Abstract
The use of dried blood spots (DBS) has many advantages over traditional plasma and serum samples such as the smaller blood volume required, storage at room temperature, and ability for sampling in remote locations. However, understanding the robustness of different analytes in DBS samples is essential, especially in older samples collected for longitudinal studies. Here we analyzed the stability of polar metabolites and lipids in DBS samples collected in 2000-2001 and stored at room temperature. The identified and statistically significant molecules were then compared to matched serum samples stored at −80°C to determine if the DBS samples could be effectively used in a longitudinal study following metabolic disease. Four hundred polar metabolites and lipids were identified in the serum and DBS samples using gas chromatograph-mass spectrometry (GC-MS), liquid chromatography-MS (LC-MS) and LC-ion mobility spectrometry-MS (LC-IMS-MS). The identified polar metabolites overlapped well between the sample types, though only one statistically significant metabolite was conserved in a case-control study of older diabetic males with high body mass indices, triacylglycerides and glucose levels, and low amounts of high density lipoproteins and non-diabetic patients with normal levels, indicating that degradation in the DBS samples affects quantitation. Differences in the lipid identifications indicated that some oxidation occurs in the DBS samples. However, thirty-six statistically significant lipids correlated in both sample types indicating that the lipids did not degrade as much as the polar metabolites in the DBS samples and quantitation was still possible.
Keywords: Lipidomics, Metabolomics, Ion Mobility Spectrometry, Mass Spectrometry
INTRODUCTION
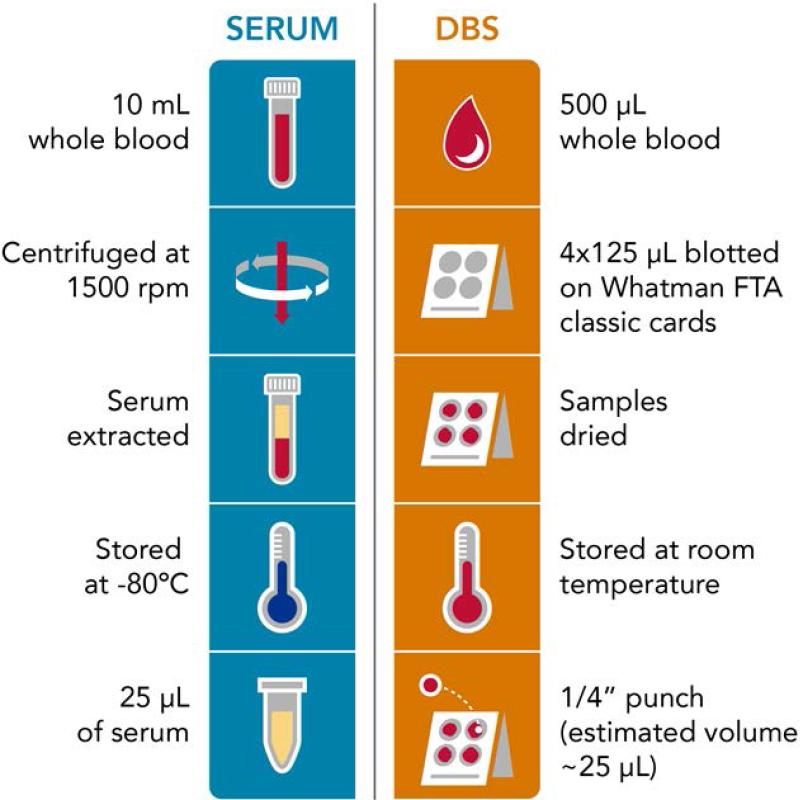
The evaluation of human disease is often aided by longitudinal studies in order to determine the occurrence of biological changes that may disrupt a system's stability. To perform longitudinal studies, biofluids are usually collected over an extended period of time to gain a more accurate understanding of the onset of disease and its progression. Blood is often used in these studies since is easy to collect and it flows through all organs, allowing the study of many molecular changes occurring in the body. Hospital and research-based studies normally use serum and plasma even though these sample types have two main challenges. First, acquiring plasma and serum requires a health professional to prepare from whole blood and second to avoid degradation these sample types must be stored at −80°C which requires valuable freezer space and energy (Figure 1). Thus to address these limitations, other blood-based sample types such as dried blood spots (DBS) are of great interest.
Figure 1.
A schematic illustrating the differences in sample collection and preparation for serum and DBS samples.
DBS offer a simpler way of storing and acquiring blood samples and have been used for disease surveillance [1-5], drug resistance [6, 7], clinical and pre-clinical pharmacokinetic studies [8, 9], population studies [10, 11] and global newborn screening of metabolic diseases since the 1960s [12]. DBS samples have many benefits compared to standard serum and plasma samples in that collection is less invasive for the patient (normally a finger or heel prick), storage and transport cost is lower, and sampling does not require a health personnel so it can be performed anywhere [13]. These benefits are of particular interest for studies in remote locations and those having large cohorts where thousands of samples are collected and must be stored. However, there is a great need to understand molecular stability in DBS samples stored over an extended period of time in order to determine how reliable their molecular results are compared to serum or plasma.
Previous studies have shown that molecular stability in DBS samples is analyte and storage condition dependent [14-17]. For example, valine was found to be stable for up to 14 years in DBS samples when stored at room temperature, while other amino acids and acylcarnitines degraded over time [18]. Lipids containing polyunsaturated fatty acids (PUFA) and oxylipins were also found to be sensitive to storage conditions and susceptible to oxidative damage. Ideal storage conditions for DBS small molecule studies were found to be in low gas-permeable zip-closure bags with desiccant, humidity indicator cards, and lower temperatures (4°C or −80°C) for enhanced stability [14, 16]. Further, lipidomics studies focusing on PUFA or oxylipins often use DBS filter paper pretreated with butylated hydroxytoluene and a chelating agent, and then stored at −80°C (ideal) indefinitely to prevent degradation or room temperature for up to 6 months [19-22]. However, due to interest in longitudinal studies, sometimes samples collected 10-20 years ago are not stored under optimal conditions or these conditions are not available in remote areas. That being said, Koulman et al. [23] investigated the rate of lipid oxidation in infant DBS and whole blood samples. They noted that only 5% of the original lipids underwent oxidation in the DBS samples when tested after 6 days storage at 40°C to expedite degradation, and no oxidized lipids were observed in the serum or plasma samples. Here, we evaluated whether DBS samples collected in 2000-2001 would be useful time points in a large cohort longitudinal study following metabolic disease progression by comparing the identified and statistically significant metabolites and lipids in serum and DBS samples for a case-control study of older diabetic males with high body mass indices (BMIs), triacylglycerides (TGs) and glucose levels, and low amounts of high density lipoproteins (HDL) compared to older non-diabetes males with normal levels. The available DBS samples were stored at room temperature after collection so there were concerns as to whether the metabolites and lipids degraded over time compared to the serum samples stored at −80°C.
