Abstract
NF-κB signaling is a central pathway of immunity and integrates signal transduction upon a wide array of inflammatory stimuli. Noncanonical NF-κB signaling is activated by a small subset of TNF family receptors and characterized by NF-κB2/p52 transcriptional activity. The medical relevance of this pathway has recently re-emerged from the discovery of primary immunodeficiency patients that have loss-of-function mutations in the MAP3K14 gene encoding NIK. Nevertheless, knowledge of protein interactions that regulate noncanonical NF-κB signaling is sparse. Here we report a detailed state-of-the-art mass spectrometry-based protein–protein interaction network including the noncanonical NF-κB signaling nodes TRAF2, TRAF3, IKKα, NIK, and NF-κB2/p100. The value of the data set was confirmed by the identification of interactions already known to regulate this pathway. In addition, a remarkable number of novel interactors were identified. We provide validation of the novel NIK and IKKα interactor FKBP8, which may regulate processes downstream of noncanonical NF-κB signaling. To understand perturbed noncanonical NF-κB signaling in the context of misregulated NIK in disease, we also provide a differential interactome of NIK mutants that cause immunodeficiency. Altogether, this data set not only provides critical insight into how protein–protein interactions can regulate immune signaling but also offers a novel resource on noncanonical NF-κB signaling.
Keywords: noncanonical NF-κB signaling, tandem affinity purification, protein−protein interaction network, NF-κB inducing kinase
Introduction
NF-κB signaling is one of the most important signaling pathways in immunity with two major branches, termed canonical and noncanonical. These pathways are characterized by activation of different transcription factor heterodimers that are members of the NF-κB family (REL-A/p65, REL-B, c-REL, NF-κB1/p50, and NF-κB2/p52). Canonical NF-κB signaling is activated by a wide array of inflammatory stimuli (e.g., cytokine receptors, pattern recognition receptors, antigen receptors). Signaling occurs via an IKKα/IKKβ/IKKγ complex and, further, via the formation of p65/p50 heterodimers.1 In contrast, noncanonical NF-κB signaling is characterized by the induction of proteolytic processing of NF-κB2/p100 to p522 (Figure 1A). REL-B and p52 form a heterodimer that translocates to the nucleus and transcriptionally activates a subset of NF-κB target genes. These are primarily involved in homeostasis, plus development and functioning of the adaptive immune system.3 The noncanonical pathway is activated by a small group of TNF family receptors including B-cell activating factor (BAFF), lymphotoxin β receptor, and CD40.2 The majority of these receptors, however, can also activate canonical NF-κB signaling. Downstream of noncanonical receptor activation, signals are relayed through the TNF receptor-associated factor (TRAF) proteins TRAF2 and TRAF3 via regulation of the serine/threonine kinase NF-κB inducing kinase (NIK, MAP3K14).4 NIK is central to noncanonical NF-κB signaling. Under steady-state conditions, a complex containing TRAF3 (a ubiquitin ligase that acts on NIK and thus represents a negative regulator) and TRAF2 continuously targets NIK for degradation.5 Upon receptor ligation, TRAF3 is degraded and NIK accumulates. This enables NIK to activate phosphorylation of the downstream effectors, kinase IKKα (IκB kinase α) and NF-κB2/p100.6 NIK is the primary kinase that can induce NF-κB2/p100 proteolytic processing to produce the active p52 transcription factor (reviewed in ref (2)). Therefore, NIK is essential for signaling via the noncanonical NF-κB branch.4 Signaling via either branch of the NF-κB pathway is flexible, as crosstalk between canonical and noncanonical NF-κB pathways is known to occur in different situations and in various cell types (reviewed in ref (7)).
Figure 1.
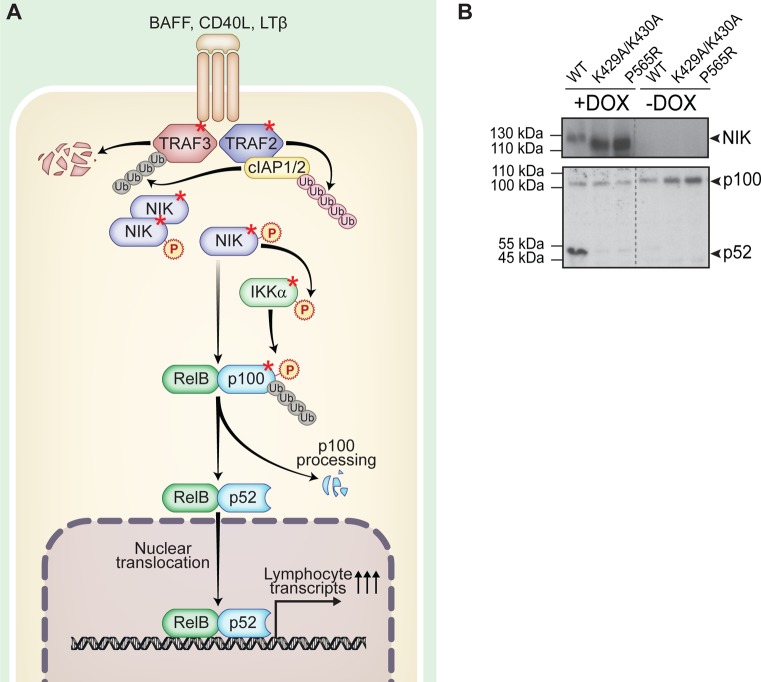
Noncanonical NF-κB signaling pathway. (A) Schematic depiction of the function and key proteins of noncanonical NF-κB signaling. Proteins used in this study are marked by a red asterisk. (B) HEK293 Flp-In stable cell line system expressing SH-tagged NIKwt, NIKK429A/K430A, and NIKP565R upon doxycycline induction. IB showing expression of NIK upon doxycycline treatment and NF-κB2/p100 processing due to expression of NIKwt. This is a reduced depiction of the IB shown in Supplementary Figure S2B. Gray dotted lines indicate IB lanes that are not adjacent to one another.
