Abstract
The gut microbiota provides essential signals for the development and appropriate function of the immune system. Through this critical contribution to immune fitness, the gut microbiota has a key role in health and disease. Recent advances in the technological applications to study microbial communities and their functions have contributed to a rapid increase in host–microbiota research. Although it still remains difficult to define a so-called ‘normal' or ‘healthy' microbial composition, alterations in the gut microbiota have been shown to influence the susceptibility of the host to different diseases. Current translational research combined with recent technological and computational advances have enabled in-depth study of the link between microbial composition and immune function, addressing the interplay between the gut microbiota and immune responses. As such, beneficial modulation of the gut microbiota is a promising clinical target for many prevalent diseases including inflammatory bowel disease, metabolic abnormalities such as obesity, reduced insulin sensitivity and low-grade inflammation, allergy and protective immunity against infections.
Introduction
Hippocrates was quoted as saying ‘all disease begins in the gut' and over 2000 years later we are beginning to appreciate his sentiment. Our body is colonised by a large number of microbes (bacteria, fungi, archaea, viruses and protozoa); which mostly reside within the gastrointestinal (GI) tract, are predominantly bacterial, and together these microbes collectively form the gut microbiota. Often denoted previously as commensal organisms, we now know the gut microbiota acts in a symbiotic manner that is also beneficial for its host.1 Interest in the gut microbiota, most notably the bacterial communities, has recently exploded, and we are beginning to uncover how crucial these microbes are to appropriate immune function and lifelong health, or conversely, susceptibility to inflammatory and infectious diseases.
Early in life the GI tract quickly becomes colonised by microbes, and the gut microbiota is purported to reach an adult state at around 3 years of age.2 Bacteroidetes and Firmicutes are the dominant phyla, making up more than 90% of the total microbial population in both mice and humans. Other major phyla present in the gut include the Proteobacteria, Tenericutes, Actinobacteria and Verrucomicrobia.3 The gut microbiota has an important role in homoeostasis by controlling metabolic pathways, nutrient metabolism and the production of vitamins.4 Furthermore, it has also been shown to be essential in the development and maturation of mucosal and systemic immune responses, and for the maintenance of intestinal epithelial barrier function.4 The importance of microbial signalling for immune development in the GI tract has also been demonstrated in germ-free (GF) mice, which have underdeveloped gut-associated lymphoid tissues (GALT) including Peyer's patches, isolated lymphoid follicles and mesenteric lymph nodes.5 Taken together, host–microbiota interactions are critical for host immunity and health.
Here, we review the recent advances in gut microbiota analysis, and define the computational approaches that can be utilised to expand our ever-growing understanding of the role of the gut microbiota in health and disease. The immense interest and impact of the gut microbiota in current research can mainly be attributed to the recent advances in these applications to study microbial communities and their functions. We also detail key microbial cross talk with the immune system, resulting in critical instruction of appropriate immunity. Finally, we have considered immune-mediated diseases where the relationship between the gut microbiota and disease susceptibility is currently most convincing, including inflammatory bowel disease (IBD), obesity-related inflammatory disorders, allergic and infectious diseases.1 Where applicable, the influence of key factors such as diet or antibiotic use on the composition of the gut microbiota is outlined. As well, we explore the use of GF mice, and gnotobiotic mouse models where GF mice are colonised with defined microbial communities, which provide a tool to elucidate the function of the gut microbiota in a disease setting.
Technological Applications for Assessing Gut Microbiota
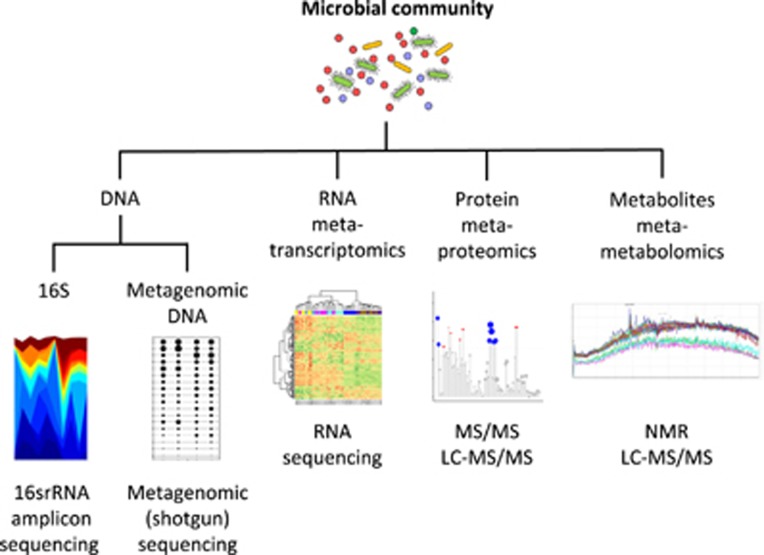
Researchers now routinely use a wide-range of culture-dependent and high-throughput culture-independent methods to assess gut microbial communities. The recent advances of the culture-independent methods analysing the DNA (metagenomics), RNA (metatranscriptomics), proteins (metaproteomics) and metabolites (metametabolomics) present within the gut will be the focus of this section (Figure 1). The development and reducing costs of next-generation sequencing, also known as high-throughput sequencing, combined with improved computational tools allowing for the analysis of large and complex data sets, have led to an enormous growth in the research field. Each tool provides unique information to interpret the different species that live within the gut; to provide understanding of how these species function; and describe why certain species may contribute to disease, or respond differently to specific stimuli such as changes in diet.
Figure 1.
Overview of current technological applications available for the assessment of gut microbial communities.