MATERIALS AND METHODS
Patient samples
Matched DBS and serum samples were simultaneously collected in 2000-2001 from 10 overnight fasted male participants having an average age of 75.5 years +/− 6.15. The institutional review boards at the participating institutions approved the protocol and the study participants provided informed consent. Case participants included 5 diabetic men with BMI, TGs and glucose levels, and low amounts of HDL. The 5 control participants had normal metabolic profiles (normal BMIs, TGs, HDL and glucose levels) as shown in Table 1. Participants having drug and medical conditions that resulted in lipid abnormalities/obesity/insulin resistance were eliminated from the study. Also those being treated for metabolic disturbances with lipid lowering agents and other drugs were removed to focus on the endogenous metabolic syndrome rather than drug/medically induced metabolic abnormalities.
Table 1.
Case & Control Patient Criteria
| Cases | Controls |
|---|---|
| BMI ≥ 30 kg/m2 | 18.5 ≤ BMI < 25 |
| HDL < 40 mg/dL | HDL ≥ 40 mg/dL |
| TG ≥ 150 mg/dL | TG < 150 mg/dL |
| Diabetes (d1fgluc≥126 mg/dL) | Normoglycemic (d1fgluc<100 mg/dL) |
The preparation scheme for each sample type is shown in Figure 1. For the serum samples, 10 mL of whole blood was drawn and centrifuged so the serum could be extracted. For the DBS samples, 500 μL of blood was collected and 125 μL of that was blotted in four areas on a Whatman FTA Classic Card. The DBS were stored at room temperature in the dark until analysis, while serum was stored at −80°C. A ¼” diameter punch was taken from a single DBS for analysis and 25 μL of serum was utilized.
Polar metabolite and total lipid extraction
Polar metabolites and lipid extracts were derived from the same sample type (DBS or serum) using a modified Folch extraction in June 2015 and analyzed with MS shortly after [24, 25]. The DBS punch from each patient was transferred into to a 2.0 mL tube where 50 μL of water and then 1200 μL of −20°C 2:1 chloroform/methanol were added. Each sample was vortexed for 30 sec then transferred into a shaker at 22°C for 60 min at 600 rpm. The samples were vortexed again for 30 sec and then 250 μL of water was added to induce a phase separation. The sample was gently inverted several times, placed at room temperature for 5 min and then centrifuged at 10,000 × g for 5 min at 4°C and put on ice to maintain the clear phase separation. Finally, 400 μL of the top polar layer was removed, dried in a speedvac, and stored at −80°C for analysis of polar metabolites, while 700 μL of the bottom nonpolar layer was removed, dried in a speedvac, and stored at −20°C in 250 μL of 2:1 chloroform/methanol for lipid analyses. Serum lipids were extracted using a similar procedure except 25 μL of serum was used and then 600 μL of −20°C 2:1 chloroform/methanol was added. After vortexing and shaking, 125 μL of water was added to induce a phase separation and 200 μL of the top polar layer and 350 μL of the bottom nonpolar lipid layer were removed and stored as outlined above. Prior to analysis, the total lipid extracts were dried down and then reconstituted in 70 μL and 100 μL of MeOH for the DBS and serum samples. To generate pooled case and control samples for LC-MS/MS and LC-IMS-MS analyses, 5 μL aliquots from each reconstituted DBS and serum sample were removed and combined.
GC-MS Instrumental Analyses
Polar metabolites were chemically derivatized and analyzed by GC-MS as reported previously [26]. Briefly, samples were derivatized by adding 20 μL of methoxyamine solution (30 mg/ml in pyridine) and agitating at 37°C for 90 min to protect the carbonyl groups and reduce carbohydrate isoforms. Then, 80 μL of N-methyl-N-(trimethylsilyl)trifluoroacetamide (MSTFA) with 1% trimethylchlorosilane (TMCS) was added to each sample to trimethylsilyate the hydroxyl and amine groups for at least 30 min. The samples were allowed to cool to room temperature prior to analysis by GC-MS. Data collected by GC-MS were processed using the MetaboliteDetector software, version 2.5 beta [27]. For these steps, Agilent .D files were converted to netCDF using Agilent Chemstation followed by conversion to binary files using Metabolite Detector. Retention indices of detected metabolites were calculated based on analysis of the FAMEs mixture, followed by chromatographic alignment across all analyses after deconvolution. Metabolites were initially identified by matching experimental spectra to a PNNL augmented version of the FiehnLib [28] containing spectra and validated retention indices for over 850 metabolites and additionally cross-checked by matching with the NIST14 GC-MS library. All metabolite identifications were manually validated to minimize deconvolution and identification errors during the automated data processing.
LC-MS/MS and LC-IMS-MS Instrumental Analyses
All extracted lipids in this manuscript were analyzed by LC-MS/MS using a Waters NanoAquity UPLC system interfaced with a Velos Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA) and an Agilent 6560 Ion Mobility QTOF MS system (Agilent, Santa Clara, CA) [29, 30]. For the LC-MS/MS analyses, 7 μL of the reconstituted total lipid extracts (TLEs) were injected onto a Waters column (HSS T3 1.0 mm × 150 mm × 1.8 μm particle size). Pooled case and control samples were injected 1 time each and individual samples were analyzed in triplicate. Lipids were separated using a 92 min gradient at a flow rate of 30 μL/min (Table S1). Mobile phase A was a 40:60 mixture of ACN/H2O containing 10 mM ammonium acetate and mobile phase B was a 10:90 mixture of ACN/IPA containing 10 mM ammonium acetate. Samples in the Velos were analyzed in both positive and negative ionization using HCD (higher-energy collision dissociation) and CID (collision-induced dissociation) to obtain high lipidome coverage. Normalized collision energies of 30 and 35 were used for CID and HCD fragmentation, respectively. Both CID and HCD were set to a maximum charge state of 2, an isolation width of 2 m/z units, and an activation Q value of 0.18 for CID. LC-MS/MS raw data files from the pooled samples were imported into an in-house developed software LIQUID (Lipid Informed Quantitation and Identification) for manual identification of lipid molecular species (available at http://github.com/PNNL-Comp-Mass-Spec/LIQUID). Confident lipid identifications were determined by examining the tandem mass spectra for diagnostic ion fragments along with associated chain fragment information. In addition, the isotopic profile, extracted ion chromatogram, and mass error of the measured precursor ions were examined for lipid species. To align the individual patient samples and gap-fill the mass spectrometry data, the identified lipid name, observed m/z, and the retention time from each analysis was used as a target database for feature identifications across all LC-MS/MS runs. To perform identification, all datasets were grouped by sample type and ionization mode, and unidentified features were matched to their identified counterparts using MZmine2 [31]. Aligned features were manually verified and peak apex intensity values were exported for statistical analysis.
