Abstract
The post-translational modification of proteins with polyubiquitin regulates virtually all aspects of cell biology. Eight distinct chain linkage types in polyubiquitin co-exist and are independently regulated in cells. This ‘ubiquitin code’ determines the fate of the modified protein1. Deubiquitinating enzymes of the Ovarian Tumour (OTU) family regulate cellular signalling by targeting distinct linkage types within polyubiquitin2, and understanding their mechanisms of linkage specificity gives fundamental insights into the ubiquitin system.
We here reveal how the deubiquitinase Cezanne/OTUD7B specifically targets Lys11-linked polyubiquitin. Crystal structures of Cezanne alone and in complex with mono- and Lys11-linked diubiquitin, in combination with hydrogen-deuterium exchange mass spectrometry, enable reconstruction of the enzymatic cycle in exquisite detail. An intricate mechanism of ubiquitin-assisted conformational changes activate the enzyme, and while all chain types interact with the enzymatic S1 site, only Lys11-linked chains can bind productively across the active site and stimulate catalytic turnover. Our work highlights the fascinating plasticity of deubiquitinases, and indicates that new conformational states can occur when a true substrate, such as diubiquitin, is bound at the active site.
The 16 human OTU family deubiquitinases (DUBs) are key regulators of the ubiquitin (Ub) code. Small OTU DUBs of the OTUD and OTUB subfamilies (catalytic core ~130-220 residues) employ distinct mechanisms to achieve linkage specificity2–5 but physiological roles are unclear for most members. In contrast, the A20-like OTU subfamily, identified by a larger catalytic domain (300-350 residues, Fig. 1a), has been well studied. A20 is a tumour suppressor and negative feedback regulator of NF-κB signalling6,7, TRABID regulates transcription8,9 by targeting Lys29- and Lys33-linked chains10, and VCPIP is associated with p97/VCP11 and cleaves Lys48 and Lys11 linkages2.
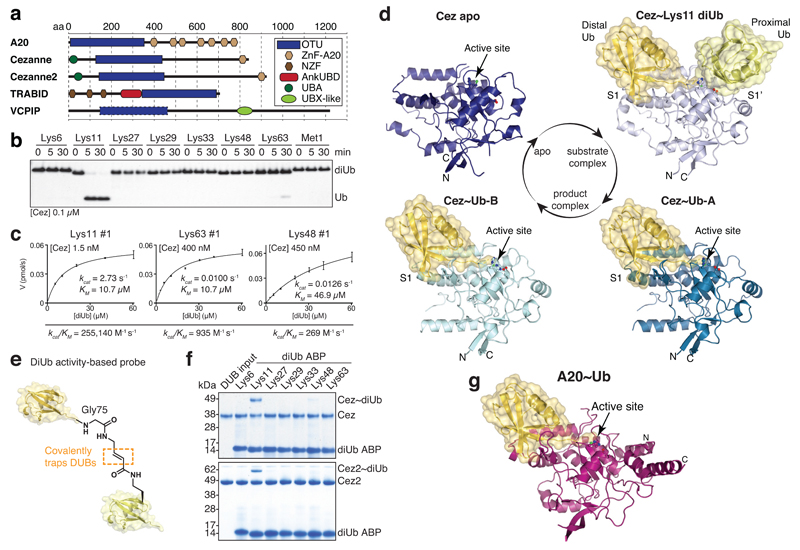
Figure 1. Cezanne biochemistry and structures.
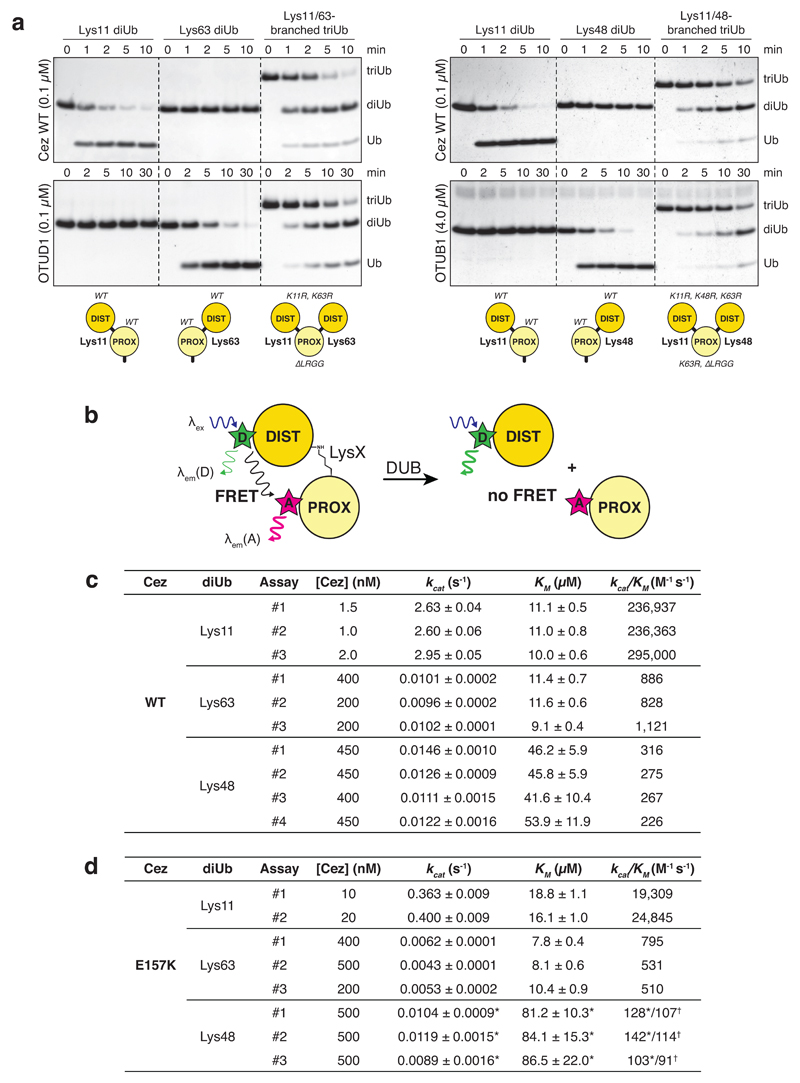
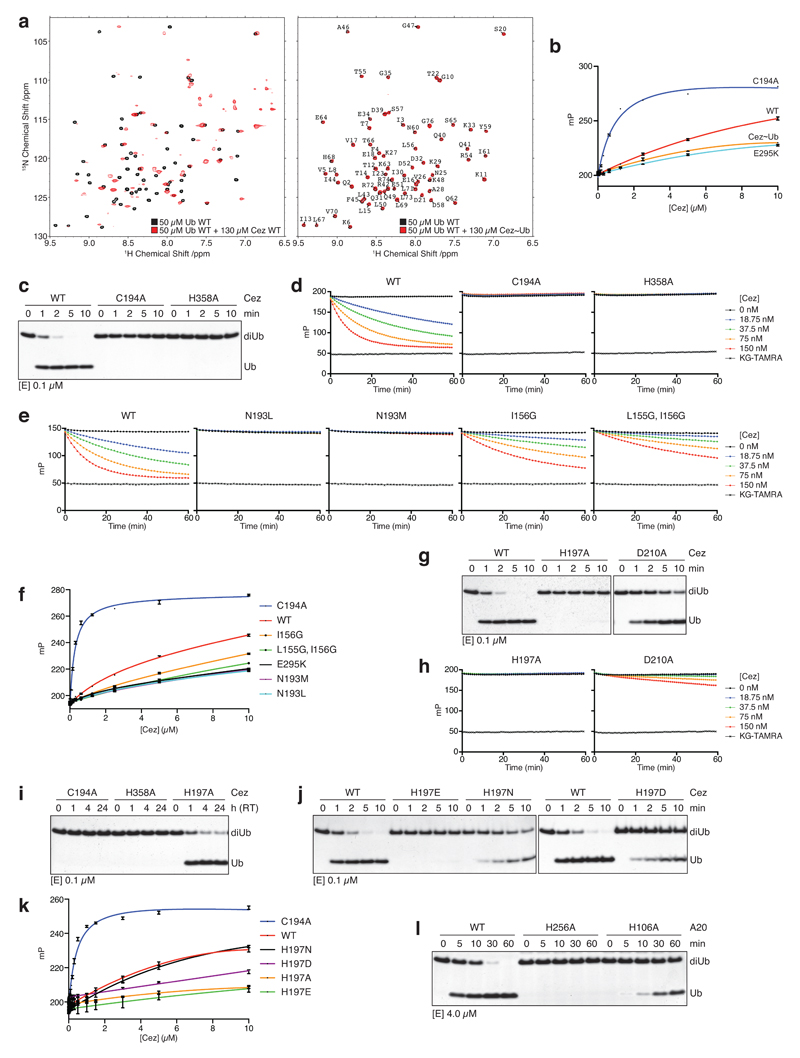
a, Domain architecture of A20-like OTU DUBs. b, Specificity analysis of the Cezanne OTU domain (residues 129-438). This experiment was performed three times. c, Representative graphs of initial rates and kinetic parameters for Lys11, Lys63 and Lys48 diUb hydrolysis by Cezanne. Assays were performed in triplicate and in at least three independent experiments (Extended Data Fig. 1c). Error bars represent standard deviation from the mean. d, Cezanne (residues 129-438) structures determined in this study: Cez apo, Cez~Lys11 diUb and Cez~Ub (‘QPG’) (see Methods). Both Cez~Ub complexes in the asymmetric unit are depicted. The OTU domain is shown as cartoon with active site residues highlighted, and Ub moieties are shown under transparent surfaces. e, Schematic of diUb ABPs. f, Probe assay of Cezanne (residues 129-438, top) and Cezanne2 (residues 1-462, bottom) with differently linked diUb ABPs. Experiments were replicated twice. g, Crystal structure of A20~Ub (residues 1-366, see Methods). For gel source data, see Supplementary Figure 1.
Cezanne/OTUD7B (Cez) regulates inflammation and NF-κB signalling12–14, T-cell activation15, EGFR trafficking16, and homeostasis of HIF-1α and HIF-2α17,18. Cezanne and Cezanne2/OTUD7A (61% identity) (Fig. 1a) are the only known DUBs specific for Lys11-linked polyUb2,17,19. Lys11-specificity is encoded in the catalytic domain2,19 (Fig. 1b), and extends to Lys11 linkages within Lys11/Lys63- and Lys11/Lys48-branched chains (Extended Data Fig. 1a). A FRET-based kinetic cleavage assay20 reveals similar KM values for Lys11-, Lys63- or Lys48-linked diUb, but a significantly higher kcat for Lys11 diUb (Fig. 1c, Extended Data Fig. 1b,c).
