Significance
High-mobility group box protein 4 (HMGB4) is a transcription repressor preferentially expressed in the testes and binds cisplatin-damaged DNA. Investigating the DNA-damage recognition potential of HMGB4 and its relevance to cancer is of clinical importance. In this study, we found that HMGB4 regulates the sensitivity of testicular germ cell tumors (TGCTs) to cisplatin treatment. HMGB4 imparts repair shielding of platinum-DNA lesions in human TGCTs, rendering lesions inaccessible to the nucleotide excision repair (NER) machinery. Our results show that cisplatin-resistant breast cancer cells complemented with HMGB4 become sensitive to cisplatin. Furthermore, the ability of HMGB4 to modulate the cell cycle response, NER, apoptosis, and MAPK suggests a critical role for this protein in conveying cisplatin hypersensitivity in TGCTs.
Keywords: platinum anticancer drug, testicular cancer, high-mobility group protein
Abstract
Cisplatin is the most commonly used anticancer drug for the treatment of testicular germ cell tumors (TGCTs). The hypersensitivity of TGCTs to cisplatin is a subject of widespread interest. Here, we show that high-mobility group box protein 4 (HMGB4), a protein preferentially expressed in testes, uniquely blocks excision repair of cisplatin-DNA adducts, 1,2-intrastrand cross-links, to potentiate the sensitivity of TGCTs to cisplatin therapy. We used CRISPR/Cas9-mediated gene editing to knockout the HMGB4 gene in a testicular human embryonic carcinoma and examined cellular responses. We find that loss of HMGB4 elicits resistance to cisplatin as evidenced by cell proliferation and apoptosis assays. We demonstrate that HMGB4 specifically inhibits repair of the major cisplatin-DNA adducts in TGCT cells by using the human TGCT excision repair system. Our findings also reveal characteristic HMGB4-dependent differences in cell cycle progression following cisplatin treatment. Collectively, these data provide convincing evidence that HMGB4 plays a major role in sensitizing TGCTs to cisplatin, consistent with shielding of platinum-DNA adducts from excision repair.
Testicular germ cell tumors (TGCTs) are among the few tumor types that can be clinically cured with chemotherapy owing to the potency of cisplatin (1). The introduction of cisplatin in combination chemotherapy helped advance cure rates from 5% to the current level of 90% (2). In the clinic, cisplatin is used with bleomycin and either vinblastine or etoposide to treat metastatic testicular neoplasms (3–6). The well-studied antitumor activity of cisplatin involves binding to DNA, inhibition of transcription, and induction of apoptosis (7, 8). Cellular events leading to apoptosis are multifactorial and begin with uptake followed by chemical transformation of cisplatin. Following aquation, or replacement of the chloride ligands in [Pt(NH3)2Cl2] by water, the activated drug binds to DNA, generating mainly 1,2-intrastrand d(GpG) cross-links that block RNA polymerase II, ultimately signaling cell death (9–11). Recently, a cisplatin-sequencing (cisplatin-seq) approach was used to confirm DNA as the target for cisplatin at the genome scale by base resolution analysis (12). High-mobility group proteins such as high-mobility group box protein 1 (HMGB1) recognize these specific cisplatin-DNA adducts and promote the toxicity of cisplatin to tumors by interfering with excision and other repair pathways (13, 14).
HMGB1 binds selectively to cisplatin 1,2-intrastrand d(GpG) and d(ApG) cross-links, which account for ∼90% of all platinum (Pt)-DNA adducts (15, 16). Studies of the interaction of HMGB1 and other cisplatin-DNA recognition proteins such as Irx1 in yeast (17, 18) with cisplatin-modified DNA revealed bending of the DNA with attendant shielding from excision repair in vitro and subsequent sensitization of cancer cells to cisplatin treatment (13, 19). Related studies showed cooperative HMGB1 and XPA–RPA binding to DNA interstrand cross-links (ICLs) induced by psolaren (20, 21) and, recently, concluded that HMGB1–XPA interactions favor the ICL repair process (22). There are three cysteine residues in HMGB1, two of which (Cys22 and Cys44) are located in the A domain directly involved in binding to cisplatin-DNA 1,2-d(GpG) intrastrand cross-links (23). These cysteines can form a disulfide bond under mild oxidizing conditions, which significantly reduces their ability to bind to cisplatin-modified DNA (24). Because the intracellular redox potential, buffered by glutathione, will generate a mixture of oxidized and reduced forms of HMGB1, HMGB1 sensitization of cancer cells to cisplatin by repair shielding can be compromised and will depend on cell type (25).
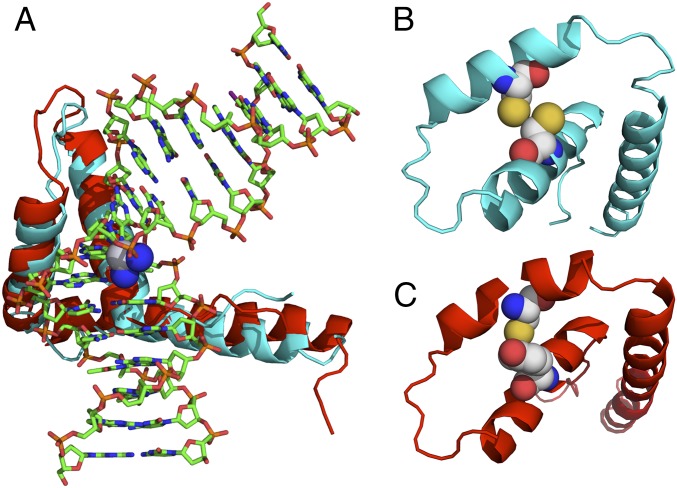
HMGB4 has two tandem HMG domains, A and B, homologous to those in HMGB1, and a shortened C-terminal tail that lacks the string of 30 acidic residues present in HMGB proteins 1–3. HMGB4 is preferentially expressed in the testes in contrast to HMGB1, which is ubiquitous (26). The amino acid sequence of HMGB4 revealed the presence of tyrosine in place of Cys22 in domain A, thereby avoiding the redox-dependent platinated-DNA binding observed for HMGB1 that we postulate to be the cause of the variable cellular response (Fig. 1). Moreover, purified HMGB4 protein at 1 μM inhibits nucleotide excision repair of 1,2-intrastrand d(GpG) cross-link damage by >90%, whereas the inhibitory property of purified HMGB1 of the same substrate is ∼45% (27). Conceivably, HMGB4 might be a critical player in promoting the hypersensitivity of TGCTs to cisplatin by blocking excision repair, thus conveying significant clinical success.