The LC-IMS-MS analyses were performed in both positive and negative ion mode and collected from 100-3200 m/z at a MS resolution of 40,000. The LC-IMS-MS data were analyzed using in-house PNNL software for deisotoping and feature finding the multidimensional LC, IMS and MS data [32]. Features were then compared to the AMT tag database generated from LC-MS/MS for identification and quantitation [33].
Statistical analysis
The triplicate runs for the serum and DBS samples from the 10 patients resulted in 60 LC-MS and 60 GC-MS datasets. Polar metabolites and lipids were statistically analyzed separately for each instrument type, allowing a thorough comparison of each group. The RMD-PAV algorithm [34] was used to identify any outlier biological samples and then further examined by Pearson correlation. One outlier was identified in the lipid serum negative ionization run and removed from further analyses. Median centering was used for normalization of the datasets prior to qualitative and quantitative statistical tests [35]. Both polar metabolites and lipids were analyzed using an ANOVA (quantitative comparison) with a Dunnett test correction and a Bonferroni-corrected g-test (qualitative comparison) to compare the patients to the associated controls, where adjusted p-values of less than 0.05 were considered statistically significant. Principal component analysis scores were also computed using sequential projection pursuit without imputing missing values [36].
RESULTS AND DISCUSSION
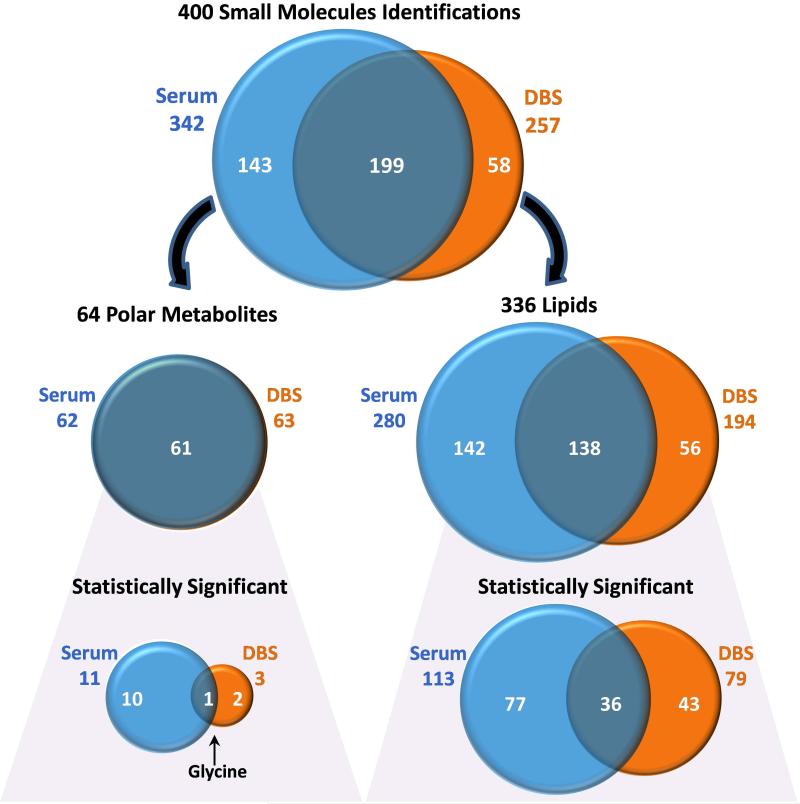
Prior to performing a large patient cohort longitudinal study with DBS samples stored at room temperature, there was great interest in understanding the degradation that occurs in the DBS samples versus matched serum. To compare the DBS and serum samples, we analyzed the overlapping metabolites and lipids in a case-control samples collected in 2000-2001 since they constitute a time point of interest for a longitudinal study. The case patients were older males with high BMI, TG and glucose levels and low HDL, while the controls were older males with normal levels (Table 1). Each patient had both serum and DBS samples taken simultaneously and triplicate GC-MS, LC-MS and LC-IMS-MS analyses were conducted on each metabolite and lipid extract. A total of 400 unique small molecules were identified in the DBS and serum samples including 336 lipids and 64 polar metabolites (Figure 2 and Tables S2-S5). The identified and statistically significant lipids and metabolites (those increasing or decreasing in the patient groups with p-values <0.05) were compared to determine degradation and assess if differences between the patient groups were similar in the two sample types. These comparisons are detailed below.
Figure 2.
Venn diagrams illustrating the number of serum and DBS small molecule identifications and the overlap between the sample types. The small molecules were then separated into polar metabolite and lipid identifications and those statistically significant among the patient groups.
Identified and Statistically Significant Polar Metabolites
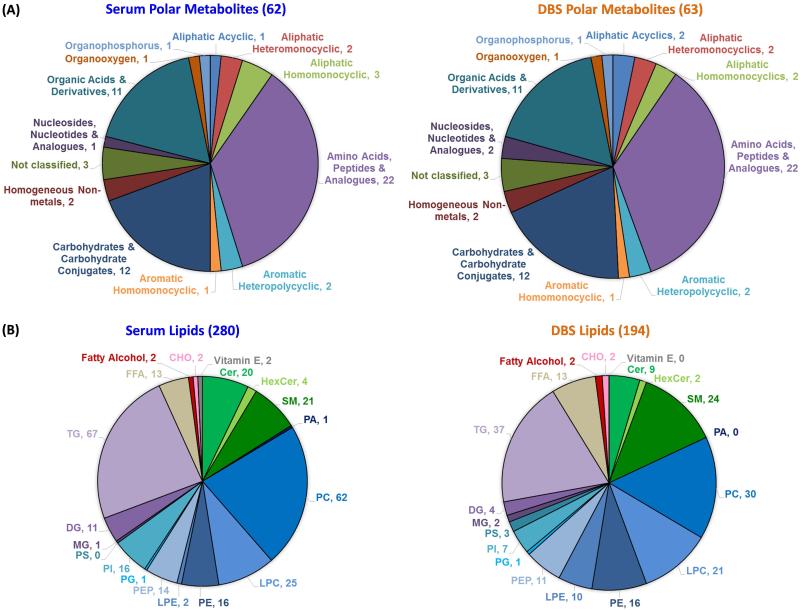
Previous studies have shown that polar metabolites can degrade in DBS studies [14-16], so their analysis in the serum and DBS sample types was of great interest to compare their stability. In the serum and DBS samples analyzed by GC-MS for the case-control patients, a total of 64 polar metabolites (Tables S2-S3) were identified across thirteen super classes (Figure 3) with most correlating to amino acids (22 identifications) and carbohydrates (12 identifications comprising monosaccharides, sugar acids and sugar alcohols). Of the 64 identified polar metabolites, 61 were in common between the serum and DBS samples, indicating that if degradation occurs in the DBS samples, the polar metabolites were not completely lost.