Recently, the medical relevance of this pathway has been elucidated by our identification of a NIK deficiency. Here patients have a catalytically inactive version of the kinase (NIKP565R).8 Patients show a life-threatening combined immunodeficiency that results in increased susceptibility to bacterial, viral, and protozoan infections and subsequent need for hematopoietic stem-cell transplantation. Lymphoid lineages are most affected by the mutation: B-cell survival is reduced in the patients, resulting in low B-cell numbers and hypogammaglobulinemia; T cells are functionally affected; and NK cells are not only reduced but also have impaired cytotoxic function. Together, this leads to a severe dysfunction of lymphoid immunity. Other kinase-deficient mutants of NIK have only been characterized in vitro.9 In mice, the NIK mutant aly(10,11) has been a long-standing subject of immunological research. This mutant preserves kinase activity but abolishes interaction with effectors such as TRAF2 and IKKα. Additionally, knockout animals12 have been described. Both models show disorganization of lymphatic tissue in the peripheral nodes, Peyer’s patches, spleen, and thymus as well as reduced B-cell numbers and immunoglobulin (Ig) serum levels concomitant to humoral immunodeficiency.10,12,13 This largely recapitulates the findings in humans. Because noncanonical NF-κB signaling is required for healthy lymphoid tissue survival, the pathway is also commonly activated in cancers, for example, B-cell neoplasms14 or multiple myelomas.15,16 Fine-tuned regulation of the noncanonical pathway and cross-talk with other immune and cellular pathways is still not fully understood. This fact is supported by the recent discovery of additional novel regulators.17,18 For example, the TRAF3 interactor and deubiquitinase OTUD7B acts via protein–protein interaction to regulate the pathway.18 Because the NF-κB pathway is of such essential interest, it has been extensively explored in large-scale proteomic studies;19 however, a detailed and technologically up-to-date interactome analysis of noncanonical NF-κB signaling has not yet been reported. Here we present the protein–protein interaction network of the noncanonical NF-κB pathway using NF-κB2/p100, IKKα, TRAF2, TRAF3, and NIK as nodes. Additionally, we chose this well-studied pathway for expanding our knowledge on a recently discovered primary immunodeficiency protein, NIK, because the effect of a given newly identified disease-causing mutation is rarely studied in the context of cellular protein machinery. Therefore, as a proof-of-principle, we compare the interactome of wild-type NIK to previously reported single gene-defect causing mutants, namely, the recently identified novel kinase inactive mutant observed in human patients (NIKP565R),8 a second catalytically inactive mutant (NIKK429A/K430A),9 and the human aly equivalent mutant (NIKG860R).11
Experimental Procedures
Constructs and Cells
Human MAP3K14 and mutant versions thereof (NIKK429A/K430A, NIKP565R, NIKG860R), GFP control, TRAF3, TRAF2, IKKα, and NFKB2 cDNA were cloned into pTO-SII-HA-GW vectors20 for N-terminal streptavidin–hemagglutinin (S-HA) tag fusion using gateway recombination as described previously.20,21 The plasmids were transfected into HEK293 Flp-In-TREx cells (Life Technologies, Carlsbad, CA) together with a Flp recombinase expression plasmid pOG44 (Life Technologies). Recombinants were selected according to the instructions supplied by the manufacturer and cell lines that have doxycycline-dependent stable transgene expression were generated. Cells were maintained in DMEM (PAA Laboratories, Germany) supplemented with 10% fetal calf serum (FCS), penicillin (100 U/mL), and streptomycin (100 μg/mL) (PAA Laboratories, Germany). The inducible expression of the SH-tagged constructs upon treatment with 1 μg/mL doxycycline (Sigma-Aldrich, Austria) for 24 h was verified by Western blotting. To validate interactions, we cloned FKBP8 and FBXW7 cDNA into pCS2-6myc-GW expression vector21 for N-terminal myc tag fusion expression using gateway recombination as described above.
Tandem Affinity Purification
Tandem affinity purifications (TAPs) were performed as previously described,22 with the following modifications. In brief, confluent 15 cm dishes of HEK293 were incubated with 1 μg/mL doxycycline for 24 h. Cells were lysed in buffer I (50 mM HEPES, pH 8.0, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF) and complete protease inhibitor cocktail (Sigma-Aldrich, Austria). Protein complexes (55–100 mg protein input) were isolated using 400 μL of Strep-Tactin agarose bead slurry (2-1201-010, IPA, Germany), followed by elution with biotin. A second purification step with 200 μL of HA agarose bead slurry (A 2095, Sigma-Aldrich, Austria) was performed, followed by elution with 100 mM formic acid. Eluates were immediately neutralized with triethylammonium bicarbonate (TEAB). After reducing cysteine bonds with 10 mM dithiothreitol (DTT) and alkylation in 55 mM iodacetamide at room temperature, the samples were digested overnight at 37 °C with 2.5 μg trypsin (Promega, Austria). The peptides were purified using C18 microspin columns (3M, USA) according to the protocol provided by the manufacturer, lyophilized, redissolved in 5% formic acid. 5% v/v of the digest of the SH-GFP control pulldown sample was used for C18 purification and subsequent MS analysis. The fraction of digested peptide of the SH-tagged bait proteins used was normalized to the SH-GFP control sample (as described in ref (22)) using anti-HA immunoblot data. Each SH-tagged protein was prepared as biological replicates and analyzed as back-to-back technical replicates by liquid chromatography mass spectrometry.