Fig. 1.
Structural representation of the ternary HMGB1/HMGB4–Pt-DNA complex. (A) Schematic representation of the crystal structure of domain A of HMGB1 in complex with platinated DNA superimposed with a computed structure of the similar domain of HMGB4. Significant homology is observed between HMGB4 and HMGB1 domain structures. SWISS-MODEL (46) with input from the human HMGB4 sequence was used to compute the HMGB4 structure. (B) Schematic representation of a simplified structure of HMGB1 protein (ribbon) highlighting Cys22 and Cys44 (space filling), critical residues implicated in disulfide bond formation under oxidizing conditions. (C) Schematic representation of a simplified structure of HMGB4 (ribbon) displaying Tyr22 and Cys44 (space filling). All structures were rendered using Pymol.
To investigate this hypothesis at the molecular level, we genetically modified an embryonic carcinoma cell line (NTera2/D1) using CRISPR/Cas9-mediated gene editing to eliminate endogenous expression of HMGB4. Following cisplatin treatment, we found differences in the DNA-repair efficacy for parental NTera2 and HMGB4 gene-edited NTera2 cells (NTera2 HMGB4−/−). In addition, different cell cycle and apoptotic induction patterns were observed. Taken together, our results point to a major role for HMGB4 in sensitizing TGCTs to cisplatin therapy.
Results
HMGB4 and Cisplatin in Testicular Cancer Cells.
The structure of the ternary HMGB1–Pt-DNA complex provides a basis for understanding HMG recognition of the platinum-induced DNA distortion (23) and led to the analysis of the protein sequence of human HMGB4 and its computed structure in complex with platinated DNA (Fig. 1). The HMGB1 phenylalanine that inserts into the hydrophobic notch created in the minor groove by the platinum 1,2-d(GpG) cross-link in the major groove is preserved in HMGB4. However, in place of two cysteine residues found at positions Cys44 and Cys22 in HMGB1, the corresponding residues in HMGB4 are Cys44 and Tyr22. This difference confers redox indifference for HMGB4 binding to cisplatin-modified DNA by eliminating the potential for disulfide bond formation that is present in HMGB1.
Because HMGB4 has not been well studied in relation to human testicular cancer, we first measured the levels of HMGB4 in a panel of TGCTs. TGCTs are classified according to their histopathology as either seminoma or nonseminoma (SI Appendix, Fig. S1A). Immunoblotting experiments revealed that different TGCTs of human origin, including the embryonic carcinomas NTera2 and Tera2 as well as the testicular teratoma GCT27, express appreciable levels HMGB4 (SI Appendix, Fig. S1B).
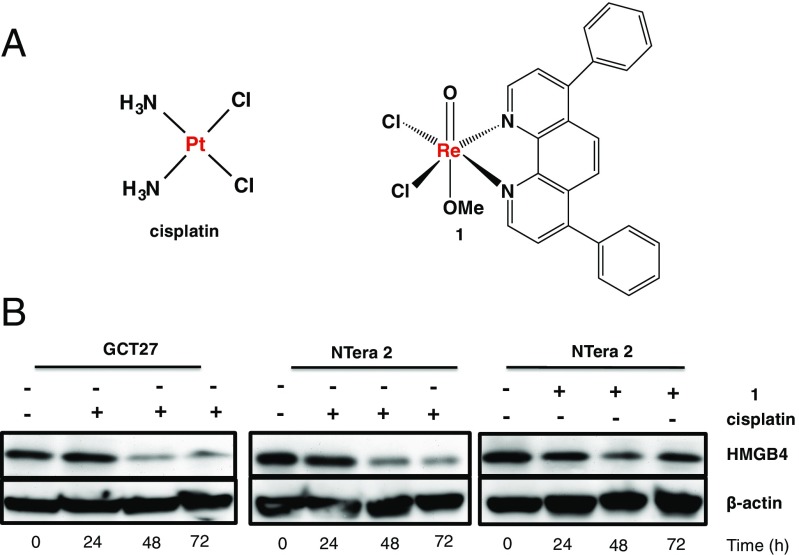
Cisplatin and its analogs, oxaliplatin and carboplatin, induce DNA damage by forming intrastrand 1,2-d(GpG), 1,2-d(ApG), and 1,3-d(GpNpG) cross-links, with fewer interstrand and DNA–protein cross-links. There is substantial evidence that HMGB4 binds 1,2-d(GpG) cross-links in vitro (27). To investigate the effect of cisplatin on HMGB4 in testes, NTera2 and GCT27 cells were treated with 2 μM of the drug for several time points and the protein level was monitored by Western blotting. Cisplatin treatment reduced HMGB4 expression over time by comparison with controls (Fig. 2), a phenomenon possibly associated with posttranslational modification of the target protein (28).
Fig. 2.
Effect of metal complexes on HMGB4. (A) Chemical structures of cisplatin (Left) and necroptosis-inducing compound 1 (Right). (B) Representative Western blot of human HMGB4 (Upper) and β-actin (Lower) at different times following treatment with 1 (2 μM) or cisplatin (2 μM), with respective untreated controls in NTera2 or GCT27 cells. Blots were probed with anti-HMGB4 or anti–β-actin antibody.
To investigate the alternative possibility that the diminution in HMGB4 recognition by the antibody after 72 h was a consequence of HMGB4 interaction with cisplatin-DNA adducts, we treated NTera2 and GCT27 cells with cisplatin and a Re(V) complex (1) that induces necroptosis but not DNA cross-link formation (Fig. 2) (29). Necroptosis is a specialized caspase-independent pathway of programmed necrosis that relies on the signaling activity of receptor interacting serine/threonine protein kinase 1 and 3 (RIP1/RIP3) to phosphorylate each other to form necrosomes (30, 31). Subsequent phosphorylation of mixed lineage kinase domain (MLKL) by necrosomes facilitates necroptosis. Characteristic features of necroptosis are swollen organelles and disintegrated plasma membrane. As seen in Fig. 2, there was no significant difference in HMGB4 recognition in NTera2 cells treated with 1.