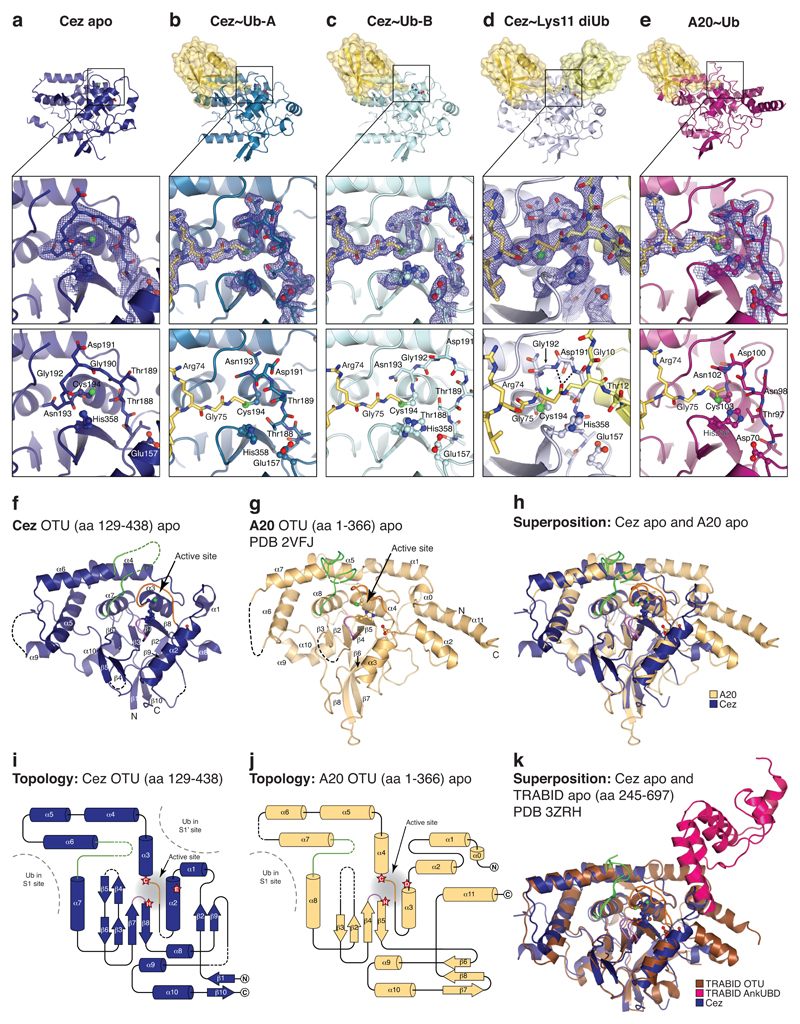
To understand the kcat-driven specificity of Cezanne, we determined crystal structures of Cezanne alone (Cez apo, 2.2 Å), in complex with monoUb (Cez~Ub, 2.0 Å, two distinct complexes in the asymmetric unit), and bound to Lys11 diUb (Cez~Lys11 diUb, 2.8 Å) (Fig. 1d, Extended Data Figs. 2,3, Extended Data Table 1, Methods). The latter utilised covalent diUb activity-based probes (ABPs)21 (Fig. 1e,f). In addition, an A20~Ub complex was determined at 2.85 Å (Fig. 1g, Extended Data Table 1, Methods). The structures of Cezanne resemble A2022,23 and TRABID10 with some topological differences (Extended Data Fig. 2).
MonoUb-bound structures have not been available for the A20-like subfamily, and the Cez~Ub and A20~Ub complexes reveal a conserved S1 Ub-binding site distinct from other OTUs (Extended Data Fig. 3). A20, Cezanne and TRABID apo states feature an unobstructed S1 site, and A20~Ub is highly similar to unliganded A20 (RMSD 0.54 Å, Extended Data Fig. 3d).
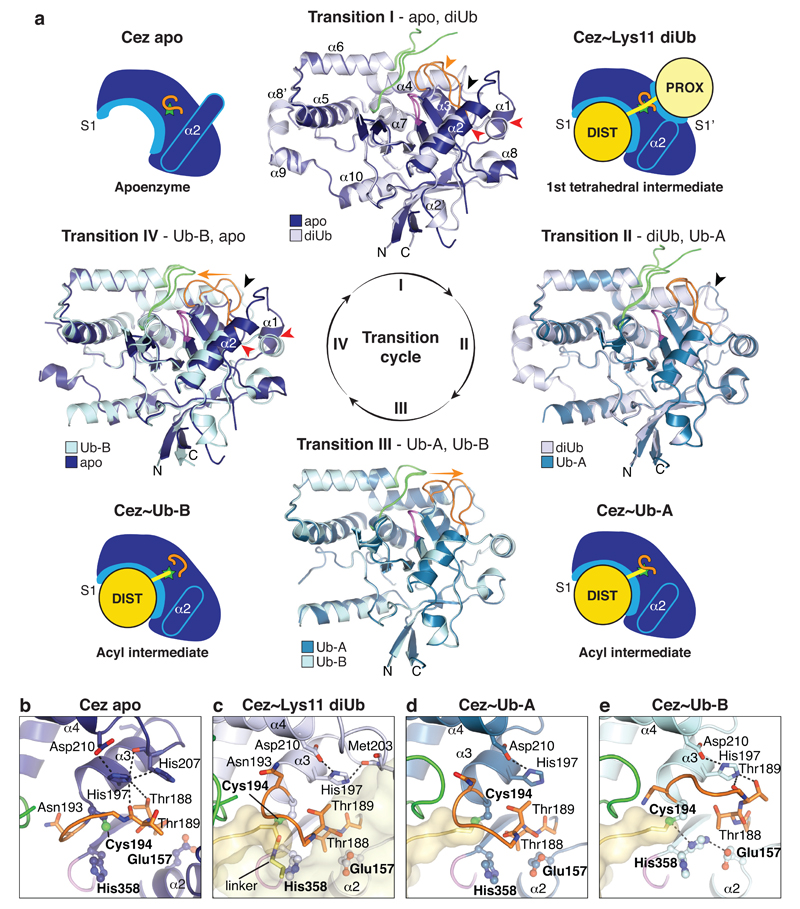
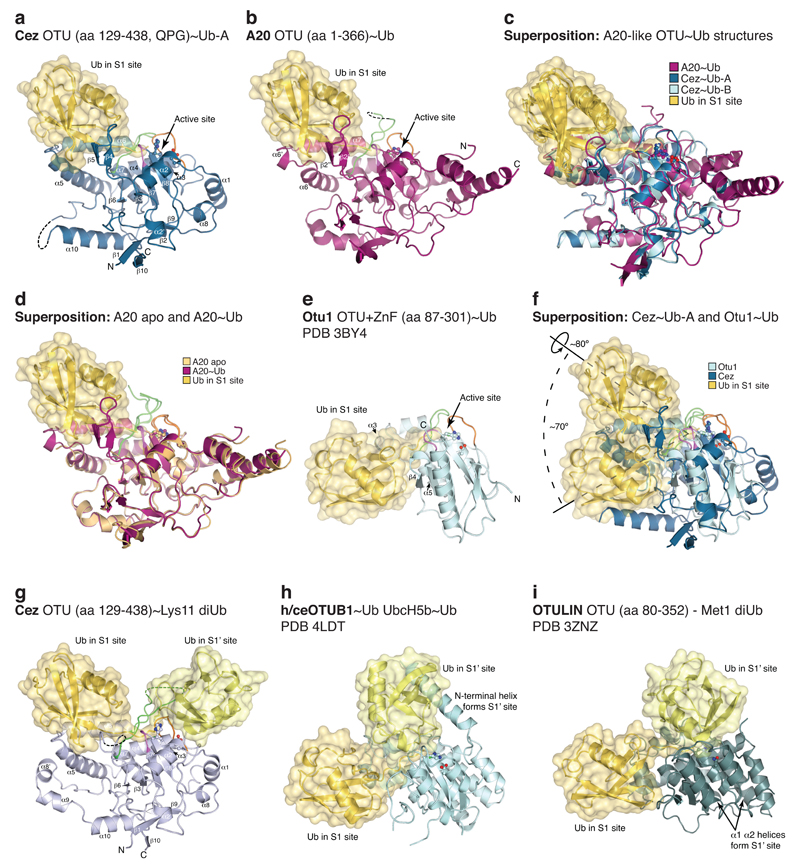
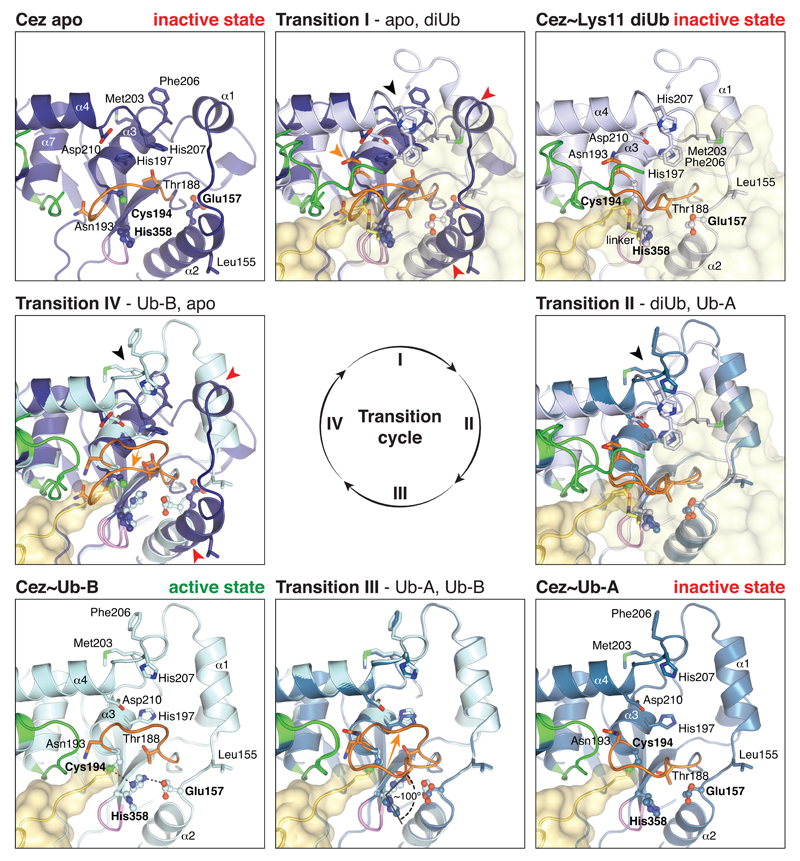
MonoUb complexes depict product-, rather than substrate-bound forms, and do not explain DUB specificity. The Cez~Lys11 diUb complex represents a substrate-bound state that reveals the S1’ site, and together, the structures explain Cezanne specificity. Indeed, large-scale conformational changes between individual structures, in which the S1’ site is transiently formed and lost, delineate the catalytic cycle for Lys11 diUb hydrolysis in Cezanne (Fig. 2, Extended Data Fig. 4, Supplementary Videos 1,2).