Liquid Chromatography Mass Spectrometry
Mass spectrometry was performed on a hybrid linear trap quadrupole (LTQ) Orbitrap Velos mass spectrometer (ThermoFisher Scientific, Waltham, MA) running the Xcalibur software (version 2.1.0). The instrument was coupled to an Agilent 1200 HPLC nanoflow system with a dual pump, one precolumn, and one analytical column (Agilent Biotechnologies, Palo Alto, CA) via a nanoelectrospray ion source with a liquid junction (Proxeon, Odense, Denmark). The peptide mixtures were automatically loaded from the thermostated autosampler (4 °C) onto a trap column (Zorbax 300SB-C18 5 μm, 5 × 0.3 mm, Agilent Biotechnologies) with the binary pump solvent that was comprised of 0.1% trifluoracetic acid (TFA) in water at a flow rate of 45 μL/min. The peptides were eluted by back-flushing from the trap column onto a 16 cm fused silica analytical column with an inner diameter of 50 μm packed with C18 reversed-phase material (ReproSil-Pur 120 C18-AQ, 3 μm, Dr. Maisch, Ammerbuch-Entringen, DE). The solvents for peptide separation were composed of 0.4% FA in water (solvent A) and 0.4% FA in 20% isopropanol, 70% methanol (solvent B). A multistep linear gradient elution of the peptides was achieved by a 27 min gradient ranging from 3 to 30% solvent B, followed by a 25 min gradient from 30 to 70% solvent B and, finally, a 7 min gradient from 70 to 100% solvent B at a constant flow rate of 100 nL/min. The analyses were performed in a data-dependent acquisition mode. The top 15 most intense ions were selected for collision-induced dissociation (CID). Normalized collision energy was 30%. Dynamic exclusion of selected ions for MS2 fragmentation was 60 s, and a single lock mass at m/z 445.120024 Si(CH3)2O)623 was used for internal mass calibration with a target loss mass abundance of 0%. Maximal ion accumulation time allowed was 50 ms for MSn in the LTQ and 500 ms in the C-trap. Overfilling of the C-trap was prevented by automatic gain control set to 106 ions for a full Fourier transform mass spectrometer (FTMS) scan and 5 × 105 ions for MSn mode. Intact peptides were detected in the Orbitrap mass analyzer at resolution of 60 000. Signal threshold for triggering MSMS fragmentation was set 2000 ion counts.24
Data Processing and Bioinformatic Analysis
Acquired raw mass spectrometry data files were converted into Mascot generic format (mgf) files using msconvert (ProteoWizard Library v2.1.2708). The resultant peak lists were compared with the human SwissProt database version v2013.01_20130110 (37 398 sequences including isoforms obtained from varsplic.pl25 and appended known contaminants) with the search engines Mascot (v2.3.02, MatrixScience, London, U.K., www.matrixscience.com) and Phenyx (v2.6, GeneBio, Geneva, Switzerland).26 Submission to the search engines was performed using a Perl script that performs an initial search with relatively broad mass tolerances (Mascot only) on both precursor and fragment ions (±10 ppm and ±0.6 Da, respectively). High-confidence peptide identifications are used to recalibrate all precursor and fragment ion masses prior to a second search with narrower mass tolerances (±4 ppm and ±0.3 Da). One missed tryptic cleavage site was allowed. Carbamidomethyl cysteine and oxidized methionine were set as variable modifications, respectively. To validate the proteins, we processed Mascot and Phenyx output files by parsers developed internally. Proteins with ≥2 unique peptides above a score T1, or with a single peptide above a score T2, were selected as unambiguous identifications. Additional peptides for these validated proteins with score > T3 were also accepted. For Mascot and Phenyx, T1, T2, and T3 were equal to 12, 45, and 10 and 5.5, 9.5, and 3.5, respectively (p value <10–3). Following the selection criteria, proteins were grouped on the basis of shared peptides, and only the group reporters are considered in the final output of identified proteins. Spectral conflicts between Mascot and Phenyx peptide identifications were discarded. The whole procedure was repeated against a reversed database to assess the protein group false discovery rate (FDR). Peptide and protein group identifications were <0.1 and <1% FDR, respectively.
All proteins identified in the SH-GFP negative control experiments as well as additional proteins from control experiments in our internal repository were removed as nonspecifically interacting proteins. Only proteins that were identified in both biological replicates and were absent in the GFP experiments were further considered. Using the SAINT27 algorithm and the Homo sapiens CRAPome28 (version 1.1) database (www.crapome.org, accessed 25 August 2015), high confidence scores were calculated. Spectral counts from technical replicates were added up, and biological replicates (n = 2) were used as the input for the analysis tools. The FC-A score was determined from internal negative control experiments (including SH-GFP) with a default background estimation, and the FC-B score was ascertained from CRAPome controls with a stringent background estimation. For calculating the FC-B score, CRAPome controls with matching experimental procedures were selected (i.e., HEK293 cells, S-HA tag, total cell lysate, 1D-LC–MS, Strep-Tactin and HA agarose affinity resins). We used the SAINT algorithm to calculate the probability score of the identified interactions via SAINT express29 with default options. This analysis included negative control experiments from our internal repository. SAINT and CRAPome scores were plotted using Ggplot2 in R. Only interactions with a SAINT score of 1 and a CRAPome FC-A score >2.5 were considered. To retrieve preannotated interactions, the STRING database (version 9.1) was used, applying a default confidence score = 0.4. Networks were generated using cytoscape (version 3.0.2). Gene set enrichment analysis was performed by uploading complete, filtered noncanonical NF-κB signaling network data to DAVID annotation database with default settings.
Co-Immunoprecipitation and Immunoblot Analysis
Co-immunoprecipitation (Co-IP) was performed using doxycycline-inducible HEK293 Flp-In cells as previously described. Confluent 15 cm dishes of cells (containing 3 × 107 cells) were induced with 1 μg/μL doxycycline for 24 h. For transfection experiments, cells were transfected with pCS2-6myc-GW expression vector using X-tremeGENE reagent (Roche, Vienna, Austria) and induced 12 h after transfection. After lysis in buffer I (composition given above in the TAP section), 7 mg of total protein together with 70 μL of HA–agarose bead slurry (Sigma-Aldrich, Austria) were used for immunoprecipitation (IP). For immunoblot (IB) analysis, protein was isolated with buffer I. SDS-PAGE and polyvinylidene difluoride or nitrocellulose membranes were prepared according to standard methods. Primary antibodies (1:1000 dilution) for IB analyses were: rabbit antihuman IKKα (2682), NIK (4994), NF-κB2/p100/p52 (4882) (Cell Signaling, Frankfurt, Germany), and rabbit antihuman FKBP8 (Pierce/ThermoFisher, Vienna, Austria). Tagged recombinant proteins were detected with antihuman c-myc (BD Biosciences, Schwechat, Austria) at 1:1000 dilution and horseradish peroxidase-coupled anti-HA (Sigma-Aldrich, Austria) at 1:3000 dilution. Mouse antihuman GAPDH (Santa Cruz Biotechnology, TX) at 1:1000 dilution was a loading control. Horseradish peroxidase-conjugated goat antirabbit (Biorad, Vienna, Austria) and goat antimouse (BD Biosciences, Schwechat, Austria) were secondary antibodies at 1:10 000 or 1:50 000 dilution. Signals were detected with a chemiluminescent substrate (Amersham ECL Prime Western Blotting Detection Reagent, GE Healthcare Life Sciences, Vienna, Austria) and Hyperfilm ECL (GE Healthcare Life Sciences).
Results and Discussion
Experimental Design
To understand the cellular context of the noncanonical NF-κB signaling pathway (Figure 1A) and in particular the central protein NIK, our aim was to map the protein interaction network of NIK, IKKα, TRAF2, TRAF3, and NF-κB2/p100 by tandem affinity purification mass spectrometry (TAP-MS). N-terminally tagged SH-fusion proteins were expressed by stable recombinant human embryonic kidney HEK293 Flp-In cells that enable doxycycline-dependent protein production.20 Doxycycline induction of NIKwt revealed that NF-κB2/p100 processing to p52 can be induced in HEK293 Flp-In cells by overexpression and subsequent accumulation of NIK (Figure 1B). This was not the case for the catalytically inactive mutants NIKP565R and NIKK429A/K430A. Thus, we were confident that all components of noncanonical NF-κB signaling are expressed and functional in the HEK293 Flp-In system, serving as a model for noncanonical NF-κB signaling in lymphocytes.