CRISPR/Cas9 KO of HMGB4 in Human Testicular Embryonic Carcinoma.
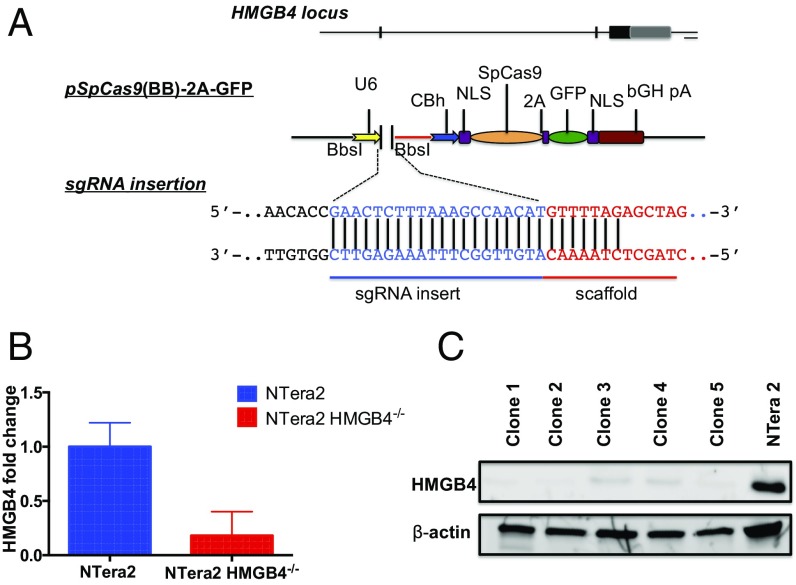
The Cas9 endonuclease of Streptococcus pyogenes (SpCas9) associated with CRISPR can be reprogrammed to target genomic loci of mammalian cells in a specific fashion using single guide RNA (sgRNA) (32–34). We used this gene-editing strategy to target HMGB4 in the human embryonic testicular cancer cell line, NTera2. Using RT-PCR, we observed a >80% change in HMGB4 mRNA expression levels in the KO relative to the parental NTera2 cells (Fig. 3B). We also confirmed a corresponding decrease in HMGB4 protein levels using Western blot analysis of the single clone HMGB4 KO NTera2 cells (Fig. 3C). The KO cell lines afford us a biological tool to explore our hypothesis that HMGB4 conveys DNA-repair shielding, which leads to sensitization of TGCTs to cisplatin treatment.
Fig. 3.
Targeting of the HMGB4 locus in mammalian embryonic carcinoma TGCT cells. (A) pSpCas9(BB)-2A-GFP expression vectors. The sgRNA cloned into the Bbs1 site of the Cas9-containing vector, encoding GFP for identification of transfected cells. GFP+ cells were sorted by FACS and single cells were expanded in culture. (B) mRNA levels of HMGB4 in parental NTera2 cells and knockout cells as determined by RT-PCR. (C) Western blot analysis of HMGB4 (Upper) protein expression 6 wk after pSpCas9(BB)-2A-GFP transfection. β-Actin (Lower) was used as loading control. Blots were probed with anti-HMGB4 or anti–β-actin antibody.
Loss of HMGB4 Desensitizes TGCTs to Cisplatin.
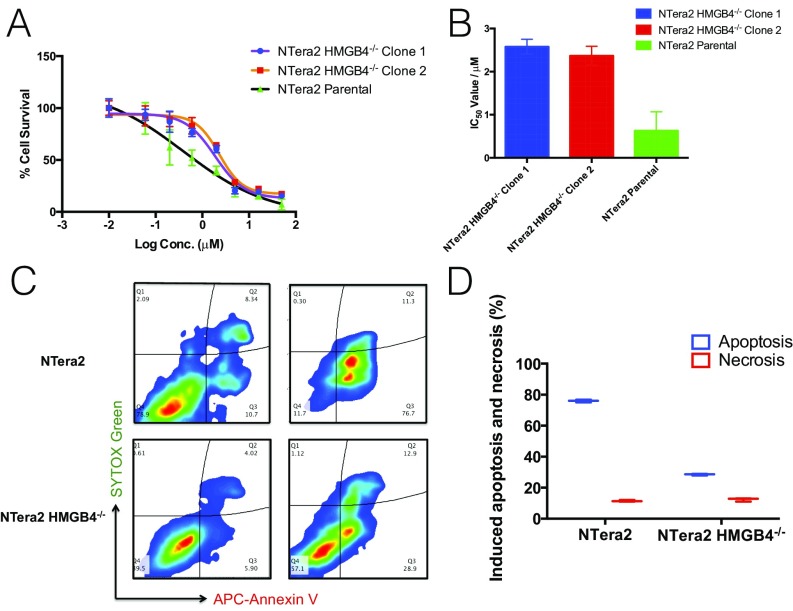
To assess the involvement of HMGB4 in mediating cellular sensitivity to cisplatin, we investigated the effect of HMGB4 on the viability of human TGCT cells treated with cisplatin using RNAi knockdown or CRISPR/Cas9 KO of HMGB4 in NTera2 cells. First, we used RNAi-mediated knockdown to obtain NTera2 cells with varying HMGB4 expression levels. As confirmed by quantitative PCR (qPCR) and Western blot, we generated NTera2 cell lines that enabled the correlation of HMGB4 expression with cisplatin sensitivity in an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) colorimetric assay (SI Appendix, Fig. S2). Notably, reduced levels of HMGB4 in silenced NTera2 cells conferred relative cisplatin resistance compared with WT cells, and a correlation was observed between cellular resistance and HMGB4 expression level. Second, we investigated cisplatin sensitivity in HMGB4 KO NTera2 cells from different clones compared with WT NTera2 cells. We observed a 4.5-fold difference in cisplatin sensitivity of the KO compared with WT NTera2 cells (Fig. 4 A and B). This result underscores the ability of HMGB4 to increase cellular sensitivity to cisplatin.
Fig. 4.
