Abstract
The rapid expansion of carbapenem-resistant Acinetobacter baumannii (CRAB) clinical isolates is a big issue. We investigated the antibiotic susceptibility, molecular epidemiology and resistance gene of A. baumannii collected at two hospitals in Shanghai, China. Besides, the A. baumannii PCR-based replicon typing method (AB-PBRT) was conducted to categorize the plasmids into homogeneous groups on the basis of replicase genes. Most CRAB isolates showed high-level resistance to almost all antibiotics but retain susceptibility to colistin and tigecycline. A total of 101 isolates carried blaOXA-51-like gene. Sequencing identified the presence of blaOXA-66 for CRAB isolates. blaOXA–23 gene were discovered in all CRAB isolates. Each CRAB isolate contained 1–3 of 19 different plasmid replicase (rep) gene homology groups (GRs) and the GR6 (repAci6) was ubiquitous. Genotyping by Multilocus Sequence Typing (MLST) showed seven defined MLST patterns and three novel STs were found. eBURST analysis indicated they were all grouped in CC92 (GCII) with the most frequent ST208 (50%). Two blaOXA–23-bearing transposons were found: Tn2006 and Tn2008. Tn2008 were detected in 54 (96.4%) isolates and Tn2006 in two remaining isolates. The blaOXA–23 carbapenem gene was vitally associated with repAci6 plasmid belong to CC92 clonal group. Our survey revealed severe drug resistance in A. baumannii isolates. Tn2008-containing CC92 A. baumannii were endemic, which may facilitate the blaoxa23 dissemination.
Keywords: carbapenem-resistant Acinetobacter baumannii (CRAB), blaOXA–23, transposon, plasmid replicase genes, multilocus sequence type (MLST)
Introduction
Acinetobacter baumannii is a significant opportunistic pathogen responsible for numerous nosocomial infections, including respiratory infections (in particular ventilator-associated pneumonia, VAP), urinary tract infections, bacteremia, soft and skin tissue infections, burn wound infections and secondary meningitis (Roca et al., 2012; Kempf et al., 2013). Carbapenem resistance in A. baumannii is an emerging problem worldwide during the last decade (Jiang et al., 2014; Guerrero-Lozano et al., 2015). One data from Taiwan involving five major hospitals showed that resistance to imipenem in intensive care units increased from 22.0% in 2000 to 66.8% in 2010 (Hsueh et al., 2001). Studies performed by Reddy also reported a disturbing tendency of sharp increase in the rates of CRAB isolates, from 1% in 2003 to 58% in 2008 (Reddy et al., 2010). Data from Wallace also found the percentage of CRAB isolates was 31% before the year 2009 followed by 99% after the year 2009 at the University of Maryland Medical Center (UMMC; Wallace et al., 2016).
Carbapenem-resistant in A. baumannii is due to combined mechanisms including production of OXA- and metallo-β-lactamases, outer membrane impermeability, increased expression of efflux pumps, and penicillin-binding proteins modification (Zarrilli et al., 2013). OXA-type enzymes related to carbapenem resistance include the natural blaOXA-51-like and three acquired genes: blaOXA–23-like, blaOXA-24-like and blaOXA-58-like (Jiang et al., 2014). These genes were integrated in the bacterial chromosome or carried by plasmids (Poirel et al., 2010). Nowadays, AB-PBRT method provides a new tool to investigate the epidemiology of plasmids in A. baumannii (Towner et al., 2011). Numerous publications reported the spread of resistance gene via transposons (Poirel et al., 2010). Four transposons harboring blaOXA–23 gene have been reported: Tn2006, Tn2007, Tn2008, and Tn2009 (Zhou et al., 2011; Espinal et al., 2013; Guerrero-Lozano et al., 2015). These types of transposons share a common region “blaOXA–23-ΔATPase.” Tn2007 owns ISAba4 promoter upstream blaOXA–23 gene. The remaining three transposons own ISAba1. The two ISAba1 copies were inversely orientated in Tn2006 compared to the same direction in Tn2009 (Guerrero-Lozano et al., 2015). Tn2006 and Tn2008 are reported globally disseminated, while Tn2009 has only been discovered in China (Zhou et al., 2011). These ISAba1-associated transposons contributed to the dissemination of blaOXA–23. Moreover, Tn2006, Tn2008, and Tn2009 all have been found in conjugative plasmids except Tn2007. It is noted that Tn2007 is immovable and is not actually considered as a transposon (Nigro and Hall, 2016).
