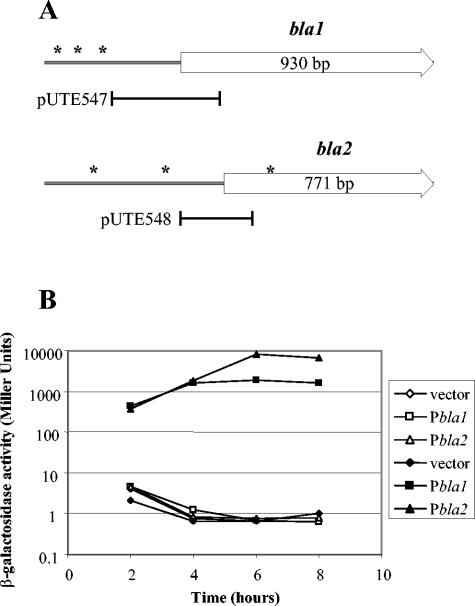
The comparison of bla1 and bla2 upstream regions (up to 500 nucleotides [nt]) in UM44 and UT223 revealed differences at specific sites (Fig. 2A). However, the first 351 nt upstream of bla1 and the first 180 nt upstream of bla2 are identical in the two strains. To determine if differential expression of the bla genes was due to sequence differences in upstream regions, common regions were cloned adjacent to a promoterless lacZ gene in pUTE546 (Table 1 and Fig. 2A). The reporter constructs were electroporated into B. anthracis strains (18), and promoter activity of the transcriptional fusions was assessed by measuring β-galactosidase activity (7). UM44 harboring either pUTE547 (Pbla1-lacZ) or pUTE548 (Pbla2-lacZ) produced a low level of β-galactosidase activity (Fig. 2B). In contrast, the enzyme activity of UT223 harboring either Pbla1-lacZ or Pbla2-lacZ was 100- to 1,000-fold higher. Both transcriptional fusions were expressed throughout growth. The bla2 promoter was more strongly expressed (about fourfold) than the bla1 promoter. These data indicate that differential expression of the bla genes in UM44 and UT223 is dependent upon strain background rather than differences in the promoter regions of the genes.
FIG. 2.
Promoter activity of bla genes in Penr and Pens B. anthracis strains. (A) Schematic representation of sequence differences in bla genes. Cloned DNA regions for lacZ fusions are indicated. Loci of nucleotide differences are shown as asterisks. (B) Promoter activities of bla genes in Penr (UT223) (filled symbols) and Pens (UM44) (open symbols) strains. Isolates harbored pUTE546 (vector), pUTE547 (Pbla1), or pUTE548 (Pbla2) as indicated. β-Galactosidase activity was determined following growth in Luria-Bertani medium at 37°C with shaking at 200 rpm. Data shown are representative of at least three experiments.
To determine the contribution of each bla gene to β-lactam antibiotic resistance in UT223, bla-null mutants were constructed by using the delivery vector pUTE583 and a method described previously (31). UT247 (bla1), UT248 (bla2), UT249 (bla1 bla2) (Table 1), and the parent strain grew at similar rates in Luria-Bertani broth, and bla gene transcripts were not detected in RNA preparations from cultures of the respective mutants (data not shown). β-Lactamase activity of UT223 was defined as 100 arbitrary units. UT247 (bla1) and UT248 (bla2) exhibited approximately one-half of the β-lactamase activity of the parent, while UT249 (bla1 bla2) produced no detectable β-lactamase activity (Fig. 3).
FIG. 3.
β-Lactamase production by B. anthracis strains. β-Lactamase activity was determined following incubation for 7 h at 37°C with shaking at 200 rpm, when the cultures were in late log phase. The strains used were UT223 (Penr), UT247 (bla1), UT248 (bla2), UT249 (bla1 bla2), and UM44 (Pens).