Initial efforts to characterise the gut microbiome using next-generation sequencing of gut microbiota involved the use of a ‘marker' gene that serves as a unique fingerprint for different bacterial genera. Amplicon sequencing is a PCR-based method that provides information about the different microorganisms present within a sample. As such, the bacterial communities within the gut can be assessed by sequencing the variable regions of the 16S ribosomal RNA (rRNA) encoding gene sequence found in bacteria and Archaea,6 whereas eukaryotic organisms (fungi, parasites and the host) can be assessed by sequencing the 18S rRNA encoding sequence.7 Today, this method still provides an inexpensive way to compare bacterial communities between patients, and assess alterations over time within a patient. However, this 16S sequencing cannot be routinely used to identify specific bacterial species as the 16S rRNA gene typically lacks sufficient phylogenetic information to differentiate between taxa at the species level,8 and some bacterial genera can share greater than 99.5% of their 16S rRNA sequences.9
Metagenomic (shotgun) sequencing was later utilised to provide strain level resolution of all the species present within a microbial community. Instead of employing an individual marker gene, a random subset of the DNA present within the sample is sequenced, and the bacterial, viral and eukaryotic species associated with the sequences can then be reconstructed using high-powered bioinformatic tools. This relatively expensive method is now becoming more widespread as costs continue to decrease. It has also been employed to provide information about the host GI tract through non-invasive sampling, as DNA from the cells within the GI tract can be detected within faecal samples.10 Despite being a very small proportion of the gut microbiome, even viruses present within the metagenome can be assessed using different sample preparation steps and metagenomic sequencing.11, 12 Using DNA hybridisation prior to sequencing, further adaptations of metagenomic sequencing have included enrichment or capture of DNA sequences specific to the species of interest.13 Within a clinical setting, this approach could be adapted to identify specific pathogens or disease-associated species from within an individual's gut microbiota without sequencing all of the DNA within a sample, decreasing costs and the bioinformatic processing time for clinicians.
In addition to identifying specific strains, metagenomic sequencing can also assess the functions that microorganisms can perform, as genes within the microbial genomes are linked to the ability of an organism to execute certain tasks. The functional capability of a bacterial community can also be predicted based on its 16S rRNA amplicon profile by comparing against databases containing fully sequenced and annotated bacterial genomes.14 However, metagenomic DNA sequencing remains the only way to identify functions from unknown species or unique microbial communities. Metatranscriptomic approaches, or the sequencing of RNA present within a sample, have also been applied to assess the current or ongoing functions of a metagenome within a specific period of time.15 Although metagenomic sequencing will divulge the total functional capacity of a microbial community, metatranscriptomics reports genes that are being actively transcribed at a given time. Further use of metatranscriptomic approaches linked with RNA sequencing of the host will reveal more information about how distinctive microbial functions are linked to disease, and how they alter human health.
Lastly, metaproteomic and metametabolomic approaches have also been developed. Metaproteomic studies examine the proteins that typically result from the microorganism,16 but can also result from inflammatory responses of the host.17 Moreover, shotgun proteomic approaches have identified unique proteins present within the gut that were not predicted by metatranscriptomic approaches,17 highlighting the yet unknown nature of many interactions between the gut microbiota and the host. Similarly, metametabolomic approaches also use a shotgun-like approach to examine a broad range of metabolic by-products and secretions of microorganisms that provide insight into active microbial functions and interplay between microbial species as well as the gut microbiota and the host.18 Many GI diseases linked to the gut microbiota may be associated with altered proteins or metabolite production, rather than a depletion of certain microbes. Once certain proteins and metabolites can be linked to microbiota-associated diseases, downstream applications will likely revolutionise our ability to quickly and efficiently detect alterations in the gut microbiota that are linked to disease phenotypes. This can provide previously unavailable insight into the metabolic pathways of the microbial community and how this intersects with the metabolism of the host.
This knowledge could provide a basis for the crucial next step and the remaining challenge for this research field, to establish causal relationships between the gut microbiota and the host. Colonisation of GF mice with mouse or human gut microbiota (referred to as humanised mice) has been used to demonstrate a causal role for the gut microbiota in immune development and disease susceptibility. It is likely that the emerging technological applications will move beyond the current focus of the bacterial communities present towards their function; which holds the most promise for the identification of microbial or metabolite based therapeutic strategies for prevention and treatment of inflammatory and infectious diseases.
Gut Microbiota and Local Immune Cell Cross-Talk
The GI tract must remain non-responsive to food antigens and the gut microbiota, while being capable of responding quickly to invading pathogens. One of the first defence mechanisms afforded by the gut microbiota is limiting the access of pathogenic bacteria to the gut epithelium by competitive exclusion. The gut microbiota also promotes secretory immunoglobulin (Ig)A production that binds to microbes at mucosal surfaces, neutralises toxins and contributes to microbial tolerance.19 Furthermore, bacterial molecular patterns are sensed by innate pattern recognition receptors (PRR) such as Toll-like receptors (TLR) or Nod-like receptors, which have a role in the maintenance of intestinal epithelial cell (IEC) homeostasis. The microbiota also contributes to the priming signal of the inflammasome pathway, leading to the transcription of cytokines such as tumour necrosis factor (TNF)-α and interleukin (IL)-6, as well as pro-IL-1β and pro-IL-18.20 The inflammasome, activated by a variety of pathogens including various viruses, can regulate the cleavage of these inactive precursors to active IL-1β and IL-18.21
The gut microbiota also helps to maintain immune homeostasis by stimulating different arms of the T-cell response. Segmented filamentous bacteria (SFB) are potent promoters of T helper (Th)17 cells in the intestine, which are important in protection against bacterial and fungal infections.22 SFB adhesion to IEC results in the activation of the IL-23 pathway, leading to IL-22 production by group 3 innate lymphoid cells (ILC3) and a subsequent Th17 response.23 Conversely, polysaccharide A from Bacteroides fragilis was shown to stimulate the generation of FoxP3+ regulatory T cells (Treg) via TLR2 activation,24 and microbial-derived butyrate has also been shown to induce colonic Treg.25
