Abstract
During asexual intraerythrocytic development, Plasmodium falciparum diverges from the paradigm of the eukaryotic cell cycles by undergoing multiple rounds of DNA replication and nuclear division without cytokinesis. A better understanding of the molecular switches that coordinate a myriad of events for the progression of the parasite through the intraerythrocytic developmental stages will be of fundamental importance for rational design of intervention strategies. To achieve this goal, we performed isobaric tag-based quantitative proteomics and phosphoproteomics analyses of three developmental stages in the Plasmodium asexual cycle and identified 2767 proteins, 1337 phosphoproteins, and 6293 phosphorylation sites. Approximately 34% of identified proteins and 75% of phosphorylation sites exhibit changes in abundance as the intraerythrocytic cycle progresses. Our study identified 43 distinct phosphorylation motifs and a range of potential MAPK/CDK substrates. Further analysis of phosphorylated kinases identified 30 protein kinases with 126 phosphorylation sites within the kinase domain or in N- or C-terminal tails. Many of these phosphorylations are likely CK2-mediated. We define the constitutive and regulated expression of the Plasmodium proteome during the intraerythrocytic developmental cycle, offering an insight into the dynamics of phosphorylation during asexual cycle progression. Our system-wide comprehensive analysis is a major step toward defining kinase–substrate pairs operative in various signaling networks in the parasite.
Keywords: malaria, Plasmodium falciparum, intraerythrocytic cycle, proteomics, phosphoproteomics, isobaric tags, phosphorylation, phosphorylation motifs, kinase–substrate pairs
Graphical Abstract
INTRODUCTION
Malaria infection presents a major global health challenge, currently afflicting over 500 million people worldwide and significantly increasing childhood mortality in the poorest nations while causing an estimated 1.3% reduction in economic productivity in affected countries.1 Of the five species of malaria parasites that infect human populations, Plasmodium falciparum is the most virulent and causes about one million deaths each year.2 Identification of novel cellular targets for therapeutic intervention requires a nuanced understanding of the idiosyncratic molecular mechanisms that underlie progression of the P. falciparum life cycle. Despite long-standing interest due to its medical and biological significance, the atypical life cycle of Plasmodium remains ill-defined at a molecular level; more generally, the signaling networks that govern all aspects of its biology, from recognition of environmental cues to cell growth and differentiation, are poorly understood. To date, our knowledge of this organism has been restricted due to challenges in applying traditional biochemical, cell biological, and genetic screens in this system. However, in recent years, a variety of technological advances have enabled a paradigm shift in biology, broadening the focus from studies restricted to individual genes or proteins to a system-based, global analysis of cellular networks. Importantly, many of these technologies, including mass spectrometry-based methods for global quantitative analysis of proteomes and phosphoproteomes, have proven to be applicable to Plasmodium,3–5 thus offering unprecedented insights into the molecular foundations of this organism’s unique and medically important life cycle.
A variety of previous proteomic studies utilizing a wide range of technologies for fractionation and quantification have explored various stages of the Plasmodium life cycle.6–9 These studies have investigated sporozoite, merozoite, trophozoite, schizont, and gametocyte stages independently of one another and typically identified several hundred to ~2500 proteins.8–12
While post-transcriptional regulation could occur from many cellular processes, reversible protein phosphorylation seems especially likely to play a key role in regulation of the Plasmodium life cycle. Throughout eukaryotes, protein kinases universally function as regulatory switches to drive cell cycle progression by phosphorylating key effector proteins. Genome-wide sequencing of P. falciparum2 has enabled us to identify the repertoire of parasite protein kinases (PKs) based on homology searches. Surprisingly, this revealed only 65 PKs related to eukaryotic PKs (ePKs), along with a novel ePK-related family of 20 members, named FIKKs after a shared Phe-Ile-Lys-Lys motif in their catalytic domain.13 This number is rather low considering that (i) Saccharomyces cerevisiae encodes approximately twice as many ePKs within its comparably sized genome and (ii) the parasite’s developmental cycle spans multiple hosts, requiring specialized regulation in each. It is expected that at least some malaria parasite ePKs are especially versatile to support the full range of cellular functions. However, it is difficult to predict physiological roles of these PKs simply based on homology because on one hand precise orthology cannot be assigned to the vast majority of these enzymes and on the other hand many of these proteins are expected to possess parasite-specific functions that orchestrate specialized features of the unique Plasmodium biology.
Phosphoproteome analysis has been performed in many organisms ranging from yeast to mouse14–16 and has proven to be an important tool for defining key proteins within cellular signaling networks. An initial attempt to analyze phosphoproteins in P. falciparum by 2D gel electrophoresis and mass spectrometry identified 170 proteins.17 More recently, reports characterizing the P. falciparum phosphoproteome focused on the schizont stage of intraerythrocytic parasitic growth.3,4,18 Notably, these previous studies did not investigate the dynamics of both protein expression and phosphorylation during the entire intraerythrocytic developmental cycle of Plasmodium. Here we present a comprehensive analysis of protein expression and phosphorylation across the three major developmental stages in the P. falciparum intraerythrocytic asexual cycle using isobaric labeling.19,20 This comparative analysis revealed stage-specific profiles of protein expression and phosphorylation. We present these quantitative protein abundance and phosphorylation profiles as a resource for the research community.
EXPERIMENTAL PROCEDURES
Plasmodium falciparum Culture
P. falciparum 3D7 were grown at a 4–10% parasitemia and 4% hematocrit in RPMI 1640 culture medium supplemented with A + erythrocytes and 5% albumax, as previously described.21 Parasites were doubly synchronized using a MACS LD (MiltenyiBiotec, Auburn, CA) column at the schizont stage and by 5% d-sorbitol in the ring stage.22,23
Tightly synchronized ring (16 ± 4 h postinvasion), trophozoites (26 ± 4 h postinvasion), and schizonts (36 ± 4 h postinvasion) were harvested following established protocols.24 Parasites were isolated from infected erythrocytes by lysing in 0.1% saponin. Parasite extracts were prepared using lysis buffer (8 M urea, 75 mM Tris, pH 8.2, 1 × HALT protease inhibitor, 1× HALT phosphatase inhibitor).25
Proteomic and Phosphoproteomic Analysis
Following cell lysis, duplicate samples for each stage (1 mg per TMT channel) were reduced, alkylated, digested with Lys-C, and then labeled with one of six TMT isobaric labeling reagents. The resulting peptides were fractionated via strong cation exchange chromatography (SCX) into 20 fractions, as previously described.14 For proteome analysis, these fractions were desalted and analyzed directly on the mass spectrometer; for phosphoproteome analysis, each SCX fraction was subjected to immobilized metal affinity chromatography to specifically enrich phosphorylated peptides.25 Both phospho-enriched and nonphospho-enriched samples were analyzed via LC-MS/MS on an LTQ-Velos-Orbitrap mass spectrometer essentially as previously described,26 except that all MS/MS spectra were acquired at high resolution in the Orbitrap and used for both peptide identification and quantification. Peptides were identified by matching individual MS/MS spectra with peptide sequences using Sequest.27 Search parameters included: 50 ppm precursor ion tolerance; 0.03 Da product ion tolerance; Lys-C digestion with up to two missed cleavages; variable modifications of Met Oxidation (+15.9949) and phosphorylation of Ser, Thr, and Tyr (+79.9663; phospho-enriched searches only); static modifications included alkylation of Cys (+57.0214); as well as TMT labeling of Lys side chains and peptide N-termini (+229.16293). Database searches employed a composite database containing P. falciparum sequences obtained from NCBI28 as well as human protein sequences derived from the International Protein Index29 and common contaminants such as Lys-C; all sequences were included in forward and reversed orientations. Peptides were filtered using a multivariate approach to remove questionable identifications based on a target-decoy strategy.14,30 Peptides were assembled into proteins, and proteins were further filtered to ensure a protein-level false discovery rate (FDR) of 1%. For phospho-enriched samples, individual phosphorylation sites on each peptide were scored to assess how confidently the modification could be localized to a single residue using AScore.31 Finally, peptides and proteins were filtered to account for protein redundancy using the principles of parsimony as previously described.32 Essentially, we report the minimal set of proteins necessary to account for all identified peptides and phosphopeptides.
For quantification, TMT reporter ion signals were extracted from MS/MS spectra and normalized assuming equal sample loading in each channel. Each protein or phosphorylation site was quantified by gathering together all matching peptides, filtering out peptides that did not meet minimum standards for reporter ion signal and isolation specificity, and rescaling reporter ion intensities so that they summed to 1.0 across the six reporter ion channels for each protein or phosphorylation site. An ANOVA model was used to identify differentially expressed proteins and sites based on the quantitation of individual peptides. The Benjamini–Hochberg approach was used to correct for multiple hypothesis testing.
Western Blot Analysis with Phospho-Specific Antibodies
For Western blot analysis, 100 µg of protein extract from 8 h time points of the intraerythrocytic developmental cycle of the parasite was resolved on a NuPAGE 4–12% Bis-Tris 1.5 mm gel with NuPAGE MOPS-SDS running buffer (Invitrogen). Phospho-tyrosine mouse monoclonal antibody (P-Tyr-100, Cell Signaling Technology) and phospho-MAPK/CDK substrates (PXSP or SPXR/K, Cell Signaling Technology) rabbit monoclonal antibody were used to detect tyrosine phosphorylated and MAPK/CDK substrates.
Immunoprecipitation with PTMScan Direct
The PTMScan Direct protocol is adapted from the PhosphoScan method developed at Cell Signaling Technology with licensed use.33
Cell Lysate Preparation
Parasite extracts were prepared in urea lysis buffer (20 mM HEPES, pH 8.0, 9 M urea, 1× HALT protease inhibitor and 1× HALT phosphatase inhibitor). Clarified lysate was reduced with 4.5 mM DTT, followed by alkylation with 55 mM iodoacetamide. Alkylated proteins were precipitated by methanol-chloroform. The protein pellet was resuspended in 4 M urea, and 50 mM Tris-HCl, pH 8.8 buffer at a concentration of 3 mg/mL. The sample was then digested with LysC (Wako 129-02451 10 AU) at a 1/100 enzyme/protein ratio in 1 M urea and 50 mM Tris-HCl, pH 8.8. Digested peptide lysates were acidified with 1% formic acid, and peptides were desalted over Sep-Pak C18 columns (Waters WAT051910). Elution of peptides was achieved with 40% acetonitrile, 1% formic acid.
Immunoprecipitation
Lyophilized peptides were resuspended in immunoprecipitation (IAP) buffer (50 mM MOPS/NaOH, pH 7.2, 10 mM Na2HPO4, 50 mM NaCl) and incubated with PTMScan Immunoaffinity beads overnight at 4 °C with agitation. The beads were then pelleted by centrifugation and peptides were eluted from the beads with 100 mM formic acid. The sequential eluates were combined and desalted as previously described.34 Finally, samples were resuspended in 5% acetonitrile/5% formic acid prior to MS analysis.
Mass Spectrometry Analysis
All samples were analyzed via LC-MS on an LTQ-Velos-Orbitrap mass spectrometer configured as previously described26 and operated in data-dependent mode with low-resolution ion trap mass spectra acquired for the top ten precursors identified in each MS survey scan. Data were then searched using Sequest,27 and resulting peptide and protein identifications were filtered to a final protein-level FDR of 1% using the target-decoy approach as previously described.14,30 Site localization was evaluated via AScore;35 finally, phosphopeptides were grouped according to the phosphorylation sites they contained.