Colistin is currently used as last-resort antibiotics against CRAB infection (Durante-Mangoni et al., 2014). It is important to analyze the local epidemiology of clinical CRAB isolates and control the dissemination. The aim of the present study was to investigate the drug-resistance and molecular characteristics of A. baumannii isolates in 101 A. baumannii clinical isolates from two hospitals in Shanghai. We also characterized the genetic environment surrounding blaOXA–23 gene. In addition, the distribution and epidemiology of plasmid replicase (rep) genes in CRAB isolates were also investigated.
Materials and Methods
Bacterial Isolates
A total of 101 non-repetitive A. baumannii clinical isolates were collected in this study from two hospitals in Shanghai, China. All subjects were anonymised in this study. Eighty-seven isolates were recovered from neurosurgery in Renji Hospital Shanghai Jiaotong University School of Medicine from July 2011 to June 2013. And the remaining 14 isolates were collected from Obstetrics and Gynecology Hospital of Fudan University from January 2015 to August 2016. All isolates were identified using the VITEK 2 Compact automatic bacteria and drug susceptibility analysis system.
Antimicrobial Susceptibility Testing
Antibiotic susceptibility testing was conducted in the present study. Fourteen antimicrobial agents were tested including piperacillin/tazobactam, cefoxitin, ceftazidime, cefotaxime, imipenem, meropenem, gentasin, amikacin, minocycline, ciprofloxacin, trimethoprim/sulfamethoxazole, cefepime, polymyxin, and tigecycline. The Minimum inhibitory concentrations (MICs) were determined by agar dilution method except polymyxin, which was performed by broth dilution method. Pseudomonas aeruginosa ATCC 27853 and Escherichia coli ATCC 25922 were used as reference strains. The susceptibility results were interpreted based on the Clinical and Laboratory standards Institute (CLSI) breakpoints (CLSI, 2011). For tigecycline, we used breakpoints recommended by the British Society for Antimicrobial Chemotherapy guidelines (BSAC, 2011). CRAB isolates were defined with both imipenem and meropenem resistance (MICs > 8 mg/L).
PCR and Sequencing of Drug Resistance Genes
PCR assay was performed to screen the presence of drug resistance genes in 101 isolates, including blaOXA–23-like, blaOXA-24-like, blaOXA-51-like, blaOXA-58-like, blaIMP-1, blaV IM-1, blaV IM-2, and blaAmpC. The entire products were sequenced and analyzed with the BLAST website1.
Multilocus Sequence Typing
Molecular typing of CRAB isolates was determined by MLST. Briefly, it was detected by sequence analysis of gltA, gyrB, gdhB, recA, cpn60, gpi, and rpoD housekeeping genes as previously described (Ying et al., 2015). Primers are available at http://pubmlst.org/abaumannii/. The results were compared with the STs databases online at http://pubmlst.org/databases/. eBURST analysis was conducted to investigate the genetic relationships and clonal complexes (CCs) of these isolates.
Genetic Environment of the blaOXA–23 Gene
PCR was performed to detect the occurrence of blaOXA–23-carrying transposons in CRAB isolates, including Tn2006, Tn2007, Tn2008, and Tn2009. Primers used in this study were shown in Table 1. Structures and primer locations were shown in Figure 1. In addition, blaOXA–23-ΔATPase was the common region of Tn2006 and Tn2008.
Table 1.
Primers used for blaOXA–23 gene detection.
| Primer name | Primer sequence (5′ –3′) | Replicase gene |
|---|---|---|
| Tn2006 Int-P3 | GTCTATCAGGAACTTGCGCG | ISAba1-blaOXA–23-ISAba1 |
| Tn2006 Int-P4 | GCAAGGCTTTAGATGCAGAAGA | |
| Tn2007 Int-P6 | ATTTGAACCCATCTATTGGC | ISAba4- blaOXA–23 |
| Tn2007 Int-P7 | ACTCTCATATTTTTTCTTGG | |
| Tn2006/8 Int-P3 | GTCTATCAGGAACTTGCGCG | blaOXA–23-ΔATPase |
| Tn2006/8 Int-P5 | GGCTCATTACAGTCAGGTACAAGT | |
| Tn2009 Int-P1 | ATCCTGATGCTCGCAATCGT | ISAba1-blaOXA–23-ISAba1 |
| Tn2009 Int-P8 | CTGTCTGCGAACACATTCAC |
FIGURE 1.