The MICs of β-lactam antibiotics were determined for the mutants by use of the broth microdilution method (NCCLS) and Mueller-Hinton broth (BD BioSciences, Sparks, Md.) (25). Results indicate that bla1 is the major contributor to resistance to ampicillin, amoxicillin-clavulanate, and piperacillin (Table 3), a finding consistent with our previous study of bla function in E. coli (7). We reported previously that Bla2 is a cephalosporinase and that E. coli harboring the cloned bla2 gene is resistant to all but one of these antibiotics (7). However, the contribution of bla2 to cephalosporin resistance is difficult to assess given the high level of intrinsic resistance in B. anthracis. UT249 lacks bla1 and bla2 but is still resistant to cefepime, ceftazidime, and cefpodoxime.
TABLE 3.
MICs of antibiotics for B. anthracis strains
| Antibiotic or inhibitor | MIC (μg/ml) for:
|
||||
|---|---|---|---|---|---|
| UT223 (Penr) | UT247 (Δbla1) | UT248 (Δbla2) | UT249 (Δbla1 Δbla2) | UM44 (Pens) | |
| Ampicillin | >64 | <0.03 | >64 | <0.03 | <0.03 |
| Amoxicillin-clavulanate | 4 | <0.5 | 4 | <0.5 | <0.5 |
| Cefazolin | <0.5 | <0.5 | <0.5 | <0.5 | <0.5 |
| Cefepime | >32 | >32 | >32 | >32 | >32 |
| Cefoxitin | 8 | 4 | 4 | 4 | 8 |
| Cefotetan | 8 | 8 | 4 | 4 | 8 |
| Ceftazidime | >128 | >128 | >128 | >128 | >128 |
| Cefpodoxime | >16 | >16 | >16 | >16 | 16 |
| Cefotaxime | 32 | 32 | 32 | 32 | 16 |
| Ceftriaxone | 16 | 16 | 16 | 16 | 8 |
| Meropenem | 0.06 | 0.06 | <0.03 | <0.03 | <0.03 |
| Piperacillin | >128 | <2 | >128 | <2 | <2 |
| Piperacillin-tazobactam (4 μg/ml) | 2 | 2 | 2 | <1 | <1 |
B. anthracis is a member of the closely related B. cereus group species. Other members of this group, B. cereus and B. thuringiensis, are generally penicillin resistant, producing inducible β-lactamases. Constitutive β-lactamase-producing mutants of B. cereus have also been reported (4). These mutants produce multiple β-lactamases, but the genetic basis has not been investigated. Our study was limited to single isolates of penicillin-resistant and -susceptible strains, but considering the monomorphic nature of B. anthracis, it is likely that other penicillin-susceptible strains harbor transcriptionally silent bla genes. It is possible that penicillin-resistant and -susceptible B. anthracis strains differ in the functions or levels of expression of their trans-acting bla regulators. Regulation of β-lactamase synthesis has been studied in certain other gram-positive bacteria (11, 36). In Streptomyces cacaoi, the expression of β-lactamase genes is controlled by a regulatory protein, BlaB (22), and mutations in blaB can lead to silencing or constitutive expression of β-lactamase genes (30). Homologs of bla regulators are present in the B. anthracis genome, but they are not linked to either bla gene. Future investigations will address the potential relationship of these genes to bla gene control in B. anthracis.
It is notable that the treatment of anthrax with penicillin is not always successful (13, 24) and the use of penicillin for prophylaxis and treatment of anthrax in experimental animals has had various outcomes (12). Considering reports of naturally occurring penicillin-resistant isolates and the increased concern regarding human anthrax, it is likely that guidelines for antibiotic treatment will continue to be revised. If the various penicillin-resistant isolates are determined to harbor a common mutation, this information may provide a rapid method for determining penicillin resistance at the DNA sequence level.
Nucleotide sequence accession numbers.
DNA sequences of bla1 and bla2 from strain UT223 were deposited in GenBank under accession numbers AY453161 and AY453162.
Acknowledgments
We thank Jasmine Chaitram for assistance with MIC testing.