HMGB4-dependent cytotoxicity and apoptosis induction in NTera2 cells. (A) Dose–response curves of cisplatin-treated cells with varying levels of HMGB4 expression obtained by MTT assay 72 h after platinum exposure. (B) IC50 values from dose–response curves shown in A. WT NTera2 cells were used as control. Experiments were performed in triplicate. Data are represented as mean ± SD. (C) Flow cytometry experiment measurement of the extent of apoptosis in NTera2 (Upper) and NTera2 HMGB4−/− (Lower), untreated or treated with cisplatin (1 μM, 72 h) using APC-annexin V and SYTOX green as the markers. (D) Quantification of apoptosis and necrosis in NTera2 and NTera2 HMGB4−/− cells as determined in C.
Next, we investigated the ability of HMGB4 to promote cisplatin-induced apoptosis in NTera2 and CRISPR/Cas9-mediated HMGB4 KO NTera2 cells. Taking advantage of the translocation of phosphatidylserine to the exterior of apoptotic cells for annexin V recognition, we used a dual-staining annexin V/SYTOX green apoptosis dead cell assay to study the apoptotic behavior of NTera2 and NTera2 HMGB4−/− after 72 h of exposure to cisplatin (Fig. 4 C and D). The apoptotic and necrotic populations in NTera2 HMGB4−/− cells were reduced by approximately twofold compared with WT cells, indicative of the role of HMGB4 in effecting cell death by cisplatin-induced apoptosis.
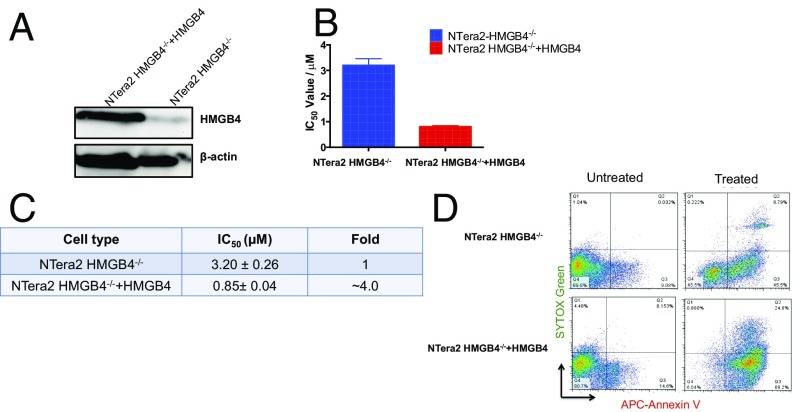
To further confirm our findings, we carried out two complementation studies to ensure that the observed desensitization of NTera2 to cisplatin is a consequence of the loss of HMGB4. We transiently transfected NTera2 HMGB4−/− cells with a plasmid encoding HMGB4 and assessed the cellular response of the transfected cells to cisplatin. The HMGB4 transfected cells restored sensitivity to cisplatin to the level of NTera2 WT cells (Fig. 5). In addition, the triple-negative breast cancer cell line MDA–MB-231, known to be highly resistant to cisplatin treatment, was transfected with the HMGB4 cDNA. HMGB4 expression was confirmed by Western blot and cisplatin sensitivity was assessed by an MTT assay (SI Appendix, Fig. S3). The IC50 (50% inhibitory concentration) values generated from dose–response curves showed twofold enhanced sensitivity for MDA–MB-231 transfected cells over WT cells. Collectively, our results establish that HMGB4 plays a role in sensitizing TGCTs to cisplatin.
Fig. 5.
HMGB4 complementation assay. (A) Western blot analysis of HMGB4 expression in NTera2 HMGB4−/− cells. Transient transfection with pcDNA3.1 encoding human HMGB4. (B) Cisplatin toxicity in NTera2 HMGB4−/− cells and NTera2 HMGB4−/− cells transiently transfected with HMGB4. (C) Quantification of IC50 and fold change extrapolated from dose–response curves. (D) APC-annexin V/SYTOX green double staining of NTera2 HMGB4−/− and NTera2 HMGB4−/− + HMGB4 after cisplatin treatment (2 μM) for 72 h.
Because HMGB4 specifically recognizes and binds 1,2-(GpG) Pt-DNA cross-links, we studied the effect of a monofunctional platinum agent on NTera2 and NTera2 HMGB4−/− cells. Monofunctional platinum adducts, unlike their bifunctional counterparts, do not significantly distort the 3D structure of duplex DNA (35). Therefore, if sensitization of TGCTs to cisplatin arises from HMGB4 recognition and repair shielding of the 1,2(GpG) cisplatin-DNA cross-link, monofunctional adducts would not elicit the same response. Indeed, phenanthriplatin, a highly potent anticancer agent developed within our laboratory (36), did not discriminate between the two different isogenic lines, at least with respect to IC50 values (SI Appendix, Fig. S4). This result supports our hypothesis that sensitivity of TGCTs to monofunctional platinum complexes is independent of HMGB4.
HMGB4 Dictates Cell-Cycle–Specific DNA-Repair Events.
Cellular DNA is under constant threat by endogenous and exogenous agents that inflict damage. In response, cells deploy sophisticated repair machinery to remove cytotoxic lesions and maintain genomic integrity. DNA repair is regulated throughout the cell cycle. We therefore examined the role of HMGB4 in cell cycle progression after exposure of both NTera2 and NTera2 HMGB4−/− cells to cisplatin. It is conceivable that lack of repair in NTera2 cells resulting from the repair-shielding role of HMGB4 will block S-phase progression because the lesions impede DNA polymerases (37). To investigate this possibility, we treated NTera2 and NTera2 HMGB4−/− cells with cisplatin (2 μM) and used flow cytometry to assay cell cycle progression using ModFit. NTera2 cells displayed a clear S-phase delay 24 and 48 h following cisplatin treatment, whereas NTera2 HMGB4−/− cells did not (SI Appendix, Fig. S5). These data support the hypothesis that HMGB4 shields repair, inducing S-phase blockage and imparting cisplatin sensitivity to testicular cancer cells. We found that the G0/G1 and S-phase checkpoints of NTera2 HMGB4−/− cells did not change significantly at various time points, suggesting that, in the absence of HMGB4, DNA damage repair is unimpeded and S-phase blockage is not observed, properties that confer cellular resistance to cisplatin.
Shielding of DNA-Excision Repair by Constitutive HMGB4 Is Important for Sensitivity of TGCTs to Cisplatin.