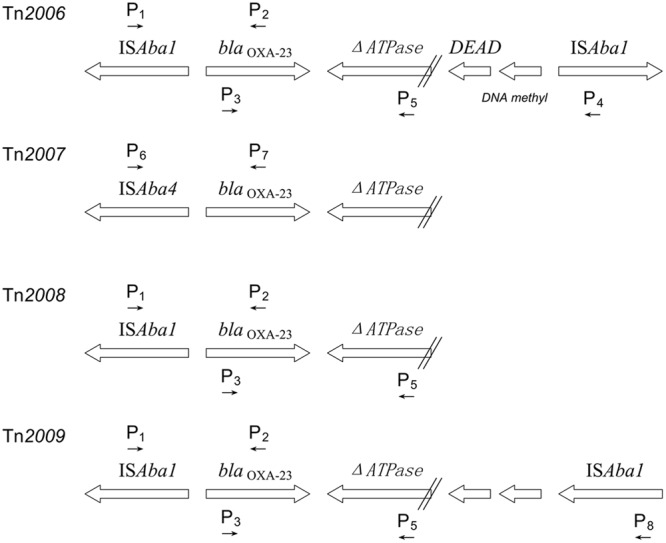
Structures and primer locations of Tn2006, Tn2007, Tn2008, and Tn2009.
AB-PBRT
Nineteen PCR amplifications were organized into six multiplexes and used to detect 27 different plasmid rep genes as described previously (Bertini et al., 2010). Each PCRs recognized three or four different homology groups (GRs). PCR products were recognized by DNA sequencing. An additional PCR was performed to identify the gene encoding the type IV secretion system protein TraC, which was found on the plasmid pACICU2 carrying the repAci6 gene (Towner et al., 2011).
Results
Antimicrobial Susceptibility Testing
The MICs of 14 antimicrobial agents were determined for all A. baumannii isolates. Sixty-five (64.4%) and 70 (69.3%) strains were resistant to imipenem and meropenem (Table 2). Fifty-six CRAB isolates were recovered and the MICs of imipenem and meropenem were both ranged from 16 to 256 mg/L. Resistance to piperacillin/tazobactam, cefoxitin, ceftazidime, cefotaxime, cefepime, gentamicin, amikacin, minocycline, ciprofloxacin, and trimethoprim/sulfamethoxazole were 71.3% (72/101), 87.1% (88/101), 75.2% (76/101), 85.1% (86/101), 69.3% (70/101), 74.3% (75/101), 62.4% (63/101), 27.7% (28/101), 74.3% (75/101), and 88.1% (89/101), respectively. However, no isolate was resistant to colistin or tigecycline. The MICs of antimicrobial agents for isolates recovered from Renji Hospital were higher than those from Fudan Obstetrics and Gynecology hospital. In addition, 14 isolates from Fudan Obstetrics and Gynecology hospital were all sensitive to imipenem and meropenem (Table 2).
Table 2.
Susceptibility analyses of 101 A. baumannii in this study.
| Antimicrobial agents | RJa (n = 87) | OGb (n = 14) | N = 101 |
|---|---|---|---|
| R (n, %c) | R (n, %) | R (n, %) | |
| Piperacillin/Tazobactam | 72, 82.8% | 0, 0 | 72, 71.3% |
| Cefoxitin | 80, 91.9% | 8, 57.1% | 83, 87.1% |
| Ceftazidime | 73, 83.9% | 3, 21.4% | 76, 75.2% |
| Cefotaxime | 83, 95.4% | 3, 21.4% | 86, 85.1% |
| Cefepime | 70, 80.5% | 0, 0 | 70, 69.3% |
| Imipenem | 65, 74.7% | 0, 0 | 65, 64.4% |
| Meropenem | 70, 80.5% | 0, 0 | 70, 69.3% |
| Gentamicin | 75, 86.2% | 0, 0 | 75, 74.3% |
| Amikacin | 63, 72.4% | 0, 0 | 63, 62.4% |
| Minocycline | 28, 32.2% | 0, 0 | 28, 27.7% |
| Ciprofloxacin | 75, 86.2% | 0, 0 | 75, 74.3% |
| Trimethoprim-sulfamethoxazole | 87, 100% | 2, 14.3% | 89, 88.1% |
| Tigecycline | 0, 0 | 0, 0 | 0, 0 |
| Colistin | 0, 0 | 0, 0 | 0, 0 |
aRJ, Renji Hospital Shanghai Jiaotong University School of Medicine; bOG, Obstetrics and Gynecology Hospital of Fudan University; c%R, Resistant percent.