Fermentation of dietary fibre in the colon by anaerobic bacteria generates short-chain fatty acids (SCFA) including butyrate, acetate and propionate.26 As such, fermentable dietary fibre is often referred to as microbiota accessible carbohydrates (MAC). SCFA signal via G-protein coupled receptors (GPCR) such as GPR41, GPR43 and GPR109A on epithelial cells and immune cells.26 When SCFA are taken up by host cells they serve as an energy source for metabolism. SCFA can also inhibit histone deacetylases that in turn affects gene transcription.26 SCFA were also shown to inhibit the transcription factor nuclear factor kappa B (NF-κB) leading to reduced inflammatory cytokine production.27 Furthermore, SCFA have been demonstrated to promote mucous production by intestinal goblet cells, induce secretory IgA and activate the inflammasome resulting in IL-18 secretion.26, 28 Importantly, systemic SCFA also reach other organs such as the bone marrow and lung via the circulation.29
When MAC beneficially affect the host's health by selectively changing the composition or activity of the gut microbiota, they can be referred to as prebiotics.30 Probiotics are defined as live microorganisms, which when administered in adequate amounts confer a health benefit on the host.31 Probiotics can interfere with the growth or survival of pathogens in the gut lumen, improve gut barrier function and immunity, or have an effect on the systemic immune system and organs.31 The molecular mechanisms by which probiotic strains act are still largely unknown and require a strain-specific approach as different probiotic strains can induce differential effects. Both pre- and probiotics can be useful for the beneficial modulation of the gut microbiota, and their capability to enhance immune function has been studied for many years.30, 31
Inflammatory Bowel Disease
No longer viewed as merely passengers, the gut microbiota has a critical role in the development and progression of IBD, and is the most extensively investigated with respect to the influence of the gut microbiota on disease susceptibility, relapse and remission. Although the specific microbial changes appear to vary from study to study, some broad patterns are apparent. These include a loss of community diversity, increased representation of some Gammaproteobacteria, and decreased relative abundance of several taxa within the Firmicutes phylum.32
Loss of faecal community diversity has been consistently shown in human IBD studies.33, 34, 35 The most prominent change in diversity associated with IBD is decreased abundance of the Firmicutes phylum, which includes a loss of Faecalibacterium prausnitzii, a bacterium with significant anti-inflammatory effects.33 Other species with decreased representation in IBD include Bacteroides fragilis, B. vulgatus, Ruminococcus albus, R. callidus, and R. bromii, which were over 5-fold more abundant in healthy subjects compared with IBD patients.34 In addition to the loss of diversity and altered composition, recent metagenomic studies showed that the overall quantity of bacteria is also reduced in IBD. On average, 25% fewer microbial genes could be detected in the faecal community of IBD patients, pointing to lower functional as well as taxonomic diversity. Indeed, some evidence suggests that the abundances of metabolic pathways are more consistently altered in IBD than the actual taxonomic abundances.32
Conversely, specific taxa that are increased in IBD patients include Enterobacteriaceae, such as Escherichia/Shigella.32 In particular, adherent-invasive E. coli strains have been identified within ileal biopsies from Crohn's disease patients,36 and are more prominent in this environment compared with faecal samples.37 The increase in Enterobacteriaceae in the small intestine may reflect their capacity to use sialic acid and fucose liberated from mucous.38 Under normal circumstances, these bacteria are likely to be outcompeted for this resource by the resident microbial community, but the alterations in the gut microbiota that occur with IBD may present the opportunity for these bacteria to flourish.38 Other mucous associated bacteria suspected of having an important role in IBD are the sulphate-reducing bacteria such as Desulfovibrio piger.39 In the gut, sulphate-reducing bacteria consume hydrogen to produce H2S by removing sulphate from sulfated mucous glycans.40 Potential links between H2S and IBD include genotoxic properties and the chemical disruption of the mucous structure, as sulphides are potent reducers of disulphide bonds.41
Specific groups of bacteria may also provide protection against IBD, by ameliorating the inflammatory response. Bacteroides thetaiotaomicron has been shown to suppress NF-κB and attenuate IEC inflammation.42 Further evidence for beneficial effects of gut microbiota in immune modulation are found in mice with impaired recognition of bacteria through deletion of TLR4 or its downstream signalling molecule myeloid differentiation primary response gene 88 (MyD88), which experience worse symptoms of GI inflammation when exposed to DSS compared with wild-type mice.43 In addition, activation of the inflammasome can occur via MAC modulation of the gut microbiota as well as SCFA administration, which promoted IL-18-mediated epithelial repair following DSS-induced GI inflammation.28 Butyrate produced by the gut microbiota, most likely by members of the Clostridia class, has also been shown to induce the expansion of Treg in mice, ameliorating intestinal inflammation in an adoptive T-cell transfer model of colitis.25 Other bacteria, such as F. prausnitzii, Bifidobacterium and Lactobacillus protect the host through a variety of mechanisms, including modulation of cytokine production33, 44 and strengthening gut barrier function.45 The evidence for the efficacy of probiotic strains like Bifidobacterium and Lactobacillus in reducing the symptoms of Crohn's disease in humans remains unclear, although some beneficial effects have been shown in patients with ulcerative colitis.46 In addition, the gut microbiota may protect the host by outcompeting pathogenic bacteria that drive GI inflammation by preventing these pathogens from occupying niches.38
The incidence and prevalence of IBD is progressively rising, making the condition a global health problem with currently no permanent therapeutic treatment. Therefore, finding alternative strategies for treating IBD is an area of increasing importance. One potential therapy of interest is faecal microbiota transplantation (FMT), which has been particularly successful in treating C. difficile infections.47 However, so far, FMT for treating IBD has been shown to have variable efficacy,48 and long-term safety for FMT is largely unknown. Risks include the possible transmission of infectious agents or antibiotic resistance genes within FMT, or the possible induction of other microbiota-associated diseases following FMT to a susceptible host. Nonetheless, gut microbiota based treatments hold promise as a future therapy for IBD patients.