Figure 2. Conformational changes in the Cezanne catalytic cycle.
a, Schematic cartoons of the determined structures (Fig. 1d) highlight four catalytic states of the reaction cycle (green star, active site; orange line, Cys-loop). In between, superpositions of the OTU domain show Transitions I-IV. Loops are coloured orange (Cys-loop), green (V-loop) and purple (His-loop). Transition I (diUb substrate binding) is characterised by conformational changes around the catalytic centre, including the Cys-loop (orange arrow), α1 and α2 helices (red arrows) and the S1’-loop (black arrow). In Transition II (proximal Ub release), a second S1’-loop rearrangement relocates S1’ site residues (black arrow). Transition III features a Cys-loop movement. Several structural changes regenerate Cez apo in Transition IV. Also see Supplementary Video 1. b-e, Active site close-up of the four states. Selected residues are shown as sticks. Hydrogen bond networks of His197 and the catalytic centre are indicated. Also see Extended Data Fig. 4 and Supplementary Video 2.
Cez apo is autoinhibited due to the conformation of the Cys-loop2 (residues 187-193) that precedes catalytic Cys194. Asn193 occupies the channel that binds the C-terminal tail of the distal Ub, and the catalytic His358 is unable to deprotonate Cys194 (Fig. 2b, Extended Data Fig. 4). A key structural residue, His197, stabilises the autoinhibited Cys-loop conformation (Fig. 2b).
Significant conformational changes take place upon substrate binding (Fig. 2a). The distal Ub binds the accessible S1 site, while the proximal Ub interacts with a new S1’ site formed by the α3-α4 linker (S1’-loop hereafter) and by helices α1 and α2 that change in position and length (Fig. 2a, Extended Data Fig. 4). These rearrangements enable hydrophobic residues (Leu155, Met203, Phe206) to come together and bind the proximal Ub (Extended Data Fig. 4). His197 no longer coordinates the Cys-loop but now binds the S1’-loop (Fig. 2c). As a result, the Cys-loop moves, forming the oxyanion hole and enabling Cys194 to form the tetrahedral intermediate mimic with diUb ABP (Extended Data Fig. 2d). Intriguingly, the catalytic His358 does not coordinate Cys194 but remains in an inactive conformation, since the proximal Ub pushes Thr188 of the Cys-loop into His358’s position in the catalytic centre (‘His358-out’ conformation) (Fig. 2c).
The next step in the cycle is illuminated by the two Cez~Ub complexes, which notably no longer feature an S1’ Ub binding site (Fig. 2a, Extended Data Fig. 4). Consistently, Cez~Ub displays no interaction with Ub in NMR or fluorescence polarisation (FP) measurements (Extended Data Fig. 5a,b). Importantly, the two Cez~Ub complexes have different catalytic centres. Cez~Ub-A features the inactive ‘His358-out’ conformation (Fig. 2d), while Cez~Ub-B displays a catalytically competent state, in which Thr188 moved out and His358 coordinates Cys194 (Fig. 2e, Extended Data Fig. 4). His197 is again a key residue that now stabilises the Cys-loop in the active state (Fig. 2e), which hydrolyses the acyl intermediate to regenerate Cez apo (Fig. 2a). The active state might also depict the initial Cys deprotonation stage prior to tetrahedral intermediate formation (within Transition I, Fig. 2a). Indeed, Cys194 and His358 are essential for hydrolysis of diUb and a monoubiquitinated fluorescent Lys-Gly (KG) dipeptide (Ub-KG*, 24) (Extended Data Fig. 5c,d).
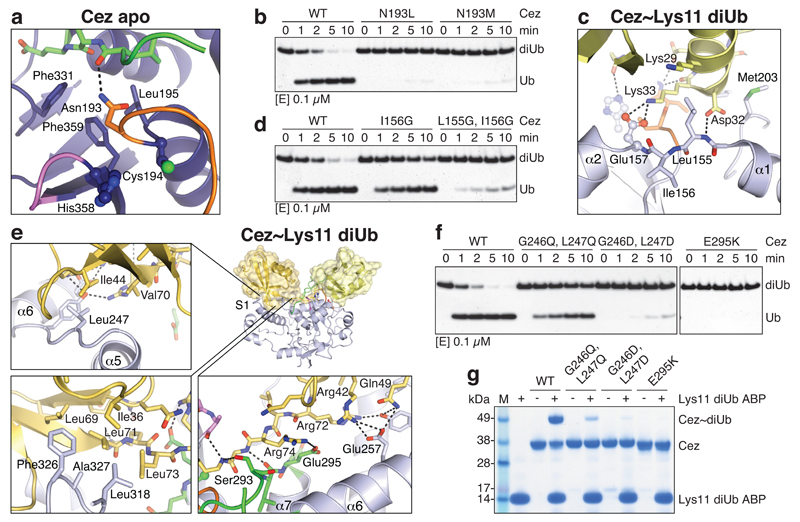
Structure-based mutagenesis confirmed key mechanistic features. Mutation of Asn193 to Leu or Met stabilised Cez apo autoinhibition by improving contacts in the Ub-binding channel and abrogated DUB activity and Ub binding (Fig. 3a,b, Extended Data Fig. 5e,f). Mutation of His197 or its coordinating residue Asp210 to Ala reduced diUb and Ub-KG* cleavage (Extended Data Fig. 5g,h). Consistent with a key structural rather than catalytic role, Cez H197A showed residual activity, and mutations maintaining its coordinating capabilities were only mildly impaired (Extended Data Fig. 5i-k). Interestingly, mutation of the corresponding residue in A20, His106, also reduced A20 DUB activity (Extended Data Fig. 5l).
Figure 3. Mutational analysis of Cezanne dynamics.
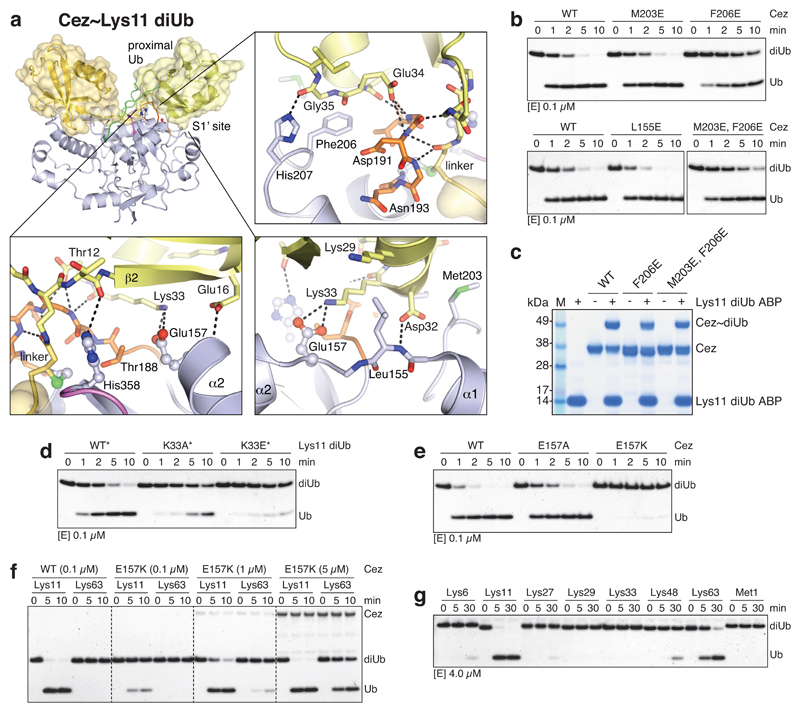
a, In Cez apo, Asn193 of the Cys-loop (orange) blocks the distal Ub-binding channel and is positioned above a hydrophobic pocket. b, DUB assay of Asn193 mutants that ‘lock’ the enzyme in the apo state. c, The α1-α2 linker in Cez~diUb contains residues Leu155 and Ile156 adjacent to Glu157. d, DUB assay of mutants with a destabilised α2-helix (see Extended Data Fig. 6c). e, Cezanne’s S1 site contacts the Ile44 patch (top left), the Ile36 patch (bottom left) and the C-terminal tail of the distal Ub (bottom right). f, DUB assay of S1 site mutants. g, Lys11 diUb ABP probe assay of S1 site mutants.All assays were performed at least twice with consistent results. For gel source data, see Supplementary Figure 1.