Detection of Drug Resistance Genes
PCR screening was performed to detect drug resistant genes in all isolates. The natural oxacillinase, blaOXA-51-like, were detected in all isolates. And blaOXA-51-like gene of CRAB strains revealed the presence of blaOXA-66 by sequencing. The prevalence of blaOXA–23 and blaAmpC gene was 60.4% (61/101) and 51.5% (52/101). Besides, all 56 CRAB isolates contained blaOXA–23 genes and 54 CRAB isolates contained blaAmpC. The genes encoding blaOXA-24/58-like, blaIMP-1 and blaV IM-1/2 were not detected in any of the 101 isolates.
Multilocus Sequence Typing
Fifty-six CRAB isolates were typed by MLST analysis. A total of seven defined STs and three novel STs were identified. ST208, which accounting for the largest proportion (28/56, 50.0%), was the major clonal type, followed by ST191 (12/56, 21.4%), ST540 (7/56, 12.5%), ST381 (2/56, 3.57%), ST643 (2/56, 3.57%), ST195 (1/56, 1.79%), and ST368 (1/56, 1.79%). Additionally, they were single locus variants of gpi gene. eBURST analysis (Figure 2) showed that the seven defined STs along with novel STs were all clustered into the same CCs (CC92), which was also commonly referred to as global/international clone II (GCII; Diancourt et al., 2010; Zarrilli et al., 2013). The three novel STs were submitted and have assigned as ST1472, ST1473, and ST1474, respectively.
FIGURE 2.
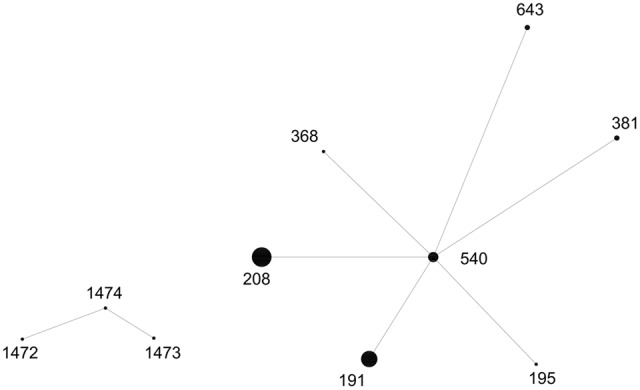
eBURST analysis results of tested CRAB isolates. One clonal complexes (CC92) were identified. The size of the dot indicates the number of CRAB isolates.
Genetic Structure of blaOXA–23 Gene
The gene encoding blaOXA–23 of 56 CRAB isolates was located exclusively within the Tn2006 or Tn2008 transposons. We found Tn2008 was almost ubiquitous (54 of 56 isolates). However, the specific region (ISAba1-blaOXA–23-ISAba1) for Tn2006 was only detected in 2 (3.6%) isolates. No ISAba4- blaOXA–23 was found, indicating there was no Tn2007 in these CRAB strains, followed by the Tn2009.
Distribution of Plasmid rep Genes
GR6 (repAci6) was the most prevalent and detected in 52 CRAB isolates along with TraC gene, which was identified on plasmid pACICU2 (Table 3). GR14 (p4AYE) was found in 29 CRAB isolates, followed by GR8 (Aci9, found in 11 isolates), GR2 (Aci1, 5 isolates), GR4 (Aci4, 3 isolates), and GR12 (p2S1, 1 isolate). The plasmid rep gene profiles and CHDL content along with the general typing of 56 CRAB isolates were summarized in Table 3.
Table 3.
Results of plasmid rep geng groups of 56 CRAB isolates.