This work was supported by Public Health Service grant AI33537 from the National Institutes of Health.
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1994. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol. Microbiol. 13:97-107. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1996. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 3.Bakici, M. Z., N. Elaldi, M. Bakir, I. Dokmetas, M. Erandac, and M. Turan. 2002. Antimicrobial susceptibility of Bacillus anthracis in an endemic area. Scand. J. Infect. Dis. 34:564-566. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, A., K. W. Nickerson, and R. A. Day. 1967. Thermal penicillinase derepression and temperature dependence of penicillinase production: inducible and constitutive strains of Bacillus cereus. Arch. Biochem. Biophys. 119:50-54. [DOI] [PubMed] [Google Scholar]
- 5.Bradaric, N., and V. Punda-Polic. 1992. Cutaneous anthrax due to penicillin-resistant Bacillus anthracis transmitted by an insect bite. Lancet 340:306-307. [DOI] [PubMed] [Google Scholar]
- 6.Cavallo, J.-D., F. Ramisse, M. Girardet, J. Vaissaire, M. Mock, and E. Hernandez. 2002. Antibiotic susceptibilities of 96 isolates of Bacillus anthracis isolated in France between 1994 and 2000. Antimicrob. Agents Chemother. 46:2307-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Y., J. Succi, F. C. Tenover, and T. M. Koehler. 2003. β-Lactamase genes of the penicillin-susceptible Bacillus anthracis Sterne strain. J. Bacteriol. 185:823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coker, P. R., K. L. Smith, and M. E. Hugh-Jones. 2002. Antimicrobial susceptibilities of diverse Bacillus anthracis isolates. Antimicrob. Agents Chemother. 46:3843-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai, Z., J.-C. Sirard, M. Mock, and T. M. Koehler. 1995. The atxA gene product activates transcription of the anthrax toxin genes and is essential for virulence. Mol. Microbiol. 16:1171-1181. [DOI] [PubMed] [Google Scholar]
- 10.Doganay, M., and N. Aydin. 1991. Antimicrobial susceptibility of Bacillus anthracis. Scand. J. Infect. Dis. 23:333-335. [DOI] [PubMed] [Google Scholar]
- 11.Filee, P., K. Benlafya, M. Delmarcelle, G. Moutzourelis, J. M. Frere, A. Brans, and B. Joris. 2002. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP beta-lactamase. Mol. Microbiol. 44:685-694. [DOI] [PubMed] [Google Scholar]
- 12.Friedlander, A. M., S. L. Welkos, M. L. Pitt, J. W. Ezzell, P. L. Worsham, K. J. Rose, B. E. Ivins, J. R. Lowe, G. B. Howe, P. Mikesell, et al. 1993. Postexposure prophylaxis against experimental inhalation anthrax. J. Infect. Dis. 167:1239-1243. [DOI] [PubMed] [Google Scholar]
- 13.Gold, H. 1967. Treatment of anthrax. Fed. Proc. 26:1563-1568. [PubMed] [Google Scholar]
- 14.Green, B. D., L. Battisti, T. M. Koehler, C. B. Thorne, and B. E. Ivins. 1985. Demonstration of a capsule plasmid in Bacillus anthracis. Infect. Immun. 49:291-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmaster, A. R., and T. M. Koehler. 1999. Autogenous regulation of the Bacillus anthracis pag operon. J. Bacteriol. 181:4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inglesby, T. V., T. O'Toole, D. A. Henderson, J. G. Bartlett, M. S. Ascher, E. Eitzen, A. M. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. T. Osterholm, G. Parker, T. M. Perl, P. K. Russell, and K. Tonat. 2002. Anthrax as a biological weapon, 2002: updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 17.Jackson, P. J., M. E. Hugh-Jones, D. M. Adair, G. Green, K. K. Hill, C. R. Kuske, L. M. Grinberg, F. A. Abramova, and P. Keim. 1998. PCR analysis of tissue samples from the 1979 Sverdlovsk anthrax victims: the presence of multiple Bacillus anthracis strains in different victims. Proc. Natl. Acad. Sci. USA 95:1224-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koshimizu, K., M. Ogata, and K. Ishida. 1972. Penicillin-resistant anthrax bacilli isolated from a dead dairy cow. Jpn. Vet. Soc. J. 25:184-189. [Google Scholar]