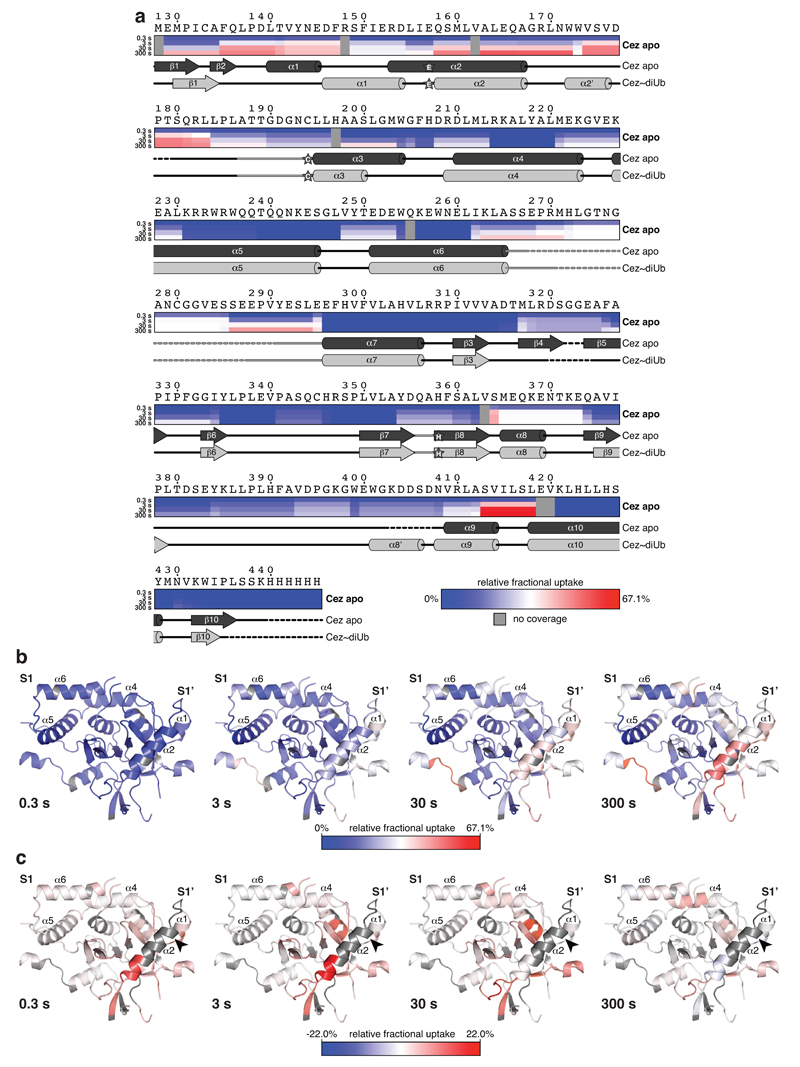
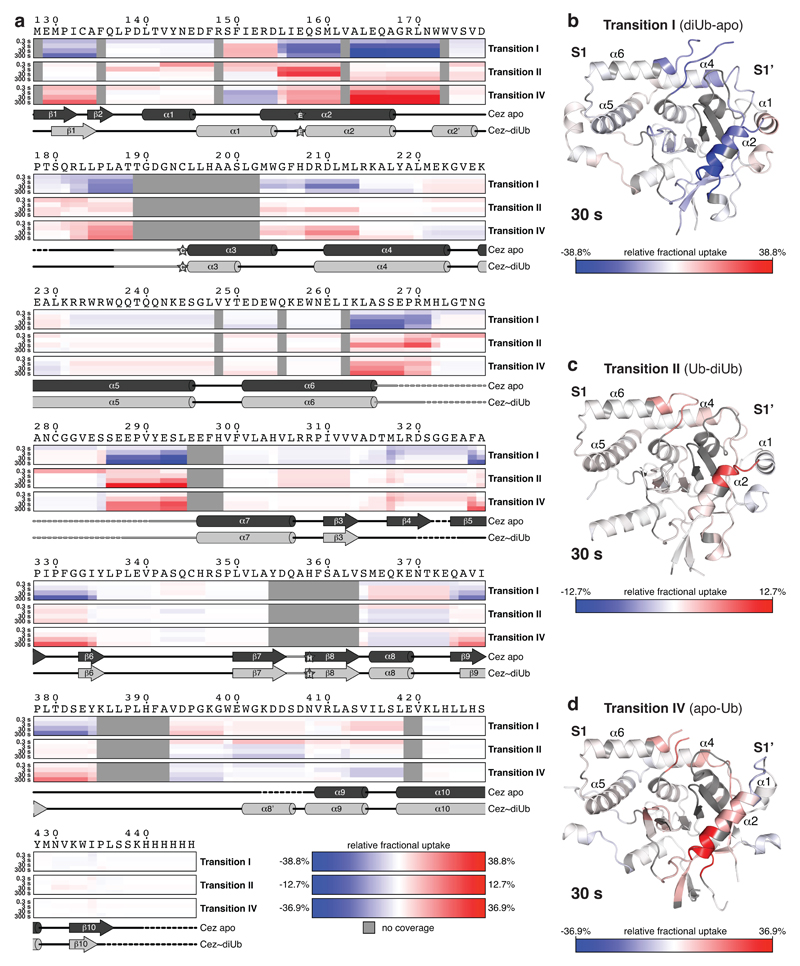
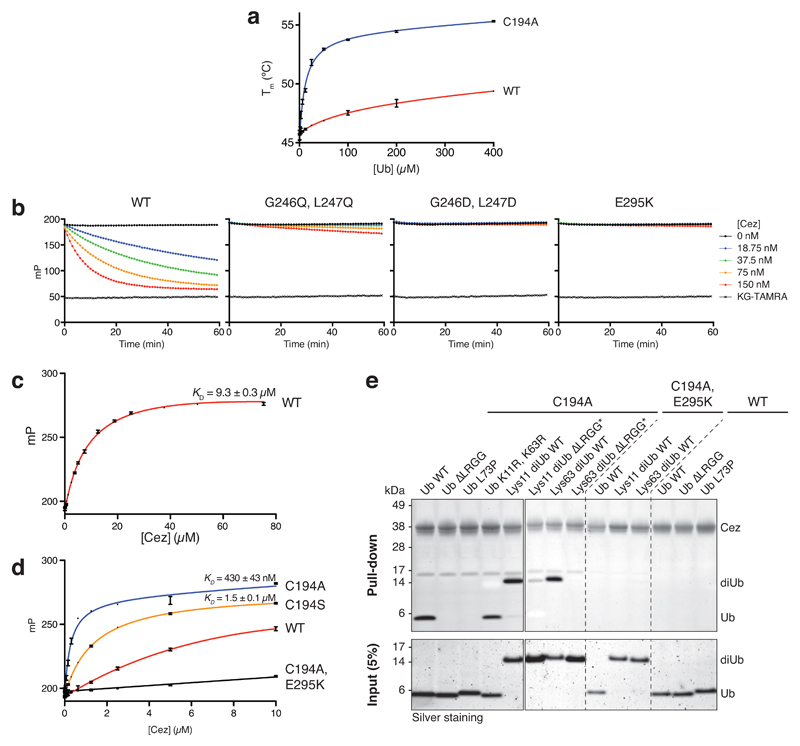
Finally, hydrogen-deuterium exchange mass spectrometry (HDX-MS) confirmed the ‘foot-prints’ of bound Ub moieties, and the conformational transitions observed crystallographically (Extended Data Figs. 6,7). The elongated α2-helix that is disrupted in complex structures (Fig. 2a) displayed an uncharacteristically high H/D exchange for helices in Cez apo, suggesting that it may be dynamic in solution (Extended Data Fig. 6a,b). Mutations that destabilise helix α2 (I156G or L155G/I156G) impaired Cezanne activity (Fig. 3c,d, Extended Data Figs. 5e,6c), indicating a required order-disorder transition in this region. Moreover, despite lack of direct contacts, these mutants reduced Ub binding to the S1 site (Extended Data Fig. 5f). Ub binding improved thermal stability of Cezanne significantly (Extended Data Fig. 8a), and an increased H/D exchange was observed in multiple elements corresponding to the S1’ site upon Ub release (Extended Data Fig. 7a,d). This reveals that S1 site Ub binding is coupled to S1’ site dynamics, and primes the enzyme for substrate discrimination and catalysis. Together, mutagenesis and HDX-MS confirm the observed conformational transitions in the catalytic cycle (Fig. 2).
We next studied how these transitions impose linkage specificity. The small difference in KM between Lys11-, Lys63- and Lys48-linked diUb (Fig. 1c) suggests that substrate engagement is primarily driven by the exposed S1 site that involves the Cezanne α5-α6 helical arm and a hydrophobic loop (Fig. 3e). These elements contact both Ile44 and Ile36 patches of Ub as well as its C-terminus. Mutations in the helical arm or in the channel for the Ub C-terminus abrogated Lys11 diUb and Ub-KG* hydrolysis (Fig. 3f, Extended Data Fig. 8b) and reactivity towards Lys11 diUb ABP (Fig. 3g). MonoUb-binding assays indicated a strong interface (KD 9.3 μM for Cez WT, 0.43 μM for Cez C194A) (Extended Data Fig. 8c,d). Moreover, monoUb or differently linked diUb bound Cez C194A similarly in pull-down assays, and this depended on a free Ub C-terminus; once removed, interactions are lost except for Lys11 diUb, as this chain type can bind across the active site (Extended Data Fig. 8e). Hence, the S1 site is solely responsible for substrate recruitment.
DUB linkage specificity relies on careful positioning of the proximal Ub, which interacts with Cezanne via an unusual surface, involving Thr12, Glu16 and its α-helix (Asp32, Lys33, Glu34, Gly35) (Fig. 3c, Extended Data Fig. 9a). A similar interface is also utilised by UBE2S, the Lys11-specific E2 enzyme25. Proximal Ub engagement transiently forms the S1’ site, enabling catalysis. Yet, consistent with the weak interface, mutations in hydrophobic S1’ site residues had little effect on DUB activity or probe reactivity (Extended Data Fig. 9b,c).
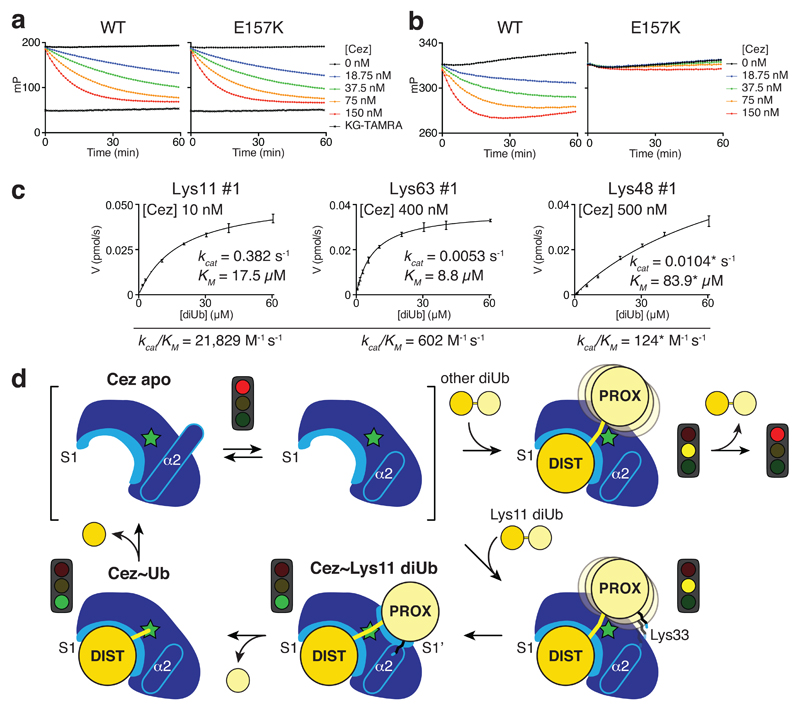
Importantly, a direct interaction between Lys33 of the proximal Ub and Glu157 of the catalytic centre (Fig. 3c) affects Cezanne catalytic turnover. Cezanne hydrolysed Lys11 diUb with proximal K33A or K33E mutations less efficiently (Extended Data Fig. 9d). Cez E157K cleaved Ub-KG* (Fig. 4a) but was severely impaired towards Lys11 diUb substrates (Fig. 4b, Extended Data Fig. 9e). Kinetic analyses of this reaction reveal a drop in kcat for Lys11 diUb hydrolysis as compared to Cez WT, while Lys63 and Lys48 cleavage was hardly affected (Fig. 4c, Extended Data Fig. 1c,d). Qualitative assays confirm that Cez E157K is less specific (Extended Data Fig. 9f,g). Lys33 binding to Glu157 requires Cezanne to be in its inactive ‘His358-out’ conformation. Hence, Glu157 is important for substrate selection but not for catalysis, and while it can coordinate His358, its presence is not essential for the catalytic dyad of Cezanne.
Figure 4. Basis of Lys11 specificity and model of Cezanne mechanism.
a, b, FP cleavage assays comparing Cez WT and E157K using Ub-KG* (a) and FlAsH-tagged Lys11-linked diUb (b). FP measurements were performed in triplicate in at least two independent experiments. c, Summary of Cez E157K diUb cleavage kinetics. Compared to Cez WT (Fig. 1c), this mutant is impaired in cleaving Lys11 linkages. Assays were performed in triplicate and in at least two independent experiments. Values marked by an asterisk (*) suffer from technical limitations (for more detail, see Extended Data Fig. 1d). Error bars represent standard deviation from the mean. d, Model of Cezanne mechanism. The apoenzyme is autoinhibited yet dynamic, and recruits a substrate with its accessible S1 site. Only Lys11-linked diUb can interact with the formed S1’ site specifically, involving an activating interaction between Cezanne and Ub Lys33. After cleavage, the S1’ site is lost and the proximal moiety expelled. Subsequent hydrolysis and distal Ub release recreates the Cezanne apo state.