Subdomain Location of Phosphorylation Sites in Protein Kinases
Putative kinases were selected from the table of all phosphory-lated proteins based on the appearance of the string “kinase” in the sequence description. From these, we identified and aligned 30 ePK domains using the program MAPGAPS36 and sequence profiles designed for apicomplexan PKs,37 while additional sequences representing atypical kinases or other kinase-associated proteins were removed from this sequence set. The aligned regions corresponding to each subdomain were identified according to the definition by Hanks and Hunter.38 An in-house Python script was used to map each observed phosphorylation site to a subdomain or tail region, count the number of phosphorylation sites in each region, and visualize the totals.
Matching Phosphorylation Sites with Kinases Using NetPhorest
Sequences of likely ePKs in the NCBI Plasmodium proteome were uploaded to the public web server for NetPhorest39 along with lists of phosphorylated residues. The probabilities associating each phosphorylation site with particular kinases and kinase families were then downloaded and sorted in order of decreasing probability. The top 2.5% of putative kinase–substrate relationships were retained and plotted as a heat map reflecting the probability that the indicated kinase or kinase family would be expected to phosphorylate the indicated residue.
RESULTS AND DISCUSSION
Identification of 2767 Proteins and 6293 Phosphorylation Sites from the Intraerythrocytic Stages of Plasmodium falciparum
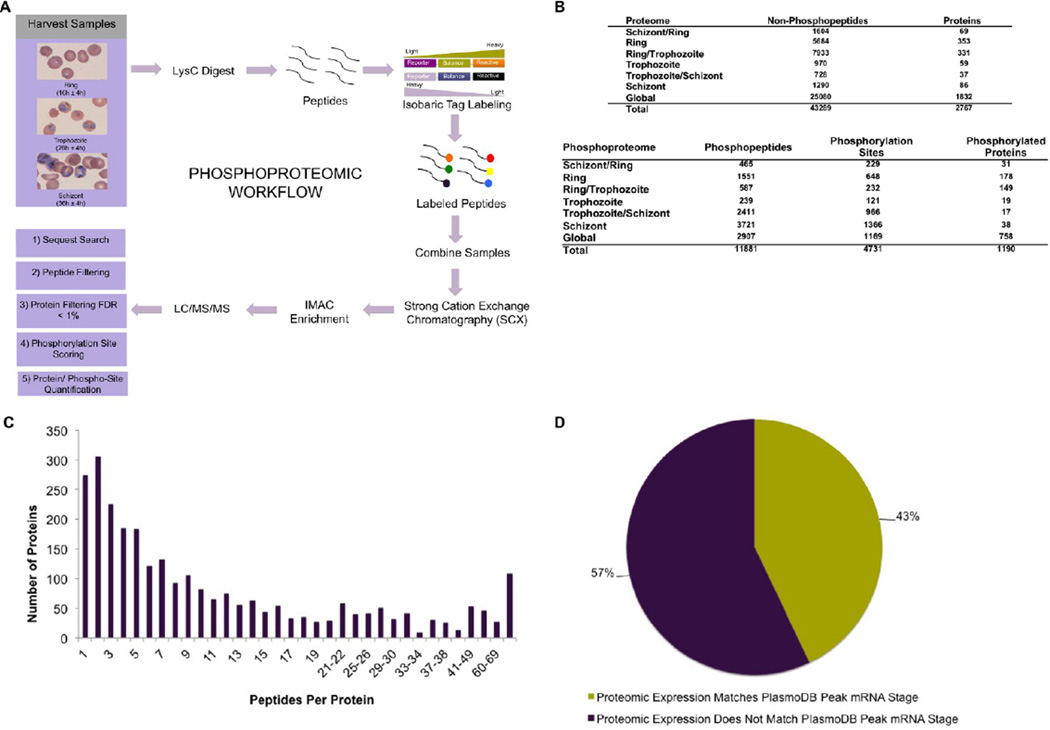
We used isobaric labeling for quantitative analysis of LysC-digested parasite extracts prepared from tightly synchronized ring (16 ± 4 h postinvasion), trophozoite (26 ± 4 h), and schizont (36 ± 4 h) stages. The combined proteomic and phosphoproteomic workflow is outlined in Figure 1A. This quantitative proteomic analysis identified a total of 3365 proteins across both proteomic and phosphoproteomic data sets. Of these proteins, 451 were human-derived, likely from contaminating RBC material, while 2914 were of malarial origin, representing 55% of the total predicted P. falciparum proteome (Supplemental Table S1 and Figure S1 in the Supporting Information).2 After human proteins were excluded, the final data set contained 2767 proteins obtained in the absence of phosphopeptide enrichment (“Proteome” table in Figure 1B) and 1337 phosphoproteins (out of which 1190 were also found in the proteome analysis; see Discussion below) containing a total of 6293 phosphorylation sites, obtained from the phosphoproteome analysis (Figure 1B and Supplemental Table S2 in the Supporting Information) at a protein-level FDR of <1%. Figure 1C displays a histogram showing how the quantified peptide count varied across quantified proteins; at least two peptides were used for quantification of 90% of the identified proteins, while the average number of peptides per identified protein was 15. The observation that the vast majority of proteins were both identified and quantified on the basis of several peptides enhances the reliability of this analysis. We identified a total of 11 881 phosphopeptides bearing 6293 phosphorylation sites from 1337 proteins. Importantly, in addition to identification, we provide quantitative analysis of phosphorylation changes spanning 4731 sites across the intraerythrocytic cell cycle (Figure 1B and Supplemental Table S2 in the Supporting Information).
Figure 1.
Identification of the phosphoproteome and proteome from the intraerythrocytic stages of Plasmodium falciparum. (A) Overview of the procedure for phosphopeptide preparation and enrichment from stage-specific P. falciparum lysates. Duplicate samples from each stage were processed to generate the lists of proteins in Supplementary Table 1 in the Supporting Information and phosphorylation sites in Supplementary Table 2 in the Supporting Information. Protein samples were digested with LysC, followed by labeling with TMT and fractionation on a SCX column. Enrichment of phosphopeptides was conducted using IMAC. The resulting phosphopeptide samples were analyzed on an LTQ-Velos Orbitrap mass spectrometer. Spectra were identified using SEQUEST, and the resulting data were filtered to a protein-level false discovery rate of 1%. AScore was used to assess phosphorylation site localization. Phosphorylation sites with AScores above 13 were considered to be localized. (B) Tables representing the total numbers and stage-specific distributions of identified peptides and phosphopeptides, phosphorylation sites (before AScore filtering), and proteins and phosphoproteins in the six stage-specific samples indicated to the left. (C) Histogram depicting the numbers of peptides used for identification and quantification of the proteins reported in this study. (D) Correlation of the transcriptome (as stated on PlasmoDB) and the proteome of identified malaria proteins.
Because many proteins of the Plasmodium proteome are exclusively expressed in the mosquito stages11,12 and many others are restricted to liver and gametocyte stages during infection of the human host, we propose that our present data set likely encompasses the majority of intraerythrocytic stage-specific proteins. Two main reasons underpin the greater depth of our analysis compared with previous reports:10,24 first, by simultaneously surveying three developmental stages, each with its own protein complement, our study encompasses a larger fraction of the proteomic landscape. Second, continuous improvements in methodology and instrument sensitivity naturally result in increased numbers of protein and phosphorylation identifications.25
An intriguing aspect of Plasmodium intraerythrocytic development is the mechanism by which parasites regulate gene expression. On the basis of transcriptome analysis, it was initially believed that the majority of genes are globally controlled in a stage-dependent manner at the transcriptional level,40,41 but it is now generally accepted that post-transcriptional and posttranslational regulatory mechanisms have dominant roles in regulation of intraerythrocytic gene expression.6,7 Indeed, the cognate mRNAs encoding 429 of the 2767 proteins identified in our study were not present in the sorbitol-synchronized ring, trophozoite, and schizont transcriptomics data set.40 For a large percentage of genes (57.1%), timing of peak mRNA expression does not coincide with maximal protein abundance (Figure 1D). In some instances, such as early transcribed membrane protein 10.1 (PF10_0019) and the putative PK PfTKL4 (PFF1145c), the delay in protein expression is significant: transcription peaks in segmenters or in rings, while the protein expression peaks in schizonts, suggesting that these mRNAs have a long half life and are not translated until the late trophozoite stage. Analysis of mRNA decay in Plasmodium supports extended mRNA half lives for these proteins.42 Overall, our analysis complements previous findings6,7 that post-transcriptional regulation of gene expression plays a major role in Plasmodium.
Developmental Stage-Specific Distribution of Proteins
To determine the dynamics of protein expression during the intraerythrocytic cell cycle, we first compared expression levels by calculating three ratios across the three stages (trophozoite:ring, schizont:ring, and schizont:trophozoite). Proteins exhibiting greater than 1.5-fold difference in abundance between any two stages and an adjusted p-value ≤ 0.05 were considered to be enriched in that particular stage, while proteins showing less than 1.5-fold changes for all pairwise comparisons or an adjusted p-value > 0.05 were considered to be globally expressed.43 Several abundant proteins enriched in a specific stage are shown in Table 1. These include merozoite surface protein 1 (enriched in the ring/schizont sample), Pfmc2-2TM Maurer’s cleft two transmembrane protein (enriched in the trophozoite sample), and ring-exported protein 1 (enriched in the trophozoite/schizont sample). Pfmc2-2TM Maurer’s cleft two transmembrane protein has been implicated in transporting proteins secreted from the parasite to the erythrocyte surface.44 Previously, its transcription has been observed during the trophozoite stage,40 and our study shows peak protein expression also at the trophozoite stage, suggesting that Pfmc2-2TM may have a role in translocation of secreted proteins during trophozoite maturation.