- 20.Lalitha, M. K., and M. K. Thomas. 1997. Penicillin resistance in Bacillus anthracis. Lancet 349:1522. [DOI] [PubMed] [Google Scholar]
- 21.Lightfoot, N. F., R. J. D. Scott, and P. C. B. Turnbull. 1990. Antimicrobial susceptibility of Bacillus anthracis: proceedings of the International Workshop on Anthrax. Salisbury Med. Bull. 68(Suppl.):95-98. [Google Scholar]
- 22.Magdalena, J., C. Gerard, B. Joris, M. Forsman, and J. Dusart. 1997. The two β-lactamase genes of Streptomyces cacaoi, blaL and blaU, are under the control of the same regulatory system. Mol. Gen. Genet. 255:187-193. [DOI] [PubMed] [Google Scholar]
- 23.Materon, I. C., A. M. Queenan, T. M. Koehler, K. Bush, and T. Palzkill. 2003. Biochemical characterization of β-lactamases Bla1 and Bla2 from Bacillus anthracis. Antimicrob. Agents Chemother. 47:2040-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McSwiggan, D. A., K. K. Hussain, and I. O. Taylor. 1974. A fatal case of cutaneous anthrax. J. Hyg. 73:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammed, M. J., C. K. Marston, T. Popovic, R. S. Weyant, and F. C. Tenover. 2002. Antimicrobial susceptibility testing of Bacillus anthracis: comparison of results obtained by using the National Committee for Clinical Laboratory Standards broth microdilution reference and Etest agar gradient diffusion methods. J. Clin. Microbiol. 40:1902-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Odendaal, M. W., P. M. Pieterson, V. de Vos, and A. D. Botha. 1991. The antibiotic sensitivity patterns of Bacillus anthracis isolated from the Kruger National Park. Onderstepoort J. Vet. Res. 58:17-19. [PubMed] [Google Scholar]
- 27.Palmer, B. R., and M. G. Marinus. 1994. The dam and dcm strains of Escherichia coli—a review. Gene 143:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Philippon, A., J. Dusart, B. Joris, and J. M. Frere. 1998. The diversity, structure and regulation of β-lactamases. Cell. Mol. Life Sci. 54:341-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin, C., C. Gerard, S. Donfut, E. Giannotta, G. Van Driessche, J. Van Beeumen, and J. Dusart. 2003. BlaB, a protein involved in the regulation of Streptomyces cacaoi β-lactamases, is a penicillin-binding protein. Cell. Mol. Life Sci. 60:1460-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saile, E., and T. M. Koehler. 2002. Control of anthrax toxin gene expression by the transition state regulator abrB. J. Bacteriol. 184:370-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Severn, M. 1976. A fatal case of pulmonary anthrax. Br. Med. J. 1:748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sterne, M. 1939. The use of anthrax vaccines prepared from avirulent (uncapsulated) variants of Bacillus anthracis. Onderstepoort J. Vet. Sci. Anim. Ind. 13:307-312. [Google Scholar]
- 34.Thorne, C. B. 1993. Bacillus anthracis, p. 113-124. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 35.Thorne, C. B. 1985. Genetics of Bacillus anthracis, p. 56-62. In L. Leive (ed.), Microbiology. American Society for Microbiology, Washington, D.C.
- 36.Zhang, H. Z., C. J. Hackbarth, K. M. Chansky, and H. F. Chambers. 2001. A proteolytic transmembrane signaling pathway and resistance to β-lactams in staphylococci. Science 291:1962-1965. [DOI] [PubMed] [Google Scholar]