Interactome Integrating the Five SH-Tagged Proteins
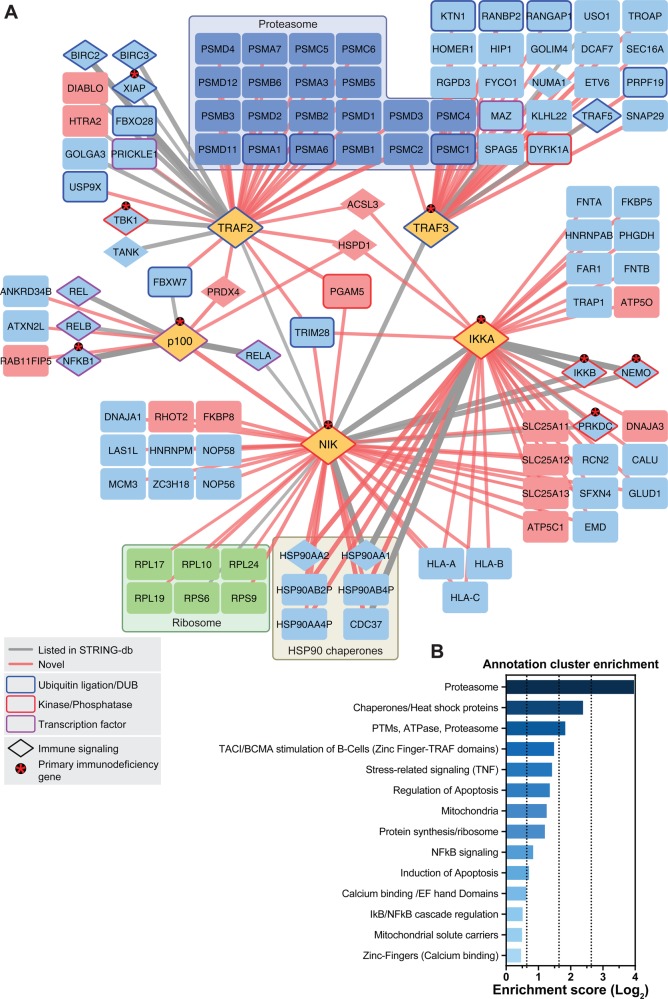
From the five SH-tagged proteins, numerous protein interactors were obtained. The data were analyzed by SAINT and the CRAPome database and filtering was applied according to the thresholds described in the material and methods section (Supplementary Figure S1A,B). Only interacting proteins that were identified in both biological replicates and that were not identified in the SH-GFP experiments were considered for the final interactome. 109 high confidence interacting proteins linked by 145 interactions were identified between all five SH-tagged baits (Figure 2, see Supplemetary Table S1 for all protein–protein interactions ranked by SAINT probability and CRAPome score). From these, we found that 30 had been previously reported with high confidence in the STRING database (Figure 2, Table S1), and 115 interactions were novel. Out of all of the previously reported (“known”) interacting proteins identified in our experiments, 85.7% had both a score of 1 in SAINT and an FC-A score of >2.5 (Supplementary Figure S1A). This result not only confirmed that known interactions were retrieved with high confidence but also allowed us to conclude that our scoring system was valid. This supported the quality of the interaction network and the potential of the interactome as a resource for novel candidate pathway regulators. In line with this, several functionally characterized regulators of canonical/noncanonical NF-κB signaling were among the identified proteins, including cIAP1, cIAP2 (encoded by BIRC2 and BIRC3, respectively), REL-A/p65, REL-B, c-REL, NF-κB1/p50, IKKβ, IKKγ, and FBXW7. These interactors are discussed below.
Figure 2.
Interaction network of the noncanonical NF-κB signaling pathway. (A) Proteins shown were identified by LC–MS. Edge (line) thickness and edge transparency represent the CRAPome FC-A score. Red edges indicate novel interactions, and gray edges indicate known interactions. SH-tagged proteins, proteasomal machinery proteins, ribosomal proteins, and mitochondrial-localizing proteins are colored yellow, dark-blue, green, and red, respectively. Ubiquitination/deubiquitination machinery proteins, kinases/phosphatases, and transcription factors are framed in blue, red, and purple, respectively. Asterisks denote proteins that cause primary immunodeficiency phenotypes in humans when mutated. (B) Gene Ontology term enrichment analysis using DAVID functional annotation clustering. Selected clusters are shown.
For instance, cIAP1 and 2 were copurified with SH-TRAF2. These proteins are known to form a complex with TRAF2 and TRAF3 that then acts as a ubiquitin ligase competent complex controlling NIK degradation.5 The association of SH-TRAF2 and SH-TRAF3 with different components of the proteasomal machinery (Figure 2A) is consistent with the role of these two proteins in the regulation of proteolytic processes in NF-κB signaling.
The most downstream member of noncanonical NF-κB signaling that was analyzed is NF-κB2/p100. The SH-tagged version of the protein revealed an association with known NF-κB family transcription factors that can heterodimerize with NF-κB2/p52 (i.e., REL-A/p65, REL-B, c-REL, NF-κB1/p50).
An only recently reported interaction between NF-κB2/p100 and FBXW717 was also observed (Figure 2A). As a substrate-targeting subunit of the SCF ubiquitin ligase complexes, FBXW7 was discovered to constitutively target NF-κB2/p100 for proteasomal degradation. Because the uncleaved nuclear p100 form of NF-κB2 is inhibitory to noncanonical NF-κB signaling,6 FBXW7 was shown to represent a novel positive regulator of noncanonical NF-κB activity.17 This interaction was validated by co-IP via the HA-tag in doxycycline-inducible HEK293 cells (Figure 3A). The data clearly indicated that our network encompasses, besides long-documented interactions, also an only recently characterized novel regulator, FBXW7, suggesting that additional candidates may also be real and potentially relevant interactions. Interestingly, in the previous study, NF-κB2/p100 was identified as an interactor of FBXW7 by immunopurification and mass spectrometry.17 In our study, the reciprocal strategy resulted in the identification of the interaction, thus strengthening previous observations. Additionally, we observed a previously undescribed interaction between FBXW7 and TRAF2, which could be confirmed using the co-IP and Western blot method (Figure 3A). This provides further evidence of the role of FBXW7 in NF-κB signaling, and it is conceivable that control of NF-κB2/p100 by FBXW7 may indeed involve TRAF complexes.