Together, our findings illuminate a complete DUB catalytic cycle and reveal the molecular basis of Cezanne’s Lys11 specificity (Fig. 4d). The dynamic Cez apo state engages polyUb at the S1 site, releasing Cys-loop mediated autoinhibition. This primes the enzyme for substrate discrimination by S1’ site remodelling. Only Lys11-linked polyUb forms favourable contacts with the S1’ site, which transiently opens to enable interactions between Lys33 and Glu157, and formation of this contact improves catalytic turnover. After isopeptide bond hydrolysis, conformational changes destroy the S1’ site and expel the proximal Ub. The remaining product-bound monoUb complex is resolved by further rearrangements that align the catalytic centre to hydrolyse the acyl intermediate. Upon distal Ub release, Cezanne regains its autoinhibited state (Fig. 4d).
Our work highlights the potential plasticity of DUBs, which in case of Cezanne results in dramatic conformational transitions along the reaction cycle (Supplementary Videos 1,2). With the rising importance of DUBs as drug targets, insights into conformational flexibility are essential. Indeed, small molecule DUB inhibitors targeting Cezanne or TRABID, which also requires conformational domain rearrangements10, may open new avenues in treatment of cancer and inflammation.
Methods
Cloning and site-directed mutagenesis
A codon-optimised human Cezanne gene (Otud7b) for bacterial expression was obtained from GeneArt Gene Synthesis (Invitrogen) and cloned into pOPIN vectors26 using the In-Fusion HD Cloning Kit (Clontech) according to manufacturer’s instructions. Site-directed mutagenesis of Cezanne, A20 and Ub was performed by using the QuikChange method.
Protein expression and purification
Proteins were expressed in E. coli Rosetta2 (DE3) pLacI (Novagen) from pOPIN-E (Cezanne) or pGEX6P1 (A20). The pOPIN-E vector introduces a C-terminal His6 tag and pGEX6P1 features a PreScission protease cleavable N-terminal GST tag. Cells were grown at 37°C to an OD600 of 0.8-1.0 and induced with 0.2-0.5 mM IPTG for 18-20 h at 18°C. Expression was performed in 2-4 L 2xTY medium supplemented with appropriate antibiotics.
All purification steps were performed at 4°C. Bacterial cells were resuspended in 40-80 mL lysis buffer (25 mM Tris [pH 8.5], 200 mM NaCl, 5 mM DTT [A20] or 2 mM β-mercaptoethanol [Cez], 1 mg/mL lysozyme, 0.1 mg/mL DNaseI, one EDTA free Complete Protease Inhibitor Cocktail tablet), sonicated and cleared by centrifugation at 20,000 x g for 35 min. His6-tagged constructs were affinity purified with TALON Superflow resin (GE Healthcare). 1-2 mL resin was incubated with the cleared lysate for 5-10 min and washed with 1 L buffer A (25 mM Tris [pH 8.5], 200 mM NaCl, 2 mM β-mercaptoethanol). Protein was eluted with 5-10 mL buffer A supplemented with 250 mM imidazole and subsequently dialysed against buffer B (25 mM Tris [pH 8.5], 5 mM DTT) plus 50 mM NaCl prior to further purification (see below). Cleared lysates of GST fusion proteins were incubated with 2-4 mL Glutathione Sepharose 4B resin (GE Healthcare) for 1 h under constant agitation. The beads were washed with 2 L buffer B supplemented with 500 mM NaCl and 0.5 L buffer B plus 50 mM NaCl. GST tag cleavage was performed overnight with 50-100 μg GST-tagged PreScission protease on the resin. Protein was eluted with buffer B plus 50 mM NaCl prior to further purification.
All proteins were subjected to anion-exchange chromatography (Resource Q, GE Healthcare) in buffer B with a salt gradient of 50-500 mM NaCl, and subsequent size-exclusion chromatography (HiLoad 16/60 Superdex 75, GE Healthcare) in buffer B supplemented with 200 mM NaCl. Peak fractions were pooled, concentrated to 2-25 mg/mL using Amicon 10 kDa MWCO Ultra Centrifugal Filters (Millipore), frozen in liquid nitrogen and stored at -80°C.
Qualitative DUB assays
Qualitative in vitro deubiquitination assays were performed as described27. In short, DUBs were diluted in DUB dilution buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 10 mM DTT) to 2x indicated concentrations and pre-incubated for 10 min at room temperature. 10 μM stocks (2x final concentration) of differently linked diUb substrates were prepared in 2x DUB reaction buffer (100 mM Tris [pH 7.5], 100 mM NaCl, 10 mM DTT). To start the hydrolysis reaction, DUB and substrate solutions were mixed in a 1:1 ratio and incubated at 37°C for indicated times. Reactions were stopped by adding 4x LDS sample buffer (Invitrogen), resolved by SDS-PAGE on 4-12% gradient gels run in MES buffer (Invitrogen) and visualised by silver staining.
Assembly of Ub chains for in vitro DUB assays and pull-down assays
Lys11-linked diUb variants were assembled using UBE2S19. For the generation of Ub moiety-specific mutations, such as Lys11 diUb with a proximal K33A or K33E mutation, chains were assembled using Ub (K11R, K63R) as the distal, and Ub (K63R, ΔLRGG) variants as proximal moiety. Lys11 diUb molecules carrying no further mutation are referred to as ‘WT’, and diUb substrates carrying additional proximal mutations are called ‘K33A’ and ‘K33E’, respectively.
In order to generate specific branched triUb molecules containing a Lys11 linkage, the UBE2S assembly system was combined with UBE2N/UBE2V1 or UBE2R1 for Lys11/63 or Lys11/48 triUb, respectively28. By using Ub (ΔLRGG) with or without K63R mutation as the proximal Ub allowed the assembly of defined branches. Furthermore, distal Ub moieties contained mutations to prevent chain elongation, i.e. Ub (K11R, K63R) and Ub (K11R, K48R, K63R) were used for Lys11/63 and Lys11/48 branched triUb, respectively.
FRET-based DUB kinetics
Recently developed FRET-based diUb substrates were used to determine Michaelis-Menten kinetics for Cezanne variants20. Two Ub moieties linked via a native isopeptide linkage feature a donor (5-carboxyrhodamine110; Rho) or an acceptor (5-carboxytetramethylrhodamine; TAMRA) fluorophore, respectively. Lys11- and Lys48-linked diUb substrates of this type were used, while Lys63 FRET diUb was purchased from Boston Biochem (cat. no. UF-330). The change in fluorescence intensity (FI) of the donor fluorophore upon diUb hydrolysis was measured on a PheraStar plate reader (BMG Labtech), equipped with an FI optic module with λex = 485 nm and λem = 520 nm (for Lys11 and Lys48 FRET diUb), or λex = 540 nm and λem = 590 nm (for Lys63 FRET diUb). Reactions were performed in black, round bottom, non-binding 384-well plates (Corning) at 25°C in a total volume of 15 μL.
Non-fluorescent Lys11, Lys63 and Lys48 diUb molecules were serially diluted in FI buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 2 mM DTT, 0.1 mg/mL BSA) and the respective FRET substrate was spiked in at a fixed concentration of 1 μM (2x final concentration). In each well, 7.5 μL substrate was mixed with 7.5 μL enzyme at 2x indicated concentrations in FI buffer. All measurements for one experiment were performed in parallel in triplicate for each substrate concentration, and the change in FI was recorded over a period of 30-45 min in 15 s intervals. The maximal FI change was determined by using 25-50 nM Cez WT (Lys11 cleavage), 5 nM USP21 (Lys63 cleavage) or 0.5 μM OTUB1 (Lys48 cleavage) as positive controls. The observed FI values were plotted against time, and initial velocities of diUb cleavage were calculated. These initial rates at a fixed Cezanne concentration were plotted against diUb substrate concentration, and Michaelis-Menten parameters were determined using Prism 6 (GraphPad Software, Inc.). In case of Lys48 diUb cleavage by Cez E157K, where the determined KM value exceeded the highest tested diUb concentration, catalytic efficiencies were also calculated from a linear fit of the lower substrate concentration range (see Extended Data Fig. 1d).
Modification of Cezanne with Ub-based suicide probes
Cezanne variants were diluted in DUB dilution buffer (25 mM Tris [pH 7.5], 150 mM NaCl, 10 mM DTT) to 11 μM stocks, and mixed with 44 μM diUb ABPs21 in a 1:1 ratio. Reactions were incubated at 37°C for 10-60 min, stopped with 4x LDS sample buffer (Invitrogen) and analysed on a Coomassie-stained SDS-PAGE gel.
Further Ub-based suicide probes used in this study include the Ub-haloalkyl probe Ub-C2Br29 and Ub propargylamide (Ub-PA)30, which were used for the generation of Cez~Ub for crystallisation (Ub-C2Br) as well as A20~Ub crystallisation, FP-based Ub-binding assays, NMR and HDX-MS (Ub-PA), respectively.
Crystallisation
Crystallisation screening was carried out in 96-well plates in a sitting-drop vapour diffusion setup using nano-liter robotics (typical drop size was 100+100 nL).
The first crystallised Cez apo construct (residues 88-438) contained an N-terminal flexible extension of 41 residues that was removed in subsequent crystallisation attempts for Cez apo and complexes. Native (9.5 mg/mL) and selenomethionine (SeMet)-substituted (7.0 mg/mL) Cez apo (residues 88-438) crystals grew at 18°C in 8% (w/v) PEG 8K, 0.1 M lithium chloride and 50 mM magnesium sulfate. Cez apo (residues 129-438, 8.0 mg/mL) crystallised in 0.1 M Bis-Tris pH 6.1 and 0.2 M magnesium formate at 18°C.
Covalent complexes of Cezanne and A20 were generated with Ub-derived suicide probes (see section above) and purified by anion-exchange chromatography and gel filtration. To obtain crystals of Cez~Ub, the long, unstructured V-loop (residues 267-291) was replaced by the corresponding sequence in TRABID (Gln-Pro-Gly; QPG). Cez (residues 129-438, QPG) was reacted with Ub-C2Br to form Cez~Ub, which was set up at a concentration of 21.7 mg/mL. Initial crystals grew from 0.1 M sodium acetate pH 4.6 and 8% (w/v) PEG 4K at 18°C, and were used for streak seeding to obtain diffraction quality crystals that grew from 0.1 M sodium acetate pH 4.8 and 6% (w/v) PEG 6K. For Cez~Lys11 diUb crystallisation, Cez (residues 129-438) was reacted with Lys11-linked diUb ABP. The complex (6.7 mg/mL) crystallised at 18°C in 0.1 M phosphate citrate pH 4.2, 20% (w/v) PEG 8K and 0.2 M sodium chloride. A20~Ub was generated from A20 (residues 1-366) and Ub-PA. Crystals were set up at a concentration of 8.0 mg/mL and grew from 0.1 M MES/imidazole pH 6.5, 7% (w/v) PEG 8K and 20% ethylene glycol at 14°C.
