Abstract
Most ribosomal proteins in Saccharomyces cerevisiae are encoded by two paralogs that additively produce the optimal protein level for cell growth. Nonetheless, deleting one paralog of most ribosomal protein gene pairs results in a variety of phenotypes not observed when the other paralog is deleted. To determine whether paralog-specific phenotypes associated with deleting RPL7A or RPL7B stem from distinct functions or different levels of the encoded isoforms, the coding region and introns of one paralog, including an intron-embedded snoRNA (small nucleolar RNA) gene, were exchanged with that of the other paralog. Among mutants harboring a single native or chimeric RPL7 allele, expression from the RPL7A locus exceeded that from the RPL7B locus, and more Rpl7a was expressed from either locus than Rpl7b. Phenotypic differences in tunicamycin sensitivity, ASH1 mRNA localization, and mobility of the Ty1 retrotransposon were strongly correlated with Rpl7 and ribosome levels, but not with the Rpl7 or snoRNA isoform expressed. Although Ty1 RNA is cotranslationally localized, depletion of Rpl7 minimally affected synthesis of Ty1 Gag protein, but strongly influenced Ty1 RNA localization. Unlike the other processes studied, Ty1 cDNA accumulation was influenced by both the level and isoform of Rpl7 or snoRNA expressed. These cellular processes had different minimal threshold values for Rpl7 and ribosome levels, but all were functional when isoforms of either paralog were expressed from the RPL7A locus or both RPL7 loci. This study illustrates the broad range of phenotypes that can result from depleting ribosomes to different levels.
Keywords: ribosomal protein, paralog, isoform, retrotransposon, RNA localization
The Saccharomyces cerevisiae ribosome comprises four ribosomal RNAs and 79 ribosomal proteins (RPs) synthesized in nearly equimolar amounts. Notably, 59 RPs are encoded by paralogous gene pairs that arose from a whole genome duplication and were selectively retained (Wolfe and Shields 1997). Paralogs may persist in the genome because they encode redundant functions that balance gene dosage, become specialized for different environmental conditions, or acquire distinct functions by subfunctionalization or neofunctionalization (Innan and Kondrashov 2010; Parenteau et al. 2015). Most RP paralogs are redundant for a ribosome-associated function that is essential for optimal cell growth or viability (Dean et al. 2008; Steffen et al. 2012; Woolford and Baserga 2013). The mRNAs transcribed from these genes, although heterogeneous in sequence, especially in untranslated regions (UTRs) (Leer et al. 1985), typically encode proteins of identical, or nearly identical, length and amino acid sequence. Fitness defects resulting from deletion of one paralog can be suppressed by ectopic expression of the coding region of the other paralog from an active promoter (Abovich and Rosbash 1984; Rotenberg et al. 1988; Simoff et al. 2009). Nonetheless, deletion of one paralog or the other of an RP gene pair often results in distinct transcriptional and phenotypic profiles (Komili et al. 2007). A single paralog of discrete subsets of RP genes is required for bud-site selection (Ni and Snyder 2001), translational repression, and bud tip localization of the ASH1 mRNA (Komili et al. 2007), actin organization (Haarer et al. 2007), vacuolar carboxypeptidase Y secretion (Bonangelino et al. 2002), propagation of the M1 satellite dsRNA of the L-A virus (Ohtake and Wickner 1995), protection from killer toxin (Page et al. 2003), telomere length control (Askree et al. 2004), replicative life span (Steffen et al. 2008, 2012), and mobility of the Ty1 retrotransposon (Dakshinamurthy et al. 2010; Risler et al. 2012). The divergent phenotypes associated with deleting one or the other paralog are not always correlated with the relative abundance of mRNA or RP produced from individual paralogs in a wild-type strain (Komili et al. 2007; Steffen et al. 2008, 2012). For example, it was argued that RPS18B is specifically required for ASH1 mRNA localization because deletion of RPS18B but not RPS18A abolished localization despite equivalent levels of epitope-tagged Rps18a and Rps18b in a wild-type strain (Komili et al. 2007). Collectively, these observations led to the “ribosome code” hypothesis, which posits that the divergent RP isoforms are incorporated in different combinations into heterogeneous ribosomes that selectively translate specific subsets of mRNAs (Mauro and Edelman 2002, 2007; Komili et al. 2007). The specialized functions of heterogeneous ribosomes may be further diversified by selective post-translational modification of RPs and ribosomal RNAs (Mauro and Edelman 2002, 2007; Gilbert 2011; Xue and Barna 2012).
Discordance between the phenotypic consequences of deleting each paralog, and the relative level of mRNA or RP produced from each paralog in a wild-type strain could result from compensatory changes in the RP level produced from one paralog when the other is deleted. In one study, the longer replicative life span of an rpl20b∆ mutant was correlated with a lower level of 60S ribosomal subunits compared to an rpl20a∆ mutant, even though RPL20A and RPL20B mRNA levels are equivalent in a wild-type strain. Thus, a disparity in Rpl20 levels in rpl20a∆ vs. rpl20b∆ mutants could explain the paralog-specific role of RPL20B in replicative life span (Steffen et al. 2008). Given that RP paralog expression levels are often altered by deletion of the other paralog (Eng and Warner 1991; Presutti et al. 1991; Paulovich et al. 1993; Li et al. 1995; Fewell and Woolford 1999; Badis et al. 2004), the question of whether paralog-specific phenotypes result from RP isoforms with disparate functions, differences in the level or activity of the RP produced from each paralog, or a combination of these mechanisms has not yet been rigorously tested.
To address this question, we have examined phenotypes associated with expressing RPL7A or RPL7B at different levels. Rpl7 isoforms participate in the earliest steps of 60S precursor rRNA processing (Jakovljevic et al. 2012), and bind to 25S and 5S rRNAs in the mature 60S ribosomal subunit (Ben-Shem et al. 2011). Five of the 244 amino acid residues in Rpl7a and Rpl7b are divergent, with four amino acid substitutions in Rpl7b relative to Rpl7a (A2S, A3T, S16T, and V26I) being in the conserved N-terminal domain that is predicted to be on the surface of the ribosome (Lin 1991; Tsai et al. 2012). The possibility that Rpl7a and Rpl7b have unique functions as components of heterogeneous ribosomes was suggested by the involvement of different ribosome biogenesis factors in assembly of ribosomes containing Rpl7a vs. Rpl7b (Komili et al. 2007). In addition, the RPL7A and RPL7B genes are unique among RP paralogs in that each contains a paralogous C/D box snoRNA gene, snR39 or snR59, that is encoded in the second intron (Figure 1A), and likely expressed from the excised intron (Vincenti et al. 2007). While divergent in sequence, snR39 and snR59 function redundantly as guide RNAs for 2′-O-methylation of residue A807 in the large subunit rRNA (Piekna-Przybylska et al. 2007). Deletion of one or both of the introns harboring these snoRNA genes has no effect on cell viability (Parenteau et al. 2011). Several paralog-specific functions have been assigned to RPL7A and RPL7B; however, they are asymmetrically expressed in a wild-type strain (Ghaemmaghami et al. 2003; Komili et al. 2007), and the rpl7a∆ mutant has a diminished level of 60S ribosomal subunits, whereas rpl7b∆ has a wild-type polysome profile (Jakovljevic et al. 2012). Thus, it remains to be determined whether Rpl7a or Rpl7b has unique functions irrespective of its expression level.
Figure 1.
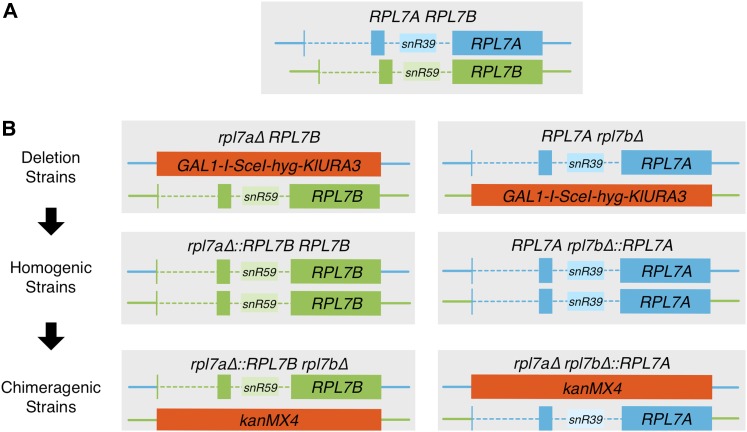
Schematic of the RPL7A and RPL7B loci used in this study. (A) RPL7A and RPL7B coding regions are interrupted by two introns. Encoded in the second intron is a snoRNA paralog, snR39 or snR59. (B) Construction of chimeragenic rpl7a∆::RPL7B rpl7b∆ (left column), and rpl7a∆ RPL7B∆::RPL7A (right column) mutants from the wild-type strain in three sequential steps. First, the RPL7A or RPL7B CRI was replaced with the GAL1-I-SceI-hygB-KlURA3 delitto perfetto cassette. Second, replacement of the delitto perfetto cassette with the RPL7B or RPL7A CRI, respectively, generated homogenic strains harboring the same CRI at both RPL7 loci. Third, the CRI of the RPL7B or RPL7A locus in homogenic strains was replaced with the kanMX4 marker to generate chimeragenic strains. Turquoise represents sequences derived from the RPL7A gene, and green represents sequences derived from the RPL7B gene. Solid, colored lines: noncoding locus DNA flanking the CRI; dashed, colored lines: introns; solid, dark-colored boxes: exons; solid, light-colored boxes: intron-encoded snoRNA genes.
Because selective pressure to suppress retrotransposon activity in the genome can drive neofunctionalization of gene paralogs involved in retrotransposon control (Münk et al. 2012), we examined the role of RPL7A and RPL7B in regulating Ty1, a long terminal repeat (LTR)-retrotransposon present in ∼30 copies in haploid S. cerevisiae strains. Ty1 elements contain two overlapping ORFs: GAG, which encodes the capsid protein, and POL, which is expressed by translational frameshifting, and encodes enzymatic mobility proteins. Retromobility involves localized translation of Ty1 RNA in cytoplasmic foci known as retrosomes, assembly of Ty1 RNA and proteins into virus-like particles (VLPs), reverse transcription of Ty1 RNA within VLPs, transport of cDNA to the nucleus, and introduction of the cDNA into the host genome by nonhomologous integration or homologous recombination [reviewed in Curcio et al. (2015)]. Deletion of RPL7A results in lower levels of Ty1 retrosomes, cDNA, and retromobility (Risler et al. 2012; Doh et al. 2014).
In this study, the coding region and introns (CRI) of RPL7A and RPL7B were exchanged to express Rpl7 and snoRNA isoforms at different levels. Expression of either RPL7 CRI from the RPL7A locus resulted in faster cell growth and higher levels of Rpl7 and 60S ribosomal subunits than expression from the RPL7B locus. Moreover, the RPL7A CRI is expressed more highly from either locus than the RPL7B CRI. Divergent growth rates, Rpl7 levels and 60S/40S ribosomal subunit ratios in strains expressing a single native or chimeric allele of RPL7 correlated strongly with differences in sensitivity to tunicamycin, efficiency of ASH1 mRNA and Ty1 retrotransposon RNA localization, and rate of Ty1 retromobility. Another phenotype, Ty1 cDNA accumulation, correlated with both the level and isoforms of Rpl7 and snoRNA expressed. These cellular processes required different minimal levels of Rpl7 or the RPL7-encoded snoRNA, but all phenotypic defects resulting from either RPL7 CRI being expressed from the RPL7B locus, or from the RPL7B CRI being expressed from the RPL7A locus were suppressed by expressing the RPL7B CRI from both loci. Thus, Rpl7 and snoRNA isoforms encoded by RPL7A and RPL7B have redundant, dosage-dependent functions. Overall, the findings suggest that cellular processes have different minimal threshold-values for the level of Rpl7 or snR39/59, and, consequently, 60S ribosomal subunits that are required, and that variations in 60S ribosomal subunit levels underlie the phenotypic diversity that results from deleting one RPL7 paralog or the other.
Materials and Methods
Yeast strains, media, and plasmids
All synthetic complete (SC) drop-out media contained 2% glucose unless otherwise noted. The strains used in this study are derivatives of strain BY4741 (Open Biosystems), and genotypes are listed in Supplemental Material, Table S1. Strain JC3212 harbors a chromosomal Ty1his3AI[∆1]-3114 element (Mou et al. 2006). Strain JC3807 is a derivative of JC3212 harboring Ty1(GAG:GFP)-3566 (Scholes et al. 2003). Oligonucleotide primers used to amplify PCR products for gene replacement are listed in Table S2. PCR products used for gene replacement were amplified with Phusion High Fidelity Polymerase (New England Biolabs). Each gene replacement was confirmed by two independent PCR analyses.