Figure 3.
Validation of interacting proteins identified by LC–MS. (A) Co-IP using SH-GFP doxycycline-inducible control cell line, SH-TRAF2 and SH-NF-κB2/p100-inducible HEK293 Flp-In lines concomitantly expressing myc-FBXW7. IP with anti-HA agarose beads confirmed the association of SH-TRAF2 and SH-NF-κB2/p100 with myc-tagged FBXW7. (B) Co-IP with SH-GFP doxycycline-inducible control cell line, SH-IKKα and SH-NIK doxycycline-inducible HEK293 Flp-In lines. IP with anti-HA agarose beads confirmed the association of SH-IKKα and SH-NIK with endogenously expressed FKBP8. Gray dotted lines indicate lanes that are not adjacent to one another.
IKKα and NIK are the kinases central to noncanonical NF-κB signaling. In our network study, well-known and studied interaction partners were identified as interactors of SH-tagged kinases. IKKα has been reported to form a trimeric IKKα/IKKβ/IKKγ complex.30 These findings are reflected in our data (Figure 2A). In turn, the kinase NIK exhibits an IKK interaction domain,31,32 and indeed the IKK complex was identified from the SH-NIK data. The downstream effector NF-κB2/p100 is also reported to participate in a NIK signaling complex,33 as evident from the data. How the NIK interactome in its entirety relates to the function of the kinase in various biological processes remains to be explored. Gene set enrichment analysis of the complete, filtered noncanonical NF-κB signaling network showed an expected enrichment of proteins involved in protein turnover, apoptosis regulation, and immune signaling groups. In addition, there was enrichment for some unexpected groups, such as proteins that are localized to the mitochondria (Figure 2B).
Because the ultimate aim of this study was to enable a systematic view of noncanonical NF-κB signaling rather than an extensive characterization of single interactions, only limited validation of single interaction candidates was initiated. To assess the predictive quality of our data set, however, we validated the identified interactions of FKBP8. FKBP8 was a high-scoring interactor that copurified with SH-NIK and was also observed as an interacting protein with SH-IKKα, albeit with a subthreshold CRAPome FC-A confidence score (2.21). FKBP8 is a cochaperone of the immunophilin family and is classified under the biological function of cell death and mitochondrial location. One of the various biological roles of FKBP8 is also the regulation of the antiapoptotic Bcl-2 protein.34 Co-IP of SH-NIK and SH-IKKα from doxycycline-inducible HEK293 cells with endogenous FKBP8 confirmed its interaction with NIK and IKKα (Figure 3B). Reciprocal co-IP corroborated the association between FKBP8 and IKKα and indicated a robust association between the proteins (Supplemetary Figure S2A). Noncanonical NF-κB signaling activity, as measured by NF-κB2/p100 processing, was not enhanced by the presence of FKBP8 (Supplementary Figure S2B). Rather than direct positive regulation of noncanonical NF-κB signaling, this observation suggested an alternative role for FKBP8 (Supplementary Figure S2B). FKBP8 is an atypical member of the immunophilin family (reviewed in ref (34)) and has been implicated as a negative regulator of Bcl-2. Bcl-2 is a pro-survival factor that protects cells by interfering with pro-apoptotic proteins at the mitochondrial membrane. Mitochondrial membrane proteins were enriched in the noncanonical NF-κB interactome network. Interestingly, an association of NF-κB signaling with mitochondria has been previously reported;35,36 however, 40% of filtered mitochondrial interactors stem from the inner mitochondrial membrane, and thus these data could also partly be explained by a postlysis association of mitochondrial proteins with the baits (Table S1). Of note, FKBP8, the protein chosen here for validation because of its role in survival signaling, is a protein that localizes to the outer mitochondrial membrane. This finding is consistent with the possibility of intracellular association of noncanonical NF-κB signaling components with mitochondria and potentially regulation of cell survival. Therefore, FKBP8 could be involved in the control of cell survival by the noncanonical NF-κB pathway.
Differential Interactome of NIK Mutants
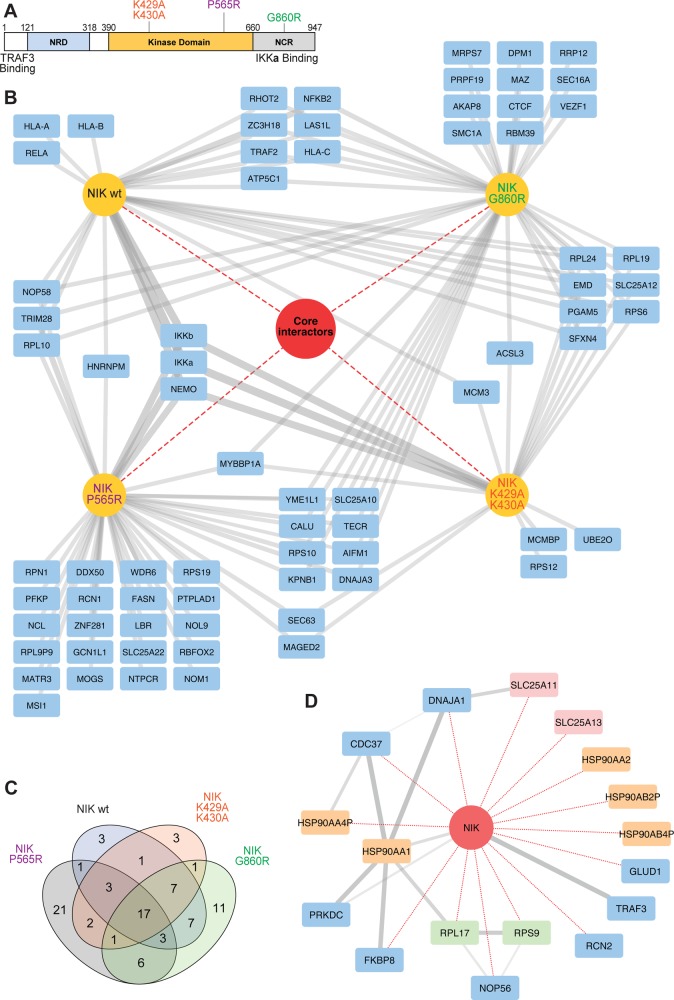
To compare the biological consequences of recently reported and long-known NIK mutations on the NIK interactome, the mutants NIKG860R (the amino acid change of the aly/aly mouse introduced into human NIK), NIKK429A/K430A (catalytically inactive), and the recently identified in human patients, novel, catalytically inactive mutant NIKP565R (Figure 4A) were expressed as N-terminal SH-tagged proteins in the doxycycline-inducible HEK293 system. Using the filtering procedure described in the material and methods section (Supplementary Figure S1B), a total of 89 interacting proteins were identified (Figure 4B). The “core interactome” was composed of 17 proteins that were common to all NIK variants (Figure 4B–D). TRAF3,5 FKBP8, various subunits of the HSP90–CDC37 protein chaperone complex, and a group of SLC25 mitochondrial membrane transporters (Figure 4D) constituted the “core interactome”. Overlaying the shared NIK proteins with public interaction data from STRING revealed interconnectivity between the HSP90 chaperone group and the cochaperone FKBP8 (Figure 4D). The data suggested that in this biological process, the HSP90–FKBP8 chaperone complex crosstalks with noncanonical NF-κB signaling. The HSP90 complex has been implicated in the maturation of other kinases of TNF-receptor associated pathways37,38 and may represent a novel NIK interaction complex. Also, the aforementioned inhibition of Bcl-2 by FKBP8 is dependent on the HSP90 complex, and FKBP8 is known to interact with this complex.39 It remains to be determined if the role of HSP90–CDC37 in kinase maturation involves the interaction with FKBP8 or if the interaction of FKBP8 with the NIK and IKKα kinases of the noncanonical NF-κB pathway has a separate role.