Table 1.
| Part A | |||||||
|---|---|---|---|---|---|---|---|
| stage | |||||||
| peak expression | accession number |
annotation | adj. p value | peptide count |
ring average |
trophozoite average |
schizont average |
| Schizont/Ring | |||||||
| A5A7B0 | merozoite surface protein 1 | 1.81472 × 10−69 | 426 | 0.19822 | 0.12845 | 0.17333 | |
| Q4AE85 | RhopH1/Clag3.1 | 1.98323 × 10−26 | 173 | 0.20033 | 0.12982 | 0.16985 | |
| Q8I2J4 | profilin | 1.94174 × 10−23 | 31 | 0.20973 | 0.12672 | 0.16355 | |
| Ring | |||||||
| Q6LFH8 | ornithine aminotransferase | 4.9966 × 10−181 | 123 | 0.26665 | 0.14896 | 0.08438 | |
| Q8IDQ9 | phosphoethanolamine N-methyltransferase | 2.9399 × 10−151 | 148 | 0.25202 | 0.14008 | 0.10789 | |
| Q8IJA9 | adenosine deaminase | 2.7421 × 10−138 | 108 | 0.25039 | 0.13255 | 0.11706 | |
| Ring/ Trophozoite |
|||||||
| Q8IC05 | heat shock protein 86 | 5.2475 × 10−178 | 403 | 0.21866 | 0.16784 | 0.11350 | |
| Q8IJN7 | enolase | 2.1139 × 10−152 | 249 | 0.22732 | 0.16188 | 0.11079 | |
| Q71T02 | l-lactate dehydrogenase | 9.3727 × 10−118 | 171 | 0.23005 | 0.15436 | 0.11559 | |
| Trophozoite | |||||||
| Q8III6 | heat shock protein 90 | 5.62519 × 10−73 | 62 | 0.14727 | 0.26304 | 0.08969 | |
| Q8I492 | mature parasite-infected erythrocyte surface antigen (MESA) or PfEMP2 |
2.44953 × 10−66 | 132 | 0.13203 | 0.25505 | 0.11292 | |
| Q8IJ11 | Pfmc-2TM Maurer’s cleft two-transmembrane protein |
2.86193 × 10−51 | 84 | 0.13169 | 0.22994 | 0.13838 | |
| Trophozoite/ Schizont |
|||||||
| Q8I2G1 | ring-exported protein 1 | 1.08717 × 10−44 | 89 | 0.07421 | 0.21638 | 0.20940 | |
| Q8ILA1 | conserved Plasmodium protein | 2.85479 × 10−44 | 190 | 0.12001 | 0.18267 | 0.19732 | |
| P13830 | RecName: full= ring-infected erythrocyte surface antigen |
4.38793 × 10−30 | 73 | 0.08755 | 0.18778 | 0.22466 | |
| Schizont | |||||||
| Q8I5M3 | conserved Plasmodium protein | 2.59184 × 10−42 | 126 | 0.13449 | 0.15767 | 0.20784 | |
| Q8IJQ4 | conserved Plasmodium protein | 5.28286 × 10−26 | 49 | 0.11808 | 0.10549 | 0.27644 | |
| Q8IDR3 | myosin A | 8.48 × 10−26 | 54 | 0.15048 | 0.08967 | 0.25985 | |
| Part B | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| stage | |||||||||
| peak phosphorylation |
accession number |
annotation | adj. p value | peptide count |
class | ring average |
trophozoite average |
schizont average |
site |
| Schizont/Ring | |||||||||
| O97239 | conserved Plasmodium protein | 5.67584 × 10−5 | 8 | A | 0.23169 | 0.14715 | 0.16912 | S406 | |
| Q8ILC8 | DNA topoisomerase II | 0.000263595 | 3 | O | 0.24748 | 0.11807 | 0.18188 | S1296 | |
| 0.001019579 | 9 | A | 0.21974 | 0.13337 | 0.19774 | S1288 | |||
| C6KT13 | conserved Plasmodium protein | 0.006320528 | 2 | O | 0.22058 | 0.13932 | 0.14850 | S25 | |
| Ring | |||||||||
| Q8I296 | ubiquitin carboxyl-terminal hydrolase | 8.91046 × 10−11 | 3 | A | 0.36738 | 0.09242 | 0.07079 | S2603 | |
| C5HEV4 | rhoptry-associated protein 1 | 1.436 × 10−9 | 8 | B | 0.35644 | 0.08576 | 0.06399 | S99 | |
| Q7K6A5 | multi-drug resistance protein | 4.05683 × 10−9 | 19 | A | 0.26736 | 0.14217 | 0.11477 | S513 | |
| Ring/ Trophozoite |
|||||||||
| Q8IDQ2 | Kelch protein | 2.50453 × 10−09 | 14 | B | 0.25089 | 0.17959 | 0.13337 | T192 | |
| P39898 | RecName: full= plasmepsin 1 | 1.201 × 10−8 | 4 | O | 0.30932 | 0.25867 | 0.05553 | T23 | |
| 5.25165 × 107 | 4 | O | 0.30742 | 0.26047 | 0.06233 | S22 | |||
| O97238 | conserved Plasmodium protein | 1.68106 × 10−6 | 9 | B | 0.24925 | 0.17173 | 0.14156 | S275 | |
| Trophozoite | |||||||||
| Q8IDG8 | membrane-associated histidin- rich protein 2 |
8.66673 × 10−11 | 7 | O | 0.12973 | 0.41704 | 0.13912 | T136 | |
| Q8IDM6 | nucleoside transporter 1 | 3.16966 × 10−7 | 6 | A | 0.16576 | 0.32821 | 0.11905 | S16 | |
| Q8I492 | mature parasite-infected erythrocyte surface antigen (MESA) or PfEMP2 |
1.52967 × 10−5 | 7 | A | 0.12416 | 0.21326 | 0.17549 | S177 | |
| Trophozoite/ Schizont |
|||||||||
| Q8IJW6 | transcription factor with AP2 domain | 2.3356 × 10−35 | 25 | A | 0.06787 | 0.23552 | 0.27157 | S913 | |
| 1.19618 × 10−24 | 16 | A | 0.06456 | 0.24616 | 0.28399 | S907 | |||
| 2.27714 × 10−23 | 14 | P | 0.05212 | 0.24595 | 0.30015 | S918 | |||
| Q8I3A3 | ubiquitin specific protease | 9.15377 × 10−22 | 16 | A | 0.08560 | 0.21405 | 0.29959 | S1457 | |
| Q8IB78 | nucleoside transporter | 1.64292 × 10−20 | 10 | O | 0.02631 | 0.35909 | 0.28085 | T336 | |
| Schizont | |||||||||
| Q8ILA9 | conserved Plasmodium protein | 1.86363 × 10−22 | 18 | P | 0.07735 | 0.16488 | 0.29758 | S121 | |
| Q8IC17 | origin recognition complex subunit 2 | 7.10986 × 10−19 | 11 | T | 0.13324 | 0.22689 | 0.37323 | S347 | |
| Q8IHR4 | dynamin-like protein | 7.40701 × 10−19 | 16 | O | 0.12588 | 0.14555 | 0.24440 | S782 | |
Stage-specific abundant proteins exhibiting a >1.5 fold difference in expression among intraerythrocytic stages. The representative proteins for each stage displayed the most significant p values. The columns corresponding to the ring average, trophozoite average, and schizont averages are the normalized intensities for peptides associated with each particular stage. Further characterization of the proteins can be found in Supplemental Table 1 in the Supporting Information.
Stage-specific abundant phosphorylation sites exhibiting a >1.5 fold difference in phosphorylation levels among intraerythrocytic stages. The proteins for each stage displayed the most significant p values. Site classes were assigned using a decision tree algorithm: A: acidic; B: basic; P: proline-directed; T: tyrosine; O: other. The columns listing the ring average, trophozoite average, and schizont averages are the normalized intensities for phosphorylated peptides associated with each stage. A more in-depth analysis of each phosphorylation event can be found in Supplemental Table 2 in the Supporting Information.
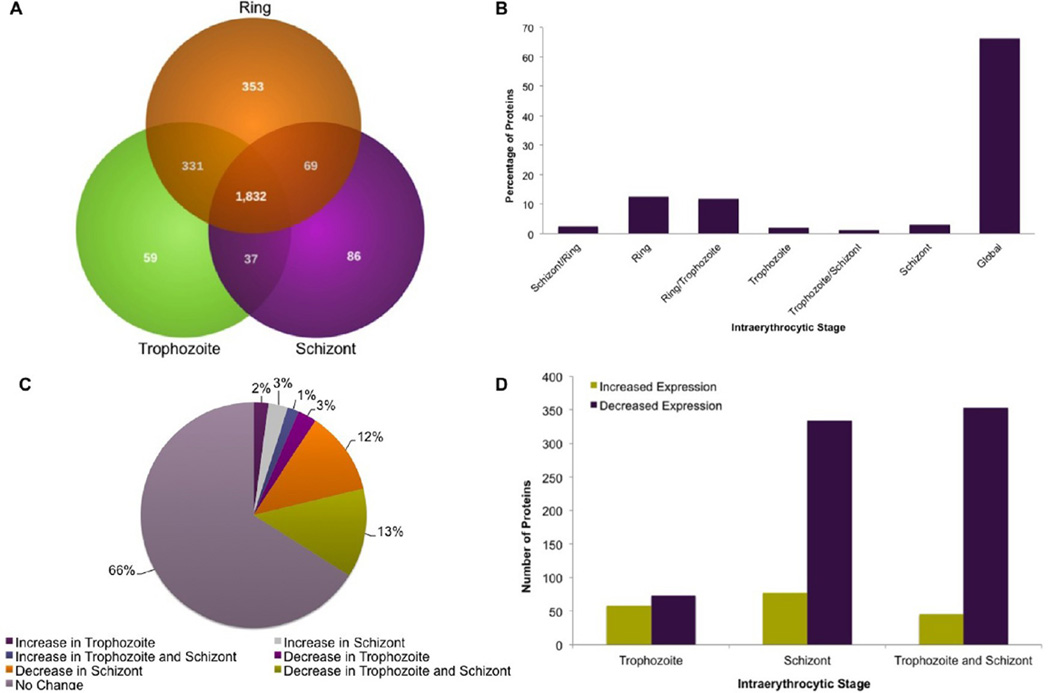
A total of 1832 proteins were globally expressed, while 935 showed a significant change in expression during development (Figure 2A). Of these, 353 (12.8%) proteins were enriched in rings, 59 (2.1%) were enriched in trophozoites, and 86 (3.1%) were enriched in schizonts (Figure 2B). The large proportion of proteins exhibiting variable expression across stages demonstrates that the P. falciparum proteome undergoes significant changes during the intraerythrocytic cell cycle. Next, we analyzed the expression pattern of the proteome that fluctuates across the intraerythrocytic cell cycle (Figure 2C,D). Among constitutively expressed proteins, ~65% are hypothetical or unclassified proteins, which is not significantly different from their prevalence in the predicted proteome.
Figure 2.
Comparative analysis of stage-specific protein expression. (A) Venn diagram depicting the distribution of identified malaria proteins over the intraerythrocytic stages. (B) Percentage of globally expressed proteins and proteins quantified at each of the intraerythrocytic stages. (C) Expression level changes as determined by a >1.5-fold difference. (D) Number of proteins up- and down-regulated in each stage in comparison with ring stage expression levels.