Plasmid pGSHU, a gift from Michael Resnick, harbors the GAL1P -I-SceI-hygB-K.l.URA3 counter-selectable delitto perfetto cassette (Storici and Resnick 2006). A PCR-amplified rpl7a∆::GAL1P -I-SceI-hygB-K.l.URA3 cassette was generated with primers PJ813 and PJ814 using plasmid pGHU as a template. The cassette was introduced into strain JC3212 by one-step gene replacement, and hygromycin-resistant (HygR) Ura+ isolates were selected and tested for the presence of the rpl7a∆::GAL1P -I-SceI-hygB-K.l.URA3 allele. An rpl7a∆::RPL7B cassette, amplified by PCR using genomic DNA of an rpl7a∆ RPL7B strain as a template, and primers PJ757 and PJ759, was introduced into rpl7a∆::GAL1P -I-SceI-hygB-K.l.URA3 strains JC5896 and JC5898 by one-step gene replacement. The RPL7B allele was deleted from this strain using an rpl7b∆::kanMX cassette that was PCR-amplified using primers PJ463 and PJ464, and genomic DNA from an rpl7b∆::kanMX derivative of strain BY4741 as a template. A rpl7b∆::GAL1P -I-SceI-hygB-K.l.URA3 cassette was amplified with primers PJ842 and PJ843 using plasmid pGHU as a template, and introduced into strain JC3212 by one-step gene replacement. An rpl7b∆::RPL7A cassette, generated by PCR with primers PJ845 and PJ846 and genomic DNA from an RPL7Arpl7b∆ strain as a template, was used to replace the rpl7b∆::GAL1P -I-SceI-hygB-K.l.URA3 allele in strains JC5900 and JC5901. The RPL7A allele was deleted from this strain using an rpl7a∆::kanMX cassette that was PCR-amplified using primers PJ459 and PJ460 and genomic DNA from the rpl7a∆::kanMX derivative of strain BY4741 as a template. An rpl31a∆::GAL1P-I-SceI-hygB-K.l.URA3 cassette, amplified with primers PJ893 and PJ894 using plasmid pGHU as a template, was introduced into strain JC3212 by gene replacement. An rpl31a∆::RPL31B allele, generated by PCR amplification with primers PJ895 and PJ896 using genomic DNA of an rpl31a∆ RPL31B strain as a template, was used to replace the rpl31a∆::GAL1P-I-SceI-hygB-K.l.URA3 allele in strains JC6130 and JC6131. The RPL31B allele in this strain was deleted by one-step gene replacement using a DNA fragment that was PCR-amplified with primers PJ471 and PJ472, and genomic DNA from the rpl31b∆::kanMX derivative of strain BY4741 as a template. The tif4631∆::kanMX derivative of strain JC3212 was made by gene replacement using a cassette that was PCR-amplified using primers PJ371 and PJ372, and the tif4631∆::kanMX derivative of strain BY4741 as a template. Strains harboring the rad52::hisG-URA3-hisG allele were constructed by one-step gene replacement using a DNA fragment isolated from plasmid pBDG542, as described previously (Curcio and Garfinkel 1994).
Derivatives of yeast strain JC3709 (Table S2) harboring 9xMYC-tagged alleles of RPL7A and RPL7B were made by using PCR-amplified RPL7A:9xMyc-K.l.TRP1 and RPL7B:9xMyc-K.l.TRP1 cassettes for one-step gene replacement. The RPL7A:9xMyc-K.l.TRP1 cassette was amplified using primers PJ564 and PJ565, and plasmid pYM6 (Knop et al. 1999) as a template. The RPL7B:9xMyc-K.l.TRP1 cassette was amplified using primers PJ566 and PJ567, and plasmid pYM6 as a template. The corresponding untagged paralog was deleted in each strain by one-step gene replacement using a PCR-amplified rpl7a∆::kanMX or rpl7b∆::kanMX cassette, respectively. Construction of these deletion cassettes is described above.
pBJC1058, a LEU2-CEN6 plasmid that contains the pLTRP:Gag1–401:GFP:ADH1TER cassette, has been described previously (Doh et al. 2014).
Plasmid pGTy1(synGAG) consists of pRS416 carrying a GAL1P-driven Ty1 element that encodes functional GAG and POL ORFs, but carries silent nucleotide substitutions in the 5′ UTR and GAG ORF, and lacks the 3′ LTR. Plasmid pGTy1(synGAG) is a derivative of plasmid pBJC838 (Stamenova et al. 2009). The transcriptional start site of Ty1 (nucleotide 241 of Ty1-H3; Boeke et al. 1988) is fused to GAL1P at a XhoI site. In addition, the 3′ LTR and his3AI marker gene were deleted by removal of a DNA fragment from the internal BglII site to the end of the element (nucleotides 5564–5918 of Ty1-H3). Finally, a sequence-optimized 577 bp XhoI–HpaI fragment has replaced the XhoI–HpaI fragment of Ty1-H3 (nucleotides 241–817 of Ty1-H3). The sequence-optimized fragment, synthesized by GenScript, harbors multiple nucleotide changes in the Ty1 RNA 5′ UTR region to reduce predicted secondary structure elements, as well as silent mutations in GAG predicted by the OptimumGene algorithm (GenScript) to optimize gene expression (Figure S1). The GAL1P:Ty1(synGAG) fragment is carried as an ApaI–EagI fragment on the LEU2-CEN6 vector pRS415.
Plasmid pBJC1198 contains the GAL1P-ASH1-MS2L(x12)-ADH1TER cassette on the LEU2-CEN6 vector, pRS415. The ASH1 ORF (from the start codon to the stop codon) was amplified from BY4741 genomic DNA with primers PJ1285, which introduced a 5′ XhoI site, and PJ1286, which introduced a 3′ BamHI site. The ApaI–XhoI fragment of plasmid pBJC838 containing the GAL1P (Stamenova et al. 2009), the XhoI–BamHI fragment containing the ASH1 ORF, and the BamHI–BglII fragment of pSL-MS2-12X (Bertrand et al. 1998) were cloned into pRS415 (Sikorski and Hieter 1989), following digestion with ApaI and BamHI. An Eag1 fragment harboring ADH1 terminator sequences (ADH1TER) was cloned into the EagI site of the resulting plasmid. ADH1TER was amplified from plasmid pBJC1058 with primers PJ1169 and PJ944.
Plasmid pMS2-CP-GFP(3X), a gift from Jeffrey Gerst, expresses the MS2 coat binding protein fused to three tandem GFP moieties, and is driven by the MET15 promoter (Haim-Vilmovsky and Gerst 2009). Basal expression of pMS2-CP-GFP(x3) in the presence of methionine was sufficient to visualize pASH1-MS2L(x12) mRNA while minimizing background signal.
Western blot analyses
Culture volumes corresponding to 2.5 OD600 units of cells grown to an OD600 of 0.6–1.0 at 20° were harvested, and protein was extracted as previously described (Yarrington et al. 2012). Proteins were separated on 10% SDS-Tris gels and transferred to an Immun-Blot PVDF membrane (Bio-Rad). To detect Gag, membranes were incubated with affinity-purified anti-VLP polyclonal antibodies (Conte and Curcio 2000), at a 1:35,000 dilution in 0.005% nonfat milk in phosphate-buffered saline and 0.05% Tween 20 (PBST), followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary polyclonal antibody (Millipore) in 0.005% nonfat milk in PBST. Rpl7a-9xMyc and Rpl7b-9xMyc were detected with a 1:5000 dilution of monoclonal anti-c-Myc antibody (clone 9E10; Sigma), followed by incubation with HRP-conjugated sheep anti-mouse secondary polyclonal antibody (GE Healthcare UK Limited; lot # 357597). Membranes were stripped in 50 mM Tris-HCl pH 7, 2% SDS, 50 mM DTT at 70°. Membranes originally used to detect Gag were incubated in incubated in a 1:2000 dilution of anti-humL7 polyclonal antibodies (Bethyl Labs, Inc.) in PBST, and then HRP-conjugated goat anti-rabbit secondary polyclonal antibodies (Millipore) in PBST. After stripping, the membrane was incubated in a 1:2000 dilution of anti-β-Actin monoclonal antibody (Abcam) in PBST with 2.5% nonfat milk, followed by incubation with a 1:4000 dilution of HRP-conjugated sheep anti-mouse secondary polyclonal antibody (GE Healthcare UK Limited; lot # 357597) in PBST with 2.5% nonfat milk. Membranes originally used to detect Rpl7a-9xMyc and Rpl7b-9xMyc were incubated in a 1:2000 dilution of anti-α-Tubulin polyclonal antibody (Chemicon International) in PBST with 2.5% nonfat milk, followed by incubation in a 1:5000 dilution of HRP-conjugated donkey anti-rat secondary polyclonal antibody (Millipore; lot # 2050222) in PBST with 2.5% nonfat milk. All bands were visualized by incubation of membranes in SuperSignal West Pico chemiluminescent substrate (Pierce) and exposure to film. Films were scanned, and the intensity of nonsaturated bands was determined using ImageJ software. Gag and Rpl7 values were individually divided by the values for β-Actin, and corrected for differences in dilution.
Growth assays
To determine the rate of cell doubling of each strain, cells were scraped from fresh patches on YPD agar, and were resuspended in YPD broth at an OD600 of <0.01. Three separate cultures of each strain were incubated with shaking at either 30 or 20° to OD600 ∼0.6. The doubling time (t2) was calculated for each culture based on the following equation (Amberg et al. 2005), where tf is the time in hours that the cultures were incubated, ODf is the OD600 of the cultures after incubation, and ODi is the OD600 of the dilutions before incubation:
For serial dilution growth assays, cells from fresh patches on YPD plates were resuspended in YPD broth at an OD600 of ∼0.3. These suspensions were diluted 10-fold serially, from 1:10 to 1:10,000. A 5 µl aliquot of each dilution was plated onto YPD agar, with or without 5 µg/ml tunicamycin (Sigma), and incubated at 30° for the indicated times.
Polysome analysis
Cells were grown to an OD600 of 0.4–0.6 at 20° in YPD broth, and 2.0 OD600 units of cells were collected. Cells were washed and lysed by bead beating in 20 mM Tris-HCl pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 1 mg/ml heparin, cOmplete Mini, EDTA-free protease inhibitor cocktail (Roche), and 200 units/ml RNasin (Promega). Debris was pelleted at 4000 rpm at 4° for 5 min. Clarified lysates were transferred to a fresh tube, and centrifuged at 10,000 rpm at 4° for 5 min. Cell extracts were separated by ultracentrifugation at 35,000 rpm in 10–50% (w/v) sucrose gradients containing 20 mM Tris-HCl pH 8.0, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, and 1 mg/ml heparin for 165 min at 4° in an SW41 rotor. Gradients were fractionated with a Teledyne/ISCO gradient fractionation system. Traces were recorded on a UA6 detector at a sensitivity setting of 0.2. In addition, traces for rpl7a∆ RPL7B and rpl7a∆ rpl7b∆::RPL7A strains were collected at a sensitivity setting of 0.1.
Fluorescence microscopy
Three independent transformants of strains JC3212, JC5987, JC5989, and JC5914 with plasmids pGAL1P-ASH1-MS2L(x12) and pMS2-CP-GFP(x3) were grown overnight in SC-HIS-LEU broth at 30°. Aliquots of each culture were inoculated into 10 ml of fresh SC-HIS-LEU broth containing 2% galactose, 2% raffinose, and 2% sucrose, and incubated at 30° to OD600 of 0.6. A 2.5 OD600 unit aliquot of each culture was harvested, resuspended in 1 ml of water, and incubated with 2.5 µl Hoechst 33258 for 20 min at room temperature. Cells were washed three times with water prior to visualization.
Detection of Ty1 RNA by fluorescence in situ hybridization was performed as previously described (Doh et al. 2014). A Zeiss Axioskop 200 M inverted microscope was used (filter set: 31 for Cy3, and 34 for DAPI) at a magnification of 63×. Photographs were taken with a Q Imagining Camera (or Hamamatsu ORCA ER), and merged and colored with Openlab 4.0.4 software (Improvision).
Gag-GFP activity assay
Two independent cultures of strain JC3807 and derivatives, which harbor the Ty1(GAG-GFP)-3566 chromosomal element (Scholes et al. 2003), and strain BY4741 and derivatives, which do not, were grown in YPD broth overnight at 30°. Duplicate cultures of each strain were diluted to an OD600 of 0.1 in YPD broth, and grown at 20° for 3 hr. The cells were diluted 1:10 in deionized H2O. The geometric mean of the GFP activity in 10,000–20,000 cells of each strain was measured by flow cytometry using a FACSCalibur (Becton Dickinson). The geometric mean of the GFP activity for each strain lacking Ty1(GAG-GFP)-3566 was subtracted from the mean GFP activity in the isogenic strain harboring Ty1(GAG-GFP)-3566 to correct for autofluorescence arising from differences in cell size.
RNA isolation and quantitative real-time PCR
Cells of each strain grown overnight in YPD broth at 30° were diluted to an OD600 of 0.3 in 50 ml YPD broth, and grown to an OD600 of 0.6 at 20°. Cell harvesting, RNA extraction, and DNase treatment was performed as previously described (Risler et al. 2012). Total RNA was used as a template for qRT-PCR, which was performed as described previously (Risler et al. 2012), with the following modifications. Ty1 and snR6 RNAs were reverse transcribed with the sequence-specific primers TY5253A and PJ751, respectively. Ty1 and snR6 single-stranded cDNAs were amplified via qPCR with PJ748 and PJ1230, and PJ750 and PJ1220, respectively. The average fold change in Ty1 RNA level was calculated according to the 2−∆∆Ct method using average Ty1 and snR6 cycle thresholds (Ct) from three biological replicates.
Ty1his3AI retrotransposition assays
To determine the retromobility rate of the Ty1his3AI-∆1-3114 element (Mou et al. 2006), three biological replicates of each strain were inoculated to a density of 1 × 10−4 cells/ml in seven cultures of YPD broth for a total of 21 cultures per genotype. Cultures were incubated with shaking at 20° for 3–4 d. A 1 µl aliquot of a 1 × 10−3 dilution of four randomly selected cultures was plated on SC agar to determine the titer. All seven cultures were plated on SC-HIS agar to determine the number of His+ prototrophs. The Ty1his3AI-∆1 retromobility rate for each genotype was determined by dividing the mean titer into the mutation rate, which was calculated by using the median number of His+ prototrophs per 1 ml culture in the Lea and Coulson median estimator (Rosche and Foster 2000).
To determine the frequency of Ty1his3AI-∆1-3114 retrotransposition during induction of expression from plasmid pGTy1(synGAG)∆3′LTR, four independent transformants of strains harboring plasmid pRS415, or pGTy1(synGAG)∆3′LTR, were inoculated into SC-LEU broth containing 2% raffinose and 2% sucrose, and incubated overnight at 30° with rolling. Cultures were diluted 1:10 in YP broth containing 2% galactose, 2% raffinose, and 2% sucrose, and incubated at 20° for 3 d. A 1 µl aliquot of each culture was plated on SC-LEU agar to determine the number of cells that retained the plasmid throughout incubation, and the remainder of each culture was plated on SC-LEU-HIS agar to determine the number of His+ prototrophs. The mean Ty1his3AI retromobility frequency is the mean of the number of His+Leu+ prototrophs per culture divided by the total number of Leu+ prototrophs per culture.