Obesity-Related Inflammatory Disorders
The world is facing an epidemic of obesity-related metabolic abnormalities, collectively known as metabolic syndrome. The hallmarks of metabolic syndrome include hyperglycaemia, hyperlipidaemia, insulin resistance, obesity and hepatic steatosis.49 Both lifestyle and genetic factors have a role in the development of obesity, and recently the gut microbiota has been suggested to be a key environmental factor that influences metabolic syndrome.50
The heritability of the gut microbiota has previously been examined linking the abundance of specific gut microbial taxa to host genetics.51 Among the heritable taxa were members of the Firmicutes phylum that are associated with leaness.51 Thus, host genetics influence the composition of the human gut microbiota, and this can impact host metabolism. Nonetheless, the diet has been suggested to be most influential in shaping the composition of the gut microbiota.51, 52 An increase in the ratio of Firmicutes to Bacteroidetes, as well as a marked reduction in the bacterial diversity of the gut microbiota, have been associated with obesity in both genetic- and diet-induced mouse models3, 53, 54 and clinical obesity.55, 56 Moreover, diets that limit weight gain led to a reduction in the relative abundance of Firmicutes and an increase in Bacteroidetes in both mice and humans.53, 55 However, other investigations have shown no association between obesity and the Bacteroidetes to Firmicutes ratio, or the diversity of the human gut microbiota, suggesting the relationship between obesity and the gut microbiota is highly complex and difficult to interpret.57
There is now a large body of evidence that supports the close link of obesity and insulin resistance with chronic, low-grade inflammation, and the gut microbiota has been implicated in the development of this inflammatory response.58 Both diet and genetic factors can alter the gut microbiota and impair intestinal integrity, leading to metabolic endotoxemia, obesity, glucose intolerance and insulin resistance via activation of TLR4 and subsequent inflammatory cytokine production.58, 59 Moreover, restoration of the gut microbiota by the MAC oligofructose resulted in amelioration of the inflammation as well as the associated metabolic abnormalities.60 Other TLR responses have been linked to obesity-induced inflammation and insulin resistance as well. For example, TLR5 sensing of bacterial flagellin is required for gut microbial homeostasis. Consequently, TLR5 deficient mice have altered gut microbiota and are prone to the development of metabolic syndrome.49
The gut microbiota provides enzymes that increase caloric harvest from MAC and the resulting monosaccharides are absorbed or metabolised to SCFA. These products get delivered to the liver, where lipogenesis results in hepatic triglyceride (TG) production. In addition, the gut microbiota acts to enhance lipoprotein lipase-directed deposition of these TG into adipocytes, promoting the storage of calories harvested from the diet into fat. Fat storage is reduced in GF compared with conventional mice, and colonisation of these GF mice with gut microbiota results in a 60% increase in body fat within two weeks.50 This body fat increase was accompanied by decreased insulin sensitivity and increased liver TG.50 GF mice also have increased host metabolism of fatty acids that protects against diet-induced obesity provoked by a prototypic high-fat, high-sugar Western-style diet.61 Furthermore, depletion of the gut microbiota using an antibiotic cocktail was sufficient to reduce adiposity and improve glucose tolerance and insulin sensitivity by promoting browning of adipose tissue in both diet and genetically induced obese mice.62
Colonisation of GF mice with the gut microbiota from mice with either genetic or diet-induced obesity increased host fat deposition compared with colonisation with microbiota from lean mice.53, 54 Humanised mice from obese donor microbiota also showed more weight gain and fat deposition than humanised mice with lean donor microbiota.63 Cohousing lean with obese humanised mice prevented the obese phenotype in these cage mates, and altered the microbiota to a more lean-like profile.63 The transfer of faeces from standard diet-fed compared with high-fat diet-fed donor mice was also sufficient to reduce the expression of inflammatory cytokines and lipogenic genes in adipose and hepatic tissue of mice with diet-induced obesity.64 These data suggest that there are available niches in the obese donor microbiota that can be filled by lean associated microbes. FMT from lean donors into humans with metabolic syndrome was also sufficient in the clinical setting to increase gut microbial diversity and improve insulin sensitivity.56 This opens the opportunity to determine specific bacterial species for transplantation and the treatment of metabolic abnormalities as a therapeutic approach in the future. Some preclinical studies have reported beneficial effects of pre- and probiotics, yet the data from human randomised controlled trials are inconsistent, and more investigations are needed to inform clinical practice.
Epidemiologic studies imply that disruption of the gut microbiota early in life, via caesarean section as well as antibiotic therapy, increases the risk of being overweight later in childhood.65, 66 Indeed, antibiotic use during the first 6 months was associated with becoming overweight later in life among children of normal weight mothers. However, early administration of antibiotics reduced the risk of becoming overweight among children of overweight and obese mothers.65 Taken together with the preclinical data, this demonstrates the complex relationship between the gut microbiota, antibiotic use and metabolic outcomes. In light of the high rates of childhood obesity, the use of antibiotics or pre- and probiotics to restore intestinal homeostasis and to potentially prevent obesity in children from overweight and obese mothers is an interesting concept.65 So far, there is no direct clinical evidence for a causal relationship between obesity and antibiotic use or delivery mode. Yet, preclinical studies imply that even transient alterations in the infant gut microbiome could potentially affect metabolic homeostasis resulting in obesity. In mice, exposure to low dose antibiotics in early life resulted in an increased risk of obesity.67, 68 Low dose antibiotics do not deplete the microbiota but cause a shift in taxonomic composition towards an increased Firmicutes to Bacteroidetes ratio.67 There was a trend towards hyperglycaemia in antibiotic treated mice and genes related to hepatic lipogenesis were upregulated, but no phenotypic signs of hepatic steatosis were observed.67 In a more recent study, Cox et al.68 showed that the period around birth was critical to alter host metabolism and adiposity. The metabolic phenotype persisted after cessation of the antibiotic treatment, even though the microbiota recovered.68 This study provides evidence for a critical window in early life when the gut microbiota can induce long-term alterations in host metabolism.68
Taken together, the gut microbiota has a substantial effect on metabolic abnormalities by influencing the efficiency of energy harvest and storage, low-grade inflammation and browning of adipose tissue. Notably, there are risk factors for these metabolic abnormalities beyond gut microbes, which include lifestyle factors such as energy intake and expenditure as well as genetic risk factors. This results in a complex interplay of these lifestyle, microbial and host factors that determine disease susceptibility. The integration of modern applications to assess microbiota in preclinical and clinical studies will help clarify this complex interplay and yield promise to identify novel treatments for beneficial modulation of the gut microbiota to improve metabolic health.
Allergic Diseases
Allergic diseases are a global health concern, affecting over half a billion people worldwide. Genetic and molecular risk factors are clearly associated with the development of allergy; however, it is unlikely genetic factors can account for the rapid rise in allergic disease over the past 50 years. This indicates that environmental factors, such as the gut microbiota, may have a key role in the development of allergic disease. For example, reduced gut microbial diversity in infants was associated with increased allergic risk in school age children,69 and the development of food allergy was associated with altered gut microbiota in these infants.70 Strikingly, a recent study has shown that lower abundance of bacteria such as Bifidobacterium, Akkermansia and Faecalibacterium, along with higher abundance of particular fungi including Candida and Rhodotorula in neonates may predispose to allergy susceptibility by influencing T-cell differentiation.71
GF mice have been an effective way to address several questions regarding the relationship between the gut microbiota and allergic inflammation. Colonisation of GF mice with gut microbiota in early life was sufficient to protect mice from allergic inflammation in the lung, whereas allergic airway inflammation (AAI) persisted in mice colonised as adults.72 Early life colonisation with gut microbiota also inhibited serum IgE responses,73 and protected against sensitisation to food allergens.74 In addition, Clostridia-induced IL-22 has also been shown to reduce the uptake of dietary antigen into the systemic circulation.74 These investigations suggest that the gut microbiota is a key environmental factor in the regulation of allergic inflammation in the lung and GI tract. Depletion of the gut microbiota using antibiotic administration in mice has also been employed to examine the impact on allergy susceptibility. Vancomycin is not absorbed from the intestine, so when administered orally affects only the gut microbiota. Oral administration of vancomycin in early life resulted in exacerbation of AAI and serum IgE,75 and exposure of neonatal mice to an oral antibiotic cocktail also supported food allergen sensitisation.74 These data show that antibiotic use increased the risk of allergy development later in life.