Prior to synchrotron data collection, crystals were vitrified in liquid nitrogen after brief soaking in mother liquor containing 27.5% glycerol (Cez apo, residues 88-438), 25% (v/v) PEG400 (Cez apo, residues 129-438), 30% (v/v) PEG400 (Cez~Ub), 28% glycerol (Cez~Lys11 diUb), or mother liquor alone (A20~Ub).
Data collection, structure determination and refinement
Diffraction data were collected at the ESRF (Grenoble, FR), beam lines ID23-1 (Cez apo; wavelength: 0.97933 Å) and ID29 (A20~Ub; 0.96863 Å), the Diamond Light Source (Harwell, UK), beam lines I02 (Cez~Ub; 0.97950 Å) and I03 (Cez~Lys11 diUb; 0.97626 Å), and the High Energy Accelerator Research Organization (KEK, Tsukuba, Japan), beam line PF BL-1A (Cez SeMet; 0.9786 Å).
Diffraction images were indexed and integrated using iMOSFLM31 or XDS32, and scaled using SCALA33 or its successor program AIMLESS34.
The structure of Cez apo (residues 88-438) was solved by SAD phasing using diffraction data collected from a SeMet-substituted crystal. The automated structure solution pipeline SHARP and autoSHARP was used for SAD phasing35,36, followed by iterative manual building using Coot37 and refinement using PHENIX38. Electron density was not visible for the first 41 residues, which were removed from the construct for subsequent crystallisation attempts.
Phases for subsequent Cez structures as well as for A20~Ub were obtained by molecular replacement in PHASER39, using Cez apo (88-438), Cez apo (129-438) or A20 apo (PDB 2VFJ, 22), and Ub (PDB 1UBQ, 40) as search models where appropriate. Models were built in Coot37 and refined in PHENIX38 in iterative rounds, using simulated annealing and TLS restraints where appropriate. Final Ramachandran statistics (favoured/allowed/outliers) were 98.5%/1.5%/0% (Cez apo), 99.1%/0.9%/0% (Cez~Ub), 98.6%/1.4%/0% (Cez~Lys11 diUb), and 97.8%/2.2%/0% (A20~Ub), respectively. Final data collection and refinement statistics are summarised in Extended Data Table 1.
Fluorescence polarisation (FP) assays
FP measurements were used to monitor the interaction between DUBs and FlAsH-tagged Ub, as well as the cleavage of a monoubiquitinated TAMRA-labelled Lys-Gly (KG) dipeptide (Ub-KG*)24 or Lys11-linked diUb-FlAsH. Measurements were performed using a PheraStar plate reader (BMG Labtech), equipped with an optic module for FlAsH (λex = 485 nm, λem = 520 nm) or TAMRA (λex = 540 nm and λem = 590 nm) detection. FP values given in millipolarisation (mP) were determined by taking the following polarisation values into account: FlAsH-Ub (195 mP), TAMRA-KG (50 mP) and Lys11 diUb-FlAsH (315 mP). Assays were performed in triplicate in black, round bottom, non-binding surface 384-well plates (Corning) at 25°C in 20 μL.
For FP binding studies, FlAsH-Ub was diluted to 300 nM in FP buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 2 mM β-mercaptoethanol, 0.1 mg/mL BSA). Serial dilutions in FP buffer were prepared of Cezanne variants, and 10 μL of this was mixed with 10 μL FlAsH-Ub in a 384-well plate. The plate was incubated in the dark for 10-15 min prior to the measurement. FP values were fitted according to a ‘one-site – total’ binding model using Prism 6 (GraphPad Software, Inc.).
FP cleavage assays were started by mixing 10 μL/well enzyme at 2x indicated concentration in FP buffer to 10 μL of predispensed FP substrate (Ub-KG* or Lys11-linked diUb-FlAsH) at 300 nM. For the TAMRA-based substrate, TAMRA-KG was included as positive control. Hydrolysis reactions were followed for 1 h in 60-90 s intervals. FP values of TAMRA-measurements were fitted according to a ‘one phase decay’ model using Prism 6 (GraphPad Software, Inc.).
NMR Ub-binding study with Cez apo WT and Cez~Ub
Cez WT (residues 129-438), a purified covalent Cez~Ub complex (assembled with Cez WT and Ub-PA), and 15N-labelled Ub WT were dialysed against NMR buffer (18 mM Na2HPO4, 7 mM NaH2PO4xH2O, 150 mM NaCl, 5 mM DTT [pH 7.2]). Samples of 50 μM 15N-labelled Ub alone or in the presence of 130 μM Cez WT or Cez~Ub were prepared in 350 μL NMR buffer containing 20 μL D2O, and were transferred into Shigemi NMR microtubes.
1H-15N BEST-TROSY (Band-selective Excitation Short-Transient Transverse Relaxation-Optimised Spectroscopy) spectra were recorded at 298 K on a Bruker AvanceII+ 700 MHz spectrometer with a TCI triple resonance probe. Data were processed in TopSpin 3.0 (Bruker Inc., Billerica, MA, USA) and analysed in Sparky (UCSF).
Thermal shift assay
Protein melting curves were recorded on a Corbett RG-6000 real-time PCR cycler. Samples contained 3 μM Cez (residues 129-438) WT or C194A, 3x SYPRO Orange, and 0-400 μM Ub. Data were recorded in triplicate and in two independent experiments. Melting temperatures (Tm) in the presence of Ub were referenced to 45.9 ± 0.2°C (WT) and 45.0 ± 0.2°C (C194A).
Pull-down assay
His6-tagged Cezanne constructs were used for in vitro pull-downs of purified Ub and diUb variants. For each reaction, 5 μL TALON Superflow resin (GE Healthcare) was equilibrated with PD buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 2 mM β-mercaptoethanol), mixed with 20 μg His6-tagged bait in 400 μL PD buffer plus 0.1 mg/mL BSA, incubated for 20 min at 4°C under constant agitation, and subsequently washed three times. Ub and diUb prey proteins were diluted in PD buffer plus BSA to 1.2 μg/mL and 2.4 μg/mL, respectively, and 400 μL were added to the immobilised bait. After incubation for 1 h at 4°C, the resin was washed three times with PD buffer prior to the addition of 50 μL of 4x LDS sample buffer (Invitrogen). Samples were boiled for 1 min, and 20 μL per sample was analysed by SDS-PAGE and silver staining.
Hydrogen-deuterium exchange mass spectrometry (HDX-MS)
Deuterium exchange reactions of Cezanne were initiated by diluting the protein in D2O (99.8% D2O ACROS, Sigma, UK) in 10 mM Tris pH 7.5 buffer to give a final D2O percentage of 95.3. For all experiments, deuterium labelling was carried out at 23°C (unless otherwise stated) at four time points (3 sec on ice (0.3 sec), 3 sec, 30 sec and 300 sec). The labelling reaction was quenched by the addition of chilled 2.4% (v/v) formic acid in 2 M guanidinium hydrochloride, and immediately frozen in liquid nitrogen. Samples were stored at -80°C prior to analysis.
The quenched protein samples were rapidly thawed and subjected to proteolytic cleavage by pepsin followed by reversed phase HPLC separation. Briefly, the protein was passed through an Enzymate BEH immobilised pepsin column, 2.1 x 30 mm, 5 μm (Waters, UK) at 200 μL/min for 2 min and the peptic peptides trapped and desalted on a 2.1 x 5 mm C18 trap column (Acquity BEH C18 Van-guard pre-column, 1.7 μm, Waters, UK). Trapped peptides were subsequently eluted over 12 min using a 5-36% gradient of acetonitrile in 0.1% (v/v) formic acid at 40 μL/min. Peptides were separated on a reverse phase column (Acquity UPLC BEH C18 column 1.7 μm, 100 mm x 1 mm (Waters, UK). Peptides were detected on a SYNAPT G2-Si HDMS mass spectrometer (Waters, UK) acquiring over a m/z of 300 to 2000, with the standard electrospray ionization (ESI) source and lock mass calibration using [Glu1]-fibrino peptide B (50 fmol/μL). The mass spectrometer was operated at a source temperature of 80°C and a spray voltage of 2.6 kV. Spectra were collected in positive ion mode.
Peptide identification was performed by MSe using an identical gradient of increasing acetonitrile in 0.1% (v/v) formic acid over 12 min. The resulting MSe data were analysed using Protein Lynx Global Server software (Waters, UK) with an MS tolerance of 5 ppm.
Mass analysis of the peptide centroids was performed using DynamX software (Waters, UK). Only peptides with a score >6.4 were considered. The first round of analysis and identification were performed automatically by the DynamX software, however, all peptides (deuterated and non-deuterated) were manually verified at every time point for the correct charge state, presence of overlapping peptides, and correct retention time. Deuterium incorporation was not corrected for back-exchange and represents relative, rather than absolute changes in deuterium levels. Changes in H/D amide exchange in any peptide may be due to a single amide or a number of amides within that peptide. All time points of a data set (i.e. data of related constructs) were prepared simultaneously and were acquired on the mass spectrometer on the same day.
Extended Data
Extended Data Figure 1. Analysis of branched triUb substrates and FRET-based diUb cleavage kinetics.
a, Branched triUb molecules with different topology were generated as shown in the schematic (bottom). Lys11 diUb, Lys63 diUb and branched Lys11/63 triUb (left panel) were treated with Cez WT (top) and OTUD1 (residues 287-481, bottom), a Lys63-specific enzyme2. Both DUBs cleaved their preferred diUb substrate as well as one linkage of the branched triUb molecule. Lys11 diUb, Lys48 diUb and branched Lys11/48 triUb (right panel) were incubated with Cez WT (top) and OTUB1 (full-length, bottom), a Lys48-specific OTU DUB2. Again, both enzymes showed similar activities towards their preferred linkage type in a diUb substrate and a branched triUb molecule. This shows that Cezanne can cleave Lys11 linkages in the context of Lys11/Lys63- and Lys11/Lys48-branched chains. For gel source data, see Supplementary Figure 1. b, Schematic of FRET-based diUb cleavage assays to derive DUB kinetics. Distal and proximal Ub moieties were modified with a donor (D) and acceptor (A) fluorophore, respectively. Upon DUB treatment, the native isopeptide bond is cleaved and the FRET signal is lost. The increase in donor intensity is measured to follow the reaction. c, Kinetic parameters for all independently performed experiments of Lys11, Lys63 and Lys48 diUb cleavage by Cez WT. Values are in good agreement with previously published parameters derived from gel-based studies41. d, Summary of kinetic parameters for Lys11-, Lys63- and Lys48-linked diUb cleavage by Cez E157K. The determined KM values for Lys48 diUb lie above the highest tested substrate concentration, so kinetic parameters marked by an asterisk (*) were calculated from experiments where substrate saturation could not be achieved due to technical limitations. Catalytic efficiencies (kcat/KM) for this substrate were also derived from a linear fit of the lower concentration range (0-20 μM, linear part of the graph). These values are marked by a cross (†). The similarity of catalytic efficiencies calculated in two different ways indicate that the kinetic parameters marked by an asterisk (*) are good estimates. See Supplementary Figure 2 for all corresponding graphs of initial rates.