Figure 3.
The small molecule distribution of (A) polar metabolites and (B) lipid subclass identifications for the serum and DBS samples. Parentheses show the number of identifications per pie chart. Abbreviations are: CHO=cholesterol; Cer=ceramide; HexCer=hexosylceramide; SM= sphingomyelin; PA=diacylglycerophosphate; PC=diacylglycerophosphocholine; LPC= monoacylglycerophosphocholine; PE=diacylglycerophosphoethanolamine; LPE= monoacylglycerophosphoethanolamine; PEP=PE plasmalogen; PG=diacylglycerophosphoglycerol; PI=diacylglycerophosphoinositol; PS=diacylglycerophosphoserine; MG=onoacylglycerol; DG=diacylglycerol, TG = triacylglycerol; FFA=free fatty acids.
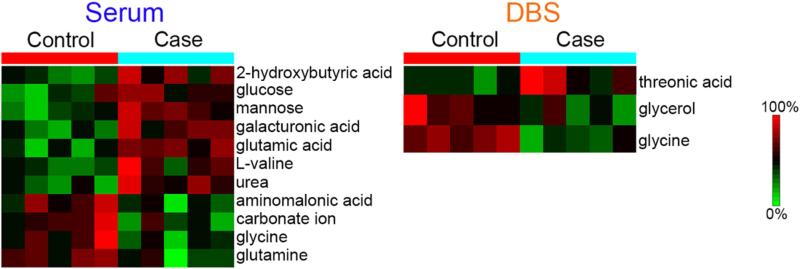
Upon comparison of the polar metabolites in the case-control patient samples, eleven were found to be statistically significant in serum and three in the DBS samples (Figure 2). Heatmaps illustrating the statistically significant polar metabolites in the two patient groups are shown in Figure 4. In serum, the statistically significant polar metabolites were made up of five amino acids (glutamic acid, L-valine, aminomalonic acid, glycine, and glutamine), three carbohydrates (glucose, mannose, and galacturonic acid), two organic acids (2-hydroxybutyric acid and carbonate ion), and one aliphatic acyclic compound (urea), and they all increased in the case patients except glycine, glutamine, aminomalonic acid, and carbonate ion. Previous studies have implicated 2-hydroxybutyric acid as an early biomarker for both insulin resistance and impaired glucose regulation [37] as well as type 2 diabetes [38]. Glucose and branched chain amino acids such as valine have also been observed to increase in prediabetes and diabetes patients, while glycine and glutamine decreased [39]. These correlations therefore showed that our serum analyses matched well with other case-control studies for similar patient types [37-39].
Figure 4.
Heatmaps illustrating the polar metabolites found to have statistically significant changes in the patient groups.
For the DBS samples, two carbohydrates (threonic acid and glycerol) and one amino acid (glycine) were identified as significant with threonic acid increasing in the case patients while the others decreased (Figure 4). Glycine was the only polar metabolite found to be statistically significant in both DBS and serum samples, and in both instances it decreased in the case patients. A previous study by Lustgarten et al. [40] showed glycine from serum was a biomarker in overweight and functionally limited older people (average BMI 27 and age 77.8 yrs) for both insulin sensitivity and regional fat deposits, and serves as a homeostasis model assessment for insulin resistance. In addition to glycine, Lustgarten et al. [40] identified 4 additional polar metabolites as biomarkers in the study, two of which (glutamine and 2-hydroxybutyric acid) were also identified as statistically significant in our serum study. The trends in the case participants (a decrease of glycine and glutamine and increase in 2-hydroxybutyric acid) also match those reported [40]. Since these polar metabolites were not observed as statistically significant in the DBS samples stored at room temperature, they appear less stable under these conditions; hence the DBS samples would not be highly informative if used as early time points for the longitudinal studies. Nonetheless, glycine being statistically significant in both sample types may be related to metabolic diseases of the case study presented here, so not all metabolite changes were lost in the DBS samples.
Identified and Statistically Significant Lipids
Due to the molecular differences between lipids and polar metabolites, lipids were analyzed separately to understand the number of identified and statistically significant lipids in the DBS and serum samples. A total of 336 lipids were identified using both LC-MS and GC-MS across 6 lipid categories (fatty acyls, glycerophospholipids, glycerolipids, sphingolipids, prenols, and sterols) and comprising nineteen lipid subclasses (Figures 2 and 3 and Tables S4-S5) [41, 42]. Of the 336 lipids, 280 were identified in the serum and 194 were identified in the DBS with 140 in common. Glycerophospholipids were found to be the most commonly identified lipids in both groups covering ~50% of the identifications, while the greatest number of identified species belonged to TGs, diacylglycerophosphocholines (PCs), sphingomyelin (SMs), and monoacylglycerophosphocholines (LPCs) (Figure 3). Further analysis showed the most abundant lipid species in the control serum and DBS samples were free fatty acids (FFAs), HexCer (galactose or glucose ceramide), and LPCs. Differences between DBS and serum samples were observed in that MG and PS subclasses were not identified in serum, and PA and vitamin E subclasses were not observed in the DBS samples (Figure 3). Some of these differences however made sense as PS lipids are present in the lipid membrane of erythrocytes [43] and platelets [44], neither of which are typically found in serum.