Figure 4.
Interactions of NIK wild-type and mutants. (A) Schematic depiction of the NIK protein indicating the domains and mutants relevant to this study.41 (B) Interaction comparison of NIKwt, NIKG860R, NIKK29R/K430A, and NIKP565R. In the network, SH-tagged proteins and interactors are colored yellow and blue, respectively. Core interactors shared by all variants are represented within the red circle. (C) Venn diagram depicting the number of interactors that overlap between the different NIK variants. (D) Network depiction of NIK core interactors. NIK is indicated as a red circle. Edges in red and gray depict interactions identified in this study and interactions retrieved from the STRING database, respectively. Interactors colored in yellow, green, and red denote HSP90 complex members, ribosomal complex members, and SLC25 family members, respectively.
Interactions specific to, or abrogated in NIK mutants, were also observed. Notably, only SH-NIKwt and the catalytically inactive mutants SH-NIKK429A/K430A and SH-NIKP565R copurified IKKα and its complex members IKKβ/IKKγ. The NIKG860ARaly mutant is reported to have the mutation within the IKKα interaction region11 and thus did not copurify IKKα (Supplementary Figure S3). Conversely, NF-κB2/p100 was only identified with high confidence in the SH-NIKwt and SH-NIKG860R experiments. NF-κB2/p100 is a target gene that is expressed upon NF-κB activation.40 Thus, this observation may be a consequence of the increased abundance of NF-κB2/p100 in NIKwt cells and subsequent functional p52 signaling, as differential binding of p100 by catalytically inactive NIK has previously not been observed.33
Impact of the Study
To enhance our knowledge on the noncanonical NF-κB signaling pathway from a systems biology point of view, we have provided here a detailed resource describing the interactome of the key nodes of noncanonical NF-κB signaling. A number of well-known interactors were verified from the data. More importantly, however, the resource provides a surprising number of novel candidate interactions that hint at unexpected regulatory links between pathway members and their interactors, such as the potential functional interaction between FKBP8 and NIK/IKKα. NF-κB signaling plays a central role in immunity and represents a convergence point for integrating diverse immune stimuli. We provide, to our knowledge, for the first time a refined interaction map of factors specific to noncanonical NF-κB signaling. This map includes mutant variants of the pathway-defining kinase NIK and provides information on the interplay with canonical NF-κB signaling components. In vivo, such interplay is crucial for immune cells to react appropriately to innate and adaptive immune stimuli. Studies in a physiological setting using immune cells will confirm whether the set of interacting proteins we found in our inducible HEK293 system is comparable in these cells. As we showed that overexpression of NIK induces processing of p100 in our system, we are confident that this system is suitable to study the pathway of interest. The purpose of this study was to build an up-to-date data set of basal, unstimulated noncanonical NF-κB signaling to provide a robust reference interactome. Further studies could compare the interactomes in stimulated versus unstimulated cells because induction of NF-κB may lead to a proteomic profile altered to the baseline reference.
Therefore, an extended network of connections including canonical and noncanonical NF-κB signaling proteins as presented here will provide a valuable platform for a deeper understanding of the nature of the pathway. Furthermore, in the light of the recently discovered rare genetic condition NIK deficiency,8 our comparison of various NIK mutants will aid in understanding this disorder further.
Taken together, this study has generated valuable novel interactome data for noncanonical NF-κB signaling. The data provide a new, unbiased and global overview on the pathway and identify unexpected candidate interactions between canonical and noncanonical signaling proteins and potential novel regulators.
Acknowledgments
We thank Jacques Colinge, André C. Müller, and Peter Májek for fruitful discussions and Elisabeth Salzer and Kate G. Ackermann for critically reading the manuscript. We thank Giulio Superti-Furga for providing pTO-SII-HA-GW plasmids. This work was supported by the Austrian Science Fund (FWF) Lise Meitner Program Fellowship (FWF M-1809, to K.L.W.), the FWF Infect-ERA framework (I-1620_B22, to S.K.), and the European Research Council (ERC StG 310857, to K.B.).
Glossary
Abbreviations:
- TAP
tandem affinity purification
- MS
mass spectrometry
- IB
immunoblot
- IP
immunoprecipitation
- S
streptavidin-binding peptide motifs
- HA
hemagglutinin
- SDS PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- 1D-LC–MS
one-dimensional gel-free liquid chromatography mass spectrometry
- CID
collision-induced dissociation
- LTQ
linear trap quadrupole
- NIK
NF-κB inducing kinase
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jproteome.5b01004.
Supplementary Figure S1: Distribution of interactors of SH-tagged proteins plotted by SAINT and CRAPome FC-A scores. Supplementary Figure S2: Data on the interactor FKBP8. Supplementary Figure S3: Interaction of NIK variants with IKKα. Supporting Information and Methods: Full plasmid sequences of the used vectors. (PDF)
Complete list of proteins identified by each purification. Proteins identified in the SH-GFP negative control experiments are removed. The list is ranked by SAINT score and CRAPome FC-A score. (XLSX)
Author Present Address
⊥ R.S.: Institute of Science and Technology Austria, 3400 Klosterneuburg, Austria.