| GR group (repAci) | Isolate(s) | Resistance genes content |
Transposons | MLST | ||
|---|---|---|---|---|---|---|
| OXA–23 | OXA-66 | AmpC | ||||
| 6 | 60, 80 | + | + | + | Tn2008 | ST208 |
| 25 | + | + | + | Tn2008 | ST191 | |
| 36 | + | + | + | Tn2006 | ST381 | |
| 56 | + | + | - | Tn2008 | ST208 | |
| 2 | 53, 69 | + | + | + | Tn2008 | ST208 |
| 23, 26 | + | + | + | Tn2008 | ST191 | |
| 6, 8 | C3, C26, 19, 21, 22, 24, 30, 33, 50, 55, 57, 61, 63, 82, 91, | + | + | + | Tn2008 | ST208 |
| C22, | + | + | + | Tn2008 | ST1472 | |
| C23, | + | + | + | Tn2008 | ST1473 | |
| 39 | + | + | + | Tn2008 | ST1474 | |
| 6, 14 | 18, 28, 32, 37, 40, 41, 59, 76, 88 | + | + | + | Tn2008 | ST191 |
| C27, 64, 65, 66, 67, 83 | + | + | + | Tn2008 | ST540 | |
| 45, 49, 51 | + | + | + | Tn2008 | ST208 | |
| 20, 35 | + | + | + | Tn2008 | ST643 | |
| C7 | + | + | + | Tn2008 | ST368 | |
| 52 | + | + | - | Tn2008 | ST208 | |
| 62 | + | + | + | Tn2006 | ST540 | |
| 34 | + | + | + | Tn2008 | ST381 | |
| 2, 6, 14 | 48 | + | + | + | Tn2008 | ST195 |
| 6, 12, 14 | C20 | + | + | + | Tn2008 | ST208 |
| 4, 6, 8 | 71, 84, 85 | + | + | + | Tn2008 | ST208 |
+, positive; -, negative.
Discussion
The increasing drug resistance of A. baumannii has raised a big challenge, especially CRAB strains reported worldwide (Durante-Mangoni et al., 2014). As observed in other literature, here we reported a high prevalence of carbapenem resistance (55.4%) in clinical A. baumannii isolates recovered from two hospitals. Most CRAB strains always exhibited high resistance to other antimicrobial agents tested.
OXA-type carbapenems resistance hydrolytic enzymes are common in A. baumannii isolates. blaOXA-51-like gene, which shares less than 60% amino acid identity with other OXA-types, was identified in all A. baumannii isolates, supporting that blaOXA-51-like are natural genes in A. baumannii (Table 3). blaOXA-66 variant might be the most prevalent and found in all 56 CRAB isolates. In general, CRAB strains contained more resistance genes. This probably explains why CRAB strains show much higher MICs than non-CRAB isolates. blaOXA-24-like β-lactamases was first identified in A. baumannii in 1997, which resulted in an outbreak in Spain (Bou et al., 2000). AndblaOXA-58-like gene was first found in France in 2003 (Poirel et al., 2005). Isolates carrying blaOXA-24/58-like genes were typically resistant to carbapenems. Fortunately, they were barely identified in China. Our results also indicated no blaOXA-24/58-like nor blaIMP-1 genes were found. However, due to their plasmid location, the distribution of these genes in A. baumannii should be early monitored.
Multilocus Sequence Typing is frequently used for strain phylogeny and global epidemiology. Fifty-six CRAB isolates with occurrence of blaOXA–23 in our study were typed. The most common ST was ST208 and CC92 was the unique CC clonal group tested. CC92 (GCII) clone outbreaks due to blaOXA–23-producing A. baumannii strains have been reported frequently (Fu et al., 2010; He et al., 2011). The discovered of three novel STs indicated that A. baumannii isolates were diverse and still clonal expansion. Our e-BURST analysis revealed clonal spread at Renji hospital during a certain period. Hence the carbapenems resistance in A. baumannii is increasing. While isolates of the same ST clone reported owning different resistance patterns, the vital spreading of CRAB isolates may have other approaches.
Acinetobacter baumannii PCR-based replicon typing assay revealed 52 CRAB isolates owning repAci6, and the gene encoding TraC was strongly linked with repAci6 plasmid. No rep genes associated with blaOXA-24/58 were found. RepAci6 was identified on plasmid pACICU2 in general and TraC protein make pACICU2 transferable (Iacono et al., 2008). In some cases (e.g., isolates Ab18 and Ab64), different MLST genotypes were found to have identical plasmid rep gene profiles. In contrast (e.g., isolates Ab18 and Ab23), isolates with the same MLST type were found to have different plasmid rep gene profiles (Table 3). Hence the plasmid rep genes were variable in the process of strain epidemics. blaOXA–23-like gene was associated with carriage of repAci6 plasmids as previously described (Towner et al., 2011). With few exceptions, strains grouped in ST208 were associated with pACICU2, pMAC02, and p4AYE. Strains grouped in ST191 and ST540 were associated with carriage of pACICU2 and p4AYE. These removable plasmids carrying CHDLs genes led to spreading among A. baumannii strains.