Extended Data Figure 2. Crystal structures determined in this study and comparison of A20-like OTU apo structures.
a-e, Active site regions Cez apo (a), Cez~Ub-A (b), Cez~Ub-B (c), Cez~Lys11 diUb (d), and A20~Ub (e). 2|FO| – |FC| electron density maps contoured at 1σ (blue) cover catalytic residues, the Cys-loop and chemical linkers in the complex structures. Hydrogen bonds between the oxyanion hole and the Lys11 diUb ABP linker carbonyl are indicated in d, and the sp3-hybridised carbon atom that is linked to the oxyanion in a native first tetrahedral intermediate is highlighted (green arrow). f, g, Cezanne OTU (as in Fig. 1d) and A20 OTU (PDB 2VFJ, 22) apo structures with labelled secondary structure elements. Catalytic residues are shown in ball-and-stick representation. Three loops surrounding the active site are coloured (Cys-loop, orange; V-loop, green; His-loop, purple). h, Superposition of f and g showing structural similarities and differences between Cezanne and A20. i, j, Topology diagrams of f and g. The catalytic centre is indicated (red stars) and Ub-binding sites are highlighted. A20 contains two additional N-terminal and one additional C-terminal helices compared to Cezanne. The β1-β10 sheet in Cezanne corresponds to the A20 β7-β8 sheet. This explains why sequence-based alignments are challenging. k, Superposition of Cez apo (f) and TRABID AnkUBD (pink) and OTU (brown) domains (residues 245-697, PDB 3ZRH, 10).
Extended Data Figure 3. Comparison of Ub and diUb complexes within the OTU family.
Ub moieties are shown in cartoon representation under transparent surfaces in shades of yellow. Secondary structure elements involved in Ub binding are labelled, and active site loops are coloured as in Extended Data Fig. 2f. a, Cez~Ub-A complex as in Fig. 1d. b, A20~Ub complex as in Fig. 1g. c, Superposition of Ub complexes reveals a conserved S1 Ub-binding mode in A20-like OTU DUBs. d, Superposition of A20 apo (Extended Data Fig. 2g) and A20~Ub (b). No large conformational changes occur upon Ub binding. However, two unstructured loops in A20 apo are stabilised by Ub, forming helix α6’ and the β2’-β2” sheet (compare Extended Data Fig. 2j). e, The structure of the yeast Otu1~Ub complex (PDB 3BY4,42) is representative for the OTUD subfamily of OTU DUBs. The Ub moiety in the S1 site is mainly bound by the short helix α3. f, The superposition of Cez~Ub (a) and Otu1 (e) reveals substantially different S1 sites between A20-like and OTUD subfamilies. Rotations around the roll axis of Ub (~80°) and the active site (~70°) would be required to align both Ub moieties. g-i, Structures of OTU domains in identical orientation bound to their respective diUb substrate. The binding mode of proximal and distal Ub differs dramatically between the here determined Cez~Lys11 diUb complex (g, as shown in Fig. 1d), the h/ceOTUB1~Ub UbcH5b~Ub structure (PDB 4LDT, 43, UbcH5b molecule is not shown), which resembles an OTUB1 Lys48 diUb complex (h), and OTULIN bound to Met1-linked diUb (PDB 3ZNZ, 3) (i).
Extended Data Figure 4. Conformational changes in the catalytic centre.
Cezanne structures according to Fig. 2a are shown in the corners, and Transitions I-IV are overlays of neighbouring structures. Side chains of catalytic residues and other selected residues are highlighted. Loops are coloured as in Extended Data Fig. 2f. Cez apo shows a catalytically incompetent state. His358 and Glu157 are in flipped-out conformations. Transition I features structural rearrangements of the Cys-loop (orange arrow), helices α1 and α2 (red arrows) and the S1’-loop (black arrow). Cez~Lys11 diUb also features an inactive state; His358 remains flipped-out, which is caused by the Cys-loop residue Thr188 that is pushed into the active site by the proximal Ub. In Transition II, another S1’-loop movement relocates S1’ site residues (black arrow). A similar inactive state is present in Cez~Ub-A, and Thr188 still resides in the active site. The absence of the proximal Ub allows the Cys-loop and Thr188 to move in Transition III (orange arrow), allowing a ~100° rotation of His358. Hence, Cez~Ub-B contains an aligned catalytic centre. Hydrogen bonds are indicated. In Transition IV, large conformational changes in various parts of the OTU domain regenerate the autoinhibited apoenzyme.
Extended Data Figure 5. Ub binding to Cezanne and mutational analysis of residues involved in catalysis and conformational dynamics.
a, NMR analysis of Ub binding to Cez WT and the covalent Cez~Ub complex. 1H-15N BEST-TROSY spectra of 50 μM 15N-labelled Ub alone (black) and in the presence of 130 μM unlabelled Cez WT (red, left) or unlabelled Cez~Ub (red, right). Strong chemical shift perturbations upon addition of Cez WT indicate binding to Ub. In contrast, no chemical shifts were detected with Cez~Ub, suggesting that all changes with Cez WT can be attributed to the S1 site (this site is occupied by unlabelled Ub in Cez~Ub). More importantly, this also indicates that a functional S1’ site is not present in the Cez~Ub complex. b, Fluorescence polarisation (FP) experiment assessing the binding of FlAsH-tagged Ub to catalytically inactive Cez (C194A), Cez WT, an S1 site mutant (E295K, see below) and the Cez~Ub complex. c, Lys11 diUb cleavage assays of catalytic Cys194 and His358 mutants. d, e, Ub-KG* cleavage by catalytic Cys194 and His358 mutants (d), as well as Asn193 and helix α2 mutants that modulate the overall dynamics of Cezanne (e). This assay follows fluorescent dye release in the reaction; the fact that Cez H358A is inactive indicates an important role in the deprotonation of the catalytic Cys at the start of the reaction (i.e. the catalytic centre transiently adopts an active state) and/or a role in resolving the first tetrahedral intermediate. In case His358 was not required for either, we would expect a single turnover of the reaction, which would stop at the thioester intermediate. The release of KG-TAMRA would still occur, but was not detected even at an enzyme concentration of 150 nM (the substrate concentration in all assays was 150 nM). The FP signal also did not increase, suggesting that no covalent first tetrahedral intermediate was formed due to impaired dye release. Hence, the data suggests a role for His358 at least in the initial Cys deprotonation in addition to the last reaction step. f, FP binding assay of Asn193 and helix α2 mutants compared to constructs used in b. g, h, Hydrolysis of Lys11-linked diUb (g) and Ub-KG* (h) by Cez H197A and D210A. i, DUB assay with Cezanne variants (extended incubation at room temperature, RT). j, Lys11 diUb cleavage assay with His197 variants. k, FP binding assay as in b testing His197 variants. l, Mutation of corresponding residues in A20 (A20 His256 corresponds to Cez His358, and A20 His106 to Cez His197) have similar effects on Lys48 diUb hydrolysis. All DUB assays are representative of at least two independent experiments for every construct. Ub-KG* cleavage experiments and FP binding assays were replicated at least twice for each variant with consistent results. FP measurements were performed in triplicate. Error bars represent standard deviation from the mean. mP, millipolarisation unit. For gel source data, see Supplementary Figure 1.
Extended Data Figure 6. HDX-MS analysis of the Cez apo state.
a, HDX-MS experiment showing the conformational dynamics of Cez WT. The relative fractional deuterium uptake is shown for four time points (0.3-300 s). Protein sequence and secondary structure elements of Cez apo (dark grey) and Cez~diUb (light grey) are aligned. Residues of the catalytic centre are indicated by stars. b, Cez apo structure coloured based upon the relative fractional deuterium uptake of Cez WT at 0.3 s, 3 s, 30 s and 300 s. The region spanning helices α1 and α2 shows a particularly high deuterium uptake, suggesting conformational flexibility in this region in solution. c, The H/D exchange of the α2-helix destabilising mutant Cez L155G/I156G compared to Cez WT. Cez apo structure coloured based upon the difference in deuterium uptake (L155G/I156G-WT) at 0.3 s, 3 s, 30 s and 300 s (heat maps are shown in Supplementary Figure 3). The data suggest that helix α2 is destabilised, as regions structurally adjacent to the mutation site (black arrow) show an increased deuterium uptake as compared to Cez WT. Peptides containing the mutations could not be analysed due to the different sequences, and are therefore coloured in grey. Notably, most differences are stronger at shorter time points, indicating increased dynamics within this time frame (0.3-30 s). At the latest time point (300 s), differences are not as pronounced, suggesting that Cez WT undergoes the same structural rearrangements at a slower speed. Importantly, the data also confirms that overall folding of the mutant was not affected by the two Gly residues introduced in helix α2.
Extended Data Figure 7. HDX-MS analysis of Transitions I, II and IV.
a, HDX-MS experiments were performed with Cez apo, Cez~Lys11 diUb and Cez~Ub. Heat maps show differences in deuterium uptake between two states in each case: Transition I (diUb-apo), Transition II (Ub-diUb) and Transition IV (apo-Ub). Hence, Cezanne regions that are stabilised or more protected upon Lys11 diUb binding (Transition I), or more flexible or exposed upon the stepwise release of the proximal Ub (Transition II) and the distal Ub (Transition IV) are highlighted. The S1 site predominantly consists of helices α5 and α6 (i.e. helical content with very low deuterium uptake in any state), and is not as easily detected as the S1’ site that features various loops and the dynamic helix α2. Cezanne sequence and secondary structure schematics are shown as in Extended Data Fig. 6a. b, Cez~Lys11 diUb structure (shown without Lys11 diUb) coloured based upon Transition I deuterium uptake at 30 s. c, Transition II deuterium uptake at 30 s plotted onto Cez~Ub-B (shown without Ub). d, Cez apo coloured based upon Transition IV deuterium uptake at 30 s.
Extended Data Figure 8. Mutational analysis of the S1 Ub-binding site.
a, Thermal shift assay of Cez WT and C194A. In the presence of Ub, the melting temperature (Tm) of Cezanne increases. Data were recorded in triplicate and in two independent experiments. b, Ub-KG* hydrolysis by S1 site mutants. c, d, FP-based affinity measurement using N-terminally FlAsH-tagged Ub. Dissociation constants (Kd values) for Cez WT (c), C194A and C194S (d) are shown. Data are representative of at least two independent experiments per construct. e, Pull-down assay with His-tagged Cez constructs (catalytically inactive C194A, S1 site mutant C194A/E295K or WT) and different Ub and diUb variants. MonoUb requires an intact C-terminus to bind to Cez C194A. To prevent unspecific binding of differently linked diUb molecules with their proximal Ub to the S1 site, the C-terminus was removed (ΔLRGG). Variants marked by an asterisk (*) were assembled by using K11R, S20C and K63R mutations in the distal Ub, as well as K63R (only for Lys11 diUb) and ΔLRGG in the proximal Ub moiety. Pull-down and input samples were analysed by SDS-PAGE and silver staining. The pull-down assay was performed in two independent experiments. For gel source data, see Supplementary Figure 1.