To determine the retromobility frequency of the Ty1his3AI-∆1-3114 element in congenic RAD52 and rad52 strains, seven independent cultures of yeast cells of each genotype were inoculated to a density of 1 × 10−4 cells/ml in YPD broth, and cultures were grown at 20° each genotype were inoculated to a density−6 dilution of each culture was plated on SC agar to determine the titer. The remaining culture was plated on SC-HIS agar to determine the number of His+ prototrophs. The retromobility frequency of each culture was calculated as the fraction of total cells that were His+. The mean retromobility frequency was determined from seven cultures of each genotype.
Quantitative Southern blot assay for Ty1 cDNA
To quantify unintegrated Ty1 cDNA in each strain, total genomic DNA was harvested from cells grown to saturation at 20°. DNA samples were digested with SphI, and separated on a 1% TBE agarose gel. Southern blot analysis was performed with an antisense Ty1 POL riboprobe in vitro-transcribed using plasmid pJC525 as a template. The intensity of bands corresponding to Ty1 cDNA, and two different genomic DNA bands, was measured with ImageQuant software.
Data availability
Strains and plasmids are available upon request. Figure S1 illustrates nucleotide substitutions in Ty1(syn-Gag) relative to Ty1-H3. Table S1 contains genotypes of strains used in this study. Table S2 contains oligonucleotide primers used in this study. Table S3 lists the chromosomal location and orientation of Ty1HIS3 transposition events. File S1 contains methods and references for Table S3. All data necessary for confirming the conclusions presented in the article are represented fully within the article and the associated supplementary files.
Results
Construction of chimeric RPL7 alleles
To determine whether the Rpl7 or snoRNA isoforms encoded by RPL7A and RPL7B have evolved distinct functions irrespective of the disparity in their expression, we created chimeric RPL7 alleles in which the CRI of one paralog, extending from the start codon to the stop codon, and including the snR39 or snR59 snoRNA gene, was substituted for the CRI of the other paralog. The resulting chimeric alleles consist of the CRI of one paralog, and the 5′ UTR, 3′ UTR, promoter, and terminator, denoted as the “locus,” of the other. Mutants harboring a chimeric allele were constructed by precisely replacing the CRI of RPL7A or RPL7B in strain JC3212 with a counterselectable delitto perfetto cassette (Storici and Resnick 2006) (Figure 1B). Each cassette was then precisely replaced with the RPL7B or RPL7A CRI, respectively. The resulting “homogenic” strains harbor the same CRI at both the native allele, and a chimeric rpl7b∆::RPL7A or rpl7a∆::RPL7B allele (Clarkson et al. 2010). Subsequently, the RPL7A or RPL7B CRI was deleted at its native locus in homogenic strains to yield “chimeragenic” strains (Figure 1B). Since one RPL7 gene is required for survival (Mizuta et al. 1992), the viability of chimeragenic strains demonstrates that both chimeric RPL7 alleles are functional.
Growth rates and 60S ribosomal subunit abundance in strains with different RPL7 alleles
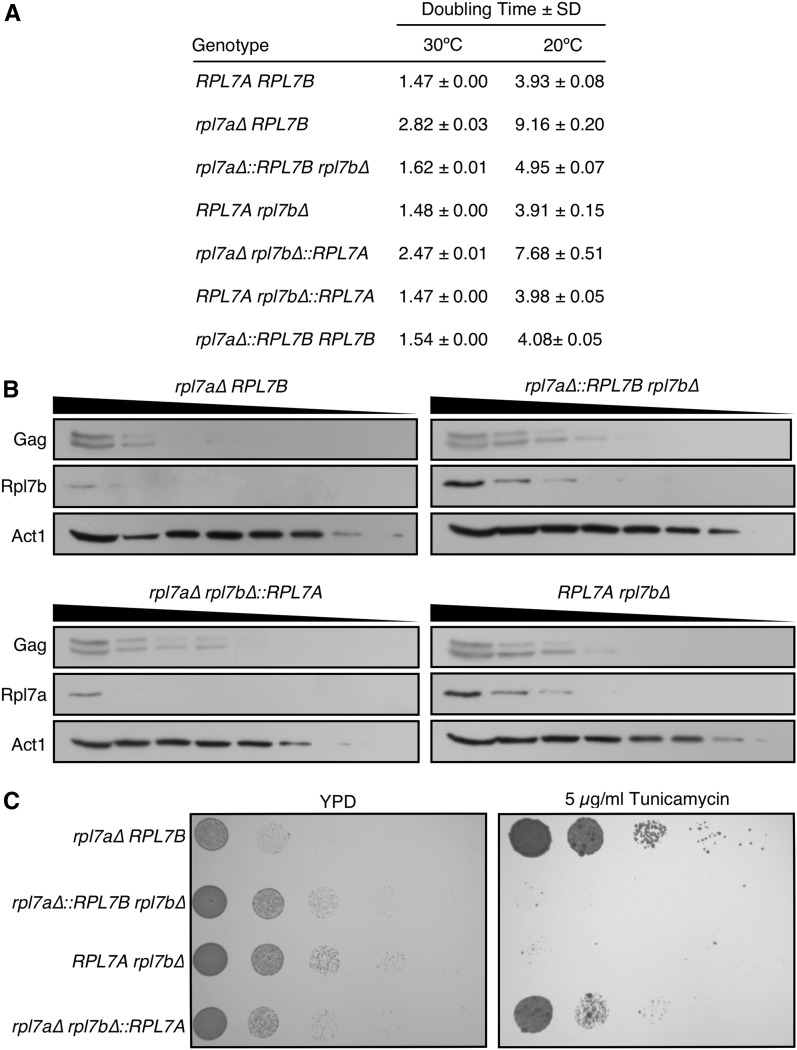
The growth rates in RPL7 deletion, chimeragenic and homogenic strains were measured in YPD broth at 30 and 20°, the latter permissive for retrotransposition of Ty1 (Figure 2A). The RPL7Arbl7b∆ mutant had a wild-type growth rate. Mutants expressing the RPL7A or RPL7B CRI from the RPL7B locus exhibited doubling times that were 1.7-fold longer at 30°, and 1.9- to 2.0-fold longer at 20°, when compared to strains expressing the corresponding CRI from the RPL7A locus. These data suggest that, in mutants harboring only one RPL7 allele, the RPL7A locus is more strongly expressed than the RPL7B locus. There was also a small increase in doubling time (1.1-fold longer at 30° and 1.2- to 1.3-fold longer at 20°) when the RPL7B vs. the RPL7A CRI was expressed from either locus, suggesting that higher levels of Rpl7a/snR39 are expressed from either RPL7 loci, or that Rpl7a/snR39 is more active than Rpl7b/snR59 in 60S ribosomal subunit biogenesis. Increasing the dosage of Rpl7b/snR59 by expressing the RPL7B CRI from both loci suppressed the major and minor growth defect of mutants expressing the RPL7B CRI from the RPL7B or RPL7A locus, respectively. In fact, the doubling times of the homogenic rpl7a∆::RPL7B strain at 20 and 30° were within 5% of those of the wild-type strain, and those of the homogenic RPL7Arpl7b∆::RPL7A strain were indistinguishable from wild type. These findings indicate that the RPL7 locus or loci expressed have a greater effect on cell doubling time than the particular RPL7 CRI that is expressed.
Figure 2.
The RPL7 locus is the primary determinant of cell growth rate, Rpl7 level and tunicamycin resistance. (A) The rate of cell doubling ± SD in three biological replicates of each strain of the indicated genotype grown in YPD broth at 20 or 30° to an OD600 of 0.6. (B) Western blot analysis of Rpl7 and Ty1 Gag, which runs as a doublet, in rpl7 deletion and chimeragenic strains. Each lane from left to right represents a twofold serial dilution of whole cell lysate. Gag was detected with anti-VLP antisera, and Rpl7 isoforms were detected with polyclonal anti-human L7 antibodies. Act1 was detected with a monoclonal anti-Actin antibody as a loading control. (C) The relative tunicamycin resistance of each strain of the indicated genotype grown to an OD600 of 0.6 in YPD broth at 30°. A 5 µl aliquot of each culture, and 1:10 serial dilutions were plated on YPD agar, with or without 5 µg/ml tunicamycin. Plates were incubated at 30° for 1 d (YPD) or 5 d (YPD with tunicamycin).
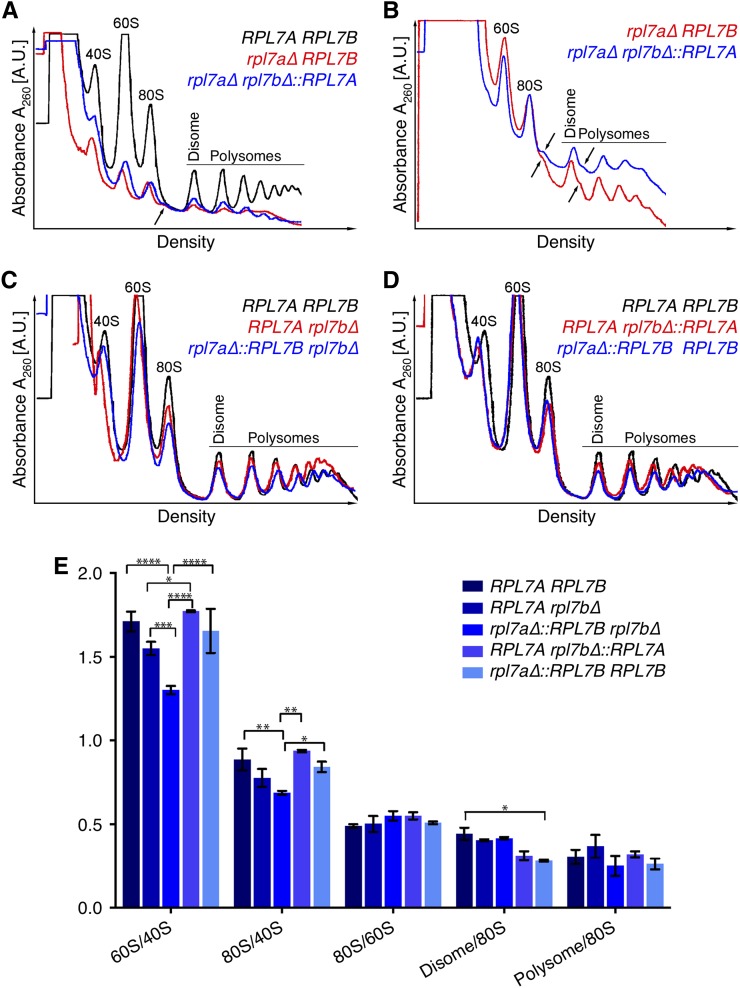
Polysome profiles of the RPL7 chimeragenic and deletion strains were compared to determine whether differences in cell doubling times are reflected in altered 60S ribosomal subunit levels (Figure 3). Cells were grown at 20° because differences in doubling time were more pronounced at this temperature (Figure 2A). Even the wild-type strain had higher 40S and 60S ribosomal subunit peaks than 80S or polysome peaks at this suboptimal temperature (Figure 3A). Nonetheless, mutants with either RPL7 CRI at the RPL7B locus only had greatly diminished levels of 60S ribosomal subunits, 80S monosomes, and polysomes relative to the wild-type strain (Figure 3A). In addition, halfmer peaks, in which 40S ribosomal subunits that are not joined with 60S subunits are associated with mRNA, were evident as shoulders on the 80S monosome and disome peaks (Figure 3B), indicating that the ratio of 60S to 40S ribosomal subunits is markedly reduced in these mutants. In contrast, the polysome profiles of mutants with either RPL7 CRI at the RPL7A locus more closely resembled that of the wild-type strain, and halfmer polysomes were not detected (Figure 3C). Compared to 40S ribosomal subunits, there was a significantly lower level of 60S ribosomal subunits, and reduced 80S monosomes and polysomes when the RPL7B CRI vs. the RPL7A CRI was present at the RPL7A locus (Figure 3, C and E). These discrepancies paralleled the modest difference in growth rate between these mutants. Expression of the RPL7B CRI from both RPL7 loci suppressed the 60S/40S ribosomal subunit imbalance of the rpl7a∆::RPL7B∆ mutant (Figure 3, C–E). The polysome profiles of homogenicRPL7Arpl7b∆::RPL7A and rpl7a∆::RPL7B strains were both similar to that of the wild-type strain, although there was a minor reduction in light polysomes in both (Figure 3D), and a decrease in the ratio of disomes to 80S monosomes in the rpl7a∆::RPL7B (Figure 3E).
Figure 3.