SCFA and diets high in MAC also influence the development of allergic diseases. Mice lacking SCFA receptors GPR4129 or GPR4376, 77 developed more severe AAI than wild-type littermates, and high MAC diet-suppressed AAI.29, 77 Dietary MAC and SCFA also promoted Treg induction, oral tolerance and protected against experimental food allergy.78 Further studies identify the lung microbiota as key player in suppressing AAI. In mice, the neonatal airways contained predominantly Firmicutes and Gammaproteobacteria, and these neonatal mice were more susceptible to AAI.79 An expansion of Bacteroidetes in the lung and the development of Treg cells with age were correlated with reduced AAI.79 Asthmatic patients have also been shown to have higher bacterial burden and diversity compared with controls.80 These data may reflect an interplay between the microbiota at these mucosal tissues. However, the function of the gut microbiota in atopic dermatitis (AD) is less clear. Similarly, the impact of the gut microbiota in eosinophilic GI diseases, such as eosinophilic esophagitis, as well as other allergic disorders including allergic rhinitis and allergic conjunctivitis, are yet to be fully elucidated.
Importantly, these findings in mice are supported by epidemiological data that early life exposure to microbes can reduce the incidence of allergic disease. Indeed, allergy is reduced in children exposed to a farm environment, and studies suggest microbial exposures in early life are a contributing factor.81 On the basis of these data, a large number of clinical studies have examined the efficacy of probiotics in the prevention and treatment of allergy. Utilising probiotics for the prevention of AD has been extensively studied, yet there is still conflicting data. As well, more studies are needed to determine the potential for prebiotics in allergy prevention at present. Two notable studies support beneficial modulation of the gut microbiota and associated metabolites in early life as an effective intervention strategy for allergic disease. The Canadian Healthy Infant Longitudinal Development (CHILD) Study identified a transient alteration in the gut microbiota during the first 100 days of life in infants at risk of asthma, which was associated with reduced faecal acetate concentrations.82 This disturbance of the microbial composition in infants at risk was characterised by significantly lower abundance in the gut of Lachnospira, Veillonella, Faecalibacterium and Rothia bacterial genera.82 Colonisation of GF mice with these specific gut microbiota ameliorated AAI in the offspring, demonstrating a causal role for the bacterial communities in reducing allergy susceptibility.82 Moreover, the gut microbiota of infants with cow's milk allergy is altered in composition and diversity as compared with non-allergic infants.83 Supplementation of extensively hydrolysed formula with a Lactobacillus probiotic has been shown to accelerate tolerance to cow's milk in infants with cow's milk allergy, and was associated with increased abundance of faecal butyrate producing bacteria and butyrate levels following Lactobacillus treatment.83
Taken together, there is mounting evidence to support gut microbiota based treatments to reduce susceptibility to allergic disease. Future research identifying the components of the gut microbiota that influence allergy avoidance or tolerance mechanisms is crucial to develop these strategies for widespread clinical use.
Infectious Diseases
Nearly one third of deaths in low-income countries are attributable to infectious diseases.84 Infection rates in the Western world have reduced dramatically over the past century due to advances in hygiene, antibiotic use and vaccination. However, infectious disease still poses significant risk to vulnerable populations such as infants and the elderly. Influenza and pneumonia remain one of the leading causes of death of elderly citizens.85 Upper respiratory tract infections are the most common illness for which people seek medical care, and the burden on healthcare services is substantial.86 In addition, C. difficile is the leading healthcare-acquired infection, which has increased in severity and morbidity over the last decade.47 The gut microbiota provide critical signals for the development and function of the immune system that provides protection from invading intestinal pathogens.87 Therefore, manipulation of the gut microbiota is an attractive therapeutic target to build immune capability to combat infectious diseases.
Interactions between epithelial cells and the gut microbiota are essential for intestinal homeostasis and barrier defences. Defined gut microbial species have been shown to maintain tight junction integrity, which in turn limits Salmonella typhimurium invasion.88 Gut microbiota promote the secretory IgA response that inactivates rotavirus and neutralises cholera toxin,87 and outcompete C. difficile colonisation thereby preventing infection.47 Key gut microbial species also prime the development of cellular immune responses in the GI tract. SFB-induced Th17 responses are sufficient to promote immunity against the intestinal pathogen Citrobacter rodentium.89 Moreover, damage induced by pathogens is limited by microbiota-induced Treg. B. fragilis-induced IL-10 producing Treg limit Helicobacter hepaticus infection, and Bifidobacterium infantis-induced Treg dampen excessive inflammation induced in S. typhimurium infection.87
The critical role for the gut microbiota in establishing immune defence against infection extends far beyond the local environment of the GI tract. In fact, recent evidence demonstrates that gut microbial signalling actively shapes the systemic immune response by controlling haematopoiesis in primary immune sites.90 Microbiota derived signals maintain granulocyte/monocyte progenitor development in the bone marrow, which influenced multiple tissue-resident innate immune populations. This in turn dictates the efficiency of early innate responses to infection. Absence of microbiota derived signals conferred susceptibility to systemic Listeria monocytogenes and Staphylococcus aureus infection, which was driven by defects in tissue-resident myeloid populations prior to infection.90
Microbiota signalling can also calibrate the activation threshold of innate immune cells, that enhances the sensitivity to detect pathogen invasion. On infection with mouse cytomegalovirus and lymphocytic choriomeningitis virus, antiviral defences including type I interferon (IFN) are stimulated through PRR ligation.91 In GF mice, type I IFN dependent priming of natural killer cells was reduced due to dampened PRR signalling, and antiviral protection was lost.91 Microbiota signalling has also been shown to enhance protection against influenza virus by facilitating inflammasome dependent IL-1β and IL-18 in the lung in a TLR dependent manner.20 Reactive oxygen species-mediated defence of alveolar macrophages in respiratory Klebsiella pneumonia infection was modulated by the gut microbiota as well.92 Interestingly, only gut and not lung microbiota derived ligands activated this respiratory defence mechanism, demonstrating the importance of the gut microbiota in establishing immune defences at other microbiota colonised sites.92
The capacity to mount adaptive immune responses is influenced by gut microbial signals as well. This was clearly demonstrated in a mouse model of seasonal trivalent influenza vaccination (TIV), where the absence of gut microbiota significantly attenuated the magnitude of vaccine-induced protective antibody response.93 A mechanistic role was elucidated, demonstrating TLR5 mediated sensing of the gut microbiota increased the magnitude of the antibody response.93 Furthermore, these investigations found a correlation between TLR5 expression in peripheral blood mononuclear cells shortly after vaccination and the magnitude of TIV-induced antibody response in a clinical cohort. The authors suggest the observed variable efficacy of TIV between vaccines may in part be attributed to the host's gut microbiota composition.93 This raises the possibility that beneficial modulation of the gut microbiota may be used as a therapeutic approach to improve the efficacy of vaccine-induced immunity.