Excision repair has long been known as the major mechanism for removing the major DNA adducts formed by cisplatin (19, 38). To study the role of NER in TGCTs, we examined the biological consequences of cisplatin treatment on the expression of genes encoding proteins involved in the repair complex. The repair mechanism comprises damage recognition by proteins, unwinding or opening of the DNA helical structure, excision of the damaged DNA strand by endonucleases, and the subsequent action of DNA polymerases and ligases to complete the repair using the undamaged strand as template (39). With the use of qPCR to interrogate selected genes within the NER pathway, we found that endonucleases XPF-ERCC1 (xeroderma pigmentosum group F-excision repair cross-complementation group 1) and XPG (xeroderma pigmentosum group G) mRNA transcripts were consistently up-regulated by approximately twofold in NTera2 HMGB4−/− relative to NTera2 cells, following treatment with cisplatin (SI Appendix, Fig. S6 A–D). Consistent with the report that mouse HMGB4 acts as a transcription repressor (26), this difference might result from HMGB4 transcriptionally regulating NER-associated proteins required to repair cisplatin-DNA adducts and that its absence in NTera2 HMGB4−/− cells derepresses excision repair. In addition, the mRNA levels of XPC (xeroderma pigmentosum group C), which is considered to function in DNA-damage recognition within the global genome repair arm of NER (GG-NER), showed a sustained increase over a 48-h period in NTera2 HMGB4−/− compared with WT NTera2 cells.
Given that XPB (xeroderma pigmentosum group B) is a helicase in transcription factor II human (TFIIH) that facilitates DNA duplex unwinding for transcription and repair within the NER machinery, we investigated the role of XPB in NTera2 and NTera2 HMGB4−/− cell lines following cisplatin treatment. We used a small molecule inhibitor, spironolactone (SI Appendix, Fig. S7A), to inactivate XPB and evaluated cisplatin-induced cytotoxicity by MTT (SI Appendix, Fig. S7B). In addition, using shRNA to target the XPB gene, we knocked down XPB (SI Appendix, Fig. S7C) and examined the response of the mutated NTera2 and NTera2 HMGB4−/− cells to cisplatin, as shown in SI Appendix, Fig. S7D. A threefold increase in sensitivity was observed in NTera2 HMGB4−/− having reduced XPB compared with the XPB-containing parental HMGB4−/− cell line (SI Appendix, Fig. S7D). Reduced XPB activity in NTera2 showed little effect on the cellular sensitivity to cisplatin (SI Appendix, Fig. S7E).
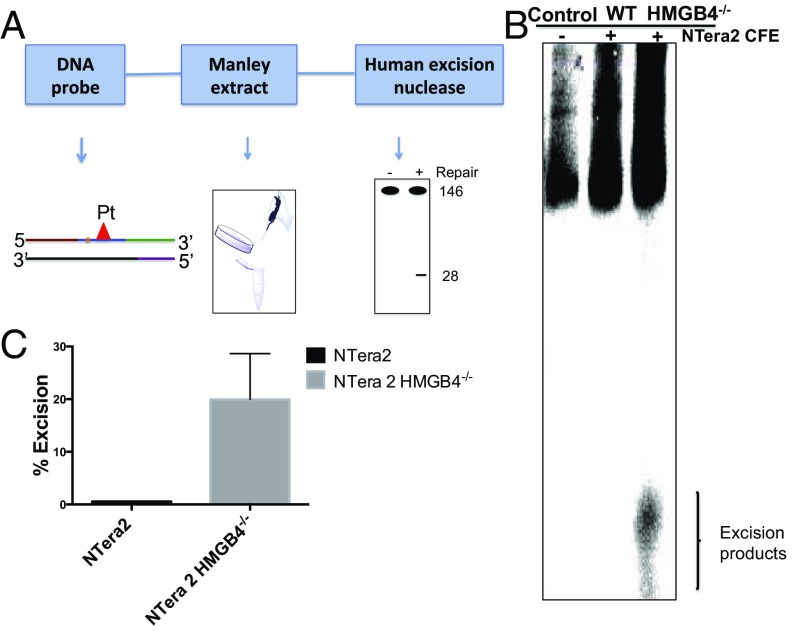
To further validate these results, we studied the impact of HMGB4 on DNA repair using a human excision nuclease assay performed with cell-free extracts (CFEs) derived from NTera2 and NTera2 HMGB4−/− cells. The assay of cisplatin-damaged DNA repair was carried out by first preparing the substrates shown in Fig. 6A, formed by annealing and ligating a short oligomer containing cisplatin 1,2-d(GpG) cross-links with four other overlapping oligomers, as previously described (19). The oligomer containing the Pt damage was labeled with [γ32P] ATP such that the 146-bp duplexes contained a radiolabel at the fifth phosphodiester bond 5′ to the cisplatin d(GpG) adduct. When these substrates were incubated with NTera2 and NTera2 HMGB4−/− CFE, the adducts were released in oligonucleotides that were mainly 25- to 30-nt long (Fig. 6B), as observed by polyacrylamide gel electrophoresis. The removal of cisplatin-modified fragments requires active nucleotide excision repair in the TGCTs with depleted HMGB4. In support of the repair-shielding hypothesis, we observed that, in NTera2 cells with normal HMGB4 levels, there was reduced 25- to 30-nt fragment removal compared with results for NTera2 HMGB4−/− cells (Fig. 6C). Site-specifically platinated control oligonucleotides prepared in an identical manner failed to produce the characteristic 25- to 30-nt fragments when incubated without CFE.
Fig. 6.
Human excision nuclease assay. (A) Schematic representation of human excision nuclease assay. (B) Excision of 1,2-d(GpG) cisplatin cross-links by excinuclease activity in testicular cancer cell extracts. The substrate (146 bp, Pt-DNA probe) was incubated with CFEs from NTera2 and NTera2 HMGB4−/− cells. The reaction was run for 75 min and the excision products were resolved on 10% (wt/vol) polyacrylamide denaturing gels (TE buffer, 55–46 mA for 5 h). The position of the main excision product, a 25–30 nt is indicated by a bracket. Replicates of this experiment with two independently prepared cell-free extract sets are reported in SI Appendix, Fig. S8. (C) Percent excision products from the NER assay using CFEs from NTera2 and NTera2 HMGB4−/− cells in B. Error bar represents SD (n = 3).