Primarily the RPL7 locus, but also the Rpl7 and snR39/59 isoforms expressed, contributes to differences in polysome profiles. Polysome profiles of cell extracts from strains of the indicated genotype grown in YPD broth at 20°, and fractionated in 10–50% (w/v) sucrose gradients. Absorbance units at 254 nm from the top to the bottom of the gradient for each genotype are shown in each graph. The same trace of the RPL7A RPL7B strain is displayed in black in (A), (C), and (D) to facilitate comparisons between panels. (A) Polysome profiles of the mutants with the RPL7B (red), or RPL7A (blue), CRI at the RPL7B locus are shown, and the genotypes indicated. Traces of the two mutants and the RPL7A RPL7B strain were overlaid by aligning the lowest point of each trace between the 80S monosome peak and the disome peak. The sensitivity setting of the UV monitor was 0.2. The arrow points to a minor peak of halfmer polyribosomes on the right shoulder of the 80S peak in rpl7a∆ RPL7B and rpl7a∆ rpl7b∆::RPL7A mutants. (B) Reanalysis of the rpl7a∆ RPL7B and rpl7a∆ rpl7b∆::RPL7A mutants, with the UV monitor at a more sensitive setting of 0.1, and profiles normalized to the 80S peak for better resolution of the halfmer polyribosome peaks, which are indicated by arrows. The 40S peak cannot be resolved at this sensitivity setting. (C, D) Polysome profiles of mutants of the indicated genotypes with the RPL7A (red), or RPL7B (blue), CRI at the RPL7A locus. In each panel, traces of three strains were overlaid by aligning the lowest point of each trace between the 80S monosome peak and disome peak. The sensitivity setting of the UV monitor was 0.2. (E) A graph of relative levels of 60S ribosomal subunits, 80S monosomes, disomes, and polysomes in three biological replicates of each strain of the indicated genotype. To normalize for differences in lysis efficiency and lysate amount, the heights of 40S, 60S, 80S, disome, and highest polysome peaks in polysome profiles were measured, and ratios indicated on the y-axis were calculated. Statistically significant differences in the indicated ratios between strains of different genotypes, as determined by a two-way ANOVA test, are indicated with asterisks. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
It was not possible to determine whether the 60S to 40S ribosomal subunit ratio differed in mutants with the RPL7B vs. RPL7A CRI at the RPL7B locus, since the level of 40S subunits was so low in both strains (Figure 3A). Nonetheless, the polysome data from mutants expressing RPL7 CRI from the RPL7A locus suggests that a reduced ratio of 60S to 40S ribosomal subunits is strongly correlated with differences in the growth rate of rpl7 deletion and chimeragenic strains.
To determine whether doubling time and 60S ribosomal subunit level differences among strains expressing only one native or chimeric RPL7 allele were reflected in differences in Rpl7 levels, western blot analysis of twofold serial dilutions of cell lysate with a human anti-L7 antibody that recognizes the identical epitope in Rpl7a and Rpl7b was performed. Rpl7 levels relative to level of a control protein, Act1 were measured by densitometry using bands at nonsaturating intensities. The Rpl7/Act1 ratio was >2-fold higher when the RPL7A CRI was expressed from the RPL7A locus vs. the RPL7B locus (0.045 vs. 0.020, respectively), or when the RPL7B CRI was expressed from the RPL7A locus vs. the RPL7B locus (0.029 vs. 0.012, respectively) (Figure 2B). In addition, there was a modestly higher level of Rpl7 when the RPL7A CRI vs. the RPL7B CRI was expressed from either the RPL7A locus (0.045 vs. 0.029) or the RPL7B locus (0.020 vs. 0.012). Therefore, differences in growth rates and relative 60S ribosomal subunit levels among strains expressing a single native or chimeric RPL7 allele may result, at least partially, from different Rpl7 levels. Together these findings indicate that RPL7Arpl7b∆ > rpl7a∆::RPL7B∆ > rpl7a∆ rpl7b∆::RPL7A > rpl7a∆ RPL7B for rate of growth, ratio of 60S to 40S ribosomal subunits, and Rpl7 level.
Negative correlation between tunicamycin resistance and Rpl7 and snR39/59 expression
Deletion of RPL7A, but not RPL7B, results in resistance to tunicamycin, an inhibitor of protein N-glycosylation in the endoplasmic reticulum (ER), and inducer of the ER stress response (Steffen et al. 2012). We asked whether resistance to tunicamycin is a function of expressing the RPL7B CRI or simply correlated with Rpl7 levels by plating serial dilutions of rpl7 deletion and chimeragenic strains on YPD agar, with or without 5 µg/ml tunicamycin (Figure 2C). Mutants with either RPL7 CRI at the RPL7A locus were sensitive to tunicamycin, while mutants with either RPL7 CRI at the RPL7B locus were resistant (Figure 2C). There was a small decrease in tunicamycin-resistance in the mutant with the RPL7A vs. the RPL7B CRI at the RPL7B locus (Figure 2C), which parallels the slightly faster growth and higher Rpl7 level in the rpl7a∆ rpl7b∆:RPL7A mutant (Figure 2, A and B). Together, the data suggest that tunicamycin resistance is correlated with Rpl7 and snR39/59 levels, and not a specific function of RPL7B isoforms.
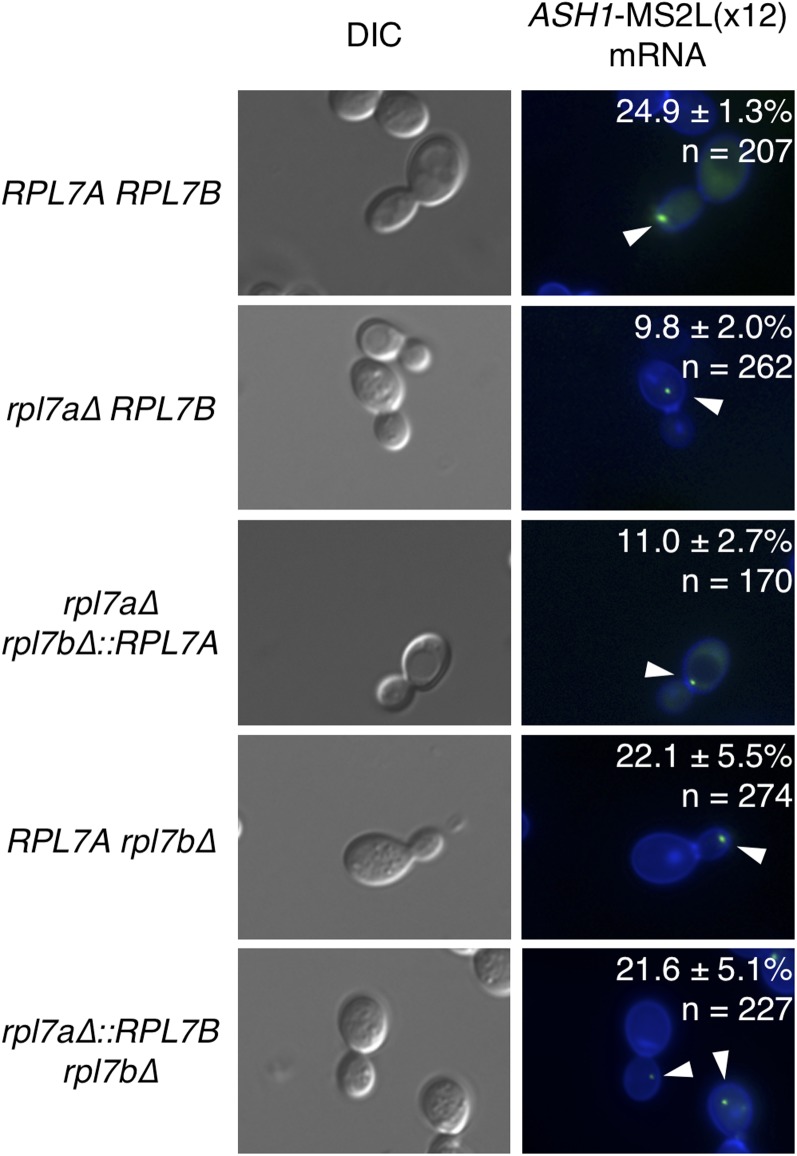
ASH1 mRNA localization in mutants with an RPL7A or rpl7a∆::RPL7B allele
The ASH1 mRNA transcript localizes to a focus at the tip of the budding daughter cell during bud growth, and migrates toward the bud neck as the cell progresses through M-phase (Beach et al. 1999). Deletion of RPL7A but not RPL7B significantly reduces the percentage of budded cells with bud-localized ASH1 mRNA foci (Komili et al. 2007). To determine whether the RPL7A isoforms or locus contribute to ASH1 mRNA localization to the bud, an mRNA consisting of the ASH1 ORF and 12 MS2 stem-loops [ASH1-MS2L(x12)] was expressed in rpl7 chimeragenic and deletion strains. Coexpression of an MS2 coat protein, which binds to the MS2L(x12) RNA sequences, fused to three GFP moieties [MS2-CP-GFP(x3)] was used to detect ASH1-MS2L(x12) RNA by fluorescent microscopy of live cells. As expected, a lower percentage of budded rpl7a∆ RPL7B cells had a bud-localized ASH1-MS2L(x12) mRNA focus compared to budded RPL7Arpl7b∆ or wild-type cells (Figure 4). An equivalently low level of bud-localized ASH1-MS2L(x12) mRNA was seen when RPL7A was expressed from the RPL7B locus (rpl7a∆ rpl7b∆::RPL7A, Figure 4), indicating that ASH1 mRNA localization is inefficient when either RPL7 CRI is expressed from the RPL7B locus. In contrast, expression of either CRI from the RPL7A locus resulted in the same percentage of budded cells exhibiting bud-localized ASH1-MS2L(x12) mRNA as in the wild-type strain. (Compare RPL7Arpl7b∆, rpl7a∆::RPL7B∆, and RPL7ARPL7B, Figure 4.) Together, these data suggest that either the RPL7A or RPL7B CRI, when expressed from the RPL7A locus results in sufficient levels of Rpl7 and snR39/59 to support wild-type levels of ASH1 mRNA localization.
Figure 4.
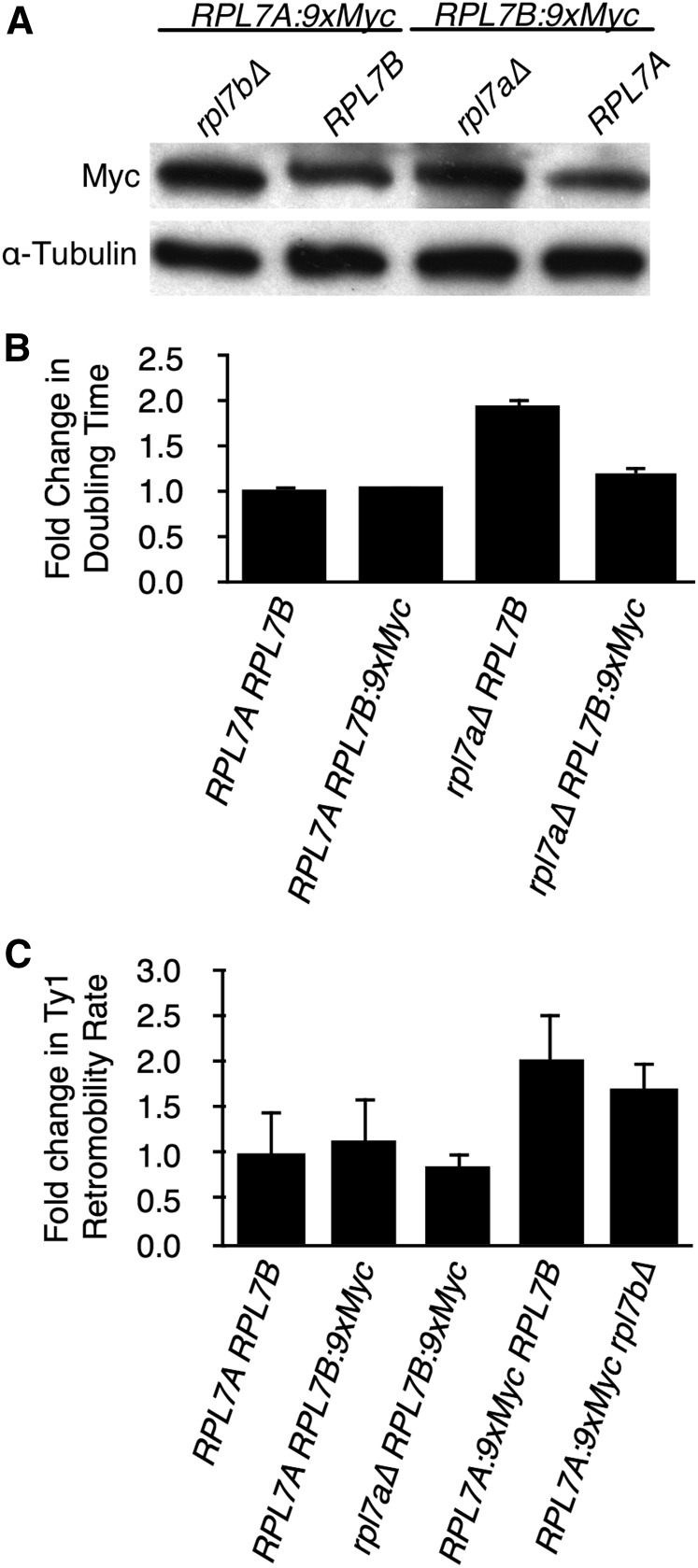
An epitope-tagged RPL7B allele suppresses the growth and Ty1 retromobility defect of an rpl7a∆ mutant. (A) Western blot analysis of Rpl7a-9xMyc or Rpl7b-9xMyc in whole cell lysate from strains, without or with deletions of RPL7B or RPL7A, respectively. Rpl7a-9xMyc and Rpl7b-9xMyc were detected with anti-c-Myc antibody, and α-Tubulin was detected with an anti-α-Tubulin antibody as a loading control. (B) The rate of cell doubling of three biological replicates of each strain of the indicated genotype grown in YPD broth at 20° to an OD600 of 0.4–0.6. Error bars represent SD. (C) The fold-change in retrotransposition rate is the retrotransposition rate of a chromosomal Ty1his3AI element in each strain of the indicated genotype relative to that in the RPL7A RPL7B strain. Error bars represent SE.