Manipulating gut microbes to enhance immune function is not a new concept to be tested in the clinic. At present the studies examining prebiotic intervention have failed to demonstrate efficacy to increase vaccine responsiveness.94 Variability observed in the effectiveness of probiotic interventions in vaccination trials needs to be acknowledged.95 However, there is strong evidence across these clinical trials to demonstrate that probiotic supplementation in adults improves vaccine-induced immunity. For example, Bifidobacterium lactis Bl-04 and Lactobacillus acidophilus La-14 significantly enhanced cholera toxin-specific IgG induced by oral cholera vaccine,95 and Lactobaccillus paracasei subsp. paracacei (L. casei 431) strain significantly increased influenza-specific IgG in response to parenteral TIV vaccination.96 Moreover, L. casei 431 along with a number of other candidate strains, have also demonstrated a protective effect against acute upper respiratory tract infections.86
These studies provide clear evidence of a role for the gut microbiota in immunity to infection as well as vaccine-induced protective antibody responses.
Discussion and Future Directions
As most gut microbes (especially anaerobes) cannot be cultured easily, the development of new culture-independent techniques have led to an increase in knowledge in the gut microbiota field. These modern technological applications are now used frequently in both pre- and clinical studies to address the role of the gut microbiota in health and disease (Figure 2), and have substantially contributed to providing understanding of the complex interplay between gut microbial composition, host genetics and biological outcomes.
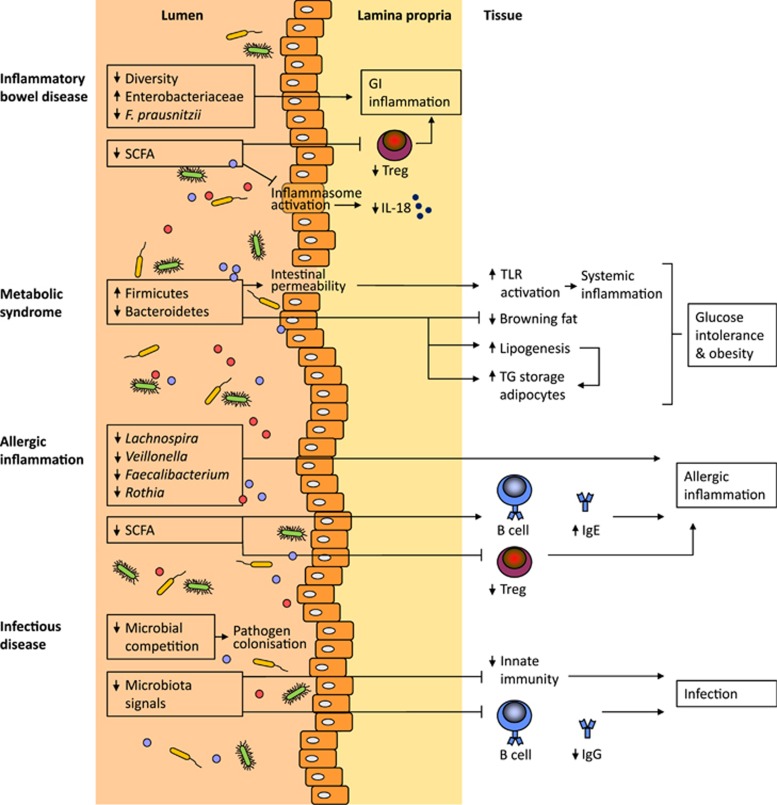
Figure 2.
The gut microbiota affects various inflammatory and infectious diseases. IBD is associated with reduced diversity of the microbial community and other specific microbial changes associated with GI inflammation. A reduction in short-chain fatty acids (SCFA) can increase GI inflammation since SCFA induce regulatory T cells (Treg) via inhibition of histone deacetylase activity. Furthermore, SCFA promote IL-18-induced epithelial repair via inflammasome activation. The microbiota profile associated with metabolic syndrome comprises an increase in Firmicutes and a reduction in Bacteroides. The obese microbiome has an increased capacity to harvest energy from the diet, leading to increased hepatic lipogenesis and storage of triglycerides (TG) in adipocytes. Furthermore, the browning of fat is regulated by the microbiota. Inflammation-associated alterations in the gut microbial composition leads to epithelial barrier disruption allowing bacterial lipopolysaccharide to circulate systemically. These metabolic changes can lead to glucose intolerance and obesity. Allergic inflammation is associated with the lower abundance of specific bacterial genera. Depleting the gut microbiota has been shown to increase IgE production, which together with reduced Treg responses can result in allergic inflammation. A lack of microbial competition in the gut lumen can lead to Clostridium difficile colonisation. Finally, a lack of microbial signals can also reduce the innate as well as adaptive immune response to microorganisms, making the host more vulnerable to infections. See the text for further details.
Studies in GF mice, which like newborns can be colonised by a diverse microbiota, demonstrate that a specific microbial composition can exert lasting effects on the immunity and disease outcomes of the host. It is only recently that investigators have begun to truly appreciate the importance of this, and translation of such findings into humans is challenging. So far there are only correlations between alterations in gut microbial composition and inflammatory and infectious diseases in humans. The humanised mouse models, when combined with current technologies, are a promising attempt to move beyond these correlations to provide causal roles for specific gut microbial functions and disease susceptibility. Colonisation with the culture collections from a human donor can be used to determine which cultured members of the microbiota can transmit or prevent a disease phenotype, and these studies may identify viable microbial-based therapeutic targets. It also opens up possibilities to study the effects of dietary components on gut microbiota composition and disease outcome, or to evaluate the ability of bacterial strains with a therapeutic potential to invade established niches in gut communities. GF and gnotobiotic models provide a great tool to study the microbiota, but as with all animal models there are limitations around the translation to the clinical setting. These models might not completely recapitulate the human disease complexity or heterogeneity of the human gut microbiota.26 Humanised mouse studies have the added complexity of the donor microbiota not having co-evolved with a mouse host, and studies have shown host-specific gut microbiota may be critical for an appropriately functioning immune system.97 Nonetheless, these model systems are invaluable for the understanding of host–microbiota interactions that contribute to disease.