Discussion
Understanding the high cure rates of TGCTs by cisplatin is of longstanding interest and could be of clinical value for treatment of refractory solid tumors. Given that cisplatin forms both intra- and interstrand cross-links on DNA that ultimately result in apoptosis if not removed, the observation that DNA-recognition proteins, including HMG-family proteins, can block the repair pathway is of potential clinical relevance. We previously demonstrated that HMG proteins potentiate sensitivity of mammalian cells to cisplatin by binding to 1,2-intrastrand d(GpG) cross-links using excision nuclease assays (40), which implies that NER proteins may be implicated in regulating cisplatin sensitivity in malignancies. Here, we expanded our investigation of HMG proteins to include HMGB4, a testes-specific protein involved in spermatogenesis.
In this study, we report an unexplored role of HMGB4 in sensitizing TGCTs to cisplatin at the molecular level. Higher IC50 and lower apoptosis induction associated with CRISPR/Cas9-mediated HMGB4 KO NTera2 cells reveals increased cisplatin resistance in the absence of HMGB4. Genome-wide screens in Saccharomyces cerevisiae identified genes that, when disrupted, confer cisplatin resistance (41). Several of the identified genes had not been previously linked to cisplatin resistance and functioned in RNA Pol II-dependent gene regulation, DNA repair, and genome stability. Complementation of individual inactivated genes eradicated cisplatin resistance. In a similar manner, we here identify the specific involvement of HMGB4 in conferring sensitivity in the cisplatin-resistant human breast cancer cell line (MDA–MB-231) by complementation with a HMGB4-encoding plasmid (SI Appendix, Fig. S3). The sensitivity doubled upon expression of the protein. Our observation that the cisplatin-resistant phenotype of the deleted HMGB4 cell line is due to the specific disruption of HMGB4 suggests that changes to HMGB4 in testicular germ cell tumors, such as posttranslational modifications, may also confer cisplatin resistance. Given that HMGB1 undergoes hyperacetylation and phosphorylation upon interaction with cisplatin-DNA adducts (28), the same or similar behavior seems highly plausible for HMGB4, considering that the two proteins share significant homology. The clinical consequence is that patients with TGCTs with cisplatin resistance are likely to have HMGB4 mutations or modifications. To explore this possibility, we are currently studying human biopsy samples from patients with TGCTs experiencing cisplatin-resistant phenotypes to determine whether they can be correlated with HMGB4 levels and associated mutations or modifications. Furthermore, the parallel of HMGB4 expression levels, as quantified by qRT-PCR, in transient knockdown cells with their cytotoxicity profiles verifies the specific involvement of HMGB4 in determining cisplatin sensitivity. For TGCTs, this report demonstrates that the platinum DNA-damage recognition protein, HMGB4, correlates with cisplatin sensitivity.
Accumulation of cells at the G2/M cell cycle transition reflects unrepaired Pt-DNA lesions in NTera2 HMGB4-proficient cells and follows delayed S phase after cisplatin treatment. The Pt lesions block DNA polymerases required for replication (42) and the transcription of the mitotic spindle apparatus needed for cell division. In cells containing HMGB4, failure to repair the damaged DNA during G1 results in replication stalling and ultimately leads to cell death. Conversely, in HMGB4-deficient cells, we propose that DNA damage is sufficiently well repaired during the G1 phase by unimpeded NER proteins to account for the unchanged S phase observed. Notably, no significant accumulation of cells at G2/M over the analysis period following cisplatin treatment was observed in NTera2 HMGB4−/−. In further support of this argument, a persistent G1 phase was observed, as indicative of growth and NER activity.
The involvement of HMGB4 in sensitizing TGCTs to cisplatin prompted our investigation of DNA repair mediated by NER in human TGCT cells. Inefficient repair of cisplatin-induced DNA damage in TGCTs has been associated with reduced XPA protein levels (43). It is possible that HMGB4 interacts with XPA to shield repair activity, although careful experiments are needed to support this presumption. Considering that cisplatin is an effective anticancer drug used to cure metastatic testicular cancer, our understanding of whether or not the predominant DNA lesion, the 1,2-d(GpG) intrastrand cross-link, is an important substrate for human excision nucleases has clinical therapeutic implications not only for testicular neoplasms but other cancer types. In this study, we used the excision nuclease assay to demonstrate that cisplatin-induced 1,2-d(GpG) intrastrand cross-links are substrates for a human excision repair system derived from the embryonic carcinoma cells, NTera2, and related genetically modified NTera2 HMGB4−/−. The detection of radiolabeled 25- to 30-nt-long products generated by excinuclease activity supports our repair-shielding hypothesis. The result demonstrates specific inhibition of repair of the 1,2-d(GpG) intrastrand cisplatin cross-link by HMGB4. This result is consistent with our previous report showing that yeast mutants lacking the HMG-domain protein Ixr1 were significantly less sensitive to cisplatin compared with WT cells (18).
Whereas numerous studies have investigated the effect of genotoxic drugs on the NER machinery (7), little work has been done in elucidating the effect of DNA-recognition proteins. We therefore, evaluated the ability of HMGB4 to impart an effect on DNA-repair processes, particularly NER. The structure-specific endonucleases, XPG and ERCC1–XPF, which cleave damaged DNA strands on the 3′ and 5′ sides of the Pt-DNA lesion, respectively, are key excision proteins within the NER pathway. ERCC1 has also been proposed as a predictive marker to assess the therapeutic benefit of cisplatin-based chemotherapy in a personalized medicine setting (44). In TGCTs, up-regulation of ERCC1 and XPF has been associated with disease progression (45). In our quest to understand how HMGB4 affects protein regulation in the NER pathway, we confirmed that the absence of HMGB4 in testicular cancer cells facilitates the action of XPG, ERCC1–XPF endonucleases, as well as XPC, consistent with their up-regulation in gene profiling studies. It is possible that cells lacking HMGB4 readily detect DNA damage and up-regulate other transcription factors for their excision repair.
As already mentioned, recruitment of NER proteins to Pt-DNA damage sites involves TFIIH, which has XPB as a principal component. Our examination of NTera2 and NTera2 HMGB4−/− cells with reduced levels of XPB shows differential sensitivity to cisplatin treatment. This result suggests that the loss of HMGB4 and NER genes, in particular those involved in the initiation steps including XPB, are required for cisplatin sensitivity. However, exclusive loss of HMGB4 in NTera2 cells is sufficient to induce significant resistance to cisplatin. It would be of interest to analyze patient genomic data from TGCTs treated with cisplatin to determine possible correlation with HMGB4 levels. Experiments of this kind are currently in progress.