As we mentioned, the spread of carbapenems resistance gene in A. baumannii are of great importance. One of the most popular gene blaOXA–23 was discovered here. Of four transposons reported, Tn2006 and Tn2008 were identified in CRAB isolates, both of which were flanked by ISAba1. However, Tn2007 and Tn2009 are not seen in agreement with previous researches. It was noted that ISAba1 facilitated the mobilization of blaOXA–23 gene hence allowed Tn2006 and Tn2008 to move. These four transposons were observed in plasmids as well as in the chromosome of A. baumannii (Nigro and Hall, 2016). Besides, Tn2006, Tn2007, and Tn2008 were found in different locations of conjugative repAci6 group plasmids (Nigro and Hall, 2015). It seemed that the blaOXA–23 dissemination might due to transposition of mentioned transposons.
Our survey suggested that both clonal spread of a GC2 strain carrying carbapenems resistance genes and the spread of conjugative plasmid among A. baumannii strains are responsible for the serious increasing carbapenems resistance. Further understanding of related plasmids may help determine their acquisition, dissemination and regulation among A. baumannii.
Author Contributions
CY conceived the work. YC performed all experiments, analyzed the results, and wrote the manuscript. JG and HZ assisted in antimicrobial sensitivity testing. All authors read and approved the final paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by National Nature Science Foundation of China (Grant 81371874).
References
- Bertini A., Poire L., Mugnier P. D., Villa L., Nordmann P., Carattoli A. (2010). Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob. Agents Chemother. 54 4168–4177. 10.1128/AAC.00542-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou G., Oliver A., Martinez-Beltran J. (2000). OXA-24, a novel class Dβ-lactamase with carbapenemase activity in an Acinetobacter baumannii clinical strain. Antimicrob. Agents Chemother. 44 1556–1561. 10.1128/AAC.44.6.1556-1561.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- BSAC (2011). BSAC Methods for Antimicrobial Susceptibility Testing. Birmingham: British Society for Antimicrobial Chemotherapy. [Google Scholar]
- CLSI (2011). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-First Informational Supplement. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Diancourt L., Passet V., Nemec A., Dijkshoorn L., Brisse S. (2010). The population structure of Acinetobacter baumannii: expanding multiresistant clones from an ancestral susceptible genetic pool. PLoS ONE 5:E10034 10.1371/journal.pone.0010034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durante-Mangoni E., Utili R., Zarrilli R. (2014). Combination therapy in severe Acinetobacter baumannii infections: an update on the evidence to date. Future Microbiol. 9 773–789. 10.2217/fmb.14.34 [DOI] [PubMed] [Google Scholar]
- Espinal P., Macià M. D., Roca I., Gato E., Ruíz E., Fernández-Cuenca F., et al. (2013). First report of an OXA–23 carbapenemase-producing Acinetobacterbaumannii clinical isolate related to Tn2006 in Spain. Antimicrob. Agents Chemother. 57 589–591. 10.1128/AAC.01157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Zhou J., Zhou H., Yang Q., Wei Z., Yu Y., et al. (2010). Wide dissemination of OXA–23-producing carbapenem-resistant Acinetobacter baumannii clonal complex 22 in multiple cities of China. J. Antimicrob. Chemother. 65 644–650. 10.1093/jac/dkq027 [DOI] [PubMed] [Google Scholar]
- Guerrero-Lozano I., Fernández-Cuenca F., Galán-Sánchez F., Egea P., Rodríguez-Iglesias M., Pascual Á. (2015). Description of the OXA–23 β-lactamase gene located within Tn2007 in a clinical isolate of Acinetobacter baumannii from Spain. Microb. Drug Resist. 21 215–217. 10.1089/mdr.2014.0155 [DOI] [PubMed] [Google Scholar]
- He C., Xie Y., Fan H., Kang M., Tao C., Zhang R., et al. (2011). Spread of imipenem-resistant Acinetobacter baumannii of European clone II in Western China. Int. J. Antimicrob. Agents 38 257–260. 10.1016/j.ijantimicag.2011.04.015 [DOI] [PubMed] [Google Scholar]