Beneficial modulation of the gut microbiota has yielded encouraging results for the prevention and treatment of various inflammatory diseases in experimental models. Further work is required to characterise strain-specific effects and efficacious dosing regimens for prebiotics and probiotics. Alongside, fundamental research needs to be conducted to define the mechanisms by which the gut microbiota can enhance immune health. Most likely non-bacterial microbes also have an important role in immune function and disease. Their contribution remains an important area for future research, as this has been relatively unexplored to date. This is a fast-paced field of research and given the positive results so far, we are cautiously optimistic it will soon yield novel therapeutics that manipulate the microbiota to prevent or treat chronic inflammatory diseases and enhance protective immunity against pathogens.
Acknowledgments
We would like to thank Anna Mooney for her contribution to the review. The contributing authors are supported by the Health Research Council (EFB), the Ministry of Business, Innovation and Employment (LvdE, HCP, WY and EFB) and LSW holds an Australian Research Council DECRA Fellowship (DE150101574).
The authors declare no conflict of interest.
References
- Zeng MY, Inohara N, Nunez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol 2016. doi:10.1038/mi.2016.75. [DOI] [PMC free article] [PubMed]
- Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 2013; 21: 167–173. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005; 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev 2010; 90: 859–904. [DOI] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 2011; 108: 4516–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 2010; 59: 518–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless CE, Guiver M, Borrow R, Edwards-Jones V, Kaczmarski EB, Fox AJ. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J Clin Microbiol 2000; 38: 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 2007; 45: 2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames SK, Gardner SN, Marti JM, Slezak TR, Gokhale MB, Allen JE. Using populations of human and microbial genomes for organism detection in metagenomes. Genome Res 2015; 25: 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray K. Gut microbiota: the gut virome and bacterial microbiome-the early years. Nat Rev Gastroenterol Hepatol 2015; 12: 609. [DOI] [PubMed] [Google Scholar]
- Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 2010; 466: 334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A et al. Target-enrichment strategies for next-generation sequencing. Nat Methods 2010; 7: 111–118. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013; 31: 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzosa EA, Morgan XC, Segata N, Waldron L, Reyes J, Earl AM et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci USA 2014; 111: E2329–E2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bergen M, Jehmlich N, Taubert M, Vogt C, Bastida F, Herbst FA et al. Insights from quantitative metaproteomics and protein-stable isotope probing into microbial ecology. ISME J 2013; 7: 1877–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberkmoes NC, Russell AL, Shah M, Godzik A, Rosenquist M, Halfvarson J et al. Shotgun metaproteomics of the human distal gut microbiota. ISME J 2009; 3: 179–189. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Segata N, Huttenhower C. Biodiversity and functional genomics in the human microbiome. Trends Genet 2013; 29: 51–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson AJ, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 2004; 303: 1662–1665. [DOI] [PubMed] [Google Scholar]
- Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA 2011; 108: 5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell 2010; 140: 821–832. [DOI] [PubMed] [Google Scholar]
- Ivanov I, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 2009; 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 2015; 163: 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA et al. The toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 2011; 332: 974–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504: 446–450. [DOI] [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16: 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami M, Kishimoto K, Ohata A, Miyoshi M, Aoyama M, Fueda Y et al. Butyrate and trichostatin A attenuate nuclear factor kappaB activation and tumor necrosis factor alpha secretion and increase prostaglandin E2 secretion in human peripheral blood mononuclear cells. Nutr Res 2008; 28: 321–328. [DOI] [PubMed] [Google Scholar]
- Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat Commun 2015, 1–15. [DOI] [PubMed]
- Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014; 20: 159–166. [DOI] [PubMed] [Google Scholar]
- Roberfroid M. Prebiotics: the concept revisited. J Nutr 2007; 137: 830S–837S. [DOI] [PubMed] [Google Scholar]
- Rijkers GT, Bengmark S, Enck P, Haller D, Herz U, Kalliomaki M et al. Guidance for substantiating the evidence for beneficial effects of probiotics: current status and recommendations for future research. J Nutr 2010; 140: 671S–676S. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13: R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008; 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M et al. Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm Bowel Dis 2010; 16: 2034–2042. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L et al. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 2006; 55: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N et al. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004; 127: 412–421. [DOI] [PubMed] [Google Scholar]
- Sokol H, Lepage P, Seksik P, Dore J, Marteau P. Temperature gradient gel electrophoresis of fecal 16S rRNA reveals active Escherichia coli in the microbiota of patients with ulcerative colitis. J Clin Microbiol 2006; 44: 3172–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013; 502: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey FE, Gonzalez MD, Cheng J, Wu M, Ahern PP, Gordon JI. Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc Natl Acad Sci USA 2013; 110: 13582–13587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitcher MC, Beatty ER, Harris RM, Waring RH, Cummings JH. Sulfur metabolism in ulcerative colitis: investigation of detoxification enzymes in peripheral blood. Dig Dis Sci 1998; 43: 2080–2085. [DOI] [PubMed] [Google Scholar]
- Ijssennagger N, Belzer C, Hooiveld GJ, Dekker J, van Mil SW, Muller M et al. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc Natl Acad Sci USA 2015; 112: 10038–10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly D, Campbell JI, King TP, Grant G, Jansson EA, Coutts AG et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol 2004; 5: 104–112. [DOI] [PubMed] [Google Scholar]
- Fukata M, Michelsen KS, Eri R, Thomas LS, Hu B, Lukasek K et al. Toll-like receptor-4 is required for intestinal response to epithelial injury and limiting bacterial translocation in a murine model of acute colitis. Am J Physiol Gastrointest Liver Physiol 2005; 288: G1055–G1065. [DOI] [PubMed] [Google Scholar]