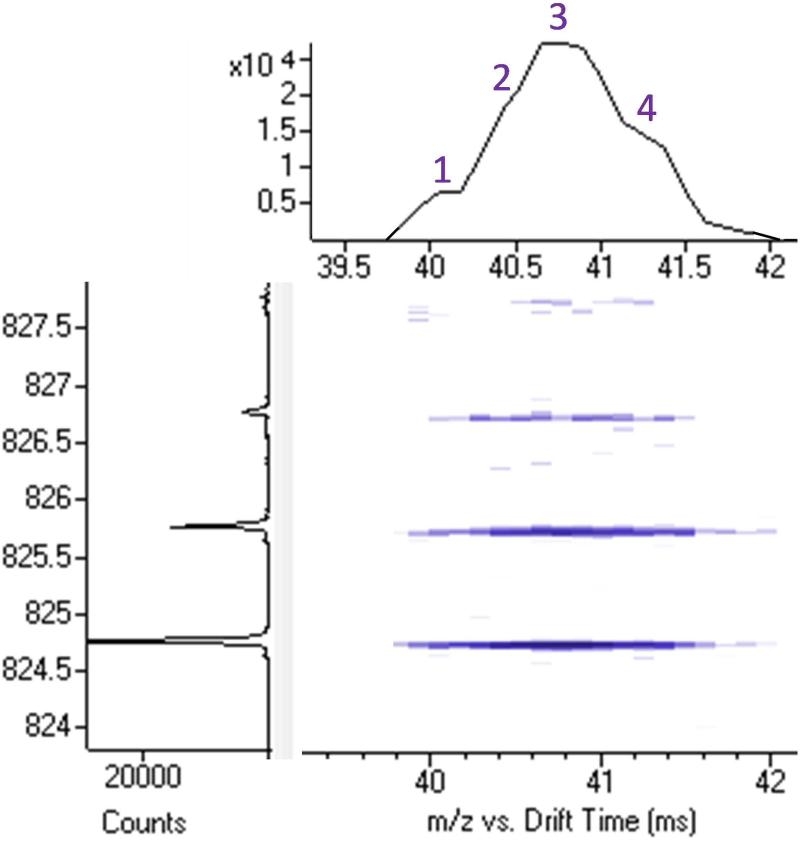
Lipid isomers were also identified in the LC-MS and LC-IMS-MS studies and each was classified and quantitated separately (Tables S4-S5). In this study, isomers were defined as those separated in LC retention time but containing the same number of carbon and double bonds. Nineteen isomer groups were identified with ten LPCs, one PC, three LPEs, three Cers (ceramide), and two SMs (sphingomyelin); however only the one PC and six of the LPCs overlapped in both sample types. Each lipid isomer was quantitated separately and several interesting trends were observed. First, when examining the two SM isomer groups identified in both the serum and DBS, SM(d18:1/24:1) and SM(d18:1/26:1), it was quickly noted that the isomers identified in the serum were different than the ones observed in the DBS sample, indicating a change in sn1/sn2 arrangement or double bond position. Another interesting observation was that in the LC-IMS-MS data, many of the TGs had multiple IMS peaks but coeluted in the LC dimension (Figure 5), and were not considered separately using our criteria as additional separation techniques were needed for their full identification. Thus, there is great interest in combining IMS with ozonolysis to understand what each TG species is and where the double bonds are located [45].
Figure 5.
The1-s nested IMS-MS spectra for (TG(48:0) + NH4)+ extracted at LC elution time = 71.8 min. Only 1 LC peak was observed for TG(48:0), but 4 IMS peaks were present indicating the co-eluting isomers.
To assess the degradation of lipids in the DBS samples, we searched for twenty-one common short-chain oxidation products of PCs in the MS/MS data. A total of nine oxidized PCs (oxPCs) were identified in the DBS samples whereas only one was identified in serum. Reis et al. [46] performed oxidation reactions with PC(16:0/18:2(9Z,12Z)) and noted that the dominant oxidized by-products were oxPC with dicarboxylic acid and aldehyde structures (i.e., PC(16:0/9:0(COOH)) and PC(16:0/9:0(CHO)). Examining the raw peak apex intensities for the PCs identified in the DBS samples, PC(16:0/18:1), PC(16:0/18:2), and PC(18:0/18:2) were the most abundant PCs with PC(16:0/9:0(COOH)) and PC(18:0/9:0(COOH)) being the most abundant oxPCs. If the oxPCs were generated from the same precursors then over the course of DBS storage, approximately 3-4% of the precursor formed the associated oxidation products. These results align nicely with the findings of Koulman et al. [23] who reported a 5% degradation upon accelerated oxidation.
Evaluation of the statistically significant lipids (those different in the case-control study by p-value <0.05) illustrated 118 in serum, 81 in DBS, and 36 in common for both sample types (Figure 2 and S1). For the most part, the statistically significant lipids decreased in the case samples except for the glycerolipids (MG [only 1 lipid detected in DBS], DGs, and most TGs) and diacylglycerophosphoethanolamines (PEs, with one exception in serum), which were higher. Also the direction of change agreed for all the overlapping statistically significant lipids except (PE(18:0/20:4) and TG(16:0/16:0/18:0)) as shown in Figure 6. The changes for (PE(18:0/20:4) were greatly significant in serum (p-value = 0.009) while it approached the cutoff in the DBS samples (p-value = 0.0488) possibly indicating its differences. The variance for TG(16:0/16:0/18:0) was less clear but could be due to TG isomers that co-elute as shown in Figure 5 by the LC-IMS-MS data.
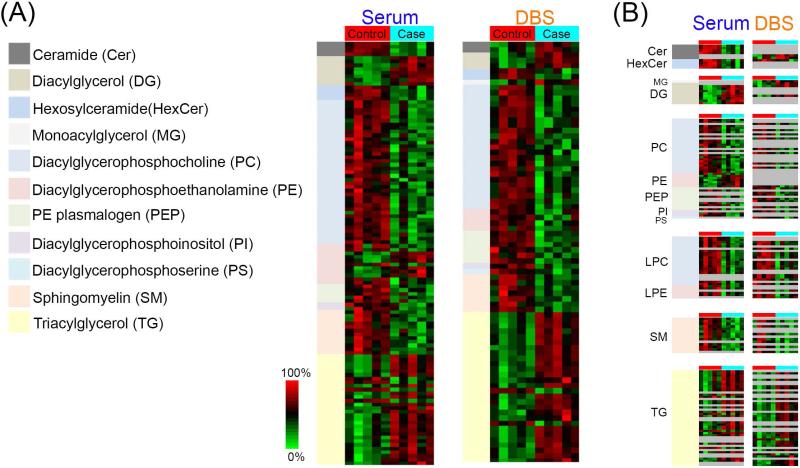
Figure 6.