Ty1 retromobility is a function of robust Rpl7 and Rpl31 expression
Ty1 retromobility is strongly reduced in the absence of RPL7A, or a specific paralog of a few other RP gene pairs (Risler et al. 2012). Retrotransposons have the potential to damage the host genome when their rate of mobility or location of insertion into the genome is not tightly controlled; therefore, we considered the possibility that differences between Rpl7b/snR59 and Rpl7a/snR39 isoforms might confer a selective advantage on the cell by repressing Ty1 retromobility while maintaining ribosome-related functions. To determine whether Ty1 retromobility is a paralog-specific function of RPL7A, we used a chromosomal Ty1his3AI element to measure retromobility in RPL7 deletion strains. The his3AI retrotranscript indicator gene contains an antisense intron interrupting the HIS3 coding sequence. The his3AI reporter is located in the 3′ UTR of the chromosomal Ty1 element in the opposite transcriptional orientation; ergo, it can be spliced from the Ty1 transcript. Cells in which Ty1his3AI RNA is spliced and reverse transcribed, and the resulting Ty1HIS3 cDNA is inserted into the genome by integration or recombination, are detected as His+ prototrophs. The rate of His+ prototroph formation per cell per generation is directly proportional to the rate of Ty1his3AI retromobility (Curcio and Garfinkel 1991). At 20°, a permissive temperature for Ty1 retrotransposition, Ty1his3AI retromobility in the rpl7a∆ RPL7B mutant was <8% of that in the wild-type strain, but in an RPL7Arpl7b∆ mutant, the rate of retromobility was equivalent to that in a wild-type strain (Table 1). Thus, Ty1 retromobility is a paralog-specific function of RPL7A.
Table 1. Effect of RPL7 and RPL31 alleles on Ty1his3AI retromobility.
| Genotype | Rate of Ty1his3AI Retromobilitya ± SE × 10−7 | Relative Ty1his3AI Retromobility Rate |
|---|---|---|
| RPL7A RPL7B | 1.01 ± 0.23 | 1.0 |
| rpl7a∆ RPL7B | <0.08 | <0.08 |
| RPL7A rpl7b∆ | 1.07 ± 0.20 | 1.1 |
| rpl7a∆ rpl7b∆::RPL7A | 0.13 ± 0.03 | 0.1 |
| rpl7a∆::RPL7B rpl7b∆ | 0.82 ± 0.18 | 0.8 |
| rpl7a∆::RPL7B RPL7B | 0.77 ± 0.26 | 0.8 |
| RPL7A rpl7b∆::RPL7A | 0.82 ± 0.21 | 0.8 |
| RPL31A RPL31B | 0.68 ± 0.29 | 1.0 |
| rpl31a∆ RPL31B | 0.03 ± 0.05 | 0.05 |
| rpl31a∆::RPL31B rpl31b∆ | 0.79 ± 0.35 | 1.2 |
Median number of His+ prototrophs per cell per generation.
The rate of Ty1his3AI retromobility in the rpl7a∆::RPL7B∆ mutant was similar to that of the RPL7Arpl7b∆ strain, suggesting that Rpl7a/snR39 isoforms do not play a specialized role in Ty1 retrotransposition. This conclusion is also supported by the observation that homogenic mutants expressing either the RPL7A or the RPL7B CRI at both loci had high levels Ty1 retromobility that were comparable to each other, and similar to the wild-type strain. In contrast, the rpl7a∆ rpl7b∆::RPL7A mutant had a strongly reduced retromobility rate similar to that of the rpl7a∆ RPL7B mutant (Table 1). Thus, expression of either Rpl7a/snR39 or Rpl7b/snR59 isoforms from the RPL7A locus, but not the RPL7B locus, resulted in efficient Ty1 retromobility.
To further test the idea that the frequency of Ty1 retromobility is correlated with the level of Rpl7, we capitalized on an interesting observation: when RPL7B is tagged with a 9xMYC epitope at the 3′ end of the CRI, the level of Rpl7b:9xMyc increased upon deletion of RPL7A (Figure 5A). Addition of the 9xMYC tag to RPL7B did not change the doubling time of cells with an RPL7A allele, but it substantially decreased the doubling time of an rpl7a∆ mutant (Figure 5B). Thus, in contrast to the ∼2-fold difference in doubling time in an rpl7a∆ RPL7B mutant relative to the RPL7ARPL7B strain (Figure 2A and Figure 5B), there was a very slight difference in doubling time between the rpl7a∆ RPL7B:9xMYC mutant and RPL7ARPL7B:9xMYC strain (Figure 5B), consistent with the compensatory increase in Rpl7b:9xMyc protein when RPL7A was deleted. Also, there was no significant difference in the frequency of Ty1his3AI retromobility between the rpl7a∆ RPL7B:9xMYC mutant and RPL7ARPL7B:9xMYC strain (Figure 5C), which contrasts starkly with the >10-fold difference in Ty1his3AI retromobility between rpl7a∆ RPL7B and RPL7ARPL7B strains (Table 1). The data suggest that increased expression of Rpl7b:9xMyc upon deletion of RPL7A suppresses the retromobility defect that normally results from deleting RPL7A, and strongly support the idea that the RPL7A paralog is required for retromobility because of the level of Rpl7 expression rather than the expression of the Rpl7a or snR39 isoform.
Figure 5.
ASH1 mRNA localization requires a high dosage of Rpl7 and snR39/59. Expression of ASH1-MS2L(x12) RNA was induced in strains of the indicated genotype cotransformed with plasmid pGAL1PASH1-MS2L(x12), and plasmid pMS2-CP-GFP(x3), by growth in galactose-containing medium. The ASH1 ORF fragment in ASH1-MS2L(x12) RNA contains three sequence motifs—E1, E2 and E3—that target the mRNA for localization to the bud tip (Beach and Bloom 2001). Cells were incubated with Hoechst 33258 dye, which stains the yeast cell wall (Hernandez et al. 1988). Cells from two or three biological replicates were analyzed for a total of three independent experiments. Numerical values represent the average percent of budded cells with bud-localized ASH1-MS2L(x12) mRNA foci; n represents the total number of budded cells counted. White arrows point to ASH1-MS2L(x12) RNA foci in budded cells.
The RPL31A gene, but not RPL31B, was also identified as essential for efficient Ty1 retromobility (Risler et al. 2012). RPL31A and RPL31B ORFs are not interrupted by introns, and encode isoforms that differ at only one amino acid residue. The rate of Ty1his3AI retromobility in an rpl31a∆ RPL31B mutant was 5% of that in a wild-type strain (Table 1). Ty1his3AI retromobility in a chimeragenic strain expressing the RPL31B ORF from the RPL31A locus was measured to determine whether Rpl31 dosage or the Rpl31a isoform is required for Ty1 retromobility. The rpl31a∆::RPL31B∆ strain had a similar rate of Ty1 retromobility to that of the wild-type strain, and a 23-fold higher rate than the rpl31a∆ RPL31B mutant (Table 1), indicating that expression of either Rpl31 isoform from the RPL31A locus is sufficient for Ty1 retromobility. Since deletion of RPL31A but not RPL31B substantially retards cell doubling (Steffen et al. 2012), a higher dosage of Rpl31b when expressed from the RPL31A vs. RPL31B locus is the most plausible cause of more efficient Ty1 retromobility.
Minor effects of Rpl7 and snR39/59 levels on Ty1 RNA translation
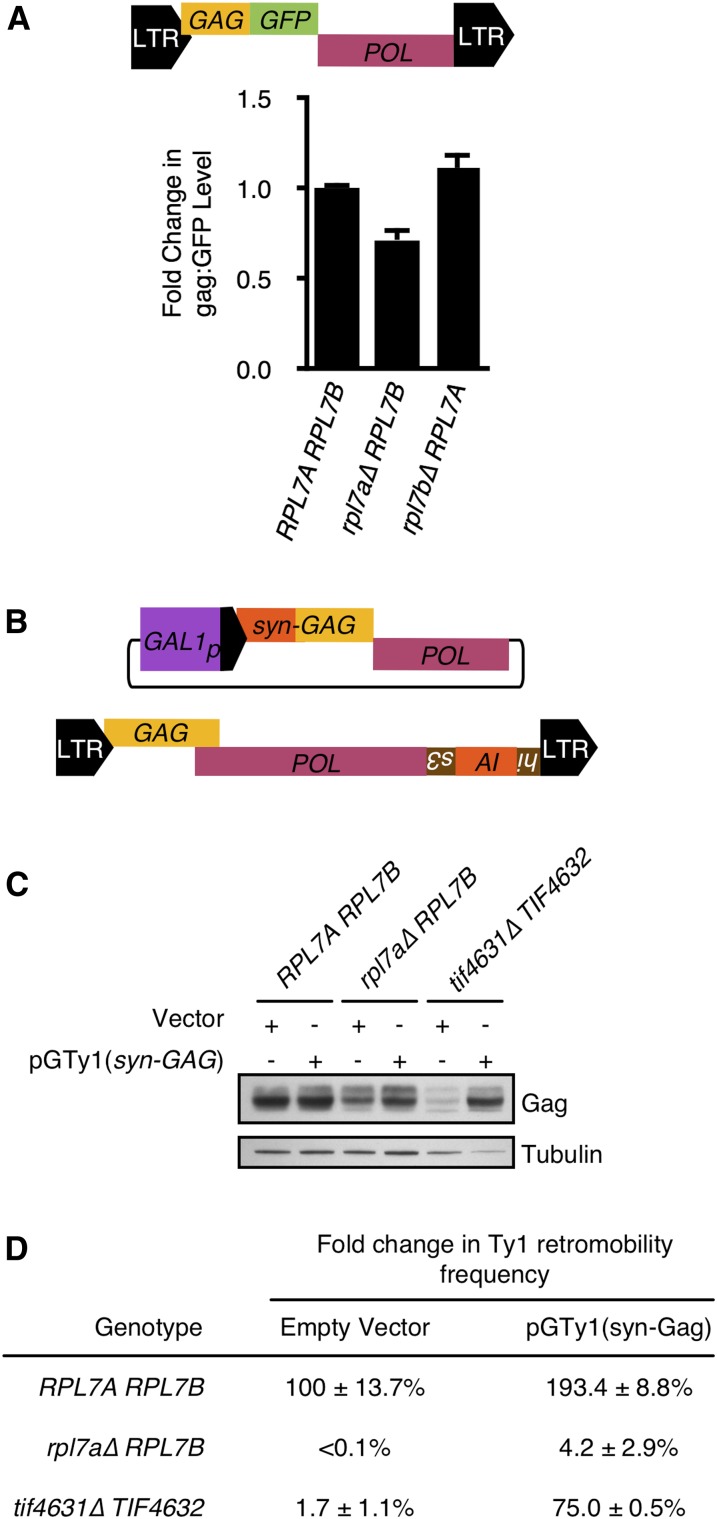
Depletion of 60S ribosomal subunits in mutants with low levels of Rpl7 could inhibit Ty1 retromobility by selectively reducing the rate of Ty1 RNA translation. However, Gag levels were not significantly different among chimeragenic and deletion mutants harboring one RPL7 allele (Figure 2B). The rpl7a∆ RPL7B mutant had 59% as much Gag as the RPL7Arpl7b∆ mutant, despite substantial differences in doubling time and 60S subunit levels between these strains (Figure 2, A and B). As an independent method of measuring relative Gag levels in the rpl7a∆ RPL7B mutant, we quantified Gag:GFP produced from a chromosomal Ty1(GAG:GFP) element (Figure 6A, top). The rpl7a∆ RPL7B mutant had ∼75% of the GFP activity in the wild-type strain or RPL7Arpl7b∆ mutant (Figure 6A, bottom). Thus, even the mutant with the strongest depletion of Rpl7 and 60S ribosomal subunits has only a minor defect in Gag accumulation, indicating that inhibition of Ty1 RNA translation may not be the major cause of the retromobility defect in the rpl7a∆ RPL7B mutant.
Figure 6.
Diminished Ty1 retromobility in the rpl7a∆ RPL7B mutant is not a result of limiting Gag levels. (A) A schematic of the chromosomal Ty1(GAG:GFP)-3566 element is shown (top). The GFP ORF is fused at the C-terminal end of the GAG ORF after codon 401 in a chromosomal Ty1 element (Scholes et al. 2003). The mean fluorescence in 10,000–20,000 cells of each genotype, corrected for autofluorescence by subtracting the mean fluorescence in each congenic strain lacking Ty1(GAG:GFP)-3566 from the mean fluorescence in each strain harboring Ty1(GAG:GFP)-3566, was determined, and the value relative to that in the RPL7A RPL7B strain is reported (bottom panel). Error bars represent the SD of mean fluorescence calculated from two independent isolates of each genotype. (B) Schematic of plasmid pGTy1(syn-GAG), which carries a modified Ty1 element that lacks the 3′ LTR, and has numerous silent nucleotide substitutions in the GAG ORF. Ty1(syn-GAG) RNA is defective for retromobility in cis, but encodes wild-type Gag and Gag-Pol proteins required in trans for retromobility of the chromosomal Ty1his3AI element, which is diagrammed below plasmid pGTy1(syn-GAG). (C) Western blot analysis of RPL7A RPL7B, rpl7a∆ RPL7B, and tif4631∆ TIF4632 strains harboring either the vector pRS415 or plasmid pGTy1(syn-GAG) and grown in galactose medium at 20°. The membrane was probed with anti-VLP antisera to detect Ty1 Gag (top), or with an anti-α-Tubulin antibody to detect α-Tubulin (bottom) as a loading control. (D) Effect of pGTy1(syn-GAG) expression on Ty1his3AI retromobility in different genetic backgrounds. RPL7A RPL7B, rpl7a∆ RPL7B and tif4631∆ TIF4632 strains harboring a chromosomal Ty1his3AI element, and the vector (pRS415), or plasmid pGTy1(syn-GAG) grown in galactose-containing medium at 20° to induce expression of Ty1 proteins, and the Ty1 retromobility frequency was determined by measuring the frequency of His+ prototrophs formed. Error represents the SD of four biological replicates.