Stage-Specific Analysis of Functional Profiles
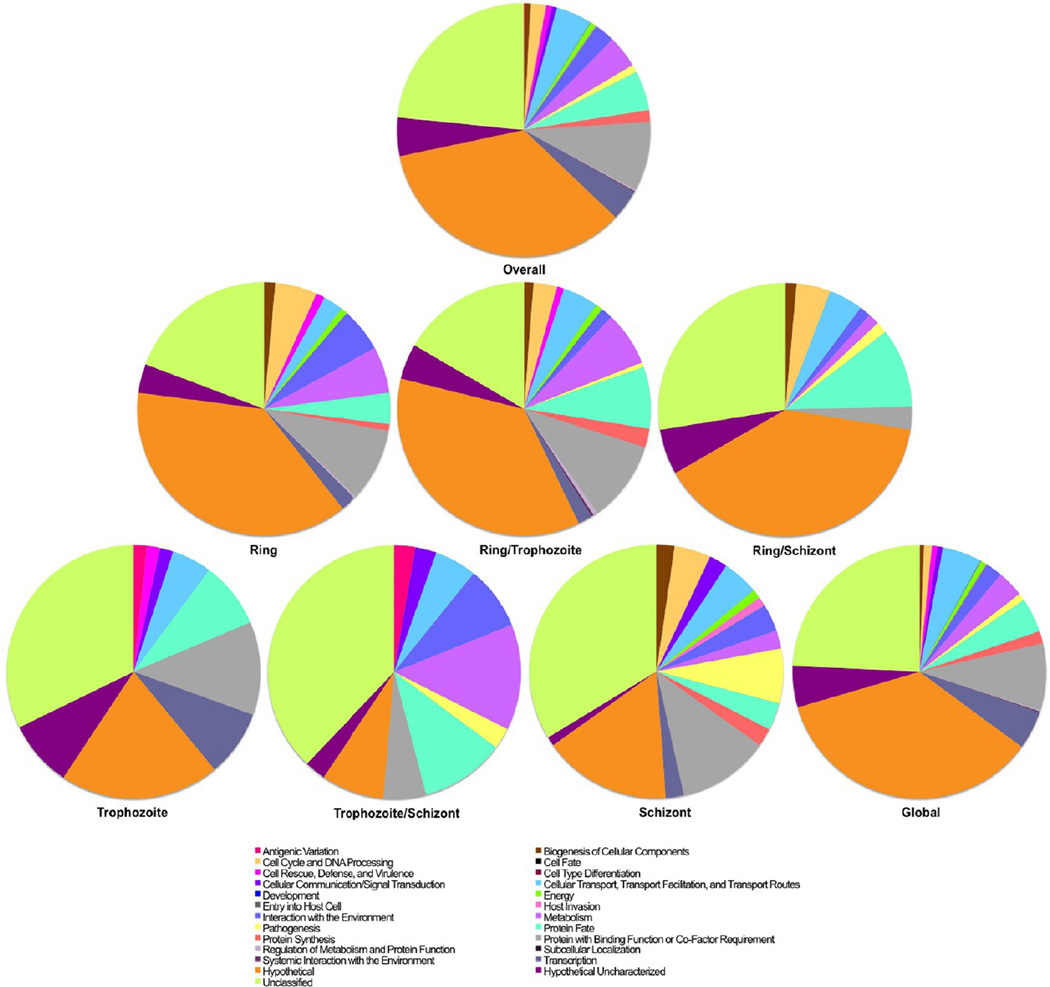
We sorted identified proteins into functional classes using the Munich Information Centre for Protein Sequences (MIPS) catalog, with some adaptations for classes specific to the parasite, such as proteins pertaining to malaria pathogenesis (Figure 3).45 The distribution of the functional profiles is summarized in Figure 3. Forty percent of the identified proteins were P. falciparum hypothetical proteins. We made an effort to categorize the hypothetical proteins into functional classes where possible based on sequence homology to orthologs in other organisms (Supplemental Figure S2 in the Supporting Information). Proteins belonging to important functional categories such as cell cycle and DNA processing (2%), cellular transport (5%), interaction with the environment (3%), metabolism (4%), protein fate (5%), protein with binding function (9%), and transcription (4%) account for 32% of the total proteins identified. Many of these protein classes are necessary for invasion, proliferation, defense, and cell communication, which are critical for pathogenesis. Functional classification revealed some marked differences between intraerythrocytic stages. More specifically, the “pathogenesis” functional category was not represented in the ring and trophozoite stage proteomes but reached ~7% in the schizont stage, although <1% of globally expressed proteins are classified in this category. This enrichment of pathogenesis-related proteins in schizonts may reflect high-level cytoadherence in this stage as well as preparation for egress from the red blood cells and invasion of new red bloods cells. Another fascinating observation involves the “transcription” functional category, which encompasses 2 and 2.3% of the ring and schizont stage proteomes, respectively, but is increased to 8.5% in trophozoites (p < 0.05, Chi-Squared Test). Interestingly, abundances of proteins implicated in “protein expression” are at their lowest during the trophozoite stage. This implies that the parasite emphasizes transcription over protein synthesis during the trophozoite stage.
Figure 3.
Stage-specific analysis of functional profiles. Functional profiles of expressed proteins using GO annotations downloaded from PlasmoDB (www.plasmodb.org) or UniProt (www.uniprot.org) as defined by the MIPS catalogue. To avoid redundancy, we assigned only one class per protein. The complete protein list is included in Supplementary Table 1 in the Supporting Information.
Proteins were further characterized by assigning GO SLIM terms using “Generic GO”12 (http://go.princeton.edu/cgi-bin/GOTermMapper). A Plasmodium-specific GO analysis for “Molecular Function” and “Cellular Component” was performed using collected PlasmoDB identifiers. The high percentage of proteins identified across multiple functional categories and cellular components demonstrates the depth and comprehensiveness of this analysis (Supplemental Figure S3a,b in the Supporting Information).
Developmental Stage-Specific Distribution of Phosphorylation Sites
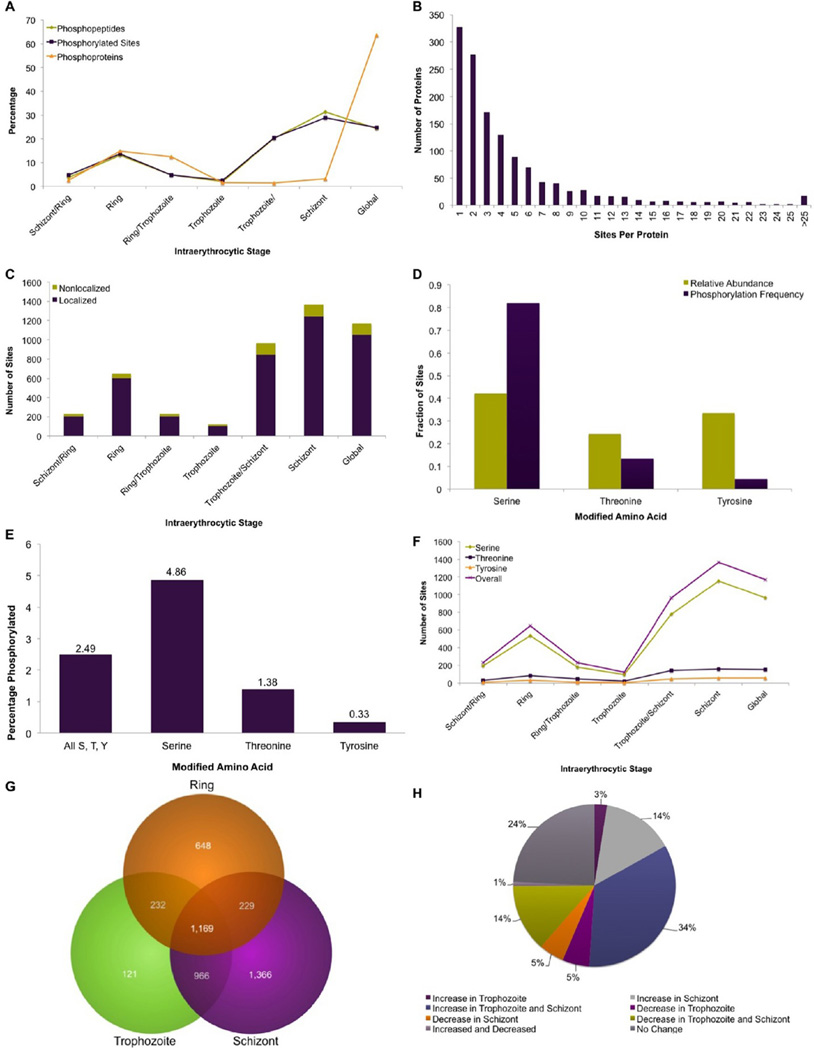
Because reversible protein phosphorylation is an important regulator of protein activity, we analyzed how protein phosphorylation fluctuates with the progression of the Plasmodium asexual life cycle. Table 1 includes examples of abundant stage-specific phosphorylation events with the most significant p values. Of the identified phosphoproteins, 452 were differentially expressed and 758 were globally expressed (Figure 1B). The numbers of phosphorylated peptides, unique phosphorylation sites, and total phosphoproteins varied among the intraerythrocytic stages (Figure 4A), reflecting the likely role of protein phosphorylation in regulating developmental progression. The highest numbers of phosphopeptides and phosphorylation sites were found in the schizont stage, while maximum expression of phosphoproteins was detected in rings. A similar trend has been found in the yeast centrosome phosphoproteome: only 54 phosphorylation sites were detected during the G1 stage, which is analogous to the ring stage in Plasmodium, but almost twice as many were detected during mitosis, which is comparable to the Plasmodium schizont stage.46
Figure 4.
Global analysis of stage-specific phosphorylation patterns. (A) Relative distributions of stage-specific phosphopeptides, phosphorylated sites, and phosphoproteins. Raw values are provided in Figure 1B. (B) Histogram depicting the numbers of phosphorylation sites identified for each phosphorylated protein. (C) Distribution of the number of localized and nonlocalized phosphorylation sites throughout the intraerythrocytic stages. (D) Relative abundance of serine, threonine, and tyrosine residues within all phosphoproteins compared with their frequency of phosphorylation. (E) Extent of phosphorylation of serine, threonine, and tyrosine residues within all phosphoproteins. (F) Stage-specific profile of the proportion of serine, threonine, and tyrosine residues phosphorylated in each of the intraerythrocytic stages. (G) Venn diagram depicting the distribution of identified malaria phosphorylation sites over the intraerythrocytic stages. (H) Phosphorylation level changes as determined by a >1.5 fold difference in comparison with observed ring stage levels.
We identified that 75% of phosphoproteins contained multiple phosphorylation sites (Figure 4B), with most being phosphorylated on three or more residues (Figure 4B). Phosphorylation on multiple residues suggests that a protein’s activity and interactions are regulated via phosphorylation at distinct sites. Phosphorylation sites were scored using the AScore algorithm to assess the confidence of site localization.35 Any site with an AScore above 13 was considered to be correctly localized. Overall, 90% of the sites were localized to a single amino acid (Figure 4C), with 87–93% of sites localized within each developmental stage.
Phosphorylation was most abundant on Ser (82%), followed by Thr (13.5%), and Tyr (4.5%). The enriched phosphorylation of Ser residues surpasses its relative abundance among all residues that are subject to phosphorylation (Figure 4D), indicating a strong bias toward Ser phosphorylation. The observed 4.5% tyrosine phosphorylation in this study is similar to the typical ~1–4% tyrosine phosphorylation seen in other organisms,14,47 which is somewhat surprising given the absence of true tyrosine kinases in Plasmodium.3,13,48–50 The observed 4.5% tyrosine phosphorylation is higher than that previously reported,3,4,18 at least in part because our data set includes the ring and trophozoite stages. Stage-specific tyrosine phosphorylation levels are moderate in rings (14.8%), decrease in trophozoites (2%), and increase to peak levels in schizonts (27.6%) (Figure 4F). Although many proteins were phosphorylated on multiple residues, only a small proportion of Ser, Thr, and Tyr residues within each phosphoprotein were modified (Figure 4E).
We detected 226 Tyr phosphorylated proteins (Supplemental Table S3 in the Supporting Information). Tyrosine phosphorylation of such a large number of Plasmodium proteins in the absence of true tyrosine kinase homologues underscores the importance of dual-specificity kinases in the parasite. Recent reports have identified tyrosine autophosphorylation in the activation loops of dual-specificity CMGC kinases PfGSK3 and PfCLK3.3,4 Tyr autophosphorylation activities by mammalian “dual-specificity tyrosine phosphorylation-regulated kinases” (DYRK) have been observed.51 Recently, the Plasmodium NIMA-like kinase Pfnek3 has also been shown to have tyrosine autophosphorylation activity.52 Characterization of Plasmodium kinases capable of Tyr phosphorylation will be an area of considerable interest.