- Smelt MJ, de Haan BJ, Bron PA, van Swam I, Meijerink M, Wells JM et al. The impact of Lactobacillus plantarum WCFS1 teichoic acid d-alanylation on the generation of effector and regulatory T-cells in healthy mice. PLoS ONE 2013; 8: e63099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakostelska Z, Kverka M, Klimesova K, Rossmann P, Mrazek J, Kopecny J et al. Lysate of probiotic Lactobacillus casei DN-114 001 ameliorates colitis by strengthening the gut barrier function and changing the gut microenvironment. PLoS ONE 2011; 6: e27961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zuo ZX, Mao AP. Effect of probiotics on inducing remission and maintaining therapy in ulcerative colitis, Crohn's disease, and pouchitis: meta-analysis of randomized controlled trials. Inflamm Bowel Dis 2014; 20: 21–35. [DOI] [PubMed] [Google Scholar]
- Seekatz AM, Young VB. Clostridium difficile and the microbiota. J Clin Invest 2014; 124: 4182–4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 2014; 8: 1569–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S et al. Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science 2010; 328: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004; 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JK, Davenport ER, Beaumont M, Jackson MA, Knight R, Ober C et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 2016; 19: 731–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody RN, Gerber GK, Luevano J, Jesus M, Gatti DM, Somes L et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015; 17: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008; 3: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1131. [DOI] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature 2006; 444: 1022–1023. [DOI] [PubMed] [Google Scholar]
- Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012; 143: 913–916.e917. [DOI] [PubMed] [Google Scholar]
- Finucane MM, Sharpton TJ, Laurent TJ, Pollard KS. A taxonomic signature of obesity in the microbiome? Getting to the guts of the matter. PLoS ONE 2014; 9: e84689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes 2008; 57: 1470–1481. [DOI] [PubMed] [Google Scholar]
- Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid–induced insulin resistance. J Clin Invest 2006; 116: 3015–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes 2011; 60: 2775–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 2007; 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-Zamorano N, Fabbiano S, Chevalier C, Ozren S, Colin DJ, Ana S et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat Med 2015; 21: 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341: 1241214–1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CJ, Lin CS, Lu CC, Martel J, Ko YF, Ojcius DM et al. Ganoderma lucidum reduces obesity in mice by modulating the composition of the gut microbiota. Nature Commun 2015; 6: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M, Sørensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes 2011; 35: 522–529. [DOI] [PubMed] [Google Scholar]
- Kuhle S, Tong OS, Woolcott CG. Association between caesarean section and childhood obesity: a systematic review and meta-analysis. Obes Rev 2015; 16: 295–303. [DOI] [PubMed] [Google Scholar]
- Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2013; 488: 621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014; 158: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H, Li N, Bonnelykke K, Chawes BL, Skov T, Paludan-Muller G et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol 2011; 128: 646–652 e641-645. [DOI] [PubMed] [Google Scholar]
- Ling Z, Li Z, Liu X, Cheng Y, Luo Y, Tong X et al. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol 2014; 80: 2546–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 2012; 336: 489–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahenzli J, Koller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe 2013; 14: 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefka AT, Feehley T, Tripathi P, Qiu J, McCoy K, Mazmanian SK et al. Commensal bacteria protect against food allergen sensitization. Proc Natl Acad Sci USA 2014; 111: 13145–13150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M et al. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 2012; 13: 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461: 1282–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun 2015; 6: 7320. [DOI] [PubMed] [Google Scholar]
- Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE et al. Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016; 15: 2809–2824. [DOI] [PubMed] [Google Scholar]
- Gollwitzer ES, Saglani S, Trompette A, Yadava K, Sherburn R, McCoy KD et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med 2014; 20: 642–647. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol 2011; 127: 372–381 e371-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mutius E, Vercelli D. Farm living: effects on childhood asthma and allergy. Nat Rev Immunol 2010; 10: 861–868. [DOI] [PubMed] [Google Scholar]
- Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 2015; 7: 307ra152. [DOI] [PubMed] [Google Scholar]
- Berni Canani R, Sangwan N, Stefka AT, Nocerino R, Paparo L, Aitoro R et al. Lactobacillus rhamnosus GG-supplemented formula expands butyrate-producing bacterial strains in food allergic infants. ISME J 2016; 10: 742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation. The top 10 causes of death. 2016. Available from http://www.who.int/mediacentre/factsheets/fs310/en/.
- Statistics NCfH Health. United States 2015. (4/2016). 2016: 1-461.
- Jespersen L, Tarnow I, Eskesen D, Morberg CM, Michelsen B, Bugel S et al. Effect of Lactobacillus paracasei subsp. paracasei, L. casei 431 on immune response to influenza vaccination and upper respiratory tract infections in healthy adult volunteers: a randomized, double-blind, placebo-controlled, parallel-group study. Am J Clin Nutr 2015; 101: 1188–1196. [DOI] [PubMed] [Google Scholar]
- Kamada N, Seo S-U, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Imm 2013; 13: 321–335. [DOI] [PubMed] [Google Scholar]
- Martz S-LE, McDonald JAK, Sun J, Zhang Y-G, Gloor GB, Noordhof C et al. Administration of defined microbiota is protective in a murine Salmonella infection model. Sci Rep 2015; 5: 16094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity 2014; 40: 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS et al. Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 2012; 37: 171–186. [DOI] [PubMed] [Google Scholar]
- Clarke TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun 2014; 82: 4596–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M et al. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 2014; 41: 478–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam J, van Stuijvenberg M, Garssen J, Knipping K, Sauer PJJ. A mixture of three prebiotics does not affect vaccine specific antibody responses in healthy term infants in the first year of life. Vaccine 2011; 29: 7766–7772. [DOI] [PubMed] [Google Scholar]
- Paineau D, Carcano D, Leyer G, Darquy S, Alyanakian M-A, Simoneau G et al. Effects of seven potential probiotic strains on specific immune responses in healthy adults: a double-blind, randomized, controlled trial. FEMS Immunol Med Microbiol 2008; 53: 107–113. [DOI] [PubMed] [Google Scholar]
- Rizzardini G, Eskesen D, Calder PC, Capetti A, Jespersen L, Clerici M. Evaluation of the immune benefits of two probiotic strains Bifidobacterium animalis ssp. lactis, BB-12 and Lactobacillus paracasei ssp. paracasei, L. casei 431 in an influenza vaccination model: a randomised, double-blind, placebo-controlled study. Br J Nutr 2012; 107: 876–884. [DOI] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 2012; 149: 1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]