To further determine whether the retromobility defect of the rpl7a∆ RPL7B mutant is a direct result of limiting levels of Ty1 proteins, the ability of Ty1 proteins supplied in trans to suppress the retromobility defect was tested. Ty1 proteins were expressed from a GAL1-driven modified Ty1 element whose transcript cannot be used as a template for retrotransposition. The plasmid-borne pGTy1(syn-GAG) element lacks a 3′ LTR, and has multiple nucleotide substitutions in the 5′ UTR and 5′ terminus of the GAG ORF that are predicted to disrupt RNA secondary structures and other cis-acting sequences required for retrotransposition (Curcio et al. 2015), but not the amino acid sequence of Gag (Figure 6B and Figure S1). As a positive control for suppression of a Ty1 RNA translation defect, pGTy1(syn-GAG) was also expressed in a tif4631∆ strain. In the absence of TIF4631, which encodes the translation initiation factor eIF4G1, the amount of Ty1 Gag is substantially reduced (Figure 6C, tif4631∆ TIF4632 strain with vector), presumably because eIF4G1 is required to initiate translation from the highly structured 5′ UTR of Ty1 RNA (Purzycka et al. 2013). In the tif4631∆ strain, galactose-mediated induction of pGTy1(syn-GAG) increased the amount of Gag (Figure 6C, compare tif4631∆ TIF4632 strain with pGTy1(syn-Gag) plasmid to that with vector). Accordingly, retromobility of the chromosomal Ty1his3AI element increased to 75% of that of the wild-type strain with a vector only (Figure 6D). In contrast, expression of pGTy1(syn-GAG) in the rpl7a∆ RPL7B strain increased Gag levels [Figure 6C, compare rpl7a∆ RPL7B strain with pGTy1(syn-GAG) to that with vector], but it only increased Ty1his3AI retromobility to 4.2% of that in the wild-type strain with a vector only (Figure 6D). Together, the data suggest that Gag is not the only limiting factor for retrotransposition in the rpl7a∆ RPL7B mutant.
High levels of Rpl7 and 60S ribosomal subunits are required for Ty1 RNA localization
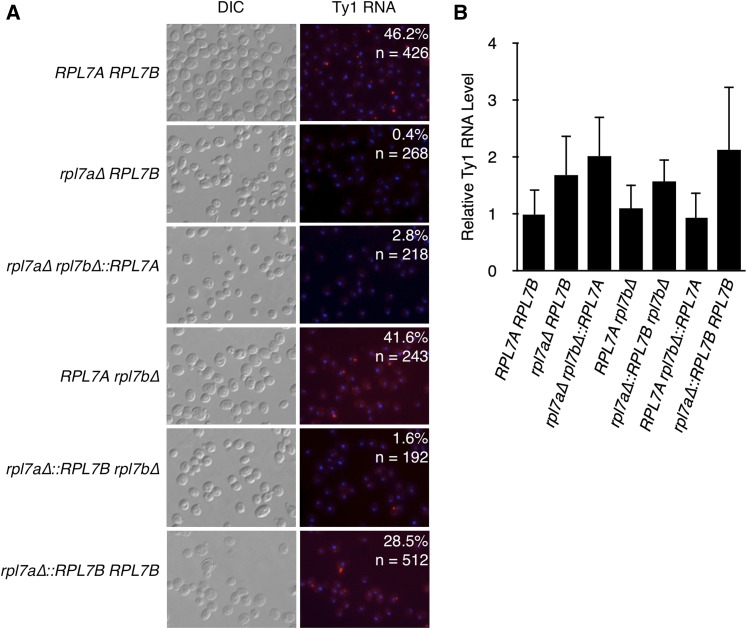
When cells are grown at the permissive temperature for Ty1 retrotransposition, Ty1 RNA is cotranslationally localized to a discrete cytoplasmic focus known as the retrosome, the presumptive site of VLP assembly (Malagon and Jensen 2008; Checkley et al. 2010; Sandmeyer and Clemens 2010; Doh et al. 2014). Ty1 RNA does not localize in retrosomes in the rpl7a∆ RPL7B mutant (Doh et al. 2014). To determine whether this localization defect is specific to deleting RPL7A, fluorescent in situ hybridization (FISH) of Ty1 RNA was performed in strains bearing a deletion of each RPL7 allele (Figure 7A). Consistent with the previous finding, the percentage of cells with at least one Ty1 RNA focus in an rpl7a∆ RPL7B strain was 0.4%; in contrast, 41.6% of RPL7Arpl7b∆ cells had a Ty1 RNA focus, which was similar to the level in the RPL7ARPL7B strain. Therefore, some feature of RPL7A, but not RPL7B, is required for Ty1 RNA localization to retrosomes. In the mutant with the RPL7A CRI expressed from the RPL7B locus, the percentage of cells with a Ty1 RNA focus was 2.8%, indicating that expression of either CRI from the RPL7B locus is not sufficient for Ty1 RNA localization. Notably, strains expressing the RPL7B CRI from the RPL7A locus also had a very low percentage of cells with a Ty1 RNA focus (1.6%). Lack of Ty1 RNA localization in these three Rpl7-deficient mutants is not a consequence of reduced Ty1 RNA levels (Figure 7B). Efficient retrosome formation in the RPL7Arpl7b∆ mutant, but not the rpl7a∆::RPL7B∆ mutant, raises the possibility that a very high level of Rpl7 or snR39/59—one which results in no growth defect or 60S ribosomal subunit deficiency—is necessary for Ty1 RNA to localize to retrosomes. Alternatively, retrosome formation could be a distinct function of the Rpl7a/snR39 isoforms, which must be expressed from the more robust RPL7A locus to perform this function efficiently. To differentiate between these possibilities, retrosomes in the homogenic strain expressing the RPL7B CRI from both RPL7 loci were quantified. The percentage of cells with a Ty1 RNA retrosome increased 18-fold relative to the strain expressing the RPL7B CRI from the RPL7A locus alone, (Figure 7A), indicating that Rpl7b/snR59 supports retrosome formation when expressed at high levels, and, therefore, that Ty1 RNA localization is not a distinct function of the Rpl7a/snR39 isoforms. The finding that Ty1 RNA efficiently localizes to retrosomes only in mutants with growth rates similar to the RPL7ARPL7B strain suggests that Ty1 RNA localization is very sensitive to reductions in 60S ribosomal subunits.
Figure 7.
A high dosage of Rpl7 or snR39/59 is required for localization of Ty1 RNA to retrosomes. (A) Fluorescent in situ hybridization was used to determine the number of cells harboring a cytoplasmic focus of Ty1 RNA, or retrosome. Ty1 RNA (red) was detected with a Cy3-labeled oligonucleotide probe. Nuclei (blue) were stained with DAPI. The percentage of cells with one or more Ty1 RNA retrosome is indicated. “n” is the number of cells scored for the presence of an RNA focus. (B) Quantitative real-time PCR of total RNA to determine the amount of Ty1 RNA relative to the amount of snR6 RNA in strains of the indicated genotypes. Error bars represent the SD of three biological replicates analyzed in triplicate.
Rpl7 or snR39/59 isoforms influence Ty1 cDNA accumulation in a dosage-dependent manner
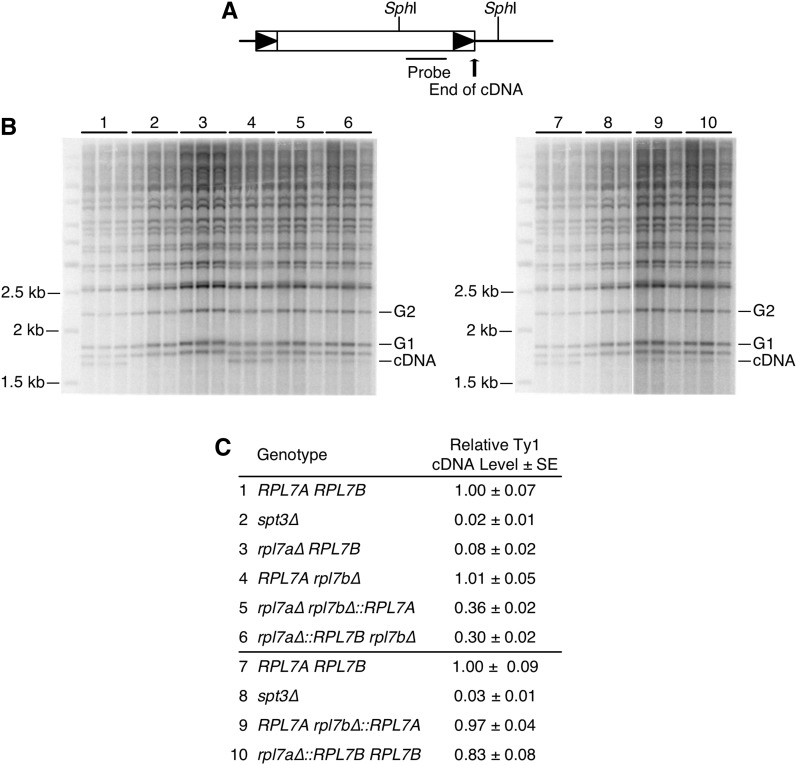
Given that Ty1 retrotransposes efficiently, but retrosomes rarely form in a mutant with modestly reduced Rpl7 and 60S subunit levels, we determined whether Ty1 cDNA accumulation, a step in retrotransposition between retrosome formation and transposition into the genome, is regulated by RPL7 paralogs. Unintegrated Ty1 cDNA levels in paralog deletion strains were measured relative to genomic copies of Ty1 via quantitative Southern blot analysis (Scholes et al. 2001). Deletion of RPL7A decreased Ty1 cDNA levels to 8% of the wild-type strain, whereas deletion of RPL7B had no effect on cDNA accumulation, demonstrating that Ty1 cDNA accumulation is a paralog-specific function of RPL7A (Figure 8). When the RPL7B CRI was substituted for the RPL7A CRI at the RPL7A locus, cDNA levels were reduced to 30% of that in the wild-type strain. On the other hand, substituting RPL7A for the RPL7B CRI at the RPL7B locus increased cDNA levels from 8 to 36% of that in the wild-type strain. Equivalent levels of Ty1 cDNA in the rpl7a∆ rpl7b∆::RPL7A and rpl7a∆::RPL7Brpl7b∆ mutants was not predicted by the substantially different Rpl7 and 60S ribosomal subunits levels between these mutants. Higher cDNA levels resulting from expression of Rpl7a/snR39, rather than Rpl7b/snR59 from both the RPL7A and RPL7B loci, suggested that Rpl7a/snR39 isoforms could have a specialized role in Ty1 cDNA accumulation. To determine whether this function is unique to Rpl7a/snR39, or can be performed by Rpl7b/snR59 at a higher dosage, cDNA levels in homogenic strains were measured (Figure 8). Expression of the RPL7B CRI from both loci increased the relative level of cDNA from 30 to 83% compared to expression from the RPL7A locus only. Thus, a mutant expressing Rpl7b/snR59 with a polysome profile that is comparable to the wild-type strain has efficient cDNA accumulation, indicating that cDNA accumulation is not a distinct function of Rpl7a/snR39. Nonetheless, equivalent levels of Ty1 cDNA in a mutant with low levels of Rpl7a, and a severe 60S ribosomal subunit defect, as well as a mutant with higher levels of Rpl7b, and a mild 60S subunit defect, suggests that Rpl7a or snR39 is more active than Rpl7b or snR59 at some aspect of Ty1 cDNA synthesis or stability.
Figure 8.
Rpl7 and snR39/59 isoforms expressed affect Ty1 cDNA accumulation in a dosage-and isoform-dependent manner. (A) Schematic of a chromosomal Ty1 element, with the positions of SphI sites in Ty1 and flanking genomic DNA indicated. The position of a riboprobe used in Southern blot analysis is indicated. (B) Southern blot analyses of three biological replicates of genomic DNA from each strain. Numbers correspond to strain genotypes indicated below in (C). The amount of unintegrated Ty1 cDNA is measured relative to genomic Ty1 elements in SphI-digested total DNA. The size of each Ty1:genomic DNA junction band is determined by the distance between the internal Ty1 SphI site and the nearest SphI site in flanking genomic DNA. The 1.7 kb Ty1 cDNA band is the fastest-migrating band because of the lack of flanking genomic DNA. The relative Ty1 cDNA level is the average ratio of the 32P-signal in the unintegrated Ty1 cDNA band (cDNA) relative to the average of two different Ty1:genomic DNA junction bands (G1 and G2) in three samples. Unmarked lanes: molecular weight marker. (C) Average (±SE) Ty1 cDNA level in each mutant relative to the RPL7A RPL7B strain. The spt3∆ strain, in which Ty1 RNA and cDNA are markedly decreased, was analyzed as a negative control.
Unaltered mechanism of cDNA integration in the rpl7a∆::RPL7B rpl7b∆ mutant
The rate of Ty1 retromobility in the rpl7a∆::RPL7Brpl7b∆ mutant was similar to that of the RPL7ARPL7B strain (Table 1), despite a low levels of Ty1 retrosomes, and a 70% reduction in unintegrated Ty1 cDNA. Therefore, we determined whether Ty1 cDNA is introduced into the genome of the rpl7a∆::RPL7Brpl7b∆ mutant via a mechanism that is different than that of the wild-type strain, which might uncover evidence for a specialized function Rpl7b/snR59 in retromobility. Ty1 cDNA integration is normally much more efficient than recombination with genomic Ty1 sequences. Accordingly, the homologous recombination factor, Rad52, is dispensable for retromobility, and deletion of RAD52 increases Ty1 retromobility by stimulating cDNA synthesis (Table 2) (Sharon et al. 1994; Rattray et al. 2000; Curcio et al. 2007). Deletion of RAD52 also increased retromobility in the rpl7a∆::RPL7Brpl7b∆ strain, suggesting that cDNA recombination is rare and integration remains the major mechanism of retromobility (Table 2). A second possibility is that the target site specificity of cDNA integration is altered when only Rpl7b/snR59 is expressed. Therefore, independent Ty1HIS3 integration sites in the rpl7a∆::RPL7Brpl7b∆ mutant were compared to those in a wild-type strain. Following selection for His+ prototrophs, the location of Ty1HIS3 was determined by sequencing Ty1HIS3 cDNA:genomic DNA junctions amplified by TAIL PCR (Table S3). Integration is usually targeted to a ∼750-bp window upstream of Pol III transcribed genes (Ji et al. 1993; Devine and Boeke 1996; Baller et al. 2012; Mularoni et al. 2012). In the RPL7ARPL7B strain, 12 of 17 Ty1HIS3 retromobility events integrated within 475 bp upstream of a Pol III-transcribed gene, and one was 150 bp downstream of a Pol III-transcribed gene. In the remaining four isolates, Ty1HIS3 cDNA was present within a tandem array of Ty1 cDNAs, so genomic sequences flanking these arrays could not be identified by TAIL PCR. A similar integration profile was observed for 15 independent Ty1HIS3 cDNA integration events in the rpl7a∆::RPL7Brpl7b∆ mutant. The two strains had equivalent fractions of Ty1HIS3 elements integrated upstream of Pol III-transcribed genes (11/15 vs. 12/17) and within multimeric arrays (4/15 vs. 4/17). One Ty1HIS3 element integrated into an ORF >6 kb away from the nearest Pol III-transcribed gene, but this low fraction of ORF disruption is not inconsistent with that observed in a study of >150,000 Ty1HIS3 insertions in a wild-type strain (Baller et al. 2012). We conclude that Ty1 target specificity is generally preserved in the rpl7a∆::RPL7Brpl7b∆ mutant, and, therefore, that expression of Rpl7b/snR59 alone does not result in an altered mechanism of Ty1 retromobility.