We examined each phosphorylation site to define those with a >1.5-fold increase or decrease between stages. Figure 4F illustrates the stage-specific distribution of serine, threonine, and tyrosine and overall phosphorylation. It is interesting to note that although the overall phosphorylation of serine, threonine, and tyrosine residues varies greatly between the stages, the distribution of phosphorylation between these residues is relatively stable as the parasite progresses through the developmental cycle. Phosphorylation of 25% of the sites did not significantly fluctuate, while 75% of the sites were stage-dependent (Figure 4G,H). Our data indicate that each stage of the cycle displays a distinct phosphoproteome (Figure 4G,H). Peak protein phosphorylation was observed in the schizont stage, suggesting that reversible protein phosphorylation is presumably a regulatory feature of nuclear division and merozoite ontogeny. The importance of phosphorylation in merozoite invasion has been demonstrated previously.53,54 More recently, the phosphorylation of glideosome motor components and other proteins involved in parasite egress and invasion has been documented.18
Distribution of Phosphorylation Site Classes and Phosphorylation Motifs Across P. falciparum Stages and Phosphoproteins
To determine the kinase classes responsible for each modification, we used a decision tree approach to classify each site as acidic, basic, proline-directed, tyrosine, or other based on the identities of surrounding amino acids.14,25 Acidic sites were the most common (36%), followed by basic (29%), while tyrosine and proline-directed sites were the least common (4% each) (Figure 5A). Approximately 60% of phosphoproteins contained phosphorylation sites from multiple kinase classes, whereas 2% of phosphoproteins were predicted to be targets for all kinase classes (Figure 5B). Variations were detected in the frequencies of these classes across developmental stages, suggesting that distinct kinases operate preferentially in various stages. Overall, sites within all classes were least abundant during the early stages (ring and trophozoite), where phosphorylation levels were the lowest, and increased drastically as the parasite progressed into the schizont stage (Figure 5C). Proline-directed sites that are utilized by CDKs and MAP kinases had the most significant increase at the schizont stage (Figure 5C).
Figure 5.
Stage-specific distribution of phosphorylation site classes and motifs. Phosphorylation sites were categorized as acidic, basic, proline-directed, tyrosine, or other as previously described.14 (A) Overall percentage of each site class and percentage of each throughout the intraerythrocytic cell cycle of P. falciparum. (B) Proportion of phosphoproteins containing phosphorylation sites from only one site class, from two, three, or four different site classes, or from all five different site classes. (C) Proportion of residues phosphorylated in each site class through the intraerythrocytic stages. (D) Comparison of phosphorylation across the P. falciparum intraerythrocytic stages for motifs containing serine, threonine, arginine, asparagine, glycine, and proline residues. (E) Table identifying potential Ser/Thr kinases responsible for observed phosphorylation on representative putative substrates.
Our analysis suggests that P. falciparum A/T codon bias has led to unique phosphorylation motifs that direct its kinase specificity. The phosphoproteomic data enabled us to identify parasite-specific phosphorylation motifs using the MotifX algorithm.55 A total of 43 distinct phosphorylation motifs were identified (Supplemental Table S4 in the Supporting Information). Of these, 33 motifs related to phosphorylated Ser residues and 10 motifs to phosphorylated Thr residues. Interestingly, some of the identified motifs are unusual. For example, there are 14 motifs with prominent Asn residues, as indicated by a previous analysis of the schizont phosphoproteome.3 The overall trends of phosphorylation for the identified Ser-, Thr-, Arg-, Asn-, Gly-, and Pro-residue-containing motifs resemble the general trend of phosphorylation. One notable difference is the substantial increase in phosphorylation at the trophozoite and schizont stages for proline-containing phosphorylation motifs (Figure 5D).
We predict possible kinase–substrate relationships by comparing phosphorylated peptide sequences to protein Ser/Thr kinase consensus phosphorylation sequences. Figure 5E lists five putative substrates with consensus phosphorylation sequences for known human Ser/Thr kinases. In addition to proline-directed CDK-like motifs, we found motifs similar to those for mammalian GSK3, TGF-βR1, and numerous other candidates. We also identified some previously undescribed motifs that are likely to be specific for as-yet unknown kinases.
Phosphorylation Profiles of Putative MAPK/CDK Substrates and Tyrosine Phosphorylated Proteins
The intraerythrocytic development of the malaria parasite diverges from the eukaryotic cell cycle paradigm that has emerged largely from studies in yeast cells. The six identified CDK-related PKs56,57 are likely key regulators of cell-cycle progression. To validate this prediction and gain insight into physiological functions of Plasmodium CDK-related kinases, it is necessary to identify their cellular substrates. CDKs are proline-directed serine/threonine kinases having a strong requirement for basic amino acids, Arg and Lys, in particular, at the +3 site.58 Filtering 206 proline-directed Ser/Thr phosphorylated proteins for those with the strong CDK consensus pS/T-P-X-K/R motif identified 45 proteins that are putative Plasmodium CDK substrates (Supplemental Table S5 in the Supporting Information). Among the proteins that may be phosphorylated by Plasmodium CDKs are homologues of origin recognition complex subunit 2 (Orc2) (MAL7P1.21), Orc4 (PF13_0189), and Orc1/CDC6 (PFE0155w), components of the prereplication complex that are known CDK substrates in other eukaryotes. Additional prereplicative complex proteins that are phosphorylated at the weak CDK consensus motif (pS/T-P) are MCM4 and MCM5 (Supplemental Table S5 in the Supporting Information). Mammalian ORC and MCM subunits are phosphorylated by CDKs to prevent helicase loading beyond G1 phase of the cell cycle, thus preventing rereplication.59,60 Other proteins of significance in potentially regulating the parasite’s DNA replication, transcription, or mitosis that are phosphorylated at the strong CDK motif are homologues of cell division cycle protein 48 (PFF0940c) and regulator of chromosome condensation (MAL7P1.38).
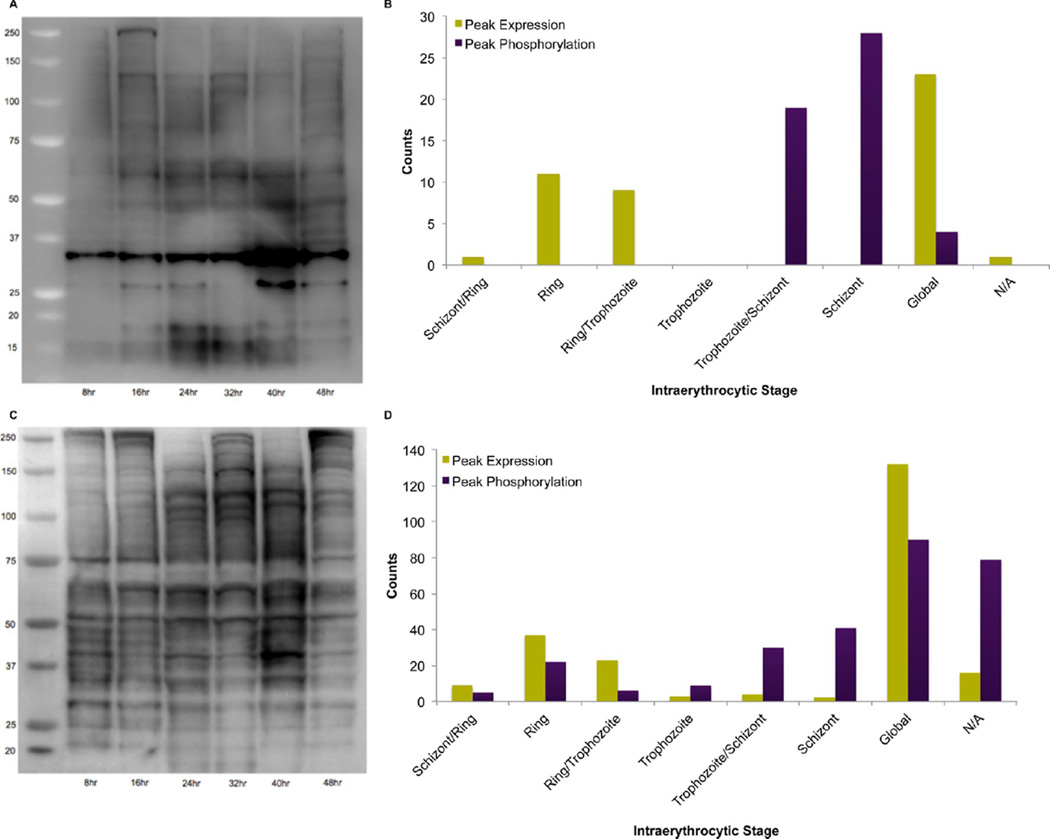
Developmental stage-specific protein phosphorylation profile analysis using phospho-MAPK/CDK substrates antibody demonstrated minimal phosphorylation in the ring stage and maximal phosphorylation in the schizont stage (Figure 6A). This correlates well with our protein phosphorylation analysis of the 45 putative MAPK/CDK substrates, concurring that prolinedirected kinase activity is predominant in late stages of the asexual cycle (Figure 6B). Using an antibody-based pull-down of phospho-motif peptides (PTMScan Direct Cell Signaling Technology) in duplicate experiments, we identified a total of 48 unique phosphorylation events (Supplemental Table S6 in the Supporting Information). Significantly, the PTMScan IAP identified two phosphorylation events, homologues of cell division cycle protein 48 (PFF0940c) (S512) and regulator of chromosome condensation (MAL7P1.38) (S602), that were predicted MAPK/CDK substrates from the global phosphoproteomic analysis. Furthermore, seven of the identified phosphopeptides identified in the PTMScan IAP were also identified in the global phosphoproteomic analysis (Supplemental Table S6 in the Supporting Information).
Figure 6.
Phosphorylation profiles of putative MAPK/CDK substrates and tyrosine phosphorylated proteins. (A) Immunoblot analysis using a MAPK/CDK phospho-specific antibody on 8 h time point lysates. (B) Analysis of 45 putative MAPK/CDK substrates revealing the number of proteins with peak expression in each stage and the number of phosphorylation events peaking at each stage. (C) Immunoblot analysis using a phospho-tyrosine antibody on 8 h time point lysates. (D) Stage-specific distribution of proteins containing tyrosine phosphorylation events.
We used a phospho-tyrosine specific monoclonal antibody (P-Tyr-100) for Western blot analysis of stage-specific extracts (Figure 6C) to demonstrate that tyrosine phosphorylation events also fluctuate with cell-cycle progression (Figure 6D). We examined the sensitivity of the parasite to the common tyrosine kinase inhibitors Genistein, Herbimycin, PP1, and PP2. The observed IC50 values were as follows: Genistein 83 uM, Herbimycin 0.3 uM, PP1 2.4 uM, and PP2 0.9 uM (unpublished data), which reveals that P. falciparum is quite sensitive to tyrosine kinase inhibitors. Herbimycin and PP2, inhibitors of Src-family kinases, exhibited submicromolar potency. Recent data implicating host erythrocyte signaling pathways in parasite survival61 suggest that some of these effects may be mediated by host tyrosine kinases. Our data suggest that tyrosine kinases may prove to be promising targets for future malaria therapeutics development.