Author Contributions
# R.S., R.M., and W.G. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Vallabhapurapu S.; Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu. Rev. Immunol. 2009, 27, 693–733. 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- Sun S. C. The noncanonical NF-kappaB pathway. Immunol Rev. 2012, 246 (1), 125–40. 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G.; Karin M. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 2004, 25 (6), 280–8. 10.1016/j.it.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Xiao G.; Harhaj E. W.; Sun S. C. NF-kappaB-inducing kinase regulates the processing of NF-kappaB2 p100. Mol. Cell 2001, 7 (2), 401–9. 10.1016/S1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- Liao G.; Zhang M.; Harhaj E. W.; Sun S. C. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J. Biol. Chem. 2004, 279 (25), 26243–50. 10.1074/jbc.M403286200. [DOI] [PubMed] [Google Scholar]
- Senftleben U.; Cao Y.; Xiao G.; Greten F. R.; Krahn G.; Bonizzi G.; Chen Y.; Hu Y.; Fong A.; Sun S. C.; Karin M. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 2001, 293 (5534), 1495–9. 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- Razani B.; Reichardt A. D.; Cheng G. Non-canonical NF-kappaB signaling activation and regulation: principles and perspectives. Immunol Rev. 2011, 244 (1), 44–54. 10.1111/j.1600-065X.2011.01059.x. [DOI] [PubMed] [Google Scholar]
- Willmann K. L.; Klaver S.; Dogu F.; Santos-Valente E.; Garncarz W.; Bilic I.; Mace E.; Salzer E.; Conde C. D.; Sic H.; Majek P.; Banerjee P. P.; Vladimer G. I.; Haskologlu S.; Bolkent M. G.; Kupesiz A.; Condino-Neto A.; Colinge J.; Superti-Furga G.; Pickl W. F.; van Zelm M. C.; Eibel H.; Orange J. S.; Ikinciogullari A.; Boztug K. Biallelic loss-of-function mutation in NIK causes a primary immunodeficiency with multifaceted aberrant lymphoid immunity. Nat. Commun. 2014, 5, 5360. 10.1038/ncomms6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinin N. L.; Boldin M. P.; Kovalenko A. V.; Wallach D. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature 1997, 385 (6616), 540–4. 10.1038/385540a0. [DOI] [PubMed] [Google Scholar]
- Miyawaki S.; Nakamura Y.; Suzuka H.; Koba M.; Yasumizu R.; Ikehara S.; Shibata Y. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur. J. Immunol. 1994, 24 (2), 429–34. 10.1002/eji.1830240224. [DOI] [PubMed] [Google Scholar]
- Shinkura R.; Kitada K.; Matsuda F.; Tashiro K.; Ikuta K.; Suzuki M.; Kogishi K.; Serikawa T.; Honjo T. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat. Genet. 1999, 22 (1), 74–7. 10.1038/8780. [DOI] [PubMed] [Google Scholar]
- Yin L.; Wu L.; Wesche H.; Arthur C. D.; White J. M.; Goeddel D. V.; Schreiber R. D. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science 2001, 291 (5511), 2162–5. 10.1126/science.1058453. [DOI] [PubMed] [Google Scholar]
- Shinkura R.; Matsuda F.; Sakiyama T.; Tsubata T.; Hial H.; Paumen M.; Miyawaki S.; Honjo T. Defects of somatic hypermutation and class switching in alymphoplasia (aly) mutant mice. Int. Immunol. 1996, 8 (7), 1067–75. 10.1093/intimm/8.7.1067. [DOI] [PubMed] [Google Scholar]
- Staudt L. M. Oncogenic activation of NF-kappaB. Cold Spring Harbor Perspect. Biol. 2010, 2 (6), a000109. 10.1101/cshperspect.a000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata C. M.; Davis R. E.; Demchenko Y.; Bellamy W.; Gabrea A.; Zhan F.; Lenz G.; Hanamura I.; Wright G.; Xiao W.; Dave S.; Hurt E. M.; Tan B.; Zhao H.; Stephens O.; Santra M.; Williams D. R.; Dang L.; Barlogie B.; Shaughnessy J. D. Jr.; Kuehl W. M.; Staudt L. M. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 2007, 12 (2), 115–30. 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats J. J.; Fonseca R.; Chesi M.; Schop R.; Baker A.; Chng W. J.; Van Wier S.; Tiedemann R.; Shi C. X.; Sebag M.; Braggio E.; Henry T.; Zhu Y. X.; Fogle H.; Price-Troska T.; Ahmann G.; Mancini C.; Brents L. A.; Kumar S.; Greipp P.; Dispenzieri A.; Bryant B.; Mulligan G.; Bruhn L.; Barrett M.; Valdez R.; Trent J.; Stewart A. K.; Carpten J.; Bergsagel P. L. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007, 12 (2), 131–44. 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L.; Millman S. E.; Scotto L.; Kyratsous C. A.; Basrur V.; O’Connor O.; Hoffmann A.; Elenitoba-Johnson K. S.; Pagano M. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 2012, 14 (4), 375–85. 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H.; Brittain G. C.; Chang J. H.; Puebla-Osorio N.; Jin J.; Zal A.; Xiao Y.; Cheng X.; Chang M.; Fu Y. X.; Zal T.; Zhu C.; Sun S. C. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature 2013, 494 (7437), 371–4. 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester T.; Bauch A.; Ruffner H.; Angrand P. O.; Bergamini G.; Croughton K.; Cruciat C.; Eberhard D.; Gagneur J.; Ghidelli S.; Hopf C.; Huhse B.; Mangano R.; Michon A. M.; Schirle M.; Schlegl J.; Schwab M.; Stein M. A.; Bauer A.; Casari G.; Drewes G.; Gavin A. C.; Jackson D. B.; Joberty G.; Neubauer G.; Rick J.; Kuster B.; Superti-Furga G. A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat. Cell Biol. 2004, 6 (2), 97–105. 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- Glatter T.; Wepf A.; Aebersold R.; Gstaiger M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 2009, 5, 237. 10.1038/msb.2008.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A.; Kandasamy K.; Alvisi G.; Mulhern O.; Sacco R.; Habjan M.; Binder M.; Stefanovic A.; Eberle C. A.; Goncalves A.; Burckstummer T.; Muller A. C.; Fauster A.; Holze C.; Lindsten K.; Goodbourn S.; Kochs G.; Weber F.; Bartenschlager R.; Bowie A. G.; Bennett K. L.; Colinge J.; Superti-Furga G. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature 2012, 487 (7408), 486–90. 10.1038/nature11289. [DOI] [PubMed] [Google Scholar]