(A) Heatmaps illustrating the lipid subclasses found to have statistically significant changes in the patient groups from the LC-MS/MS and LC-IMS-MS analyses. (B) A direct comparison of each statistically significant lipid subclass and individual lipid species in the serum and DBS samples.
It is not surprising that glycerolipids were upregulated in the case patients since they have well-established relationships with obesity, insulin resistance, and diabetes [39, 47-49]. Both MGs and DGs have been identified as signaling molecules with regards to insulin activity and obesity [48], and plasma TGs are not only a signature for insulin resistance but predictors of type 2 diabetes [47]. The other lipid subclass shown to be higher in the case participants was PE, but this signature was not retained in the DBS samples as only one significantly changing PE was observed versus five in serum. Four LPEs however did increase in the serum and three in the DBS samples. Increased PEs has also been positively correlated with type 2 diabetes and prediabetes [50], possibly due to more lipoprotein production in particular VLDL which have high levels of PE [51, 52] or the increase of PEs in membranes enhancing the binding of amyloid fibers of the islet amyloid polypeptide [53]. Thirteen free fatty acids were also identified in both sample types; however palmitoleic acid (16:1n-9) and hexanoic acid (6:0) were only found to be statistically significant in the DBS samples. Palmitoleic acid increased in the case patients and has been associated with the risk of type 2 diabetes and insulin resistance in men [54]; however, lifestyle can influence its levels [55]. Hexanoic acid was found to decrease in the case patients and while its role is less clear, a recent study noted that the relative abundance of hexanoic acid in plasma from obese individuals is associated with the levels of certain fungi in the human gut [56]. Other exceptions where lipids were noted to increase include Cer(d18:1/26:0) and Cer(d18:1/26:1), which may be due to the degradation of SM lipids (the loss of the choline head group forms Cer) especially since SM(d18:1/26:1) was noted to be lower in the cases versus controls. Other SM lipids, however, trended the same direction as the ceramides with the same chains lengths (e.g., (d18:1/23:0) and (d18:1/24:0)). All of these findings show that the statistically significant lipids found in both the DBS and serum samples correlate with current literature on the case-control study. Further, they support that lipids could be analyzed in older DBS samples and provide important information in longitudinal studies.
SUMMARY
The analyses of patient-matched serum and DBS samples collected in 2000-2001 from older male diabetic patients with high BMIs, TGs and glucose levels, and low HDL levels and non-diabetic male patients with normal levels, illustrated that the number of identified polar metabolites in each sample type overlapped extremely well. However, when the statistically significant polar metabolites were evaluated in the case-control study, only glycine was conserved in both. The lipidomic analyses showed much better agreement between the sample types with thirty-six statistically significant lipids in common for the sample types, indicating lipids are more robust in the older DBS samples. Thus, this study indicates that serum samples are needed for longitudinal study of polar metabolites if DBS samples are stored at room temperature. Conversely, because most lipid changes are preserved, they could be used in patient analyses of older DBS samples. Results from this study indicate that further analyses are needed to address questions relating to polar metabolites and lipids in fresh DBS and serum samples, or DBS samples stored at cooler temperatures for long time periods. The robustness of the lipids in the older samples nonetheless indicate they will probably correlate well in fresh samples and those stored at cooler temperatures, but the stability of polar metabolites cannot be predicted from this study.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Nathan Johnson for assistance in preparing the figures. Portions of this research were supported by grants from the National Institute of Environmental Health Sciences of the NIH (R01 ES022190), National Institute of General Medical Sciences (P41 GM103493), the Laboratory Directed Research and Development Program at Pacific Northwest National Laboratory. This research utilized capabilities developed by the Pan-omics program (funded by the U.S. Department of Energy Office of Biological and Environmental Research Genome Sciences Program) and by the National Institute of Allergy and Infectious Diseases under grant U19 AI106772. The Osteoporotic Fractures in Men (MrOS) Study in the US was supported by the National Institute on Aging (NIA), the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. This work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory (EMSL), a DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the DOE under contract DE-AC05-76RL0 1830.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
REFERENCES
- 1.Solomon SS, Solomon S, Rodriguez, McGarvey ST, Ganesh AK, Thyagarajan SP, et al. Dried blood spots (DBS): a valuable tool for HIV surveillance in developing/tropical countries. Int J STD AIDS. 2002;13(1):25–8. doi: 10.1258/0956462021924578. [DOI] [PubMed] [Google Scholar]
- 2.Ross RS, Stambouli O, Gruner N, Marcus U, Cai W, Zhang W, et al. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots--performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J. 2013;10:72. doi: 10.1186/1743-422X-10-72. PMCID: PMC3599381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smit PW, Elliott I, Peeling RW, Mabey D, Newton PN. An overview of the clinical use of filter paper in the diagnosis of tropical diseases. Am J Trop Med Hyg. 2014;90(2):195–210. doi: 10.4269/ajtmh.13-0463. PMCID: PMC3919219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snijdewind IJ, van Kampen JJ, Fraaij PL, van der Ende ME, Osterhaus AD, Gruters RA. Current and future applications of dried blood spots in viral disease management. Antiviral Res. 2012;93(3):309–21. doi: 10.1016/j.antiviral.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 5.El Mubarak HS, Yuksel S, Mustafa OM, Ibrahim SA, Osterhaus AD, de Swart RL. Surveillance of measles in the Sudan using filter paper blood samples. J Med Virol. 2004;73(4):624–30. doi: 10.1002/jmv.20136. [DOI] [PubMed] [Google Scholar]