Subdomain Distribution of Phosphorylation Sites within Protein Kinases
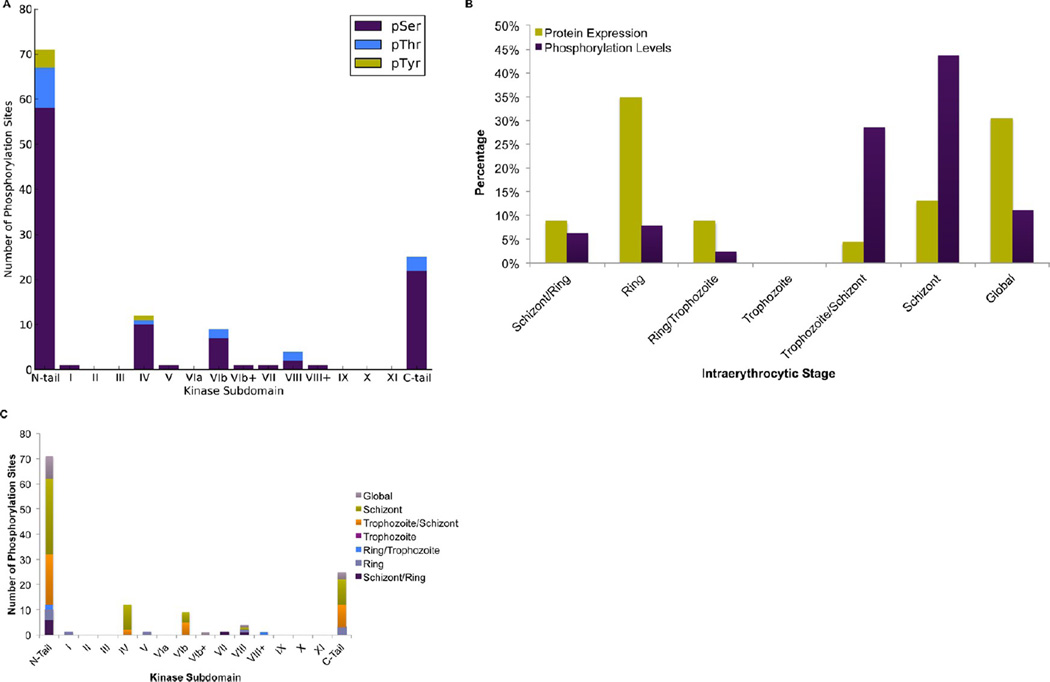
Because a common regulatory mechanism of eukaryotic phospho-signaling pathways is the autophosphorylation of key regions of the kinase domain itself, we investigated the subdomain distribution of phosphorylation in Plasmodium kinases. Thirty of the phosphorylated proteins from our phosphoproteomic data set were identified as ePKs. In these proteins, we identified the location of each of the 126 observed phosphorylation sites, whether in the kinase domain, the N-terminal tail, or the C-terminal tail (Supplemental Table S7 in the Supporting Information). Those in the kinase domain were further assigned to a subdomain location (Figure 7A). The majority of phosphorylation sites, 71 in total, appear in the N-terminal extension of the kinase domain, while 25 appear in the C-terminal tail and 30 appear in the kinase domain itself.
Figure 7.
Subdomain location of phosphorylation sites in protein kinases. (A) Subdomain location of 126 phosphorylation sites observed on 30 eukaryotic protein kinases. Bar height indicates the number of observed phosphorylation sites relative to the kinase domain, whether in the domain, the N-terminal tail, or the C-terminal tail. Those in the kinase domain were further assigned to a subdomain location. (B) Stage-specific comparison of eukaryotic protein kinase expression levels and phosphorylation levels. (C) Histogram depicting the stage-specific distribution of the number of phosphorylation sites at a given time during the intraerythrocytic cell cycle for each of the kinase subdomains as well as the N-terminal tail and C-terminal tail.
When examining the stage-specific expression profile of the 30 identified ePKs, we observed peak expression in the ring stage, whereas their phosphorylation peaks in schizonts (Figure 7B). Figure 7C depicts the stage-specific distribution of the number of phosphorylation sites during the cycle for each of the kinase subdomains as well as the N-terminal and C-terminal tails.
The PK subdomains VII and VIII contain the activation loop, a structural region whose phosphorylation is a key step in the activation of many kinases.62 We observed activation loop phosphorylation in three kinases: PfCDPK1 (PFB0815w), PfCRK3 (PFD0740w), and an orphan kinase (PF11_0227) (Supplemental Table S7 in the Supporting Information). Of these, the single phosphorylation sites on PfCDPK1 and PfCRK3 are consistent with a typical mechanism of kinase activation by phosphorylation of the activation loop, while the orphan kinase PF11_0227 contains a greatly extended activation loop of 135 amino acids encompassing three phosphorylation sites (Supplemental Table S7 in the Supporting Information), suggesting the possibility of a novel regulatory elaboration.
Phosphorylation in sequence regions between the defined subdomains was observed in two kinases, FIKK4.1 (PFD1165w)63 and PfPK1 (PF08_0044), which are related to glycogen synthase kinases (GSK3)37,64 (Figure 7A). The N-terminal region of subdomain VIb, which includes the catalytic loop and beta-7 strand, was phosphorylated in two other kinases, PfSRPK1 (PFC0105w) at six sites and the orphan kinase PF14_0392 at three sites (Supplemental Table S8 in the Supporting Information). In PfSRPK1, this is the site of a large insert known as the spacer domain, which influences localization of the protein in yeast and human SRPKs.65 Given that the spacer region itself is not required for PfSRPK1 activity,66 its extensive phosphorylation within PfSRPK1 may instead be a mechanism controlling localization or binding.
While tyrosine phosphorylation is rare overall in the P. falciparum phosphoproteome,3–5 those occurrences are noteworthy because they may point to novel parasitic signaling pathways or mechanisms. We observed only one phosphorylated tyrosine site within the kinase domain, in the β-4 sheet of subdomain IV of PfCRK4 (PFC0755c) (Supplemental Table S7 in the Supporting Information). PfCRK4 was also phosphorylated on serine or threonine residues at six other sites in subdomain IV. Four more instances of tyrosine phosphorylation were observed in the N-terminal tails of PfCRK4, PfCRK1 (PFD0865c), PfPKG (PF14_0346), and the orphan kinase PF14_0392.
Preliminary Identification of Kinases Responsible for Kinase Domain Phosphorylation
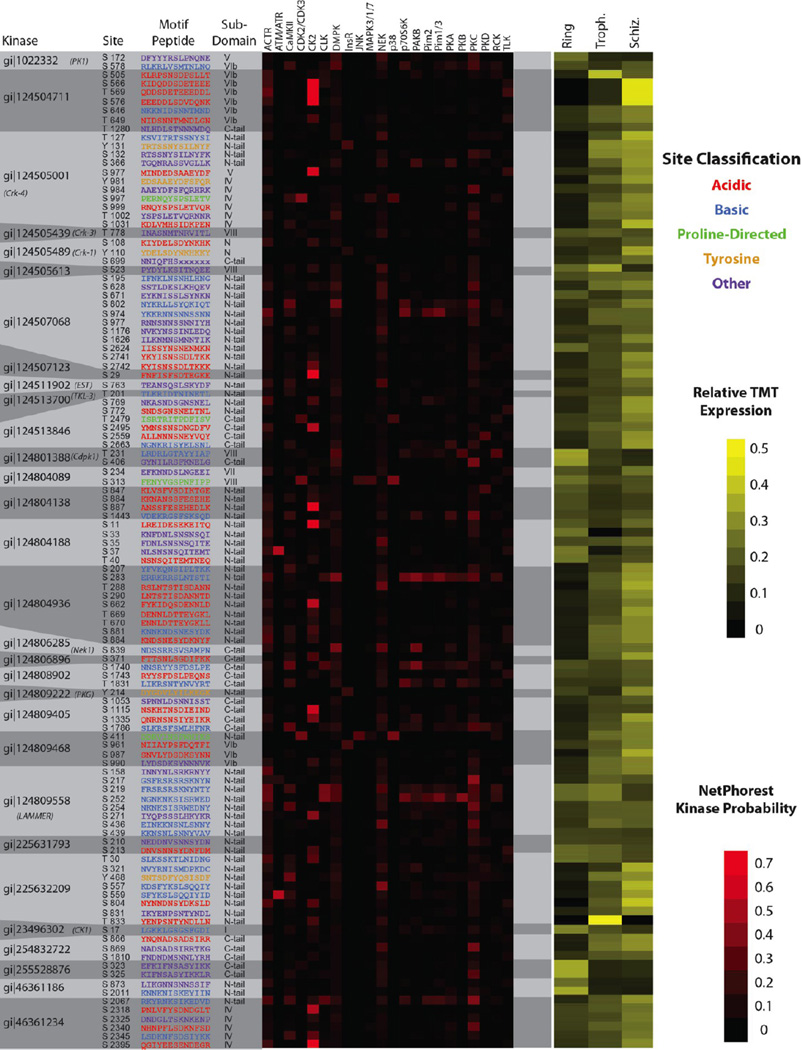
Because phosphorylation events govern the activities of many kinases, identifying the kinases responsible can reveal the phosphorylation networks that regulate Plasmodium biology. We used the NetPhorest algorithm39 to identify candidate kinases that may be responsible for the 126 phosphorylation events we observed upon 30 Plasmodium kinases. The 2.5% most likely kinase–substrate pairings are depicted in Figure 8 for all phosphorylation sites observed on Plasmodium kinases in conjunction with structural and quantitative data. Although such predictions on their own do not prove particular kinase–substrate relationships, they nevertheless provide working hypotheses that can be tested experimentally.
Figure 8.
Associating kinase subdomain phosphorylation events with probable kinases Those localized phosphorylation events that mapped to known kinase subdomains were analyzed using NetPhorest to identify the kinase or kinase groups most likely to phosphorylate each site based on neighboring sequences.39 A total of 22 kinases and kinase groups accounted for the most confident 2.5% of all predicted kinase-site relationships and are mapped above. The red heat map displays the predicted confidence that each kinase or kinase family would phosphorylate the sequence in question; the yellow heat map displays relative levels of each phosphorylation site throughout three stages of the Plasmodium life cycle. Kinase abbreviations: ACTR, ACTR2_ACTR2B_TGFbR2_group; ATM/ATR, ATM_ATR_group; CaMKII, CaMKIIbeta_CaMKIIgamma_group; CDK2/CDK3, CDK2_CDK3_group; CK2, CK2_group; CLK, CLK_group; DMPK, DMPK_group; InsR, InsR_group; JNK, JNK_group; MAPK3/1/7, MAPK3_MAPK1_MAPK7_NLK_group; NEK, NEK1_NEK5_NEK3_NEK4_NEK11_NEK2_group; p38, p38_group; p70S6K, p70S6K_group; PAKB, PAKB_group; Pim2, Pim2; Pim1/3, Pim3_Pim1_group; PKA, PKA_group; PKB, PKB_group; PKC, PKC_group; PKD, PKD_group; RCK, RCK_group; TLK, TLK_group.
Although 22 kinases and kinase families were associated with the 126 kinase phosphorylation sites, the most confident predictions implicated kinases of the CK2 family (similar to casein kinase 2). Sites on nearly a dozen kinases were located in highly acidic regions favorable for CK2 activity. Finally, we observed a single phosphorylation site on NEK1, which was scored as a candidate substrate for DMPK, PAKB, or a NEK-family kinase: thus, this site could potentially be a case of autophosphorylation.