Extended Data Figure 9. Biochemical analysis of S1’ site mutations.
a, The interface between Cezanne and the proximal Ub in the Cez~Lys11 diUb complex. An unusual surface of Ub comprising Glu16, Asp32, Lys33 and Glu34 is contacted by the S1’ site (Leu155, Glu157, Met203, Phe206 and His207). b, c, Lys11 diUb cleavage (b) and Lys11 diUb ABP reactivity (c) assays with S1’ site mutants. d, DUB assays with Cez WT and Ub variants. Lys11 diUb substrates were assembled to specifically mutate the proximal Ub by using K11R, K63R mutations in the distal, and K63R, ΔLRGG in the proximal Ub moiety. No further mutations were introduced in WT*, while K33A* and K33E* variants additionally contained respective mutations in their proximal Ub only. e, Lys11 diUb cleavage assay with Glu157 variants. f, g, Gel-based specificity analysis of Cez E157K. The mutant shows a reduced activity towards Lys11-linked diUb and therefore specificity compared to Cez WT (compare Fig. 1b). Assays with each variant were performed at least twice with consistent results. For gel source data, see Supplementary Figure 1.
Extended Data Table 1. a Data collection and refinement statistics.
| Cez apo SeMet (88-438) | Cez apo (129-438) | Cez~Ub (129-438, QPG) | Cez~Lys11 diUb (129-438) | A20~Ub (1-366) | ||
|---|---|---|---|---|---|---|
| Data collection | ||||||
| Space Group | P41212 | P41212 | H3 | P212121 | P1211 | |
| Cell dimensions | ||||||
| a, b, c (Å) | 96.72, 96.72, 83.37 | 103.35, 103.35, 90.20 | 157.56, 157.56, 75.6 | 92.03, 56.52, 91.75 | 64.20, 71.95, 203.93 | |
| α, β, γ (Å) | 90, 90, 90 | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 | 90, 94.64, 90 | |
| Resolution (Å) | 50.00 - 3.70 (3.83 - 3.70) | 90.20 - 2.20 (2.27 - 2.20) | 66.13 - 2.00 (2.05 - 2.00) | 56.52 - 2.80 (2.87 - 2.80) | 49.33 - 2.85 (2.96 - 2.85) | |
| Rmerge | 0.212 (0.718) | 0.071 (0.829) | 0.125 (0.854) | 0.145 (0.689) | 0.070 (0.384) | |
| < I/σI > | 11.7 (4.3) | 13.1 (1.9) | 8.6 (1.9) | 9.5 (2.0) | 9.6 (2.1) | |
| CC1/2 | 0.985 (0.437) | 0.994 (0.515) | 0.990 (0.691) | 0.995 (0.867) | ||
| Completeness (%) | 99.8 (100) | 100 (99.9) | 100 (100) | 94.6 (95.5) | 99.4 (99.8) | |
| Redundancy | 7.7 (7.6) | 7.9 (5.8) | 4.2 (4.1) | 4.6 (4.6) | 3.2 (3.3) | |
| Refinement | ||||||
| Resolution (Å) | 73.08 - 2.20 | 45.48 - 2.00 | 48.16 - 2.80 | 49.33 - 2.85 | ||
| No. reflections | 25351 | 47215 | 11478 | 43393 | ||
| Rwork / Rfree | 19.8 / 21.9 | 17.6 / 21.7 | 20.7 / 24.4 | 19.5/ 24.6 | ||
| No. atoms | ||||||
| Protein | 2143 | 5599 | 3318 | 11558 | ||
| Ligand/ion | - | 18 | 35 | 8 | ||
| Water | 90 | 621 | 3 | - | ||
| B factors | ||||||
| Protein | 72.3 | 29.0 | 46.8 | 67.1 | ||
| Ligand/ion | - | 36.9 | 56.3 | 67.6 | ||
| Water | 58.8 | 34.2 | 17.2 | - | ||
| R.m.s deviations | ||||||
| Bond length (Å) | 0.002 | 0.002 | 0.002 | 0.002 | ||
| Bond angles (°) | 0.59 | 0.62 | 0.52 | 0.54 | ||
Values in parentheses are for the highest resolution shell. All datasets were collected from a single crystal each.
Supplementary Material
Acknowledgments
We thank Soichi Wakatsuki and Naohiro Matsugaki for access to KEK PF BL-1A, and beam-line scientists at ESRF ID23-1, ID29 and Diamond I02 and I03; Roger Williams and Mark Skehel for help and discussion on HDX-MS; Chris Johnson and Stephen McLaughlin for help with biophysics. Access to DLS was supported in part by the EU FP7 infrastructure grant BIOSTRUCT-X (contract no. 283570).
This work was supported by the Medical Research Council [U105192732] (DK), the European Research Council [309756 (DK); 281699 (HO)], the Lister Institute for Preventive Medicine (DK), the British Heart Foundation [PG11/109/29247] (JEB), and a Netherlands Organization for Scientific Research VICI grant [724.013.002] (HO). TETM was supported by the Marie Curie ITN UPStream and YK by Marie Curie and EMBO Long Term Fellowships.
Footnotes
Accession numbers
Coordinates and structure factors have been deposited with the protein data bank accession codes 5LRU (Cez apo), 5LRV (Cez~Lys11 diUb), 5LRW (Cez~Ub) and 5LRX (A20~Ub).
Author contribution
DK directed the research. TETM performed all biochemical experiments, crystallised and determined the structure of the Cez~Lys11 diUb complex, and refined and analysed all structures. YK crystallised and determined the structure of Cez, Cez~Ub and A20~Ub, and performed preliminary biochemistry. MPCM and FEO designed and generated diUb ABP probes, PPG designed and generated (with BDMvT) diUb based FRET probes, SLM performed HDX-MS, JEB performed preliminary HDX-MS, MG performed TSA assays, PRE, MA and MK helped with data collection and structure determination, SMVF performed NMR analysis, HO guided chemical biology efforts. TETM and DK analysed the data and wrote the manuscript with help from all authors.
Conflict of Interest Statement
D.K. and H.O. are part of the DUB Alliance that includes Cancer Research Technology and FORMA Therapeutics. H.O. and F.E. are co-founders and shareholders of UbiQ Bio BV.
References
- 1.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 2.Mevissen TET, et al. OTU Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiener R, Zhang X, Wang T, Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483:618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Juang Y-C, et al. OTUB1 Co-opts Lys48-Linked Ubiquitin Recognition to Suppress E2 Enzyme Function. Mol Cell. 2012;45:384–397. doi: 10.1016/j.molcel.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hymowitz SG, Wertz IE. A20: from ubiquitin editing to tumour suppression. Nat Rev Cancer. 2010;10:332–341. doi: 10.1038/nrc2775. [DOI] [PubMed] [Google Scholar]
- 7.Wertz IE, et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature. 2015;528:370–375. doi: 10.1038/nature16165. [DOI] [PubMed] [Google Scholar]
- 8.Tran H, Hamada F, Schwarz-Romond T, Bienz M. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 2008;22:528–542. doi: 10.1101/gad.463208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin J, et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat Immunol. 2016;17:259–268. doi: 10.1038/ni.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licchesi JDF, et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID's specificity for atypical ubiquitin chains. Nat Struct Mol Biol. 2012;19:62–71. doi: 10.1038/nsmb.2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat Struct Mol Biol. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu H, et al. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature. 2013;494:371–374. doi: 10.1038/nature11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enesa K, et al. NF-kappaB suppression by the deubiquitinating enzyme Cezanne: a novel negative feedback loop in pro-inflammatory signaling. J Biol Chem. 2008;283:7036–7045. doi: 10.1074/jbc.M708690200. [DOI] [PubMed] [Google Scholar]
- 14.Luong LA, et al. Cezanne regulates inflammatory responses to hypoxia in endothelial cells by targeting TRAF6 for deubiquitination. Circ Res. 2013;112:1583–1591. doi: 10.1161/CIRCRESAHA.111.300119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu H, et al. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. J Exp Med. 2016;213:399–414. doi: 10.1084/jem.20151426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pareja F, et al. Deubiquitination of EGFR by Cezanne-1 contributes to cancer progression. Oncogene. 2012;31:4599–4608. doi: 10.1038/onc.2011.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bremm A, Moniz S, Mader J, Rocha S, Komander D. Cezanne (OTUD7B) regulates HIF-1α homeostasis in a proteasome-independent manner. EMBO Rep. 2014;15:1268–1277. doi: 10.15252/embr.201438850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moniz S, et al. Cezanne regulates E2F1-dependent HIF2α expression. J Cell Sci. 2015;128:3082–3093. doi: 10.1242/jcs.168864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bremm A, Freund SMV, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geurink PP, et al. Development of Diubiquitin-Based FRET Probes To Quantify Ubiquitin Linkage Specificity of Deubiquitinating Enzymes. ChemBioChem. 2016;17:816–820. doi: 10.1002/cbic.201600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulder MPC, El Oualid F, ter Beek J, Ovaa H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. ChemBioChem. 2014;15:946–949. doi: 10.1002/cbic.201402012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Komander D, Barford D. Structure of the A20 OTU domain and mechanistic insights into deubiquitination. Biochem J. 2008;409:77–85. doi: 10.1042/BJ20071399. [DOI] [PubMed] [Google Scholar]
- 23.Lin S-C, et al. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol. 2008;376:526–540. doi: 10.1016/j.jmb.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geurink PP, Oualid ElF, Jonker A, Hameed DS, Ovaa H. A general chemical ligation approach towards isopeptide-linked ubiquitin and ubiquitin-like assay reagents. ChemBioChem. 2012;13:293–297. doi: 10.1002/cbic.201100706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berrow NS, et al. A versatile ligation-independent cloning method suitable for high-throughput expression screening applications. Nucleic Acids Res. 2007;35:e45. doi: 10.1093/nar/gkm047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hospenthal MK, Mevissen TET, Komander D. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest) Nat Protoc. 2015;10:349–361. doi: 10.1038/nprot.2015.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faggiano S, Alfano C, Pastore A. The missing links to link ubiquitin: Methods for the enzymatic production of polyubiquitin chains. Anal Biochem. 2016;492:82–90. doi: 10.1016/j.ab.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 29.Borodovsky A, et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9:1149–1159. doi: 10.1016/s1074-5521(02)00248-x. [DOI] [PubMed] [Google Scholar]
- 30.Ekkebus R, et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J Am Chem Soc. 2013;135:2867–2870. doi: 10.1021/ja309802n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 34.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 36.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams PD, et al. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 41.Pham GH, et al. Comparison of native and non-native ubiquitin oligomers reveals analogous structures and reactivities. Protein Sci. 2016;25:456–471. doi: 10.1002/pro.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Messick TE, et al. Structural basis for ubiquitin recognition by the otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiener R, et al. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol. 2013;20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.