- 6.Kimaro J, Shao E, Nyombi B, Kifaro E, Maruapula D, Gaseitsiwe S, et al. Using dried blood spots collected under field condition to determine HIV-1 diversity and drug resistance mutations in resource limited Tanzania. J Int AIDS Soc. 2014;17(4 Suppl 3):19686. doi: 10.7448/IAS.17.4.19686. PMCID: PMC4225344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garrido C, Zahonero N, Fernandes D, Serrano D, Silva AR, Ferraria N, et al. Subtype variability, virological response and drug resistance assessed on dried blood spots collected from HIV patients on antiretroviral therapy in Angola. J Antimicrob Chemother. 2008;61(3):694–8. doi: 10.1093/jac/dkm515. [DOI] [PubMed] [Google Scholar]
- 8.Kothare PA, Bateman KP, Dockendorf M, Stone J, Xu Y, Woolf E, et al. An Integrated Strategy for Implementation of Dried Blood Spots in Clinical Development Programs. AAPS J. 2016;18(2):519–27. doi: 10.1208/s12248-015-9860-3. PMCID: PMC4779096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Henion J, Abbott R, Wang P. Dried blood spots as a sampling technique for the quanti tative determination of guanfacine in clinical studies. Bioanalysis. 2011;3(22):2501–14. doi: 10.4155/bio.11.262. [DOI] [PubMed] [Google Scholar]
- 10.McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44(4):899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- 11.Crimmins E, Kim JK, McCreath H, Faul J, Weir D, Seeman T. Validation of blood-based assays using dried blood spots for use in large population studies. Biodemography Soc Biol. 2014;60(1):38–48. doi: 10.1080/19485565.2014.901885. PMCID: PMC4117354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guthrie R, Susi A. A Simple Phenylalanine Method for Detecting Phenylketonuria in Large Populations of Newborn Infants. Pediatrics. 1963;32:338–43. [PubMed] [Google Scholar]
- 13.Holen T, Norheim F, Gundersen TE, Mitry P, Linseisen J, Iversen PO, et al. Biomarkers for nutrient intake with focus on alternative sampling techniques. Genes & Nutrition. 2016;11(1):1–20. doi: 10.1186/s12263-016-0527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner M, Tonoli D, Varesio E, Hopfgartner G. The use of mass spectrometry to analyze dried blood spots. Mass Spectrom Rev. 2016;35(3):361–438. doi: 10.1002/mas.21441. [DOI] [PubMed] [Google Scholar]
- 15.Therrell BL, Hannon WH, Pass KA, Lorey F, Brokopp C, Eckman J, et al. Guidelines for the retention, storage, and use of residual dried blood spot samples after newborn screening analysis: statement of the Council of Regional Networks for Genetic Services. Biochem Mol Med. 1996;57(2):116–24. doi: 10.1006/bmme.1996.0017. [DOI] [PubMed] [Google Scholar]
- 16.Li W, Tse FLS. Dried blood spot sampling in combination with LC-MS/MS for quantitative analysis of small molecules. Biomed Chromatogr. 2010:24. doi: 10.1002/bmc.1367. [DOI] [PubMed] [Google Scholar]
- 17.Prentice P, Turner C, Wong MC, Dalton RN. Stability of metabolites in dried blood spots stored at different temperatures over a 2-year period. Bioanalysis. 2013;5(12):1507–14. doi: 10.4155/bio.13.121. [DOI] [PubMed] [Google Scholar]
- 18.Strnadova KA, Holub M, Muhl A, Heinze G, Ratschmann R, Mascher H, et al. Long-term stability of amino acids and acylcarnitines in dried blood spots. Clin Chem. 2007;53(4):717–22. doi: 10.1373/clinchem.2006.076679. [DOI] [PubMed] [Google Scholar]
- 19.Bastani NE, Gundersen TE, Blomhoff R. Dried blood spot (DBS) sample collection for determination of the oxidative stress biomarker 8-epi-PGF(2alpha) in humans using liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2012:26. doi: 10.1002/rcm.6149. [DOI] [PubMed] [Google Scholar]
- 20.Metherel AH, Aristizabal Henao JJ, Stark KD. EPA and DHA levels in whole blood decrease more rapidly when stored at -20 °C as compared with room temperature, 4 and −75 °C. Lipids. 2013:48. doi: 10.1007/s11745-013-3827-x. [DOI] [PubMed] [Google Scholar]
- 21.Liu G, Muhlhausler BS, Gibson RA. A method for long term stabilisation of long chain polyunsaturated fatty acids in dried blood spots and its clinical application. Prostaglandins Leukot Essent Fatty Acids. 2014;91(6):251–60. doi: 10.1016/j.plefa.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Pupillo D, Simonato M, Cogo PE, Lapillonne A, Carnielli VP. Short-Term Stability of Whole Blood Polyunsaturated Fatty Acid Content on Filter Paper During Storage at −28 degrees C. Lipids. 2016;51(2):193–8. doi: 10.1007/s11745-015-4111-z. [DOI] [PubMed] [Google Scholar]
- 23.Koulman A, Prentice P, Wong MC, Matthews L, Bond NJ, Eiden M, et al. The development and validation of a fast and robust dried blood spot based lipid profiling method to study infant metabolism. Metabolomics. 2014;10(5):1018–25. doi: 10.1007/s11306-014-0628-z. PMCID: PMC4145199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim Y-M, Kyle JE, et al. MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems. 2016;1(3) doi: 10.1128/mSystems.00043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folch J, Lees M, Stanley GHS. A Simple Method for the Isolation and Purification of Total Lipides from Animal Tissues. Journal of Biological Chemistry. 1957;226(1):497–509. [PubMed] [Google Scholar]
- 26.Kim YM, Nowack S, Olsen MT, Becraft ED, Wood JM, Thiel V, et al. Diel metabolomics analysis of a hot spring chlorophototrophic microbial mat leads to new hypotheses of community member metabolisms. Front Microbiol. 2015;6:209. doi: 10.3389/fmicb.2015.00209. PMCID: PMC4400912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiller K, Hangebrauk J, Jager C, Spura J, Schreiber K, Schomburg D. MetaboliteDetector: comprehensive analysis tool for targeted and nontargeted GC/MS based metabolome analysis. Anal Chem. 2009;81(9):3429–39. doi: 10.1021/ac802689c. [DOI] [PubMed] [Google Scholar]