Peak Protein Expression and Phosphorylation are Unlinked
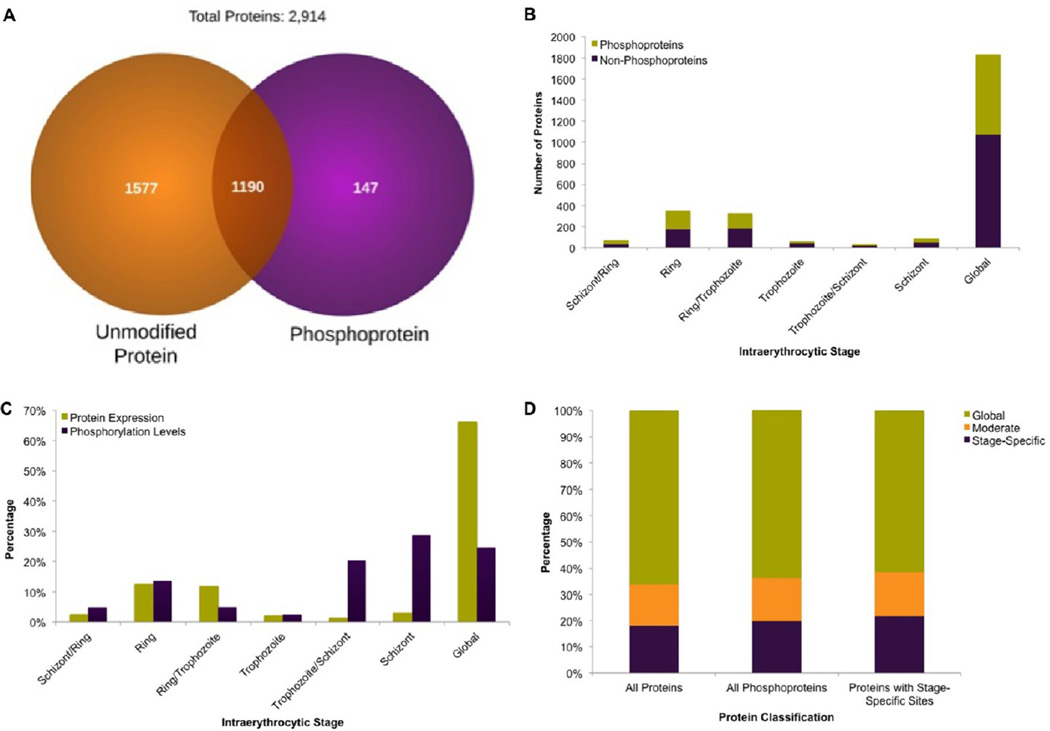
Differential levels of phosphopeptide detection can arise from changes in protein abundance or from genuine changes in the phosphorylation occupancy at the target site. To differentiate these two possibilities, we compared the proteomic data of the intraerythrocytic stages to our phosphoproteomic analysis. Overall, we identified 2914 proteins, 40% of which were identified both with and without phosphopeptide enrichment (Figure 9A). A similar overlap was observed across developmental stages (Figure 9B). The proteins that are identified solely on the basis of phosphorylation data are those of low abundance.
Figure 9.
Comparison of protein and phosphoprotein expression across P. falciparum intraerythrocytic stages. (A) Comparison of proteins identified with and without phosphopeptide enrichment. (B) Numbers of proteins and phosphoproteins with peak expression at each of the intraerythrocytic stages. (C) Stage-specific comparison of protein expression and phosphorylation levels. (D) Bar chart reflecting proportions of proteins with global, moderate. and stage-specific expression in an unmodified form, modified form, and those containing stage-specific phosphorylation sites.
Phosphorylated and nonphosphorylated protein profiles varied throughout the asexual cell cycle (Figure 9C). 75% of proteins with stage-specific phosphorylation sites were expressed across multiple intraerythrocytic stages in nonphosphorylated form (Figure 9D). In general, protein expression and phosphorylation display obvious differences throughout the P. falciparum intraerythrocytic cycle. Malaria parasite protein expression can be classified as a global event (66.2%) more frequently than is observed for protein phosphorylation (25%). Among those proteins that are variably expressed across developmental stages, a significant number attained peak expression during ring stage; in contrast, phosphorylation peaks in the schizont stage, demonstrating that peak phosphorylation events are generally not due to stage-specific protein expression.
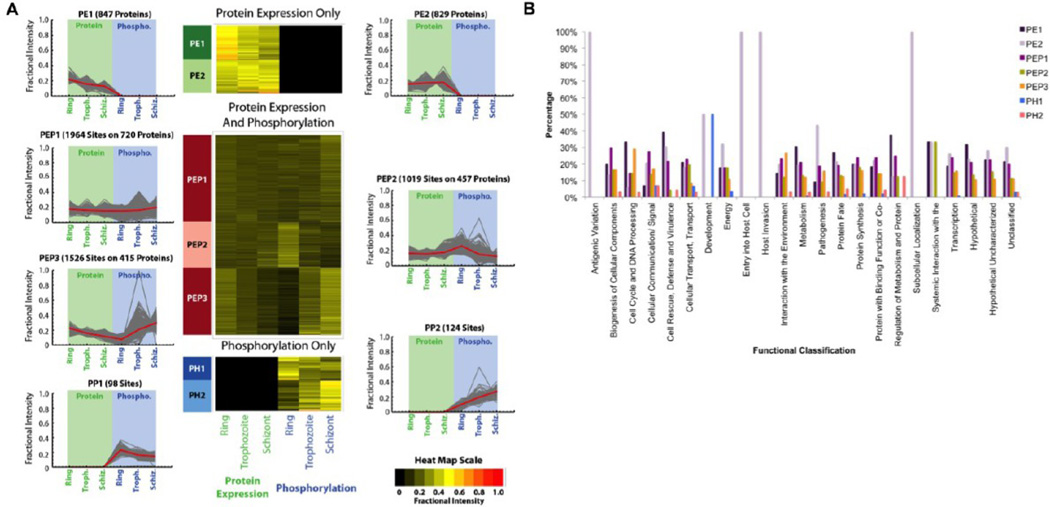
Next, proteins and phosphorylation sites that showed statistically significant differences in protein abundance, phosphorylation levels, or both across the three growth stages were classified via K-medoids clustering into groups with similar patterns of both protein expression and phosphorylation. The silhouette method was then used to determine the optimal number of clusters for the data, seven clusters in our case. Figure 10A summarizes numbers of proteins and phosphorylation sites that were assigned to each of the 7 clusters, accompanied by trends observed for proteins and sites in each group. Also, heat maps were generated for each cluster to illustrate changes in protein expression and phosphorylation. The results confirm that protein abundance and phosphorylation levels change independently. There are relatively few cases where both change in tandem, while there are many cases where phosphorylation increases while protein abundance stays the same or decreases. For example, in cluster PEP3, there are 1526 phosphorylation sites with associated protein measurements. Members of this cluster generally showed increasing levels of phosphorylation, while protein levels decreased (Figure 10A).
Figure 10.
K-medoids cluster analysis of differential proteins. (A) Proteins and phosphorylation sites that showed statistically significant differences across the three growth stages in protein abundance, phosphorylation levels, or both were classified via K-medoids clustering. The numbers of proteins and phosphorylation sites that were assigned to each cluster as well as the trends observed for protein expression and phosphorylation are depicted. (B) Functional profiles of proteins in each cluster using GO annotations downloaded from PlasmoDB (www.plasmodb.org) or UniProt (www.uniprot.org) as defined by the MIPS catalogue. The complete protein list is included in Supplementary Table 7 in the Supporting Information.
To further characterize the biological significance of the observed protein clustering, we examined the functional classes of all of the proteins in each cluster using the MIPS catalogue (Supplemental Table S8 in the Supporting Information). Interestingly, some of the seven clusters show a significant enrichment in functional classes. When applying a chi-squared test and determining a two-tailed p value for each cluster, the distribution of functional classes proved to be highly nonrandom. This signifies that proteins following similar expression and phosphorylation trends also possess similar functional roles (Figure 10B). A remarkable trend can be seen for Cluster PE2 functional analysis. Cluster PE2 proteins show a slight increase in protein expression in the schizont stage, while the parasite prepares for egress from the erythrocyte and invasion into a new host cell. Supporting this is the fact that 100% of proteins in the host invasion, entry into host cell, and antigenic variation functional categories are members of Cluster PE2. Another interesting trend can be seen for Cluster PEP3, which comprises a significant number of cell-cycle regulatory proteins. Cluster PEP3 shows a dramatic increase in protein phosphorylation in the schizont stage. This is the period in the Plasmodium developmental cycle where multiple rounds of DNA replication are ongoing. The fact that ~29% of proteins in the cell cycle and DNA processing functional category are members of Cluster PEP3 demonstrates the involvement of phosphorylation in the control of P. falciparum cell-cycle progression.
CONCLUSIONS
A major challenge in the postgenome sequencing era is to understand the cellular roles of the vast number of proteins whose function is not deducible from sequence analysis. Achieving a comprehensive functional understanding of the Plasmodium falciparum proteome is particularly challenging given that 60% of putative proteins have no known orthologs in other systems. Many Plasmodium proteins, including PKs, are atypical and possess hybrid features;67 thus, this work brings a significant contribution to the ongoing global and multipronged efforts at implementing functional genomics studies of Plasmodium falciparum, the end goal of which is to facilitate the discovery of novel targets for intervention. Our phosphoproteomic analysis also represents a step toward elucidation of kinase–substrate pairs that will eventually form the basis of a comprehensive picture of Plasmodium signaling networks. In view of the phylogenetic divergence of malaria parasites relative to model eukaryotes such as yeast and mammalian cells, this is likely to shed light from a different angle on a domain that is of central importance in biology. Data from this study are available at http://plasmodb.org/plasmo/.
Supplementary Material
Acknowledgments
This work is supported by an NIH/NIAID grant AI73795 (to DC and RC).