In NTera2 cells, which are cisplatin sensitive relative to NTera2 HMGB4−/−, p-ERK1/2 (phosphorylated extracellular signal-regulated kinase) and p-c-Jun (phosphorylated c-Jun) expression decreased over a 72-h period following cisplatin treatment. Interestingly, basal levels of p-ERK1/2 and p-c-Jun were relatively diminished in NTera2 HMGB4−/− compared with normal NTera2 cells (SI Appendix, Fig. S9). Additionally, the proapoptotic protein expression for NTera2 HMGB4−/− cells treated with cisplatin was significantly lower than that of the WT cells, whereas the antiapoptotic Bcl-xL protein expression was higher in NTera2 HMGB4−/− than in NTera2 cells (SI Appendix, Fig. S10). This imbalance supports the differential apoptosis response observed for the TGCT cells under study.
In conclusion, our results are consistent with a repair-shielding model in which HMGB4 recognizes and binds cisplatin-DNA intrastrand d(GpG) cross-links, stalling the NER machinery recruited to the damage site, which otherwise would have excised and ultimately repaired the damage. Although we do not rule out transcriptional activity imposed by HMGB4 to regulate genes in the NER pathway, we conclude, based on the present findings, that a significant component of the hypersensitivity of TGCTs to cisplatin is a result of repair shielding by HMGB4. We speculate that HMGB4 may also impart transcriptional activity by regulating NER genes based on the evaluation of mRNA transcripts in NTera2 or NTera2 HMGB4−/− treated with cisplatin. Consistent with this suggestion is the G2/M accumulation in HMGB4-proficient NTera2 cells and the relatively lower levels of XPC, XPG, ERCC1, and XPF mRNA transcripts in NTera2 cells compared with HMGB4 NTera2-deficient cells, which leads to significant sensitivity of the former to cisplatin.
Materials and Methods
All cell lines were grown at 37 °C under a humidified atmosphere of 5% (vol/vol) CO2. The TGCT, NTera2, and the breast cancer cell line MDA–MB-231 were purchased from American Type Culture Collection. An NTera2 HMGB4−/− cell line was generated via CRISPR/Cas9-mediated KO. Detailed experimental procedures, including gene editing, transfection, cell cycle, apoptosis, and human excision nuclease assays, are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Kogularamanan Suntharalingam for providing the control Re(V) complex (1), Dr. Peter Bruno for helpful discussions and biological reagents, Dr. Amanda Hussey for assistance with figures, and the staff of the Koch Institute flow cytometry core facility. This work was supported by National Cancer Institute Grant CA034992 (to S.J.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615327114/-/DCSupplemental.
References
- 1.Masters JR, Köberle B. Curing metastatic cancer: Lessons from testicular germ-cell tumours. Nat Rev Cancer. 2003;3(7):517–525. doi: 10.1038/nrc1120. [DOI] [PubMed] [Google Scholar]
- 2.Einhorn LH. Curing metastatic testicular cancer. Proc Natl Acad Sci USA. 2002;99(7):4592–4595. doi: 10.1073/pnas.072067999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einhorn LH, Donohue J. Cis-diamminedichloroplatinum, vinblastine, and bleomycin combination chemotherapy in disseminated testicular cancer. Ann Intern Med. 1977;87(3):293–298. doi: 10.7326/0003-4819-87-3-293. [DOI] [PubMed] [Google Scholar]
- 4.Schabel FM, Jr, Trader MW, Laster WR, Jr, Corbett TH, Griswold DP., Jr cis-Dichlorodiammineplatinum(II): Combination chemotherapy and cross-resistance studies with tumors of mice. Cancer Treat Rep. 1979;63(9–10):1459–1473. [PubMed] [Google Scholar]
- 5.Corbett TH, Griswold DP, Jr, Wolpert MK, Venditti JM, Schabel FM., Jr Design and evaluation of combination chemotherapy trials in experimental animal tumor systems. Cancer Treat Rep. 1979;63(5):799–801. [PubMed] [Google Scholar]
- 6.Williams SD, et al. Treatment of disseminated germ-cell tumors with cisplatin, bleomycin, and either vinblastine or etoposide. N Engl J Med. 1987;316(23):1435–1440. doi: 10.1056/NEJM198706043162302. [DOI] [PubMed] [Google Scholar]
- 7.Siddik ZH. Cisplatin: Mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4(4):307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 9.Jamieson ER, Lippard SJ. Structure, recognition, and processing of cisplatin-DNA adducts. Chem Rev. 1999;99(9):2467–2498. doi: 10.1021/cr980421n. [DOI] [PubMed] [Google Scholar]
- 10.Todd RC, Lippard SJ. Structure of duplex DNA containing the cisplatin 1,2-{Pt(NH3)2}2+-d(GpG) cross-link at 1.77 A resolution. J Inorg Biochem. 2010;104(9):902–908. doi: 10.1016/j.jinorgbio.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Todd RC, Lippard SJ. Consequences of cisplatin binding on nucleosome structure and dynamics. Chem Biol. 2010;17(12):1334–1343. doi: 10.1016/j.chembiol.2010.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shu X, Xiong X, Song J, He C, Yi C. Base-resolution analysis of cisplatin-DNA adducts at the genome scale. Angew Chem Int Ed Engl. 2016;55(46):14246–14249. doi: 10.1002/anie.201607380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung Y, Lippard SJ. Direct cellular responses to platinum-induced DNA damage. Chem Rev. 2007;107(5):1387–1407. doi: 10.1021/cr068207j. [DOI] [PubMed] [Google Scholar]
- 14.Yamada M, O’Regan E, Brown R, Karran P. Selective recognition of a cisplatin-DNA adduct by human mismatch repair proteins. Nucleic Acids Res. 1997;25(3):491–496. doi: 10.1093/nar/25.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kartalou M, Essigmann JM. Recognition of cisplatin adducts by cellular proteins. Mutat Res. 2001;478(1–2):1–21. doi: 10.1016/s0027-5107(01)00142-7. [DOI] [PubMed] [Google Scholar]
- 16.Kartalou M, Essigmann JM. Mechanisms of resistance to cisplatin. Mutat Res. 2001;478(1–2):23–43. doi: 10.1016/s0027-5107(01)00141-5. [DOI] [PubMed] [Google Scholar]