- 28.Kind T, Wohlgemuth G, Lee DY, Lu Y, Palazoglu M, Shahbaz S, et al. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal Chem. 2009;81(24):10038–48. doi: 10.1021/ac9019522. PMCID: PMC2805091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.May JC, Goodwin CR, Lareau NM, Leaptrot KL, Morris CB, Kurulugama RT, et al. Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem. 2014;86(4):2107–16. doi: 10.1021/ac4038448. PMCID: PMC3931330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibrahim YM, Baker ES, Danielson WF, 3rd, Norheim RV, Prior DC, Anderson GA, et al. Development of a New Ion Mobility (Quadrupole) Time-of-Flight Mass Spectrometer. Int J Mass Spectrom. 2015;377:655–62. doi: 10.1016/j.ijms.2014.07.034. PMCID: PMC4501404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. PMCID: PMC2918584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowell KL, Slysz GW, Baker ES, LaMarche BL, Monroe ME, Ibrahim YM, et al. LC-IMS-MS Feature Finder: detecting multidimensional liquid chromatography, ion mobility and mass spectrometry features in complex datasets. Bioinformatics. 2013;29(21):2804–5. doi: 10.1093/bioinformatics/btt465. PMCID: 3799467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25(3):450–82. doi: 10.1002/mas.20071. PMCID: PMC1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matzke MM, Waters KM, Metz TO, Jacobs JM, Sims AC, Baric RS, et al. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics. 2011;27(20):2866–72. doi: 10.1093/bioinformatics/btr479. PMCID: PMC3187650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Webb-Robertson BJ, McCue LA, Waters KM, Matzke MM, Jacobs JM, Metz TO, et al. Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J Proteome Res. 2010;9(11):5748–56. doi: 10.1021/pr1005247. PMCID: PMC2974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb-Robertson BJ, Matzke MM, Metz TO, McDermott JE, Walker H, Rodland KD, et al. Sequential projection pursuit principal component analysis--dealing with missing data associated with new -omics technologies. Biotechniques. 2013;54(3):165–8. doi: 10.2144/000113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gall WE, Beebe K, Lawton KA, Adam KP, Mitchell MW, Nakhle PJ, et al. alpha-hydroxybutyrate is an early biomarker of insulin resistance and glucose intolerance in a nondiabetic population. PLoS One. 2010;5(5):e10883. doi: 10.1371/journal.pone.0010883. PMCID: PMC2878333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Xu Z, Lu X, Yang X, Yin P, Kong H, et al. Comprehensive two-dimensional gas chromatography/time-of-flight mass spectrometry for metabonomics: Biomarker discovery for diabetes mellitus. Anal Chim Acta. 2009;633(2):257–62. doi: 10.1016/j.aca.2008.11.058. [DOI] [PubMed] [Google Scholar]
- 39.Guasch-Ferre M, Hruby A, Toledo E, Clish CB, Martinez-Gonzalez MA, Salas-Salvado J, et al. Metabolomics in Prediabetes and Diabetes: A Systematic Review and Meta-analysis. Diabetes Care. 2016;39(5):833–46. doi: 10.2337/dc15-2251. PMCID: PMC4839172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lustgarten MS, Price LL, Phillips EM, Fielding RA. Serum glycine is associated with regional body fat and insulin resistance in functionally-limited older adults. PLoS One. 2013;8(12):e84034. doi: 10.1371/journal.pone.0084034. PMCID: Pmc3877144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Jr., Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46(5):839–61. doi: 10.1194/jlr.E400004-JLR200. [DOI] [PubMed] [Google Scholar]
- 42.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, et al. Update of the LIPID MAPS comprehensive classification system for lipids. Journal of Lipid Research. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodge JT, Phillips GB. Composition of phospholipids and of phospholipid fatty acids and aldehydes in human red cells. J Lipid Res. 1967;8(6):667–75. [PubMed] [Google Scholar]
- 44.Clark SR, Thomas CP, Hammond VJ, Aldrovandi M, Wilkinson GW, Hart KW, et al. Characterization of platelet aminophospholipid externalization reveals fatty acids as molecular determinants that regulate coagulation. Proc Natl Acad Sci U S A. 2013;110(15):5875–80. doi: 10.1073/pnas.1222419110. PMCID: Pmc3625294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas MC, Mitchell TW, Harman DG, Deeley JM, Murphy RC, Blanksby SJ. Elucidation of double bond position in unsaturated lipids by ozone electrospray ionization mass spectrometry. Analytical Chemistry. 2007;79(13):5013–22. doi: 10.1021/ac0702185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reis A, Domingues P, Ferrer-Correia AJ, Domingues MR. Fragmentation study of short-chain products derived from oxidation of diacylphosphatidylcholines by electrospray tandem mass spectrometry: identification of novel short-chain products. Rapid Commun Mass Spectrom. 2004;18(23):2849–58. doi: 10.1002/rcm.1686. [DOI] [PubMed] [Google Scholar]
- 47.Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, et al. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. Journal of Clinical Investigation. 2011;121(4):1402–11. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen HS, Rosenkilde MM, Holst JJ, Schwartz TW. GPR119 as a fat sensor. Trends Pharmacol Sci. 2012;33(7):374–81. doi: 10.1016/j.tips.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Erion DM, Shulman GI. Diacylglycerol-mediated insulin resistance. Nature Medicine. 2010;16(4):400–2. doi: 10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meikle PJ, Wong G, Barlow CK, Weir JM, Greeve MA, MacIntosh GL, et al. Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS One. 2013;8(9):e74341. doi: 10.1371/journal.pone.0074341. PMCID: Pmc3785490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adiels M, Olofsson SO, Taskinen MR, Boren J. Overproduction of very low-density lipoproteins is the hallmark of the dyslipidemia in the metabolic syndrome. Arterioscler Thromb Vasc Biol. 2008;28(7):1225–36. doi: 10.1161/ATVBAHA.107.160192. [DOI] [PubMed] [Google Scholar]
- 52.Dashti M, Kulik W, Hoek F, Veerman EC, Peppelenbosch MP, Rezaee F. A phospholipidomic analysis of all defined human plasma lipoproteins. Sci Rep. 2011;1:139. doi: 10.1038/srep00139. PMCID: Pmc3216620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sciacca MF, Kotler SA, Brender JR, Chen J, Lee DK, Ramamoorthy A. Two-step mechanism of membrane disruption by Abeta through membrane fragmentation and pore formation. Biophys J. 2012;103(4):702–10. doi: 10.1016/j.bpj.2012.06.045. PMCID: PMC3443794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mozaffarian D, Cao H, King IB, Lemaitre RN, Song X, Siscovick DS, et al. Trans-palmitoleic acid, metabolic risk factors, and new-onset diabetes in U.S. adults: a cohort study. Ann Intern Med. 2010;153(12):790–9. doi: 10.1059/0003-4819-153-12-201012210-00005. PMCID: PMC3056495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mozaffarian D, Jacobson MF, Greenstein JS. Food reformulations to reduce trans fatty acids. N Engl J Med. 2010;362(21):2037–9. doi: 10.1056/NEJMc1001841. [DOI] [PubMed] [Google Scholar]
- 56.Rodriguez MM, Perez D, Chaves FJ, Esteve E, Marin-Garcia P, Xifra G, et al. Obesity changes the human gut mycobiome. Sci Rep-Uk. 2015:5. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.