- Rudashevskaya E. L.; Sacco R.; Kratochwill K.; Huber M. L.; Gstaiger M.; Superti-Furga G.; Bennett K. L. A method to resolve the composition of heterogeneous affinity-purified protein complexes assembled around a common protein by chemical cross-linking, gel electrophoresis and mass spectrometry. Nat. Protoc. 2012, 8 (1), 75–97. 10.1038/nprot.2012.133. [DOI] [PubMed] [Google Scholar]
- Olsen J. V.; de Godoy L. M.; Li G.; Macek B.; Mortensen P.; Pesch R.; Makarov A.; Lange O.; Horning S.; Mann M. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 2005, 4 (12), 2010–21. 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- Bennett K. L.; Funk M.; Tschernutter M.; Breitwieser F. P.; Planyavsky M.; Ubaida Mohien C.; Muller A.; Trajanoski Z.; Colinge J.; Superti-Furga G.; Schmidt-Erfurth U. Proteomic analysis of human cataract aqueous humour: Comparison of one-dimensional gel LCMS with two-dimensional LCMS of unlabelled and iTRAQ(R)-labelled specimens. J. Proteomics 2011, 74 (2), 151–66. 10.1016/j.jprot.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Kersey P.; Hermjakob H.; Apweiler R. VARSPLIC: alternatively-spliced protein sequences derived from SWISS-PROT and TrEMBL. Bioinformatics 2000, 16 (11), 1048–9. 10.1093/bioinformatics/16.11.1048. [DOI] [PubMed] [Google Scholar]
- Colinge J.; Masselot A.; Giron M.; Dessingy T.; Magnin J. OLAV: towards high-throughput tandem mass spectrometry data identification. Proteomics 2003, 3 (8), 1454–63. 10.1002/pmic.200300485. [DOI] [PubMed] [Google Scholar]
- Choi H.; Larsen B.; Lin Z. Y.; Breitkreutz A.; Mellacheruvu D.; Fermin D.; Qin Z. S.; Tyers M.; Gingras A. C.; Nesvizhskii A. I. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 2011, 8 (1), 70–3. 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D.; Wright Z.; Couzens A. L.; Lambert J. P.; St-Denis N. A.; Li T.; Miteva Y. V.; Hauri S.; Sardiu M. E.; Low T. Y.; Halim V. A.; Bagshaw R. D.; Hubner N. C.; Al-Hakim A.; Bouchard A.; Faubert D.; Fermin D.; Dunham W. H.; Goudreault M.; Lin Z. Y.; Badillo B. G.; Pawson T.; Durocher D.; Coulombe B.; Aebersold R.; Superti-Furga G.; Colinge J.; Heck A. J.; Choi H.; Gstaiger M.; Mohammed S.; Cristea I. M.; Bennett K. L.; Washburn M. P.; Raught B.; Ewing R. M.; Gingras A. C.; Nesvizhskii A. I. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 2013, 10 (8), 730–6. 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G.; Liu G.; Zhang J.; Nesvizhskii A. I.; Gingras A. C.; Choi H. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 2014, 100, 37–43. 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi E.; Karin M. Bridging the gap: composition, regulation, and physiological function of the IkappaB kinase complex. Mol. Cell. Biol. 1999, 19 (7), 4547–51. 10.1128/MCB.19.7.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier C. H.; Song H. Y.; Gao X.; Goeddel D. V.; Cao Z.; Rothe M. Identification and characterization of an IkappaB kinase. Cell 1997, 90 (2), 373–83. 10.1016/S0092-8674(00)80344-X. [DOI] [PubMed] [Google Scholar]
- Woronicz J. D.; Gao X.; Cao Z.; Rothe M.; Goeddel D. V. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 1997, 278 (5339), 866–9. [DOI] [PubMed] [Google Scholar]
- Xiao G.; Fong A.; Sun S. C. Induction of p100 processing by NF-kappaB-inducing kinase involves docking IkappaB kinase alpha (IKKalpha) to p100 and IKKalpha-mediated phosphorylation. J. Biol. Chem. 2004, 279 (29), 30099–105. 10.1074/jbc.M401428200. [DOI] [PubMed] [Google Scholar]
- Edlich F.; Lucke C. From cell death to viral replication: the diverse functions of the membrane-associated FKBP38. Curr. Opin. Pharmacol. 2011, 11 (4), 348–53. 10.1016/j.coph.2011.03.011. [DOI] [PubMed] [Google Scholar]
- Johnson R. F.; Witzel II; Perkins N. D. p53-dependent regulation of mitochondrial energy production by the RelA subunit of NF-kappaB. Cancer Res. 2011, 71 (16), 5588–97. 10.1158/0008-5472.CAN-10-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazarentzos E.; Mahul-Mellier A. L.; Datler C.; Chaisaklert W.; Hwang M. S.; Kroon J.; Qize D.; Osborne F.; Al-Rubaish A.; Al-Ali A.; Mazarakis N. D.; Aboagye E. O.; Grimm S. IkappaBetaalpha inhibits apoptosis at the outer mitochondrial membrane independently of NF-kappaB retention. EMBO J. 2014, 33 (23), 2814–28. 10.15252/embj.201488183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.; Cao P.; Goeddel D. V. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol. Cell 2002, 9 (2), 401–10. 10.1016/S1097-2765(02)00450-1. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Moghaddas Gholami A.; Kuster B. Systematic identification of the HSP90 candidate regulated proteome. Mol. Cell. Proteomics 2012, 11 (6), M111 016675. 10.1074/mcp.M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlich F.; Erdmann F.; Jarczowski F.; Moutty M. C.; Weiwad M.; Fischer G. The Bcl-2 regulator FKBP38-calmodulin-Ca2+ is inhibited by Hsp90. J. Biol. Chem. 2007, 282 (21), 15341–8. 10.1074/jbc.M611594200. [DOI] [PubMed] [Google Scholar]
- Dejardin E.; Droin N. M.; Delhase M.; Haas E.; Cao Y.; Makris C.; Li Z. W.; Karin M.; Ware C. F.; Green D. R. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 2002, 17 (4), 525–35. 10.1016/S1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- Liu J.; Sudom A.; Min X.; Cao Z.; Gao X.; Ayres M.; Lee F.; Cao P.; Johnstone S.; Plotnikova O.; Walker N.; Chen G.; Wang Z. Structure of the nuclear factor kappaB-inducing kinase (NIK) kinase domain reveals a constitutively active conformation. J. Biol. Chem. 2012, 287 (33), 27326–34. 10.1074/jbc.M112.366658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.