Footnotes
ASSOCIATED CONTENT
Supporting Information
Summary table of proteins identified in the intraerythrocytic stages of P. falciparum. Distribution of identified human versus malaria proteins and phosphorylation sites. Phosphorylated peptides discovered during the P. falciparum intraerythrocytic stages. Functional profile of hypothetical proteins Gene ontology term analysis of intraerythrocytic stages: Enrichment for GO “Molecular Function” terms of protein detected in P. falciparum intraerythrocytic stages and enrichment for GO “Cellular Component” terms of proteins detected in the intraerythrocytic stages. Tyrosine phosphorylation events. MotifX analysis of phosphorylation events. Putative CDK substrates identified in phosphoproteomic data set. MAPK/CDK PTMScan immunoprecipitation. Distribution of phosphorylation events within eukaryotic protein kinases. K-Medoids clustering of protein and phosphorylation events. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Gallup JL, Sachs JD. The economic burden of malaria. Am. J. Trop. Med. Hyg. 2001;64(1–2 Suppl):85–96. doi: 10.4269/ajtmh.2001.64.85. [DOI] [PubMed] [Google Scholar]
- 2.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe. 2011;10(4):410–419. doi: 10.1016/j.chom.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Solyakov L, Halbert J, Alam MM, Semblat JP, Dorin-Semblat D, Reininger L, Bottrill AR, Mistry S, Abdi A, Fennell C, Holland Z, Demarta C, Bouza Y, Sicard A, Nivez MP, Eschenlauer S, Lama T, Thomas DC, Sharma P, Agarwal S, Kern S, Pradel G, Graciotti M, Tobin AB, Doerig C. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat. Commun. 2011;2:565. doi: 10.1038/ncomms1558. [DOI] [PubMed] [Google Scholar]
- 5.Lasonder E, Treeck M, Alam M, Tobin AB. Insights into the Plasmodium falciparum schizont phospho-proteome. Microbes Infect. 2012;14:811–819. doi: 10.1016/j.micinf.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, Yates JR, 3rd, Winzeler EA. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14(11):2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foth BJ, Zhang N, Mok S, Preiser PR, Bozdech Z. Quantitative protein expression profiling reveals extensive posttranscriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol. 2008;9(12):R177. doi: 10.1186/gb-2008-9-12-r177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuss C, Gan CS, Gunalan K, Bozdech Z, Sze SK, Preiser PR. Quantitative proteomics reveals new insights into erythrocyte invasion by Plasmodium falciparum. Mol. Cell. Proteomics. 2011;11 doi: 10.1074/mcp.M111.010645. 10.1074/mcp.M111.010645–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oehring SC, Woodcroft BJ, Moes S, Wetzel J, Dietz O, Pulfer A, Dekiwadia C, Maeser P, Flueck C, Witmer K, Brancucci NM, Niederwieser I, Jenoe P, Ralph SA, Voss TS. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 2012;13(11):R108. doi: 10.1186/gb-2012-13-11-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ. A proteomic view of the Plasmodium falciparum life cycle. Nature. 2002;419(6906):520–526. doi: 10.1038/nature01107. [DOI] [PubMed] [Google Scholar]
- 11.Lasonder E, Ishihama Y, Andersen JS, Vermunt AM, Pain A, Sauerwein RW, Eling WM, Hall N, Waters AP, Stunnenberg HG, Mann M. Analysis of the Plasmodium falciparum proteome by high-accuracy mass spectrometry. Nature. 2002;419(6906):537–542. doi: 10.1038/nature01111. [DOI] [PubMed] [Google Scholar]
- 12.Lasonder E, Janse CJ, van Gemert GJ, Mair GR, Vermunt AM, Douradinha BG, van Noort V, Huynen MA, Luty AJ, Kroeze H, Khan SM, Sauerwein RW, Waters AP, Mann M, Stunnenberg HG. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog. 2008;4(10):e1000195. doi: 10.1371/journal.ppat.1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson-Grady JT, Villen J, Gygi SP. Phosphoproteome analysis of fission yeast. J. Proteome Res. 2008;7(3):1088–1097. doi: 10.1021/pr7006335. [DOI] [PubMed] [Google Scholar]
- 16.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 2002;20(3):301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 17.Wu Y, Nelson MM, Quaile A, Xia D, Wastling JM, Craig A. Identification of phosphorylated proteins in erythrocytes infected by the human malaria parasite Plasmodium falciparum. Malar. J. 2009;8:105. doi: 10.1186/1475-2875-8-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lasonder E, Green JL, Camarda G, Talabani H, Holder AA, Langsley G, Alano P. The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase A signaling. J. Proteome Res. 2012;11(11):5323–5337. doi: 10.1021/pr300557m. [DOI] [PubMed] [Google Scholar]
- 19.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75(8):1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 20.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3(12):1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Nirmalan N, Sims PF, Hyde JE. Quantitative proteomics of the human malaria parasite Plasmodium falciparum and its application to studies of development and inhibition. Mol. Microbiol. 2004;52(4):1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim CC, Wilson EB, DeRisi JL. Improved methods for magnetic purification of malaria parasites and haemozoin. Malar. J. 2010;9:17. doi: 10.1186/1475-2875-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 1979;65(3):418–420. [PubMed] [Google Scholar]
- 24.Prieto JH, Koncarevic S, Park SK, Yates J, 3rd, Becker K. Large-scale differential proteome analysis in Plasmodium falciparum under drug treatment. PLoS One. 2008;3(12):e4098. doi: 10.1371/journal.pone.0004098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Villen J, Gygi SP. The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 2008;3(10):1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8(11):937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 28.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38(Database issue):D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4(7):1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 30.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4(3):207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 31.Beausoleil A, Desjardins P, Rochefort A. Effects of long jumps, reversible aggregation, and Meyer-Neldel rule on submonolayer epitaxial growth. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2008;78(2 Pt 1):021604. doi: 10.1103/PhysRevE.78.021604. [DOI] [PubMed] [Google Scholar]
- 32.Nesvizhskii AI, Aebersold R. Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 2005;4(10):1419–1440. doi: 10.1074/mcp.R500012-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23(1):94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 34.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 35.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 36.Neuwald AF. Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics. 2009;25(15):1869–1875. doi: 10.1093/bioinformatics/btp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talevich E, Mirza A, Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol. Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–596. [PubMed] [Google Scholar]
- 39.Miller ML, Jensen LJ, Diella F, Jorgensen C, Tinti M, Li L, Hsiung M, Parker SA, Bordeaux J, Sicheritz-Ponten T, Olhovsky M, Pasculescu A, Alexander J, Knapp S, Blom N, Bork P, Li S, Cesareni G, Pawson T, Turk BE, Yaffe MB, Brunak S, Linding R. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signaling. 2008;1(35):ra2. doi: 10.1126/scisignal.1159433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, Winzeler EA. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- 41.Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1(1):E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shock JL, Fischer KF, DeRisi JL. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 2007;8(7):R134. doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H, Zhao Q, Singla LD, Min J, He S, Cong H, Li Y, Su C. Differential proteomic profiles from distinct Toxoplasma gondii strains revealed by 2D-difference gel electrophoresis. Exp. Parasitol. 2013;133:376–382. doi: 10.1016/j.exppara.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Sam-Yellowe TY, Florens L, Johnson JR, Wang T, Drazba JA, Le Roch KG, Zhou Y, Batalov S, Carucci DJ, Winzeler EA, Yates JR., 3rd A Plasmodium gene family encoding Maurer’s cleft membrane proteins: structural properties and expression profiling. Genome Res. 2004;14(6):1052–1059. doi: 10.1101/gr.2126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30(1):31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keck JM, Jones MH, Wong CC, Binkley J, Chen D, Jaspersen SL, Holinger EP, Xu T, Niepel M, Rout MP, Vogel J, Sidow A, Yates JR, 3rd, Winey M. A cell cycle phosphoproteome of the yeast centrosome. Science. 2011;332(6037):1557–1561. doi: 10.1126/science.1205193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan CS, Bodenmiller B, Pasculescu A, Jovanovic M, Hengartner MO, Jorgensen C, Bader GD, Aebersold R, Pawson T, Linding R. Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signaling. 2009;2(81):ra39. doi: 10.1126/scisignal.2000316. [DOI] [PubMed] [Google Scholar]
- 48.Peixoto L, Chen F, Harb OS, Davis PH, Beiting DP, Brownback CS, Ouloguem D, Roos DS. Integrative genomic approaches highlight a family of parasite-specific kinases that regulate host responses. Cell Host Microbe. 2010;8(2):208–218. doi: 10.1016/j.chom.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tewari R, Straschil U, Bateman A, Bohme U, Cherevach I, Gong P, Pain A, Billker O. The systematic functional analysis of Plasmodium protein kinases identifies essential regulators of mosquito transmission. Cell Host Microbe. 2010;8(4):377–387. doi: 10.1016/j.chom.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkes JM, Doerig C. The protein-phosphatome of the human malaria parasite Plasmodium falciparum. BMC Genomics. 2008;9:412. doi: 10.1186/1471-2164-9-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aranda S, Laguna A, de la Luna S. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J. 2011;25(2):449–462. doi: 10.1096/fj.10-165837. [DOI] [PubMed] [Google Scholar]
- 52.Low H, Chua CS, Sim TS. Plasmodium falciparum possesses a unique dual-specificity serine/threonine and tyrosine kinase, Pfnek3. Cell. Mol. Life Sci. 2012;69:1523–1535. doi: 10.1007/s00018-011-0888-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamaoki T. Use and specificity of staurosporine, UCN-01, and calphostin C as protein kinase inhibitors. Methods Enzymol. 1991;201:340–347. doi: 10.1016/0076-6879(91)01030-6. [DOI] [PubMed] [Google Scholar]
- 54.Ward GE, Fujioka H, Aikawa M, Miller LH. Staurosporine inhibits invasion of erythrocytes by malarial merozoites. Exp. Parasitol. 1994;79(3):480–487. doi: 10.1006/expr.1994.1109. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005;23(11):1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 56.Doerig C, Endicott J, Chakrabarti D. Cyclin-dependent kinase homologues of Plasmodium falciparum. Int. J. Parasitol. 2002;32(13):1575–1585. doi: 10.1016/s0020-7519(02)00186-8. [DOI] [PubMed] [Google Scholar]
- 57.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5(1):79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang EJ, Begum R, Chait BT, Gaasterland T. Prediction of cyclin-dependent kinase phosphorylation substrates. PloS One. 2007;2(7):e656. doi: 10.1371/journal.pone.0000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vas A, Mok W, Leatherwood J. Control of DNA rereplication via Cdc2 phosphorylation sites in the origin recognition complex. Mol. Cell. Biol. 2001;21(17):5767–5777. doi: 10.1128/MCB.21.17.5767-5777.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen S, Bell SP. CDK prevents Mcm2–7 helicase loading by inhibiting Cdt1 interaction with Orc6. Genes Dev. 2011;25(4):363–372. doi: 10.1101/gad.2011511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sicard A, Semblat JP, Doerig C, Hamelin R, Moniatte M, Dorin-Semblat D, Spicer JA, Srivastava A, Retzlaff S, Heussler V, Waters AP, Doerig C. Activation of a PAK-MEK signalling pathway in malaria parasite-infected erythrocytes. Cell Microbiol. 2011;13(6):836–845. doi: 10.1111/j.1462-5822.2011.01582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85(2):149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 63.Schneider AG, Mercereau-Puijalon O. A new Apicomplexa-specific protein kinase family: multiple members in Plasmodium falciparum, all with an export signature. BMC Genomics. 2005;6:30. doi: 10.1186/1471-2164-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kappes B, Yang J, Suetterlin BW, Rathgeb-Szabo K, Lindt MJ, Franklin RM. A Plasmodium falciparum protein kinase with two unusually large kinase inserts. Mol. Biochem. Parasitol. 1995;72(1–2):163–278. doi: 10.1016/0166-6851(95)00075-c. [DOI] [PubMed] [Google Scholar]
- 65.Siebel CW, Feng L, Guthrie C, Fu XD. Conservation in budding yeast of a kinase specific for SR splicing factors. Proc. Natl. Acad. Sci. U. S. A. 1999;96(10):5440–5445. doi: 10.1073/pnas.96.10.5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dixit A, Singh PK, Sharma GP, Malhotra P, Sharma P. PfSRPK1, a novel splicing-related kinase from Plasmodium falciparum. J. Biol. Chem. 2010;285(49):38315–38323. doi: 10.1074/jbc.M110.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Doerig C. Protein kinases as targets for anti-parasitic chemotherapy. Biochim. Biophys. Acta. 2004;1697(1–2):155–168. doi: 10.1016/j.bbapap.2003.11.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.