- 17.McA’Nulty MM, Whitehead JP, Lippard SJ. Binding of Ixr1, a yeast HMG-domain protein, to cisplatin-DNA adducts in vitro and in vivo. Biochemistry. 1996;35(19):6089–6099. doi: 10.1021/bi952877u. [DOI] [PubMed] [Google Scholar]
- 18.McA’Nulty MM, Lippard SJ. The HMG-domain protein Ixr1 blocks excision repair of cisplatin-DNA adducts in yeast. Mutat Res. 1996;362(1):75–86. doi: 10.1016/0921-8777(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 19.Huang JC, Zamble DB, Reardon JT, Lippard SJ, Sancar A. HMG-domain proteins specifically inhibit the repair of the major DNA adduct of the anticancer drug cisplatin by human excision nuclease. Proc Natl Acad Sci USA. 1994;91(22):10394–10398. doi: 10.1073/pnas.91.22.10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy MC, Christensen J, Vasquez KM. Interplay between human high mobility group protein 1 and replication protein A on psoralen-cross-linked DNA. Biochemistry. 2005;44(11):4188–4195. doi: 10.1021/bi047902n. [DOI] [PubMed] [Google Scholar]
- 21.Lange SS, Mitchell DL, Vasquez KM. High mobility group protein B1 enhances DNA repair and chromatin modification after DNA damage. Proc Natl Acad Sci USA. 2008;105(30):10320–10325. doi: 10.1073/pnas.0803181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee A, Vasquez KM. HMGB1 interacts with XPA to facilitate the processing of DNA interstrand crosslinks in human cells. Nucleic Acids Res. 2016;44(3):1151–1160. doi: 10.1093/nar/gkv1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohndorf UM, Rould MA, He Q, Pabo CO, Lippard SJ. Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature. 1999;399(6737):708–712. doi: 10.1038/21460. [DOI] [PubMed] [Google Scholar]
- 24.Park S, Lippard SJ. Redox state-dependent interaction of HMGB1 and cisplatin-modified DNA. Biochemistry. 2011;50(13):2567–2574. doi: 10.1021/bi2000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, et al. Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA. Biochem Biophys Res Commun. 2013;441(4):701–706. [PubMed] [Google Scholar]
- 26.Catena R, et al. HMGB4, a novel member of the HMGB family, is preferentially expressed in the mouse testis and localizes to the basal pole of elongating spermatids. Biol Reprod. 2009;80(2):358–366. doi: 10.1095/biolreprod.108.070243. [DOI] [PubMed] [Google Scholar]
- 27.Park S, Lippard SJ. Binding interaction of HMGB4 with cisplatin-modified DNA. Biochemistry. 2012;51(34):6728–6737. doi: 10.1021/bi300649v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, et al. HMGB1 bound to cisplatin-DNA adducts undergoes extensive acetylation and phosphorylation in vivo. Chem Sci. 2015;6(3):2074–2078. doi: 10.1039/c4sc03650f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suntharalingam K, et al. Necroptosis-inducing rhenium(V) oxo complexes. J Am Chem Soc. 2015;137(8):2967–2974. doi: 10.1021/ja511978y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho YS, et al. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137(6):1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hitomi J, et al. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135(7):1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyzocha NK, Ran FA, Hsu PD, Zhang F. RNA-guided genome editing of mammalian cells. Methods Mol Biol. 2014;1114:269–277. doi: 10.1007/978-1-62703-761-7_17. [DOI] [PubMed] [Google Scholar]
- 34.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lovejoy KS, et al. cis-Diammine(pyridine)chloroplatinum(II), a monofunctional platinum(II) antitumor agent: Uptake, structure, function, and prospects. Proc Natl Acad Sci USA. 2008;105(26):8902–8907. doi: 10.1073/pnas.0803441105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park GY, Wilson JJ, Song Y, Lippard SJ. Phenanthriplatin, a monofunctional DNA-binding platinum anticancer drug candidate with unusual potency and cellular activity profile. Proc Natl Acad Sci USA. 2012;109(30):11987–11992. doi: 10.1073/pnas.1207670109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Novarina D, Amara F, Lazzaro F, Plevani P, Muzi-Falconi M. Mind the gap: Keeping UV lesions in check. DNA Repair (Amst) 2011;10(7):751–759. doi: 10.1016/j.dnarep.2011.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reardon JT, Sancar A. Molecular mechanism of nucleotide excision repair in mammalian cells. In: Dizdaroglu M, Karakaya AE, editors. Advances in DNA Damage and Repair. Spinger; New York: 1999. pp. 377–393. [Google Scholar]
- 39.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 40.Zamble DB, Mu D, Reardon JT, Sancar A, Lippard SJ. Repair of cisplatin--DNA adducts by the mammalian excision nuclease. Biochemistry. 1996;35(31):10004–10013. doi: 10.1021/bi960453+. [DOI] [PubMed] [Google Scholar]
- 41.Huang RY, Eddy M, Vujcic M, Kowalski D. Genome-wide screen identifies genes whose inactivation confer resistance to cisplatin in Saccharomyces cerevisiae. Cancer Res. 2005;65(13):5890–5897. doi: 10.1158/0008-5472.CAN-04-4093. [DOI] [PubMed] [Google Scholar]
- 42.Chaney SG, Campbell SL, Bassett E, Wu Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit Rev Oncol Hematol. 2005;53(1):3–11. doi: 10.1016/j.critrevonc.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 43.Köberle B, Masters JR, Hartley JA, Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr Biol. 1999;9(5):273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 44.Jun HJ, et al. ERCC1 expression as a predictive marker of squamous cell carcinoma of the head and neck treated with cisplatin-based concurrent chemoradiation. Br J Cancer. 2008;99(1):167–172. doi: 10.1038/sj.bjc.6604464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Köberle B, et al. ERCC1 and XPF expression in human testicular germ cell tumors. Oncol Rep. 2010;23(1):223–227. [PubMed] [Google Scholar]
- 46.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31(13):3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.