Table 2. Effect of RAD52 disruption on Ty1his3AI retromobility.
| Frequency of Ty1his3AI Retromobilitya ± SE × 10−7 | |||
|---|---|---|---|
| Genotype | RAD52 | rad52 | rad52/RAD52 Ratio |
| RPL7A RPL7B | 3.78 ± 0.13 | 102.63 ± 23.55 | 27 |
| RPL7A rpl7b∆ | 3.41 ± 0.48 | 88.10 ± 11.21 | 26 |
| rpl7a∆::RPL7B rpl7b∆ | 3.63 ± 0.74 | 57.30 ± 4.18 | 16 |
The mean of the number of His+ prototrophs per culture divided by the total number of cells plated.
Discussion
Different Rpl7 or snoRNA levels underlie paralog-specific functions of RPL7A and RPL7B
The question of whether the diverse phenotypic consequences of deleting one paralog or the other of an RP gene pair can be explained by differences in RP gene dosage, or constitute evidence of “specialized” ribosomes containing distinct subsets of RP isoforms has been widely debated (Komili et al. 2007; Parenteau et al. 2011, 2015; Steffen et al. 2012; Xue and Barna 2012; Slavov et al. 2015; Dinman 2016). Based on the mislocalization of Rpl7b, but not Rpl7a, when certain ribosome biogenesis factors were depleted, it was proposed that Rpl7 isoforms are incorporated into heterogenous ribosomes that selectively translate specific mRNAs, and give rise to paralog-specific functions of RPL7A and RPL7B (Mauro and Edelman 2002, 2007; Komili et al. 2007; Xue and Barna 2012). However, phenotypic comparisons of strains expressing either Rpl7a/snR39 or Rpl7b/snR59 from the robust RPL7A locus, or weaker RPL7B locus, suggest that paralog-specific functions of RPL7A and RPL7B can be explained by differences in the level of Rpl7 or RPL7-encoded snoRNA expression, and the effect of depleting these gene products on 60S ribosomal subunit levels. For example, the efficiencies of ASH1 mRNA localization, and Ty1 retromobility in mutants expressing either RPL7 CRI from the RPL7A locus, were similar to each other, and to the wild-type strain, whereas mutants with either RPL7 CRI at the RPL7B locus had marked defects in these processes. Because the rpl7a∆ RPL7B and rpl7a∆ rpl7b∆::RPL7A mutants have severe reductions in 60S ribosomal subunits that lead to halfmer ribosomes, and the rpl7a∆::RPL7Brpl7b∆ and RPL7Arpl7b∆ mutants have minor reductions in 60S ribosomal subunits; these observations indicate that ASH1 mRNA localization and Ty1 retromobility are correlated with the dosage of Rpl7 and snR39/59, and not with the specific isoforms expressed. The Ty1 retromobility phenotypes of strains expressing an epitope-tagged Rpl7b, the level of which increased when RPL7A is deleted, support this view, and further suggest that only the dosage of single Rpl7 isoform need be altered to produce different phenotypic outcomes. Dosage of the encoded product(s) also explains the paralog-specific regulation of Ty1 retromobility by RPL31A. Together, these findings support the hypothesis that phenotypic variation in complex cellular processes can result from different degrees of ribosomal subunit imbalance.
Other phenotypes examined here differed depending on which CRI was expressed from a particular locus. There was greater resistance to tunicamycin when Rpl7b/snR59 vs. Rpl7a/snR39 was expressed from the RPL7B locus, and Ty1 RNA localization was significantly more robust when Rpl7a/snR39 was expressed from the RPL7A locus. These phenotypic differences do not necessarily result from distinct, or even specialized, functions of the Rpl7 or snoRNA isoforms, because Rpl7a is more highly expressed from either locus than Rpl7b. The reason for this expression difference was not investigated, but it likely reflects the negative regulatory effects that RPL7B introns have on RPL7B mRNA expression (Parenteau et al. 2011). Growth rates and polysome profiles indicated that mutants expressing Rpl7a/snR39 have a less severe deficiency in 60S ribosomal subunits that parallels their higher Rpl7 levels. Thus, a different extent of 60S subunit depletion is the likely explanation for the phenotypic differences associated with disparate RPL7 CRI at the same locus. The importance of Rpl7 and snoRNA dosage and irrelevance of isoform in these phenotypes was further illustrated by the effects of increasing the dosage of Rpl7b/snR59. Tunicamycin resistance was suppressed by expressing Rpl7b/snR59 from the RPL7A locus, and the Ty1 RNA localization defect was rescued by expressing Rpl7b/snR59 from both RPL7 loci. The dependence of the phenotypes above on the dosage of Rpl7 or snoRNA strongly suggests that neither Rpl7 nor snR39/59 isoforms have distinct functions in the relevant cellular processes.
Ty1 cDNA accumulation was the single phenotype studied here that was not strictly correlated with the extent of 60S ribosomal subunit depletion. Similar levels of Ty1 cDNA in a rpl7a∆ rpl7b∆::RPL7A mutant, which has a severe 60S ribosomal subunit biogenesis defect, and an rpl7a∆::RPL7Brpl7b∆ mutant, which has only a modest reduction in 60S ribosomal subunits, suggests that Rpl7a or snR39 could have a specialized role in promoting cDNA synthesis or stability. Nonetheless, this is not a unique role, since expressing Rpl7b/snR59 from both RPL7 loci rescued the defect in cDNA accumulation in mutants expressing Rpl7b/snR59 from either single locus. The finding that defective cDNA accumulation is correlated with a depletion of Rpl7a/snR39 or Rpl7b/snR59, albeit to substantially different levels depending on the isoforms, argues against the idea that control of cDNA accumulation is an extraribosomal function of Rpl7 isoforms. Rather, these observations are consistent with Rpl7a being more active than Rpl7b in a specific ribosome-associated function. Numerous host cofactors and signaling pathways influence Ty1 cDNA levels (Curcio et al. 2015), and, thus, the translation of an mRNA that encodes a Ty1 cofactor could be more efficient in a strain expressing Rpl7a/snR39. The N-terminal domain of Rpl7, which contains four of the five amino acid substitutions that differentiate the Rpl7 isoforms and forms an α-helix located on the surface of the ribosome (Wilson and Doudna Cate 2012), could differentially recruit a factor that influences the efficiency of translation of specific mRNAs. Three of the four N-terminal substitutions in Rpl7a are serine and threonine residues, suggesting the possible involvement of signaling via post-translational modification of Rpl7a.
Although evidence for neofunctionalization of Rpl7, snR39/59 or Rpl31 isoforms was not uncovered, it remains possible that these isoforms have distinct, dosage-independent functions in other processes not tested here, and that RPL7 and RPL31 paralogs are differentially expressed as environmental conditions vary (Parenteau et al. 2011, 2015). The chimeragenic and homogenic RPL7 strains described here provide a useful tool for testing the isoform specificity of other cellular processes under a variety of environmental conditions. It is also possible that isoforms encoded by RP gene pairs other than RPL7 paralogs have unique functions in specialized ribosomes. Nonetheless, this study highlights the importance of considering the relative levels of each paralog’s protein products in the absence of the corresponding paralog when assigning RP isoform-specific functions. Other studies have used the rate of cell growth and polysome profiling in paralog-deletion strains as informative proxies of relative RP levels that are incorporated into functional ribosomes (Steffen et al. 2008, 2012), and these findings also support the idea that RP dosage and levels of functional ribosomes underlie many complex cellular phenotypes in yeast.
The effects of Rpl7 depletion on intermediate steps in Ty1 retromobility
Uncovering the mechanism by which Rpl7 or the RPL7-encoded snoRNAs regulate Ty1 retromobility was not the primary goal of this study, but several mechanistic conclusions can be drawn from our findings. First, Ty1 mRNA is not likely to be translated preferentially on ribosomes containing Rpl7a or snR39-modified rRNA, since equivalent levels of Gag accumulate when either the RPL7 CRI was expressed from the RPL7A locus. Second, depletion of Rpl7, even to the point of a severe deficiency in 60S ribosomal subunits, caused a <2-fold decrease in the level of Gag, and overexpressing Ty1 proteins only weakly rescued the Ty1 retromobility defect of an rpl7a∆ RPL7B mutant; therefore, reduced Gag is not likely to be the major cause of the decrease in retromobility in rpl7a∆ mutants. Many RP genes influence Ty1 retromobility (Harger et al. 2001; Griffith et al. 2003; Dakshinamurthy et al. 2010; Risler et al. 2012; Suresh et al. 2015), and several others that are superfluous for Ty1 Gag accumulation have been characterized (Suresh et al. 2015). Third, Ty1 retromobility was more sensitive to Rpl7 or snR39/59 depletion than Gag accumulation, suggesting that Rpl7 or snR39/59 might regulate retrotransposition at a post-translational level. However, it is important to note that our studies have not addressed the effect of Rpl7 depletion on translational frameshifting from the GAG to POL ORF, the efficiency of which impacts the level of Ty1 retrotransposition (Curcio et al. 2015). Fourth, localization of Ty1 RNA to retrosomes, and cDNA accumulation, had very low thresholds for Rpl7 and snR39/59 levels, suggesting that Rpl7 or RPL7-encoded snoRNAs might directly influence Ty1 RNA localization. While retrotransposition was efficient even in a mutant with reduced retrosomes and cDNA, the findings suggest a model in which Rpl7 or snR39/59 control the cotranslational localization of Ty1 RNA in retrosomes. Rpl7 or ribosomal RNA modifications made by snR39/59 could facilitate the interaction between Gag and Ty1 RNA on translating ribosomes, which is critical for retrosome formation (Doh et al. 2014). Another possibility supported by our findings is that depletion of Rpl7, like that of a few other 60S ribosomal subunit proteins, increases expression of a truncated form of Gag (p22-Gag) that potently inhibits retromobility and reduces retrosome formation (Saha et al. 2015; Suresh et al. 2015). It is not obvious how a shortage of 60S ribosomal subunits would increase the expression of the internal Ty1 transcript that encodes p22-Gag, but it could be via an indirect effect on cell metabolism or the cell cycle.
Variable phenotypic consequences of reduced expression of Rpl7
The results presented here suggest that different cellular pathways have varied thresholds for 60S ribosomal subunit depletion. For example, the dosage of Rpl7b/snR59 produced from the RPL7A locus, which results in a moderately reduced level of 60S ribosomal subunits, and slower growth relative to the wild-type strain, is sufficient for wild-type levels of ASH1 mRNA localization, and Ty1 retromobility, but insufficient for Ty1 RNA localization. Depletion of other 60S ribosomal subunits by deletion of different RP gene paralogs, or 60S subunit biogenesis factors, also quantitatively alters distinct cellular processes, including replicative life span and translation of the L-A virus mRNA (Ohtake and Wickner 1995; Steffen et al. 2008). Therefore, phenotypic variation in many cellular processes might be linked to varying degrees of ribosomal subunit imbalance. The asymmetric regulation and expression of many RP gene pairs also supports the idea that differences in ribosome levels could underlie paralog-specific phenotypes (Parenteau et al. 2011, 2015).
In humans, mutations in RP genes, most of which are present in a single copy, have been linked to a plethora of inherited developmental abnormalities, and somatic mutations in RP genes have been found in specific cancers. Tissue-specific defects that characterize specific mutations in various RP genes may reflect unique functions of ribosomes lacking a particular RP, or imbalances in ribosomal subunits that affect some cellular pathways more strongly than others, and each of these cellular pathways may be more or less critical for differentiation in specific cell types (Xue and Barna 2012; McCann and Baserga 2013; De Keersmaecker et al. 2015). In mice, ribosomopathy-associated tissue-specific abnormalities are, in some cases, linked to impaired translation of specific subsets of mRNAs (Xue and Barna 2012; McCann and Baserga 2013). For example, depletion of Rpl38 in certain tissues blocks translation of the mRNAs of a specific subset of homeobox genes during mouse embryogenesis; these mRNAs have specific cis-acting regulatory sequences in their 5′ UTRs that govern their dependence on Rpl38 (Kondrashov et al. 2011; Xue et al. 2015). What remains to be determined in human ribosomopathies and mouse models is whether the depletion of a specific RP changes the composition or level of functional ribosomes (McCann and Baserga 2013). Because Rpl7 is required for an early step in ribosome biogenesis in yeast, and there is a tight correlation between Rpl7 and 60S subunit levels (Jakovljevic et al. 2012), it is likely that the phenotypes associated with Rpl7 and snoRNA depletion in yeast are a result of a reduced level of functional ribosomes, rather than an increased percentage of ribosomes lacking Rpl7. Thus, this study supports a model in which subtle differences in ribosomal subunit biogenesis caused by depletion of a single RP result in varied phenotypic consequences, perhaps because translation of specific subsets of mRNAs have different sensitivities to imbalances between ribosomal subunits (Ohtake and Wickner 1995; Jakovljevic et al. 2012; Steffen et al. 2012). Characterization of global translation patterns in yeast strains that are homogenic or chimeragenic for a single RP paralog, or harbor a single native allele, could provide a systematic means of identifying classes of mRNAs that are differentially sensitive to ribosomal subunit imbalances.