- Hsueh P. R., Liu Y. C., Yang D., Yan J. J., Wu T. L., Huang W. K., et al. (2001). Multicenter surveillance of antimicrobial resistance of major bacterial pathogens in intensive care units in 2000 in Taiwan. Microb. Drug Resist. 7 373–382. 10.1089/10766290152773383 [DOI] [PubMed] [Google Scholar]
- Iacono M., Villa L., Fortini D., Bordoni R., Imperi F., Bonnal R. J., et al. (2008). Whole-genome pyrosequencing of an epidemic multidrug-resistant Acinetobacter baumannii strain belonging to the European clone II group. Antimicrob. Agents Chemother. 52 2616–2625. 10.1128/AAC.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Zhang Z., Zhao S. (2014). Epidemiological characteristics and drug resistance analysis of multidrug-resistant Acinetobacter baumannii in a China hospital at a certain time. Pol. J. Microbiol. 63 275–281. [PubMed] [Google Scholar]
- Kempf M., Rolain J. M., Azza S., Diene S., Joly-Guillou M. L., Dubourg G., et al. (2013). Investigation of Acinetobacter baumannii resistance to carbapenems in Marseille hospitals, south of France: a transition from an epidemic to an endemic situation. APMIS 121 64–71. 10.1111/j.1600-0463.2012.02935.x [DOI] [PubMed] [Google Scholar]
- Nigro S. J., Hall R. M. (2015). Distribution of the blaOXA–23-containing transposons Tn2006 and Tn2008 in Australian carbapenem-resistant Acinetobacter baumannii isolates. J. Antimicrob. Chemother. 70 2409–2411. 10.1093/jac/dkv102 [DOI] [PubMed] [Google Scholar]
- Nigro S. J., Hall R. M. (2016). Structure and context of Acinetobacter transposons carrying the oxa23 carbapenemase gene. J. Antimicrob. Chemother. 71 1135–1147. 10.1093/jac/dkv440 [DOI] [PubMed] [Google Scholar]
- Poirel L., Marque S., Heritier C., Segonds C., Chabanon G., Nordmann P. (2005). OXA-58 a novel class D β-lactamase involved in resistance to carbapenems in Acinetobacter baumannii. Antimicrob. Agents Chemother. 49 202–208. 10.1128/AAC.49.1.202-208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Naas T., Nordmann P. (2010). Diversity, epidemiology, and genetics of class D β-lactamases. Antimicrob. Agents Chemother. 54 24–38. 10.1128/AAC.01512-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy T., Chopra T., Marchaim D., Pogue J. M., Alangaden G., Salimnia H., et al. (2010). Trends in antimicrobial resistance of Acinetobacter baumannii isolates from a metropolitan Detroit health system. Antimicrob. Agents Chemother. 54 2235–2238. 10.1128/AAC.01665-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca I., Espinal P., Vila-Farrés X., Vila J. (2012). The Acinetobacter baumannii oxymoron: commensal hospital dweller turned pan-drug-resistant menace. Front. Microbiol. 3:148 10.3389/fmicb.2012.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner K. J., Evans B., Villa L., Levi K., Hamouda A., Amyes S. G., et al. (2011). Distribution of intrinsic plasmid replicase genes and their association with carbapenem-hydrolyzing cass Dβ-Lactamase genes in European clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 55 2154–2159. 10.1128/AAC.01661-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace L., Daugherty S. C., Nagaraj S., Johnson J. K., Harris A. D., Rasko D. A. (2016). Use of comparative genomics to characterize the diversity of Acinetobacter baumannii surveillance isolates in a health care institution. Antimicrob. Agents Chemother. 60 5933–5941. 10.1128/AAC.00477-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying C., Li Y., Wang Y., Zheng B., Yang C. (2015). Investigation of the molecular epidemiology of Acinetobacter baumannii isolated from patients and environmental contamination. J. Antibiot. 68 562–567. 10.1016/j.meegid.2015.02.017 [DOI] [PubMed] [Google Scholar]
- Zarrilli R., Pournaras S., Giannouli M., Tsakris A. (2013). Global evolution of multidrug-resistant Acinetobacter baumannii clonal lineages. Int. J. Antimicrob. Agents 41 11–19. 10.1016/j.ijantimicag.2012.09.008 [DOI] [PubMed] [Google Scholar]
- Zhou H., Zhang T., Yu D., Pi B., Yang Q., Zhou J., et al. (2011). Genomic analysis of the multidrug-resistant Acinetobacter baumannii strain MDR-ZJ06 widely spread in China. Antimicrob. Agents Chemother. 55 4506–4512. 10.1128/AAC.01134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]