Supplementary Material
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.035931/-/DC1.
Acknowledgments
We are grateful to Patrick Maxwell for sharing experimental findings that initiated this project, to Eric Gamache for technical contributions, to Pooja Flora for figure preparation, to Michael Resnick and Jeffrey Gerst for providing plasmids, and to Randy Morse and Dieter Kressler for thoughtful comments on the manuscript. We thank the Wadsworth Center Applied Genomic Technologies Core for DNA sequencing, the Wadsworth Center DAI Microscope facility for use of fluorescent microscopes, and the Wadsworth Immunology Core for use of the flow cytometer.
Footnotes
Communicating editor: J. Rine
Literature Cited
- Abovich N., Rosbash M., 1984. Two genes for ribosomal protein 51 of Saccharomyces cerevisiae complement and contribute to the ribosomes. Mol. Cell. Biol. 4: 1871–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D. C., Burke D. J., Strathern J. N., 2005. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Askree S. H., Yehuda T., Smolikov S., Gurevich R., Hawk J., et al. , 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101: 8658–8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badis G., Saveanu C., Fromont-Racine M., Jacquier A., 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15: 5–15. [DOI] [PubMed] [Google Scholar]
- Baller J. A., Gao J., Stamenova R., Curcio M. J., Voytas D. F., 2012. A nucleosomal surface defines an integration hotspot for the Saccharomyces cerevisiae Ty1 retrotransposon. Genome Res. 22: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. L., Bloom K., 2001. ASH1 mRNA localization in three acts. Mol. Biol. Cell 12: 2567–2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach D. L., Salmon E. D., Bloom K., 1999. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 9: 569–578. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Garreau de Loubresse N., Melnikov S., Jenner L., Yusupova G., et al. , 2011. The structure of the eukaryotic ribosome at 3.0 A resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S. M., Singer R. H., et al. , 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 2: 437–445. [DOI] [PubMed] [Google Scholar]
- Boeke J. D., Eichinger D., Castrillon D., Fink G. R., 1988. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol. Cell. Biol. 8: 1432–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino C. J., Chavez E. M., Bonifacino J. S., 2002. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley M. A., Nagashima K., Lockett S. J., Nyswaner K. M., Garfinkel D. J., 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30: 382–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson B. K., Gilbert W. V., Doudna J. A., 2010. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoS One 5: e9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte D., Curcio M. J., 2000. Fus3 controls Ty1 transpositional dormancy through the invasive growth MAPK pathway. Mol. Microbiol. 35: 415–427. [DOI] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. USA 88: 936–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Garfinkel D. J., 1994. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136: 1245–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Lutz S., Lesage P., 2015. The Ty1 LTR-retrotransposon of budding yeast, Saccharomyces cerevisiae. Microbiol. Spectr. 2: 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio M. J., Kenny A. E., Moore S., Garfinkel D. J., Weintraub M., et al. , 2007. S-phase checkpoint pathways stimulate the mobility of the retrovirus-like transposon Ty1. Mol. Cell. Biol. 27: 8874–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakshinamurthy A., Nyswaner K. M., Farabaugh P. J., Garfinkel D. J., 2010. BUD22 affects Ty1 retrotransposition and ribosome biogenesis in Saccharomyces cerevisiae. Genetics 185: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean E. J., Davis J. C., Davis R. W., Petrov D. A., 2008. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 4: e1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K., Sulima S. O., Dinman J. D., 2015. Ribosomopathies and the paradox of cellular hypo- to hyperproliferation. Blood 125: 1377–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devine S. E., Boeke J. D., 1996. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 10: 620–633. [DOI] [PubMed] [Google Scholar]
- Dinman J. D., 2016. Pathways to specialized ribosomes: the Brussels lecture. J. Mol. Biol. 428: 2186–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh J. H., Lutz S., Curcio M. J., 2014. Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet. 10: e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng F. J., Warner J. R., 1991. Structural basis for the regulation of splicing of a yeast messenger RNA. Cell 65: 797–804. [DOI] [PubMed] [Google Scholar]
- Fewell S. W., Woolford J. L., Jr, 1999. Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 19: 826–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., et al. , 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. [DOI] [PubMed] [Google Scholar]
- Gilbert W. V., 2011. Functional specialization of ribosomes? Trends Biochem. Sci. 36: 127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J. L., Coleman L. E., Raymond A. S., Goodson S. G., Pittard W. S., et al. , 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haarer B., Viggiano S., Hibbs M. A., Troyanskaya O. G., Amberg D. C., 2007. Modeling complex genetic interactions in a simple eukaryotic genome: actin displays a rich spectrum of complex haploinsufficiencies. Genes Dev. 21: 148–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim-Vilmovsky L., Gerst J. E., 2009. m-TAG: a PCR-based genomic integration method to visualize the localization of specific endogenous mRNAs in vivo in yeast. Nat. Protoc. 4: 1274–1284. [DOI] [PubMed] [Google Scholar]
- Harger J. W. J. W., Meskauskas A. A., Nielsen J. J., Justice M. C. M. C., Dinman J. D. J. D., 2001. Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology 286: 216–224. [DOI] [PubMed] [Google Scholar]
- Hernandez R., Diaz-de Leon F., Castaneda M., 1988. Molecular cloning and partial characterization of ribosomal RNA genes from Trypanosoma cruzi. Mol. Biochem. Parasitol. 27: 275–279. [DOI] [PubMed] [Google Scholar]
- Innan H., Kondrashov F., 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108. [DOI] [PubMed] [Google Scholar]
- Jakovljevic J., Ohmayer U., Gamalinda M., Talkish J., Alexander L., et al. , 2012. Ribosomal proteins L7 and L8 function in concert with six A(3) assembly factors to propagate assembly of domains I and II of 25S rRNA in yeast 60S ribosomal subunits. RNA 18: 1805–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Moore D. P., Blomberg M. A., Braiterman L. T., Voytas D. F., et al. , 1993. Hotspots for unselected Ty1 transposition events on yeast chromosome III are near tRNA genes and LTR sequences. Cell 73: 1007–1018. [DOI] [PubMed] [Google Scholar]
- Knop M., Siegers K., Pereira G., Zachariae W., Winsor B., et al. , 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15: 963–972. [DOI] [PubMed] [Google Scholar]
- Komili S., Farny N. G., Roth F. P., Silver P. A., 2007. Functional specificity among ribosomal proteins regulates gene expression. Cell 131: 557–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov N., Pusic A., Stumpf C. R., Shimizu K., Hsieh A. C., et al. , 2011. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 145: 383–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Molenaar C. M., Witsenboer H. M., Mager W. H., et al. , 1985. Yeast contains two functional genes coding for ribosomal protein S10. Nucleic Acids Res. 13: 5027–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Paulovich A. G., Woolford J. L., Woolford J. L., Jr, 1995. Feedback inhibition of the yeast ribosomal protein gene CRY2 is mediated by the nucleotide sequence and secondary structure of CRY2 pre-mRNA. Mol. Cell. Biol. 15: 6454–6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A., 1991. Localization of surface peptide from ribosomal protein L7 on 80 S ribosome by biotinylation. FEBS Lett. 287: 121–124. [DOI] [PubMed] [Google Scholar]
- Malagon F., Jensen T. H., 2008. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 28: 6022–6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V. P., Edelman G. M., 2002. The ribosome filter hypothesis. Proc. Natl. Acad. Sci. USA 99: 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro V. P., Edelman G. M., 2007. The ribosome filter redux. Cell Cycle 6: 2246–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann K. L., Baserga S. J., 2013. Mysterious ribosomopathies. Science 341: 849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta K., Hashimoto T., Otaka E., 1992. Yeast ribosomal proteins: XIII. Saccharomyces cerevisiae YL8A gene, interrupted with two introns, encodes a homolog of mammalian L7. Nucleic Acids Res. 20: 1011–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z., Kenny A. E., Curcio M. J., 2006. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172: 2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mularoni L., Zhou Y., Bowen T., Gangadharan S., Wheelan S. J., et al. , 2012. Retrotransposon Ty1 integration targets specifically positioned asymmetric nucleosomal DNA segments in tRNA hotspots. Genome Res. 22: 693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münk C., Willemsen A., Bravo I. G., 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol. Biol. 12: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Snyder M., 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol. Biol. Cell 12: 2147–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake Y., Wickner R. B., 1995. Yeast virus propagation depends critically on free 60S ribosomal subunit concentration. Mol. Cell. Biol. 15: 2772–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page N., Gerard-Vincent M., Menard P., Beaulieu M., Azuma M., et al. , 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J., Durand M., Morin G., Gagnon J., Lucier J. F., et al. , 2011. Introns within ribosomal protein genes regulate the production and function of yeast ribosomes. Cell 147: 320–331. [DOI] [PubMed] [Google Scholar]
- Parenteau J., Lavoie M., Catala M., Malik-Ghulam M., Gagnon J., et al. , 2015. Preservation of gene duplication increases the regulatory spectrum of ribosomal protein genes and enhances growth under stress. Cell Rep. 13: 2516–2526. [DOI] [PubMed] [Google Scholar]
- Paulovich A. G., Thompson J. R., Larkin J. C., Li Z., Woolford J. L., Jr, 1993. Molecular genetics of cryptopleurine resistance in Saccharomyces cerevisiae: expression of a ribosomal protein gene family. Genetics 135: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D., Decatur W. A., Fournier M. J., 2007. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA 13: 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presutti C., Ciafre S. A., Bozzoni I., 1991. The ribosomal protein L2 in S. cerevisiae controls the level of accumulation of its own mRNA. EMBO J. 10: 2215–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purzycka K. J., Legiewicz M., Matsuda E., Eizentstat L. D., Lusvarghi S., et al. , 2013. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 41: 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A. J., Shafer B. K., Garfinkel D. J., 2000. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics 154: 543–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risler J. K., Kenny A. E., Palumbo R. J., Gamache E. R., Curcio M. J., 2012. Host co-factors of the retrovirus-like transposon Ty1. Mob. DNA 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosche W. A., Foster P. L., 2000. Determining mutation rates in bacterial populations. Methods 20: 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg M. O., Moritz M., Woolford J. L., Jr, 1988. Depletion of Saccharomyces cerevisiae ribosomal protein L16 causes a decrease in 60S ribosomal subunits and formation of half-mer polyribosomes. Genes Dev. 2: 160–172. [DOI] [PubMed] [Google Scholar]
- Saha A., Mitchell J. A., Nishida Y., Hildreth J. E., Ariberre J. A., et al. , 2015. A trans-dominant form of Gag restricts Ty1 retrotransposition and mediates copy number control. J. Virol. 89: 3922–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeyer S. B., Clemens K. A., 2010. Function of a retrotransposon nucleocapsid protein. RNA Biol. 7: 642–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D. T., Banerjee M., Bowen B., Curcio M. J., 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D. T., Kenny A. E., Gamache E. R., Mou Z., Curcio M. J., 2003. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. USA 100: 15736–15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Burkett T. J., Garfinkel D. J., 1994. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol. Cell. Biol. 14: 6540–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P., 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoff I., Moradi H., Nygard O., 2009. Functional characterization of ribosomal protein L15 from Saccharomyces cerevisiae. Curr. Genet. 55: 111–125. [DOI] [PubMed] [Google Scholar]
- Slavov N., Semrau S., Airoldi E., Budnik B., van Oudenaarden A., 2015. Differential stoichiometry among core ribosomal proteins. Cell Rep. 13: 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamenova R., Maxwell P. H., Kenny A. E., Curcio M. J., 2009. Rrm3 protects the Saccharomyces cerevisiae genome from instability at nascent sites of retrotransposition. Genetics 182: 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K. K., MacKay V. L., Kerr E. O., Tsuchiya M., Hu D., et al. , 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen K. K., McCormick M. A., Pham K. M., MacKay V. L., Delaney J. R., et al. , 2012. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 191: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storici F., Resnick M. A., 2006. The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol. 409: 329–345. [DOI] [PubMed] [Google Scholar]
- Suresh S., Ahn H. W., Joshi K., Dakshinamurthy A., Kananganat A., et al. , 2015. Ribosomal protein and biogenesis factors affect multiple steps during movement of the Saccharomyces cerevisiae Ty1 retrotransposon. Mob. DNA 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. J., Lee H. I., Lin A., 2012. Ribosome distribution in HeLa cells during the cell cycle. PLoS One 7: e32820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenti S., De Chiara V., Bozzoni I., Presutti C., 2007. The position of yeast snoRNA-coding regions within host introns is essential for their biosynthesis and for efficient splicing of the host pre-mRNA. RNA 13: 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N., Doudna Cate J. H., 2012. The structure and function of the eukaryotic ribosome. Cold Spring Harb. Perspect. Biol. 4: a011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K. H., Shields D. C., 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708–713. [DOI] [PubMed] [Google Scholar]
- Woolford J. L., Jr, Baserga S. J., 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Barna M., 2012. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S., Tian S., Fujii K., Kladwang W., Das R., et al. , 2015. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 517: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrington R. M., Richardson S. M., Lisa Huang C. R., Boeke J. D., 2012. Novel transcript truncating function of Rap1p revealed by synthetic codon-optimized Ty1 retrotransposon. Genetics 190: 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains and plasmids are available upon request. Figure S1 illustrates nucleotide substitutions in Ty1(syn-Gag) relative to Ty1-H3. Table S1 contains genotypes of strains used in this study. Table S2 contains oligonucleotide primers used in this study. Table S3 lists the chromosomal location and orientation of Ty1HIS3 transposition events. File S1 contains methods and references for Table S3. All data necessary for confirming the conclusions presented in the article are represented fully within the article and the associated supplementary files.